Significance
DNA damage can lead to chromosome instability and loss of genetic information, resulting in cell death or diseases such as cancer. One source of damage is DNA:RNA hybrids that form when RNA transcripts hybridize to homologous sequences in the genome. Cells from all branches of life possess two enzymes that can degrade DNA:RNA hybrids: RNase H1 and RNase H2. In this study we explore the differential functions of the RNase H1 and H2 in preventing chromosome instability in the model organism Saccharomyces cerevisiae. We present evidence that RNase H2 acts on hybrids genome-wide to prevent chromosome instability. In contrast, RNase H1 acts at a subset of hybrids to repress locus-restricted chromosome instability.
Keywords: RNase H, R-loops, DNA:RNA hybrids, chromosome instability, genome instability
Abstract
DNA:RNA hybrids can lead to DNA damage and genome instability. This damage can be prevented by degradation of the RNA in the hybrid by two evolutionarily conserved enzymes, RNase H1 and H2. Indeed, RNase H-deficient cells have increased chromosomal rearrangements. However, the quantitative and spatial contributions of the individual enzymes to hybrid removal have been unclear. Additionally, RNase H2 can remove single ribonucleotides misincorporated into DNA during replication. The relative contribution of DNA:RNA hybrids and misincorporated ribonucleotides to chromosome instability also was uncertain. To address these issues, we studied the frequency and location of loss-of-heterozygosity (LOH) events on chromosome III in Saccharomyces cerevisiae strains that were defective for RNase H1, H2, or both. We showed that RNase H2 plays the major role in preventing chromosome III instability through its hybrid-removal activity. Furthermore, RNase H2 acts pervasively at many hybrids along the chromosome. In contrast, RNase H1 acts to prevent LOH within a small region of chromosome III where the instability is dependent upon two hybrid-prone sequences. This restriction of RNase H1 activity to a subset of hybrids is not the result of its constrained localization, because we found it at hybrids genome-wide. This result suggests that the genome-protection activity of RNase H1 is regulated at a step after hybrid recognition. The global function of RNase H2 and the region-specific function of RNase H1 provide insight into why these enzymes with overlapping hybrid-removal activities have been conserved throughout evolution.
Preventing chromosome instability is an essential process for maintaining genetic information. A source of chromosome instability is the accumulation of R-loops, which form when an RNA molecule hybridizes with a portion of genomic DNA, creating a DNA:RNA hybrid and a displaced single-stranded DNA (reviewed in ref. 1). One mechanism to prevent hybrid-mediated damage involves RNase H1 and RNase H2, two endogenous enzymes, conserved from bacteria to humans, that can degrade the RNA in R-loops (reviewed in ref. 2). RNase H2 also functions in the removal of single ribonucleotides that are inappropriately incorporated into DNA by DNA polymerases during replication. Why RNase H1 and H2, which appear to have overlapping functions, remain highly conserved across many branches of life has been an outstanding question. Two areas of inquiry that will help address this conundrum are (i) does one of these RNases carry the major burden of preventing spontaneous R-loop–mediated chromosome instability, and, if so which, and (ii) do RNase H1 and H2 protect the same or different regions of the genome from R-loop–mediated damage?
Whether RNase H1 and H2 contribute differentially to protecting against hybrid-mediated genome instability has been controversial. Studies of the inactivation of RNase H1 in yeast have shown little effect on chromosome instability (3, 4), whereas inactivation of RNase H2 has been shown to increase chromosome instability (4, 5). However, results from the Conover study (5) suggested that the elevated instability in the RNase H2-deficient cells is caused by elevated misincorporation of ribonucleotides rather than by the failure to remove hybrids. In contrast, results from the O’Connell study (4) suggested the opposite: that the elevated instability is caused by hybrids rather than by misincorporated ribonucleotides. The failure in these studies to reveal a prominent role for RNase H1 in protecting against hybrid-mediated chromosome instability also was surprising. Previous studies had shown that RNase H1, when constitutively overexpressed, can suppress genome-wide hybrid formation and hybrid-mediated genome instability induced by mutations in the RNA biogenesis machinery (3, 6, 7). These results suggested that RNase H1 had the ability to remove many hybrids within the cell but could do so only under artificial conditions of constitutive overexpression, not addressing the roles that RNase H1 and H2 play in physiological conditions.
Whether RNase H1 and H2 protect different regions of the genome from hybrid-mediated instability also remains unanswered. A recent study mapped the mitotic recombination events genome-wide in cells deficient for both RNase H1 and H2 (4). These mitotic recombination events presumably marked the sites of repair from damage induced by RNase H deficiency. No correlation was observed between the positions of these mitotic events and the positions of R-loop–prone regions that had been defined by a genome-wide tiling array map (8). This failure in correlation could be attributed both to the low number of events resulting from the genome-wide nature of study and to the low resolution of the R-loop map.
In this study we have used a number of strategies and tools to address these questions. First we used diploid strains of Saccharomyces cerevisiae deficient for RNase H1 (rnh1∆), RNase H2 (rnh201∆), or both (rnh1∆ rnh201∆) with different markers on the two copies of chromosome III that allowed quantitative analyses of loss-of-heterozygosity (LOH) events. These strains also allowed the mapping of the junctions of LOH as a means to localize sites of damage to potential hybrid-prone regions. We also exploited a new high-resolution map of hybrid-prone regions throughout the genome (9) to identify hybrids that are potentially causative for LOH. With these tools, we showed that RNase H2 is the predominant effector, through its hybrid-removal activity, for preventing hybrid-induced instability at many distinct sites on chromosome III. Moreover, the existence of distinct sites suggests that many hybrids are capable of inducing damage. In contrast, we showed that RNase H1 acts preferentially to prevent hybrid-mediated instability within a single mapping interval. This instability is correlated specifically with two hybrid-prone sequences within the region. Further analyses of RNase H1 localization suggests that this enzyme binds to hybrids within this interval as well as to most, if not all, hybrids in the genome. This result suggests that RNase H1 specificity for hybrid removal occurs at a step after hybrid recognition. Thus, the hybrid-removal activities of RNases H1 and H2 have distinct quantitative and spatial functions in vivo.
Results
RNase H Deficiency Increases LOH.
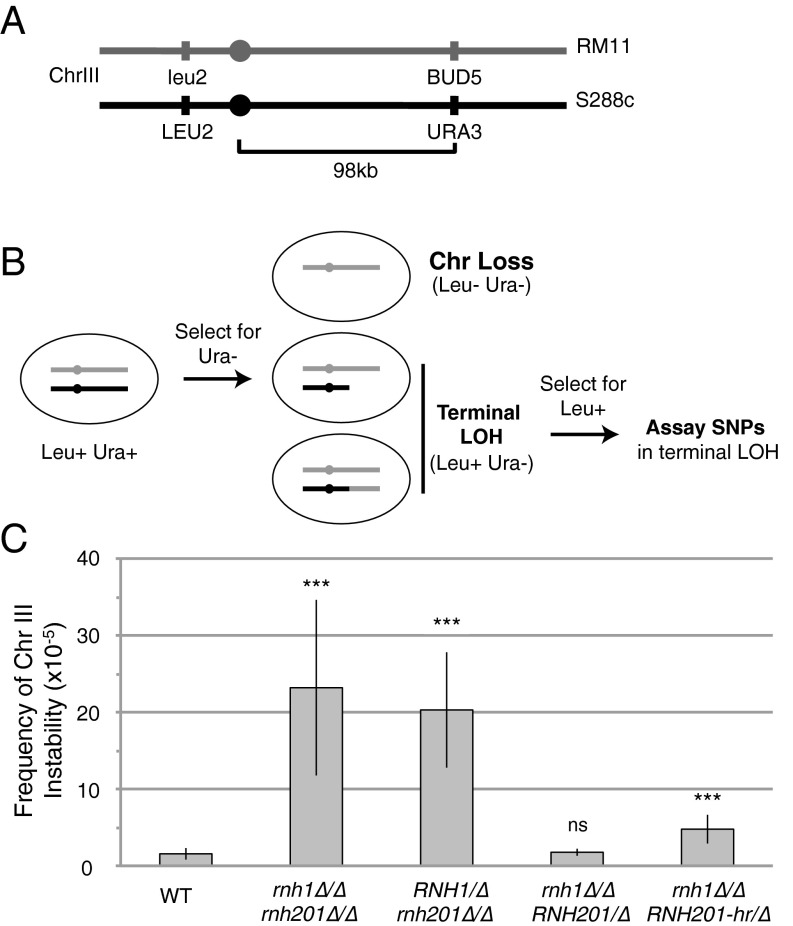
To investigate the links among RNase H1 and H2, DNA:RNA hybrids, and chromosome instability, we designed an assay to measure LOH in wild-type and RNase H-deficient diploid yeast. We marked the right arm of one homolog of chromosome III with the URA3 gene and the left arm with LEU2 (Fig. 1A). These genetic markers allowed us to monitor LOH from both arms of the chromosome (Fig. 1B). We report the frequency of LOH as the fraction of 5-fluoroorotic acid (5-FOA)–resistant colonies compared with the total number of viable cells, thus providing an approximation of the rate of LOH (Materials and Methods).
Fig. 1.
Assay for LOH of chromosome III in RNase H mutants. (A) Diagram of chromosome III genetic markers relevant for the LOH assay. (B) Schematic of the workflow of the LOH assay and possible outcomes for chromosome III. The marked S288c-derived chromosome is diagramed in black, and the RM11-derived chromosome is diagrammed in gray. Diploid cells (Leu+ Ura+) are propagated on medium lacking uracil. Individual colonies then are plated onto medium containing 5-FOA, selecting for loss of the URA3 marker. Resultant colonies may have complete chromosome loss or terminal LOH, shown here as a de novo telomere addition and a recombination repair event. Colonies then are replica plated to medium lacking leucine to select for Leu+ colonies. The SNPs of resultant colonies are assayed for heterozygosity by Sanger sequencing. (C) Frequency of LOH in wild-type cells and RNase H mutants. The mean of 40 parent colonies is shown with error bars indicating ±1 SD. Statistical analysis comparing mutants to wild-type using an unpaired t test: ***P < 0.001; ns, not significant at P < 0.1.
Using this assay, we investigated the chromosome instability phenotypes of wild-type and RNase H mutants. Deficiency of both RNase H1 and H2 (rnh1∆/∆ rh201∆/∆) elevated chromosome instability 14.9-fold over wild-type levels (Fig. 1C). These events in both wild-type and RNase H-deficient cells were about half terminal LOH and half chromosome loss (Fig. S1). These results corroborate previous reports that deletion of both RNase H1 and H2 elevates chromosome instability (3, 4). We then generated RNase H single mutants by taking the double mutant and adding back a single copy of either RNH1 or RNH201 to its endogenous locus to confirm linkage between the observed phenotypes and the presence or absence of a specific RNase H. A strain deficient in RNase H2 but harboring a wild-type copy of RNH1 (RNH1/∆ rnh201∆/∆) showed instability levels only slightly lower than those in the double mutant (Fig. 1C). In contrast, a strain deficient in RNase H1 but harboring one wild-type copy of RNH201 (rnh1∆/∆ RNH201/∆) showed instability at wild-type levels (Fig. 1C). Therefore, the majority of the chromosome instability phenotype in RNase H-deficient cells is caused by the lack of RNase H2.
Fig. S1.
Percent terminal LOH. The percent of colonies that underwent LOH that had terminal LOH events (Leu+, Ura−) as opposed to whole chromosome loss (Leu−, Ura−) in wild-type and RNase H mutants.
RNase H2 can perform two enzymatic activities: removal of R-loops and removal of single ribonucleotides misincorporated into DNA. The relative contribution of these activities to the maintenance of chromosome stability has remained unclear. To address this controversy, we used a mutant of Rnh201 that lacks the ability to remove single ribonucleotides but retains the ability to degrade R-loops (10). This separation-of-function allele (RNH201-P45D,Y219A) was shown to have undetectable levels of single ribonucleotide removal activity in multiple in vitro and in vivo assays. Here, we refer to this allele as “RNH201-hr” (hybrid removal) because it retains hybrid-removal activity. In strains harboring a copy of RNH201-hr at the endogenous RNH201 locus (rnh1∆/∆ RNH201-hr/∆), chromosome III instability was reduced to just threefold over wild-type levels (Fig. 1C). This suppression indicates that hybrids are the major cause of the LOH events and that the hybrid-removal activity of RNase H2 is the main protector against hybrid-mediated chromosome instability.
Mapping LOH Events in RNase H-Deficient Strains.
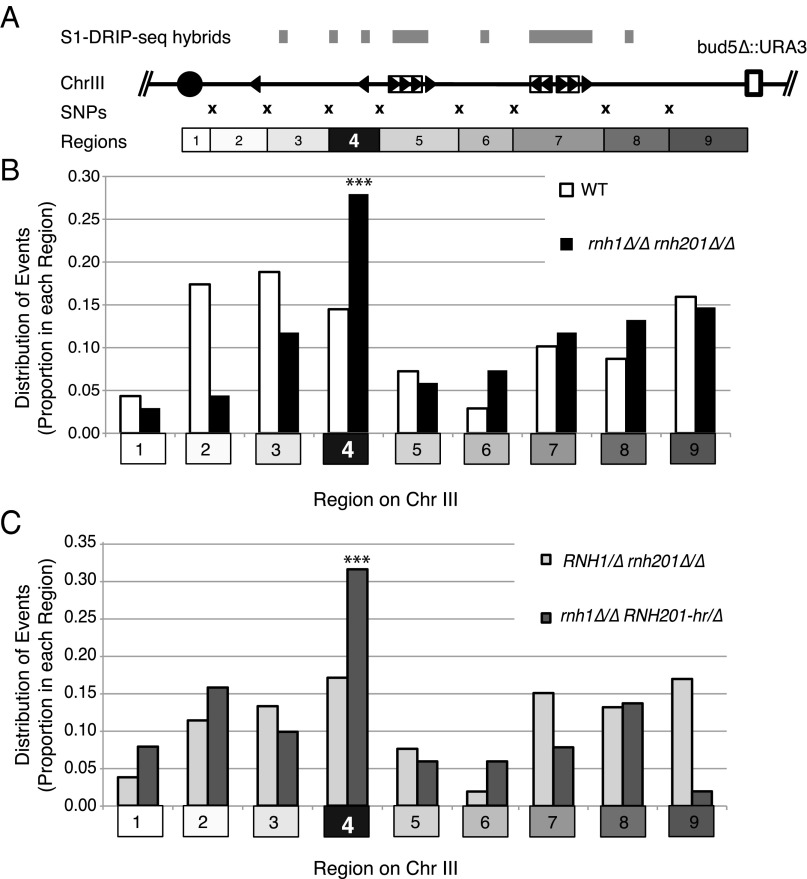
We previously mapped hybrid-prone regions genome-wide in wild-type and RNase H mutants of haploid S288c yeast using a high-resolution technique termed “S1 nuclease DNA:RNA immunoprecipitation with deep sequencing” (S1-DRIP-seq) (9) and identified hybid-prone regions between CEN3 and our URA3 marker, a 98-kb region of the right arm of chromosome III (Fig. 2A). These hybrid-prone regions were candidates for causing chromosome III LOH induced by RNase H deficiency. To assess the potential implication of these hybrid regions in LOH events, we asked three related questions: (i) Did the pattern of LOH in this region suggest that multiple hybrids were capable of inducing LOH events? (ii) Were some hybrids more likely to induce LOH events? (iii) Did RNase H1 and H2 play different roles in protecting against hybrid-induced LOH?
Fig. 2.
Distribution of LOH junctions. (A) Diagram of the region of the right arm of chromosome III assayed for LOH junctions. The first row shows the locations of hybrid-prone sequences mapped in ref. 9. The second line shows chromosome III with the centromere diagramed as a circle, Ty elements diagrammed as boxed triangles, solo delta elements as triangles, and the location of the URA3 marker inserted at the BUD5 locus shown as a square. The third row shows the locations of the SNPs (marked as “X”) assayed for heterozygosity. The fourth row shows the nine regions in which LOH junctions may occur. (B and C) Locations of LOH boundaries. Boundaries in wild-type cells (n = 69) and RNase H double-mutant cells (n = 68) (B) and RNase H2-deficient cells (n = 53) and H1-deficient cells (n = 51) (C) were mapped. The proportion of LOH boundaries occurring in each of the nine regions is plotted. ***P < 0.001; χ2 test.
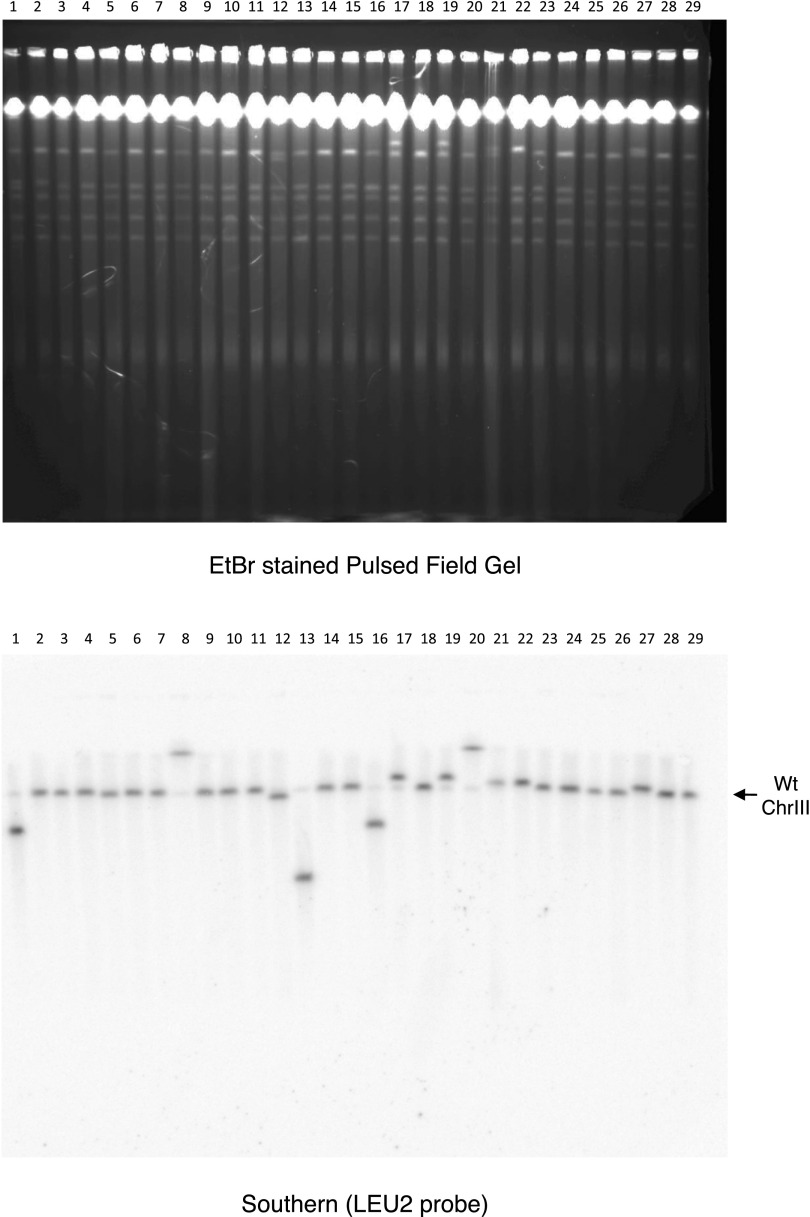
To map the LOH events on the right arm of chromosome III, we constructed diploids deriving one set of parental chromosomes from the haploid S288c strain used to map hybrids and the second set of parental chromosomes from the haploid RM11 strain. The RM11 strain differed from the S288c strain by about 46,000 SNPs (11). We first identified progeny that had undergone terminal LOH on the right arm of chromosome III by their growth phenotype (Ura− Leu+) (Fig. 1B). We then mapped the LOH in these progeny by monitoring the heterozygosity of the SNPs along the chromosome arm at ∼10-kb intervals (Fig. 2A). The junctions between the regions of retention of heterozygosity and LOH marked the sites of resolution of damage induced by RNase H deficiency, i.e., sites of crossover, initiation of break-induced replication, or de novo telomere addition. Indeed, when we examined the size of the chromosomes from colonies with terminal LOH events, many had a wild-type karyotype consistent with LOH by a mechanism of homologous recombination, but in some colonies chromosome III was smaller or larger, indicating the occurrence of de novo telomere addition and other complex chromosome rearrangements (Fig. S2).
Fig. S2.
Chromosome III size in colonies that have undergone terminal LOH. (Upper) An ethidium bromide-stained pulsed-field electrophoresis gel. (Lower) A Southern blot of that gel with a probe against LEU2, which probes for the S288c-derived chromosome III homolog. Chromosomes are shown for wild-type (lanes 1–2 and 27–28), rnh1∆/∆ rnh201∆/∆ (lanes 3–12 and 25–26), rnh1∆/∆ RNH201/∆ (lanes 13–17 and 21–24), and RNH1/∆ rnh201∆/∆ (lanes 17–20). The parental size of chromosome III in a non-rearranged cell is shown in the rightmost lane 29.
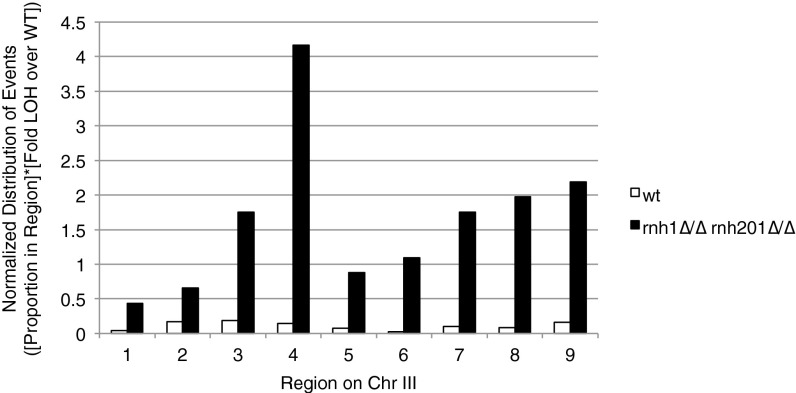
The junctions of spontaneous LOH events in the wild-type strain were distributed along the 98-kb segment, with no region showing any more or any fewer events than would be predicted by the interval length (χ2 test) (Fig. 2B, white bars). Although the intervals are ∼10 kb, the length between intervals varies, and the distribution of events in the wild-type strain did not differ significantly from the expected number in each interval. In contrast, the junctions of LOH events mapped in the RNase H-deficient strain (rnh1∆/∆ rh201∆/∆) were not uniformly distributed. Junctions were overrepresented in region 4, occurring at a higher proportion than would be expected by the length of the interval (P < 0.001, χ2 test) (Fig. 2B, black bars). However, this hotspot represented only 25% of the total LOH events, indicating that most junctions of LOH occurred in other intervals in rnh1∆ rnh201∆ cells. The overall frequency of LOH was 14.9-fold higher in the RNase H-deficient cells than in wild-type cells, so LOH events were elevated in all the intervals and were even elevated in interval 4. The distribution of events normalized to the overall frequency of LOH is shown in Fig. S3.
Fig. S3.
Normalized distribution of LOH boundaries. The distribution of LOH boundary events in each SNP-defined interval was normalized to the fold frequency of LOH in RNase H-deficient cells over wild-type cells.
To understand the contributions of the individual loss of Rnh1 and Rnh201 to the pattern of LOH on chromosome III, we examined the pattern of LOH in rnh201∆ and rnh1∆ single mutants. Unlike the RNase H double mutants, strains deficient in RNase H2 (RNH1/∆ rnh201∆/∆) showed a more uniform distribution of events, similar to the distribution in the wild-type strain (Fig. 2C, light gray bars). Given that the frequency of LOH in these cells was 13-fold higher than in wild-type cells, this result indicates that loss of Rnh201 led to the induction of damage in most, if not all, regions on chromosome III. Because hybrid-forming regions were dispersed along the chromosome, these results are consistent with multiple hybrid-prone regions inducing the damage that led to LOH and with RNase H2 suppressing this damage by removing these hybrids.
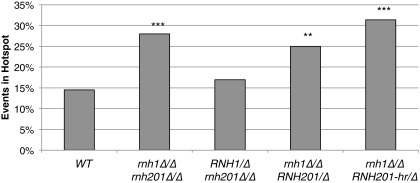
In contrast, cells lacking RNH1 but expressing RNH201-hr (rnh1∆/∆ RNH201-hr/∆) had a hotspot of LOH junctions mapping to region 4, similar to the hotspot in the RNase H double mutant (Fig. 2C, dark gray bars). Cells lacking RNH1 but expressing a wild-type copy of RNH201 (rnh1∆/∆ RNH201/∆) also had a hotspot in region 4 (Fig. S4). Therefore, the hotspot of junctions in region 4 was dependent on RNase H1 but not on RNase H2. These results demonstrate a specific role of RNase H1 in targeting chromosome instability at a particular region of the chromosome.
Fig. S4.
LOH boundary events in the region 4 hotspot. The percent of LOH boundary events in the region 4 hotspot in wild-type and RNase H mutants. RNase H1-deficient cells with either wild-type RNH201 or RNH201-hr display an instability hotspot. ***P < 0.001; **P < 0.01; χ2 test.
Defining Instability-Prone Hybrid-Forming Regions.
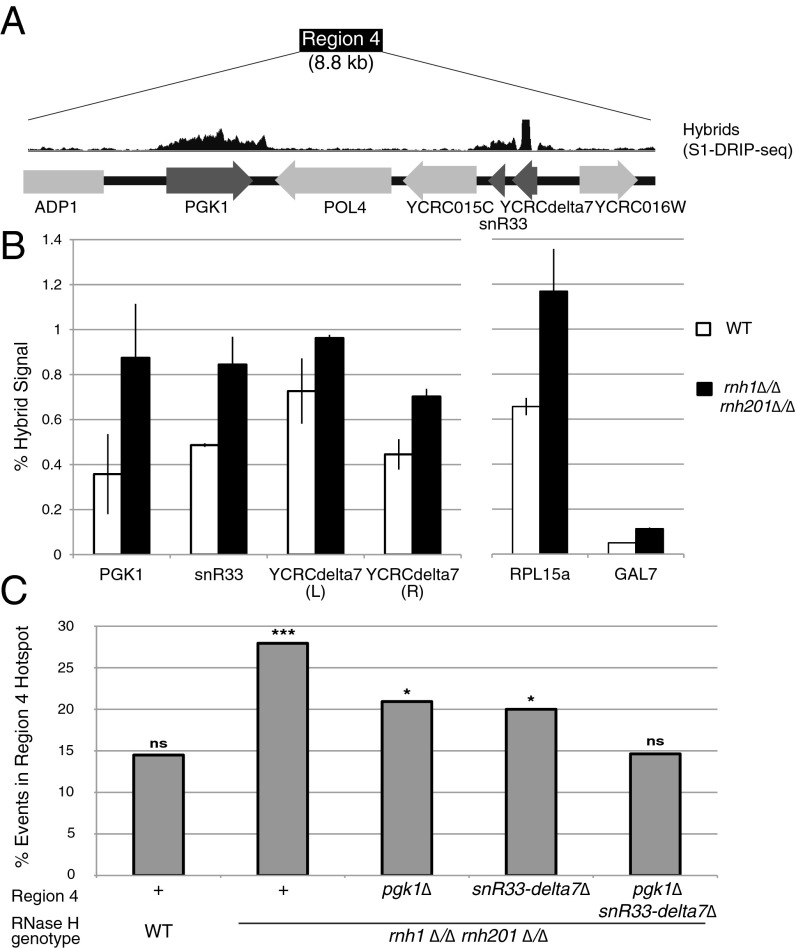
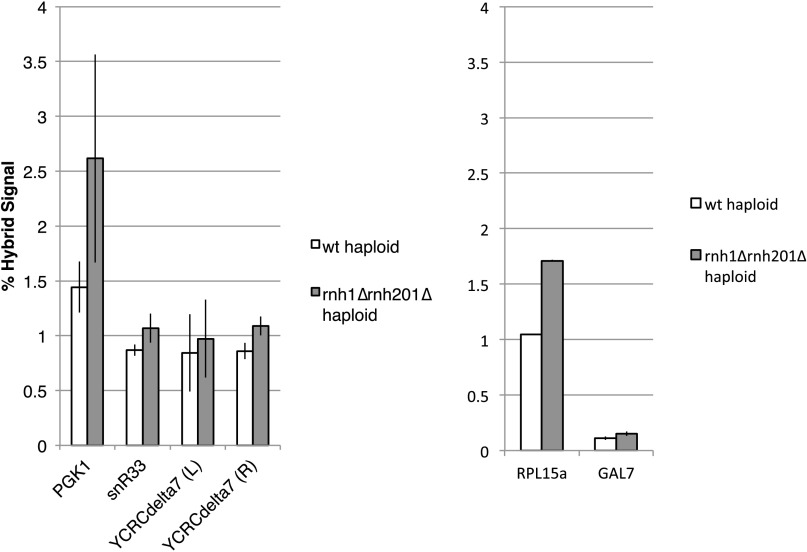
Region 4 contains multiple hybrid-prone regions, suggesting that the RNase H1-dependent chromosome instability in this region may be caused by hybrid formation. Two clusters of hybrid formation were identified in region 4 in haploid wild-type and RNase H-deficient cells by S1-DRIP-seq. (9): PGK1 and a region containing snR33 and YCRCdelta7 (Fig. 3A). These hybrid-prone loci can be categorized into known hybrid-prone families as a highly transcribed gene, a small nucleolar RNA, and a repetitive solo delta element, respectively (9).To confirm that these hybrid-prone regions form hybrids in the diploid cells used in this study, we performed DRIP followed by quantitative PCR (qPCR) at the loci of interest (Fig. 3B). We found that all three regions formed hybrids in the diploid strains, and, like all the hybrid regions identified by our S1-DRIP-seq study, these regions were hybrid forming in both wild-type and rnh1∆/∆ rnh201∆/∆ strains. Additionally, hybrids at these sites, as in most hybrid regions genome-wide, formed at higher levels in RNase H-deficient cells than in wild-type cells. We verified that these loci also form hybrids on the genetically marked chromosome III homolog by performing DRIP on the haploid S288c parents of the diploids assayed in this study (Fig. S5).
Fig. 3.
Characterization of hybrid-prone sequences in the region 4 hotspot. (A) Diagram of the region 4 hotspot. Hybrids by S1-DRIP-seq reads (9) are shown above sequence features. (B) Hybrid signal by DRIP-qPCR. Hybrid signals (as percent input) at hybrid-prone sequences in the hotspot [PGK1, snR33, unique sequences just left (L) and right (R) of YCRCdelta7, another known hybrid-prone sequence (RPL15a), and a non–hybrid-prone sequence (GAL7)] are shown. Error bars indicate ±1 SD. (C) Percent of LOH boundary events in the region 4 hotspot in wild-type and RNase H double-mutants with deletions of hybrid-forming sequences. ***P < 0.001; *P < 0.01; ns, not significant at P < 0.1 using the χ2 test.
Fig. S5.
Hybrid signal in haploids. Hybrid signal by DRIP-qPCR in S288c haploids. Hybrid signals are shown at hybrid-prone sequences in the hotspot (Left) and at another known hybrid-prone sequence (RPL15a) and at a non–hybrid-prone sequence (GAL7) (Right). Error bars indicate ±1 SD.
We next generated deletions within the hotspot interval to determine whether hybrid-prone sequences were necessary for instability at this hotspot. After deleting hybrid-prone sequences from the homolog bearing the genetic markers, we assayed the location of LOH junctions in an rnh1∆/∆ rnh201∆/∆ strain. Deleting either PGK1 or a region encompassing snR33 and YCRCdelta7 and replacing these loci with a similarly sized HIS3 marker led to a significant reduction in LOH events mapping to the hotspot (Fig. 3C). Although the proportion of LOH events occurring at the hotspot was smaller when these sequences were deleted, more events still occurred at the hotspot than would be expected based on the size of the interval (P < 0.05, χ2 test). Deletion of both PGK1 (with HIS3) and the region encompassing snR33 and YCRCdelta7 (with TRP1) led to the elimination of the LOH hotspot. In these strains, the proportion of events occurring in the hotspot was the same as in the wild-type strain, at the level expected based on the interval length. In total, these experiments showed that the hotspot for LOH events resulted from the contribution of multiple hybrid-forming sequences within the region. The proximity of these causative hybrid-prone regions to the LOH events in the hotspot interval suggests that hybrid-induced damage and its repair occur proximal to the causative hybrids.
RNases H1 and H2 Localize to Hybrid-Forming Regions.
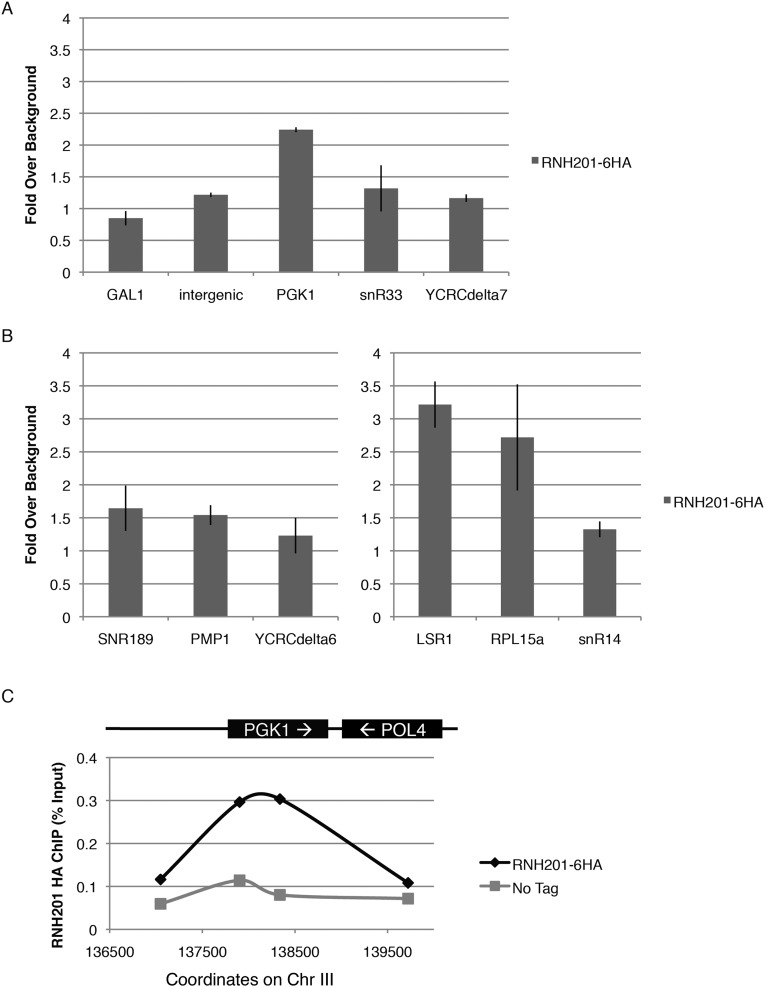
The level and distribution of LOH events in RNase H2-deficient cells (Figs. 1 and 2) suggested that RNase H2 protects against hybrid-induced damage at multiple distinct intervals. To test whether this broad mode of action was reflected at the level of localization, we performed ChIP studies of Rnh201. Rnh201 was enriched at the PGK1 locus but was not significantly enriched at the other region 4 hybrids (Fig. S6 A and C). It was enriched only weakly at most other tested hybrid-prone regions, with stronger enrichment detected at LSR1 and RPL15a (Fig. S6B). This weak enrichment could be caused by a weak signal because of potential transient association of RNase H2 with most of the hybrid regions at which it acts or by an elevated background because of its binding to single misincorporated ribonucleotides dispersed throughout the genome. Whatever the cause of the weak enrichment might be, we were unable to identify the localization of RNase H2 confidently.
Fig. S6.
Localization of RNase H2. (A and B) Enrichment of Rnh201 with the 6×HA tag at the region 4 hotspot (A) and at other hybrid-prone loci (B). Fold enrichment over nonhybrid background loci is shown. Two of these background loci (GAL7 and an intergenic sequence upstream of RGS2) are shown. Error bars indicate ±1 SD. (C) Percent enrichment of HA in HA-tagged Rnh201 and untagged cells at primers in the hybrid-prone PGK1 locus and in up- and downstream non–hybrid-prone loci.
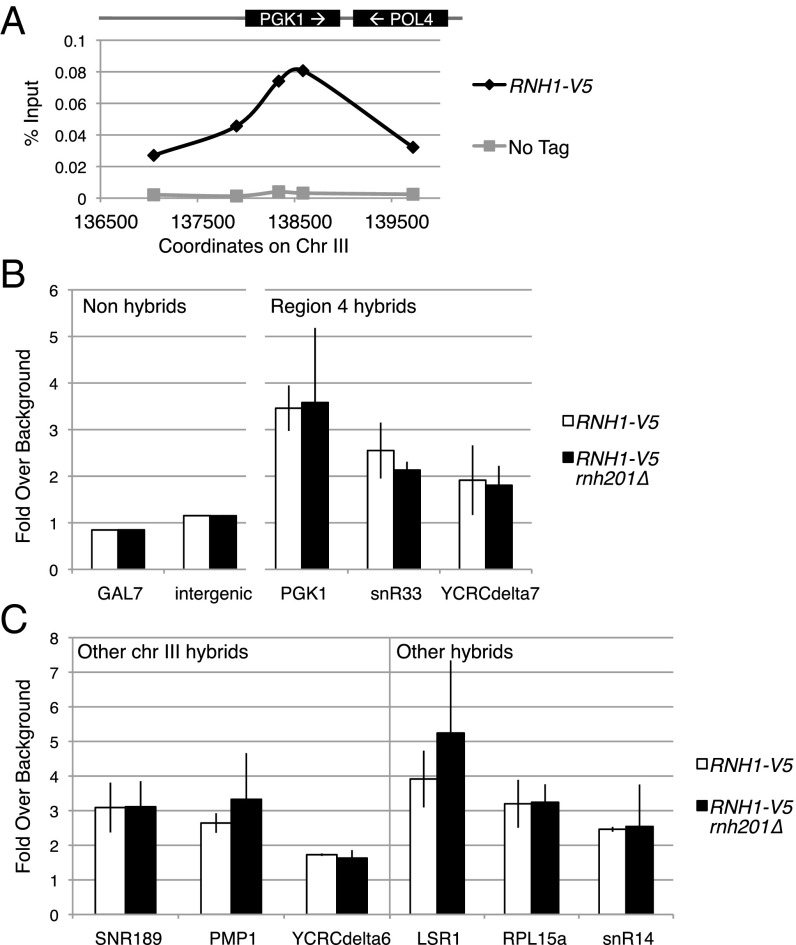
The LOH SNP-mapping assay revealed that RNase H1 protected against LOH within a specific interval while having a limited role for protection in adjacent intervals. We asked whether this interval-specific function of RNase H1 reflected its preferred localization to this region. To address this question, we performed ChIP studies of Rnh1 localization using an internal 3×V5 tag after P85 (RNH1-V5). The location of the tag in a less evolutionarily conserved region of the protein was chosen to disrupt protein function minimally (Materials and Methods).
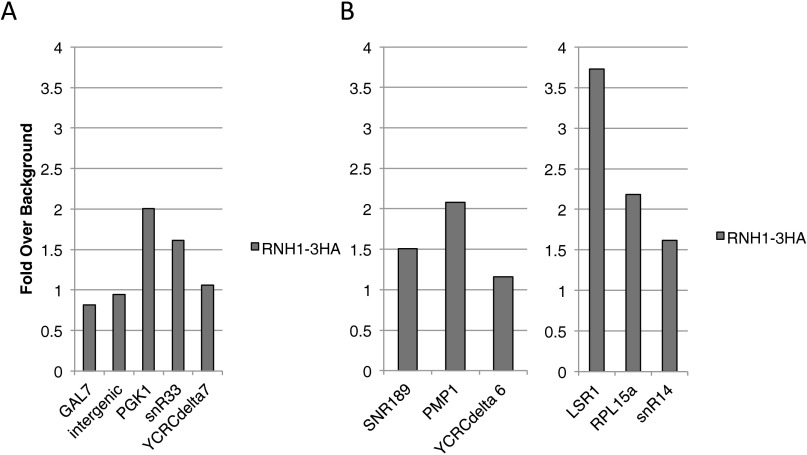
We were able to detect Rnh1 localization specifically to hybrid-forming regions. As shown in Fig. 4A, Rnh1 localized to the hybrid-forming ORF of PGK1 and not to upstream or downstream non–hybrid-forming sequences. Our ChIP studies had very little nonspecific signal, with a strain containing no V5 tag showing very low signal (Fig. 4A, gray line). We found that Rnh1 localized to all the hybrid-forming loci in the instability hotspot. Rnh1 ChIP signal was enriched 3.5-fold over background at the PGK1 locus, 2.6-fold at snR33, and 1.9-fold at YCRCdelta7 (Fig. 4B). Rnh1 also localized to other hybrid-forming loci along the right arm of chromosome III: 3.1-fold at SNR189, 2.6-fold at PMP1, and 1.7-fold at YCRCdelta6 (Fig. 4C). In fact, Rnh1 localized to all tested hybrid-forming loci. Other strong hybrid-forming loci—LSR1, RPL15a, and snR14—all showed enrichment of Rnh1 (Fig. 4C). We additionally performed the ChIP using Rnh1 tagged with a 3×HA tag. This ChIP showed a pattern of localization similar to that of the 3×V5-tagged protein (Fig. S7).
Fig. 4.
RNase H1 localization by ChIP. (A) Percent enrichment of V5 in V5-tagged Rnh1 cells and untagged cells at primers in the hybrid-prone PGK1 locus and up- and downstream non–hybrid-prone loci. (B and C) Enrichment of Rnh1 at the region 4 hotspot (B) and at other hybrid-prone loci (C) in wild-type and rnh201∆ cells. Fold enrichment over nonhybrid background loci is shown. Two of these background loci (GAL7 and an intergenic sequence upstream of RGS2) are shown in B. Error bars indicate ±1 SD.
Fig. S7.
Localization of RNase H1 with an HA tag. (A and B) Enrichment of Rnh1 with 3×HA at the region 4 hotspot (A) and at other hybrid-prone loci (B). Fold enrichment over nonhybrid background loci is shown. Two of these background loci (GAL7 and an intergenic sequence upstream of RGS2) are shown.
RNase H1 and RNase H2 have overlapping enzymatic functions. To determine if RNase H2 has an effect on RNase H1 localization, we deleted RNH201 and performed ChIP of Rnh1. We found that Rnh1 localization was similar at all tested loci in the wild-type and rnh201∆ background (Fig. 4 B and C), indicating that Rnh201 does not affect Rnh1 localization.
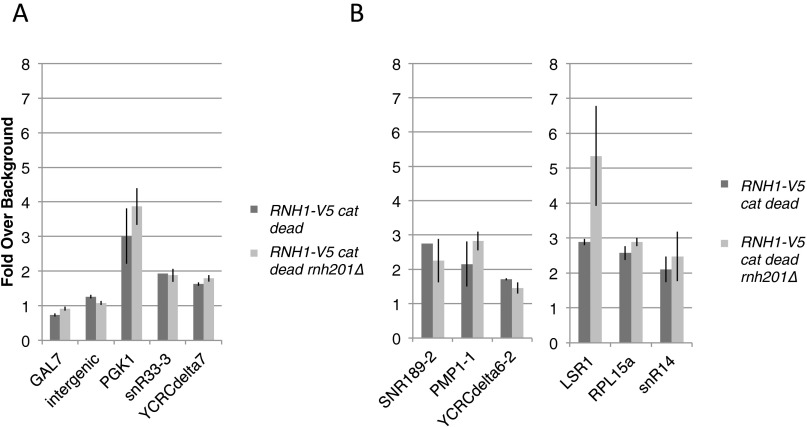
A previous study in human cells used a catalytically dead RNase H1 to immunoprecipitate chromatin (12). To determine whether a catalytically dead RNase H1 has the same localization pattern as the wild-type enzyme, we performed ChIP of the catalytically dead Rnh1-D193N in both a wild-type and rnh201∆ background. The enrichment of the catalytically dead Rnh1 in both backgrounds was similar to that of wild-type Rnh1 (Fig. S8), indicating that the catalytically dead enzyme does not have altered localization or level of binding to hybrids. This similarity suggests that RNase H1 binding to hybrids is independent of its enzymatic activity in degrading hybrids. These results coupled with the global localization of RNase H1 to many hybrids indicates that RNase H1 binds broadly to hybrids but does not remove them or disassociate from them rapidly.
Fig. S8.
Localization of RNase H1 catalytically dead mutant. (A and B) Enrichment of the Rnh1 catalytically dead mutant at the region 4 hotspot (A) and other hybrid-prone loci (B) in wild-type and rnh201∆ cells. Fold enrichment over nonhybrid background loci is shown. Two of these background loci (GAL7 and an intergenic sequence upstream of RGS2) are shown. Error bars indicate ±1 SD.
Discussion
Why organisms from bacteria to humans harbor two distinct RNases H, both with the ability to remove DNA:RNA hybrids, has been an enigma. In this study, we investigated the in vivo roles of RNases H1 and H2 in budding yeast through the lens of R-loop–induced chromosome instability. We found that the frequency of LOH on the right arm of chromosome III is 15-fold greater in cells lacking both RNase H1 and H2. These events could occur through chromosome loss, chromosome rearrangement, or repair events that result in long tracts of LOH-like mitotic recombination or break-induced replication. More than 90% of these large-scale LOH events could be suppressed by restoring RNase H2, but not RNase H1, activity. Furthermore, the LOH could be suppressed by introducing a mutant of RNase H2 that retained its R-loop degradation activity but lacked the ability to remove single ribonucleotides. Taken together, these results suggest that most spontaneous R-loops are inhibited from inducing large-scale LOH because they are removed by the hybrid-degrading activity of RNase H2 before they can induce DNA damage.
The conclusion that RNase H2, but not H1, carries the major load of protecting cells against large-scale LOH corroborates a previous study (4). Although our work and that of O’Connell and colleagues (4) concluded that this protection results from RNase H2’s removing R-loops, a different study of RNase H2’s activities by Conover and colleagues (5) suggested that this protection is afforded by RNase H2’s ability to remove misincorporated nucleotides. In fact, many studies have shown that misincorporated ribonucleotides can lead to DNA damage and mutation in a topoisomerase I-dependent manner (13–16). However, it has remained unclear if misincorporated ribonucleotides are a major contributor to larger-scale chromosome instability. These recent large-scale LOH studies that reached different conclusions (4, 5) both relied on results from mutants of DNA polymerases that incorporated greater or fewer ribonucleotides into DNA. By using an orthogonal approach with an RNase H2 variant defective in the removal of misincorporated nucleotides (RNH201-hr), we provide strong evidence supporting the Conover study (5), i.e., that RNase H2 protects against large-scale chromosome instability mostly by removing R-loops. The RNH201-hr mutant greatly suppresses LOH; however, we do note that it does not suppress LOH to wild-type levels. This finding indicates that removal of misincorporated ribonucleotides by RNase H2 does play a role, albeit a minor one, in preventing chromosome instability.
Further insight into the distinct functions of these two enzymes came when we mapped the position of the junctions between the regions of heterozygosity and LOH along a 98-kb region of chromosome III. These junctions presumably map the repair sites of lesions induced by specific R-loops in the intervals. Our map suggests that RNase H2 and H1 have distinct spatial specificity. A comparison of RNase H2-deficient and wild-type cells revealed no significant difference in the distribution of the LOH junctions. Given the 13-fold induction of overall LOH in the RNase H2-defective mutants, the LOH events induced by RNase H2 deficiency were equally distributed among all intervals. Therefore these results suggest that RNase H2 acts as the major protector against LOH by degrading DNA:RNA hybrids in R-loops at many, if not all, sites in the genome. This spatially unconstrained function of RNase H2 in hybrid removal fits with its genome-wide activity in removing random ribonucleotide misincorporation. Consistent with this ubiquitous genomic function of RNase H2 in both single ribonucleotide and hybrid removal, it exhibited very weak binding to many sites on chromosomes as assayed by ChIP (this study). One might have imagined that the misincorporated ribonucleotide removal activity of this enzyme would compete with its R-loop removal activity, thereby explaining the need for a second enzyme, such as RNase H1, dedicated to R-loop removal. However, the genome-wide load of single-ribonucleotide misincorporation in wild-type cells seems to be insufficient to generate this competition. Such a preoccupation with the global removal of misincorporated nucleotides may occur in a stress condition that increases misincorporation, thereby explaining the synergistic increase in LOH in DNA polymerase mutants lacking RNase H2.
In contrast, the mapping of the junctions of LOH events in RNase H1-deficient cells showed elevated LOH in only the fourth of the nine contiguous intervals. This restricted spatial impact on LOH explains why RNase H1-deficient cells did not exhibit an increase in total LOH of this chromosome III arm (i.e., the sum of the LOH in all nine intervals). Intriguingly, the ability of RNase H1 to protect primarily the fourth interval but not the other eight intervals from hybrid-induced LOH did not reflect its preferred access to the fourth interval. This conclusion was based on our RNase H1 ChIP, which showed that RNase H1 localized equally well to hybrid-prone regions within the fourth interval and to representative hybrid-prone regions outside this interval on chromosome III and elsewhere in the genome. This equal distribution suggests a model in which RNase H1 uses its hybrid-recognition activity to bind to spontaneous R-loops but in which its nuclease activity normally is suppressed except at subsets such as those in the fourth interval.
Three additional observations are consistent with this hypothesis. The ChIP signal was the same at R-loops from wild-type and catalytically dead RNase H1. If RNase H1 were active on most hybrids where it was bound, one might expect it to degrade the hybrids and interact transiently, whereas the catalytically dead RNase H1, being unable to degrade the hybrids, would have a prolonged interaction and generate a higher ChIP signal (17, 18). However, this was not the case. Second, constitutive overexpression of RNase H1 can suppress the instability of a yeast artificial chromosome in RNase H2-deficient cells (3). Overexpression of RNase H1 may allow it to escape repression or regulation, perhaps by titrating out a repressor, and therefore to degrade spontaneous R-loops normally degraded by RNase H2. Third, biochemical characterization of the RNase H activity of yeast extracts found that almost all RNase H activity was derived from RNase H2, not from RNase H1, again reflecting a possible repression of H1 activity in yeast (19). Why is the RNase H1 nuclease repressed at most sites of spontaneous hybrid formation? That the inactivation of RNase H1 causes a synergistic increase in hybrid-induced LOH when transcription is perturbed by defects in RNA biogenesis machinery may offer a clue (3). RNase H1 may be a stress-induced factor that is unleashed at sites that accumulate hybrids resulting from aberrant transcription in a few loci under normal conditions but at many loci when the cells are stressed. Another possibility is that RNase H1 resolves hybrids only during distinct cell-cycle stages. Previous studies have suggested that expression of RNase H2, but not RNase H1, is cell-cycle regulated, with two bursts of expression in S and G2 (19). Intriguingly, in that same study RNase H activity appeared to cycle in rnh201∆ cell extracts, perhaps indicating some posttranscriptional regulation of RNase H1 activity. Given our results, additional characterization of the expression, protein levels, activity, and binding of RNase H1 and H2 through the cell cycle would be very interesting.
Finally, our study provides important insights into the relationship of specific R-loops with R-loop–induced LOH. Previous studies identified LOH events induced by RNase H deficiency, but they lacked a high-resolution map of hybrids to correlate these events with specific hybrids. Our recent map of hybrid-prone regions (9) allowed us to assess various models of how R-loops lead to LOH. One possibility is that all R-loops have the potential to induce LOH because they induce damage, independent of their context. Alternatively, R-loops may differ in their ability to induce damage because of unique features such as their position in genes or nucleotide content. We showed that RNase H2-deficient cells exhibit elevated LOH at multiple intervals. This broad effect suggests that, if hybrids are allowed to persist, many, if not all, have the potential to generate damage that leads to LOH. However, we observed a hotspot for LOH in one interval upon RNase H1 deficiency, suggesting that some hybrids may be more prone to cause damage than others. Three hybrid-prone regions lie within the fourth interval, one on the PGK1 gene and two clustered on snR33 and YCRCdelta7. We showed that elevated LOH in this interval was partially reduced by deleting either PGK1 or the cluster and was eliminated completely when both were deleted. Engineered hybrids have been shown to cause chromosome instability (20), but our results link specific natural hybrids with LOH in yeast. Elevated LOH in this interval appears to result from the sum of the events induced by each hybrid, suggesting that a feature in this region makes hybrids more prone to LOH.
In summary, the experiments presented in this study provide important examples of functional differences for the hybrid-removal activities of RNase H1 and H2. Understanding the molecular basis for these differences may provide important insights into why these two enzymes have been so highly conserved in evolution.
Materials and Methods
Yeast Strains, Media, and Methods.
Yeast strain construction, pulsed-field gel electrophoresis, ChIP, and DNA/RNA immunoprecipitation were performed according to standard procedures as described in SI Materials and Methods. Strains are listed in Table S1 and primers are listed in Table S2.
Table S1.
Strains used in this study
| Description | Strain | Strain background | Genotype |
| LOH assay diploids | |||
| WT | AZ88 | S288c/RM11 | MATa/α LEU2/leu2∆his3/HIS3 ura3/ura3-Δ0 TRP1/TRP1 lys2/LYS2 HO::KanMX BUD5/bud5Δ::URA3 |
| rnh1∆/∆ rnh201∆/∆ | AZ92 | S288c/RM11 | MATa/α rnh1∆::HYG/rnh1∆::HYG rnh201∆::NAT/rnh201∆::NAT LEU2/leu2∆ his3/HIS3 ura3/ura3-Δ0 TRP1/TRP1 lys2/LYS2 HO::KanMX BUD5/bud5Δ::URA3 |
| RNH1+/∆ rnh201∆/∆ | AZ174 | S288c/RM11 | MATa/α RNH1:HIS3/rnh1∆::HYG rnh201∆::NAT/rnh201∆::NAT LEU2/leu2∆ his3/HIS3 ura3/ura3-Δ0 TRP1/TRP1 lys2/LYS2 HO::KanMX BUD5/bud5Δ::URA3 |
| rnh1∆/∆ RNH201+/∆ | AZ203a | S288c/RM11 | MATa/α rnh1∆::HYG/rnh1∆::HYG RNH201:HIS3/rnh201∆::NAT LEU2/leu2∆ his3/HIS3 ura3/ura3-Δ0 TRP1/TRP1 lys2/LYS2 HO::KanMX BUD5/bud5Δ::URA3 |
| rnh1∆/∆ RNH201 h/∆ | AZ204a | S288c/RM11 | MATa/α rnh1∆::HYG/rnh1∆::HYG RNH201-P45D-Y219A:HIS3/rnh201∆::NAT LEU2/leu2∆ his3/HIS3 ura3/ura3-Δ0 TRP1/TRP1 lys2/LYS2 HO::KanMX BUD5/bud5Δ::URA3 |
| Hotspot hybrid deletion diploids | |||
| rnh1∆/∆ rnh201∆/∆ pgk1∆ | AZ207a | S288c/RM11 | MATa/α PGK1/pgk1∆::HIS3 rnh1∆::HYG/rnh1∆::HYG rnh201∆::NAT/rnh201∆::NAT LEU2/leu2∆ his3/HIS3 ura3/ura3-Δ0 TRP1/TRP1 lys2/LYS2 HO::KanMX BUD5/bud5Δ::URA3 |
| rnh1∆/∆ rnh201∆/∆ snRNA/δ∆ | AZ217a | S288c/RM11 | MATa/α SNR33-YCRCdelta7/snr33-ycrcdelta7∆::HIS3 rnh1∆::HYG/rnh1∆::HYG rnh201∆::NAT/rnh201∆::NAT LEU2/leu2∆ his3/HIS3 ura3/ura3-Δ0 TRP1/TRP1 lys2/LYS2 HO::KanMX BUD5/bud5Δ::URA3 |
| rnh1∆/∆ rnh201∆/∆ pgk1∆ snRNA/δ∆ | AZ221a | S288c/RM11 | MATa/α PGK1/pgk1∆::TRP1 SNR33-YCRCdelta7/snr33-ycrcdelta7∆::HIS3 rnh1∆::HYG/rnh1∆::HYG rnh201∆::NAT/rnh201∆::NAT LEU2/leu2∆ his3/HIS3 ura3/ura3-Δ0 TRP1/trp1∆::KanMX lys2/LYS2 HO::KanMX BUD5/bud5Δ::URA3 |
| Haploids for DRIP | |||
| WT | AZ61c | S288c | MATa LEU2 bud5Δ::URA3 his3 |
| rnh1∆ rnh201∆ | AZ62d | S288c | MATa rnh1∆::HYG rnh201∆::NAT LEU2 bud5Δ::URA3 his3 ade- |
| RNase H1 ChIP | |||
| RNH1-V5 | JA151 | S288c | Mata RNH1-P85-3xV5 MIF2-3xV5:G418 his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| RNH1-V5 rnh201∆ | JA183 | S288c | Mata RNH1-P85-3xV5 rnh201∆::HYG his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| RNH1-V5 Cat Dead | JA172 | S288c | Mata RNH1-P85-3xV5-D193N MIF2-3xV5:G418 his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| RNH1-V5 Cat Dead rnh201∆ | JA184 | S288c | Mata RNH1-P85-3xV5-D193N rnh201∆::HYG his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| WT, No Tag | LW6836a | S288c | MATa leu2 his3 ura3 TRP1 ade- |
| RNH1-HA | AZ205c | RM11 | MATα RNH1-F95-3HA lys2 ura3-Δ0 HO::KanMX leu2 |
| RNase H2 ChIP | |||
| RNH201-HA | AZ218b | S288c | MATa RNH201-6HA:HIS3MX LEU2 bud5Δ::URA3 |
Table S2.
Primers
| Primer name | Sequence |
| PGK1_F | TTT CGA CTT GCC ACA ACG TG |
| PGK1_R | ATC TTG TCA GCA ACC TTG GC |
| snR33_F | GCA AAT CGA TTG TCC ACA CAC |
| snR33_R | GCC TAG CTT TTA CAC CGG TTT G |
| YCRCdelta7(L)_F | CCG TTC CGC CAA ATT TTT CAT G |
| YCRCdelta7(L)_R | TAG CGC AAG TGG TTT AGT GG |
| YCRCdelta7(R)_F | AGA ACA TCC TTG AAA GGT CGA C |
| YCRCdelta7(R)_R | AGC TGA ATA ATG CCG TGG TG |
| RPL15a_F | ACC GCT GAA GAA AGA GTT GG |
| RPL15a_R | TGT TGA GGG TCG ACC AAG AT |
| GAL7_F | CCA ACC AAG AAT TTC CGA AC |
| GAL7_R | CGC CTC GAT TTT AAA GCA AC |
| PGK1_5′_F1 | GAC TTC AAC TCA AGA CGC ACA G |
| PGK1_5′_R1 | AAA GGA TTC GCG CCC AAA TC |
| PGK1_5′_F2 | TTG CTG CTT TGC CAA CCA TC |
| PGK1_5′_R2 | CAA GTG AGA AGC CAA GAC AAC G |
| PGK1_F2 | TTG ATG GAA AAG GCC AAG GC |
| PGK1_R2 | TTG GCA TCA GCA GAG AAA GC |
| POL4_F1 | CCC AAC AAT CTT CGC TGT ACG |
| POL4_R1 | CGA CCG AGT TGG CAA AAA TC |
| RGS2_intergenic_F | CGT GTC TGG CTC GGA AGT AT |
| RGS2_intergenic_R | CCG CAA TAA CGT ACA CAT CG |
| GAL1_F | GAG CTT TAC TGC CGA CGA AG |
| GAL1_R | CGG GAA CCA TAT GAT CCA TT |
| UBC6_F | GAT ACT TGG AAT CCT GGC TGG TCT GTC TC |
| UBC6_R | AAA GGG TCT TCT GTT TCA TCA CCT GTA TTT GC |
| CYC1_F | TGA ATT CAA GGC CGG TTC TG |
| CYC1_R | TTA TGT GGG CCA CCC TTT TC |
| DYN2_F | ACA TTG CTG GGA CGG TAA AG |
| DYN2_R | AAT GGC CCT TTT CGT GTG TC |
| SNR189_F | CGT AAG TAC TCC AAA GCA GTC TC |
| SNR189_R | ACG GGC CTG ACA TCT CTA TTC |
| PMP1_F | AAA GGG TAT CGC ACA CAC AC |
| PMP1_R | CGG AGC GAG CCA TTT TAT TTC C |
| YCRCdelta6_F | GTG AGG AAT TAT CGG GCA TCT TG |
| YCRCdelta6_R | GCC ATT TCA TGA GGA CGG AAT AC |
| LSR1_F | TTT TGG TTT GCA AGG AAA GG |
| LSR1_R | TGT AGA CCA ACC CCA CCC TA |
| snR14_F | CCT TAT GCA CGG GAA ATA CG |
| snR14_R | ATT CAA AAG CGA ACA CCG AAT |
LOH Assay.
Diploid cells were dilution-streaked on synthetic complete uracil (SC-URA) plates grown at 30 °C. Single colonies were resuspended in 0.25 mL of water, diluted, and plated onto 5-FOA–containing plates (Zymo Research); 107 cells were plated for wild-type and rnh1∆ mutants, and 106 cells were plated for rnh201∆ and RNase H double mutants. Plating efficiency was monitored by plating 200 cells onto yeast peptone dextrose (YPD) plates. Plates were incubated at 30 °C for 2–3 d; then the number of colonies forming on each plate was counted. The number of colonies that grow on 5-FOA, normalized for plating efficiency, is a measure of the frequency of events. To determine the proportion of terminal LOH versus chromosomes loss, the colonies grown on 5-FOA plates were replica plated onto synthetic complete leucine (SC-LEU) medium. The number of colonies that grew on SC-LEU divided by the number of colonies that grew on 5-FOA represented the percentage of terminal LOH.
Mapping Terminal LOH Events on Chromosome III by SNPs.
Diploid cells with terminal LOH events on chromosome III were isolated on SC-LEU according to the LOH assay described above. Each colony to be mapped was derived from an independent starting colony on SC-URA. SNP-containing regions (SNP locations are listed in Table S3) were amplified by optimized yeast colony PCR and subjected to Sanger sequencing.
Table S3.
Locations of SNPs assayed in chromosome III
| Position in SGD | Allele in S288C | Allele in RM11 |
| 114,986 | G | T |
| 125,079 | A | G |
| 135,857 | T | C |
| 145,017 | C | T |
| 156,868 | A | G |
| 165,025 | T | C |
| 175,359 | T | C |
| 185,113 | T | C |
SGD, Saccharomyces Genome Database.
SI Materials and Methods
Yeast Strains, Media, and Reagents.
Full genotypes of strains used in this study are listed in Table S1. All strains are derived from the S288c or RM11 strains, as noted in the table. Genetic markers on chromosome III and gene knockouts were introduced by standard yeast transformation. Yeast strains were grown in yeast extract peptone (YEP) or synthetic complete medium supplemented with 2% glucose at 30 °C according to standard yeast protocols. 5-FOA was purchased from Zymo Research.
Mapping Terminal LOH Events on Chromosome III by SNPs.
Diploid cells with terminal LOH events on chromosome III were isolated on SC-LEU according to the LOH assay described in Materials and Methods. Each colony to be mapped was derived from an independent starting colony on SC-URA. Regions containing SNPs (SNP locations are listed in Table S3) were amplified by optimized yeast colony PCR as follows: ∼5 μL of the yeast colony was resuspended in 20 μL of 0.02 M NaOH and was boiled for 10 min. Then 1 μL of this boiled yeast colony was used as a template for PCR amplification in the following reaction conditions: 1× Q buffer (Qiagen), 1× standard Taq buffer (New England BioLabs), 0.2 M dNTP, 1 μM of each primer, and 0.5 U Taq polymerase (New England BioLabs). Touchdown PCR cycles were used: 94 °C for 1 min; nine cycles of 94 °C for 20 s, 62 °C decreasing by 1 °C each cycle for 45 s, 68 °C for 45 s; and 24 cycles of 94 °C for 20 s, 52 °C for 45 s, 68 °C for 45 s. The PCR-amplified DNA was cleaned using standard enzymatic cleanup conditions [1× CutSmart buffer (New England BioLabs), 1 U rSAP (New England BioLabs), 1.8 U ExoI (Thermo Scientific)] and was incubated at 37 °C for 1 h followed by heat inactivation at 80 °C for 15 min. This reaction was diluted, and the equivalent of 1/20th of the reaction was used for standard Sanger sequencing. Heterozygosity or homozygosity of the SNP was determined by the relative intensity of each base on sequencing chromatographs. For the deletion studies of hybrid-forming sequences at the hotspot (Fig. 3), only the two SNPs flanking the hotspot were sequenced.
Pulsed-Field Gel Electrophoresis and Southern Blot Analysis.
Yeast genomic DNA was prepared in 1% pulsed-field–grade agarose plugs (SeaPlaque 50100; Lonza) and resolved as previously described (21) with a Bio-Rad CHEF-DR III system. The following parameters were used: 1.3% agarose gel in 0.5× Tris/borate/EDTA, 6 V/cm, 120° angle, 15- to 25-s switch times, 24 h at 14 °C. For Southern analysis, gels were transferred onto a GeneScreen Plus membrane (PerkinElmer NEF988) and probed with a 1.8-kb fragment containing the LEU2 sequence.
DRIP.
DRIP experiments were performed as described previously (9). Briefly, genomic DNA was isolated from ∼1010 log-phase cells. Then 100 μg of genomic DNA was digested overnight at 37 °C with SpeI-HF, HindIII-HF, BsrGI, and XbaI. Digested DNA was immunoprecipitated with S9.6 antibody. The percentage of the hybrid signal was quantified using qPCR on DNA from immunoprecipitation and total input with the DyNAmo HS SYBR Green qPCR kit (Thermo Scientific).
Construction of Tagged Rnh1 and Rnh201 Strains.
The RNH201-hr allele was amplified off the ycNPH2-FL2 plasmid harboring a C-terminally FLAG-tagged RNH201 gene with the P45D-Y219A mutations from ref. 10 and was integrated into the endogenous RNH201 locus with a HIS3 marker by standard PCR and recombination in yeast. Rnh1 was internally tagged with a 3×V5 or 3×HA tag placed after proline 85 in a region of low conservation within the Saccharomyces clade. The catalytically dead Rnh1 harbors the point mutation D193N, which was chosen as the evolutionarily conserved residue from the human RNase H1 protein of D145N (12).
ChIP.
ChIP was performed as described previously (22). Briefly, a 50-mL YPD culture of asynchronous cells was grown to OD 0.6. Cells were fixed for 2 h in a final concentration of 1% formaldehyde. After mechanical cell lysis with glass beads, chromatin was sheared 24 times for 45 s each (settings at duty cycle, 20%; intensity, 10; cycles per burst, 200; 30-s rest between cycles) using a Covaris S2 ultrasonicator. Immunoprecipitation of epitope-tagged proteins was isolated using 5 μL of anti-V5 (R960-25; Invitrogen) or 1 μL of anti-HA (12CA5; Roche Life Sciences) monoclonal antibodies. ChIP using a no-primary-antibody control and a no-tag strain was also performed to ensure specificity. Appropriate dilutions of input and immunoprecipitated DNA samples were used for qPCR analysis to ensure linearity of the PCR signal. A background level of no enrichment was established by averaging six sets of background primers chosen as having very low hybrid signal in S1-DRIP-seq (9): GAL7, RGS2 intergenic, GAL1, UCB6, CYC1, and DYN2. The fold increase over this average background was calculated for all loci tested. All experiments were performed at least twice, and an average of the fold increase over background is presented. ChIP primers are listed in Table S2.
Acknowledgments
We thank Lorenzo Costantino, Lamia Wahba, Nicholas Ingolia, and Vincent Guacci for suggestions and helpful comments on the manuscript; Jeremy Amon for helpful discussions, technical assistance, and strains for ChIP; Jeremy Roop from Rachel Brem’s laboratory for an RM-11 isolate; and all members of the D.K. laboratory for support and discussion. This work was supported by NIH Grant 1R35GM118189-01 (to D.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613448113/-/DCSupplemental.
References
- 1.Santos-Pereira JM, Aguilera A. R loops: New modulators of genome dynamics and function. Nat Rev Genet. 2015;16(10):583–597. doi: 10.1038/nrg3961. [DOI] [PubMed] [Google Scholar]
- 2.Cerritelli SM, Crouch RJ. Ribonuclease H: The enzymes in eukaryotes. FEBS J. 2009;276(6):1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wahba L, Amon JD, Koshland D, Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell. 2011;44(6):978–988. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Connell K, Jinks-Robertson S, Petes TD. Elevated genome-wide instability in yeast mutants lacking RNase H activity. Genetics. 2015;201(3):963–975. doi: 10.1534/genetics.115.182725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conover HN, et al. Stimulation of chromosomal rearrangements by ribonucleotides. Genetics. 2015;201(3):951–961. doi: 10.1534/genetics.115.181149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12(3):711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 7.Stirling PC, et al. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 2012;26(2):163–175. doi: 10.1101/gad.179721.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan YA, et al. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet. 2014;10(4):e1004288. doi: 10.1371/journal.pgen.1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wahba L, Costantino L, Tan FJ, Zimmer A, Koshland D. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 2016;30(11):1327–1338. doi: 10.1101/gad.280834.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chon H, et al. RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Res. 2013;41(5):3130–3143. doi: 10.1093/nar/gkt027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi J, et al. Characterization of meiotic crossovers and gene conversion by whole-genome sequencing in Saccharomyces cerevisiae. BMC Genomics. 2009;10(1):475. doi: 10.1186/1471-2164-10-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ginno PA, Lott PL, Christensen HC, Korf I, Chédin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45(6):814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nick McElhinny SA, et al. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010;6(10):774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark AB, Lujan SA, Kissling GE, Kunkel TA. Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase ε. DNA Repair (Amst) 2011;10(5):476–482. doi: 10.1016/j.dnarep.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim N, et al. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332(6037):1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams JS, et al. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol Cell. 2013;49(5):1010–1015. doi: 10.1016/j.molcel.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Britton S, et al. DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res. 2014;42(14):9047–9062. doi: 10.1093/nar/gku601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 2005;19(8):942–954. doi: 10.1101/gad.1298905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arudchandran A, et al. The absence of ribonuclease H1 or H2 alters the sensitivity of Saccharomyces cerevisiae to hydroxyurea, caffeine and ethyl methanesulphonate: Implications for roles of RNases H in DNA replication and repair. Genes Cells. 2000;5(10):789–802. doi: 10.1046/j.1365-2443.2000.00373.x. [DOI] [PubMed] [Google Scholar]
- 20.Wahba L, Gore SK, Koshland D. The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. eLife. 2013;2:e00505. doi: 10.7554/eLife.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwartz DC, Cantor CR. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- 22.Eng T, Guacci V, Koshland D. Interallelic complementation provides functional evidence for cohesin–cohesin interactions on DNA. Mol Biol Cell. 2015;26(23):4224–4235. doi: 10.1091/mbc.E15-06-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]