Significance
The pathogenic trypanosomatids—Trypanosoma cruzi, Trypanosoma brucei, and Leishmania spp.—are the causative agents of Chagas disease, African trypanosomiasis, and leishmaniasis, respectively. These diseases, with high morbidity and mortality rates, affect millions of people worldwide. Current treatments typically use drugs with high toxicity and marginal efficacy. Here we present, a 2.5-Å structure of the T. cruzi ribosome large subunit by single-particle cryo-EM. Our structure highlights distinctive trypanosome interactions and has allowed us to propose a tentative model for assembly of the 60S large ribosomal subunit. These atomic details highlighting trypanosome-specific interactions and the differences between T. cruzi and the human ribosome can be used directly for structure-based drug design of antitrypanosome drugs.
Keywords: ribosome structure, Trypanosoma cruzi, biogenesis, multiply fragmented rRNA, antitrypanosome drug design
Abstract
Ribosomes of trypanosomatids, a family of protozoan parasites causing debilitating human diseases, possess multiply fragmented rRNAs that together are analogous to 28S rRNA, unusually large rRNA expansion segments, and r-protein variations compared with other eukaryotic ribosomes. To investigate the architecture of the trypanosomatid ribosomes, we determined the 2.5-Å structure of the Trypanosoma cruzi ribosome large subunit by single-particle cryo-EM. Examination of this structure and comparative analysis of the yeast ribosomal assembly pathway allowed us to develop a stepwise assembly model for the eight pieces of the large subunit rRNAs and a number of ancillary “glue” proteins. This model can be applied to the characterization of Trypanosoma brucei and Leishmania spp. ribosomes as well. Together with other details, our atomic-level structure may provide a foundation for structure-based design of antitrypanosome drugs.
Ribosomes share a universally conserved core that carries out the fundamental processes of protein synthesis (1). Outside of this core, ribosome composition varies considerably. The main differences among eukaryotic ribosomes are due to rRNA expansion segments (ESs) and variations of ribosomal proteins (r-proteins) (2). An extreme case is presented by the ribosome of trypanosomatids, a family of kinetoplastid protozoans the members of which cause human diseases such as Chagas disease (Trypanosoma cruzi), sleeping sickness (Trypanosoma brucei), and Leishmaniasis (Leishmania spp.) (3–5). Trypanosomatid ribosomes have unusually large ESs and r-protein variations (6). Moreover, their large subunit contains eight pieces of rRNA, six of which result from unique cleavages of a precursor rRNA. Upon assembly, these rRNAs fulfill the functions of the 28S rRNA found in other eukaryotes (7, 8). This unique multiple fragmentation strongly suggests that trypanosomatid ribosomes have pronounced differences that could be exploited for drug design.
Results and Discussion
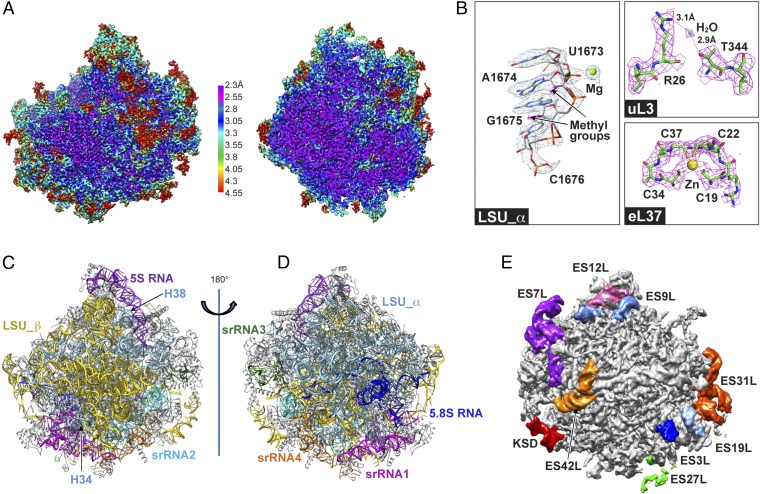
Here, we report the structure of the 60S large ribosomal subunit of T. cruzi at a 2.5-Å resolution (Fig. 1; SI Appendix, Fig. S2). Our large subunit structure represents a significant improvement over earlier trypanosomatid ribosome models (6, 9) (SI Appendix, Fig. S14) and provides extensive details of the constitutive molecular interactions that stabilize the eight pieces of rRNA (Fig. 1 C and D; SI Appendix, Fig. S4). The quality of the EM map is indicated by the clearly resolved density of the nucleotides and the base-pairing and base-stacking in the helical segments of rRNA (Fig. 1B; SI Appendix, Fig. S3 A–D). Purine bases can be readily distinguished from pyrimidine bases, and it is also routinely possible to distinguish between G and A (Fig. 1B; SI Appendix, Fig. S3). Well-defined EM density is also observed for most protein side chains (SI Appendix, Fig. S7 and Table S7). In addition, the high resolution allowed us to identify magnesium ions as well as water molecules (Fig. 1B; SI Appendix, Fig. S3). Importantly, we located many rRNA covalent modifications (Fig. 1B). The number of 2′-O methylations (66 sites) remarkably exceeds that in other eukaryotes, which is consistent with bioinformatics analysis (9) and the modifications on the Leishmania ribosome structure recently revealed (10).
Fig. 1.
Structure of the T. cruzi large subunit ribosome. (A) Cryo-EM map of the 60S subunit after sharpening, colored by local resolution and viewed from the subunit interface. (Left) Surface view. (Right) Central cut-away view. (B) Selected views of density for rRNA and proteins with associated ions and water molecule. (C) Some expansion segments in the unsharpened map of a large subunit, viewed from the solvent side. (C and D) rRNA architecture of the large subunit: interface (C) and solvent (D) view. (E) Some expansion segments in the unsharpened map of a large subunit, viewed from the solvent side.
In general outline, the 60S ribosomal subunit of T. cruzi resembles that of yeast. However, the total size of the six rRNA pieces is about one-fourth larger than the 25S rRNA in yeast (4,225 vs. 3,392 nts) (11, 12). This increase is accounted for by the larger size of ESs and the addition of trypanosome-specific ESs. We refer to the six pieces analogous to 28S rRNA as LSU-α, LSU-β, and srRNAs1–4 (13). The corresponding six pieces across the three pathogenic trypanosomatids are shown in SI Appendix, Fig. S15. LSU-α roughly corresponds to domains I and II of the yeast 25S RNA, srRNA1 to domain III, LSU-β to domains IV and V, and srRNA2–4 to domain VI (SI Appendix, Fig. S6). LSU-α and LSU-β are located on the solvent and interface side, respectively, whereas srRNA1 is located at the bottom of the large subunit (solvent-side view) (Fig. 1D; SI Appendix, Fig. S4). The srRNAs2–4 are in close mutual contact and situated on the left side of the large subunit under the P stalk (solvent-side view) (Fig. 1; SI Appendix, Fig. S4). Most r-proteins are conserved between yeast and T. cruzi with exceptions typically occurring near the srRNAs. Comparative analysis of trypanosome-specific rRNA–rRNA and r-protein–rRNA interactions with those of other eukaryotes, especially yeast, has allowed us to identify the interactions stabilizing the eight pieces of rRNA to form a mature large subunit. Using our structure as the endpoint of assembly in combination with this comparative analysis has allowed us to arrive at a plausible model for assembly of the eight trypanosomal rRNA pieces.
The rRNA Scaffold.
Examination of rRNA–rRNA interactions in our structure reveals that the 5.8S rRNA and the conserved LSU-α and LSU-β domains form the backbone of the rRNA scaffold with two rigid blocks acting as stabilizing core. The first block is formed by the whole 5.8S rRNA and nucleotides in the 5′-end of LSU-α. The second rigid block is constructed by nucleotides in the 3′-end of LSU-α and the 5′-end of LSU-β. Both of the rigid blocks are conserved in yeast; however, in trypanosomes they interact with each other via ES3L of 5.8S rRNA (Fig. 2A). Clearly, the 5.8S serves as a critical organizing center for rRNA folding and assembly. The stability of the LSU-α–LSU-β junction is further reinforced by the insertion of two helices, H38 and H34 from LSU-α, into the interface from the solvent side anchoring LSU-α to LSU-β. Mg2+ ions and modified nucleotides also mediate contacts in the scaffold (SI Appendix, Fig. S11). Along with rRNA–rRNA interactions, r-proteins play important roles in stabilizing this scaffold (SI Appendix, Fig. S7).
Fig. 2.
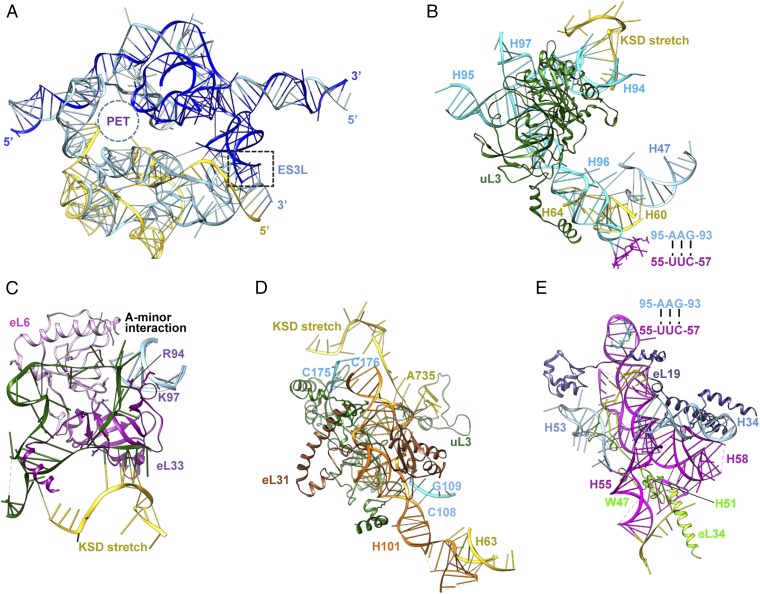
Interactions stabilizing the srRNAs. (A) Scaffold formed by 5.8S rRNA, LSU-α, and LSU-β. PET, peptide exit tunnel. (B) srRNA2 (cyan). The components interacting with srRNA2 include the KSD stretch, H60, H64, H47, and uL3. (C) srRNA3 (forest green). The rRNA-contacting residues of the anchoring protein, eL6 and eL33, are shown. R94 and K97 are in the trypanosome-specific insertion of eL33. (D) srRNA4 (orange). The interacting residues are from the KSD stretch, H63, srRNA2, eL31, and uL3. (E) srRNA1 (magenta). W47 and H51 are from the trypanosome-specific insertion of eL34.
Structure and Interactions Stabilizing the Small rRNAs.
The srRNA1 consists of five helices corresponding to yeast H55-59 and a trypanosome-specific ES, named srRNA1-ES, which adopts a similar position in T. brucei (13) and shows highly dynamic behavior, preventing us from modeling it (SI Appendix, Fig. S9A). The srRNA1 is anchored to the scaffold with the help of srRNA2 via base-pairing interactions. In addition to the contacts with srRNA2, two anchoring proteins, eL19 and eL34, pin H58 and H55 of srRNA1 to the scaffold (Fig. 2E). Protein eL34 has adopted an insertion and protein eL19 has a C-terminal extension of about 170 aa, crossing through to the SSU, as already revealed in the structure of T. brucei, which suggests that the small subunit bears a role in further stabilizing srRNA1 (13).
The srRNA2 has a scissor-shape architecture and is composed of four helices corresponding to H94-97 in yeast (SI Appendix, Fig. S9B). H94-95 and H96-97 line up to form the two prongs of the scissor. We found in our structure that srRNA2 has five contact sites with LSU-β. In addition, srRNA2 forms various contacts with LSU-α (Fig. 2B). Apart from direct interactions with the scaffold, we found srRNA2 to be further anchored by uL3 onto the scaffold. Specifically, protein uL3 forms a C-shaped cavity, which facilitates the stable assembly of srRNA2 onto the scaffold by accommodating H94 and H96 (Fig. 2B; SI Appendix, Figs. S8B and S9B).
The srRNA3 of T. cruzi corresponds to yeast ES39L, which is made up of three helices, ES39L1–3, threaded together by single rRNA stretches (SI Appendix, Fig. S9C). In contrast, srRNA3 is composed of a stretch of single-stranded rRNA plus a helix corresponding to helix ES39L-h2 (SI Appendix, Fig. S12A). The vacancy resulting from the missing ES39L-h1 adjacent to the sRNA2–4 cleavage sites is filled partially by ES42L, a trypanosome-specific ES. The absence of ES39L-h3 makes room for the kinetoplastid-specific domain (KSD) stretch, the extension of the C-terminal portion of eL14, and the N-terminal domain of eL33. Protein eL33 anchors srRNA3 to the scaffold and contains an insertion in its globular domain that provides extra binding sites for both srRNA3 and LSU-α (Fig. 2C; SI Appendix, Figs. S8D, S9C, and S11A). Moreover, srRNA3 forms an A-minor interaction with ES7L (Fig. 2C) (14), indicating that ES7L also facilitates the assembly of srRNA3 onto the scaffold.
Finally, the srRNA4 is composed of a long helix, corresponding to H101 in yeast, and two short helices (H99 and H100 in yeast) located on one end of H101, as well as an ES, named srRN4-ES, emerging from H101 (SI Appendix, Fig. S9D). Nucleotides 106–108 of srRNA2 and the exposed positively charged residues of uL3 establish a binding surface for srRNA4 to stack onto (Fig. 2D). Furthermore, several residues of the N-terminal extension of eL31 cover H101 of srRNA4. These additions on the N-terminal portion of eL31 exhibit more flexibility compared with the C-terminal region of eL31, which results in only C-A modeling for this region. Also, T. cruzi srRNA4 adopts a shorter loop at the base of H100 than its counterpart in yeast, thus avoiding conflict with the N terminus of eL31 (SI Appendix, Fig. S9D). In addition, residues 1561–1564 of the KSD stretch provide two binding sites with H99 from srRNA4 (Fig. 2D).
From the arrangement of the srRNAs revealed in our structure, we see that both srRNA1 and srRNA4 require srRNA2 to be in its final position to provide binding sites for their stable assembly onto the scaffold (Fig. 2 B, D, and E). Interestingly, the known relative time points of assembly of the r-proteins anchoring these small pieces correlates with this arrangement. Studies in yeast have shown that r-proteins, localized in specific positions within the mature 60S subunit, can be classified as early, middle-, or late-acting proteins, thus providing a temporal hierarchy of 60S assembly (15–17). As described earlier, we found in our structure that srRNA2 is anchored by uL3 and srRNA3 is anchored by eL6 and eL33 (Fig. 2 B and C), all of which are early acting proteins whereas proteins eL19, eL34, and eL31 (Fig. 2 D and E; SI Appendix, Fig. S8 E–G), which are identified as middle-acting proteins in yeast, interact with srRNA1 and srRNA4 (Fig. 2 D and E) (17, 18). Thus, the arrangement of the srRNAs as well as their anchoring proteins help define the likely order of assembly of the srRNAs, such that stable association of srRNA2 and srRNA3 occurs before that of srRNA1 and srRNA4. Indeed, deletion of the counterpart of srRNA3 in yeast (ES39L) leads to an early processing defect (19). Strikingly, although srRNA1 is the first small rRNA transcribed, it is stably assembled after srRNA2 and srRNA3, suggesting that its RNA processing occurs during or after assembly of the ribosome instead of the 5′ to 3′ direction following transcription. Furthermore, this assembly order is also consistent with the proposed order of assembly of the three focal points on the Leishmania ribosome structure (10) (SI Appendix, Fig. S6).
Prominent Expansion Segments and the 5S rRNA.
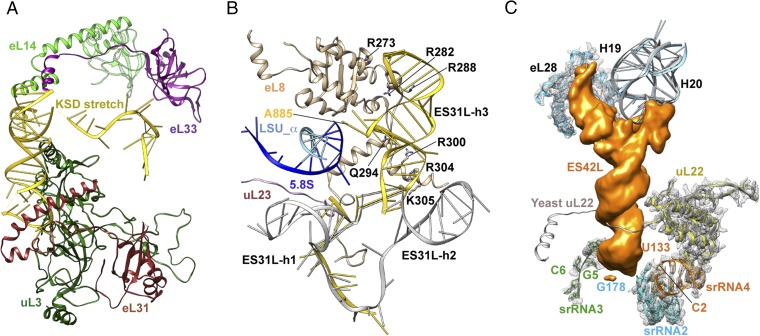
The trypanosome-specific ESs, ES42L and the KSD, both located on the surface of the ribosome, bundle srRNAs2–4. The KSD is a binding partner of the sRNA2- and srRNA4-anchoring proteins, uL3 and eL31, respectively (Fig. 3A). This places KSD in a pivotal position interacting with ribosomal functional centers (SI Appendix, Table S6) (20, 21). ES42L covers the hub region encompassing the cleavage sites of srRNAs2–4 (Fig. 3C; SI Appendix, Fig. S12), where the extension of uL22, a component of the peptide exit tunnel (PET), would reach toward ES39L in yeast. The location of ES42L suggests that it may also play a role in the remote regulation of PET by uL22 (22) (Fig. 3C; SI Appendix, Fig. S12). Both ES42L and KSD envelope small rRNAs2–4 and make additional contacts to stabilize these small rRNA pieces (Figs. 1E and 3 A and C), which suggests that their assembly occurs after that of srRNAs2–4.
Fig. 3.
T. cruzi-specific expansion segments. (A) KSD contacts uL3, eL31, eL33, and eL14. (B) ES31L (yellow) in T. cruzi has the specific helix ES31L-h3 contacting eL8 and the 5′ end of 5.8S rRNA. ES31L-h1 and -h2 are only partially modeled. For comparison, ES31L from yeast is also shown here (in gray). (C) ES42L, emerging from H19, covers the cleavage sites of srRNA2–4 and blocks the extension of uL22.
Another prominent ES, ES31L, emerges from the base of the L1 stalk and contains an additional helix compared with yeast (Fig. 3B). This helix contacts extensions of eL8 and interacts with the 3′-end of the 5.8S rRNA. In yeast, deleting ES31L or the N-terminal extension of eL8 blocks ribosome assembly at later stages, suggesting that ES31L may be involved in subsequent stabilization steps of the previously established scaffold (23). Also associated with the later stages is the final positioning of the 5S rRNA. As in other eukaryotes, the T. cruzi 5S rRNA is independently transcribed and associates with uL5 and uL18 forming the 5S ribonucleoprotein particle (RNP) (24). In yeast, this 5S RNP associates with the body of the large subunit during early steps of biogenesis, but is essential only for much later pre-rRNA processing. During this late nuclear stage, the 5S RNP undergoes a 180° rotation before nuclear export (25, 26). The conservation of 5S RNA and its associated proteins uL5 and uL18 (SI Appendix, Figs. S10 and S13), as well as identification of trypanosome homologs for 5S RNP assembly factors (27), lead us to hypothesize that similar pathways may exist for maturation of the 5S RNP in trypanosomes at late stages.
Implications for Trypanosomatid Ribosome Assembly.
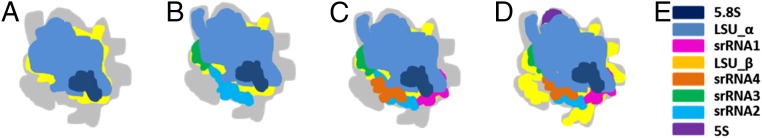
Ribosome biogenesis is a complex, multistep process, requiring the actions of hundreds of assembly factors and small nucleolar RNAs (26, 28). The multiply fragmented rRNAs increase the complexity of T. cruzi ribosome biogenesis (27). Based on the above analysis of interactions of individual rRNA components with r-proteins, and on a comparison with the hierarchy of yeast ribosome biogenesis, we propose here a step-wise assembly model for rRNAs of the large subunit (Fig. 4). First, the earlier transcribed components, 5.8S rRNA, LSU-α (domains I, II and part of domain III in yeast), and LSU-β (domains IV, V, and part of domain III), form two large intermingled blocks, and with the addition of the associated r-proteins, the scaffold is constructed. Next, srRNA2 and srRNA3 (both part of domain VI) are positioned in part by their anchoring r-proteins, which are already bound to the scaffold. ES7L is stably assembled concurrently with srRNA3. Subsequently, srRNA1 (the rest of domain III) and srRNA4 (the rest of domain VI) are stabilized with the aid of strong binding interactions with srRNA2. ES31L stabilizes the previously established scaffold, and the trypanosome-specific ESs (ES42L and the KSD) stabilize srRNAs2–4. The 5S rRNP containing 5S RNA, having assembled with the body of the large subunit earlier, is then rotated to its final position, enabling subsequent maturation steps, concurrent with proper folding of the large ESs on the periphery.
Fig. 4.
A model of the proposed T. cruzi large subunit rRNAs assembly pathway. (A) 5.8S, LSU-α, and LSU-β form a scaffold. The gray color indicates the outline of the completely assembled large subunit. (B) srRNA2, -3, and ES7L are assembled. (C) srRNA4 and srRNA1 are assembled next. (D) 5S rRNA, ES42L, ES31L, and the KSD properly assemble to complete the large subunit. (E) Color key for the eight pieces of rRNA.
Conclusion.
Our high-resolution cryo-EM density map has revealed the structure of the 60S large ribosomal subunit in T. cruzi in unprecedented detail, including the rRNA nucleotides, amino acid side chains, and rRNA modifications as well as small molecules (metal ions, and water molecules). We present an analysis of distinctive trypanosome interactions and propose a tentative model for assembly of the 60S large ribosomal subunit. These results from trypanosomatid ribosomes can be used to further understand how different rRNA domains are compacted as assembly proceeds in yeast and other eukaryotes. The multiple fragmentation of the T. cruzi ribosome is conserved within the whole trypanosomatid family (SI Appendix, Fig. S15 and Table S1), and thus our assembly model can be applied to the characterization of T. brucei and Leishmania spp. ribosomes as well. Moreover, trypanosomatids, as a group of protozoa early diverged from the eukaryotic lineage, are a compelling model to study the variability of highly conserved processes such as protein translation. Our structure thus serves as a basis for future studies of trypanosome-specific features in protein synthesis. Most importantly, the atomic details highlighting the differences between the T. cruzi and the human ribosome can be directly used for structure-based design of antitrypanosome drugs (SI Appendix, Tables S9 and S10).
Materials and Methods
T. cruzi Culture and Ribosome Purification.
T. cruzi Y-strain epimastigote cultures were grown at 27 °C in American Type Culture Collection 1029 liver infusion trypsose medium (9 mg/ml liver infusion broth, 5 mg/ml tryptose, 17 mM NaCl, 56 mM Na2HPO4, 5 mM KCl, 5 mM glucose, 10% heat inactivated fetal bovine serum, 0.01 mg/ml hemin) to a concentration of 3 × 107/mL in 1-L glass bottles. A total of 12–18 L of culture was chilled on ice, and cells were harvested by centrifugation at 1,000 × g for 20 min. Cell pellets from 5 L of culture were pooled and washed three times in ice-cold buffer containing 0.25 M sucrose and 5 mM KCl. Washed pellets were resuspended in 30 mL of buffer containing 0.25 M sucrose, 5 mM KCl, 5 mM EGTA, 1× protease inhibitor mixture (Sigma P2714), and 0.5% Triton X-100. Lysis was achieved by five freeze–thaw cycles using a dry ice/ethanol bath and warm water. Following lysis the sample was frozen at −80 °C.
Ribosomes were prepared based on previous purification protocols (29) with slight modifications. Cell lysate was loaded onto a 30% (wt/vol) sucrose cushion prepared in buffer I (containing 20 mM Hepes⋅KOH, pH 7.5), 100 mM KOAc, 4 mM Mg(OAc)2, 20% glycerol, 2 mM DTT, and Roch EDTA-free protease inhibitor mixture) and was centrifuged at 33,800 × g for 18 h at 4 °C. The resulting ribosome-enriched pellets were further purified using the 20-K PEG precipitation methods described previously (29). The final pellets were suspended in buffer II (10 mM Hepes⋅KOH, pH 7.4, 50 mM KOAc, 10 mM NH4Cl, 5 mM Mg(OAc)2, 2 mM DTT, and protease inhibitor mixture) and kept at −80 °C for further use.
Electron Microscopy.
Three microliters of ∼80 nM (0.3 mg/mL) purified ribosomes were applied onto a holey carbon grid (carbon-coated Quantifoil 2/2 grid, Quantifoil Micro Tools). The grids were coated by an additional prefloated continuous thin layer of carbon and glow-discharged before use. The sample-loaded grids were blotted for 3 s at 4 °C in 100% humidity and vitrified by plunging rapidly into liquid ethane cooled by liquid nitrogen at −180 °C with a Vitrobot (FEI). Prepared grids were quickly transferred to a storage box and stored in liquid nitrogen for imaging.
Micrographs were acquired with a back-thinned FEI Falcon II detector on a TITAN KRIOS (FEI) operating at 300 kV with the automatic imaging software FEI EPU. The defocus ranged from −1.0 to −3.0 µm, and the nominal magnification was 143,000× (133,970 after calibration), resulting in a final pixel size of 1.045 Å. Sixteen frames per second were collected with a total dose of about 32e-/Å2 in integrating mode—the only mode available for this camera. To select the grid for imaging, four grids were loaded into the microscope column and subjected to careful screening. Only grids presenting thin ice and good particle distribution were used for data collection. Before data collection, the microscope was carefully aligned including the Cs corrector to minimize electron optical aberrations, linear geometrical distortions, and coma. During the imaging session, micrographs were monitored for evidence of bad pixels, unusually large drift, beam shift, or astigmatism, in which case corrective actions were taken including regaining reference, skipping the squares exhibiting drift, and realigning the microscope. About 11,000 micrographs were collected from a total of two grids during 5 consecutive days.
Image Processing.
Dose-fractionated frame stacks were aligned using the program dosefgpu_driftcorr (30). The averaged micrographs were first screened with Arachnid (31), a python version of SPIDER (32), to exclude micrographs with obvious drift, ice contamination, or sparse coverage with particles. The contrast transfer function (CTF) parameters of the remaining good micrographs were calculated using CTFFIND3 (33). Particle selection was performed using the program e2boxer.py (34). About 700,000 selected ribosome-like single particles were subjected to 3D classifications with RELION (35) using a 70S ribosome map (EMD-6315) (36) filtered to 60 Å as the starting reference (SI Appendix, Fig. S1). About 400,000 particles were kept after excluding bad particles. One further classification with 10 classes was performed to separate ribosomes in different binding states in the dataset, yielding 60S subunits, E-tRNA–bound ribosomes, and empty ribosomes as the only relevant distinctions. No other tRNAs or factors such as eEF2 or eEF1A were found bound to the ribosomes. One resulting dominant class of 80S-E-tRNA including 79,000 particles was auto-refined and yielded a 3.1-Å resolution map showing a rigid 60S subunit and a less ordered 40S subunit, which indicated residual heterogeneity in the dataset.
To improve the resolution of the reconstruction, we used several image-processing strategies. First, all of the 60S subunit-containing particles from the first classification were recombined into a pool of about 345,000 particles. These were subjected to focused classification using a large-subunit mask. Before the refinement, the particle dataset was subjected to 2D reference-free classification to eliminate heterogeneous particles. Focused classification and reconstruction were used to overcome heterogeneity in the dataset and even in the classified subset (37). To further improve the homogeneity, three sets of particle positional parameters were obtained by refining each particle with three independent references, which were generated from 3D classification, and only the particles having consistent views were kept (38). In this way, about 4% of the particles were excluded. Second, the micrographs and their power spectra were visually reassessed to exclude those with contamination by ice and ethane, uncorrected drift, and astigmatism, ensuring that only particles extracted from high-quality micrographs were kept. Third, CTF values were recalculated using another program, S2focus (39), and the resultant values were cross-validated with those previously determined by CTFFIND3. Micrographs with discrepancies in defocus larger than 500 Å were excluded—in our case, a total of 480 micrographs (4.4%). Fourth, particles for final reconstruction were extracted from good micrographs averaged with weighting from frames 3–10, taking into account the fact that the earlier frames have more drift and the later frames experience more radiation damage.
In this manner, about 235,000 particles were used to reconstruct the large subunit to 2.5 Å. Resolution was estimated using the 0.143 Fourier shell correlation criterion computed from two independently reconstructed maps, following the “gold standard” protocol (40) (SI Appendix, Fig. S2). In addition, the two independently reconstructed maps were also used to estimate the distribution of local resolution using Resmap (41) (Fig. 1A and SI Appendix, Fig. S2 B–D). The resolution estimates were in agreement with visual assessment of map quality for secondary structure elements (e.g., appearance of α-helices and β-sheets), individual amino acid residues and nucleotides, small molecules such as water, and methyl groups (SI Appendix, Fig. S2 B–D). The accuracies of rotation and translation from final reconstruction reported by RELION were 0.52° and 0.49 Å, respectively. The resolution of the final reconstruction without masking was 3.0 Å. RELION postprocessing with auto_mask, auto_bfac, and the inimask_threshold 0.035 (which yields a masked map of the 60S subunit approximately enclosing the atomic model) determined the B-factor as 58.5 Å2 and the resolution of the masked region as 2.5 Å. The resolution of the whole map of the 80S ribosome including the disordered small subunit was determined as 2.8 Å (SI Appendix, Fig. S2).
Modeling.
A multimap modeling strategy was used in model building (42). The crystal structure of the yeast large ribosomal subunit (PDB: 4V88) (12) and the high-resolution cryo-EM model of the T. brucei large ribosomal subunit (PDB: 4V8M) (13) were used as references for building atomic models of rRNA and ribosomal proteins, respectively. The references were initially fitted as rigid bodies into the negative B-factor–sharpened density map using Chimera (43). The rRNA sequences were obtained from TriTrypDB Kinetoplastid Genomics Resource (tritrypdb.org/tritrypdb/) (11). Regions of rRNA conserved between T. cruzi and yeast were used as starting points, followed by extensive manual building using Coot (44). The high-resolution density map and the base-pairing principle allowed us to identify most of the bases. The polypeptide chains in T. brucei were mutated to match their sequences in T. cruzi taken from the National Center for Biotechnology Information protein databases (www.ncbi.nlm.nih.gov) and manually adjusted to fit the density map. The atomic model of the large ribosomal subunit was refined in real space using PHENIX (45). In addition to Mg2+ and Zn2+ ions, a number of water molecules were identified as density globules in the distance range of 2.8–3.4 Å from neighboring amino acids, a typical range for a hydrogen bond, occurring reproducibly in two independent reconstructions. The statistics for reconstruction and model refinement are summarized in SI Appendix, Table S2. The modeled parts and unmodeled residues are summarized in SI Appendix, Tables S3–S5.
Supplementary Material
Acknowledgments
We thank Amédée des Georges for helpful suggestions on image processing; Harry Kao for assistance with computer hardware; Nadia Severina for help in ribosome purification; Melissa Thomas-Baum (Buckyball Design) for assistance with the preparation of figures; and Z. Yu, C. Hong, R. Huang, and H. He for their assistance in the data collection at the Janelia Farm Research Campus of Howard Hughes Medical Institute (HHMI). This study was supported by HHMI and NIH Grants R01 GM29169 (to J.F.), R35GM118093 (to L.T.), and R01 GM028301 (to J.L.W.) and by National Science Foundaton Grant GRFP DGE-11-44155 (to C.G.-V).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this article have been deposited in The Electron Microscopy Data Bank (accession no. EMD-8361) and the Protein Data Bank (PDB ID code: 5T5H).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614594113/-/DCSupplemental.
References
- 1.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461(7268):1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 2.Yusupova G, Yusupov M. High-resolution structure of the eukaryotic 80S ribosome. Annu Rev Biochem. 2014;83:467–486. doi: 10.1146/annurev-biochem-060713-035445. [DOI] [PubMed] [Google Scholar]
- 3.El-Sayed NM, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309(5733):409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 4.El-Sayed NM, et al. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309(5733):404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- 5.Ivens AC, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309(5733):436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao H, Ayub MJ, Levin MJ, Frank J. The structure of the 80S ribosome from Trypanosoma cruzi reveals unique rRNA components. Proc Natl Acad Sci USA. 2005;102(29):10206–10211. doi: 10.1073/pnas.0500926102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray MW. Unusual pattern of ribonucleic acid components in the ribosome of Crithidia fasciculata, a trypanosomatid protozoan. Mol Cell Biol. 1981;1(4):347–357. doi: 10.1128/mcb.1.4.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White TC, Rudenko G, Borst P. Three small RNAs within the 10 kb trypanosome rRNA transcription unit are analogous to domain VII of other eukaryotic 28S rRNAs. Nucleic Acids Res. 1986;14(23):9471–9489. doi: 10.1093/nar/14.23.9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang XH, et al. A genome-wide analysis of C/D and H/ACA-like small nucleolar RNAs in Trypanosoma brucei reveals a trypanosome-specific pattern of rRNA modification. RNA. 2005;11(5):619–645. doi: 10.1261/rna.7174805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shalev-Benami M, et al. 2.8-Å Cryo-EM ctructure of the large ribosomal subunit from the eukaryotic parasite Leishmania. Cell Reports. 2016;16(2):288–294. doi: 10.1016/j.celrep.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aslett M, et al. TriTrypDB: A functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38(Database issue):D457–D462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334(6062):1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 13.Hashem Y, et al. High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome. Nature. 2013;494(7437):385–389. doi: 10.1038/nature11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc Natl Acad Sci USA. 2001;98(9):4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woolford JL, Jr, Baserga SJ. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195(3):643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de la Cruz J, Karbstein K, Woolford JL., Jr Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu Rev Biochem. 2015;84:93–129. doi: 10.1146/annurev-biochem-060614-033917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamalinda M, et al. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev. 2014;28(2):198–210. doi: 10.1101/gad.228825.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pöll G, et al. rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PLoS One. 2009;4(12):e8249. doi: 10.1371/journal.pone.0008249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramesh M, Woolford JL., Jr Eukaryote-specific rRNA expansion segments function in ribosome biogenesis. RNA. 2016;22(8):1153–1162. doi: 10.1261/rna.056705.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García-Ortega L, Alvarez-García E, Gavilanes JG, Martínez-del-Pozo A, Joseph S. Cleavage of the sarcin-ricin loop of 23S rRNA differentially affects EF-G and EF-Tu binding. Nucleic Acids Res. 2010;38(12):4108–4119. doi: 10.1093/nar/gkq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mailliot J, et al. Crystal structures of the uL3 mutant ribosome: Illustration of the importance of ribosomal proteins for translation efficiency. J Mol Biol. 2016;428(10 Pt B):2195–2202. doi: 10.1016/j.jmb.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilsson OB, et al. Cotranslational protein folding inside the ribosome exit tunnel. Cell Reports. 2015;12(10):1533–1540. doi: 10.1016/j.celrep.2015.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramesh M, Woolford JL, Jr (2016) Eukaryote-specific rRNA expansion segments function in ribosome biogenesis. RNA 22(8):1153–1162. [DOI] [PMC free article] [PubMed]
- 24.Michaeli S, Agabian N. A Trypanosoma brucei small RNP particle containing the 5S rRNA. Mol Biochem Parasitol. 1990;41(1):7–15. doi: 10.1016/0166-6851(90)90091-y. [DOI] [PubMed] [Google Scholar]
- 25.Leidig C, et al. 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle. Nat Commun. 2014;5:3491. doi: 10.1038/ncomms4491. [DOI] [PubMed] [Google Scholar]
- 26.Wu S, et al. Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature. 2016;534(7605):133–137. doi: 10.1038/nature17942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michaeli S. rRNA biogenesis in Trypanosomes. In: Bindereif A, editor. RNA Metabolism in Trypanosomes (Nucleic Acids and Molecular Biology) Vol 28. Springer; Berlin: 2011. pp. 123–148. [Google Scholar]
- 28.Gamalinda M, Woolford JL., Jr Paradigms of ribosome synthesis: Lessons learned from ribosomal proteins. Translation (Austin) 2015;3(1):e975018. doi: 10.4161/21690731.2014.975018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khatter H, et al. Purification, characterization and crystallization of the human 80S ribosome. Nucleic Acids Res. 2014;42(6):e49. doi: 10.1093/nar/gkt1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10(6):584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langlois R, et al. Automated particle picking for low-contrast macromolecules in cryo-electron microscopy. J Struct Biol. 2014;186(1):1–7. doi: 10.1016/j.jsb.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 33.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142(3):334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 34.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Scheres SH. Semi-automated selection of cryo-EM particles in RELION-1.3. J Struct Biol. 2015;189(2):114–122. doi: 10.1016/j.jsb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W, et al. Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G. Sci Adv. 2015;1(4):e1500169. doi: 10.1126/sciadv.1500169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown A, et al. Structure of the large ribosomal subunit from human mitochondria. Science. 2014;346(6210):718–722. doi: 10.1126/science.1258026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Z, Guo F, Wang F, Li TC, Jiang W. 2.9 Å resolution Cryo-EM 3D reconstruction of close-packed virus particles. Structure. 2016;24(2):319–328. doi: 10.1016/j.str.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang W, Guo F, Liu Z. A graph theory method for determination of cryo-EM image focuses. J Struct Biol. 2012;180(2):343–351. doi: 10.1016/j.jsb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henderson R, et al. Outcome of the first electron microscopy validation task force meeting. Structure. 2012;20(2):205–214. doi: 10.1016/j.str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11(1):63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2015;71(Pt 1):136–153. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettersen EF, et al. UCSF chimera: A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.