Significance
Mammalian mitoribosomes are idiosyncratic. They reversed the typical 70% RNA:30% protein ratio to generate a protein-rich ribosome, which lacks the conventional 5S transcript as a third rRNA component. Recent subnanometer-resolution structures revealed a further unexpected eccentricity: integration of a mitochondrially encoded tRNA into the large-subunit structure. However, mitochondrially encoded tRNA (mt-tRNA) selection varies with mt-tRNAVal integration into human mitoribosomes compared with porcine mitoribosomes favoring mt-tRNAPhe. Our analysis of mitoribosomes from multiple tissues isolated from different mammals found that of 22 mitochondrially encoded tRNAs, only mt-tRNAVal or mt-tRNAPhe is incorporated. Further, there was no evidence of tissue-specific selectivity. Remarkably, human mitoribosomes can exhibit adaptive plasticity when mt-tRNAVal levels are severely depleted by selectively integrating mt-tRNAPhe instead, to generate translationally competent mitoribosomes.
Keywords: mammalian mitochondria, ribosomes, rRNA, tRNA, mitochondrial protein synthesis
Abstract
The recent developments in cryo-EM have revolutionized our access to previously refractory structures. In particular, such studies of mammalian mitoribosomes have confirmed the absence of any 5S rRNA species and revealed the unexpected presence of a mitochondrially encoded tRNA (mt-tRNA) that usurps this position. Although the cryo-EM structures resolved the conundrum of whether mammalian mitoribosomes contain a 5S rRNA, they introduced a new dilemma: Why do human and porcine mitoribosomes integrate contrasting mt-tRNAs? Human mitoribosomes have been shown to integrate mt-tRNAVal compared with the porcine use of mt-tRNAPhe. We have explored this observation further. Our studies examine whether a range of mt-tRNAs are used by different mammals, or whether the mt-tRNA selection is strictly limited to only these two species of the 22 tRNAs encoded by the mitochondrial genome (mtDNA); whether there is tissue-specific variation within a single organism; and what happens to the human mitoribosome when levels of the mt-tRNAVal are depleted. Our data demonstrate that only mt-tRNAVal or mt-tRNAPhe are found in the mitoribosomes of five different mammals, each mammal favors the same mt-tRNA in all tissue types, and strikingly, when steady-state levels of mt-tRNAVal are reduced, human mitoribosome biogenesis displays an adaptive response by switching to the incorporation of mt-tRNAPhe to generate translationally competent machinery.
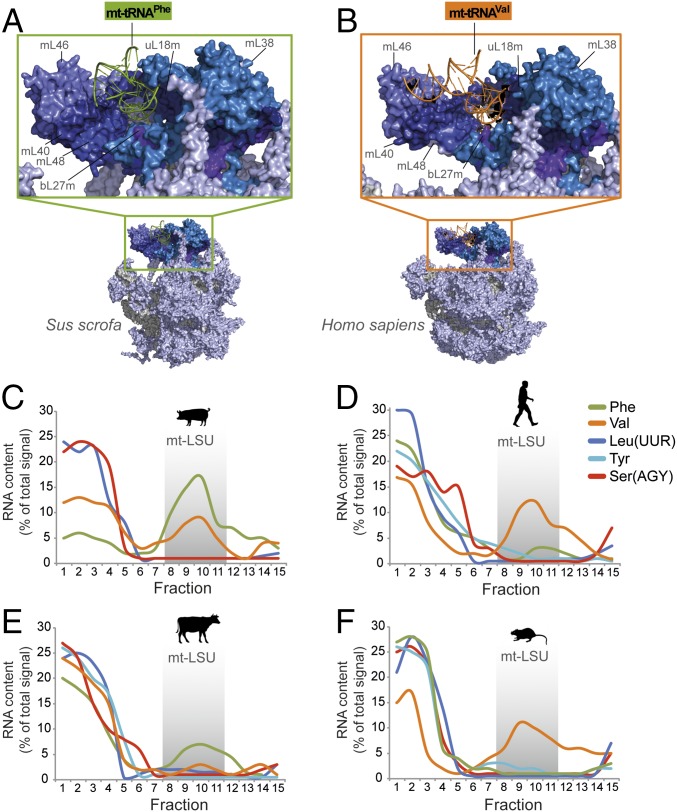
Mammalian mitoribosomes differ considerably from other ribosomes, primarily because of a reversed protein:RNA mass ratio, such that proteins predominate. Of the 80 known mitoribosomal proteins, 36 are mitospecific and lack bacterial orthologs, and those with orthologs often carry mitospecific extensions (1). Further idiosyncrasies have been revealed by recent subnanometer-resolution cryo-EM structures, which simultaneously resolved a long-debated question concerning mitoribosomal rRNA content (1, 2). Until recently, mammalian mitoribosomes were considered divergent by containing only one rRNA species per subunit, although the potential presence of the 5S species in the mitochondrial large subunit (mt-LSU) remained a contentious issue (3). Recent data have shown that there is indeed a third RNA moiety present in mammalian mitoribosomes. It is not, however, a 5S species, but a tRNA encoded by the mitochondrial genome (mtDNA) (1, 2, 4–6). Interestingly, which tRNA was selected differed between mammalian species, with mt-tRNAVal predominating in human mt-LSU, in contrast to mt-tRNAPhe being selected for incorporation into the porcine mt-LSU (Fig. 1 A and B).
Fig. 1.
Species- and tissue-specific variation in mt-tRNA incorporation into the mt-LSU was analyzed. Depicted are the overall structures of porcine (A) and human (B) mt-LSUs, detailing the region of the central protuberance containing mt-tRNAPhe (porcine) or mt-tRNAVal (human). The mitoribosomal proteins in direct contact with the mt-LSU-incorporated mt-tRNA are indicated in dark blue. Human (PDB: 3J9M); Porcine (PDB: 5AJ4). (C–F) To analyze and quantify species-specific mt-tRNA distributions, high-resolution Northern blots of sucrose gradients were performed. Analysis of porcine dermal fibroblasts (C), human osteosarcoma 143B cells (D), bovine cardiac tissue (E), and rat hepatic tissue (F) depicts the relative abundance of each mt-tRNA across fractions (ImageQuant software); represented as a percentage of the total signal.
Because mammalian mtDNA encodes 22 tRNAs (7), we sought to clarify whether incorporation into the mt-LSU is restricted to mt-tRNAVal and mt-tRNAPhe, or whether there is a more promiscuous use of mt-tRNAs in either a species- or tissue-specific manner. Furthermore, we investigated the consequence of reduced mt-tRNAVal levels on human mitoribosomes.
Results and Discussion
Do Other Mammals Integrate Other Different Mitochondrially Encoded tRNAs into Their Mitoribosomes?
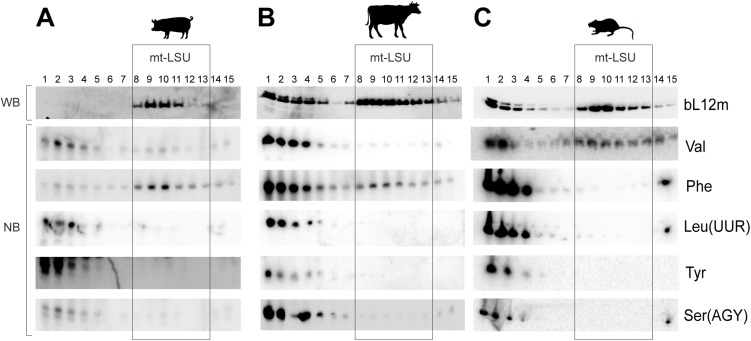
Because the overall structure of tRNAs is generically considered to be similar, the level of discrimination displayed in the selection of mt-tRNA was perhaps surprising. To identify whether there was a process that integrated mitochondrially encoded tRNAs, other than mt-tRNAVal or mt-tRNAPhe, into the central protuberance of mammalian mitoribosomes, we investigated the mt-tRNA content of porcine, human, bovine, and rat mt-LSUs. Sucrose gradient separation of cell lysates facilitated analysis of the distribution of RNA and mt-LSU proteins within different fractions (Fig. S1 A–C). The predominant mt-tRNA comigrating with porcine and bovine mt-LSUs was mt-tRNAPhe (Fig. 1 C and E), whereas mt-tRNAVal was dominant in humans and rats (Fig. 1 D and F). The migration of other mt-tRNAs was determined, the positions of which are widely distributed across the encoding polycistronic transcript. None of these was substantially enriched among mt-LSU-specific fractions in any of the mammalian samples tested (Fig. 1 C–F). Our results indicate that only mt-tRNAVal or mt-tRNAPhe are integrated into mammalian mitoribosomes.
Fig. S1.
Determination of the mt-LSU migration within sucrose gradients (A–C). Western blotting (WB) was used to determine the migration profile of mt-LSU (bL12m) in pig dermal fibroblasts (A), bovine cardiac tissue (B), and rat hepatic tissue (C) within sucrose gradients using antibodies against bL12m. High-resolution Northern blotting (NB) was performed with probes specific to mt-tRNAs Val, Phe, LeuUUR, Tyr and SerAGY.
Is There Tissue-Specific Variation in the Selection of mt-tRNA Integration?
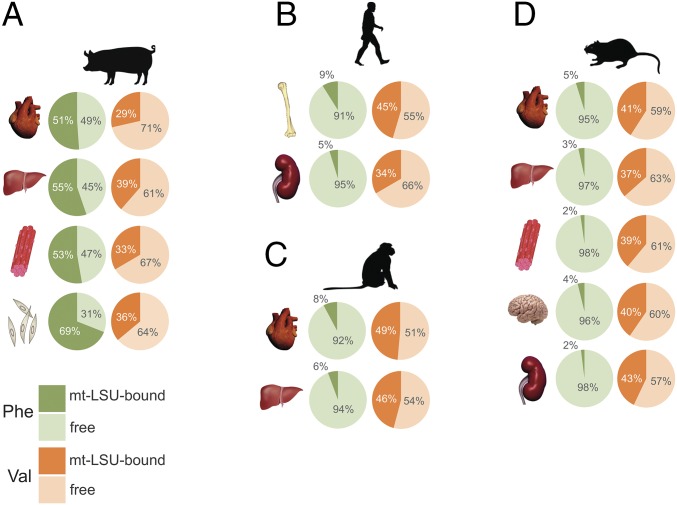
Having demonstrated species-specific incorporation of either mt-tRNAVal or mt-tRNAPhe into mammalian mitoribosomes, we addressed whether within a single organism there existed any tissue-specific variation. Mitoribosome sedimentation was performed for several mammalian tissues, and the ratio of free versus mt-LSU-bound mt-tRNA was quantified. Results indicated that mt-tRNAPhe was the principal tRNA in porcine mt-LSU of cardiac, hepatic, skeletal muscle, and dermal epithelium (Fig. 2A). Notably, a fraction of mt-tRNAVal was also detected in porcine mt-LSU across the tissues analyzed. In contrast, mt-tRNAVal was consistently the dominant tRNA in the human, macaque, and rat mt-LSU across all tissues analyzed (Fig. 2 B–D). These data suggest there is no tissue-specific incorporation of different mt-tRNAs into mitoribosomes.
Fig. 2.
Determination of tissue-specific variation in mt-tRNA incorporation into the mammalian mt-LSU. Selected tissues and cultured cells isolated from (A) pig (heart, liver, skeletal muscle, and dermal fibroblasts), (B) human (osteoblasts and embryonic kidney cells), (C) macaque (heart and liver), and (D) rat (heart, liver, skeletal muscle, brain, and kidney) were analyzed to determine any tissue-specific variation in mt-tRNA incorporation into the mt-LSU. Upon lysis, the samples were sedimented through sucrose gradients, followed by RNA extraction. RNA samples from fractions 1 and 2 (free pool) and fractions 10 and 11 (mt-LSU-associated) were subjected to Northern blotting. Abundance of mt-tRNAPhe (green) and mt-tRNAVal (orange) in free (pale green/orange), and mt-LSU-associated fractions (dark green/orange) is represented as a percentage of their combined signal.
Are the mt-tRNAs That Are Integrated as Structural Components Posttranscriptionally Modified?
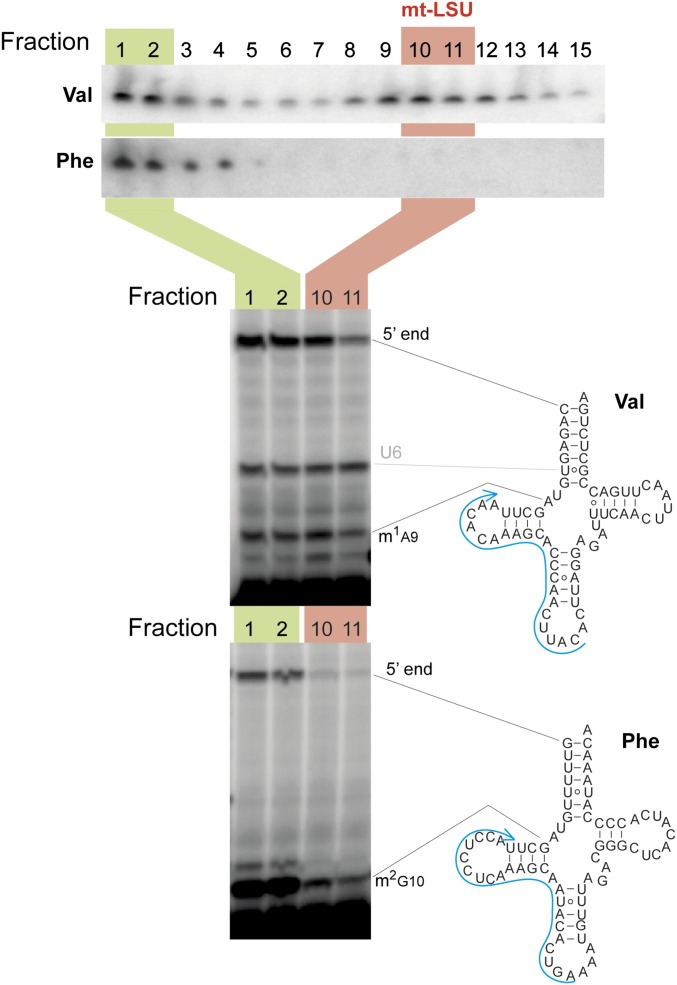
Mitochondrial tRNAs undergo extensive posttranscriptional nucleotide modification. The structural data identified that the mt-LSU-integrated mt-tRNAs were matured by the addition of CCA on the 3′ terminus (6). We therefore sought to determine whether any differences in nucleotide modification status existed between these free and mt-LSU-bound pools. No changes in m1A9 modification were detected between the free versus integrated pools of human mt-tRNAVal (Fig. S2). This is consistent with the mt-LSU structures (Fig. 1), where to integrate into the mt-LSU, mt-tRNAs adopt an l-shaped tertiary structure dependent on m1A9 (8).
Fig. S2.
Analysis of maturation and aminoacylation status of different pools of mt-tRNAs. Human osteoblast RNA isolated from sucrose gradient fractions 1–2 (green) and 10–11 (red) was subjected to run-off reverse transcription primer extension (RT-Ex) with a radioactively labeled, complementary primer (blue) to detect modified nucleotides in mt-tRNAVal and mt-tRNAPhe. The presence of m1A9 (mt-tRNAVal) or m2G10 (mt-tRNAVal) results in RT-Ex pausing producing a shorter product. Pausing of RT-Ex detected at U6 of mt-tRNAVal is likely to correspond to yet-unidentified nucleotide posttranscriptional modification in this mt-tRNA.
What Happens to Mitoribosome Biogenesis If Levels of mt-tRNAVal Are Depleted?
Despite mt-tRNAs making up only ∼10% of the mitochondrial genome, more than 200 disease-causing mutations have been reported in these genes (9, 10). Among these pathogenic changes is a homoplasmic m.1624C > T mutation in the gene encoding mt-tRNAVal (MTTV). The consequence of this particular mutation is a destabilization of the tRNA structure, which results in significantly reduced steady-state levels (11, 12). These depleted levels of mt-tRNAVal were assumed to be the cause of compromised mitochondrial translation in patient tissue and cell lines (12). However, in light of the observation that mt-tRNAVal is a component of the mitoribosome, we sought to determine the effect of reduced mt-tRNAVal levels on mitoribosome formation.
Individual clonal lines were derived from a cybrid population that harbored the homoplasmic m.1624C > T mutation. To eliminate any conflicting contribution resulting from the aneuploid nature of 143B cells (13), five clones were analyzed. All shared a consistent biochemical and molecular phenotype, including decreased steady-state levels of mt-tRNAVal compared with the parental 143B cell line (Fig. 3 depicts a representative clone). Experiments were designed to determine whether the protein synthesis defect was predominantly caused by decreased amounts of mitoribosome resulting from the reduced levels of mt-tRNAVal, or whether both mitoribosome biogenesis and translation elongation were compromised by the available mt-tRNAVal being distributed between integration into the mitoribosome and elongation, thus compromising both aspects of translation.
Fig. 3.
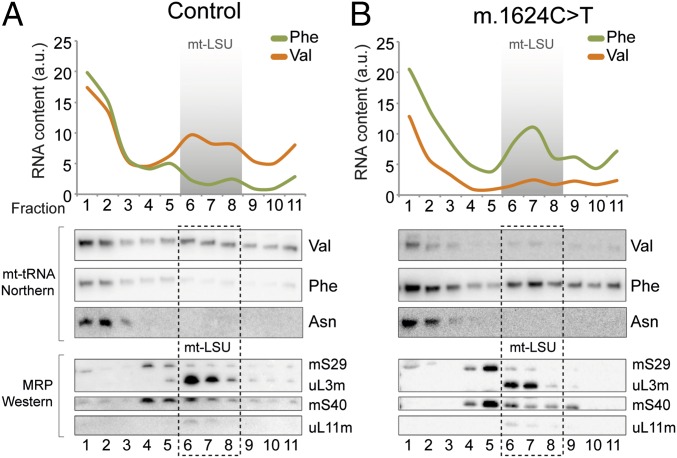
The integration of mt-tRNAVal or mt-tRNAPhe into mitoribosomes of control cells or those harboring the MTTV mutation m.1624C > T was analyzed by sucrose gradient. Cell lysates from control 143B parental (A) and m.1624C > T (B) lines were separated through 10–30% (wt/vol) sucrose gradients and fractions collected (1 = top; 11 = bottom). Both RNA (Upper) and proteins (Lower) were extracted and analyzed to determine the relative positions of mitoribosomal proteins (MRP) and mt-tRNAVal, mt-tRNAPhe with mt-tRNAAsn as a control. The representative images are, in each case, the same membranes sequentially probed for different mt-tRNAs or MRPs.
The redistribution of mt-tRNAVal within sucrose gradients of the m.1624C > T cybrid lines was striking. The detectable majority was shifted to the free fractions compared with mt-LSU association in the 143B parental controls (Fig. 3, lanes 1–3; Fig. S3). This was consistent with the majority of the available mt-tRNAVal being preferentially retained for use in elongation. Western blotting of the sucrose gradient fractions showed comparable levels and distribution of mitoribosomal proteins between the mutant and control lines. This indicated that the levels of mitoribosome were not directly proportional to the levels of mt-tRNAVal, despite this element normally being required as a structural component.
Fig. S3.
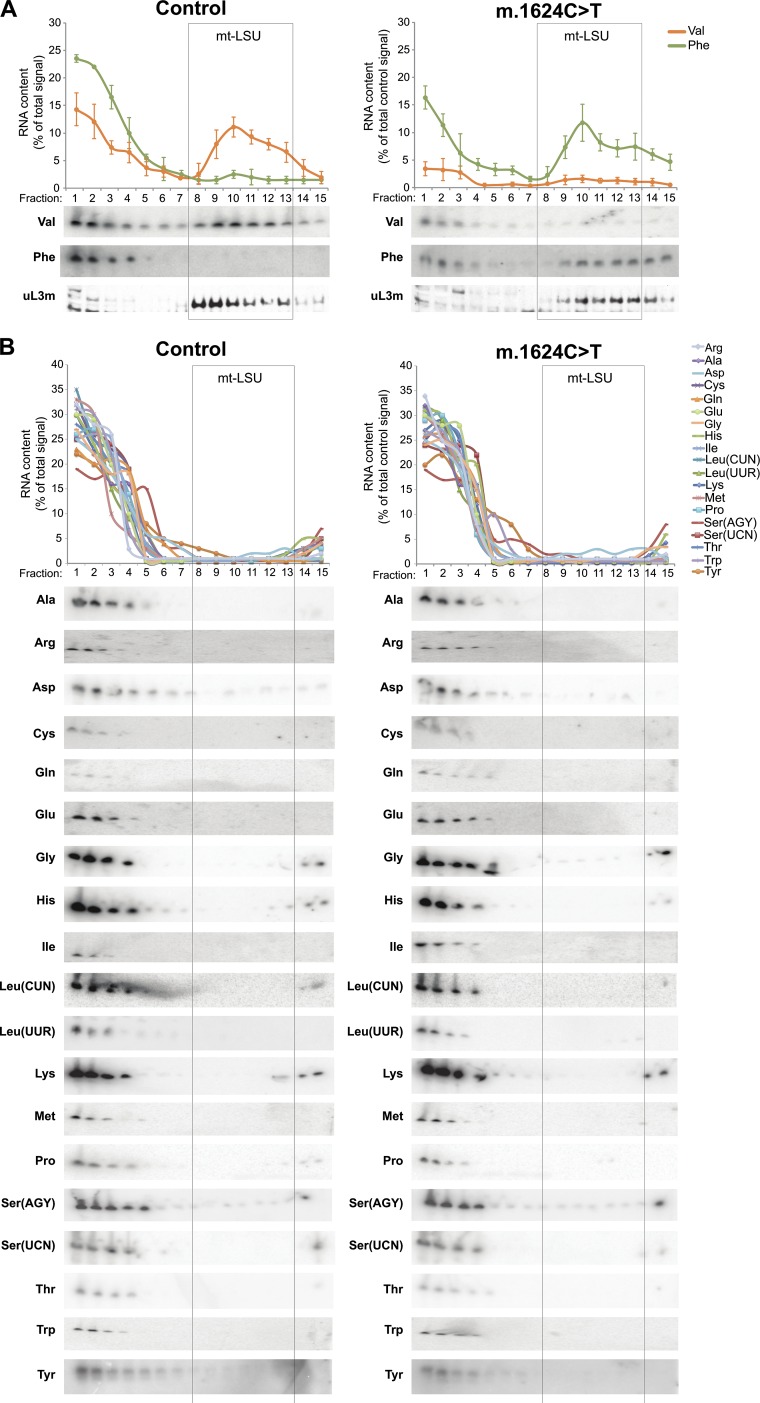
Comprehensive analysis of human mt-tRNAs to determine association with the large subunit of the mitoribosome. Cell lysates prepared from human osteosarcoma 143B parental (Control; Left) and a cybrid clone harboring the m.1624C > T mutation in mt-tRNAVal (Right) were separated through 10–30% (wt/vol) sucrose gradients. RNA was isolated from each of 15 fractions, and high-resolution northern analyses were performed to determine the migration of the mt-tRNAs. (A) The migration of mt-tRNAVal and mt-tRNAPhe were determined and compared with the position of the mt-LSU, verified by Western blotting with antibodies against uL3m. For the control samples, relative abundance of mt-tRNAVal and mt-tRNAPhe in each fraction was quantified, using ImageQuant software, and is presented as a percentage of its total signal. For the m.1624C > T samples, relative abundance of mt-tRNAs was quantified in the same way, but expressed as a percentage of total control signal for each mt-tRNA. (B) The remaining mt-tRNAs were analyzed and the relative abundance of mt-tRNA in each fraction was quantified and presented as a percentage of the total signal.
To examine whether the mitoribosomes had formed in the absence of any mt-tRNA component, we probed the distribution of the remaining mt-tRNAs. All mt-tRNAs were analyzed and retained the same profile in control and m.1624C > T cells, with the majority present in the fractions corresponding to the free pool, available for aminoacylation and elongation. The notable exception was mt-tRNAPhe, which, in contrast to 143B parental controls, now showed a significant proportion that partitioned with the mt-LSU (Fig. 3, lanes 6–8; Fig. S3). These data revealed that when the steady-state levels of mt-tRNAVal are limiting, the human mitoribosome displays adaptive plasticity by integrating mt-tRNAPhe into the mt-LSU.
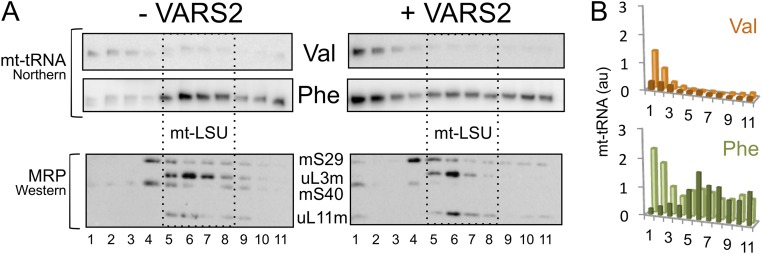
We have shown previously that the molecular and biochemical defects in m.1624C > T-cell lines (11, 12) can be partially overcome by increasing levels of the cognate aminoacyl tRNA synthetase, VARS2, which stabilizes the mt-tRNAVal (12). Therefore, to investigate whether suppression of the defect, observed with VARS2 overexpression, was caused by the expected redistribution of mt-tRNAVal back into the mitoribosome, we analyzed independently generated lines with or without exogenous VARS2 induction (Fig. S4). Despite the increased availability of mt-tRNAVal, biogenesis of the mt-LSU appeared to be sensitive to the structural abnormality in the mutant mt-tRNAVal, as mt-tRNAPhe was preferentially retained in the mt-LSU (Fig. S4). These data are consistent with the partial restoration of intraorganellar protein synthesis resulting from an increased pool of charged mt-tRNAVal being available for elongation. High-resolution structures of mt-tRNAs, either in isolation or in complex with the mt-LSU, are currently lacking. It is, therefore, not possible to determine whether the preferential incorporation of mt-tRNAPhe into the mt-LSU results from a better physicochemical fit than for the mutated mt-tRNAVal. However, translationally active mitoribosomes are generated, irrespective of which mt-tRNA is integrated (12).
Fig. S4.
Relative distribution of mt-tRNAval and mt-tRNAPhe in cell lines harboring MTTV mutation with and without overexpression of mitochondrial valyl-tRNA synthetase. (A) Individual clonal cybrid lines carrying the MTTV mutation were transfected to be able to inducibly express VARS2. Cell lysates prepared from uninduced (−) and induced (+; 14 d) were separated through 10–30% (wt/vol) sucrose gradients. RNA and protein samples from each fraction were analyzed by high-resolution northern or Western blot (dashed box indicates mt-LSU). (B) Distribution of the mt-tRNA was analyzed (ImageJ), and densitometric signals (mt-tRNAVal, orange; mt-tRNAPhe , green) within the gradient before and after induction are presented in histograms (induced, pale; uninduced, dark).
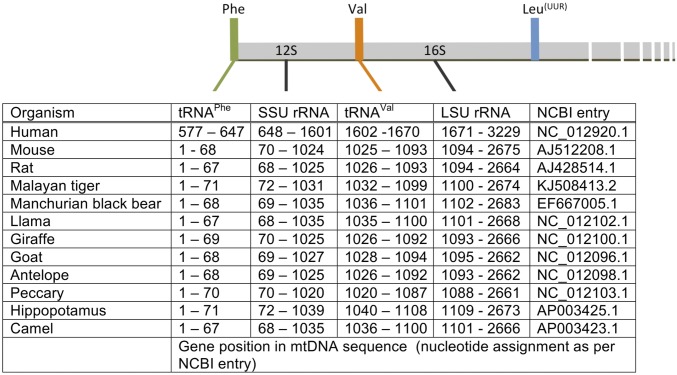
Mammalian mitoribosomes have many features that make them distinct from other ribosomes, even compared with mitochondrial ribosomes from other organisms. Until recently, the apparent lack of a 5S rRNA was actively debated, particularly considering the level of conservation in its sequence, structure, and presence within ribosomes (14). Although the increased proportion of protein components in mammalian mitoribosomes is unusual, the substitution of the 5S for a tRNA is currently without precedent. The 5S rRNA conservation also extends to the location of its gene relative to other rRNA genes in the genome (15). Intriguingly, an analogous gene arrangement is found in most mammalian mitochondrial genomes, such that the two mt-tRNAs, which substitute for the 5S moiety in the mitoribosome, flank the two mt-rRNA genes (Fig. S5). This gene arrangement in mammalian mtDNA means that the polycistronic transcription unit encompassing both the 12S and 16S mt-rRNA, which is reportedly synthesized in a major excess over other units (16), would always include both mt-tRNAPhe and mt-tRNAVal (17). This infers stoichiometric availability of these RNA species for incorporation into mt-LSU. Our understanding of mitoribosome biogenesis is too rudimentary to identify when the mt-tRNA is incorporated, but on the basis of the positioning of the cleaved termini of the 16S and the mt-tRNAVal, it seems unlikely that a polycistronic element is incorporated into the mt-LSU and subsequently cleaved. The context for CCA addition once integrated in the mt-LSU would be very different to all of the other mt-tRNAs, perhaps suggesting this occurs before incorporation, and therefore not when the mt-tRNA is part of a polycistronic unit. Because we know that certain mammals and, under certain circumstances, human mitoribosomes incorporate mt-tRNAPhe in preference to mt-tRNAVal, it seems unlikely that a ∼2.5-knt RNA would be associated with the assembling mt-LSU before a cleavage event that would release the required mt-tRNAPhe.
Fig. S5.
Genetic map of mtDNA, indicating the conservation of gene positions of mammalian mt-rRNAs and their upstream mt-tRNAs. Schematic representation of the primary polycistronic H-strand transcript (Upper), indicating the location of mitochondrial rRNAs, mt-tRNAPhe and mt-tRNAVal in a range of mammals (Lower).
The mammalian mitoribosome has substituted an mtDNA-encoded -tRNA for the 5S rRNA as a third structural RNA component. It has been well documented that mitochondrial RNase P is a protein-only complex that does not require RNA import into the mitochondrial matrix (18). Together, these observations support the absence of a functional requirement for RNA import into mammalian mitochondria.
In summary, the structural evidence for mt-tRNA incorporation, and the conserved positions of mammalian mtDNA genes, suggests an evolutionary significance of gene arrangement that would promote coordinated transcription of the RNA species destined to be structural components of the mt-LSU. The inclusion of two mt-tRNAs within this highly transcribed unit provides a degree of plasticity to the process of mitoribosome biogenesis, as seen by the differing use between organisms and the adaptive response observed under conditions of limiting mt-tRNAVal. What controls the selection of a specific mt-tRNA, despite the relative similarities in structure, or whether other mt-tRNAs can be used when levels of both these species are limiting is yet to be determined.
Materials and Methods
Cell Culture.
All cell lines used in this study were standard, commercially available cell lines except for the m.1624C > T transmitochondrial cybrid 143B lines, which used cytoplasts from patient cell lines that were derived from donated tissue samples. Access to samples that were excess to diagnostic requirements and were approved for research was covered by the license “Role of mitochondrial abnormalities in disease” (REC ref 2002/205) issued by Newcastle and North Tyneside Local Research Ethics Committee. Human 143B.206 Rho+ cells and cybrid derivatives were cultured [37 °C, humidified 5% (vol/vol) CO2] in DMEM (Sigma) supplemented with 10% (vol/vol) FCS, 1× nonessential amino acids, and 2 mM l-glutamine. Where appropriate, BlasticidinS (10 µg/mL) and HygromycinB (100 µg/mL) were added to select for successful integration and retention of the VARS2 gene. VARS2 induction was achieved by addition of 1 µg/mL tetracycline for 14 d.
Mitochondrial Transcript Analysis.
Total RNA from the relevant tissues was extracted using TRIzol (ThermoFisher Scientific) and treated with Turbo DNase, following manufacturer’s protocols. Northern blot analysis was essentially as previously performed (19), total RNA was resolved on 6% or 15% (wt/vol) UREA-PAGE (mt-tRNAs), subjected to wet transfer in 2× SSC (150 mM sodium chloride and 15 mM sodium citrate at pH 7.0) to a nylon membrane (MAGNEPROBE, 0.45 µ; GE Healthcare or Genescreenplus, NEN DuPont). After UV-crosslinking (0.120 J), membranes were hybridized with radioactive probes, corresponding to mt-tRNAs, overnight at 65 °C in 7% (wt/vol) SDS and 0.25 M sodium phosphate buffer (pH 7.6), washed with 1× SSC three times for 20 min and then three times with 1× SSC containing 0.1% SDS (20 min; 65 °C). The membranes were exposed to a storage phosphor screen (GE Healthcare) visualized using a Typhoon phosphorimaging system and quantified using ImageQuant software (Molecular Dynamics, GE Healthcare) or ImageJ (https://imagej.nih.gov/ij).
To analyze modifications, RNA was extracted using TRIzol reagent (ThermoFisher Scientific), following the manufacturer’s instructions, from sucrose gradient fractions as indicated. The reverse transcription primer extension (RT-PEx) was as in ref. 20, the specific 5′ 32P-labeled primer (0.1 pmol) was incubated with 0.5–1 μg of the RNA isolated from sucrose gradient fractions and subjected to reverse transcription reactions, as described by ref. 21. The reaction mixture was separated by 12.5% (vol/vol) PAGE containing 7 M urea, and dried gels were exposed to a storage phosphor screen and scanned as earlier. The following primers were used in RT-PEx: mt-tRNAVal: 5′-GTAAGTTGGGTGCTTTGTGTT, and mt-tRNAPhe: 5′-TCAGTGTATTGCTTTGAGGAGGT.
Analysis of Mitochondrial Ribosomal Protein Profile on Isokinetic Sucrose Density Gradients.
Mitochondria were isolated from indicated tissues, as previously described (22). Cultured cells or mitochondria isolated from the tissues were lysed in buffer containing 50 mM Tris⋅HCl (pH 7.2), 1% Triton X-100, 10 mM Mg(OAc)2, 100 mM NaCl EDTA-free complete protease inhibitor (Roche), and 0.08 U/mL RNAsin (Promega). The lysates (0.7 and 0.1 mg of cell and mitochondrial lysates, respectively) were loaded on a linear sucrose gradient [2 mL 10–30% (vol/vol)] in 50 mM Tris⋅HCl (pH 7.2), 10 mM Mg(OAc)2, 80 mM NH4Cl, 0.1 M KCl, 1 mM PMSF, and centrifuged for 2 h 15 min at 100,000 g max at 4 °C (39,000 rpm; Beckman Coulter TLS-55 rotor). Fractions (15 × 100 μL) were collected and 10 μL aliquots were analyzed directly by Western blotting with the following antibodies; bL12m (PTG laboratories 14795–1-AP), uL3m (Abcam ab39268), mS29 (Abcam ab11928), mS40 (PTG laboratories 16139–1-AP), uL11m (CST D68F2).
Acknowledgments
Bovine, rat, and macaque tissues were kindly provided by James Blaza, Hannah Bridges, and Judy Hirst. Porcine tissues and cultured cells were obtained from Dario Brunetti and Massimo Zeviani. This work was supported by the Wellcome Trust (096919/Z/11/Z to R.N.L. and Z.M.C.-L.); Medical Research Council UK (MC_U105697135 to J.R., C.A.P., A.D., and M.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609338113/-/DCSupplemental.
References
- 1.Amunts A, Brown A, Toots J, Scheres SH, Ramakrishnan V. Ribosome. The structure of the human mitochondrial ribosome. Science. 2015;348(6230):95–98. doi: 10.1126/science.aaa1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greber BJ, et al. Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science. 2015;348(6232):303–308. doi: 10.1126/science.aaa3872. [DOI] [PubMed] [Google Scholar]
- 3.Smirnov A, Entelis N, Martin RP, Tarassov I. Biological significance of 5S rRNA import into human mitochondria: Role of ribosomal protein MRP-L18. Genes Dev. 2011;25(12):1289–1305. doi: 10.1101/gad.624711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown A, et al. Structure of the large ribosomal subunit from human mitochondria. Science. 2014;346(6210):718–722. doi: 10.1126/science.1258026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greber BJ, et al. Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature. 2014;505(7484):515–519. doi: 10.1038/nature12890. [DOI] [PubMed] [Google Scholar]
- 6.Greber BJ, et al. The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature. 2014;515(7526):283–286. doi: 10.1038/nature13895. [DOI] [PubMed] [Google Scholar]
- 7.Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 8.Helm M, et al. The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA. Nucleic Acids Res. 1998;26(7):1636–1643. doi: 10.1093/nar/26.7.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor RW, et al. Use of whole-exome sequencing to determine the genetic basis of multiple mitochondrial respiratory chain complex deficiencies. JAMA. 2014;312(1):68–77. doi: 10.1001/jama.2014.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yarham JW, Elson JL, Blakely EL, McFarland R, Taylor RW. Mitochondrial tRNA mutations and disease. Wiley Interdiscip Rev RNA. 2010;1(2):304–324. doi: 10.1002/wrna.27. [DOI] [PubMed] [Google Scholar]
- 11.Rorbach J, et al. Overexpression of human mitochondrial valyl tRNA synthetase can partially restore levels of cognate mt-tRNAVal carrying the pathogenic C25U mutation. Nucleic Acids Res. 2008;36(9):3065–3074. doi: 10.1093/nar/gkn147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hornig-Do HT, et al. Human mitochondrial leucyl tRNA synthetase can suppress non cognate pathogenic mt-tRNA mutations. EMBO Mol Med. 2014;6(2):183–193. doi: 10.1002/emmm.201303202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao H, Morrison LE, Moraes CT. Suppression of a mitochondrial tRNA gene mutation phenotype associated with changes in the nuclear background. Hum Mol Genet. 1999;8(6):1117–1124. doi: 10.1093/hmg/8.6.1117. [DOI] [PubMed] [Google Scholar]
- 14.Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. 5S ribosomal RNA database. Nucleic Acids Res. 2002;30(1):176–178. doi: 10.1093/nar/30.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srivastava AK, Schlessinger D. Mechanism and regulation of bacterial ribosomal RNA processing. Annu Rev Microbiol. 1990;44:105–129. doi: 10.1146/annurev.mi.44.100190.000541. [DOI] [PubMed] [Google Scholar]
- 16.Gelfand R, Attardi G. Synthesis and turnover of mitochondrial ribonucleic acid in HeLa cells: The mature ribosomal and messenger ribonucleic acid species are metabolically unstable. Mol Cell Biol. 1981;1(6):497–511. doi: 10.1128/mcb.1.6.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 18.Holzmann J, et al. RNase P without RNA: Identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135(3):462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Minczuk M, et al. TEFM (c17orf42) is necessary for transcription of human mtDNA. Nucleic Acids Res. 2011;39(10):4284–4299. doi: 10.1093/nar/gkq1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powell CA, et al. TRMT5 mutations cause a defect in post-transcriptional modification of mitochondrial tRNA associated with multiple respiratory-chain deficiencies. Am J Hum Genet. 2015;97(2):319–328. doi: 10.1016/j.ajhg.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rorbach J, et al. MRM2 and MRM3 are involved in biogenesis of the large subunit of the mitochondrial ribosome. Mol Biol Cell. 2014;25(17):2542–2555. doi: 10.1091/mbc.E14-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koulintchenko M, Temperley RJ, Mason PA, Dietrich A, Lightowlers RN. Natural competence of mammalian mitochondria allows the molecular investigation of mitochondrial gene expression. Hum Mol Genet. 2006;15(1):143–154. doi: 10.1093/hmg/ddi435. [DOI] [PubMed] [Google Scholar]