Summary
Nono is a component of the para-speckle, which stores and processes RNA. Mouse embryonic stem cells (mESCs) lack para-speckles, leaving the function of Nono in mESCs unclear. Here we find that Nono functions as a chromatin regulator cooperating with Erk to regulate mESC pluripotency. We report that Nono loss results in robust self-renewing mESCs with epigenomic and transcriptomic features resembling the 2i (GSK and Erk inhibitors)-induced “ground state”. Erk interacts with and is required for Nono localization to a subset of bivalent genes that have high levels of poised RNA polymerase. Nono loss compromises Erk activation and RNA polymerase poising at its target bivalent genes in undifferentiated mESCs; thus disrupting target gene activation and differentiation. These findings argue that Nono collaborates with Erk signaling to regulate the integrity of bivalent domains and mESC pluripotency.
Graphical Abstract
Introduction
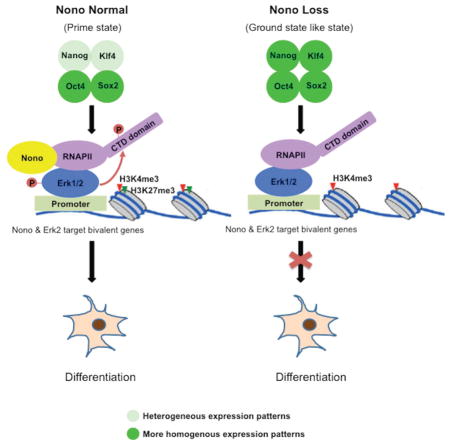
Mouse embryonic stem cells (mESCs) cultured under traditional serum/LIF conditions are naïve but “metastable”, a phenomenon characterized by heterogeneous expression of pluripotency factors, such as Nanog, Klf4 and Tbx3 (Chambers et al., 2007; Dan et al., 2013; Festuccia et al., 2012; Filipczyk et al., 2015; Hatano et al., 2005; Kalmar et al., 2009; Niwa et al., 2009; Singh et al., 2007; Torres-Padilla and Chambers, 2014). This heterogeneity reflects distinct differentiation potentials of individual mESCs. Those expressing lower levels of pluripotency factors are considered more prone to differentiation (primed) while cells expressing higher levels of these factors have more robust self-renewal capability. However, when mESCs are cultured with inhibitors of the Mek/Erk and Gsk3β signaling pathways (2i, PD03 for Mek/Erk, and CHIR for Gsk3β), this heterogeneity is reduced, resulting in mESCs with more homogeneous and higher expression levels of pluripotency factors, such as Nanog and Klf4, thus resembling the in vivo, “ground state” state (Martello and Smith, 2014; Nichols and Smith, 2009; Wray et al., 2010; Ying et al., 2008). Despite extensive epigenomic and gene expression analyses of mESCs under different culture conditions, mechanisms by which the ERK pathway regulates stem cell pluripotency remain incompletely understood.
Recently, Erk, an important kinase downstream of Mek, has been identified as associating with a subset of bivalent genes in mESCs under traditional serum/LIF cultures (Tee et al., 2014). At these bivalent regions, activated Erk (phophorylated) mediates phosphorylation of Serine 5 in the C-terminal domain (CTD) of RNA polymerase II (RNAPIICTD), and helps keep RNAPII in a poised state (Tee et al., 2014). Mek inhibition using PD03 results in a loss of Erk activation, Erk dissociation from chromatin and altered RNAPII activity at bivalent targets. This is accompanied by a phenotypic switch to a more robust self-renewing state, similar to the ground state pluripotency (Kunath et al., 2007; Tee et al., 2014; Ying et al., 2008). These findings suggest an intrinsic connection between the chromatin states of the Erk-bound bivalent regions and the differential pluripotency potentials of mESCs. Therefore, understanding how inactivation of Erk leads to more robust self-renewal and reduced differentiation potentials of mESCs will give significant insight into stem cell biology.
Nono (aka Nrb54 and P54nrb) was initially identified as a non-POU-domain-containing, octamer-binding protein (Dong et al., 1993; Yang et al., 1993). Nono binds both DNA and RNA, possibly via its helix-turn-helix (HTH) and the RNA recognition motif (RRM) domains, and has been suggested to regulate transcription (Dong et al., 1993; Park et al., 2013; Yang et al., 1993). In differentiated cells, Nono, together with Pspc1, Psf and a scaffolding non-coding RNA, Neat1, forms the nuclear para-speckle structure, known to regulate RNA processing, nuclear retention of hyperedited mRNAs and stress responses induced by viral infection and DNA damage (Clemson et al., 2009; Fox et al., 2002; Fox and Lamond, 2010; Hutchinson et al., 2007; Li et al., 2009; Sunwoo et al., 2009). However, the para-speckle structure is absent in hESC and mESCs, possibly due to the lack of sufficient Neat1 (long isoform) expression (Ghosal et al., 2013), suggesting that Nono may play a para-speckle-independent role in undifferentiated ESCs. Here, we report that Nono acts as an Erk cofactor to regulate mESC self-renewal and differentiation. Specifically, we show that Nono physically interacts with Erk and co-localizes with Erk to a subset of development-related, bivalent genes. Loss of Nono leads to impaired Erk activation and RNA Polymerase II C-Terminal Domain Serine 5 phosphorylation (RNAPIIS5P) at Nono/Erk bound bivalent genes, and compromised activation of these genes during retinoic acid (RA) induced differentiation. Inactivation of Erk evicts Nono from chromatin. Nono null cells behave similarly to mESCs cultured under 2i conditions or with PD03, i.e., they exhibit increased expression of pluripotency-associated factors, are more self-renewing and more resistant to differentiation. Molecular analyses also revealed substantial epigenomic and transcriptomic similarities between Nono KO and 2i “ground state” or PD03 treated mESCs. Taken together, our findings suggest that Nono is a critical regulator of bivalent domains and mESC pluripotency, providing insight into the molecular mechanisms that balance self-renewal and differentiation.
Results
Nono KO mESCs show increased expression of Nanog and KLF4 with enhanced self-renewal capacity
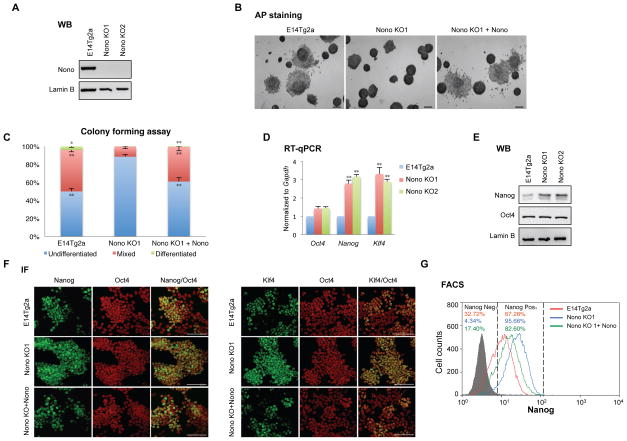
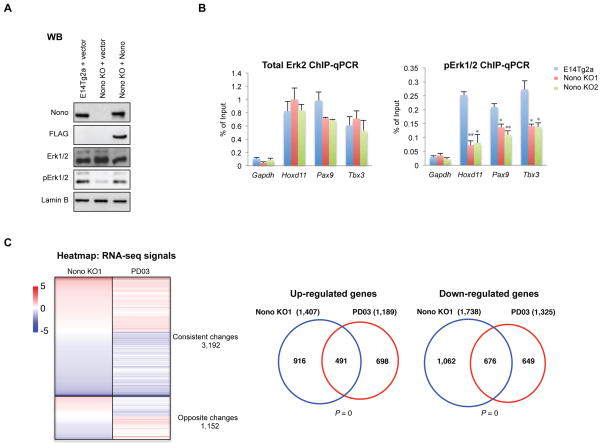
As discussed, the para-speckle structure is absent in mESCs but Nono nonetheless is expressed and has been suggested to regulate transcription of Oct4 (Park et al., 2013), raising the possibility that Nono may play a role in regulating mESC pluripotency. To investigate this possibility, we first generated two independent Nono KO (KO1 and KO2) mESC lines using the CRISPR technology (Figure 1A and S1A). Interestingly, Nono KO cells formed more compacted alkaline phosphatase-positive colonies with a greatly reduced number of differentiated cells (Figure S1B, middle and right panels; Figure 1B and 1C, middle panels), suggesting an enhanced clonal self-renewal capacity. Importantly, such phenotypic changes were largely rescued by reintroduction of wildtype Nono, indicating that the phenotype is due to the loss of Nono (Figure 1B and 1C, right panels).
Figure 1. Nono loss results in enhanced self-renewal.
(A) Immunoblot analyses of Nono and Lamin B (as control) in WT (E14Tg2a) and Nono KO ESC cell lines.
(B) Alkaline phosphatase (AP) staining of colonies formed 7 days post plating (100 Units/ml LIF) from the WT (E14Tg2a), Nono KO1 and Nono KO rescued cells. The scale bar represents 200 μm.
(C) Quantitation of colony types from the experiments shown in (B).
(D) RT-qPCR analyses of Oct4, Nanog and Klf4 levels in WT (E14Tg2a) and two Nono KO cell lines.
(E) Immunoblot analyses of Nanog, Oct4 and Lamin B (as control) in WT (E14Tg2a) and two Nono KO cell lines.
(F–G) Immunofluorescence analyses (F) of Nanog (green, panel Left), Klf4 (green, panel Right) and Oct4 (red) expression, and quantitative flow cytometry analysis (G) of Nanog expression, in WT (E14Tg2a), Nono KO1 and Nono KO rescued cells. The scale bar represents 100 μm.
In panel C–D, all data are represented as mean ± SD (n=3), * p < 0.05; ** p < 0.01, T test.
mESC self-renewal is governed by a network of pluripotency-associated factors, including Nanog and Oct4. The Nono KO mESC phenotype prompted us to investigate whether Nono regulates the expression of these factors. As shown in Figure 1D and 1E, while Oct4 displayed only a modest increase at mRNA level, Nanog expression, surprisingly, was significantly elevated in Nono KO mESCs at both the mRNA (Figure 1D) and protein (Figure 1E) levels. Furthermore, while Nanog expression levels vary (heterogeneously expressed) among individual wildtype mESCs, Nono KO cells express increased Nanog and have a reduced Nanog-negative population as shown by both immunofluorescence and FACS analyses (Figure 1F, left, and 1G). Consistently, the expression of another pluripotent factor, Klf4, a downstream target of Nanog, also showed a similar change (Figure 1D and 1F, right). Importantly, the normal expression pattern of Nanog and Klf4 was by and large rescued by re-expression of wildtype Nono (Figure 1F lower panels and 1G), indicating that the altered Nanog and Klf4 expression patterns were due to the loss of Nono.
Previous studies showed that individual mESCs transition between a high Nanog state with an enhanced self-renewal capacity and a low Nanog state, in which cells are primed for lineage specification (Abranches et al., 2014; Chambers et al., 2007; Kalmar et al., 2009; Torres-Padilla and Chambers, 2014). Our finding that loss of Nono reduces the population of Nanog-negative cells is consistent with the enhanced self-renewal capability of Nono KO mESCs. Importantly, these features resemble the “ground state” mESCs cultured with inhibitors of MEK and GSK3β (2i), which display increased self-renewal capacity associated with increased and more homogeneous expression of Nanog (Abranches et al., 2014; Ying et al., 2008).
Nono KO mESCs show an epigenomic and transcriptomic signature similar to “ground-state” pluripotency
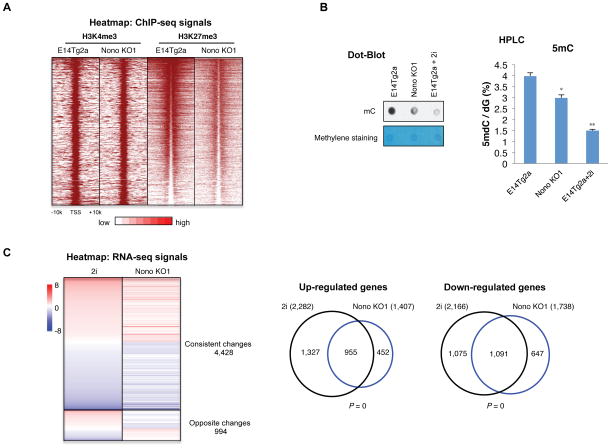
Since the “2i ground state” has been linked to global epigenomic and transcriptomic changes, including a reduction of H3K27me3 at bivalent genes, as well as a global reduction of DNA methylation (Marks et al., 2012; Tee et al., 2014), we next asked whether Nono KO cells also show these changes. Through genome-wide ChIP-seq analyses, we found a significant reduction of H3K27me3 at bivalent genes, while H3K4me3 was largely unaffected, similar to 2i mESCs (Figure 2A). Along the same line, we also found a global reduction of DNA methylation in Nono KO cells, resembling 2i “ground state” mESCs (Figure 2B), although the effect is milder (Figure 2B). Consistent with the epigenomic similarities, Nono KO cells also shared a similar gene expression profile as 2i mESCs (Figure 2C). Collectively, our findings suggest that Nono loss leads to robust self-renewal and epigenomic and transcriptomic re-patterning, resembling those observed for mESCs cultured with 2i.
Figure 2. Nono loss results in transcriptomic signature resembling “ground-state” pluripotency.
(A) Heatmap analysis of H3K4me3 and H3K27me3 ChIP-seq data from E14Tg2a and Nono KO1 ESCs at 3,104 annotated bivalent genes. The ChIP-seq signals are displayed in the distance of ±10 kb to TSS. Color scale represents normalized read density.
(B) Global DNA methylation in the parental E14Tg2a, Nono KO1 and 2i treated E14Tg2a mESCs measured by dot-blot (100ng per dot, left) and quantitative HPLC (right).
Data are represented as mean ± SD (n=3), * p < 0.05; ** p < 0.01, T test.
(C) Left: Heatmap analysis of total differentially regulated genes (fold >= 1.5, FPKM > 0.5) in 2i treated E14Tg2a mESCs and Nono KO1 mESCs compared with E14Tg2a, ranked by genes with altered expression in 2i treated E14Tg2a (from up to down). Right: Venn diagram illustrating a significant overlap between 2i and Nono KO1 up- and down-regulated genes. P values are determined by the Hypergeometric Distribution test.
Genomic distribution of Nono in mESCs
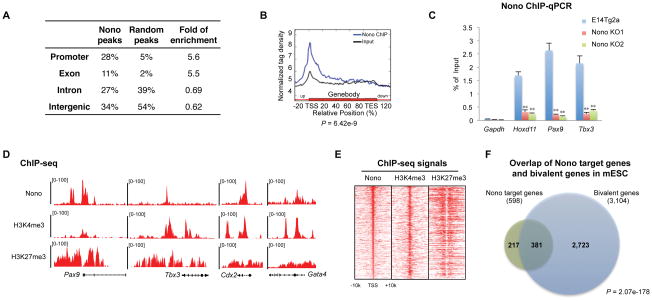
To address the mechanism of action of Nono in mESCs, we sought to identify endogenous Nono bound genomic regions by ChIP-seq. This effort yielded 1,193 potential Nono binding events, covering 598 genes, which appear to be enriched at promoter regions (Figure 3A). Consistently, analyses of the genomic distributions of Nono ChIP-seq signals across an average Reference Sequence database gene and around TSSs (Transcription Start Sites) also showed Nono enrichment at promoters, with the highest signal density around TSSs (Figure 3B). That these are genuine Nono binding events was further confirmed using the two Nono KO mESC lines as negative controls (Figure 1A and 3C). Importantly, GO analysis identified a significant enrichment of Nono bound genes in the category of transcription regulation and developmental genes (Figure S2A and S2B), including Pax9, Tbx3, Cdx2, Gata4 as well as all Hox clusters of genes, which are known to be decorated and regulated by bivalent domains (Bernstein et al., 2006; Vastenhouw and Schier, 2012) (Figure 3D). Significantly, further bioinformatics analyses showed that 63.7% (381/598) of Nono bound genes are associated with bivalent domains (Figure 3E–F and S2C), suggesting Nono may directly regulate mESC pluripotency through regulating this subset of bivalent genes.
Figure 3. Nono localizes to a subset of bivalent domains in mESCs.
(A) Genome-wide distribution of Nono binding events in mES cells. Random peaks were generated as control to show fold of enrichment.
(B) Average Nono ChIP-seq and Input signals on Nono bound genes. P values by ANOVA test.
(C) Nono ChIP-qPCR analyses at 3 selected bivalent genes and Gapdh locus (as control) in WT (E14Tg2a) and two Nono KO ESC lines. Data are represented as mean ± SD (n=3), ** p < 0.01, T test.
(D) UCSC snapshots of Nono, H3K4me3 and H3K27me3 ChIP-seq signals at four representative bivalent regions.
(E) Heatmap analyses of Nono, H3K4me3 and H3K27me3 ChIP-seq signals ranked by Nono ChIP-seq intensities over target TSSs.
(F) Venn diagrams showing significant overlap between Nono bound genes and bivalent genes in mESCs. P values by Hypergeometric Distribution test.
Erk2 interacts and is required for Nono localization to a subgroup of bivalent genes
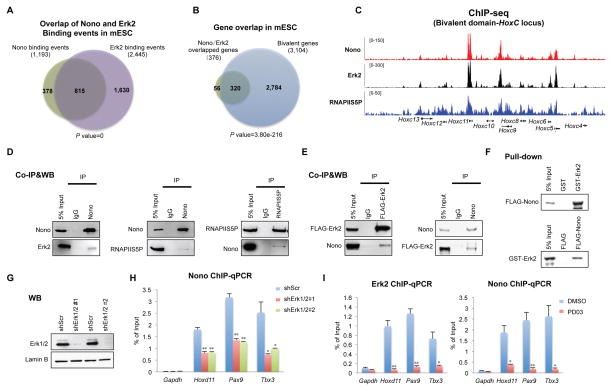
Although most Nono bound genes carry bivalent modifications, only a small portion (~12%, 381/3,104) of total mESC bivalent genes are bound by Nono (Figure 3F and S2C), suggesting that bivalent state alone does not define Nono chromatin association. Interestingly, further bioinformatics analyses found that more than two thirds of Nono binding events (68.3%, 815/1,193) are shared by Erk2, while reciprocally one third (33.3%, 815/2,445) of Erk2 binding events are shared by Nono (Figure 4A). However, the actual degree of overlap might be higher as our ChIP-seq is limited by the detection sensitivity of the Nono antibody. Supporting this, we were able to detect Nono binding by the more sensitive ChIP-qPCR at all 6 randomly selected Erk2 targets, which were otherwise negative for Nono binding based on the ChIP-seq signals (Figure S2D). Nonetheless, these data revealed a significantly more extensive overlap between Nono and Erk2 bound chromatin locations, as opposed to the general pool of bivalent domains, in mESCs. Our further analyses showed that the majority of the 815 Nono and Erk2 co-bound events are located in bivalent genes (85.1%, 320/376) (Figure 4B and S2C). Importantly, Erk signaling is critical for mESC self-renewal and differentiation, and activated Erk has recently been shown to phosphorylate Serine 5 in the CTD of RNAPII to keep RNAPII at the Erk bound bivalent regions at a poised state (Kunath et al., 2007; Tee et al., 2014; Ying et al., 2008). Consistently, most of Nono bound genes also tracked with RNAPIIS5P (Figure 4C and S2C), and the interactions among Nono, Erk2 and RNAPII5SP in mESC extracts were readily detectable by co-immunoprecipitation (Figure 4D for endogenous interactions, and 4E for interactions between Nono and ectopic FLAG-tagged Erk2). To exclude the possibility of chromatin mediated interactions between Nono and Erk2, we turned to in vitro pull-down assays. As shown in Figure 4F, recombinant GST-tagged Erk2, and FLAG-tagged Nono purified from E. coli and Sf9 insect cells, respectively, interacted with each other in vitro, suggesting a direct interaction. Taken together, these results suggest that Nono co-localizes and interacts with Erk2, thus identifying Nono as a bivalent domain component.
Figure 4. Nono interacts with and requires Erk2 for its localization at a subgroup of bivalent genes.
(A–B) Venn diagrams showing significant overlap between Nono and Erk2 binding events in mESCs (A), and that the majority of Nono/Erk2 co-bound genes are bivalent genes (B). P values by Fisher Exact test.
(C) UCSC snapshots of Nono, Erk2 and RNAPIIS5P ChIP-seq signals at the Hox C cluster. Erk2 data are adapted from published data (Tee et al., 2014).
(D–E) Co-IP analyses showing the interaction between endogenous Nono, RNAPIIS5P and Erk2 (D), FLAG-Erk2 and Nono (E).
(F) In vitro pull-down analyses using recombinant GST-Erk2 and FLAG-Nono purified from E. coli and insect Sf9 cells, respectively.
(G) Western blot showing Erk1/2 levels for shScr and Erk1/2 depleted (shErk1/2#1 and shErk1/2#2) ESCs.
(H) Nono ChIP qPCR on a few selected target genes (indicated at the bottom) in scramble control (shScr) and Erk1/2 depleted ESCs. Error bars represent SD; n=3.
(I) Erk2 and Nono ChIP qPCR in control and PD03 treated mESCs. Error bars represent SD; n=3.
In panel H and I, all q-PCR data are represented as mean ± SD (n=3), * p < 0.05; ** p < 0.01, T test.
To understand how Nono is localized to bivalent chromatin, we asked whether Nono chromatin occupancy is dependent on Erk. Importantly, we found that inhibition of Erk1/2 by two independent shRNA pairs (Figure 4G) significantly reduced Nono chromatin association, as shown by ChIP-qPCR at three target regions (Figure 4H). We further asked whether activation of Mek/Erk signaling is required for Nono bivalent domain localization. As shown in Figure 4I (left), PD03 treatment not only caused a reduction of Erk2 binding as previously reported (Tee et al., 2014), but also a significant reduction of Nono binding at these bivalent genes (Figure 4I, right). These data indicate that Erk and activated Mek/Erk signaling play a critical role in Nono chromatin localization.
Nono loss results in impaired Erk activation
The fact that Nono and Erk2 interact and co-localize at a subset of bivalent genes prompted us to investigate whether Nono plays a role in the regulation of Erk in mESCs. Interestingly, we found that although global levels of Erk1/2 were not altered in the Nono KO cells, the active form of Erk1/2 (pErk1/2) was significantly reduced (Figure 5A), indicating that Erk activation was impaired in the absence of Nono. Consistently, we further found a substantial local reduction of pErk1/2, but not total Erk2, at bivalent genes (Figure 5B), suggesting that the activation but not chromatin occupancy of Erk is regulated by Nono. This is similar but not identical to Mek/Erk inactivation mediated by PD03, which results in an impaired chromatin occupancy of both phosphorylated Erk as well as total Erk protein (Tee et al., 2014). Consistently, we found that Nono KO mESCs showed a similar expression profile to that of the PD03 treated mESCs (Figure 5C), with significant overlaps of both up- and down-regulated genes. We were able to confirm by RT-qPCR this same trend of regulation by Nono versus PD03 at a few selected Nono and Erk co-bound bivalent genes (Figure S3A). Interestingly, we found that PD03 further increased the expression levels of Nanog and Klf4 in the Nono KO cells and further reduced Nanog-negative population as shown by FACS analyses, suggesting additive effects of Nono loss and PD03 treatment (Figure S3B and S3C).
Figure 5. Nono loss results in impaired Erk activity.
(A) Western blot analyses of Nono, Erk1/2, phospho-Erk1/2 (pErk1/2) and Lamin B (as control) levels in the indicated cell lines.
(B) ChIP-qPCR analyses of Erk2 and pErk1/2 chromatin occupancies at selected genes in WT (E14Tg2a) and two Nono KO mESCs. Data are represented as mean ± SD (n=3), * p < 0.05; ** p < 0.01, T test.
(C) Left: Heatmap analysis of total differentially regulated genes in Nono KO1 and PD03 treated E14Tg2a mESCs compared with E14Tg2a (fold >= 1.5, FPKM > 0.5), ranked by genes with altered expression in Nono KO1 (from up to down). Right: Venn diagram illustrating the overlap between Nono KO1 and PD03 up- and down-regulated genes. P values are determined by the Hypergeometric Distribution test.
Nono is required for RNAPIIS5 phosphorylation at its target bivalent genes and their activation during differentiation
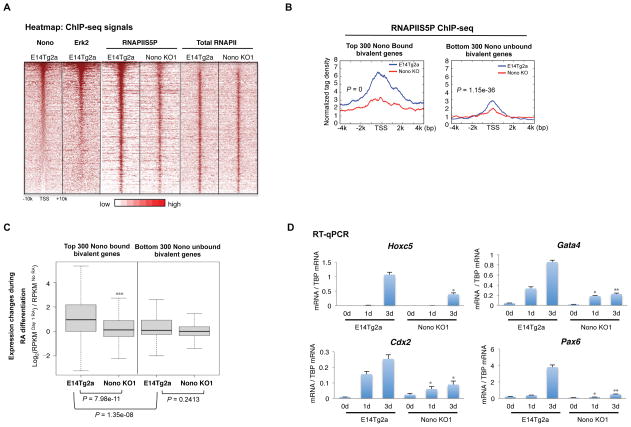
As Nono co-localizes with Erk2 and is required for Erk activation at a subset of bivalent genes, we next asked whether Nono loss leads to an impaired RNAPIIS5P at its bivalent target genes due to a defect in Erk activation. Through genome-wide ChIP-seq analyses, we found the highest level of RNAPIIS5P at Nono/Erk bound bivalent genes, as well as a significant reduction of RNAPIIS5P but not total RNAPII at bivalent genes (Figure 6A), similar to Erk inhibited mESCs (Tee et al., 2014). Interestingly, a more significant reduction of RNAPIIS5P was observed at the top 300 Nono bound bivalent genes, most of which are co-bound by Erk, when compared to the unbound ones (Figure 6B), supporting a direct mode of action of Nono at its target bivalent genes. However, a much milder reduction of RNAPIIS5P at the Nono/Erk unbound bivalent genes was also detected, suggesting a possible involvement of additional mechanisms.
Figure 6. Nono is required for RNAPIIS5P at its target bivalent genes and their activation during differentiation.
(A) Heatmap analysis of Nono, Erk2, RNAPIIS5P and Total RNAPII ChIP-seq data from E14Tg2a and Nono KO1 ESCs ranked by Nono ChIP-seq signals at 3,104 annotated bivalent genes. The ChIP-seq signals are displayed in the distance of ±10 kb to TSS. Color scale represents normalized read density.
(B) Signal plot analyses showing the reduction of RNAPIIS5P at the top 300 Nono bound (left) and 300 unbound bivalent genes. P values by ANOVA test.
(C) Fold of bivalent gene activation (presented as Log2) in WT (E14Tg2a) and Nono KO1 cells one-day post RA induced differentiation. Expression data from RNA-seq. Left: Top 300 bivalent genes bound by Nono; right: a control set of 300 bivalent genes with the lowest Nono binding intensities. P values by Student’s T-test (two sample T-tests).
(D) RT-qPCR analyses of selected Nono target gene expression in the indicated cell lines at different time points after the initiation of RA induced differentiation. Data are represented as mean ± SD (n=3), * p < 0.05; ** p < 0.01, T test.
Since the expression of many bivalent genes needs to be activated upon differentiation, we further investigated Nono function in bivalent gene activation upon RA-induced differentiation. By RNA-seq, we found that the fold of activation of the top 300 Nono bound bivalent genes, most of which are also co-bound by Erk, is significantly higher than the unbound controls (Figure 6C, compare column 1 to 3, P = 1.35e-08), possibly due to the already higher level of RNAPIIS5P before differentiation (Figure 6A and 6B). Interestingly, compared to the unbound ones, the activation of Nono bound bivalent genes is more significantly compromised upon differentiation in Nono KO cells (Figure 6C, compare left panel, P = 7.98e-11, to right panel, P = 0.2413). The activation of some of these target genes remained impaired even at day 3 after the initiation of differentiation, suggesting a sustained defect in transcriptional activation in Nono KO cells during differentiation (Figure 6D).
Nono loss compromises mESC differentiation
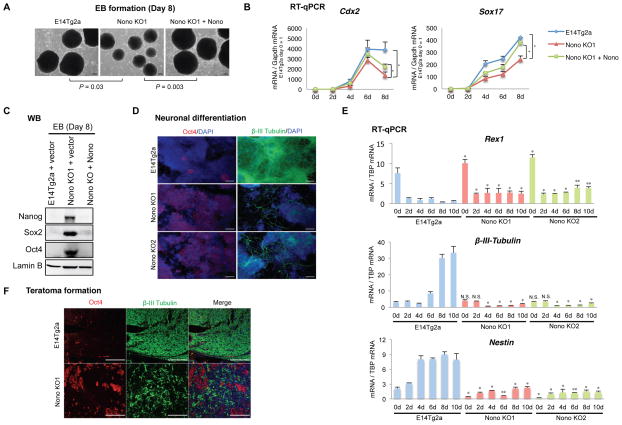
We next investigated whether Nono null cells also exhibit compromised ability to undergo differentiation as a result of an impaired Erk activity and compromised activation of Nono/Erk co-bound bivalent genes. We first used embryonic body (EB) formation assay to investigate the differentiation potential of the parental, the Nono KO and the Nono KO with a Nono rescuing construct. Consistently, we found that EBs developed from the Nono KO mESC cells were often much smaller and displayed disorganized structures, compared to the parental control (Figure 7A, compare the middle to the left, P = 0.03). In addition to the compromised activation of some developmental genes such as Cdx2 and Sox17 (Figure 7B), Nono KO cells also showed a compromised repression of the pluripotent genes, including Nanog, Sox2 and Oct4 (Figure 7C), suggesting a comprised differentiation. Importantly, these defects can be largely rescued by reintroducing wildtype Nono (Figure 7A–C), indicating on-targeting effects.
Figure 7. Nono loss leads to differentiation defect.
(A) Morphological analyses of EBs formed from the indicated cell lines at day 8 post-induction. The scale bar represents 100 μm. P values by T test.
(B) RT-qPCR analyses of Cdx2 and Sox17 expressions in the indicated cell lines at different time points after the initiation of EB differentiation. Data are represented as mean ± SD (n=3), * p < 0.05, T test.
(C) Immunoblot analyses of pluripotency markers, Nanog, Sox2 and Oct4, from day 8 EBs formed from the indicated cell lines.
(D–E) Immunofluorescence analyses (D) of Oct4 (red) and β-III Tubulin (green) proteins at day 10, and RT-qPCR analyses (E) of Rex1, β-III Tubulin and Nestin mRNAs at indicated point points, in mESCs induced toward NSC differentiation for 10 days. Cells were counterstained with Dapi (blue, panel D). The scale bar represents 100 μm. Data are represented as mean ± SD (n=3) in panel E, * p < 0.05; ** p < 0.01, T test.
(F) Immunofluorescence analysis of Oct4 and β-III Tubulin in the teratomas from E14Tg2a and Nono KO1 cells. The scale bar represents 100 μm.
We further investigated the role of Nono in a monolayer neuronal stem cell (NSC) differentiation process. As shown in Figure 7D, Nono loss significantly compromised neural differentiation, as evidenced by the presence of much fewer cells with βIII-Tubulin-positive axonal processes. Consistently, most Nono KO cells retained high and persistent expression of Oct4 even at day 10-post differentiation induction while in most of the control mESC cells at this time point Oct4 should have already been largely silenced (Figure 7D). Supporting the phenotypic analysis, we also found that suppression of the expression of pluripotency factors and activation of neuronal markers were significantly compromised in Nono KO cells during differentiation (Figure 7E).
To further confirm Nono function in differentiation in vivo, we employed mouse teratoma model. Although both WT and Nono KO cells formed teratomas in 5 weeks, the Nono KO teratomas were much bigger than the controls (Figures S4A), and with significantly more Oct4-positive and fewer βIII-Tubulin-positive cells (Figure 7F) suggesting a compromised differentiation. Additionally, although Nono KO teratomas were able to develop all three germ layers (Figure S4B), they by and large gave rise to much smaller ectoderm-derived, keratinized epithelium tissues (Figure S4B, left) and fewer mesoderm-derived muscle tissues (Figure S4B, middle), but no obvious alterations of endoderm-derived intestinal epithelium tissues (Figure S4B, right).
It’s important to note that, consistent with our molecular findings that Nono and Erk reciprocally regulate each other, Nono KO mESCs behaved similarly as the PD03 treated (Figure S4C and S4D) and Erk2 KO mESCs (Kunath T et al., 2007), in our EB and monolayer NSC differentiation assays.
Taken together, we find that Nono KO mESCs displayed many similarities to 2i “ground state” and PD03 treated mESC in terms of the robust self-renewal capability, resistance to differentiation as well as epigenomic and transcriptomic features. Mechanistically, Nono localization to the Erk2-marked bivalent domains requires Erk, and on the other hand, Nono is also involved in Erk activation and bivalency integrity, thus revealing a reciprocal regulation between Nono and Erk. Importantly, Nono/Erk bound bivalent genes undergo robust activation upon differentiation, which is likely to be dependent on Nono/Erk mediated RNAPIIS5P at the pluripotent state.
Discussion
This study identifies Nono as a regulator of mESC pluripotency. Loss of Nono in mESCs results in a ground state-like pluripotency, characterized by robust self-renewal with epigenomic and transcriptomic features resembling those of 2i treatment or Erk inhibition alone. Mechanistic analyses reveal that Nono functions in Erk signaling, i.e., Nono interacts with and co-localizes with Erk at a subset of bivalent genes with the highest level of RNAPIIS5P. Loss of Nono blocks Erk activation and results in reduced poised RNAPII at its target bivalent genes in undifferentiated mESCs, compromised target gene activation upon differentiation, and differentiation itself. Loss of Erk protein or inhibition of Mek/Erk signaling leads to Nono dissociation from chromatin. Our data thus reveal a role for Nono at bivalent chromatin, where it works with Erk to regulate mESC pluripotency.
Nono and Erk in the regulation of mESC bivalent domain and pluripotency
Our data indicate that Nono is necessary for Erk activation, which would place Erk downstream of Nono and explain the similarity of the phenotypic changes upon loss of Nono or Erk inactivation. However, both chemical inhibition and genetic ablation of Erk also result in Nono dissociation from chromatin, loss of bivalent domain integrity and up-regulation of a subset of shared target genes. Thus these data collectively reveal a reciprocal and positive reinforcement loop, and suggest that Nono and Erk function together rather than hierarchically.
Our data also reveal that Nono/Erk co-bound bivalent genes show the highest RNAPIIS5P comparing to the unbound ones (Figure 6A). Importantly, this subset of bivalent genes are also the ones that undergo stronger activation during differentiation, and loss of Nono leads to compromised activation of these genes (Figure 6C and 6D), perhaps due to loss of Erk-mediated RNAPIIS5P before differentiation (Figure 6A and 6B). Although we cannot fully exclude the possibility of indirect effects due to compromised differentiation of Nono KO mESCs, we only observed the impaired gene activation for Nono-bound bivalent genes not unbound ones, favoring a specific mode of action. These data indicate that Nono and Erk are required for RNAPIIS5P at a subset of bivalent genes in mESCs, and this process sets up the poised transcriptional machinery needed for later gene activation. This is consistent with the idea that bivalent genes are subject to dynamic regulation, different from fully silent genes. However, why the Nono/Erk co-bound but not all bivalent genes are selected for such regulation and how the specificity is achieved remain interesting questions for future investigation.
Nono and para-speckle in self-renewing and differentiated cells
As discussed earlier, Nono is an RNA binding protein and a major component of para-speckles. The main functions of para-speckles are RNA processing, nuclear retention of mRNA and stress responses (Chen and Carmichael, 2009; Fox and Lamond, 2010). However, para-speckles are thought to exist in differentiated cells, but not in ESCs, due to insufficient expression of the scaffolding non-coding RNA, Neat1, suggesting that Nono may function differently in mESCs (Ghosal et al., 2013; Mercer et al., 2010; Standaert et al., 2014; West et al., 2014; Zeng et al., 2014). Our finding that Nono functions as a bivalent chromatin factor in mESCs is consistent with this prediction.
Our study shows that Nono interacts and co-localizes with Erk2 and RNAPIIS5P at bivalent genes in mESCs, while a previous study also reported that Nono binds both phosphorylated and unphosphorylated RNAPII CTD at Ser5 (Emili et al., 2002). This raises the possibility that Nono may not only interact with poised RNAPII in mESCs, but also play a role in gene activation and mRNA processing upon differentiation. Supporting this, we observed compromised activation of Nono/Erk co-bound bivalent genes in Nono KO cells during RA induced differentiation. Since Nono is reported to bind mRNAs and required for mRNA processing in differentiation cells (Clemson et al., 2009; Fox et al., 2002; Fox and Lamond, 2010; Hutchinson et al., 2007; Li et al., 2009; Sunwoo et al., 2009), it is conceivable that such a mechanism may be critical for Nono-bound bivalent genes to produce mature mRNAs once mESCs leave the primed state to differentiate. Whether such a mechanism is dependent on functional para-speckles that are assembled after differentiation will be another interesting question to address (Emili et al., 2002; Kaneko et al., 2007; West et al., 2014). It will also be of importance to evaluate whether this mechanism is conserved in the regulation of ESCs of human and other species.
In summary, our findings highlight a function for Nono in mESCs where it plays a critical role at the chromatin interface to regulate Erk signaling, impacting the integrity of bivalent chromatin at developmental genes and mESC pluripotency.
Experimental Procedures
Cell culture and cell differentiation
mESCs were grown in standard ESC medium containing serum and 100 U/ml LIF unless otherwise stated. For the generation of Nono rescued mESCs, Nono open reading frame was cloned into pPB Flag-HA puromycin expression vector. The Nono rescued and vector control mESCs were maintained in 2mg/ml puromycin.
Immunofluorescence, flow cytometry analysis and co-Immunoprecipitation
mESC cells were seeded in 12-well plates at a density of 50K cells/well at 48 hours before immunofluorescence examination. Flow cytometry analysis of Nanog expression was carried out as previously described (Festuccia and Chambers, 2011). Briefly, 2×105 cells were stained with Nanog antibody (1:300, eBioMLc-51) and anti-rat conjugated to Cy3 (1:500), and 20K cells were analyzed for each sample.
Nuclear extracts used for co-immunoprecipitation were prepared from mESC cells and co-Immunoprecipation analyses were carried out as described previously (Mendez and Stillman, 2000).
In Vitro pull-down assay
FLAG tagged Nono was purified from insect cells, and GST-tagged Erk2 proteins were purified from E. coli. A total of 5 μg FLAG-Nono and 5 μg GST-Erk2 proteins were incubated in 200 ul reaction system in binding buffer (20 mM TrisCl pH 7.4, 150 mM NaCl and 0.1% Triton X100) for 3 hours at 4° C. FLAG-Nono and GST-Erk2 complex was captured using FLAG or GST beads and then subjected to Western blot analyses.
ChIP, ChIP-seq and data analyses
ChIP assay was performed as described elsewhere (Lan et al., 2007). The precipitated DNA samples were analyzed using real-time quantitative PCR (qPCR) and prepared for deep sequencing according to manufacture’s guidelines (Guo et al., 2014 and Illumina). DNA deep sequencing analyses were performed at the Epigenetics Key Laboratory at IBS of Fudan University, Shanghai, China.
For ChIP-seq data processing, the FASTQ data were mapped to mouse genome(mm9) using Bowtie, and significant enrichments (peaks) were identified by MACS 1.4 using Broad Peak mode with P value≤1×10−5, FDR≤0.01 as a cutoff from the mapped reads (Zhang et al., 2008).
A total of 3,104 bivalent genes are classified by the overlap of H3K4me3 and H3K27me3 with mapping tags > 50 and fold enrichment > 8 compared to input (Marks et al., 2012).
Supplementary Material
Acknowledgments
We thank Dr. Danny Reinberg and Wee-Wei Tee for providing the Erk2 expression construct. We thank Drs. Jennifer Nichols, Valerie Wilson, Tilo Kunath and William Hamilton for valuable suggestions on teratoma formation assays and ERK signaling. We thank Drs. Emily Brookes, Bo Wen, Jiekai Chen, Jing Liu, Ning Sun, Ting Ni and Bisi Miao for critical reading of the manuscript and for valuable suggestions. F.L is supported in part by State Key Development Programs (2016YFA0101800 and 2014CB943103), China “Thousand Youth Talents” (KHH1340001), NSFC grant (91419306) and International Science & Technology Cooperation Program of China (2014DFB30020). This work was also supported in part by a grant (CA118487) from the NIH and Boston Children’s Hospital funds to Y.S., who is an American Cancer Society Research Professor.
Footnotes
Conflict of Interest Statement
F.L. is a shareholder of Constellation Pharmaceuticals. Y.S. is a co-founder of Constellation Pharmaceuticals, Inc., as well as a member of its scientific advisory board. F.L. and Y.S. are also consultants for Active Motif, Inc.
Author Contributions
C.M. carried out most of the experiments with V.K-N. and H.T. described in this manuscript. V.K-N. and H. T. carried out part of the pluripotency examination. H.T. carried out part of the biochemical analyses, including molecular cloning, protein-protein interaction analysis and the gene expression study. Z.S. and L.T. assisted in Nono KO and KD experiments. W.L., W.X., R.L. and F.W. carried out bioinformatics analyses. Z.W. and L.K. prepared recombinant proteins from Sf9. W.Z. and P.Y contributed to DNA deep sequencing and preliminary data processing. Y.S., and F.L. directed all the experiments with the input from Y.G.S.; Y.S. and F.L. conceived the project and co-wrote the manuscript with C.M. and V.K-N.
ACCESSION NUMBERS
All data are deposited at the GEO database. All ChIP-Seq and mRNA-seq data are under accession number GSE73426.
Further experimental details can be found in Extended Experimental Procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abranches E, Guedes AM, Moravec M, Maamar H, Svoboda P, Raj A, Henrique D. Stochastic NANOG fluctuations allow mouse embryonic stem cells to explore pluripotency. Development. 2014;141:2770–2779. doi: 10.1242/dev.108910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brookes E, de Santiago I, Hebenstreit D, Morris KJ, Carroll T, Xie SQ, Stock JK, Heidemann M, Eick D, Nozaki N, et al. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell. 2012;10:157–170. doi: 10.1016/j.stem.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J, Li M, Yang J, Li J, Okuka M, Ye X, Liu L. Roles for Tbx3 in regulation of two-cell state and telomere elongation in mouse ES cells. Sci Rep. 2013;3:3492. doi: 10.1038/srep03492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Horowitz DS, Kobayashi R, Krainer AR. Purification and cDNA cloning of HeLa cell p54nrb, a nuclear protein with two RNA recognition motifs and extensive homology to human splicing factor PSF and Drosophila NONA/BJ6. Nucleic Acids Res. 1993;21:4085–4092. doi: 10.1093/nar/21.17.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili A, Shales M, McCracken S, Xie W, Tucker PW, Kobayashi R, Blencowe BJ, Ingles CJ. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA. 2002;8:1102–1111. doi: 10.1017/s1355838202025037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festuccia N, Osorno R, Halbritter F, Karwacki-Neisius V, Navarro P, Colby D, Wong F, Yates A, Tomlinson SR, Chambers I. Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell Stem Cell. 2012;11:477–490. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipczyk A, Marr C, Hastreiter S, Feigelman J, Schwarzfischer M, Hoppe PS, Loeffler D, Kokkaliaris KD, Endele M, Schauberger B, et al. Network plasticity of pluripotency transcription factors in embryonic stem cells. Nat Cell Biol. 2015;17:1235–1246. doi: 10.1038/ncb3237. [DOI] [PubMed] [Google Scholar]
- Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI. Paraspeckles: a novel nuclear domain. Curr Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb Perspect Biol. 2010;2:a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal S, Das S, Chakrabarti J. Long noncoding RNAs: new players in the molecular mechanism for maintenance and differentiation of pluripotent stem cells. Stem Cells Dev. 2013;22:2240–2253. doi: 10.1089/scd.2013.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano SY, Tada M, Kimura H, Yamaguchi S, Kono T, Nakano T, Suemori H, Nakatsuji N, Tada T. Pluripotential competence of cells associated with Nanog activity. Mech Dev. 2005;122:67–79. doi: 10.1016/j.mod.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmar T, Lim C, Hayward P, Munoz-Descalzo S, Nichols J, Garcia-Ojalvo J, Martinez AA. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev. 2007;21:1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath T, Saba-El-Leil MK, Almousailleakh M, Wray J, Meloche S, Smith A. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 2007;134:2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- Landeira D, Bagci H, Malinowski AR, Brown KE, Soza-Ried J, Feytout A, Webster Z, Ndjetehe E, Cantone I, Asenjo HG, et al. Jarid2 Coordinates Nanog Expression and PCP/Wnt Signaling Required for Efficient ESC Differentiation and Early Embryo Development. Cell Rep. 2015;12:573–586. doi: 10.1016/j.celrep.2015.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Kuhne WW, Kulharya A, Hudson FZ, Ha K, Cao Z, Dynan WS. Involvement of p54(nrb), a PSF partner protein, in DNA double-strand break repair and radioresistance. Nucleic Acids Res. 2009;37:6746–6753. doi: 10.1093/nar/gkp741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Smith A. The nature of embryonic stem cells. Annu Rev Cell Dev Biol. 2014;30:647–675. doi: 10.1146/annurev-cellbio-100913-013116. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- Obier N, Lin Q, Cauchy P, Hornich V, Zenke M, Becker M, Muller AM. Polycomb protein EED is required for silencing of pluripotency genes upon ESC differentiation. Stem Cell Rev. 2015;11:50–61. doi: 10.1007/s12015-014-9550-z. [DOI] [PubMed] [Google Scholar]
- Park Y, Lee JM, Hwang MY, Son GH, Geum D. NonO binds to the CpG island of oct4 promoter and functions as a transcriptional activator of oct4 gene expression. Mol Cells. 2013;35:61–69. doi: 10.1007/s10059-013-2273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AM, Hamazaki T, Hankowski KE, Terada N. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells. 2007;25:2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- Singh AM, Sun Y, Li L, Zhang W, Wu T, Zhao S, Qin Z, Dalton S. Cell-Cycle Control of Bivalent Epigenetic Domains Regulates the Exit from Pluripotency. Stem Cell Reports. 2015;5:323–336. doi: 10.1016/j.stemcr.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standaert L, Adriaens C, Radaelli E, Van Keymeulen A, Blanpain C, Hirose T, Nakagawa S, Marine JC. The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA. 2014;20:1844–1849. doi: 10.1261/rna.047332.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee WW, Shen SS, Oksuz O, Narendra V, Reinberg D. Erk1/2 activity promotes chromatin features and RNAPII phosphorylation at developmental promoters in mouse ESCs. Cell. 2014;156:678–690. doi: 10.1016/j.cell.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Padilla ME, Chambers I. Transcription factor heterogeneity in pluripotent stem cells: a stochastic advantage. Development. 2014;141:2173–2181. doi: 10.1242/dev.102624. [DOI] [PubMed] [Google Scholar]
- Vastenhouw NL, Schier AF. Bivalent histone modifications in early embryogenesis. Curr Opin Cell Biol. 2012;24:374–386. doi: 10.1016/j.ceb.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villasante A, Piazzolla D, Li H, Gomez-Lopez G, Djabali M, Serrano M. Epigenetic regulation of Nanog expression by Ezh2 in pluripotent stem cells. Cell Cycle. 2011;10:1488–1498. doi: 10.4161/cc.10.9.15658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, Tolstorukov MY, Kingston RE. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray J, Kalkan T, Smith AG. The ground state of pluripotency. Biochem Soc Trans. 2010;38:1027–1032. doi: 10.1042/BST0381027. [DOI] [PubMed] [Google Scholar]
- Yang YS, Hanke JH, Carayannopoulos L, Craft CM, Capra JD, Tucker PW. NonO, a non-POU-domain-containing, octamer-binding protein, is the mammalian homolog of Drosophila nonAdiss. Mol Cell Biol. 1993;13:5593–5603. doi: 10.1128/mcb.13.9.5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, Xu Y, Xu L, Yu X, Cheng J, Yang L, Chen S, Li Y. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer. 2014;14:693. doi: 10.1186/1471-2407-14-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.