Significance
Terpenoids are ubiquitous products made by land plants with diverse biological functions. Their formation in seed plants is catalyzed by typical plant terpene synthases (TPSs), a well-characterized group of enzymes. In contrast, our knowledge of terpenoid biosynthesis in nonseed plants is very limited. By systematically analyzing the transcriptomes and/or genomes of more than 1000 plant species, we report that microbial terpene synthase-like genes, which are only distantly related to typical plant TPS genes, are widely distributed in nonseed plants, but virtually absent in seed plants. The study provides insights into the evolution of TPS genes in early land plants and opens the door to investigating the diversity and functions of terpenoids in nonseed plants.
Keywords: terpene synthase, specialized metabolism, nonseed plant, gene evolution
Abstract
The vast abundance of terpene natural products in nature is due to enzymes known as terpene synthases (TPSs) that convert acyclic prenyl diphosphate precursors into a multitude of cyclic and acyclic carbon skeletons. Yet the evolution of TPSs is not well understood at higher levels of classification. Microbial TPSs from bacteria and fungi are only distantly related to typical plant TPSs, whereas genes similar to microbial TPS genes have been recently identified in the lycophyte Selaginella moellendorffii. The goal of this study was to investigate the distribution, evolution, and biochemical functions of microbial terpene synthase-like (MTPSL) genes in other plants. By analyzing the transcriptomes of 1,103 plant species ranging from green algae to flowering plants, putative MTPSL genes were identified predominantly from nonseed plants, including liverworts, mosses, hornworts, lycophytes, and monilophytes. Directed searching for MTPSL genes in the sequenced genomes of a wide range of seed plants confirmed their general absence in this group. Among themselves, MTPSL proteins from nonseed plants form four major groups, with two of these more closely related to bacterial TPSs and the other two to fungal TPSs. Two of the four groups contain a canonical aspartate-rich “DDxxD” motif. The third group has a “DDxxxD” motif, and the fourth group has only the first two “DD” conserved in this motif. Upon heterologous expression, representative members from each of the four groups displayed diverse catalytic functions as monoterpene and sesquiterpene synthases, suggesting these are important for terpene formation in nonseed plants.
Terpenoids are the largest class of land plant secondary metabolites, but they are not uniformly distributed in the plant kingdom (1). Many seed plants (angiosperms and gymnosperms) produce terpenoids of diverse types in large quantities. However, among nonseed plants, only liverworts are known as copious producers of terpenoids (2). Terpenoids have diverse biological and ecological functions with many serving as chemical defenses against herbivores and pathogens (3, 4). Some have lineage-specific functions, such as the volatile terpenoids in flowers that are involved in attracting pollinators (5). Characterizing the biosynthesis of terpenoids in all plant lineages is therefore an important avenue to understanding their roles in the adaptation of various lineages of terrestrial plants.
Terpene synthases (TPSs) are pivotal enzymes for terpenoid biosynthesis, forming a distinctive superfamily based on both sequence identity and structure classification. However, within this group, typical plant and microbial (bacterial and fungal) TPSs share very low sequence similarity and are therefore only distantly related (6). The typical plant TPSs form subfamilies with individual subfamilies generally associated with specific biochemical functions, such as monoterpene, sesquiterpene, or diterpene biosynthesis (7, 8). Monoterpene synthases and sesquiterpene synthases have been proposed to have evolved independently in gymnosperms and angiosperms from diterpene synthase ancestors (7, 9). Interestingly, the typical plant TPSs in the moss Physcomitrella patens (10) and the lycophyte Selaginella moellendorffii (11–13), two nonseed plants, were found to be of the diterpene synthase type. Therefore, the molecular basis underlying the biosynthesis of monoterpenes and sesquiterpenes identified in nonseed plants has long been unclear.
Recently, microbial terpene synthase-like (MTPSL) genes were identified in S. moellendorffii that encode monoterpene and sesquiterpene synthases (13). Unlike typical plant TPSs, which are composed of either three domains (αβγ) or two domains (αβ) (14, 15), MTPSLs contain only an α-domain. Phylogenetic analysis indicated that MTPSLs from S. moellendorffii are more closely related to microbial TPSs, in particular fungal TPSs, than to typical plant TPSs (13). So far, MTPSLs have only been identified in S. moellendorffii (13), raising intriguing questions about the origin, evolution, and function of this type of plant terpene synthase genes. The goal of this study was to investigate the distribution of MTPSL genes in the green plants, infer their evolution, and determine their biochemical functions.
Results and Discussion
Terpene Synthase Genes of the Microbial Type Are Widespread in the Transcriptomes of Nonseed Land Plants, but Not in Green Algae and Seed Plants.
To determine the distribution of MTPSL genes in the green plants, the transcriptomes of 1,103 species (SI Appendix, Table S1) of green plants (779 species of seed plants, 166 species of nonseed land plants, 47 species of charophytes, and 111 species of chlorophytes) generated from the 1,000 Plant (OneKP) initiative (https://sites.google.com/a/ualberta.ca/onekp/) were searched for microbial type terpene synthase genes using a HMMER method as previously described (13). A total of 712 MTPSL genes were identified from the transcriptomes of 146 species. Strikingly, the vast majority of MTPSL genes (706 of the 712 MTPSL genes or 99.2%) were found in the transcriptomes of nonseed land plants (Fig. 1).
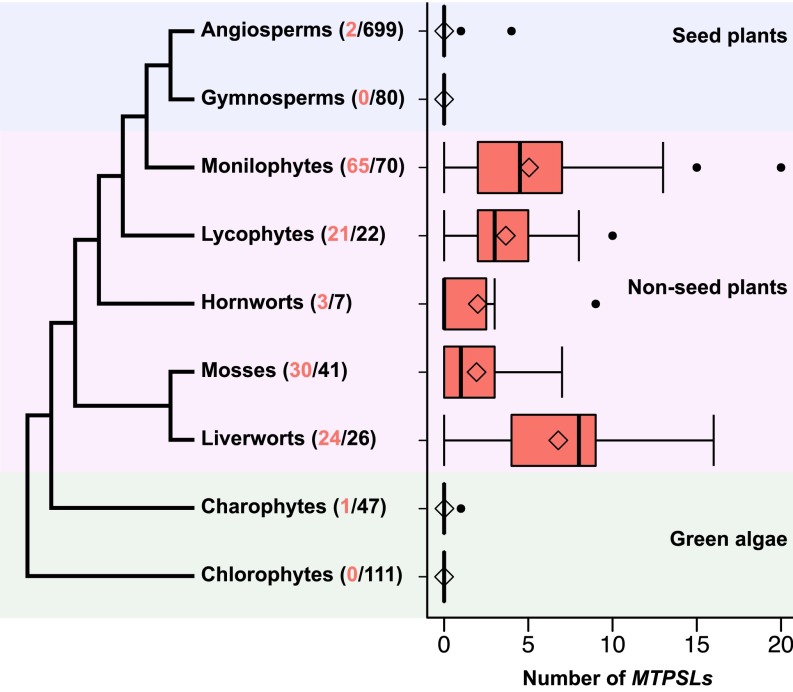
Fig. 1.
Distribution of MTPSL genes identified from the transcriptomes of 1,103 plant species. The numbers in parentheses represent the number of transcriptomes containing putative MTPSLs (in red) and total transcriptomes analyzed in each lineage (in black). The phylogeny of green plants presented was modified from refs. 25 and 26. Each boxplot represents the number of MTPSLs found for individual species in each plant lineage. The solid black lines denote the median number of MTPSLs from each species. Whiskers represent 1.5 times the quantile of the data. Points outside of the range of the whiskers are outliers.
Bryophytes consist of three lineages: hornworts, mosses, and liverworts, which have 7, 41, and 26 species in the OneKP dataset, respectively. The number of hornwort, moss, and liverwort species whose transcriptomes contain MTPSL genes was 3, 30, and 24, respectively. Among the 22 lycophyte species, 21 possessed MTPSL genes in their transcriptomes. For monilophytes, 65 of the 70 species were found to contain MTPSL genes in their transcriptomes. The median number of MTPSL genes from the transcriptome of each species for hornworts, mosses, liverworts, lycophytes, and monilophytes was 0, 1, 8, 3, and 4.5, respectively (Fig. 1 and SI Appendix, Table S2). Among all species, the monilophyte Cystopteris utahensis (a tetraploid) was found to contain the most MTPSL genes with 20 members (SI Appendix, Table S1).
On the other hand, extremely low occurrences of MTPSL genes were found in the transcriptomes of seed plants and charophytes. Among the 779 species of seed plants, only two species, Phytolacca bogotensis and Opuntia sp., both members of the Caryophyllales, were found to contain MTPSL genes in their transcriptomes with one and four members, respectively. Among the 47 species of charophytes, only one species, Micrasterias fimbriata, contained a MTPSL gene (one member) in its transcriptome. No MTPSL genes were found in the transcriptomes of 111 species of chlorophytes.
The Majority of MTPSL Genes Identified in Plant Transcriptomes Belong to Four Groups Clustered with Either Fungal or Bacterial Terpene Synthases.
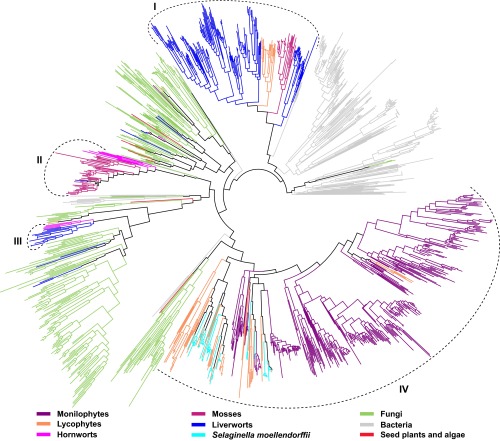
Phylogenetic analysis was performed for the 712 MTPSL genes identified from plant transcriptomes together with the 48 known MTPSL genes from S. moellendorffii (13) and selected terpene synthase genes from bacteria and fungi. The resulting phylogenetic tree indicates that the distribution of MTPSL genes in nonseed plants exhibits lineage-specific characteristics and that the majority of them (690 of 712) were clustered into four major groups with either similarity to bacterial TPS (groups I and II) or fungal TPS (groups III and IV) (Fig. 2).
Fig. 2.
Phylogeny of MTPSLs identified from OneKP with known MTPSLs from S. moellendorffii, bacterial TPSs, and fungal TPSs. Genes are color-coded based on their source. The majority of the newly identified MTPSLs are clustered into four major groups (I–IV).
Group I, the second largest MTPSL gene group, contains about 86% of MTPSL genes (152 of 177) from 23 species of liverworts, 34% of MTPSL genes (27 of 79) from 10 species of mosses, and 28% of MTPSL genes (23 of 83) from 9 species of lycophytes. Group II was composed of MTPSL genes primarily from mosses (about 66% of MTPSL genes from 24 species) and hornworts (50% of MTPSL genes from all three species in which MTPSL genes have been found). There was also one group II MTPSL gene found in a liverwort species, Scapania nemorea. Members of MTPSL genes in this species were also present in groups I and III. Group III, the smallest group, contains 4 MTPSL genes from 3 species of hornworts and 14 MTPSL genes from 7 species of liverworts. These 18 genes were clustered with the fungal trichodiene synthase (Tri5) genes. Group IV contains almost all MTPSL genes found from 65 species of monilophytes (352 of 353) and about 70% of MTPSL genes in lycophytes (58 of 83). The known MTPSL genes from S. moellendorffii were closely related to MTPSL genes from the transcriptomes of other lycophyte species.
Twenty-two MTPSL genes that lie outside of the four major groups were designated as “unclassified” (SI Appendix, Table S3). For example, the MTPSL genes found in two seed plant species, Opuntia sp. (four members) and P. bogotensis (one member), and one green alga, M. fimbriata (one member), were included in this list (Fig. 2 and SI Appendix, Table S3).
The Majority of MTPSL Genes Identified from Plant Transcriptomes Are Genuine Plant Genes.
The putative MTPSL genes identified in the plant transcriptomes could have one of two possible origins: from plants or from plant-associated microbes. Three lines of evidence were used to judge that the vast majority, if not all, of these genes in the four major MTPSL clades are plant genes.
The first line of evidence comes from the analysis of putative MTPSL genes from axenic culture. The liverwort S. nemorea was selected for this purpose. A total of eight putative MTPSL genes were identified in the transcriptome of S. nemorea (SI Appendix, Table S4), belonging to group I (five genes), group II (one gene), group III (one gene), and unclassified (one gene). An axenic culture of S. nemorea was initiated by germinating isolated spores, using sterile culture methods. This culture was therefore free of contamination of endophytic microbes. We extracted genomic DNA from axenically cultured S. nemorea and used PCR to amplify DNA fragments for each of the eight putative MTPSL genes, and the results were compared with those obtained from the transcriptome analysis. Six of the eight MTPSL genes (five from group I and one from group III) were amplified and confirmed by sequencing. However, the amplification of the members from group II and the unclassified group failed. The member from the unclassified group was suggested to be a contaminant from endophytic fungi (SI Appendix, Fig. S1). Analysis of the OneKP transcriptomes has suggested that S. nemorea was contaminated with an unknown source of plant material (https://pods.iplantcollaborative.org/wiki/display/iptol/Sample+source+and+purity). The putative MTPSL from S. nemorea in group II is the only MTPSL from liverworts to be assigned to this group, suggesting that it was also derived from contamination by other plant material. Overall, this experimental study confirms that the group I and group III MTPSLs from the S. nemorea transcriptome are endogenous S. nemorea genes.
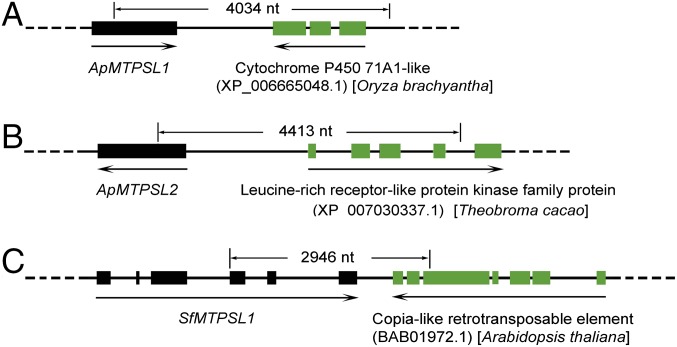
The second line of evidence for the plant origin of the majority of MTPSL genes reported is the identification of their immediate genomic neighbors as bona fide plant genes. We obtained such evidence for group II and III genes of the hornwort Anthoceros punctatus, whose genome has been recently sequenced (13). Assembling the raw genome sequence and then searching for MTPSL genes using the HMMER search, seven putative full-length MTPSL genes were identified; two of the seven genes belong to group II and the remaining five to group III (SI Appendix, Table S5). ApMTPSL1 from group II and ApMTPSL2 from group III were then selected as representatives for the following study. In the assembled genome of A. punctatus, ApMTPSL1 is the immediate neighbor to a cytochrome P450 gene, for which the most similar gene is from the plant Oryza brachyantha (Fig. 3A). Similarly, ApMTPSL2 is the immediate neighbor to a leucine-rich receptor-like kinase gene, for which the most similar gene is from the plant Theobroma cacao (Fig. 3B). We extracted genomic DNA from A. punctatus grown in axenic culture and performed PCR to amplify the coding sequence of ApMTPSL1 and -2 and their respective neighbors. The amplified DNA fragment was cloned and fully sequenced confirming that both ApMTPSL1 (Fig. 3A) and ApMTPSL2 genes (Fig. 3B) reside in the A. punctatus genome and are neighbors to a plant gene. Similar evidence for a group I gene was obtained from the moss Sphagnum fallax. The sequenced S. fallax genome contains 21 MTPSL genes, all of which belong to group I (SI Appendix, Table S6). A representative MTPSL gene (SfMTPSL1) from S. fallax was confirmed to reside in its genome and neighbor with a Copia-like retrotransposable element, to which the most similar sequence is from the plant Arabidopsis thaliana (Fig. 3C). The genes in group IV form a clade with SmMTPSL genes from S. moellendorffii, which have already been determined to be plant genes (13). This evidence that representative MTPSL genes of groups I–IV originated from plant genomes supports that their apparent orthologs/homologs in each group are also of plant origin.
Fig. 3.
Validation that representative MTPSL genes are of plant origin. Schematic genomic organization of representative MTPSL genes with their neighboring genes is shown. The genomic region spanning each MTPSL gene, its neighboring gene, and the intergenic region were amplified using PCR from genomic DNA and confirmed by sequencing. (A) ApMTPSL1 with its neighboring gene annotated as a cytochrome P450 71A1-like from the hornwort A. punctatus. (B) ApMTPSL2 and its neighboring gene annotated as a leucine-rich receptor-like kinase family protein from the hornwort A. punctatus. (C) SfMTPSL1 with its neighboring gene annotated as a Copia-like retrotransposon element from the moss S. fallax. In each schematic figure, underneath the neighboring gene, the ID in parentheses represents the accession number of the best hit of this gene by searching the nonredundant database of the National Center for Biotechnology Information. The plant species indicates from which species the best hit was identified.
The third line of evidence for plant origin is that the overall evolutionary relationships of MTPSLs are largely congruent with the relationships of the land plant species from which the MTPSL genes are identified. The MTPSL genes in each of the four groups are inferred to share a common evolutionary origin (Fig. 2). Within each group, the MTPSLs from the same plant lineage showed higher sequence similarity than with MTPSLs from a different plant lineage. For instance, in group I the MTPSLs from three plant groups, liverworts, mosses, and lycophytes, form three distinct subclades (Fig. 2). The MTPSLs from closely related species are often also most closely related. For instance, the analyzed mosses included three species from the same genus Sphagnum: Sphagnum lescurii, Sphagnum palustre, and Sphagnum recurvatum. In group I, the MTPSLs from these three species reside in a clade (Fig. 2). Such fine-scale correlations of MTPSL sequence similarity and plant phylogeny also support a plant origin for most of these genes.
On the other hand, the 22 unclassified MTPSL genes (SI Appendix, Table S3) have a high probability of being derived from plant-associated microbes because their similarities with microbial TPS genes are extremely high (SI Appendix, Table S3), as in the example of the S. nemorea gene mentioned above, and so were not considered further in this study. Nevertheless, some of these genes may have been obtained from microbes very recently through horizontal gene transfer (HGT), which will be a subject of future investigation.
MTPSL Genes Are Patchily Distributed in Green Plants: Evolutionary Implications.
The confirmation that the vast majority of MTPSL genes identified from the OneKP transcriptomes are plant genes indicates that MTPSL genes occur widely in nonseed plants. Group I contains MTPSL genes from liverworts, mosses, and lycophytes (Fig. 2), which implies the presence of MTPSL genes in the common ancestor of land plants. However, our survey found that MTPSL genes are generally absent from the transcriptomes of green algae (Fig. 1). To provide further evidence on the presence/absence of MTPSL genes in green algae, we conducted a focused search on sequenced genomes for six species of chlorophytes and one species of charophyte (SI Appendix, Table S7). No MTPSL genes were detected in these sequenced green algae. The absence of MTPSL genes in the transcriptomes of a wide range of chlorophytes and the genome of the charophyte, Klebsormidium flaccidum (16) suggests that MTPSL genes may have their origin in an ancestral land plant rather than an algal ancestor. Broader genome sampling from green algae, especially charophytes, is needed to test this hypothesis. Nonetheless, the evolution of MTPSL genes may have been associated with the transition of plants from aquatic to terrestrial habitats. The pioneer land plants faced a harsh environment replete with many new biotic and abiotic stresses. Many products of TPSs are volatile hydrocarbons that may be more useful in a terrestrial environment than in an aquatic one.
MTPSLs from nonseed land plants exhibited different degrees of relatedness to bacterial TPSs and fungal TPSs. Group I is most closely related to bacterial TPSs, whereas groups III and IV are most closely related to fungal TPSs. Group II is most closely related to a number of bacterial TPSs, which, however, reside within a fungal clade (Fig. 2). These patterns suggest a complex evolutionary history of microbial type TPSs. Whereas it is possible that microbial-type TPS genes are ancestral in all kingdoms of life and took different evolutionary trajectories, their confinement to bacteria, fungi, and plants implies HGT. Assuming that bacterial and fungal TPS genes are ancestral to MTPSLs, the distribution pattern of MTPSLs can be explained by multiple HGT events from bacteria and fungi. However, it is premature to make strong claims about the donors and recipients of such transfer events because our understanding of phylogenetic relationships of TPSs in bacteria and fungi is still very limited. A better understanding of relationships among TPS genes in bacteria and fungi will allow testing of this hypothesis.
The absence of MTPSL genes in seed plants is also notable (Fig. 1). To gain additional evidence about the presence/absence of MTPSL genes in seed plants, we analyzed the genomic sequences of 48 species of seed plants (SI Appendix, Table S7): no MTPSL genes were identified in them. As mentioned previously, land plants contain typical plant TPSs, which catalyze similar biochemical reactions for the production of terpenoids as do MTPSLs, but typical plant TPSs are only distantly related to MTPSLs (13). However, in the nonseed plants that have been studied, the typical plant TPSs function as diterpene synthases (7) rather than the full range of monoterpene synthases, sesquiterpene synthases, and diterpene synthases found in seed plants. Several MTPSL genes from S. moellendorffii have been demonstrated to encode monoterpene synthases and sesquiterpene synthases (13), and we hypothesized that most MTPSL genes in nonseed plants function in this way.
Representative MTPSL Genes Encode Active TPSs with Diverse Catalytic Activities.
The presence of MTPSL genes only in nonseed land plants poses an intriguing question about their functions. In seed plants, TPSs are responsible for the production of a diversity of terpenoids important for ecological interactions, especially as defenses against herbivores and pathogens. Some nonseed plants, such as liverworts (17) and mosses (18), also produce a vast diversity of terpenoids. However, little is known about how such terpenoids are synthesized or about their biological functions.
In general, TPSs contain two highly conserved motifs: the DDxxD and NSD/DTE motifs, which are both involved in substrate binding (14, 15, 19). Whereas the NSD/DTE motif is highly conserved in the MTPSLs, the aspartate-rich DDxxD motif exhibits variations (SI Appendix, Fig. S2). Group I and IV proteins contain the canonical DDxxD motif, but group II proteins displayed a conserved DDxxxD motif. In the group III proteins, only the first two aspartates (DD) are conserved.
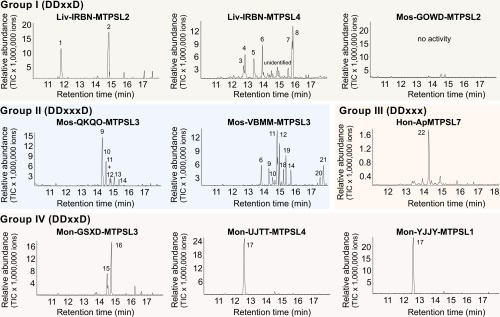
To gain an initial assessment of the biochemical functions of MTPSLs, a total of nine genes representing the four MTPSL groups (SI Appendix, Table S8) were selected for experimental work. Recombinant MTPSLs produced from Escherichia coli were tested for TPS activity with geranyl diphosphate [(E)-GPP], farnesyl diphosphate [(E,E)-FPP], and geranylgeranyl diphosphate [(E,E,E)-GGPP], the substrates for monoterpenes, sesquiterpenes, and diterpenes, respectively. With the exception of Mos-GOWD-MTPSL2, all tested enzymes were able to convert (E,E)-FPP into individual sesquiterpenes or complex sesquiterpene mixtures (Fig. 4). Whereas most of the MTPSL sesquiterpene products are also known as products from typical plant TPSs, Mon-UJTT-MTPSL4 and Mon-YJJY-MTPSL1, which are closely related to fungal TPS, produced protoillud-6-ene, a sesquiterpene that has only been reported from a fungus so far (20). Mos-GOWD-MTPSL2 showed exclusively monoterpene synthase activity and converted (E)-GPP into (Z)-β-ocimene and some minor monoterpene products (SI Appendix, Fig. S3). In addition to sesquiterpene synthase activity, Liv-IRBN-MTPSL2, Mon-UJTT-MTPSL4, and Hon-Ap-MTPSL7 were able to produce monoterpenes from (E)-GPP and Liv-IRBN-MTPSL4 and Mos-VBMM-MTPSL3 were able to convert (E,E,E)-GGPP into diterpenes (SI Appendix, Fig. S4). Recently, it was shown that certain typical plant TPSs naturally use cis-prenyl diphosphates as substrates for terpene production (21). Because most of the representative MTPSLs tested showed sesquiterpene synthase activity using (E,E)-FPP (Fig. 4), we performed additional assays to determine whether (Z,E)-FPP and (Z,Z)-FPP, the cis-isomers of FPP, could serve as substrates for these enzymes. Mos-GOWD-MTPSL2 was inactive with either substrate, whereas all of the other eight characterized MTPSLs showed activity with either (Z,E)-FPP or (Z,Z)-FPP or both (SI Appendix, Fig. S5). It is interesting to observe that some enzymes, such as Mos-QKQO-MTPSL3, produced the same products using the cis-FPP isomers as with (E,E)-FPP, whereas others such as Mon-UJTT-MTPSL4 produced different products (Fig. 4 and SI Appendix, Fig. S5). Nevertheless, the fact that the Myriopteris eatonii UJTT-MTPSL4 forms a single distinctive product from (E,E)-FPP and broad mixtures of over 10 products from each of the cis-FPP isomers suggests that (E,E)-FPP is the natural substrate. However, more information about the actual occurrence of these assay products in M. eatonii is needed before the natural substrates of these enzymes can be determined with certainty.
Fig. 4.
Representative MTPSL genes from four groups (I–IV) encode active TPSs. The aspartate-rich motif associated with each group is indicated. MTPSL genes were heterologously expressed in E. coli and crude protein extracts were incubated with the potential substrate farnesyl diphosphate [(E,E)-FPP]. Enzyme products were collected using solid-phase microextraction and analyzed with gas chromatography/mass spectrometry. 1, bicycloelemene; 2, bicyclogermacrene; 3, α-isocomene; 4, β-elemene*; 5, (E)-β-caryophyllene*; 6, (E)-β-farnesene*; 7, nerolidol*; 8, dactylol; 9, γ-curcumene; 10, α-zingiberene; 11, β-bisabolene*; 12, β-curcumene; 13, sesquiphellandrene*; 14, (E)-α-bisabolene; 15, (Z,E)-α-farnesene; 16, (E,E)-α-farnesene; 17, protoillud-6-ene*; 18, (Z)-γ-bisabolene; 19, (E)-γ-bisabolene; 20, β-bisabolol; 21, α-bisabolol; and 22, β-acoradiene. Compounds marked with an asterisk (*) were identified using authentic standards. The origin of each MTPSL gene is listed in SI Appendix, Table S8. Ap, A. punctatus; GOWD, S. lescurii; GSXD, M. eatonii; Hon, hornwort; IRBN, S. nemorea; Liv, liverwort; Mon, monilophyte; Mos, moss; QKQO, Pseudotaxiphyllum elegans; UJTT, Pityrogramma trifoliate; VBMM, Anomodon rostratus; and YJJY, Woodsia scopulina.
The substrates actually used by seed plant TPSs depend on their subcellular locations because the various prenyl diphosphate substrates are restricted to different subcellular compartments (GPP and GGPP to plastids and FPP to the cytosol) (22). Thus, in seed plants, mono- and diterpene synthases are present in plastids and sesquiterpene synthases in the cytosol. However, information on TPS enzyme and substrate localization is not yet available for nonseed plants. To learn more, we used an in silico protein-targeting program (Target P, www.cbs.dtu.dk/services/TargetP) for Mon-UJTT-MTPSL4, an enzyme that produced a single sesquiterpene from (E,E)-FPP but also formed monoterpenes from (E)-GPP. Because the program suggests a cytosolic, nonplastid location for Mon-UJTT-MTPSL4, and FPP is known to be cytosolic in seed plants, this enzyme is likely to act as a sesquiterpene synthase in planta. Under steady-state conditions, the apparent KM and kcat values of Mon-UJTT-MTPSL4 using (E,E)-FPP as substrate were determined to be 2.13 ± 0.23 µM and 0.15⋅s−1, respectively. Such kinetic parameters are very comparable to those of typical plant TPSs (23), suggesting that MTPSL enzymes function in almost the same way as typical plant TPSs.
Based on the in vitro biochemical activities of representative MTPSLs, one could speculate that the MTPSLs have been the primary enzymes to make mono- and sesquiterpenes in early land plants and that the evolution of monoterpene and sesquiterpene synthases among the typical plant TPS family allowed the eventual loss of MTPSL genes in seed plants.
Conclusions
In this study, microbial-type terpene synthase genes, once thought to be confined to bacteria and fungi, were systematically mined from large-scale plant transcriptomes. Of 779 seed plant species, only 5 MTPSL genes were found in 2 species, whereas 706 MTPSL genes were found in 143 nonseed land plant species. So, in addition to the previous report on S. moellendorffii (13), MTPSL genes are widely distributed in nonseed land plants, but generally absent from seed plants and green algae. Although these genes are also found in fungi and bacteria, their occurrence in plants is in most cases not due to microbial contamination of the plant samples used for sequencing, based on experiments with axenic cultures, phylogenetic analyses, and their embedment in plant genomes with bona fide plant genes as neighbors. MTPSL genes form four lineage-specific groups that exhibit diverse structural features, which implies multiple evolutionary origins. Biochemical studies of selected MTPSL genes showed that they encode sesquiterpene and monoterpene synthases. However, much more remains to be done to investigate the biological functions of their products and how they have influenced the evolution of the MTPSL gene family in nonseed land plants.
Materials and Methods
MTPSL genes were searched against the assembled transcriptomes for 1,103 nonmodel plant species derived from the OneKP (sites.google.com/a/ualberta.ca/onekp/) (24). Fresh materials of three axenically cultured plants, A. punctatus, S. nemorea, and S. fallax, were used for the extraction of genomic DNA that served as template for PCR analysis. For TPS activity assays, crude proteins extracted from E. coli expressing individual representative MTPSL genes were assayed with individual prenyl diphosphates. The kinetic properties of Mon-UJTT-MTPSL4 were measured with its purified recombinant enzyme following a radiochemical protocol as previously described (23). Details on transcriptome and genome assembly, sequence searches, phylogenetic reconstruction, plant cultures, and biochemical analysis of MTPSLs are provided in SI Appendix.
Supplementary Material
Acknowledgments
We acknowledge the contributions of Dr. James Leebens-Mack’s laboratory at the University of Georgia in obtaining material for RNA sequencing and developing the infrastructure for distribution of the OneKP data. We thank Natascha Rauch (Max Planck Institute for Chemical Ecology) for technical assistance. This project was supported by a University of Tennessee (UT) AgResearch Innovation Grant (to F.C.), the Max Planck Society (to T.G.K. and J.G.), and the Program for New Century Excellent Talents in University (NCET-12-0896 to G.L.). The OneKP initiative led by G.K.-S.W. was funded by the Alberta Ministry of Advanced Education, Alberta Innovates Technology Futures, Innovates Centres of Research Excellence, Musea Ventures, and the China National Genebank. The BGI-Shenzhen group led by Y.Z. was funded by the Shenzhen Supporting Projects Program under Grant CXZZ20140421112021913. S. fallax plant and genomic resources were supported by the Laboratory-Directed Research and Development Program of Oak Ridge National Laboratory, managed by UT–Battelle Memorial Institute, for the US Department of Energy under contract DE-AC05-000R22725.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences for the biochemically characterized MTPSLs reported in this paper have been deposited in the GenBank database (accession nos. KX230835–KX230843).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1607973113/-/DCSupplemental.
References
- 1.Connolly JD, Hill RA. Dictionary of Terpenoids. Chapman & Hall; London: 1991. [Google Scholar]
- 2.Asakawa Y. Chemosystematics of the hepaticae. Phytochemistry. 2004;65(6):623–669. doi: 10.1016/j.phytochem.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Gershenzon J, Dudareva N. The function of terpene natural products in the natural world. Nat Chem Biol. 2007;3(7):408–414. doi: 10.1038/nchembio.2007.5. [DOI] [PubMed] [Google Scholar]
- 4.Tholl D. Biosynthesis and biological functions of terpenoids in plants. Adv Biochem Eng Biotechnol. 2015;148:63–106. doi: 10.1007/10_2014_295. [DOI] [PubMed] [Google Scholar]
- 5.Pichersky E, Gershenzon J. The formation and function of plant volatiles: Perfumes for pollinator attraction and defense. Curr Opin Plant Biol. 2002;5(3):237–243. doi: 10.1016/s1369-5266(02)00251-0. [DOI] [PubMed] [Google Scholar]
- 6.Cao R, et al. Diterpene cyclases and the nature of the isoprene fold. Proteins. 2010;78(11):2417–2432. doi: 10.1002/prot.22751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen F, Tholl D, Bohlmann J, Pichersky E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011;66(1):212–229. doi: 10.1111/j.1365-313X.2011.04520.x. [DOI] [PubMed] [Google Scholar]
- 8.Bohlmann J, Meyer-Gauen G, Croteau R. Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA. 1998;95(8):4126–4133. doi: 10.1073/pnas.95.8.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trapp SC, Croteau RB. Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics. 2001;158(2):811–832. doi: 10.1093/genetics/158.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi K, et al. Identification and functional analysis of bifunctional ent-kaurene synthase from the moss Physcomitrella patens. FEBS Lett. 2006;580(26):6175–6181. doi: 10.1016/j.febslet.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 11.Mafu S, Hillwig ML, Peters RJ. A novel labda-7,13e-dien-15-ol-producing bifunctional diterpene synthase from Selaginella moellendorffii. ChemBioChem. 2011;12(13):1984–1987. doi: 10.1002/cbic.201100336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugai Y, et al. Enzymatic (13)C labeling and multidimensional NMR analysis of miltiradiene synthesized by bifunctional diterpene cyclase in Selaginella moellendorffii. J Biol Chem. 2011;286(50):42840–42847. doi: 10.1074/jbc.M111.302703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li G, et al. Nonseed plant Selaginella moellendorffi [corrected] has both seed plant and microbial types of terpene synthases. Proc Natl Acad Sci USA. 2012;109(36):14711–14715. doi: 10.1073/pnas.1204300109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Starks CM, Back K, Chappell J, Noel JP. Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science. 1997;277(5333):1815–1820. doi: 10.1126/science.277.5333.1815. [DOI] [PubMed] [Google Scholar]
- 15.Köksal M, Jin Y, Coates RM, Croteau R, Christianson DW. Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis. Nature. 2011;469(7328):116–120. doi: 10.1038/nature09628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hori K, et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun. 2014;5:3978. doi: 10.1038/ncomms4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asakawa Y. Liverworts-potential source of medicinal compounds. Curr Pharm Des. 2008;14(29):3067–3088. doi: 10.2174/138161208786404272. [DOI] [PubMed] [Google Scholar]
- 18.Saritas Y, et al. Volatile constituents in mosses (Musci) Phytochemistry. 2001;57(3):443–457. doi: 10.1016/s0031-9422(01)00069-3. [DOI] [PubMed] [Google Scholar]
- 19.Hyatt DC, et al. Structure of limonene synthase, a simple model for terpenoid cyclase catalysis. Proc Natl Acad Sci USA. 2007;104(13):5360–5365. doi: 10.1073/pnas.0700915104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanssen HP, Sprecher E, Abraham WR. 6-Protoilludene, the major molatile metabolite from ceratocystis piceae liquid cultures. Phytochemistry. 1986;25(8):1979–1980. [Google Scholar]
- 21.Matsuba Y, et al. Evolution of a complex locus for terpene biosynthesis in solanum. Plant Cell. 2013;25(6):2022–2036. doi: 10.1105/tpc.113.111013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemmerlin A, Harwood JL, Bach TJ. A raison d’être for two distinct pathways in the early steps of plant isoprenoid biosynthesis? Prog Lipid Res. 2012;51(2):95–148. doi: 10.1016/j.plipres.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 23.Tholl D, Chen F, Petri J, Gershenzon J, Pichersky E. Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers. Plant J. 2005;42(5):757–771. doi: 10.1111/j.1365-313X.2005.02417.x. [DOI] [PubMed] [Google Scholar]
- 24.Matasci N, et al. Data access for the 1,000 Plants (1KP) project. Gigascience. 2014;3:17. doi: 10.1186/2047-217X-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu YL, et al. The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci USA. 2006;103(42):15511–15516. doi: 10.1073/pnas.0603335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wickett NJ, et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci USA. 2014;111(45):E4859–E4868. doi: 10.1073/pnas.1323926111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.