Abstract
On record, there are 17 species in the Yersinia genus, of which three are known to be pathogenic to human. While the chromosomal and pYV (or pCD1) plasmid-borne virulence genes as well as pathogenesis of these three species are well studied, their genomic evolution is poorly understood. Our study aims to predict the key evolutionary events that led to the emergence of pathogenic Yersinia species by analyzing gene gain-and-loss, virulence genes, and “Clustered regularly-interspaced short palindromic repeats”. Our results suggest that the most recent ancestor shared by the human pathogenic Yersinia was most probably an environmental species that had adapted to the human body. This might have led to ecological specialization that diverged Yersinia into ecotypes and distinct lineages based on differential gene gain-and-loss in different niches. Our data also suggest that Y. pseudotuberculosis group might be the donor of the ail virulence gene to Y. enterocolitica. Hence, we postulate that evolution of human pathogenic Yersinia might not be totally in parallel, but instead, there were lateral gene transfer events. Furthermore, the presence of virulence genes seems to be important for the positive selection of virulence plasmid. Our studies provide better insights into the evolutionary biology of these bacteria.
Yersinia is a genus of Gram-negative bacteria consisting of at least 17 known species1. Among these, Y. pestis, Y. pseudotuberculosis and Y. enterocolitica are pathogenic to human, Y. ruckeri is pathogenic to salmonids2,3, while the other Yersinia species are apathogenic3. Both Y. pseudotuberculosis and Y. enterocolitica are enteropathogens that cause gastrointestinal infection and are distantly related to each other2. Y. pestis, which diverged from Y. pseudotuberculosis at least 2,000 years ago, can be transmitted by flea into the bloodstream of mammals, causing three pandemics of plague4.
The human pathogenic Yersinia species carry the virulence plasmid, called pYV in Y. enterocolitica or pCD1 in Y. pseudotuberculosis and Y. pestis, which encodes the Ysc-Yop type three secretion system (T3SS). T3SS allows pathogenic Yersinia to escape phagocytosis and takes control of the signaling systems of the host cells5. Other known virulence genes in the Yersinia species that cause pathogenesis are the chromosome-borne invasin (inv), the attachment-invasion gene (ail), pH 6 antigen and the virulence plasmid-borne yadA6. They encode proteins that mediate adhesion and entry into the host cell lining6.
While the virulence genes and pathogenesis of human pathogenic Yersinia are well studied, the evolution of the genus and emergence of pathogenic species are poorly understood5,6,7,8. A previous model proposed that all human pathogenic Yersinia descended from a pathogenic Yersinia, without regard to apathogenic species2. Later, other studies showed incongruence with the previous model, proposing that both Y. pseudotuberculosis group (comprising Y. pseudotuberculosis and Y. pestis) and Y. enterocolitica have evolved independently but acquired similar set of virulence genes9,10.
In view of the contradictory concepts, we further examine the evolution of Yersinia to elucidate (1) the role of the most recent ancestor shared by the human pathogenic species before their divergence, and (2) factors that mediate the acquisition of the virulence genes and virulence plasmid to transform into pathogenic species.
Results
Properties of Yersinia genomes
A total of 15 complete Yersinia genomes were used in this study (Supplementary Table 2). Six of them were human pathogenic strains. The size of these genomes ranged from approximately 3.7 Mbp to 4.9 Mbp, while the average GC content was about 47%. All Yersinia had seven rRNA operons, except Y. pestis CO92 which had six.
Yersinia phylogeny
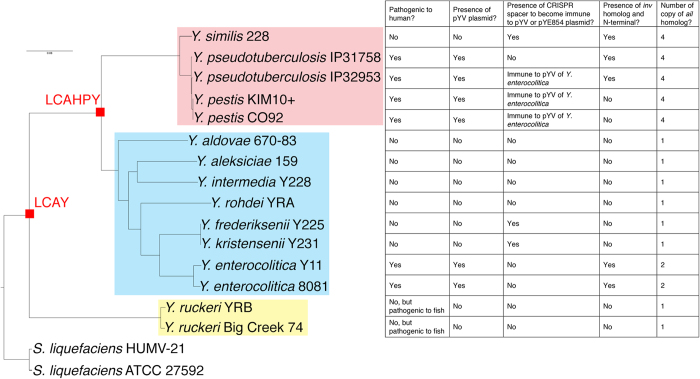
To study the phylogenetic relationship of the 15 Yersinia strains, we constructed a phylogenetic tree using a set of concatenated core protein coding sequences with 245,662 nucleotides. Our rooted supermatrix tree clearly showed that the Yersinia species could be clustered into three phylogroups that descended from Last Common Ancestor of all Yersinia (LCAY): phylogroup-P, phylogroup-E, and phylogroup-R (Fig. 1). Human pathogenic Y. pseudotuberculosis group (consisting of Y. pseudotuberculosis and Y. pestis) and Y. enterocolitica belonged to phylogroup-P and phylogroup-E respectively. Besides, the Y. pseudotuberculosis group and Y. enterocolitica appeared to be at the basal position of the supermatrix tree and closer to the apathogenic species in their respective phylogroups, suggesting that they might have evolved from different apathogenic populations (Fig. 1). We found that the gene content-based phylogenetic tree (Supplementary Fig. 1) had highly similar phyletic patterns with the supermatrix tree (Fig. 1), indicating that different genes are likely borne by the Yersinia species of different phylogroups. Thus, we suggest that lateral gene transfer is unlikely to be the major force in shaping the composition of Yersinia genomes11.
Figure 1. Yersinia supermatrix tree rooted using Serratia liquefaciens with 100 bootstrap value in every internal node.
Phylogroup-P, phylogroup-E, and phylogroup-R are highlighted in magenta, cyan, and yellow respectively. Last Common Ancestor of all Yersinia (LCAY) is hypothesized as the most recent hypothetical ancestor shared by all Yersinia species while Last Common Ancestor of Human Pathogenic Yersinia (LCAHPY) is hypothesized as the most recent hypothetical ancestor shared by human pathogenic Y. enterocolitica, Y. pseudotuberculosis, and Y. pestis. Important properties of each Yersinia genome are tabulated on the table to the right of supermatrix tree.
The average relative rate of recombination (R) to mutation (θ) of Yersinia genus was estimated to be R/θ = 0.011, mean DNA import length was δ = 603 base pair (bp), mean divergence of imported DNA was ν = 0.041. As R/θ was smaller than 1, mutation is likely a dominant occurrence in the genus, taking place at 90 (1 /0.011 = 90) times more often than recombination. It is possible that recombination across different species would decrease due to the increase of nucleotide divergence between Yersinia species12.
Gene gain-and-loss in Yersinia
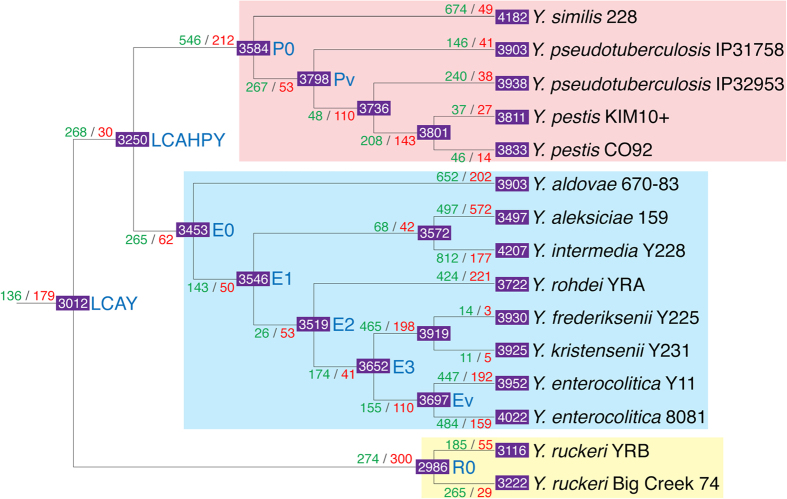
To understand how gene content of Yersinia changed since their emergence, we performed gene gain-and-loss analysis to identify acquired and lost genes. Reconstruction of gene gain-and-loss suggests that gene gain was dominant in the evolution of Yersinia (Fig. 2). In the next sections, we will discuss the hypothetical ancestors leading to the emergence of pathogenic Yersinia in more details.
Figure 2. Cladogram shows reconstruction of gene gain-and-loss in Yersinia.
Green, red, white color numbers indicate gene gain, gene loss and estimated number of gene respectively. Hypothetical ancestors of interest discussed in the main text are labelled in blue color text. Magenta, cyan and yellow backgrounds indicate phylogroup-P, phylogroup-E, and phylogroup-R respectively.
Emergence of LCAY
LCAY is considered the most recent hypothetical ancestor shared by all Yersinia species. LCAY might have preferred an aerobic environment due to the acquisition of aerobic citrate transporter genes (tctABCDE)13, and might have been able to extract heme from the host organism as indicated by the gain of heme receptor gene (hasR) and hemophore gene (hasA)14.
Our data showed that LCAY had lost the genes dsdAXC which are important for tolerance to D-serine, an anti-microbial compound abundant in the brain and urinary tract, which inhibits growth of enterohemorrhagic Escherichia coli15. Besides, 3-hydroxy-phenylacetate (3HPA) and 4HPA catabolism genes (hpaBCGEDHIAX) were lost, suggesting LCAY no longer used 3HPA and 4HPA in its new niche.
Emergence of R0 ancestor
R0 ancestor descended from LCAY and was the first hypothetical ancestor in phylogroup-R. R0 ancestor was found to gain several putative virulence loci including ysa-T3SS locus, yts1-type two secretion system (T2SS) locus, ent locus16,17. The ent locus consists of entABCES genes which synthesize ruckerbactin and are up regulated when Y. ruckeri infects fish18. R0 ancestor had also gained genes encoding for anti-sigma factor (rsbW) and anti-anti-sigma factor (rsbV) that play an important role in osmoprotection of Streptomyces coelicolor19, probably reflecting the importance of these genes to Y. ruckeri since it lives in freshwater.
It should be noted that Y. ruckeri has narrower niche as it mainly associates with and infects fishes3. This could explain our observation that several metabolic genes and transporters were lost in the R0 ancestor, probably because they were unneeded by Y. ruckeri in the more restricted niche. For instance, myo-inositol degradation genes (iolABCDEG) that encode enzymes to degrade myo-inositol, an abundant compound in soil, would probably be no longer useful in freshwater. Another locus, efeUOB encoding a ferrous transporter induced under acidic environment, was also lost in the R0 ancestor. This loss might have been due to the shift to freshwater which has more neutral pH20.
Emergence of LCAHPY
LCAHPY was the last common ancestor shared by human pathogenic Y. pseudotuberculosis/Y. pestis and Y. enterocolitica (Figs 1 and 2). We found that LCAHPY had acquired pgaABCD (poly-beta-1,6-N-acetyl-D-glucosamine synthesis and transport genes), pel and pelW (pectate lyases), togBANM and togT (oligogalacturonide transporter genes). Previous studies have shown that these genes allow human enteric pathogen, such as Escherichia coli EDL933, to persist and proliferate on vegetables21,22,23,24. Hence, the acquisition of pga, pel and tog loci suggests that LCAHPY may have the capability to grow on vegetables and be introduced into the human gastrointestinal tract after consumption of vegetables.
Besides the above-mentioned genes which facilitated survival outside human intestines, we found LCAHPY ancestor had also acquired yut and urtABCDE (urea transporter genes), yut and urtABCDE (nickel transporter genes), ureABCEFGD (urease genes). Previous study showed that these genes allowed Helicobacter pylori to colonize and cause infections in stomach, suggesting similar role in LCAHPY25. The survival of LCAHPY in human gastrointestinal tract could be further enhanced through the acquisition of lsrABCD (autoinducer-2 transporter genes) and lsrABCD (autoinducer-2 processing enzymes genes). Previous study proposed that enteric bacteria may use Lsr proteins to interrupt intercellular communication among competing bacterial cells26.
Emergence of E0, E1, E2, E3 ancestors
Both phylogroup-E and phylogroup-P descended from LCAHPY. Within phylogroup-E, many hypothetical ancestors (designated E0, E1, E2 and E3 in this study) existed before emergence of human pathogenic Y. enterocolitica (Fig. 2). We found that these hypothetical ancestors had acquired hyb and hyf loci (hydrogenase genes), cbiABCDETFGHJKLMNOQP and cobSTU (cobalamin biosynthesis genes), pduBCELPQW (1,2-propanediol degradation genes) and ttrABCRS locus (tetrathionate reduction genes). Previous study showed that these loci provided growth advantage to Salmonella enterica serotype Typhimurium in the gastrointestinal tract and to outcompete other enteric bacteria27. This suggests similar role of these acquired genes during emergence of phylogroup-E species. Besides, our data suggest that cellobiose is important to phylogroup-E as the ancestor had gained second copy of celABC (cellobiose phosphotransferase system).
Emergence of Ev ancestor
Ev was the most recent ancestor shared by human pathogenic Y. enterocolitica and it was a descendant of above-mentioned E0, E1 and E3 ancestors in phylogroup-E. Cellobiose seemed to be important to the lifestyle of Ev ancestor because we found Ev ancestor had acquired the third copy of celABC genes. The pyrimidine catabolic genes (rutRABCDEFG) were also acquired by Ev ancestor but their physiological role in bacteria is not yet understood. The absence of rut locus in all apathogenic species within phylogroup-E suggests that it might play an important role in the virulence traits of Y. enterocolitica. Most importantly, we found that the Ev ancestor had acquired pYV plasmids and several other virulence genes, such as mucoid Yersinia factor (Myf) genes and ail.
Emergence of P0 ancestor
P0 ancestor was the first hypothetical ancestor of phylogroup-P, as well as the direct descendant of LCAHPY. We found P0 ancestor had gained different types of metabolic genes compared to the phylogroup-E species. It gained tellurite resistance genes (terZABCD) and itaconate catabolic genes (ripABC), which had been shown by previous studies as adaptive strategies to survive inside macrophages28,29,30. Besides, P0 ancestor had gained several virulence genes, including pilWVUSRQPONML (type IV pilus gene cluster which resides in Yersinia adhesion pathogenicity island), psaABCEF (pH 6 antigen genes). All of these virulence genes had been shown to be important in pathogenicity of human pathogenic Yersinia31,32.
We found P0 had lost bcsGFE and bcsQABZC which are cellulose synthesis genes. A recent study had demonstrated that repression of cellulose biosynthesis in Salmonella when it was inside a macrophage could increase its virulence33. It is possible that the loss of cellulose biosynthesis genes and gain of itaconate (antimicrobial compound secreted by macrophage) catabolic genes could enhance survival of phylogroup-P species inside the macrophage.
Emergence of Pv ancestor
Pv was the most recent ancestor shared by human pathogenic Y. pseudotuberculosis and Y. pestis in phylogroup-P. We found that Pv ancestor had acquired mqsR and mqsA, which are a pair of toxin-antitoxin genes. Previous study has showed that mqsR and mqsA are the most highly up regulated gene in persistent E. coli cells and they regulated other physiological genes34. This suggests that the mqs toxin-antitoxin gene pair may be important for the pathogenic phylogroup-P species to overcome stresses from the host immune mechanisms.
Genes exclusive to pathogenic Yersinia
We attempted to search for genes exclusive to pathogenic Yersinia from different phylogroups. These genes included pYV (or pCD1 in Y. pseudotuberculosis and Y. pestis) virulence plasmid-borne yadA and ysc-yop T3SS, chromosomal ybt locus (yersiniabactin synthesis and transport system genes) and yts1-T2SS. Previous studies have demonstrated that both ybt and yts1-T2SS loci are important to highly human pathogenic Yersinia16,17,35.
inv and ail homologs within Yersinia
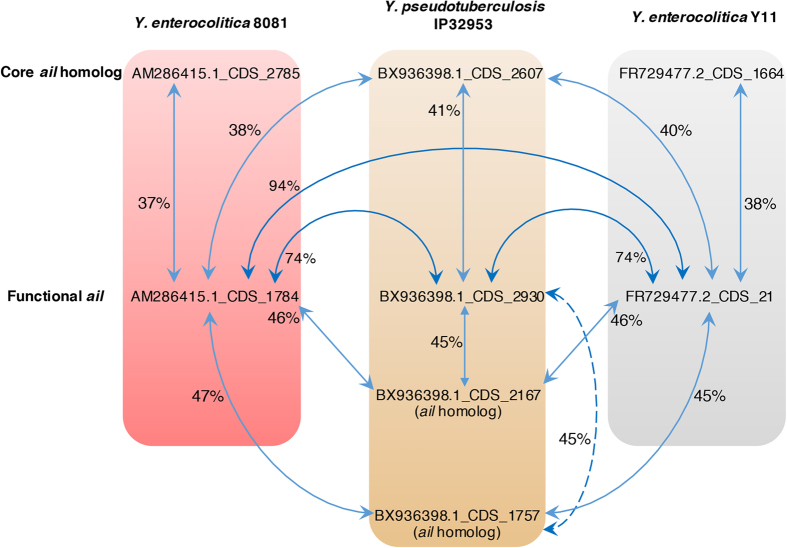
Both Ail and Inv are important virulence factors in human pathogenic Yersinia to mediate adhesion and invasion into host cells6. Therefore, we attempted to analyze ail and inv homologs in Yersinia. We found a total of 32 genes in Yersinia homologous to the functional ail from the pathogenic Y. pestis CO92, which we used as a reference gene for comparison in this analysis. BLASTP searches of all 32 ail homologs against the NCBI NR database showed that Yersinia species were always in the top significant hits for each homolog, suggesting that these putative homologs are likely from the Yersinia genus36. In another BLASTP search for functional ail of Y. enterocolitica 8081 against all 32 ail homologs of Yersinia, we found that phylogroup-P species were in the top significant hits (Supplementary Table 4). We further calculated the pairwise sequence identity between all functional ail genes and homologs from Yersinia. We found that the functional ail genes of Y. enterocolitica Y11 and 8081 were closer to the ail and ail homologs of the Y. pseudotuberculosis IP32953 (reference of phylogroup-P species) than their own ail homologs (Fig. 3). As the top hits returned by the BLAST program can be used to predict the donor of laterally transferred gene36, we thus hypothesize that the ail of Y. enterocolitica might be originated from the ail of Y. pseudotuberculosis, for example through lateral gene transfer.
Figure 3. Pairwise percentage of identity between ail protein sequences homologs of Y. pseudotuberculosis IP32953, Y. enterocolitica 8081 and Y11.
Pairwise relationships are indicated by blue double arrow pointing to two locus tags while percentage of identity is labeled next to the arrow.
Clustering of the 32 homologs based on protein sequence similarity clearly showed three separate gene clusters (Supplementary Table 3): Cluster 1 consists of known ail from both Y. pestis and Y. pseudotuberculosis and one ail-homolog from the Y. similis; the Cluster 2 consists of known ail from Y. enterocolitica and two ail-homologs from the Y. similis; Cluster 3 consists of the core ail-homologs present in all Yersinia. In the Cluster 1, we found that all pathogenic Yersinia species from the phylogroup-P had their own known ail with two copies of ail-homologs in each genome, suggesting that these genes are in-paralogs that were likely acquired through duplication events before the divergence of these species from their Pv ancestor.
We found that apathogenic Yersinia species generally had inv homologs. However, our data showed that there is a difference between inv homolog of apathogenic Yersinia and the functional inv of human pathogenic Yersinia. For instance, the aligned region between inv homologs of all apathogenic Yersinia (except Y. similis) and the functional inv of the pathogenic species did not start at first amino acid (Supplementary Table 5). We believe that this might account for different expression of the protein transcribed from inv homolog in apathogenic species as the N-terminal of Inv is required for proper localization in the outer membrane of Yersinia37.
CRISPR-Cas system in Yersinia
The CRISPR-Cas system is known to be a defense mechanism for bacteria to become immune to phage and plasmid38. Spacers which located in CRISPR array can provide resistance to foreign DNA if there is sequence homology between them. Analyses on the spacers found in Yersinia revealed that they could provide immunity against pYE854 conjugative and pYV virulence plasmid (Supplementary Table 6). pYE854 has been demonstrated to be self-transmissible, and is able to mobilize pYV plasmid39. The loss of CRISPR-Cas system in Y. enterocolitica suggests the event might be one of the key factors to acquire the pYV plasmid. Besides, apathogenic Y. similis 228 had spacers that were similar to virulence plasmids of pathogenic Y. enterocolitica and Y. pseudotuberculosis group. We suggest that these spacers inhibited the acquisition of pYV (or pCD1) plasmids by Y. similis after its divergence from P0 ancestor. The spacers in Y. pseudotuberculosis IP32953 and Y. pestis were found to be similar to the pYV plasmid from Y. enterocolitica (Supplementary Table 6), suggesting the phylogroup-P species could fragment and fail to maintain the pYV plasmid from Y. enterocolitica if the plasmid is transferred laterally.
Analyses of pYV (or pCD1) virulence plasmid
We further analyzed the pYV (or pCD1 in Y. pseudotuberculosis and Y. pestis) virulence plasmids because they are the key to pathogenesis in Yersinia species. The virulence plasmids encode for the hostile Ysc-Yop T3SS that takes over the host cell signaling system5. We found that the virulence plasmids borne by both phylogroup-P and phylogroup-E species had very high pairwise average nucleotide identity (ANI) values (e.g. >95%) and highly similar GC contents, suggesting that these virulence plasmids are likely closely related even though they were borne by different human pathogenic Yersinia species40 (Supplementary Tables 7 and 8).
The clustering of protein sequences encoded by these virulence plasmids showed that 53 out of a total of 128 gene families (<50%) were core genes or conserved across all species. The number of accessory genes and strain-specific genes in the plasmid gene families were 30 and 45 respectively, suggesting that these genes are not conserved across all pathogenic Yersinia species. The core genes mainly encoded for Ysc-Yop T3SS, whereas several transposase and integrase genes were found in the accessory gene pool. Among the strain-specific genes in the pYV plasmid, we found arsenic detoxification genes (arsCBRH) borne by Y. enterocolitica Y11, which is consistent with a previous study suggesting that the presence of the ars locus might be important for the spread of low pathogenic Y. enterocolitica41.
Discussions
Ecological speciation has been proposed to be a major mechanism in explaining diversification of prokaryotes, and lateral gene transfer is recognized as an important force to acquire niche-specific genes, yielding nascent populations in new niches42. In this study, the highly similar topology between the supermatrix tree (Fig. 1) and gene content phylogenetic tree (Supplementary Fig. 2) suggests that the lateral gene transfer between phylogroups might not be extensive11.
Our data suggest that LCAHPY, the most recent ancestor of human pathogenic Yersinia, had adapted to live in the human gastrointestinal tracts. This could be an important milestone in the evolution of Yersinia since the environment can provide a wide range of nutrients and niches to the bacterial populations, allowing subpopulations to exploit different food in new niches relative to the ancestral one. As a result, different metabolic genes have been gained and lost in the P0 and E0-3 ancestors throughout the evolution time42.
We found that the phylogroup-P species seemed to have expanded their ecological niche to macrophages, probably due to the acquisition of putative genes such as ter and rip loci, and the loss of two cellulose biosynthesis loci (bcs), which could also increase virulence inside the macrophages28,29,30,33. This might allow the phylogroup-P species to occupy the macrophage compared to its predecessor and phylogroup-E, which usually adapt to the intestinal tracts. This adaptation could be another efficient way to divide resources for utilization between the two different phylogroups and add weight to the ecological speciation.
During the ecological speciation process, the genetic recombination and gene flow between bacterial populations of different niches might still be possible, preventing them to diverge into distinct lineages42. However, in our estimation of the rate of recombination analysis, we clearly showed that the mutations play a major role in causing elevated nucleotide divergence in these Yersinia phylogroups. Therefore, this could be a barrier for sexual mating between these Yersinia species12.
Differentiation of sub-populations in response to new ecological niches may not fully explain nor justify the transformation into pathogenic species. Our analysis suggests that the loss of the CRISPR-Cas system might be critical in mediating the acquisition of the pYV (or pCD1) virulence plasmid in Yersinia. However, this might also introduce two enigmatic questions: (1) Most Yersinia species have lost the CRISPR-Cas system, but why do they have no virulence plasmids? (2) Some apathogenic Yersinia species had CRISPR-Cas system and spacer, but the spacer might be mutated and could decrease the efficiency of their CRISPR-Cas system. Will this allow the bacteria to acquire the virulence plasmid? Answers to these questions pertain to the redundancy of virulence plasmids in the human apathogenic Yersinia. For instance, Y. rukeri is known to be only pathogenic to salmonids and does not have the pYV plasmid. However, genes in the pYV (or pCD1) plasmids are usually induced at 37 °C5, but the salmon bodies do not reach such high temperature. In this case, the pYV is unlikely to be beneficial to and could be redundant or costly for the Y. ruckeri to bear it. Moreover, the pYV (or pCD1)-encoded Ysc-Yop T3SS proteins require direct physical contact between the bacteria and host cells for the effector proteins to be injected, and also require several virulence loci to assist in delivery of Yop proteins5,6. We found that none of the human apathogenic Yersinia had functional virulence genes, e.g. inv and ail. For example, although human apathogenic Yersinia species have inv homologs, but they are nonfunctional. It could be due to lack of proper N-terminal at the beginning of its protein product. Therefore, the apathogenic Yersinia are unlikely to be able to adhere to and invade the cell lining of the host if they accidentally acquire the virulence plasmids. If the physical contact and invasion are not established, the acquisition of virulence plasmid would be redundant for the human apathogenic species. In summary, we believe that the loss or mutation of CRISPR-Cas system might increase the chance of the acquisition of pYV (or pCD1) virulence plasmid by Yersinia species. However, to maintain the virulence plasmid, it must first be favored for selection because it is costly for bacteria to bear plasmid43. Thus, the presence of the important functional virulence genes, as well as the ability of Ysc-Yop T3SS to express at 37 °C environment could also become important factors determining the successful acquisition of pYV (or pCD1) virulence plasmid.
Our data support the view that gene duplication may play important evolutionary role in the ail of human pathogenic Yersinia. The ail genes in the pathogenic phylogroup-P species might have been aroused from gene duplication. Multiple copies of such ail paralogs might have rendered one (or some) of the duplicated genes to have weaker purifying selection and experienced multiple mutations44. This could have caused non-silent changes in the outer membrane receptor and increased efficiency in interaction between bacterial and mammalian receptors. As a result, neofunctionalization of paralog could have happened and facilitated the emergence of ail.
Our study suggests that there is a possibility of lateral transfer of the ail gene from Y. pseudotuberculosis to Y. enterocolitica, supported by the higher percentage of protein sequence identity between ail from Y. enterocolitica and ail (and ail homologs) from Y. pseudotuberculosis compared to ail homolog from Y. enterocolitica. To the best of our knowledge, pYV (or pCD1) virulence plasmid is only present in human pathogenic Yersinia, but not apathogenic Yersinia species. As our data clearly showed that the virulence plasmids borne by the human pathogenic Y. pseudotuberculosis and Y. enterocolitica are generally highly similar, they might have the same origin. Since both Y. pseudotuberculosis and Y. enterocolitica are distantly related to each other and do not share the same direct ancestor, we propose that the virulence plasmids might have been transferred laterally, for example, from the Y. pseudotuberculosis to Y. enterocolitica. We believe that the transfer of the virulence plasmid from the Y. enterocolitica to Y. pseudotuberculosis is unlikely to happen. It is because the spacer in the CRISPR array of the Y. pseudotuberculosis are highly similar to the spacer-recognized region in the pYV plasmid of Y. enterocolitica, therefore Y. pseudotuberculosis could recognize and fragment the pYV plasmid from the Y. enterocolitica.
Our study suggests that the evolution of human pathogenic Yersinia species might not be completely in parallel or independent to each other9,10, but instead, there might be also some lateral gene transfer events. The evolution of pathogenic Yersinia might reach another milestone when Y. pseudotuberculosis evolved into Y. pestis, which is transmitted by flea4. This breakthrough was accompanied by the acquisition of pFra and pPst plasmids in the Y. pestis. The pFra and pPst plasmids are known to be important for transmission of flea-borne infection rather than food-borne4. The pFra plasmid encodes ymt, which enables Y. pestis to survive inside flea and ensure successful transmission to the infected hosts, while the pPst plasmid encodes for a Pla protein, which is an important virulence factor that causes systematic dissemination after Y. pestis is injected subcutaneously4.
Last but not least, our data showed that the metabolism genes and virulence genes known to be involved in pathogenicity, are also conserved in the apathogenic Y. similis45. These genes (ripABC, terZABCD, pil locus, ail and inv homologs) are present in Y. pseudotuberculosis group and function to metabolize anti-microbial compounds (rip), persist in macrophages (ter) and contribute to pathogenicity29,30. The presence of these genes in the apathogenic Y. similis may make it prudent to monitor this species and its potential pathogenicity in different environmental situations in future.
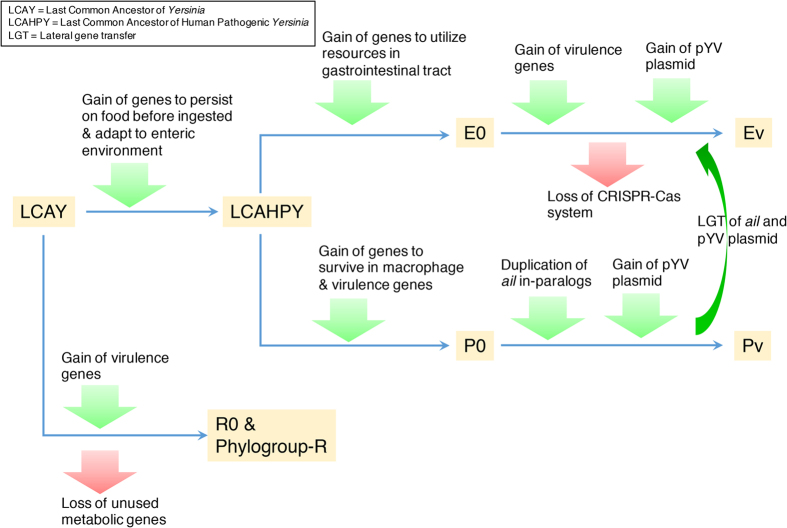
Based on our analyses, we hypothesize five possible main evolutionary events, in chorological order, to explain the emergence of human pathogenic Yersinia (Fig. 4).
LCAHPY might have, through gene gains, developed the ability to persist on food before being ingested, and adapted to gastrointestinal environment.
Diversification of Yersinia was likely to have occurred in new ecological niches by developing abilities to metabolize different nutrients available in different niches and body parts.
The gain of ail and inv genes might help the evolved species to adhere to and invade intestinal cell lining. Y. pseudotuberculosis might have gained ail through gene duplication and became donor of the gene to Y. enterocolitica.
Loss of CRISPR-Cas system and immunity against pYV virulence plasmid in some hypothetical ancestors.
Acquisition and maintenance of pYV virulence plasmid, followed by transformation into pathogenic species.
Figure 4. Proposed key evolutionary events that occurred in Yersinia and led to the emergence of pathogenic species.
All hypothetical ancestors are highlighted in orange color and correspond to node in Fig. 2. Green and red colors indicate gene gain and gene loss respectively.
Conclusion
Here we present an evolutionary study of human pathogenic Yersinia species. In contrast to previous studies9,10, we found that the evolution of the Y. enterocolitica and Y. pseudotuberculosis/Y. pestis might not be totally parallel. Instead, some of the virulence loci might have been transferred laterally from Y. pseudotuberculosis/Y. pestis to Y. enterocolitica. In summary, our study provides better insights into the evolution of human pathogenic Yersinia.
Method
Genome sequences and annotation
A total of 124 complete genome sequences of Enterobacteriaceae (including 15 Yersinia species) and 2 Haemophilus influenza were downloaded from National Center for Biotechnology Information (NCBI) database1. Details of Yersinia genomes are tabulated in Supplementary Table 1. For consistency, all genomes were annotated using Rapid Annotation using Subsystem Technology (RAST) online server to generate a list of open reading frames (ORFs) and protein sequences (see Supplementary Table 2 for summary)46. Then, the function of each protein sequence was predicted by using BLASTP to search for homolog in COG (E-value cutoff: 1E-5), KOBAS (default cutoff) and Virulence Factors Database (E-value cutoff: 1E-5) while HMMER was used to search against TIGRFAM47,48,49,50,51,52.
JSpecies was used to calculate average nucleotide identity (ANI) value between Yersinia pYV plasmids53.
Protein sequences clustering
All protein sequences were clustered thrice by using ProteinOrtho with default parameters54 (E-value cutoff: 1E-5; minimum percentage of identity: 25%; and minimum percentage of coverage: 50%). The first dataset consisted of all Enterobacteriaceae and Haemophilus influenza (hereinafter named the Enterobacteriaceae dataset), the second dataset consisted of Serratia liquefaciens and all Yersinia (hereinafter named the Yersinia-Serratia dataset) and the third dataset consisted of all Yersinia (hereinafter named the Yersinia dataset).
Multiple sequence alignment
Protein sequences of single copy core genes from all datasets were aligned using L-INS-i algorithm implemented in Multiple Alignment using Fast Fourier Transform (MAFFT) program55. Then, aligned protein sequences of each gene family were translated back to codon alignment using PAL2NAL56, and poorly aligned region was removed using GBlocks57.
Recombination testing
Codon alignments of the Yersinia-Serratia and Enterobacteriaceae datasets were used as input to PHI to test for recombination with 10,000 iterations and 0.05 as p-value cutoff 58. Next, alignments without recombination were concatenated together to form a “super-sequence” in the two dataset independently. ClonalFrameML was used to estimate rate of recombination to mutation in Yersinia dataset59.
Phylogenetic tree construction
Super-sequence of the Yersinia-Serratia dataset was used to infer phylogenetic trees using RAxML60, with maximum likelihood method, GTR + GAMMA model and 1,000 bootstrap iterations. Due to the large Enterobacteriaceae dataset, the Enterobacteriaceae phylogenetic tree was constructed by maximum likelihood in FastTree2 with 1,000 bootstrap iterations61. A matrix consisting of presence and absence of gene family in Yersinia-Serratia dataset was used to construct the gene content phylogenetic tree using neighbor-joining implemented in MEGA611,62.
Gene gain-and-loss analysis
Reconstruction of gene gain-and-loss in Yersinia genus was performed using Enterobacteriaceae dataset and COUNT with 1.5 as gain penalty63. Then, acquired and lost pathways and genes in ancestors of interest were inspected manually.
CRISPR analysis
CRISPR was predicted using CRT64. The spacer within CRISPR array was then searched against NCBI database using BLASTN to look for closely related plasmid sequence.
Virulence ail and inv genes analysis
Protein sequences of functional ail from Y. pestis CO92 and inv from Y. enterocolitica 8081 were used to search for their respective homologs in Yersinia using BLASTP47. The BLASTP outputs were further filtered by 1E-7 for E-value, 50% sequence completeness for subject and query sequences. All putative ail homologs were searched against NCBI NR database, and functional ail of Y. enterocolitica was searched against ail homologs of Yersinia using BLASTP47.
Additional Information
How to cite this article: Tan, S. Y. et al. Evolutionary study of Yersinia genomes deciphers emergence of human pathogenic species. Sci. Rep. 6, 36116; doi: 10.1038/srep36116 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This project is supported by High Impact Research (HIR) grant (Grant number: UM.C/625/HIR/MOHE/CHAN-08) from the Ministry of Higher Education of Malaysia.
Footnotes
Author Contributions S.Y.T. and S.W.C. conceived and designed the analyses. S.Y.T. and M.F.T. performed the analyses and collected data. S.Y.T., I.K.P.T., A.D. and S.W.C. wrote the manuscript. All authors read and approved the final manuscript.
References
- Benson D. A. et al. GenBank. Nucleic Acids Res 43, D30–D35, 10.1093/nar/gku1216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren B. W. The yersiniae—a model genus to study the rapid evolution of bacterial pathogens. Nat Rev Microbiol 1, 55–64, 10.1038/nrmicro730 (2003). [DOI] [PubMed] [Google Scholar]
- Sulakvelidze A. Yersiniae other than Y. enterocolitica, Y. pseudotuberculosis, and Y. pestis: the ignored species. Microbes Infect 2, 497–513 (2000). [DOI] [PubMed] [Google Scholar]
- Achtman M. et al. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci USA 96, 14043–14048 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G. R. The Yersinia Ysc-Yop ‘type III’ weaponry. Nat Rev Mol Cell Biol 3, 742–752, 10.1038/nrm932 (2002). [DOI] [PubMed] [Google Scholar]
- Mikula K. M., Kolodziejczyk R. & Goldman A. Yersinia infection tools-characterization of structure and function of adhesins. Front Cell Infect Microbiol 2, 169, 10.3389/fcimb.2012.00169 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. D. & Fetherston J. D. Yersinia pestis--etiologic agent of plague. Clin Microbiol Rev 10, 35–66 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottone E. J. Yersinia enterocolitica: the charisma continues. Clin Microbiol Rev 10, 257–276 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter S. et al. Parallel independent evolution of pathogenicity within the genus Yersinia. Proc Natl Acad Sci USA 111, 6768–6773, 10.1073/pnas.1317161111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally A., Thomson N. R., Reuter S. & Wren B. W. ‘Add, stir and reduce’: Yersinia spp. as model bacteria for pathogen evolution. Nat Rev Microbiol 14, 177–190, 10.1038/nrmicro.2015.29 (2016). [DOI] [PubMed] [Google Scholar]
- Snel B., Bork P. & Huynen M. A. Genome phylogeny based on gene content. Nat Genet 21, 108–110, 10.1038/5052 (1999). [DOI] [PubMed] [Google Scholar]
- Majewski J., Zawadzki P., Pickerill P., Cohan F. M. & Dowson C. G. Barriers to genetic exchange between bacterial species: Streptococcus pneumoniae transformation. J Bacteriol 182, 1016–1023 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocker M., Schaffer S., Mack C. & Bott M. Citrate utilization by Corynebacterium glutamicum is controlled by the CitAB two-component system through positive regulation of the citrate transport genes citH and tctCBA. J Bacteriol 191, 3869–3880, 10.1128/JB.00113-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letoffe S., Nato F., Goldberg M. E. & Wandersman C. Interactions of HasA, a bacterial haemophore, with haemoglobin and with its outer membrane receptor HasR. Mol Microbiol 33, 546–555 (1999). [DOI] [PubMed] [Google Scholar]
- Connolly J. P. et al. A Highly Conserved Bacterial D-Serine Uptake System Links Host Metabolism and Virulence. PLoS Pathog 12, e1005359, 10.1371/journal.ppat.1005359 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller J. C., Carlson S., Pederson K. J. & Pierson D. E. A chromosomally encoded type III secretion pathway in Yersinia enterocolitica is important in virulence. Mol Microbiol 36, 1436–1446 (2000). [DOI] [PubMed] [Google Scholar]
- Iwobi A. et al. Novel virulence-associated type II secretion system unique to high-pathogenicity Yersinia enterocolitica. Infect Immun 71, 1872–1879 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez L., Marquez I. & Guijarro J. A. Identification of specific in vivo-induced (ivi) genes in Yersinia ruckeri and analysis of ruckerbactin, a catecholate siderophore iron acquisition system. Appl Environ Microbiol 70, 5199–5207, 10.1128/AEM.70.9.5199-5207.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. J., Cho Y. H., Kim H. S., Ahn B. E. & Roe J. H. Regulation of sigmaB by an anti- and an anti-anti-sigma factor in Streptomyces coelicolor in response to osmotic stress. J Bacteriol 186, 8490–8498, 10.1128/JB.186.24.8490-8498.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Woodhall M. R., Alvarez J., Cartron M. L. & Andrews S. C. EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in Escherichia coli O157:H7. Mol Microbiol 65, 857–875, 10.1111/j.1365-2958.2007.05802.x (2007). [DOI] [PubMed] [Google Scholar]
- Yaron S. & Romling U. Biofilm formation by enteric pathogens and its role in plant colonization and persistence. Microb Biotechnol 7, 496–516, 10.1111/1751-7915.12186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy C. et al. Modes of action of five different endopectate lyases from Erwinia chrysanthemi 3937. J Bacteriol 181, 3705–3709 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux-Cotte-Pattat N. & Reverchon S. Two transporters, TogT and TogMNAB, are responsible for oligogalacturonide uptake in Erwinia chrysanthemi 3937. Mol Microbiol 41, 1125–1132 (2001). [DOI] [PubMed] [Google Scholar]
- Yamazaki A. et al. Commensal effect of pectate lyases secreted from Dickeya dadantii on proliferation of Escherichia coli O157:H7 EDL933 on lettuce leaves. Appl Environ Microbiol 77, 156–162, 10.1128/AEM.01079-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley H. L. The role of Helicobacter pylori urease in the pathogenesis of gastritis and peptic ulceration. Aliment Pharmacol Ther 10 Suppl 1, 57–64 (1996). [DOI] [PubMed] [Google Scholar]
- Xavier K. B. et al. Phosphorylation and processing of the quorum-sensing molecule autoinducer-2 in enteric bacteria. ACS Chem Biol 2, 128–136, 10.1021/cb600444h (2007). [DOI] [PubMed] [Google Scholar]
- Rohmer L., Hocquet D. & Miller S. I. Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol 19, 341–348, 10.1016/j.tim.2011.04.003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnusamy D. & Clinkenbeard K. D. Role of Tellurite Resistance Operon in Filamentous Growth of Yersinia pestis in Macrophages. PLoS One 10, e0141984, 10.1371/journal.pone.0141984 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnusamy D., Hartson S. D. & Clinkenbeard K. D. Intracellular Yersinia pestis expresses general stress response and tellurite resistance proteins in mouse macrophages. Vet Microbiol 150, 146–151, 10.1016/j.vetmic.2010.12.025 (2011). [DOI] [PubMed] [Google Scholar]
- Sasikaran J., Ziemski M., Zadora P. K., Fleig A. & Berg I. A. Bacterial itaconate degradation promotes pathogenicity. Nat Chem Biol 10, 371–377, 10.1038/nchembio.1482 (2014). [DOI] [PubMed] [Google Scholar]
- Collyn F. et al. Yersinia pseudotuberculosis harbors a type IV pilus gene cluster that contributes to pathogenicity. Infect Immun 70, 6196–6205 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Merriam J. J., Mueller J. P. & Isberg R. R. The psa locus is responsible for thermoinducible binding of Yersinia pseudotuberculosis to cultured cells. Infect Immun 64, 2483–2489 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes M. H., Lee E. J., Choi J. & Groisman E. A. Salmonella promotes virulence by repressing cellulose production. Proc Natl Acad Sci USA 112, 5183–5188, 10.1073/pnas.1500989112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B. L. et al. Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog 5, e1000706, 10.1371/journal.ppat.1000706 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert S., Rakin A. & Heesemann J. The Yersinia high-pathogenicity island (HPI): evolutionary and functional aspects. Int J Med Microbiol 294, 83–94, 10.1016/j.ijmm.2004.06.026 (2004). [DOI] [PubMed] [Google Scholar]
- Ravenhall M., Skunca N., Lassalle F. & Dessimoz C. Inferring horizontal gene transfer. PLoS Comput Biol 11, e1004095, 10.1371/journal.pcbi.1004095 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong J. M., Fournier R. S. & Isberg R. R. Identification of the integrin binding domain of the Yersinia pseudotuberculosis invasin protein. EMBO J 9, 1979–1989 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft D. H., Selengut J., Mongodin E. F. & Nelson K. E. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol 1, e60, 10.1371/journal.pcbi.0010060 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerl J. A., Klein I., Lanka E., Appel B. & Hertwig S. Genetic and functional properties of the self-transmissible Yersinia enterocolitica plasmid pYE854, which mobilizes the virulence plasmid pYV. J Bacteriol 190, 991–1010, 10.1128/JB.01467-07 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfield J., Fram N. R. & Ely B. Across bacterial phyla, distantly-related genomes with similar genomic GC content have similar patterns of amino acid usage. PLoS One 6, e17677, 10.1371/journal.pone.0017677 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyt C., Iriarte M., Thi V. H. & Cornelis G. R. Virulence and arsenic resistance in Yersiniae. J Bacteriol 179, 612–619 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassalle F., Muller D. & Nesme X. Ecological speciation in bacteria: reverse ecology approaches reveal the adaptive part of bacterial cladogenesis. Res Microbiol 166, 729–741, 10.1016/j.resmic.2015.06.008 (2015). [DOI] [PubMed] [Google Scholar]
- San Millan A. et al. Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat Commun 5, 5208, 10.1038/ncomms6208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov F. A., Rogozin I. B., Wolf Y. I. & Koonin E. V. Selection in the evolution of gene duplications. Genome Biol 3, RESEARCH0008 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague L. D. & Neubauer H. Genome Sequence of Yersinia similis Y228T, a Member of the Yersinia pseudotuberculosis Complex. Genome Announc 2, 10.1128/genomeA.00216-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R. K. et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75, 10.1186/1471-2164-9-75 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. J Mol Biol 215, 403–410, 10.1016/S0022-2836(05)80360-2 (1990). [DOI] [PubMed] [Google Scholar]
- Galperin M. Y., Makarova K. S., Wolf Y. I. & Koonin E. V. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43, D261–D269, 10.1093/nar/gku1223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C. et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39, W316–W322, 10.1093/nar/gkr483 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xiong Z., Sun L., Yang J. & Jin Q. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res 40, D641–D645, 10.1093/nar/gkr989 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S. R. A new generation of homology search tools based on probabilistic inference. Genome Inform 23, 205–211 (2009). [PubMed] [Google Scholar]
- Haft D. H., Selengut J. D. & White O. The TIGRFAMs database of protein families. Nucleic Acids Res 31, 371–373 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M. & Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106, 19126–19131, 10.1073/pnas.0906412106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M. et al. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics 12, 124, 10.1186/1471-2105-12-124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K. & Standley D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30, 772–780, 10.1093/molbev/mst010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M., Torrents D. & Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34, W609–W612, 10.1093/nar/gkl315 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G. & Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56, 564–577, 10.1080/10635150701472164 (2007). [DOI] [PubMed] [Google Scholar]
- Bruen T. C., Philippe H. & Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics 172, 2665–2681, 10.1534/genetics.105.048975 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X. & Wilson D. J. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 11, e1004041, 10.1371/journal.pcbi.1004041 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313, 10.1093/bioinformatics/btu033 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S. & Arkin A. P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490, 10.1371/journal.pone.0009490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30, 2725–2729, 10.1093/molbev/mst197 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csuros M. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 26, 1910–1912, 10.1093/bioinformatics/btq315 (2010). [DOI] [PubMed] [Google Scholar]
- Bland C. et al. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 8, 209, 10.1186/1471-2105-8-209 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.