Significance
Clinical outcomes in cholangiocarcinoma (CC) are poor; few patients are candidates for curative resection, and palliative chemotherapy produces only modest effects on survival. With an increasing incidence, new targets are urgently needed. Notch has been identified as having potential to induce CC when transgenically overexpressed, and this study aimed to characterize how endogenous Notch might drive tumorigenesis. We identify the atypical receptor Notch3 as differentially overactivated in CCs in humans, rats, and mice, with genetic deletion significantly reducing CC growth. Notch3 sustains tumor cell survival through PI3k/Akt activation via a noncanonical mechanism independent of Recombinant Signal Binding Protein for Immunoglobulin Kappa J Region (RBPJ), presenting an opportunity to target the pathway without disrupting classical Notch and bypassing toxicities associated with γ-secretase inhibitors.
Keywords: cholangiocarcinoma, Notch, noncanonical, bile duct, cancer
Abstract
The prognosis of cholangiocarcinoma (CC) is dismal. Notch has been identified as a potential driver; forced exogenous overexpression of Notch1 in hepatocytes results in the formation of biliary tumors. In human disease, however, it is unknown which components of the endogenously signaling pathway are required for tumorigenesis, how these orchestrate cancer, and how they can be targeted for therapy. Here we characterize Notch in human-resected CC, a toxin-driven model in rats, and a transgenic mouse model in which p53 deletion is targeted to biliary epithelia and CC induced using the hepatocarcinogen thioacetamide. We find that across species, the atypical receptor NOTCH3 is differentially overexpressed; it is progressively up-regulated with disease development and promotes tumor cell survival via activation of PI3k-Akt. We use genetic KO studies to show that tumor growth significantly attenuates after Notch3 deletion and demonstrate signaling occurs via a noncanonical pathway independent of the mediator of classical Notch, Recombinant Signal Binding Protein for Immunoglobulin Kappa J Region (RBPJ). These data present an opportunity in this aggressive cancer to selectively target Notch, bypassing toxicities known to be RBPJ dependent.
Cholangiocarcinoma (CC) is an aggressive primary liver malignancy with an increasing global incidence. Surgery remains the only potential cure, but few patients present with operable disease. New adjuvant treatments are urgently required; however, few targets have been put forward and none have been shown to have efficacy.
Notch is a master regulator of cell fate in the mammalian liver. In the embryo, hepatoblast specification to a biliary fate and tubulogenesis are dependent on Recombinant Signal Binding Protein for Immunoglobulin Kappa J Region (RBPJ)-driven effector transcription, i.e., canonical Notch signaling (1). Furthermore niche-derived ligand reactivates Notch during biliary injury in the adult to expand the hepatic progenitor cell (HPC) pool for repair (2). The four receptors play distinct roles, as evidenced by the spectrum of phenotypes seen after transgenic KO, as well in vitro and in vivo studies of HPC differentiation (3). Aberrant activation of Notch paralogs results in a spectrum of cancer phenotypes (4), implying differing potentials for therapeutic targeting. Their individual contribution to biliary carcinogenesis remains unclear.
A population of periportal hepatocytes has been identified enriched for biliary gene expression, with special replicative capacity and potential for parenchymal regeneration during hepatocyte injury (5). Introduction of transgenically activated, supraphysiological levels of Notch1 intracellular domain (N1-ICD) in hepatocytes can redirect cell identity to a ductular lineage, activating the cancer program (6, 7). There is further evidence that after damage, hepatocytes can contribute to the HPC pool, adopting biliary-specific functions, and that this reverses during recovery (8). This potential for hepatocyte plasticity may explain the appearance of perivenular CC in chronic hepatitis C virus (HCV) infection. We used lineage tracing to demonstrate CC can arise from CK19+ ductular cells; however, the contribution from biliary vs. hepatocyte-derived HPCs and the CC cell of origin is still hotly debated.
Oncogenic Notch1 is a driver of a proportion of T-cell acute lymphoblastic leukemias, whereas other tumors rarely exhibit mutated Notch; rather, WT signaling is dysregulated. Sequencing of CC has failed to identify NOTCH mutations, and therefore we sought to evaluate the contribution of endogenous WT Notch. As the role of Notch in cancer depends on somatic context, we aimed to use a range of models to reflect the mutational heterogeneity of CC. Both pan-receptor and Notch1 inhibition are associated with off-target effects, so we hypothesized that characterizing the signal might identify specific drivers to enable targeting to bypass toxicity.
Results
Notch3 Is Differentially Activated in Human CC.
We used a targeted NOTCH PCR array in five surgically resected samples paired with matched noncancerous liver (Fig. 1A and Table S1; four perihilar and one mass-forming intrahepatic CC, all moderately differentiated adenocarcinoma). NOTCH3 was highly up-regulated: 18.2-fold (P ≤ 0.000025); NOTCH1, 1.9-fold (P = 0.105153); NOTCH2, 1.8-fold (P = 0.076917), and NOTCH4, 1.6-fold (P = 0.076371). Up-regulation of JAG1 (8.4-fold, P = 0.000426) and JAG2 (12.6-fold, P = 0.003088) indicated that signaling may be triggered by nearby ligand. This preliminary screen suggested pathway activity, with up-regulation of the Hes/Hey family of effectors: HEY1, 10.25-fold, (P = 0.016558); and HEYL, 6.0-fold (P = 0.000829), although in this cohort, there was no change in the archetypal effector of classical Notch, HES1 (0.9-fold, P = 0.687197; Fig. 1A). We expanded the analysis to a larger cohort of 48 CC cases and compared them with healthy livers using quantitative RT-PCR (qRT-PCR; n = 42). NOTCH3 was again up-regulated 38-fold (P ≤ 0.0001), with NOTCH1, 1.1-fold (P ≤ 0.0001); NOTCH2, 7.5-fold (P ≤ 0.0001); NOTCH4, 2.0 fold (P ≤ 0.0001); JAG1, 363.3-fold (P ≤ 0.0001); JAG2, 938.6-fold (P ≤ 0.0001); HES1, 483.7-fold (P ≤ 0.0001); HES4, 304.2-fold (P ≤ 0.0001); HEY1, 46.8-fold (P ≤ 0.0001); HEY2, 384.4-fold (P = 0.0005), HEYL, 160.6-fold (P ≤ 0.0001) (Fig. 1B). We stained the cohort and a tissue CC microarray for Notch receptors with a panel of cell-specific markers. In the healthy liver, we observed little expression of NOTCH1 (Fig. 1C and Fig. S1 A and B) in contrast to NOTCH3, which was consistently seen on vascular smooth muscle (Fig. S1A) and on many, although not all, bile ducts (Fig. 1C and Fig. S1A). Large regions of almost all tumors stained positively for NOTCH3 (19 ± 0.77% displayed >10% coverage; 31 ± 0.84% displayed >20%; 1.6 ± 5.40% displayed >40%). Pixel analysis showed mean coverage of each core was 56.2% greater in tumors compared with noncancerous controls (Fig. 1C). NOTCH1 positivity was also greater in tumors, but not to the same extent (mean coverage, 4.49 ± 3.17% tumors vs. 2.03 ± 0.43% nontumors). In all CC samples, positivity colocalized with CK19, and a subset of tumors also exhibited stromal positivity, colocalizing with the myofibroblast marker α-SMA (Fig. 1D and Fig. S1D). NOTCH3 did not colocalize with endothelial or inflammatory cell markers (CD31 and CD68) (Fig. 1D). In malignant ductules, NOTCH3 was frequently nuclear; reactivity of the intracellular domain (N3-ICD) suggested functionality (Fig. 1E). To corroborate this, we performed N3-ICD immunoblotting: the mean signal of N3-ICD (normalized to β-actin) was 95 ± 74.66 times greater in tumor vs. matched nontumor lysates (P = 0.0286; Fig. S1E). Almost all tumors exhibited stromal expression of JAGGED1 (Fig. S1F).
Fig. 1.
Notch3 is differentially activated in human CC. (A) Volcano plot of rt-PCR Notch array in human CC and patient-matched liver (n = 5, n = 5). Gray line represents P value of 0.05. Red labels, up-regulation at least fourfold; green, down-regulation at least fourfold. (B) Notch expression in human CC (n = 48) and healthy liver (n = 42) (RT-PCR). Medians compared with Mann–Whitney U test. (C) Tissue microarray human CC (n = 77) and noncancerous liver (n = 47). Representative Notch1 and Notch3 immunostaining (positive and isotype controls; Fig. S1 B and C). Filled arrowheads, Notch3+ ductules and vascular smooth muscle in healthy liver. Pixel analysis of CC and controls compared with Mann–Whitney U test. (D) Dual fluorescence of Notch3 (green) in human CC with αSMA, CD31+, or CD68+ (red) (Scale bar, 100 µm.) (E) N3-ICD (green) in human CC (white filled arrowheads). (Scale bar, 50 µm.) Data are means ± SEM. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Table S1.
Notch pathway and pathway target expression in human CC compared with non-CC liver
| Unigene | Refseq | Gene symbol | Fold change | P value |
| Hs.8546 | NM_000435 | NOTCH3 | 18.202 | 0.000025 |
| Hs.433445 | NM_002226 | JAG2 | 12.5846 | 0.003088 |
| Hs.653700 | NM_007129 | ZIC2 | 12.2213 | 0.014216 |
| Hs.234434 | NM_012258 | HEY1 | 10.2518 | 0.016558 |
| Hs.244723 | NM_001238 | CCNE1 | 9.1662 | 0.08289 |
| Hs.728907 | NM_000214 | JAG1 | 8.4314 | 0.000426 |
| Hs.173859 | NM_003507 | FZD7 | 7.5751 | 0.01193 |
| Hs.472566 | NM_014571 | HEYL | 6.0241 | 0.000829 |
| Hs.525198 | NM_003035 | STIL | 5.9598 | 0.005442 |
| Hs.563344 | NM_006186 | NR4A2 | 5.0864 | 0.178302 |
| Hs.159142 | NM_001040167 | LFNG | 4.5722 | 0.172785 |
| Hs.142912 | NM_001466 | FZD2 | 4.5159 | 0.125445 |
| Hs.502328 | NM_000610 | CD44 | 3.3175 | 0.058305 |
| Hs.386567 | NM_004120 | GBP2 | 3.1529 | 0.020033 |
| Hs.492974 | NM_003882 | WISP1 | 2.8781 | 0.669151 |
| Hs.137510 | NM_006312 | NCOR2 | 2.8553 | 0.078323 |
| Hs.94234 | NM_003505 | FZD1 | 2.5179 | 0.251392 |
| Hs.517603 | NM_002405 | MFNG | 2.4929 | 0.086738 |
| Hs.591863 | NM_003506 | FZD6 | 2.3132 | 0.155446 |
| Hs.664706 | NM_024015 | HOXB4 | 2.264 | 0.078899 |
| Hs.149261 | NM_001754 | RUNX1 | 2.1419 | 0.279567 |
| Hs.446352 | NM_004448 | ERBB2 | 1.9708 | 0.099765 |
| Hs.495473 | NM_017617 | NOTCH1 | 1.9332 | 0.105153 |
| Hs.504096 | NM_005188 | CBL | 1.8568 | 0.068651 |
| Hs.487360 | NM_024408 | NOTCH2 | 1.8422 | 0.136808 |
| Hs.2256 | NM_002423 | MMP7 | 1.8389 | 0.318428 |
| Hs.728902 | NM_203458 | NOTCH2NL | 1.838 | 0.076917 |
| Hs.437922 | NM_005376 | MYCL1 | 1.7638 | 0.372321 |
| Hs.390736 | NM_003879 | CFLAR | 1.7396 | 0.122128 |
| Hs.728776 | NM_012423 | RPL13A | 1.6804 | 0.123115 |
| Hs.716382 | NM_004210 | NEURL | 1.6387 | 0.309506 |
| Hs.404914 | NM_003183 | ADAM17 | 1.6198 | 0.142189 |
| Hs.436100 | NM_004557 | NOTCH4 | 1.5761 | 0.076371 |
| Hs.73090 | NM_002502 | NFKB2 | 1.546 | 0.503839 |
| Hs.404089 | NM_016169 | SUFU | 1.5366 | 0.575025 |
| Hs.349094 | NM_002351 | SH2D1A | 1.4657 | 0.332305 |
| Hs.517517 | NM_001429 | EP300 | 1.3542 | 0.953128 |
| Hs.592082 | NM_003502 | AXIN1 | 1.329 | 0.521975 |
| Hs.251680 | NM_000427 | LOR | 1.2918 | 0.284098 |
| Hs.445498 | NM_012245 | SNW1 | 1.2895 | 0.300704 |
| Hs.578508 | NM_001110 | ADAM10 | 1.214 | 0.384552 |
| Hs.88556 | NM_004964 | HDAC1 | 1.2073 | 0.401968 |
| Hs.34560 | NM_005574 | LMO2 | 1.2055 | 0.58369 |
| Hs.515053 | NM_001130 | AES | 1.1931 | 0.320549 |
| Hs.283565 | NM_005438 | FOSL1 | 1.0615 | 0.468063 |
| Hs.654408 | NM_003998 | NFKB1 | 1.0525 | 0.878895 |
| Hs.445733 | NM_002093 | GSK3B | 0.9903 | 0.997244 |
| Hs.520640 | NM_001101 | ACTB | 0.9579 | 0.676272 |
| Hs.272367 | NM_018411 | HR | 0.9513 | 0.745327 |
| Hs.534255 | NM_004048 | B2M | 0.9409 | 0.754207 |
| Hs.3260 | NM_000021 | PSEN1 | 0.9259 | 0.957733 |
| Hs.25363 | NM_000447 | PSEN2 | 0.9211 | 0.719164 |
| Hs.437846 | NM_005631 | SMO | 0.9192 | 0.917649 |
| Hs.250666 | NM_005524 | HES1 | 0.9042 | 0.687197 |
| Hs.632702 | NM_005269 | GLI1 | 0.9019 | 0.726693 |
| Hs.534465 | NM_172341 | PSENEN | 0.8563 | 0.643321 |
| Hs.654609 | NM_003744 | NUMB | 0.8241 | 0.916316 |
| Hs.655331 | NM_021961 | TEAD1 | 0.7914 | 0.665451 |
| Hs.459691 | NM_002613 | PDPK1 | 0.7708 | 0.465492 |
| Hs.379912 | NM_005618 | DLL1 | 0.7449 | 0.389426 |
| Hs.476018 | NM_001904 | CTNNB1 | 0.7301 | 0.341752 |
| Hs.374127 | NM_003903 | CDC16 | 0.718 | 0.168774 |
| Hs.654464 | NM_016734 | PAX5 | 0.7176 | 0.503075 |
| Hs.531754 | NM_145185 | MAP2K7 | 0.7174 | 0.605296 |
| Hs.728789 | NM_005252 | FOS | 0.6793 | 0.425548 |
| Hs.472409 | NM_172236 | POFUT1 | 0.6762 | 0.402127 |
| Hs.523852 | NM_053056 | CCND1 | 0.6041 | 0.178436 |
| Hs.198998 | NM_001278 | CHU.K. | 0.589 | 0.010609 |
| Hs.412707 | NM_000194 | HPRT1 | 0.5764 | 0.23416 |
| Hs.11392 | NM_004469 | FIGF | 0.5699 | 0.213409 |
| Hs.569700 | NM_002917 | RFNG | 0.539 | 0.264641 |
| Hs.40735 | NM_017412 | FZD3 | 0.4341 | 0.35265 |
| Hs.6347 | NM_002335 | LRP5 | 0.4239 | 0.358288 |
| Hs.197320 | NM_005077 | TLE1 | 0.4125 | 0.084711 |
| Hs.80828 | NM_006121 | KRT1 | 0.4108 | 0.178684 |
| Hs.370771 | NM_000389 | CDKN1A | 0.396 | 0.150495 |
| Hs.156979 | NM_014443 | IL17B | 0.3786 | 0.3182 |
| Hs.162646 | NM_015869 | PPARG | 0.3637 | 0.001923 |
| Hs.181300 | NM_005065 | SEL1L | 0.3378 | 0.201228 |
| Hs.856 | NM_000619 | IFNG | 0.3377 | 0.903416 |
| Hs.19545 | NM_012193 | FZD4 | 0.3148 | 0.242607 |
| Hs.533055 | NM_003884 | KAT2B | 0.2946 | 0.049651 |
| Hs.108219 | NM_004626 | WNT11 | 0.176 | 0.251588 |
| Hs.372152 | NM_004416 | DTX1 | 0.1495 | 0.011557 |
| Hs.164537 | NM_000193 | SHH | 0.0625 | 0.180074 |
| Hs.524518 | NM_003153 | STAT6 | 1.1651 | 0.155658 |
| Hs.592355 | NM_002046 | GAPDH | 1.1455 | 0.476017 |
| Hs.231367 | NM_000417 | IL2RA | 1.0872 | 0.71911 |
| Hs.169002 | NM_138296 | PTCRA | 1.0858 | 0.453129 |
Resected human CC vs. patient-matched noncancerous liver controls (n = 5). Data are represented as fold change and P value.
Fig. S1.
Supporting data for Notch expression in human liver and cholangiocarcinoma. (A) Immunofluorescent staining of Notch1 (red) and Notch3 (red) in healthy human liver (counterstained with DAPI). (Scale bars, 100 µm.) (B) Positive control for Notch1 staining: liver from AhCre+Mdm2fl/fl mice (day 3 after induction) that demonstrate a florid Notch1-driven progenitor reaction in response to large-scale hepatocyte senescence (27). Notch1 is observed around blood vessels and ductules. (C) Rabbit isotype controls for immunohistochemical staining of Notch1 and Notch3 in Fig. 1C (antibodies to Notch1 and Notch3 both raised in rabbit). (D) Notch3 immunostaining of tissue microarray of human CC and noncancerous liver controls. Filled arrowheads indicate stromal Notch3 positivity. (E) Immunoblot for Notch3 intracellular domain (N3-ICD) in human CC and patient-matched noncancerous liver lysates (samples as in Fig. 1A). Blots were probed for β-actin as loading control to which N3-ICD signal was standardized for densitometric analysis. (F) Jagged1 (red) expression in human CC in the tumor-associated stroma. (Scale bars, 100 µm.) All data are expressed as mean ± SEM. *P ≤ 0.05.
Notch3 Is Differentially Up-Regulated During CC Development.
To determine the contribution of Notch to CC development, we used a well-characterized toxin-induced model in rat using the hepatocarcinogen thioacetamide (TAA) to induce injury followed by cancer (9). After 16 wk, multifocal foci of the invasive CC are seen with mucin production and desmoplasia. The model has a penetrance of 100% at 20 wk, when tumors are numerous, large, and coalescent (Fig. S2A). We used a Notch PCR array to compare expression in uninjured animals to those with inflammation (8- to 10-wk TAA) (Fig. 2A, Left), fibrosis (12–14 wk), early malignancy (20 wk), and invasive adenocarcinoma (26 wk) (Fig. 2A, Right, and Table S2). An induction in transcription was observed in line with tumor development as confirmed with qPCR (Fig. 2B); Notch3 was a highly up-regulated receptor at 26 wk (52.01-fold by qRT-PCR; P = 0.0022), contrasted by modest up-regulation of Notch1 (5.32-fold, P = 0.0411), Notch2 (4.75-fold, P = 0.0022), and Notch4 (9.67-fold, P = 0.0022). Jag1 was up-regulated 24.00-fold (P = 0.0022). We saw nonsignificant up-regulation of Jag2 (2.35-fold, P = 0.3095), and unlike in human disease, no change in effector transcription: Hes1, 0.67-fold (P = 0.3095); Hey1, 0.70-fold (P = 0.3095); Hey2, 0.77-fold (P = 0.3939); and HeyL, 2.10-fold (P = 00649). Immunostaining the time course mirrored these data; up-regulation occurred in line with tumor expansion, with Jagged1 and Notch3 in stroma and malignant ducts (Fig. 2C).
Fig. S2.
Time course of rat model of TAA-induced liver injury with bile duct carcinogenesis. Representative H&E-stained liver sections from the TAA time course (at 10, 14, 16, 22, and 26 wk) and uninjured rat controls. (Scale bars, 100 µm.)
Fig. 2.
Notch3 is differentially up-regulated during CC development. (A) PCR Notch pathway array in rats after 600 mg/L TAA for 8–10 wk (inflamed) (n = 6) vs. control (Left) and 24–26 wk (adenocarcinoma) (n = 6) vs. control (n = 6) (Right). Red labels, at least fourfold up-regulation; green, at leat fourfold down-regulation. (B) qRT-PCR of Notch expression in TAA rat liver normalized to uninjured controls at 8–10 (inflamed), 12–14 (fibrotic), 20 (early malignant), and 26 wk (late malignant) (n = 3; n = 6 control). (C) IHC of TAA time course. CK19 (DAB). Notch3, green; Jagged1, red; αSMA, green. (Scale bar, 100 µm.)
Table S2.
Notch pathway and pathway target expression in TAA-induced CC in rats compared with matched non-CC liver
| Gene symbol | Fold change | |||
| Inflamed (8–10 wk) | Fibrotic (12–14 wk) | Early malignancy (16–20 wk) | Advanced malignancy (21–26 wk) | |
| Mmp7 | 0.9658 | 2.4933 | 9.5454 | 104.8512 |
| Heyl | 0.3739 | 1.4555 | 2.4446 | 6.5823 |
| Cd44 | 0.2347 | 0.9341 | 1.4094 | 6.3304 |
| Ccne1 | 9.4719 | 12.3632 | 6.122 | 5.8848 |
| Notch3 | 0.131 | 2.8114 | 1.269 | 5.7307 |
| Jag1 | 0.2286 | 1.4154 | 1.1896 | 4.0773 |
| Fosl1 | 1.1151 | 1.5161 | 0.9916 | 4.062 |
| Il17b | 3.1246 | 4.4388 | 3.7048 | 3.0122 |
| Ncor2 | 2.355 | 4.9988 | 2.7228 | 2.6779 |
| hr | 0.3141 | 2.3182 | 1.4387 | 2.5085 |
| Neurl | 0.5173 | 1.9776 | 1.1683 | 2.4982 |
| Fzd1 | 0.2892 | 0.8235 | 0.7968 | 2.475 |
| Fos | 0.3056 | 1.0761 | 0.6038 | 2.4218 |
| Nfkb1 | 0.8177 | 1.2992 | 1.3289 | 2.3251 |
| Mycl1 | 0.1105 | 0.5385 | 0.7352 | 2.1658 |
| Map1b | 0.5557 | 2.1614 | 1.4026 | 2.0793 |
| Dll1 | 0.7969 | 3.0818 | 1.445 | 1.7991 |
| Ccnd1 | 1.4515 | 2.4019 | 1.4937 | 1.7516 |
| Psen1 | 0.5517 | 1.013 | 1.1565 | 1.7456 |
| Tle1 | 1.7781 | 3.357 | 2.2917 | 1.7171 |
| Nfkb2 | 0.8156 | 1.5006 | 1.05 | 1.6006 |
| Hprt1 | 1.3644 | 1.1395 | 1.8796 | 1.566 |
| Gli1 | 0.131 | 1.2467 | 0.358 | 1.5506 |
| Smo | 0.184 | 2.9473 | 0.6858 | 1.5199 |
| Cdkn1a | 2.755 | 3.1557 | 2.9396 | 1.4803 |
| Gbp2 | 0.2465 | 0.265 | 0.3446 | 1.4688 |
| Ctnnb1 | 0.8229 | 1.1203 | 0.8046 | 1.4456 |
| Fzd3 | 0.6346 | 0.5179 | 0.7967 | 1.4208 |
| Il2ra | 0.0963 | 2.3574 | 0.8523 | 1.3875 |
| Hoxb4 | 0.141 | 0.8742 | 0.7024 | 1.3791 |
| Fzd7 | 0.9339 | 1.3145 | 1.5592 | 1.3746 |
| Hdac1 | 0.4119 | 0.7379 | 0.9512 | 1.3587 |
| Runx1 | 0.0638 | 0.1163 | 0.2109 | 1.3097 |
| Cdc16 | 1.1533 | 1.8708 | 1.228 | 1.2955 |
| Ep300 | 0.6419 | 1.5713 | 0.8289 | 1.2646 |
| Erbb2 | 0.0945 | 0.5952 | 0.522 | 1.2505 |
| Gsk3b | 0.5827 | 0.7904 | 1.4263 | 1.2334 |
| Supt6h | 1.179 | 2.7661 | 1.5787 | 1.1925 |
| Pdpk1 | 0.9428 | 1.4195 | 1.0594 | 1.1796 |
| Chuk | 1.6998 | 2.7608 | 1.3433 | 1.1771 |
| Map2k7 | 0.6696 | 2.5015 | 0.9001 | 1.1423 |
| Nr4a2 | 0.049 | 0.599 | 0.9597 | 1.1419 |
| Ldha | 3.7554 | 2.616 | 1.4337 | 1.1323 |
| Stat6 | 0.3678 | 0.7935 | 1.0264 | 1.1321 |
| Sufu | 0.3248 | 0.912 | 0.9442 | 1.0668 |
| Axin1 | 0.7851 | 2.291 | 0.9233 | 1.0499 |
| Lmo2 | 0.1582 | 0.2616 | 0.491 | 1.0446 |
| Jag2 | 0.0048 | 0.0455 | 0.2216 | 0.9878 |
| Rplp1 | 2.7966 | 2.9224 | 0.9466 | 0.9875 |
| Lrp5 | 0.6323 | 2.3288 | 0.71 | 0.9696 |
| Rpl13a | 1.2381 | 1.2937 | 0.8515 | 0.935 |
| Wisp1 | 0.0671 | 0.6386 | 0.1834 | 0.9306 |
| Figf | 0.1737 | 1.0062 | 0.4602 | 0.9263 |
| Notch1 | 0.5341 | 0.8383 | 0.7734 | 0.9254 |
| Aes | 0.4479 | 1.1824 | 1.1767 | 0.9229 |
| Fzd6 | 0.3298 | 1.1423 | 0.7095 | 0.9089 |
| Lfng | 0.132 | 0.227 | 0.3168 | 0.863 |
| Notch2 | 0.5683 | 2.0499 | 0.8686 | 0.8612 |
| Cflar | 0.35 | 0.7758 | 0.4303 | 0.845 |
| Psen2 | 0.4754 | 1.5348 | 0.5839 | 0.8249 |
| Rfng | 0.6476 | 1.2304 | 0.6965 | 0.811 |
| Cbl | 0.3906 | 0.3242 | 0.5618 | 0.8054 |
| Notch4 | 0.0778 | 0.1924 | 0.3379 | 0.7989 |
| Adam17 | 0.3842 | 0.4657 | 0.4597 | 0.7486 |
| Numb | 0.5473 | 1.531 | 0.7815 | 0.732 |
| Fzd5 | 0.6628 | 1.6935 | 1.1146 | 0.7159 |
| Tead1 | 0.3454 | 0.6381 | 0.6177 | 0.7157 |
| Actb | 0.1576 | 0.2593 | 0.4358 | 0.6032 |
| Ptcra | 0.1411 | 1.2467 | 0.3787 | 0.5943 |
| Wnt11 | 0.0351 | 0.0488 | 0.1646 | 0.5863 |
| Pcaf | 0.2744 | 0.6128 | 0.5401 | 0.5769 |
| Rbpjl | 0.225 | 0.4805 | 0.3876 | 0.4806 |
| Krt1 | 0.1442 | 0.6017 | 0.6005 | 0.4756 |
| Pofut1 | 0.2798 | 1.7287 | 0.1935 | 0.4483 |
| Mfng | 0.0504 | 0.2298 | 0.2023 | 0.4287 |
| Ifng | 0.131 | 1.2467 | 0.358 | 0.4251 |
| Zic2 | 0.131 | 1.2467 | 0.358 | 0.4251 |
| Fzd4 | 0.1043 | 0.3493 | 0.2346 | 0.3815 |
| Fzd2 | 0.018 | 0.1664 | 0.1184 | 0.3046 |
| Hes5 | 0.0827 | 0.6452 | 0.1198 | 0.1818 |
| Hey1 | 0.0151 | 0.0254 | 0.0484 | 0.1194 |
| Pparg | 0.1967 | 0.1586 | 0.2157 | 0.1053 |
| Sel1l | 0.3418 | 1.1946 | 0.5706 | 0.0606 |
| Shh | 0.0052 | 0.0496 | 0.0142 | 0.0169 |
| Adam10 | 0.7499 | 0.763 | 0.3217 | 0.0052 |
| Psenen | 0.7293 | 0.9997 | 1.1626 | 1.1192 |
| Hes1 | 0.6774 | 0.7625 | 1.1288 | 1.1116 |
| Ncstn | 0.9243 | 1.7537 | 0.787 | 1.0836 |
| Il6st | 0.5737 | 1.0788 | 0.7305 | 1.0779 |
Toxin-induced CC vs. matched noncancerous liver controls (n = 6). Data are represented as fold change and P value.
Reports demonstrated an inhibitory effect using γ-secretase inhibitors (GSIs) in CC cell lines and xenograft models. We aimed to evaluate efficacy on in vivo CC growth in a model where desmoplastic CC arises from the liver without transgenic overactivation of Notch. We administered N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT) to rats on the TAA protocol, treating animals during the last 5 wk of injury, i.e., once tumors had established (Fig. S3A). TAA damage was equivalent in the two groups (Fig. S3B). Following DAPT, liver-to-body weight ratio was reduced by 19 ± 0.53% (P = 0.0121; Fig. S3C), and the proportion of liver infiltrated by the tumor was reduced by 78 ± 0.84% (P = 0.0148; Fig. 3B). There was no apparent difference in the microscopic appearance of DAPT-treated tumors; all cancerous foci exhibited features of well-differentiated adenocarcinoma with mucin production and desmoplasia, with no apparent difference in cell death or necrosis histologically. Moreover, tumor number was unchanged, consistent with the observation that by 21 wk, tumors are established and DAPT after this point slows CC growth. To establish that inhibition of the γ-secretase complex resulted in a reduction in signaling via Notch3, we stained for the Notch3 protein and looked for nuclear positivity, i.e., Notch3 intracellular domain (Fig. S3D). Immunostaining for the proliferation marker Ki67 demonstrated a 38.15% reduction in cycling cells in tumor cells (P = 0.0005; 244.14 ± 10.03 vehicle vs. 150.99 ± 20.40 DAPT; Fig. 3D).
Fig. S3.
Supporting data for rat CC model treated with Notch inhibitor. (A) Rats had 26 wk of injury with TAA and thrice weekly 10 mg/kg DAPT (n = 8) or vehicle (n = 10) during weeks 21–26. (B) Serum biochemical markers of liver injury and function in rats treated with DAPT or vehicle [bilirubin, P = 0.2848; alanine aminotransferase (ALT), P = 0.3494; aspartate aminotransferase (AST), P = 0.0676; AlkPhos, P = 0.5148; albumin, P = 0.7528] (data are represented as means ± SEM). (C) Liver-to-body weight ratio of rats treated with DAPT or vehicle (P = 0.0121). (D) Representative Notch3 DAB immunostaining in TAA + vehicle– and TAA + DAPT–treated rats. Filled arrows denote N3-ICD positivity. (Scale bars, 100 µm.)
Fig. 3.
Pan-inhibition of Notch reduces CC progression. (A) Tiled low power photomicrographs of rat liver after TAA with DAPT or vehicle during weeks 21–26. (Scale bar, 100 mm.) (B) (Left) Proportion of liver infiltrated by CC after DAPT (n = 8) vs. vehicle (n = 10) (P = 0.0148). (Right) Tumor number in rats treated with DAPT or vehicle (P = 0.2856). Data are means ± SEM. Medians compared with Mann–Whitney U test (*P ≤ 0.05). (C) High- and low-power H&E sections of rat liver after vehicle (Upper) or TAA (Lower). Dashed lines are tumor boundary. (D) Ki67 immunostaining and quantification of rat liver sections after vehicle or TAA. (Scale bar, 100 µm.) Number of Ki67-positive tumor cells per ×40 field (30 fields per rat) compared using Student t test. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Genetic Deletion of Notch3 Reduces CC Formation and Progression.
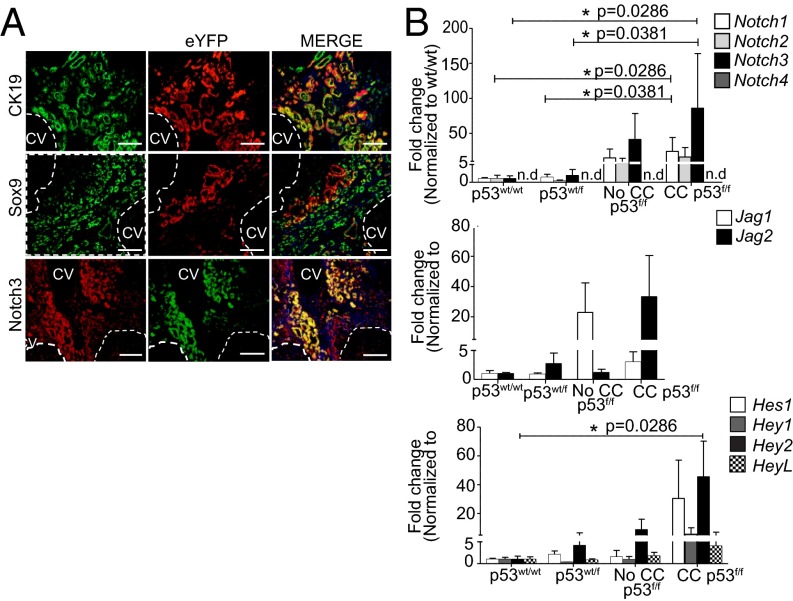
γ-Secretase is a large protease complex, and, although blockade results in total loss of Notch signal (single point mutation causes embryonic lethality) (10), Notch is only one of its substrates. Notch3 is an atypical receptor with structural differences to Notch1 and 2 and can be targeted without disrupting normal development (11). We therefore aimed to evaluate its potential as a nonredundant CC driver using genetic Notch3 deletion. Loss of the tumor suppressor p53 is a common occurrence in CC (12). CC arises following chronic inflammation as in primary sclerosing cholangitis. We therefore used a mouse model in which loss of Tp53 is conditionally targeted to enhanced yellow fluorescent protein (eYFP)-labeled CK19+ epithelia using tamoxifen inducible Cre recombinase (CK19CreERTeYFPp53f/f) followed by injury with TAA to induce oncogenic stress (13). At 26 wk, multifocal invasive CC was observed in livers of CK19CreYFPp53f/f mice at 80% penetrance, but not CK19CreYFPp53wt/f or CK19CreYFPp53wt/wt mice (Fig. S4A). Tumors stained for ductular markers CK19 and Sox9, and these frequently but not exclusively colocalized with eYFP (Fig. 4A), in line with the weak efficiency of Cre recombination in this model (14). In tumors, eYFP+ epithelia were almost always positive for NOTCH3, although not all NOTCH3+ cells carried the heritable eYFP label, indicating p53 loss is not required for Notch3 induction. In mice, we observed apparently less stromal Notch3 positivity (Fig. 4A, Bottom).
Fig. S4.
Characterization of CK19CreERT eYFPp53 CC mouse model. Representative photomicrographs of H&E-stained livers from CK19CreERT eYFPp53wt/wt, CK19CreERT eYFPp53wt/f, and CK19CreERT eYFPp53f/f mice following 26 wk of TAA or vehicle (n = 5/group). (Scale bars, 100 µm.)
Fig. 4.
Notch3 is overexpressed in a transgenic mouse model of CC. (A) Cofluorescence of CK19 (green) and eYFP (red) (Top); Sox9 (green) and eYFP (red) (Middle), and Notch3 (red) with eYFP (green) (Bottom) in CC foci from CK19CreERTR26ReYFPp53f/f mice after 26-wk TAA (CV, central vein; dashed line tumor boundary) (Scale bar, 100 µm.) (B) qRT-PCR of whole liver from CK19CreYFPp53f/f mice after 26-wk TAA. Comparisons between single groups are represented on a single graph for clarity; however, individual Mann–Whitney U tests used to compare individual genes between genotypes (p53wt/wt, n = 4; p53wt/f, n = 6; p53f/f no CC, n = 3; p53f/f with CC, n = 4) Data are means ± SEM.
Notch3 mRNA and to a lesser degree Notch2, but not Notch1 or Notch4 (undetectable), was overexpressed in CC in CK19CreYFPp53f/f mice compared with CK19CreYFPp53wt/f and CK19CreYFPp53wt/wt mice, as well as CK19CreYFPp53f/f mice without CC (Fig. 4B). When normalized to CK19CreYFPp53wt/wt mice with 26 wk of TAA, Notch3 is up-regulated 85.92-fold (P = 0.0286) in CK19CreYFPp53f/f mice with CC, compared with Notch1 at 24.28-fold (P = 0.0286). In CK19CreYFPp53f/f mice that did not develop CC, Notch3 was up-regulated 41.35-fold (P = 0.0286), compared with Notch1 at 14.94-fold (P = 0.0381). Nonsignificant increases in Jag1 and Jag2 were observed and the only effector to reach significance was Hey2: 45.47-fold (P = 0.0286; Fig. 4B).
We then compared tumor burden in CK19CreERTeYFPp53f/f mice on the TAA protocol to mice carrying constitutive deletion of the Notch3 gene (CK19CreERTp53f/fN3). A difference in livers in N3+/+ mice compared with N3+/− and N3−/− animals was seen at 26 wk (Fig. 5A). Although macroscopic cancerous nodules were not numerous on the liver surface of mice of any genotype, microscopic foci of invasive CC were clearly evident in all groups (Fig. 5 A and C). A 99.14 ± 0.48% reduction was seen in liver infiltrated by tumor in N3+/− mice, as well as a reduction in the mean tumor number [28.78 ± 15.37 N3+/+ mice (n = 9) vs. 0.875 ± 0.38 N3+/− mice (n = 8)], indicating single copy loss of Notch3 is sufficient to inhibit CC formation (Fig. 5B and Fig. S5A). N3−/− mice exhibited a similar phenotype; there was no statistical difference in tumor burden to N3+/− animals (N3+/− mice, 0.035 ± 0.01 mean% tumor area vs. 0.086 ± 0.05 N3−/−). Staining for pan-cytokeratin and pERK demonstrated an apparent reduction in proliferating malignant ductules in mice with Notch3 deletion (Fig. 5C). No significant compensatory up-regulation of Notch1, Notch2, or Notch4 was observed in response to Notch3 deletion (Fig. S5B). To evaluate the role of Notch1 in CC development, CK19CreERTeYFPp53f/fN1f/f mice were induced with tamoxifen and given TAA. These animals did not tolerate injury; they exhibited weight loss and signs of hepatic failure (jaundice and ascites), suggesting a failure of liver regeneration (Fig. S5C).
Fig. 5.
Genetic deletion of Notch3 reduces CC formation and progression. (A) Photographs and tiled low-power photomicrographs of livers from CK19CreYFPp53f/fN3+/+ (n = 9) and CK19CreYFPp53f/fN3+/− mice (n = 8) after 26-wk TAA. (Scale bar, 100 mm.) (B) Tumor number and total and % infiltrated liver area in N3+/+, N3+/−, and N3−/− mice after 26-wk TAA. Comparisons made with one-way ANOVA and Dunn’s multiple comparison test for post hoc analyses. (C) Representative H&E-, pan-CK–, and pERK-stained sections from N3+/+, N3+/−, and N3−/− mice after 26-wk TAA. Dashed line, tumor boundary. (Scale bar, 100 µm.) (D) Tumor mass and volume of NOTCH3 shRNA xenografts (n = 6) vs. scrambled control (n = 11). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Fig. S5.
Supporting data for in vivo and in vitro Notch3 inhibition experiments. (A) Area of sampled liver from CK19CreYFPp53f/fN3+/+ and CK19CreYFPp53f/fN3+/− mice for analysis of tumor burden following 26 wk of TAA. (B) qRT-PCR data for Notch receptor expression in CK19CreYFPp53f/fN3+/+, N3+/−, and N3−/− mice after 26-wk TAA. Data are mean ± SEM. (C) Kaplan–Meier survival analyses of CK19CreYFPp53f/fN3+/+-, CK19CreYFPp53f/fN3+/−, CK19CreYFPp53f/fN3−/−, and CK19CreYFPp53f/fN1f/f mice during 26 wk of TAA injury. (D) Immunocytochemistry of Notch 1, 2, 3, and 4 and Jagged1 in human CC lines; CC-LP-1, CC-SW-1, and SNU-1079 with isotype controls. Counterstained with DAPI. (Scale bars, 50 µm.) (E) NOTCH3 gene expression in human CC cells (CC-LP-1) following transfection and puromycin selection of NOTCH3 shRNA and scrambled sequence and untransfected controls. (F) NOTCH3 and N3-ICD immunoblots of CC cells (CC-LP-1) following transfection and puromycin selection of NOTCH3 shRNA and scrambled sequence and untransfected controls. Clone 1 was taken forward for further analysis as a Notch3 knockdown cell line. (G) HES/HEY expression in NOTCH3 knockdown human CC cells (clone1 from C and D; fold change: HES1, +1.4-fold, P = 0.7922; HES4, −5.1-fold, P = 0.0303; HEY1, −15.58-fold, P = 0.1775; HEY2, −9.16-fold, P = 0.0043; HEYL, −6.03-fold, P = 0.0823). (H) MTT viability assessment of Notch3 WT, scrambled control, and Notch3 knockdown human CC cells following 72 h of culture at a cell density of 1.0 × 105 cells/mL (n = 6 per group; comparison made between N3 KD cells and scrambled shRNA control). (I) Assessment of neoangiogenesis using CD31+ immunostaining and quantification in human CC xenografts from NOTCH3 KD cells compared with scrambled controls (n = 6/group). Data are mean ± SEM. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
To assess whether this role for Notch3 was reproducible in a human system, we stably inhibited the gene using shRNA in cultured human CC cells and xenografts. Immunofluorescence of receptors was performed on three lines, and one was selected (CC-LP-1) (Fig. S5D). Cells were transfected with four independent shRNA with puromycin resistance cassettes for stable selection or scrambled sequences for comparison (15). Across multiple colonies, transfection inhibited NOTCH3 expression (Fig. S5 E and F). Almost total ablation of effectors was observed, suggesting functional signaling inhibition (Fig. S5G). Clone 1 exhibited efficient knockdown and was used for further experiments. In vitro, a modest attenuation in proliferation was observed [19.42 ± 2.87% reduction in 3-(4,5-dimethylthiazol-2-yl)-diphenyltetrazolium bromide (MTT) absorbance; P = 0.0765; Fig. S5H], and when xenografted, a 62 ± 28.74% reduction in size (P = 0.0237) and 76 ± 28.44% reduction in mass (P = 0.0237) was seen in Notch3 KD xenografts (Fig. 5D). We confirmed this was not due to reduced neoangiogenesis by quantification of CD31 (mean signal CD31 to DAPI, 0.0506 ± 0.0056 scrambled vs. 0.0285 ± 0.0079 N3shRNA xenografts; P = 0.329; Fig. S5I).
Genetic Silencing of Notch3 but Not RBPJ Reduces Signaling Through the PI3k-AKT Cascade.
We then sought to identify potential targets preferentially activated by Notch3 that might drive cell survival or proliferation. To compare the immediate effects of knockdown on downstream signaling, we transfected human CC cells (CC-LP-1) with siRNA against either NOTCH3 or the canonical effector RBPJ. Inhibition was confirmed with qRT-PCR and immunoblotting (Fig. S6 A and B). Eighty-four known drivers of hepatic carcinogenesis were screened with a PCR array (Tables S3 and S4). Almost all genes exhibiting changes in transcription (defined as at least fourfold) were either upstream mediators or downstream targets of the AKT cascade including MET, IRS1, and XIAP and the death receptors FAS and FADD. Surprisingly no changes were observed in response to RBPJ inhibition (Fig. 6A and Tables S3 and S4).
Fig. S6.
Data supporting efficacy of NOTCH3 and RBPJ knockdown in siRNA experiments. (A) qRT-PCR and immunoblots of NOTCH3 and RBPJ expression in human CC cells (CC-LP-1) 48 h following transfection with NOTCH3 and RBPJ siRNA, respectively, with comparison with scrambled sequences and untransfected controls. Comparisons between groups performed using Kruskal–Wallis test. (B) qRT-PCR analysis of HES/HEY gene family expression in CC cells 48 h following transfection with NOTCH3 or RBPJ siRNA or scrambled and untransfected controls. Data are expressed as mean ± SEM in A and as single measurements of gene expression (qRT-PCR performed in triplicate) in C. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. (C) AKT target gene expression in xenografted Notch3 shRNA CC cells and scrambled controls. *P ≤ 0.05, **P ≤ 0.01.
Table S3.
Liver oncogene and cell cycle gene expression in human CC cells (CC-LP-1) treated with siRNA targeted to NOTCH3
| Gene symbol | Compared with scrambled control | ||||||||
| NOTCH3_1 | NOTCH3_2 | NOTCH3_3 | Untransfected | Scrambled | |||||
| Ct | Fold change | Ct | Fold change | Ct | Fold change | Ct | Fold change | Ct | |
| ADAM17 | 26.13 | 1.31 | 26.5 | 1.47 | 26.57 | 1.09 | 26 | 1.21 | 25.37 |
| AKT1 | 20.04 | 0.75 | 20 | 0.63 | 20.47 | 0.62 | 20.57 | 1.09 | 20.09 |
| ANGPT2 | 20.82 | 0.67 | 20.8 | 0.57 | 21.09 | 0.49 | 21.17 | 0.86 | 21.03 |
| BAX | 19.06 | 0.76 | 19.08 | 0.67 | 19.58 | 0.67 | 19.53 | 1.07 | 19.08 |
| BCL2 | 26.15 | 1 | 26.16 | 0.88 | 26.51 | 0.79 | 26.25 | 1.09 | 25.77 |
| BCL2L1 | 22.71 | 0.83 | 22.84 | 0.79 | 23.59 | 0.93 | 23.49 | 1.44 | 22.61 |
| BID | 23.06 | 0.89 | 23.55 | 1.08 | 23.78 | 0.9 | 23.13 | 0.94 | 22.86 |
| BIRC2 | 25.66 | 2.4 | 25.86 | 2.4 | 26.26 | 2.23 | 23.5 | 0.54 | 24.02 |
| BIRC5 | 25.21 | 2.04 | 25.6 | 2.32 | 26.31 | 2.68 | 23.58 | 0.67 | 23.81 |
| CASP8 | 29.79 | 0.82 | 29.8 | 0.71 | 30.16 | 0.65 | 29.79 | 0.83 | 29.71 |
| CCL5 | 31.81 | 0.78 | 31.59 | 0.58 | 31.13 | 0.3 | 31.37 | 0.58 | 31.8 |
| CCND1 | 19.16 | 1.54 | 19.2 | 1.38 | 19.43 | 1.14 | 18.52 | 1 | 18.16 |
| CCND2 | 27.99 | 0.65 | 28.06 | 0.6 | 28.46 | 0.55 | 28.67 | 1.06 | 28.23 |
| CDH1 | 0 | 0.77 | 40 | 7.39E+11 | 0 | 0.47 | 40 | 8.59E+11 | 0 |
| CDH13 | 36.52 | 7.60E+10 | 0 | 0.67 | 0 | 0.47 | 0 | 0.78 | 0 |
| CDKN1A | 20.37 | 0.84 | 20.36 | 0.73 | 20.74 | 0.67 | 21.53 | 1.91 | 20.24 |
| CDKN1B | 24.23 | 1.42 | 24.23 | 1.24 | 24.68 | 1.19 | 24.61 | 1.87 | 23.35 |
| CDKN2A | 22.18 | 1.25 | 22.17 | 1.08 | 22.69 | 1.09 | 22.08 | 1.18 | 21.48 |
| CFLAR | 23.9 | 1.15 | 23.18 | 0.61 | 23.97 | 0.74 | 24.15 | 1.38 | 23.33 |
| CTNNB1 | 22.74 | 0.89 | 22.75 | 0.78 | 22.48 | 0.45 | 23.08 | 1.14 | 22.54 |
| CXCR4 | 33.34 | 1.79 | 32.77 | 1.05 | 32.89 | 0.8 | 31.38 | 0.46 | 32.13 |
| DAB2IP | 23.7 | 0.52 | 23.88 | 0.51 | 24.04 | 0.4 | 23.78 | 0.56 | 24.27 |
| DLC1 | 20.3 | 1.08 | 20.67 | 1.21 | 20.63 | 0.83 | 20.2 | 1.02 | 19.82 |
| E2F1 | 21.51 | 0.89 | 21.22 | 0.64 | 21.77 | 0.66 | 21.66 | 1 | 21.3 |
| EGF | 0 | 0 | 0 | 0 | 40 | 42.22 | 0 | 0 | 33.52 |
| EGFR | 20.55 | 0.82 | 20.35 | 0.62 | 20.8 | 0.6 | 21.36 | 1.46 | 20.46 |
| EP300 | 21.92 | 0.95 | 22.12 | 0.95 | 22.2 | 0.71 | 22.63 | 1.57 | 21.62 |
| FADD | 26.02 | 3.09 | 26.54 | 3.85 | 26.46 | 2.57 | 24.95 | 1.49 | 24.02 |
| FAS | 25.34 | 3.85 | 25.32 | 3.31 | 25.61 | 2.85 | 23.28 | 0.94 | 23.02 |
| FHIT | 34.96 | 1.35 | 36.92 | 4.58 | 37.64 | 5.31 | 34 | 0.7 | 34.15 |
| FLT1 | 0 | 0.77 | 38.4 | 2.44E+11 | 0 | 0.47 | 0 | 0.78 | 0 |
| FZD7 | 23.85 | 0.66 | 23.8 | 0.55 | 23.98 | 0.44 | 24.13 | 0.81 | 24.08 |
| GADD45B | 20.2 | 0.76 | 20.46 | 0.79 | 20.96 | 0.79 | 20.63 | 1.04 | 20.22 |
| GSTP1 | 20.01 | 1.8 | 19.58 | 1.16 | 19.8 | 0.95 | 19.18 | 1.02 | 18.79 |
| HGF | 24.63 | 1.42 | 24.27 | 0.96 | 24.6 | 0.85 | 24.55 | 1.36 | 23.75 |
| HHIP | 34.92 | 2.78 | 34.66 | 2.02 | 34.69 | 1.45 | 34.66 | 2.35 | 33.07 |
| HRAS | 20.15 | 1.08 | 19.97 | 0.83 | 20.51 | 0.85 | 21.23 | 2.3 | 19.67 |
| IGF2 | 40 | 1.19 | 40 | 1.03 | 40 | 0.73 | 39.95 | 1.16 | 39.38 |
| IRS1 | 30.93 | 3.72 | 31.35 | 4.33 | 31.01 | 2.41 | 28.59 | 0.74 | 28.66 |
| ITGB1 | 21.63 | 2.24 | 21.82 | 2.23 | 21.86 | 1.61 | 20.14 | 0.81 | 20.09 |
| KDR | 0 | 0.77 | 0 | 0.67 | 0 | 0.47 | 0 | 0.78 | 0 |
| LEF1 | 30.61 | 1.16 | 30.84 | 1.19 | 30.89 | 0.86 | 30.48 | 1.07 | 30.02 |
| MCL1 | 20.02 | 1.29 | 19.62 | 0.85 | 20.46 | 1.07 | 19.67 | 1.02 | 19.28 |
| MET | 22.98 | 4.28 | 23.29 | 4.61 | 23.34 | 3.36 | 20.83 | 0.98 | 20.51 |
| MSH2 | 23.49 | 2.18 | 23.96 | 2.63 | 23.95 | 1.84 | 21.86 | 0.71 | 21.99 |
| MSH3 | 23.24 | 1 | 23.66 | 1.16 | 23.68 | 0.83 | 23.48 | 1.19 | 22.87 |
| MTDH | 19.77 | 1.03 | 19.75 | 0.88 | 20.02 | 0.75 | 19.78 | 1.05 | 19.36 |
| MYC | 19.06 | 0.92 | 18.86 | 0.7 | 19.31 | 0.67 | 19.42 | 1.19 | 18.81 |
| NFKB1 | 22.81 | 0.79 | 23.02 | 0.8 | 23.31 | 0.69 | 23.62 | 1.41 | 22.77 |
| NRAS | 20.73 | 0.71 | 20.84 | 0.67 | 20.97 | 0.52 | 20.91 | 0.82 | 20.84 |
| OPCML | 0 | 0.77 | 0 | 0.67 | 0 | 0.47 | 0 | 0.78 | 0 |
| PDGFRA | 33.9 | 0.19 | 0 | 0 | 35.69 | 0.4 | 0 | 0 | 35.94 |
| PIN1 | 20.37 | 0.56 | 20.22 | 0.44 | 20.7 | 0.43 | 20.62 | 0.67 | 20.84 |
| PTEN | 25.6 | 4.58 | 25.33 | 3.31 | 26.25 | 4.41 | 24 | 1.53 | 23.03 |
| PTGS2 | 36.09 | 1.99 | 0 | 0 | 0 | 0 | 34.88 | 0.87 | 34.72 |
| PTK2 | 21.89 | 1.04 | 22.05 | 1.01 | 22.14 | 0.76 | 21.85 | 1.02 | 21.46 |
| PYCARD | 31.72 | 1.08 | 31.12 | 0.62 | 31.81 | 0.7 | 31.51 | 0.94 | 31.24 |
| RAC1 | 21.62 | 2.78 | 22.34 | 3.99 | 22.13 | 2.43 | 19.94 | 0.88 | 19.77 |
| RASSF1 | 27.21 | 2.24 | 27.6 | 2.56 | 27.1 | 1.27 | 26.63 | 1.52 | 25.67 |
| RHOA | 20.83 | 0.75 | 21.16 | 0.82 | 21.55 | 0.75 | 20.37 | 0.55 | 20.88 |
| RUNX3 | 30.48 | 3.65 | 30.36 | 2.92 | 30.82 | 2.83 | 28.97 | 1.3 | 28.24 |
| SFRP2 | 0 | 0.77 | 0 | 0.67 | 0 | 0.47 | 0 | 0.78 | 0 |
| SMAD4 | 24.18 | 2.26 | 23.87 | 1.59 | 24.05 | 1.27 | 23.22 | 1.18 | 22.63 |
| SMAD7 | 29.03 | 0.96 | 29.6 | 1.24 | 28.93 | 0.55 | 29.48 | 1.33 | 28.71 |
| SOCS1 | 28.03 | 0.84 | 28.22 | 0.84 | 28.54 | 0.74 | 28.29 | 1.02 | 27.9 |
| SOCS3 | 24.3 | 1.51 | 24.35 | 1.36 | 24.83 | 1.34 | 23.52 | 0.89 | 23.33 |
| STAT3 | 21.75 | 1.2 | 21.96 | 1.21 | 21.74 | 0.73 | 21.83 | 1.29 | 21.11 |
| TCF4 | 21.53 | 0.71 | 21.66 | 0.68 | 22.2 | 0.69 | 21.74 | 0.83 | 21.65 |
| TERT | 25.79 | 0.73 | 25.75 | 0.62 | 25.88 | 0.48 | 25.91 | 0.8 | 25.87 |
| TGFA | 33.85 | 1.44 | 33.25 | 0.83 | 32.33 | 0.31 | 33.58 | 1.21 | 32.95 |
| TGFB1 | 18.9 | 0.46 | 18.61 | 0.32 | 19.2 | 0.34 | 19.06 | 0.52 | 19.66 |
| TGFBR2 | 20.01 | 0.76 | 19.69 | 0.53 | 20.21 | 0.54 | 20.29 | 0.94 | 20.03 |
| TLR4 | 40 | 8.48E+11 | 0 | 0.67 | 0 | 0.47 | 0 | 0.78 | 0 |
| TNFRSF10B | 24.41 | 0.82 | 24.71 | 0.88 | 24.74 | 0.63 | 24.89 | 1.16 | 24.32 |
| TNFSF10 | 35.78 | 2.94 | 35.77 | 2.54 | 33.29 | 0.32 | 27.63 | 0.01 | 33.85 |
| TP53 | 24.28 | 1.01 | 24.28 | 0.88 | 24.67 | 0.81 | 24.19 | 0.96 | 23.89 |
| VEGFA | 22.24 | 0.91 | 22.36 | 0.86 | 23 | 0.94 | 22.37 | 1 | 22.01 |
| WT1 | 35.94 | 5.09E+10 | 0 | 0.67 | 0 | 0.47 | 0 | 0.78 | 0 |
| XIAP | 30.69 | 7.24 | 30.74 | 6.53 | 32.21 | 12.73 | 27.09 | 0.6 | 27.46 |
| YAP1 | 21.47 | 0.72 | 21.62 | 0.7 | 22.03 | 0.66 | 21.5 | 0.75 | 21.56 |
Three independent siRNA sequences target to the NOTCH3 gene were used (NOTCH3_1; NOTCH3_2; NOTCH3_3) and RNA was pooled from three replicate wells for each sequence. Gene expression was measured 48 h following transfection and compared with scrambled sequence controls. Data are expressed as fold change and P value.
Table S4.
Liver oncogene and cell cycle gene expression in human CC cells (CC-LP-1) treated with siRNA targeted to RBPJ
| Gene symbol | Compared with scrambled control | ||||||||
| RBPJ_1 | RBPJ_2 | RBPJ_3 | Untransfected | Scrambled | |||||
| Ct | Fold change | Ct | Fold change | Ct | Fold change | Ct | Fold change | Ct | |
| ADAM17 | 24.32 | 1.77 | 26.06 | 1.21 | 24.81 | 1.17 | 25.19 | 0.98 | 24.29 |
| AKT1 | 22.3 | 1.62 | 23.09 | 2.13 | 23.17 | 0.82 | 22.85 | 1.12 | 22.14 |
| ANGPT2 | 23.12 | 0.98 | 24.66 | 0.77 | 23.04 | 0.96 | 22.83 | 1.22 | 22.24 |
| BAX | 21.31 | 1.3 | 22.52 | 1.27 | 21.59 | 0.99 | 21.78 | 0.95 | 20.83 |
| BCL2 | 28.82 | 2.21 | 30.99 | 1.12 | 30.29 | 0.74 | 29.67 | 1.24 | 29.11 |
| BCL2L1 | 24.74 | 1.54 | 26.46 | 1.06 | 25.52 | 0.83 | 25.53 | 0.91 | 24.51 |
| BID | 22.57 | 2.47 | 24.58 | 1.39 | 23.31 | 1.37 | 23.3 | 1.51 | 23.02 |
| BIRC2 | 22.56 | 1.03 | 24.02 | 0.85 | 23.43 | 0.52 | 22.78 | 0.89 | 21.74 |
| BIRC5 | 24.19 | 0.72 | 25.92 | 0.5 | 24.18 | 0.68 | 24.46 | 0.61 | 22.87 |
| CASP8 | 32.57 | 0.83 | 33.47 | 1.01 | 31.82 | 1.3 | 32.32 | 1 | 31.45 |
| CCL5 | 33.26 | 1.03 | 33.72 | 1.71 | 32.45 | 1.68 | 32.48 | 1.8 | 32.45 |
| CCND1 | 18.85 | 2.09 | 20.96 | 1.1 | 19.76 | 1.03 | 19.97 | 0.98 | 19.06 |
| CCND2 | 29.2 | 1.59 | 30.25 | 1.74 | 29.64 | 1.08 | 30.1 | 0.86 | 29.01 |
| CDH1 | 37.3 | 1.81 | 37.62 | 4.11 | 36.74 | 1.68 | 37.07 | 1.84 | 36.18 |
| CDH13 | 40 | 1.81 | 40 | 4.11 | 40 | 1.68 | 40 | 1.84 | 39.97 |
| CDKN1A | 22.52 | 1.76 | 24.68 | 0.9 | 22.8 | 1.34 | 22.87 | 1.4 | 22.48 |
| CDKN1B | 24.04 | 1.97 | 25.99 | 1.16 | 24.7 | 1.15 | 25 | 1.03 | 24.16 |
| CDKN2A | 23.76 | 1.14 | 25.62 | 0.71 | 23.66 | 1.13 | 23.88 | 1.06 | 23.09 |
| CFLAR | 25.48 | 1 | 26.47 | 1.15 | 25.52 | 0.91 | 25.87 | 0.78 | 24.63 |
| CTNNB1 | 24.35 | 1.44 | 25.14 | 1.89 | 25.5 | 0.6 | 25.63 | 0.6 | 24.02 |
| CXCR4 | 32.53 | 1.56 | 34.54 | 0.88 | 33.53 | 0.72 | 36.32 | 0.29 | 32.32 |
| DAB2IP | 26.94 | 1.36 | 27.98 | 1.51 | 27.44 | 0.89 | 27.65 | 0.84 | 26.53 |
| DLC1 | 21.7 | 1.17 | 22.8 | 1.24 | 21.79 | 1.02 | 22.3 | 0.78 | 21.07 |
| E2F1 | 22.96 | 2.28 | 24.64 | 1.61 | 23.95 | 1.06 | 24.58 | 0.75 | 23.29 |
| EGF | 34.76 | 1.36 | 37.16 | 2.62 | 33.79 | 2.47 | 35.87 | 1.17 | 34.35 |
| EGFR | 21.57 | 2.23 | 23.47 | 1.36 | 22.58 | 1.03 | 22.92 | 0.89 | 21.87 |
| EP300 | 23.05 | 1.39 | 24.55 | 1.12 | 23.71 | 0.82 | 24.16 | 0.65 | 22.67 |
| FADD | 23.81 | 2.49 | 26.11 | 1.15 | 24.78 | 1.18 | 25.04 | 1.08 | 24.27 |
| FAS | 22.87 | 1.9 | 25.17 | 0.88 | 23.59 | 1.07 | 23.91 | 0.94 | 22.94 |
| FHIT | 35.77 | 1.81 | 37.26 | 4.11 | 34.99 | 1.69 | 37.17 | 1.84 | 35.83 |
| FLT1 | 0 | 1.81 | 0 | 4.11 | 0 | 1.68 | 0 | 1.84 | 0 |
| FZD7 | 27.12 | 1.3 | 28.63 | 1.04 | 27.18 | 1.15 | 27.66 | 0.91 | 26.64 |
| GADD45B | 22.5 | 1.81 | 24.63 | 0.94 | 22.92 | 1.25 | 23.1 | 1.21 | 22.5 |
| GSTP1 | 19.22 | 2.12 | 20.96 | 1.44 | 19.69 | 1.42 | 19.92 | 1.33 | 19.45 |
| HGF | 25.99 | 1.87 | 27.88 | 1.15 | 26.98 | 0.87 | 26.99 | 0.95 | 26.04 |
| HHIP | 34.67 | 0.94 | 36.76 | 1.71 | 35.35 | 0.7 | 36.78 | 0.76 | 33.73 |
| HRAS | 22.18 | 1.77 | 24.23 | 0.97 | 22.57 | 1.25 | 22.63 | 1.32 | 22.15 |
| IGF2 | 40 | 1.81 | 40 | 4.11 | 40 | 1.68 | 40 | 1.84 | 40 |
| IGFBP1 | 40 | 1.81 | 40 | 4.11 | 40 | 1.68 | 0 | 1.84 | 40 |
| IGFBP3 | 27.72 | 2.04 | 29.67 | 1.2 | 29.01 | 0.77 | 28.72 | 1.03 | 27.89 |
| IRS1 | 26.61 | 2.56 | 29.04 | 1.08 | 28.3 | 0.74 | 28.45 | 0.72 | 27.11 |
| ITGB1 | 18.53 | 3.96 | 21.93 | 0.85 | 20.61 | 0.87 | 20.79 | 0.84 | 19.66 |
| KDR | 38.12 | 1.81 | 40 | 4.11 | 38.42 | 1.68 | 40 | 1.84 | 38.39 |
| LEF1 | 32.19 | 0.68 | 33.11 | 0.82 | 31.78 | 0.84 | 32.14 | 0.71 | 30.78 |
| MCL1 | 22.44 | 0.93 | 23.94 | 0.75 | 22.2 | 1.02 | 22.08 | 1.21 | 21.48 |
| MET | 19.89 | 1.77 | 21.93 | 0.98 | 21.1 | 0.71 | 21.52 | 0.58 | 19.86 |
| MSH2 | 21.94 | 1.51 | 23.59 | 1.09 | 21.94 | 1.4 | 22.01 | 1.46 | 21.68 |
| MSH3 | 24.01 | 1.76 | 25.86 | 1.11 | 24.57 | 1.11 | 24.88 | 0.98 | 23.97 |
| MTDH | 21.59 | 1.52 | 23.33 | 1.04 | 21.85 | 1.18 | 21.66 | 1.47 | 21.34 |
| MYC | 20.91 | 1.71 | 22.65 | 1.16 | 21.16 | 1.33 | 21.15 | 1.47 | 20.83 |
| NFKB1 | 25.92 | 1.03 | 27.24 | 0.94 | 26.04 | 0.88 | 26.05 | 0.96 | 25.11 |
| NRAS | 22.86 | 1.75 | 25.07 | 0.86 | 23.6 | 0.97 | 23.07 | 1.53 | 22.81 |
| OPCML | 40 | 1.81 | 40 | 4.11 | 40 | 1.68 | 39.5 | 1.84 | 40 |
| PDGFRA | 35.99 | 1.81 | 40 | 4.11 | 36.64 | 1.68 | 37.86 | 1.84 | 35.98 |
| PIN1 | 22.96 | 1.2 | 25.05 | 0.64 | 24.19 | 0.48 | 23.71 | 0.72 | 22.37 |
| PTEN | 23.49 | 2.08 | 26.65 | 0.53 | 24.99 | 0.68 | 24.55 | 1.01 | 23.69 |
| PTGS2 | 0 | 1.81 | 26.34 | 1663.49 | 0 | 1.68 | 0 | 1.84 | 0 |
| PTK2 | 22.31 | 1.8 | 23.86 | 1.39 | 23.06 | 0.99 | 23.33 | 0.9 | 22.3 |
| PYCARD | 32.77 | 2.08 | 33.88 | 2.19 | 33.1 | 1.53 | 33.47 | 1.3 | 32.97 |
| RAC1 | 19.95 | 1.86 | 22.5 | 0.72 | 20.83 | 0.94 | 20.6 | 1.2 | 19.99 |
| RASSF1 | 26.53 | 1.94 | 28.71 | 0.97 | 28.12 | 0.6 | 28.51 | 0.5 | 26.63 |
| RHOA | 21.11 | 1.27 | 23.06 | 0.75 | 21.59 | 0.84 | 20.53 | 1.93 | 20.6 |
| RUNX3 | 30.26 | 1.33 | 32.15 | 0.81 | 31.07 | 0.7 | 30.96 | 0.83 | 29.81 |
| SFRP2 | 36.7 | 1.81 | 40 | 4.11 | 33.18 | 5.92 | 37.88 | 1.84 | 36.54 |
| SMAD4 | 23.84 | 1.6 | 25.92 | 0.86 | 25.11 | 0.61 | 24.7 | 0.89 | 23.66 |
| SMAD7 | 29.46 | 1.51 | 31.02 | 1.16 | 29.81 | 1.1 | 30.59 | 0.7 | 29.2 |
| SOCS1 | 30.53 | 1.68 | 31.85 | 1.53 | 31.76 | 0.66 | 31.47 | 0.89 | 30.42 |
| SOCS3 | 23.92 | 1.8 | 25.13 | 1.77 | 24.92 | 0.83 | 25.26 | 0.72 | 23.91 |
| STAT3 | 22.99 | 1.32 | 23.66 | 1.88 | 23.25 | 1.02 | 23 | 1.33 | 22.53 |
| TCF4 | 23.63 | 1.1 | 24.74 | 1.16 | 23.83 | 0.89 | 23.53 | 1.19 | 22.91 |
| TERT | 28.67 | 1.91 | 29.67 | 2.17 | 30.24 | 0.6 | 30.14 | 0.7 | 28.75 |
| TGFA | 35.79 | 1.53 | 38.68 | 3.48 | 35.55 | 1.42 | 34.55 | 2.12 | 34.76 |
| TGFB1 | 21.07 | 1.74 | 22.94 | 1.08 | 21.77 | 0.99 | 21.55 | 1.26 | 21.01 |
| TGFBR2 | 21.82 | 1 | 23.08 | 0.95 | 22.17 | 0.72 | 22.26 | 0.75 | 20.96 |
| TLR4 | 34.47 | 2.61 | 36.03 | 4.11 | 34.57 | 2.26 | 35.05 | 1.84 | 35.64 |
| TNFRSF10B | 24.75 | 1.84 | 26.38 | 1.35 | 25.79 | 0.83 | 26.66 | 0.5 | 24.77 |
| TNFSF10 | 34.53 | 2.51 | 37.86 | 4.11 | 34.54 | 2.31 | 37.62 | 1.84 | 35.91 |
| TP53 | 27.61 | 1.2 | 28.16 | 1.87 | 27.83 | 0.96 | 28.2 | 0.81 | 27.02 |
| VEGFA | 24.99 | 1.46 | 26.66 | 1.04 | 25.29 | 1.1 | 25.94 | 0.77 | 24.68 |
| WT1 | 37.28 | 1.81 | 40 | 4.11 | 36.12 | 1.68 | 40 | 1.84 | 40 |
| XIAP | 24.89 | 1.75 | 27.07 | 0.88 | 25.73 | 0.91 | 25.71 | 1 | 24.84 |
| YAP1 | 20.82 | 2.08 | 22.75 | 1.24 | 22.08 | 0.8 | 22.1 | 0.87 | 21.02 |
Three independent siRNA sequences targeted to RBPJ were used (RBPJ_1; RBPJ_2; RBPJ_3) and RNA was pooled from three replicate wells for each sequence. Gene expression was measured 48 h following transfection and compared with scrambled sequence controls. Data are expressed as fold change and P value.
Fig. 6.
Genetic silencing of Notch3, but not RBPJ, reduces PI3k-AKT transcription. (A) Human (CC-LP-1) cells transfected with NOTCH3 or RBPJ siRNA and analyzed with oncogene PCR array. Three independent siRNA sequences were used, and RNA was pooled from three replicate wells for each sequence. Gene expression measured 48 h after transfection and compared with scrambled controls (dotted line represents no change in transcription). Genes in color are at least fourfold down-regulated. (B) IHC of pAKT(Thr308), pmTor, and pS6 with pixel analysis in Notch3 shRNA/scrambled CC-LP-1 xenografts. (Scale bar, 100 µm.)
We therefore returned to previous models to assess whether induction of AKT by Notch3 held true in these systems. In shRNA Notch3 KD CC xenografts, pixel analysis revealed reduced phosphorylated AKT(Thr308) (0.537 ± 0.078 rodamine:DAPI signal scrambled vs. 0.346 ± 0.115 N3shRNA tumors), as well as phosphorylated downstream targets p-mTor (1.465 ± 0.675 scrambled vs. 0.606 ± 0.211 N3 shRNA) and pS6 (1.194 ± 0.322 scrambled vs. 0.379 ± 0.996 N3 shRNA; Fig. 6B). At the gene level, qPCR results mirrored the reduced transcription of targets identified in the siRNA-treated cells using the PCR array: MET, IRS1, FAS, and RAC1 (Fig. S6C).
To confirm this phenomenon was not an off-target effect of RNAi, we looked at Akt in CK19CreERTeYFPp53f/f mice on the TAA protocol with (n = 8) and without (n = 9) Notch3 deletion. A reduction in Fas, Fadd, and Rac1 gene expression was seen, although this did not reach significance (Fig. S7A). Immunoblots, however, revealed a 72% reduction in pAKT protein (N3+/− 0.41 ± 0.10 vs. N3−/− 0.12 ± 0.06; P = 0.0426), a 30% reduction in pmTor (N3+/− 1.55 ± 0.13 vs. N3−/− 1.08 ± 0.13; P = 0.0426), a 54% reduction in pS6 (N3+/− 1.19 ± 0.13 vs. N3−/− 0.54 ± 0.16; P = 0.0127) and an 88% reduction in p70-S6 (N3+/− 0.91 ± 0.34 vs. N3−/− 0.11 ± 0.06; P = 0.0593; Fig. 7A). Finally, to independently verify Akt blockade reduces CC growth, we xenografted nude mice with WT CC cells, allowed tumors to establish, and systemically treated them with a small molecule inhibitor of PI3K, PI-103. At 28 d, we observed a 60.87% reduction in tumor size (mean volume, 228.07 ± 48.68 vs. 89.25 ± 32.54 mm3 vehicle; P = 0.0288; Fig. S7B).
Fig. S7.
Supporting data for the role of PI3K/AKT pathway in Notch3 driven cholangiocarcinogenesis. (A) AKT target expression in CK19CreYFPp53f/fN3+/+ and CK19CreYFPp53f/fN3+/− mice after 26 wk of TAA. (B) CC xenograft size after 14-d treatment with the PI3k inhibitor PI-103 (30 mg/kg) or vehicle. *P ≤ 0.05.
Fig. 7.
Genetic silencing of Notch3 reduces activity through the PI3k-AKT cascade. Immunoblots and corresponding densitometry for p-Akt, p-mTor, p-S6, and p-p70 S6k from CK19CreYFPp53f/fN3+/+ and CK19CreYFPp53f/fN3+/− livers after TAA. *P ≤ 0.05.
Discussion
Exogenous oncogene activation in mice can initiate carcinogenesis in many tissues and indeed often in tissues where these oncogenes are not overexpressed or mutated in human cancer. Consistent with the role of Notch as a cell fate determinant, transgenic overactivation of Notch1 (N1-ICD) in albumin-expressing cells results in biliary tumor formation (6, 7). In an almost identical model, however, N1-ICD expression under albumin and α-fetoprotein promoters produce HCC at 100% penetrance (16). Studies of KRAS and MYC show precise expression levels are critical to biological outcome. Because genomic analyses of CC conclude transforming Notch mutations are infrequent (17) and antibodies blocking Notch1 increase the number and extent of tumors (18), we aimed to elucidate the contribution of endogenous WT Notch to CC and identify components with potential for targeting.
We used CC models in three species not reliant on any one oncogenic alteration, and Notch3 is consistently overexpressed. As reported by others, Notch1 is barely detectable in the healthy adult liver (19). Conversely, Notch3 is consistently present around the vasculature, making the up-regulation observed in tumors all of the more striking. Notch3 up-regulation occurs with disease; the greatest increase occurs late during expansion and invasion. Overexpression is associated with functional activity as evidenced by consistent nuclear visualization of the intracellular domain. Inhibition in xenografted cells with shRNA or genetic KO in mice both result in attenuated tumor growth. This target, with many functions and interactions distinct from canonical signaling, offers an attractive prospect for therapy. Past work suggests antibody-mediated Notch3 inhibition has no effect on liver cancer; however, evidence of Notch3 activity in the model and antibody efficacy was lacking (18). In contrast, other work acknowledges that, in addition to Notch1, Notch3 is strongly expressed in human CC compared with the liver (7).
Notch3 drives 40% of non–small-cell lung cancers (NSCLCs) and almost all T-cell acute lymphoblastic leukemia. Tumor-inhibiting effects of GSIs are lost after Notch3 silencing in NSCLCs, suggesting cell survival is mediated via Notch3 (20). Serial transplantation studies indicate Notch3 is a regulator of self-renewal in tumor-propagating cells, and with no essential function in development or homeostasis (Notch3-null mice have no liver phenotype), Notch3 inhibition appears a safe strategy (11). GSIs have been pursued as therapy in a range of cancers, but translation has been hampered by toxicity. Such effects arise not due to disrupting the GS complex; the same phenotype occurs in RBPJ- or Hes1- deficient mice (21). Therefore, the possibility of a tumor-forming role via an RBPJ-independent mechanism is appealing. Our data suggest activation of AKT by Notch3 might be one such route.
Using independent techniques of blockade, we identify the PI3K/AKT pathway as one route of Notch3-driven cell survival; these data in line with Fan et al. who showed enhanced biliary tumorigenesis with transgenic activation of Notch and AKT (6). Many studies show the PI3K/AKT/mTor axis is dysregulated in CC, with AKT phosphorylation correlating with poor survival, and dual treatment with AKT and mTor inhibitors synergistically slowing tumor growth (22).
Although N-ICD translocation via RBPJ to drive Hes/Hey transcription is the most studied pathway, alternative modes of signaling are described including GS activation independent of ligand; N-ICD activity independent of RBPJ; or activation by membrane-tethered receptors without GS cleavage (23). RBPJ-independent signaling is characterized in T cells where N3-ICD interacts with IKKα to stimulate NF-κB and drive leukemia (24). Indeed, noncanonical Notch signaling is not uncommonly described in cancer, triggering cascades including PI3K/AKT, Wnt, and HIF1-α (25). Our data in rats of profound receptor overexpression without concomitant effector up-regulation further suggest Notch-driven CC can arise via an RBPJ-independent route, given the restriction of tumor growth after GSI.
The stimuli for Notch3 up-regulation are as yet unknown. In our rat time course, early ligand up-regulation by fibroblasts tempts speculation that stroma-derived factors might be a trigger. However, as tumors evolve, Jagged1 appears on ductules, suggesting a switch to autonomous signaling or activation of an alternative pathway. In ovarian carcinoma where Notch3 gene amplification is common, Jagged1 is itself dependent on Notch3 activity; deletion and ectopic expression inhibit and promote Jagged1, respectively, implementing a self-sustaining signaling loop (26). This role for Jagged1 is an important question as ligands are attractive alternative therapeutic targets. In Drosophila, cis interactions (receptor stimulated by ligand from the same cell) inhibit receptor activity within the cell while stimulating activity in neighboring cells. Potential for Jagged1 to exert differential effects on Notch1 and Notch3 here is intriguing.
Stimulation of the ductular response by Notch1 in biliary regeneration requires classical signaling via Hes1. Further work is needed to understand whether this signal required in CC, how it is affected by Notch3, if at all (we see no change in Notch1 following Notch3 inhibition), and how Hes/Hey are involved. Our data suggest this role is complex: we observe Hes/Hey up-regulation in human disease and mice but to a much lesser extent in rats. The fact we observe reduced Hes/Hey expression with Notch3 silencing and yet the observed changes in Akt-related components do not occur with RBPJ inhibition suggests that at least two signaling routes are active downstream of the receptor, and further mechanistic work is needed to understand this better. Taken together, however, our data suggest Notch3 is an important driver in CC and drives cell survival independently of RBPJ, opening up new therapeutic targets for this largely untreatable cancer.
Materials and Methods
Human Tissue.
Human CC and liver were collected prospectively from patients undergoing resection at the Royal Infirmary Edinburgh with informed consent. The study was reviewed and approved by the Tayside Committee in Medical Research Ethics B. Retrospectively collected specimens were obtained from the National Health Service Lothian Scottish Academic Health Sciences Collaboration BioResource and healthy liver from the Edinburgh Medical Research Council Sudden Death Tissue bank. Tissue CC microarrays were purchased from Pantomics.
Animal Models and Xenografts.
CK19CreERTR26ReYFP mice (14) were a kind gift from Guoquaing Gu (Vanderbilt Medical Center, Nashville, TN). These mice were cross-bred with Trp53tm1Brn mice (p53flox/flox) (ref. B6.129P2-Trp53tm1Brn/J), Notch3tm1Grid (N3−/−) mice (ref. B6.129S1-Notch3tm1Grid/J) (11), or or Notch1fl/fl (Notch1tmRko/Grid) from Jackson Laboratories. Trp53tm1Brn (p53fl/fl) and Notch3tm1Grid (N3−/−) mice were on a C57BL/6;C129 background; Notch1fl/fl mice were on a 129 background. Before experimental use, animals were cross-bred with the CK19CreERTR26ReYFP line, which carried a CD1;C57BL/6 background. Progeny were subsequently on a mixed background and used for experimental comparison. In studies where Notch3 is altered, all experimental mice were from the same colony and had a consistent mixed background (CD1/C57BL/6/129). Throughout, littermates were included as controls where possible. All animals used were male and aged matched. Mice were genotyped by Transnetyx. LoxP recombination was induced with three doses of 4 mg tamoxifen (Sigma) in corn oil i.p. on alternate days at 6 wk of age. CC was induced using oral sweetened TAA (Sigma; 600 mg/mL) or vehicle for 26 wk (n = 8). Eight-week-old male Sprague–Dawley rats were given 600 mg/L sweetened oral TAA or vehicle for 26 wk (9). Animals were killed at 10, 12, 14, 16, 18, 20, 22, 24, and 26 wk (n = 3). Rats received 10 mg/kg DAPT (Tocris) in olive oil s.c. (n = 8) or olive oil alone (n = 12) thrice weekly during weeks 21–26.
Xenografts were performed on 6-wk-old CD1-nude mice with bilateral s.c. flank injection of the commercial CC line CC-LP-1 (5 × 105) (15) or CC-LP-1 cells transfected and stably selected for NOTCH3 targeted shRNA or scrambled sequence control. Tumors were allowed to engraft for 28 d before mice were either killed or exposed to one of the following treatment regimes: DAPT (10 mg/kg), PI-103 (30 mg/kg, Selleckchem), or equivalent dose of vehicle for 14 d (n > 5 per group). Tumor volume was calculated using the modified ellipsoid formula: 0.5(l × w2). Animal studies were conducted in accordance with UK Home Office regulations under procedural guidelines, severity protocols, and with approval from the Animal Welfare and Ethical Approval Review Body (AWERB).
Quantification of in Vivo Tumor Burden.
Rat and mouse livers were cut into 3-mm slices before embedding and sectioning. Limits of malignancy were defined on H&E sections from each block (five per liver) by a histopathologist blinded to the regime. Tumor area as a proportion of liver area was quantified with Image J (NIH).
Statistical Analyses.
Analyses were performed with Prism (GraphPad v5). Data are presented as mean ± SEM. Data distribution was assessed using the D’Agostino & Pearson normality test and comparisons between two groups using the Student t test or Mann–Whitney U test; multiple groups were compared with the one-way ANOVA or Kruskal–Wallis test. Post hoc testing groups of nonparametric data were performed using Dunn’s multiple comparison test.
SI Materials and Methods
Cell Culture and RNA Interference Studies.
The human CC cell line CC-LP-1 was purchased from the Korean cell line bank (cellbank.snu.ac.kr/) and cultured in high glucose DMEM (Gibco) with 2.5% (12.5 mL/500 mL) FCS, 2 mM glutamine, 10 U/mL penicillin, and 100 µg/mL streptomycin at 37 °C and 5% CO2. The MTT assay was used for assessment of cell viability. Cells were plated at a density of 1 × 105/mL in 96-well plates in triplicate and cultured for 48 h before 20 µL MTT was added. Cells were incubated at 37 °C for 4 h before medium was aspirated, and crystals were dissolved in DMSO and read by a spectrophotometer at a wavelength of 570 nm with a reference wavelength of 630 nm. siRNA was purchased from Qiagen in the 1-nmol FlexiTube format along with scrambled sequence control (Negative Allstars siRNA sense sequence GCAAGCUGACCCUGAAGUUCAU). The following siRNA sense sequences were used: Hs_Notch3_1, CAGCGTGACCGAGATAGGTCA; Hs_Notch3_2, ATGCCTAGACCTGGTGGACAA; Hs_Notch3_3, AAGAATAGTTAACACTCAAA; Hs_Notch3_5, CTGCGATTAATGAGGATGA; Hs_RBPJ_1, TAGGGAAGCTATGCGAAATTA; Hs_RBPJ_2, GTGGCTGGAATACAAGTTGAA; Hs_RBPJ_3, TCCAGTAACTTTGGTCCGAAA; and Hs_RBPJ_SUH_7, CACGCTATTATAGTACACCTT. Human CC cells (CC-LP-1) were transfected using 5 nM HiPerfect reagent (Qiagen) according to the manufacturer’s instructions. Cells were washed in PBS the following day, and media were replaced. Wells were lysed 48 h later for RNA and protein extraction to assess gene knockdown efficiency. A stable NOTCH3 knockdown CC line was generated as previously described using four unique SureSilencing shRNA plasmids (pGeneClip), each containing an shRNA under the U1 promoter and a puromycin resistance cassette (SA Biosciences). Plasmids contained the following insert sequences: clone 1, GGATGGCGTCAACACCTATAA; clone 2, GCAGTCGCCTGAGAATGATCA; clone 3, GGTGGACATTGACGAGTGTCA; clone 4, CAGTCACGGCATCTGCTATGA; and control, GGAATCTCATTCGATGCATAC. The full sequence of the entire vector is available at www.sabiosciences.com/RNAiResource.php. Transfection of the human CC line CC-LP-1 was performed with 0.5 µg pDNA using Effectene transfection reagent in conjunction with buffer EC and Enhanceer (Qiagen). Stable selection was achieved by the addition of 5 µg/mL puromycin. Three colonies per shRNA sequence were picked and transferred to individual wells for expansion. Knockdown efficiency was assessed using qRT-PCR and Western blotting.
Gene Expression Analysis and PCR Arrays.
For quantitative real-time PCR analysis, 50 mg whole liver tissue was dissociated with a tissue homogenizer in 500 µL TRIzol reagent (Amersham), and 100 µL chloroform was added to separate the aqueous phase, which was precipitated with isopropanol. A Qiagen RNEasy mini kit was used for RNA purification according to the manufacturer’s instructions, which included treatment with DNase, and concentration and purity were determined using a Nanodrop spectrophotometer (ThermoScientific). For extraction of RNA from formalin-fixed, paraffin-embedded archival samples, an RNEasy FFPE extraction kit was used (Qiagen). One microgram or 100 ng RNA was used for cDNA strand synthesis using a Quantitect reverse transcription kit (Qiagen). qRT-PCR was performed in triplicate wells using SYBR green master mix (Qiagen) and Quantitect primers (Qiagen) (see Table S4 for complete primer list). cDNA from scrambled control duplex and siRNA-transfected CCLP-1 cells were applied to a human liver cancer RT2 profiler PCR array (PAHS-133Z); cDNA from human CC samples and patient-matched noncancerous liver controls was applied to a human NOTCH pathway RT2 profiler PCR array (PAHS-059Y); and cDNA from the TAA rat time course with uninjured control was applied to a rat Notch pathway RT2 profiler array (PARN-059Z) (all SABiosciences). All reactions were performed using a Lightcycler 480-II (Roche). Data analysis for PCR arrays was carried out using an integrated web-based software package. The 2−∆∆CT method was used with normalization of data to the mean CT values of five independent housekeeping genes. Data are expressed as the fold regulation compared with control samples.
Immunohistochemistry.
At the time of death, livers were perfused with 5 mL sterile PBS via the inferior vena cava, fixed overnight in 4% buffered formalin or methacarn, and paraffin embedded with sections cut at 4 µm. Sections were dewaxed by immersion in xylene and rehydrated through serial ethanol solutions to PBS. Heat-mediated antigen retrieval was used before endogenous peroxidase activity was blocked with 3% hydrogen peroxide and sections were permeabilized with 0.01% Tween20 (Sigma). For peroxidase detection, endogenous avidin and biotin were blocked (Avidin-Biotin blocking kit; Dako) before a universal protein block was applied (Spring Bioscience). Sections were then incubated for 1 h or overnight with the following primary antibodies: goat anti-NOTCH1 (Abcam), goat anti-NOTCH2 (Santa Cruz), rabbit anti-NOTCH3 (Abcam), goat anti-NOTCH4 (Santa Cruz), rabbit anti-JAGGED1 (Santa Cruz, Abcam), rat anti-CK19 (DSHB Hybridoma Product TROMA-III: TROMA-III was previously deposited in The Developmental Studies Hybridoma Bank by Rolf Kemler), rabbit anti-SOX9 (Millipore), mouse anti-αSMA (Sigma), rabbit anti-pAKT(Thr308) (Cell Signaling), rabbit anti-pAKT (Ser473) (Cell Signaling), rabbit anti-pS6 ribosomal protein (Cell Signaling), rabbit anti-pmTOR (Cell Signaling), rabbit anti-CD31 (Abcam), chicken anti-GFP (Abcam), and rabbit anti-Ki67 (abcam). Sections were washed in PBS before incubation with the appropriate biotinylated secondary antibodies (Dako) or Alexa Fluor-conjugated antibodies (Invitrogen). Counterstaining was performed with Harris hematoxylin and Scott’s tap water and mounted with Pertex. All bright field and fluorescent microscopy was performed with a Nikon Eclipse E600 microscope.
Cell Counting and Pixel Analysis.
For manual counting of ductular cells, 30 high-power (×20) fields with liver parenchyma at all boundaries were photographed, and Image J software (NIH) was used for quantification. Cells were identified on morphology (small round/oval appearance with high nuclear to cytoplasmic ratio), and only cells adjacent to a ductular lumen were counted. For pixel analysis of Notch staining of CC tissue microarrays, ImageProPremier software version 9.1 (MediaCybernetics) was used to select regions of positivity and automatically analyzed using a macroinstruction. For fluorescent staining of CC xenografts, Adobe Photoshop (version CS3) was used to select single color intensity that was then standardized to DAPI nuclear stain to control for cell number. As the total amount of tissue available for staining was more limited for xenografts, staining was quantified across the entire section. Results are expressed as the mean percentage of positive pixels per field. All counts and pixel analyses were performed blinded to reduce bias.
Immunoblotting.
Cell extracts were prepared from 30 mg whole liver or, in the case of siRNA experiments, one well of cells growing in monolayer on plastic from a 6-well plate 48 h following transfection. Protein content was quantified using a BSA standard and Pierce reagent before being resolved using SDS/PAGE and blotted onto nitrocellulose membrane. Immunoblots were incubated at 4 °C overnight with primary antibody [mouse anti-β actin (Sigma), rabbit anti-Notch3 (Abcam), rat anti-RBP1F1(RBPJ) (Ascension), rabbit anti-phospo AKT (Thr308; CST), rabbit anti-phospho S6 ribosomal protein (S235/236; CST), rabbit anti-phospho p70 S6 kinase (T389; CST), or rabbit anti-phospho mTOR (S2448; Abcam)]. Appropriate HRP-labeled secondary antibodies were used (Dako or CST) and signal detected with ECL reagent (Amersham).
Acknowledgments
CK19CreERTR26ReYFP mice were a gift from G. Gu (Vanderbilt University Medical Center). R.V.G. and T.J.K. are funded by Wellcome Trust fellowships. L.B., W.-Y.L., A.R., and S.J.F. are funded by the Medical Research Council, Cancer Research UK (CRUK), and the Alan Morement Memorial Fund (AMMF) charity. A.J.R. and S.E.M.-L. are funded by Medical Research Council fellowships. J.P.M. is funded by CRUK. O.J.S. is funded by the European Research Council and CRUK.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.T. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600067113/-/DCSupplemental.
References
- 1.Zong Y, et al. Notch signaling controls liver development by regulating biliary differentiation. Development. 2009;136(10):1727–1739. doi: 10.1242/dev.029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boulter L, et al. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat Med. 2012;18(4):572–579. doi: 10.1038/nm.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ortica S, Tarantino N, Aulner N, Israel A, Gupta-Rossi N. The 4 Notch receptors play distinct and antagonistic roles in the proliferation and hepatocytic differentiation of liver progenitors. FASEB J. 2014;28(2):603–614. doi: 10.1096/fj.13-235903. [DOI] [PubMed] [Google Scholar]
- 4.Mazur PKEH, et al. Notch2 is required for progression of pancreatic intraepithelial neoplasia and development of pancreatic ductal adenocarcinoma. Proc Natl Acad Sci USA. 2010;107(30):13438–13443. doi: 10.1073/pnas.1002423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Font-Burgada J, et al. Hybrid periportal hepatocytes regenerate the injured liver without giving rise to cancer. Cell. 2015;162(4):766–779. doi: 10.1016/j.cell.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan B, et al. Cholangiocarcinomas can originate from hepatocytes in mice. J Clin Invest. 2012;122(8):2911–2915. doi: 10.1172/JCI63212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zender S, et al. A critical role for notch signaling in the formation of cholangiocellular carcinomas. Cancer Cell. 2013;23(6):784–795. doi: 10.1016/j.ccr.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 8.Tarlow BD, et al. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell. 2014;15(5):605–618. doi: 10.1016/j.stem.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeh C-NMA, Maitra A, Lee KF, Jan YY, Chen MF. Thioacetamide-induced intestinal-type cholangiocarcinoma in rat: An animal model recapitulating the multi-stage progression of human cholangiocarcinoma. Carcinogenesis. 2004;25(4):631–636. doi: 10.1093/carcin/bgh037. [DOI] [PubMed] [Google Scholar]
- 10.Huppert SS, et al. Embryonic lethality in mice homozygous for a processing-deficient allele of Notch1. Nature. 2000;405(6789):966–970. doi: 10.1038/35016111. [DOI] [PubMed] [Google Scholar]
- 11.Krebs LT, et al. Characterization of Notch3-deficient mice: Normal embryonic development and absence of genetic interactions with a Notch1 mutation. Genesis. 2003;37(3):139–143. doi: 10.1002/gene.10241. [DOI] [PubMed] [Google Scholar]
- 12.Khan SATH, Thomas HC, Toledano MB, Cox IJ, Taylor-Robinson SD. p53 Mutations in human cholangiocarcinoma: A review. Liver Int. 2005;25(4):704–716. doi: 10.1111/j.1478-3231.2005.01106.x. [DOI] [PubMed] [Google Scholar]
- 13.Guest RV, et al. Cell lineage tracing reveals a biliary origin of intrahepatic cholangiocarcinoma. Cancer Res. 2014;74(4):1005–1010. doi: 10.1158/0008-5472.CAN-13-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Means ALXYX, Xu Y, Zhao A, Ray KC, Gu G. A CK19(CreERT) knockin mouse line allows for conditional DNA recombination in epithelial cells in multiple endodermal organs. Genesis. 2008;46(6):318–323. doi: 10.1002/dvg.20397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimizu Y, et al. Two new human cholangiocarcinoma cell lines and their cytogenetics and responses to growth factors, hormones, cytokines or immunologic effector cells. Int J Cancer. 1992;52(2):252–260. doi: 10.1002/ijc.2910520217. [DOI] [PubMed] [Google Scholar]
- 16.Villanueva A, et al. Notch signaling is activated in human hepatocellular carcinoma and induces tumor formation in mice. Gastroenterology. 2012;143(6):1660–1669. doi: 10.1053/j.gastro.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen JBSB, et al. Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology. 2012;142(4):1021–1031.e15. doi: 10.1053/j.gastro.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huntzicker EG, et al. Differential effects of targeting Notch receptors in a mouse model of liver cancer. Hepatology. 2015;61(3):942–952. doi: 10.1002/hep.27566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahn S, Hyeon J, Park CK. Notch1 and Notch4 are markers for poor prognosis of hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2013;12(3):286–294. doi: 10.1016/s1499-3872(13)60046-6. [DOI] [PubMed] [Google Scholar]
- 20.Bellavia D, et al. Combined expression of pTalpha and Notch3 in T cell leukemia identifies the requirement of preTCR for leukemogenesis. Proc Natl Acad Sci USA. 2002;99(6):3788–3793. doi: 10.1073/pnas.062050599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Es JH, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435(7044):959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 22.Ewald F, et al. Combined targeting of AKT and mTOR using MK-2206 and RAD001 is synergistic in the treatment of cholangiocarcinoma. Int J Cancer. 2013;133(9):2065–2076. doi: 10.1002/ijc.28214. [DOI] [PubMed] [Google Scholar]
- 23.Ayaz F, Osborne BA. Non-canonical notch signaling in cancer and immunity. Front Oncol. 2014;4:345. doi: 10.3389/fonc.2014.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vacca A, et al. Notch3 and pre-TCR interaction unveils distinct NF-kappaB pathways in T-cell development and leukemia. EMBO J. 2006;25(5):1000–1008. doi: 10.1038/sj.emboj.7600996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee KS, et al. Roles of PINK1, mTORC2, and mitochondria in preserving brain tumor-forming stem cells in a noncanonical Notch signaling pathway. Genes Dev. 2013;27(24):2642–2647. doi: 10.1101/gad.225169.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, et al. Jagged1 expression regulated by Notch3 and Wnt/β-catenin signaling pathways in ovarian cancer. Oncotarget. 2010;1(3):210–218. doi: 10.18632/oncotarget.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu WY, et al. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat Cell Biol. 2015;17(8):971–983. doi: 10.1038/ncb3203. [DOI] [PMC free article] [PubMed] [Google Scholar]