Significance
Hybrid vigor is an important phenomenon in basic genetics and in agricultural practice, but the bases of the superior performance of the hybrid relative to its parents in biomass and seed production remain elusive. In recent years, it has been suggested that epigenetic controls on levels of gene action are involved. Using mutants of genes involved in DNA methylation, we show that RNA polymerase IV or methyltransferase I do not contribute to the generation of the heterotic phenotype but that decrease in DNA methylation 1, a nucleosome remodeller with an effect on DNA methylation level, is required to produce a full level of hybrid vigor.
Keywords: heterosis, hybrid vigor, Arabidopsis, DNA methylation, DDM1
Abstract
Hybrid vigor or heterosis refers to the superior performance of F1 hybrid plants over their parents. Heterosis is particularly important in the production systems of major crops. Recent studies have suggested that epigenetic regulation such as DNA methylation is involved in heterosis, but the molecular mechanism of heterosis is still unclear. To address the epigenetic contribution to heterosis in Arabidopsis thaliana, we used mutant genes that have roles in DNA methylation. Hybrids between C24 and Columbia-0 (Col) without RNA polymerase IV (Pol IV) or methyltransferase I (MET1) function did not reduce the level of biomass heterosis (as evaluated by rosette diameter). Hybrids with a mutation in decrease in dna methylation 1 (ddm1) showed a decreased heterosis level. Vegetative heterosis in the ddm1 mutant hybrid was reduced but not eliminated; a complete reduction could result if there was a change in methylation at all loci critical for generating the level of heterosis, whereas if only a proportion of the loci have methylation changes there may only be a partial reduction in heterosis.
Heterosis or hybrid vigor describes the phenomenon where hybrids exhibit superior performance in many traits relative to their parental inbred lines. This phenomenon has been widely used in the breeding of crop and vegetable cultivars through F1 hybrid seed production systems.
Genetic approaches using quantitative trait locus analysis have been applied to some crop species and revealed that a large number of genes contribute to heterotic phenotypes by dominant, overdominant, or epistatic changes in gene activity (1, 2). Because heterosis requires genetic variation between parental inbred lines, parental genetic distance could be a useful indicator for hybrid performance. However, the relationship between genetic distance and heterosis is not straightforward (3). Recently developed molecular analyses such as transcriptomics, proteomics, metabolomics, and epigenomics (including DNA methylomes, small RNAomes, and genomewide distribution of histone modifications) allow us to study the molecular basis of heterosis in more detail (4–7).
The plant phenotype is controlled both genetically and epigenetically. In plants, epigenetic marks such as DNA methylation or histone modification vary among species or accessions (8, 9). Recently, the possibility that epigenetic regulation might contribute to heterosis has been suggested (10–14). There are two types of DNA methylation, de novo and maintenance DNA methylation. The process of de novo DNA methylation is triggered by 24-nt siRNAs produced by the RNA interference (RNAi) pathway, RNA-directed DNA methylation (RdDM). Two plant-specific RNA polymerases, Pol IV and Pol V, along with RDR2 (RNA-dependent RNA polymerase 2), DCL3 (Dicer-like 3), and AGO4 (Argonaute 4) function in this RdDM pathway in Arabidopsis thaliana. In maintenance DNA methylation, CG context methylation is maintained by MET1 (Methyltransferase 1), and non-CG contexts are maintained by two Domains Rearranged Methyltransferase (DRM) enzymes, Chromomethylase 2 (CMT2) and CMT3 (15, 16). In addition to DNA methyltransferases, a chromatin remodeling factor, Decrease in DNA Methylation 1 (DDM1), is involved in the maintenance of DNA methylation (15).
In A. thaliana, Columbia-0 (Col) × C24 and Landsberg erecta (Ler) × C24 hybrids show heterosis in vegetative biomass (17, 18), and the hybrids with different parental combinations show different levels of heterosis throughout their lifecycle (19–21). The heterosis phenotype is seen at an early developmental stage where hybrids have an increased cotyledon size relative to the parents at only a few days after sowing (18, 22). The efficiency of the photosynthetic process is the same in parents and C24 × Col hybrids, but the total amount of photosynthesis is greater in the hybrids than in parents because of the larger leaves; this increased total amount of photosynthesis may be important for the heterosis phenotype (18).
In this study, using plant size selection in backcross generations, we showed that two backcrosses enabled sufficient recovery of the genetic background to produce heterosis when crossed to another parental line. We also examined whether Pol IV, MET1, or DDM1, all important for DNA methylation, are involved in heterosis in A. thaliana.
Results
Three F1 Hybrid Combinations Show Heterosis in Seedling Size.
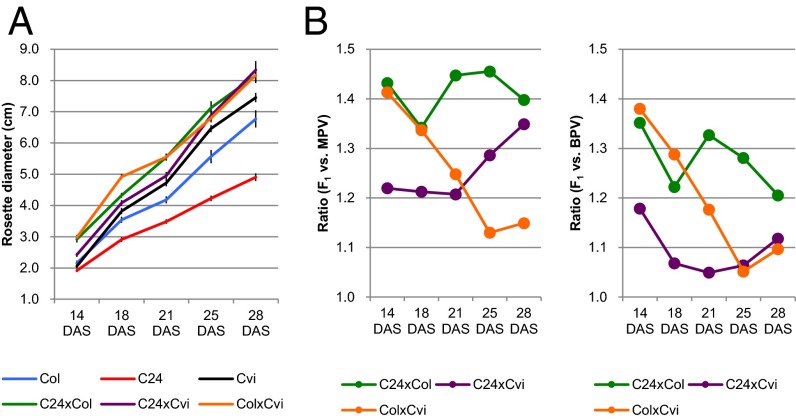
Hybrids from different parental combinations show different patterns of vegetative growth (21). Biomass heterosis evaluated by rosette diameter (RD) is observed in C24 × Col hybrids at early seedling stages (18). In this study, we compared the RD in C24 × Cvi and Col × Cvi hybrids with their parental lines at 14, 18, 21, 25, and 28 days after sowing (DAS) in addition to the comparison between C24 × Col and its parental lines (Fig. 1 A and B). The RD in C24 × Col hybrids is larger than that of the better parent value (BPV) from 14 to 28 DAS (Fig. 1B). At 14 DAS, the RD in C24 × Cvi hybrids was smaller than that of C24 × Col hybrids, whereas the RD in Col × Cvi hybrids was the same as that in C24 × Col hybrids (Fig. 1A). The ratio of RD between the F1 and the BPV (termed rBPV) in the C24 × Cvi hybrid is smaller than that in C24 × Col hybrids, and rBPV in Col × Cvi hybrids was the same as that in C24 × Col hybrids at 14 DAS (Fig. 1B). This result indicated that Col × Cvi hybrids have obvious heterosis, whereas C24 × Cvi hybrids have moderate heterosis at 14 DAS. At 28 DAS, the plant sizes of the three hybrids were the same (Fig. 1A), but rBPVs in C24 × Cvi and Col × Cvi hybrids were lower than that in the C24 × Col hybrid (Fig. 1B). These results indicate that the heterosis in C24 × Cvi and Col × Cvi hybrids is only moderate at 28 DAS, due to the large RD in the better parental line; the RD in Cvi was larger than that in Col or C24 at 28 DAS (Fig. 1A). The parental ecotypes have different alleles of the two flowering time genes, FRI and FLC, (Materials and Methods) that determine the flowering time of the hybrids in the different parental crosses. Flowering time in C24 × Col and C24 × Cvi hybrids was later than in the parental lines, whereas flowering time in Col × Cvi hybrids was the same as the parental lines (SI Appendix, Fig. S1). We measured heterosis before and after flowering for each line and found that all three hybrids had heterotic growth before the flowering time of their parents; the Col × Cvi early flowering hybrid had decreased heterosis after flowering.
Fig. 1.
Heterosis in three F1 hybrids. (A) Time course of rosette diameter in F1 hybrids and their parents. Data represent mean values ± SE obtained from more than 20 plants. (B) Levels of heterosis measured by the ratio of rosette diameter of the hybrids to MPV (Left). Levels of heterosis calculated from the ratio of the rosette diameter of the hybrids to BPV (Right). DAS, days after sowing. Germination efficiency was the same in all lines. Flowering time of Col was 20–25 DAS, Cvi 20–25 DAS, C24 28–35 DAS, C24 x Col > 35 DAS, C24 x Cvi >35 DAS, Col x Cvi 22–25 DAS.
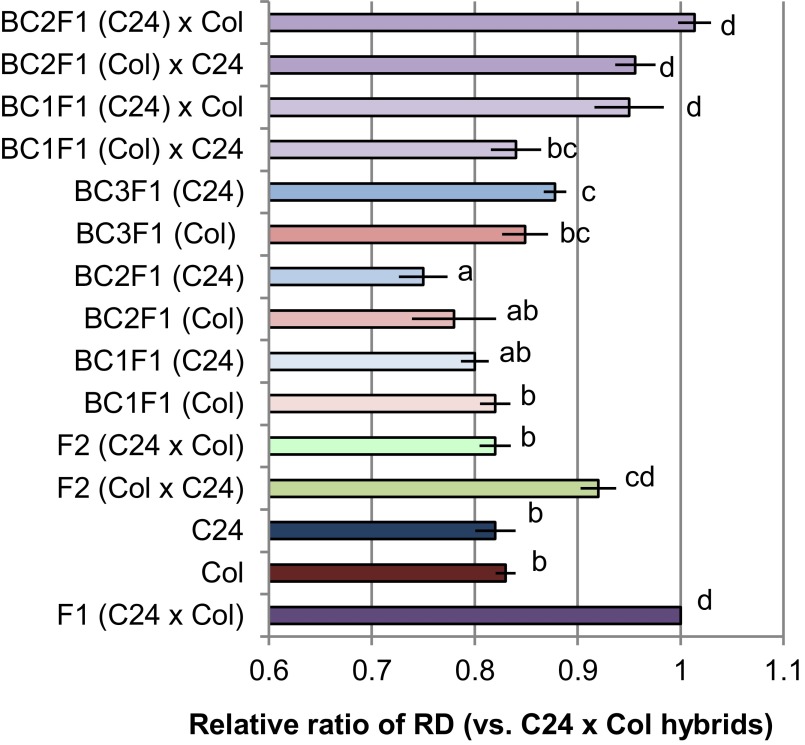
We measured rosette diameter in F2 generations derived from C24 × Col and Col × C24 hybrids. The averages of the ratio of the RD in F2 plants derived from C24 × Col and Col × C24 at 14 DAS compared with the C24 × Col hybrid were 0.82 or 0.92, respectively (Fig. 2). We produced a BC1F1 population by crossing the C24 × Col hybrid and Col [BC1F1 (Col)], or the C24 × Col hybrid and C24 [BC1F1 (C24)]. The averages of the ratio of RD in BC1F1 (Col) and BC1F1 (C24) compared with that of C24 × Col hybrids (F1) were 0.82 and 0.80, respectively, at 14 DAS (Fig. 2). These scores are similar to the parental lines: 0.83 in Col and 0.82 in C24 (Fig. 2). We produced BC2F1 plants by crossing BC1F1 plants with each parental line. The averages of the ratio of RD in BC2F1 (Col) and BC2F1 (C24) compared with the F1 hybrid were 0.78 and 0.75, respectively, at 14 DAS (Fig. 2). We crossed BC2F1 plants with the parents to produce BC3F1 plants. The averages of the ratio of RD in BC3F1 (Col) and BC3F1 (C24) compared with the F1 hybrid were 0.85 and 0.88, respectively, at 14 DAS (Fig. 2).
Fig. 2.
The ratio of rosette diameter in F2, backcrossed lines, and the F1 hybrid between backcrossed lines and the other parental line compared with C24 x Col hybrids. Data represent mean values ± SE obtained from more than 50 plants. Different letters indicate significant differences at P < 0.05 (Student’s t test).
We crossed BC1F1 plants with the alternative parent. The averages of the ratio of RD in BC1F1 (Col) × C24 and BC1F1 (C24) × Col hybrids compared with F1 were 0.84 and 0.95, respectively, at 14 DAS. Again, we made the hybrids between BC2F1 (Col) and C24 or between BC2F1 (C24) and Col. The averages of the ratio of RD in BC2F1 (Col) × C24 and BC2F1 (C24) × Col hybrids compared with F1 was 0.96 and 1.01, respectively, at 14 DAS (Fig. 2). These results show that two backcrosses were sufficient to generate F1 equivalent heterosis in crosses of BC2 plants with the alternative parent plant.
Establishment of Seedling Heterosis Is Independent of Pol IV Activity.
There is nonadditive 24nt-siRNA content in heterotic F1 hybrids, suggesting that a change in 24nt-siRNA levels with associated changes in the pattern of gene expression in the F1 hybrids may contribute to the heterotic phenotype (17, 23). Pol IV is a key enzyme for generating 24nt-siRNAs (16, 24). To identify the contribution of 24nt-siRNAs to seedling heterosis, we crossed pol IV mutants in both parental accessions [nrpd1a-3 (SALK_128428) in Col, and sde4-2 in C24; ref. 25]. We confirmed the reduction of the 24nt-siRNAs, siRNA02 and siRNA1003, by Northern blot analysis (Fig. 3B), decrease of DNA methylation levels in AtSN1 by chop-PCR (Fig. 3C), and transcriptional activation of the AtSN1 and Solo LTR by RT-PCR in the pol IV mutant hybrids (Fig. 3D). The reciprocal F1 hybrids between nrpd1a-3 and sde4-2 developed heterosis at the same level as the C24 × Col wild-type hybrids (Fig. 3 and SI Appendix, Fig. S2).
Fig. 3.
Normal levels of heterosis were observed in pol IV mutant hybrids. (A) Ratio of rosette diameter of pol IV mutant hybrids compared with that of Col at 10 DAS. Data represent mean values ± SE obtained from more than 20 plants. (B) Northern blot analysis in pol IV mutant hybrids. Reduction of 24nt-siRNA expression (siR02 and 1003) in the nrpd1a-3 (nrpd1) and sde4-2 x nrpd1a-3 (F1) was confirmed, but accumulation of 21nt-miRNA (miR171) was not changed. (C) DNA methylation status examined by chop-PCR in the pol IV mutant. No PCR amplification in nrpd1a-3 and sde4-2 x nrpd1a-3 indicated decreased DNA methylation in the endogenous RdDM target AtSN1 (a SINE-like retroelement). The actin gene that does not contain a Hae III site was used as nonmethylated control. (D) RT-PCR analysis of AtSN1 and Solo LTR in leaves. For each locus, −RT shows control lacking reverse transcriptase. GAP (glyceraldehyde-3-phosphate dehydrogenase C subunit) was used as a control.
We examined the expression levels of 27 genes in the pol IV mutant hybrids, and these genes had shown nonadditive expression in wild-type C24 × Col hybrids; 16 genes had expression levels greater than the midparent value (MPV) and 11 genes had expression levels less than the MPV (18). Most of the genes have the same differential expression in the mutant and wild-type hybrids. Two of the 27 genes showed differential expression between the pol IV mutant hybrids and wild-type hybrids (SI Appendix, Fig. S3). We compared the expression level of these two genes in nrpd1a-3 and wild-type Col or between sde4-2 and wild-type C24; they showed differential expression only between nrpd1a-3 and Col, indicating that the expression in the pol IV mutant hybrids relative to wild-type hybrids was due to an increased expression level of the Col allele in the presence of the pol IV mutation (SI Appendix, Fig. S3).
Maintenance of CG Methylation by MET1 Is Not Involved in Heterosis.
We used two approaches to examine the involvement of MET1, or maintenance CG methylation, in heterosis. We made met1-RNAi knockdown lines in a C24 background (SI Appendix, Fig. S4A). Hybrids between met1-RNAi (C24) and met1-1 (Col) had RDs the same as in the C24 × Col wild-type hybrids (SI Appendix, Fig. S4 B and C). The expected decreased CG methylation of the promoter region of FIS2 in the hybrids between met1-RNAi (C24) and met1-1 (Col) was confirmed by bisulfite sequencing (SI Appendix, Fig. S5).
We crossed met1-1 (Col) to C24 and generated the BC2F1 (C24), which was heterozygous for the met1-1 mutation. We crossed met1-1 to BC2F1 (C24) (MET1/met1) and obtained hybrids being homozygous or heterozygous met1-1; there was no size difference between the plants with these two genotypes (Fig. 4). The RD in the BC2F1 (C24) × met1-1 hybrid was larger than in the parents, BC2F1 (C24) and met1-1 (Col) (Fig. 4). Decreased CG methylation of the homozygous met1 hybrid plants from the cross between BC2F1 (C24) (MET1/met1) and met1-1 was confirmed by bisulfite sequencing of three control loci (SI Appendix, Fig. S5).
Fig. 4.
(A) Flowchart of met1 mutation backcrossing into C24. (B) Rosette diameter in the hybrids between met1 backcrossed plants in C24 background and met1-1 at 14 DAS (Left) and 24 DAS (Right). Data represent mean values ± SE obtained from more than 20 plants.
DDM1 Is Involved in the Maintenance of Heterosis.
The first generation of the ddm1 mutation in both the Col and C24 has rosette diameters closely similar to the wild type (Table 1). Using these mutants, we made a hybrid between ddm1-9 (C24) and ddm1-1 (Col). We confirmed the decrease of DNA methylation in the ddm1 homozygous hybrids by MeDIP-qPCR (SI Appendix, Fig. S6). The RD in the ddm1 mutant hybrids was smaller throughout development than that in C24 × Col wild-type hybrids (Table 1 and Figs. 5 and 6). The reduction in the level of heterosis was greater at 18 DAS than at 14 DAS.
Table 1.
Rosette diameter of wild-type, homozygous ddm1 mutants, and hybrids
| Sample | Date, DAS | DDM1/DDM1, cm | ddm1/ddm1, cm | Ratio of homozygous ddm1 relative to DDM wild type | Ratio of heterosis vs. MPV between to ddm1/ddm1 and DDM1/DDM1 hybrids |
| Col | 14 | 1.73 ± 0.02 | 1.67 ± 0.03 | 0.97 | |
| 18 | 2.81 ± 0.03 | 2.69 ± 0.05 | 0.96 | ||
| C24 | 14 | 1.55 ± 0.02 | 1.56 ± 0.02 | 1.01 | |
| 18 | 2.33 ± 0.04 | 2.37 ± 0.05 | 1.02 | ||
| C24 × Col | 14 | 2.33 ± 0.01 | 2.11 ± 0.03 | 0.91 | 0.92 |
| 18 | 3.46 ± 0.03 | 2.75 ± 0.04 | 0.79 | 0.81 |
Fig. 5.
Phenotype of the ddm1 mutant hybrids. (A) Time course of rosette diameter. Data represent mean values ± SE obtained from more than 20 plants. (B) Relative ratio of rosette diameter in ddm1 mutant hybrids compared with that in C24 x Col hybrids. Seedlings of 21 (C) and 28 (D) DAS plants. d, ddm1 mutant; D, wild type DDM1. Asterisk indicates P < 0.05 (Student’s t test). Flowering time of the ddm1 lines was ddm1 (C24) 18–20 DAS, ddm1 (Col) 22–25 DAS, ddm1 C24 x ddm1 Col >35 DAS, ddm1 C24 x ddm1 Cvi >35 DAS.
Fig. 6.
Ratio of rosette diameter (RD) in three ddm1 mutant hybrids relative to wild-type hybrids at 14 and 30 DAS. d, ddm1; D, DDM1.
Using the backcrossing procedure (26), ddm1-1 in a Cvi background was made. We again used the first generation of the homozygous ddm1 mutation. The C24 × Cvi and Col × Cvi hybrids had heterotic RDs (Fig. 1). We produced the ddm1-9 (C24) × ddm1-1 (Cvi) and ddm1-1 (Col) × ddm1-1 (Cvi) hybrids. The rosette size of both these two hybrids was smaller than the wild-type hybrids (Fig. 6 and SI Appendix, Figs. S7 and S8), indicating that the DDM1 protein is involved in seedling heterosis. Crosses using the second generation of ddm1 mutants showed the same plant size as first-generation ddm1 mutant hybrids.
Distribution of the ddm1 Mutation Effects and the Level of Heterosis.
To examine whether the decrease in heterosis in ddm1 hybrids is linked to the absence of DDM1 function, we made an F1 hybrid between a heterozygote of the ddm1-9 mutation in C24 (DDM1/ddm1) and a ddm1-1 homozygous mutant in Col (ddm1/ddm1). From 14 to 26 DAS, the average RD in the F1 hybrid plants homozygous for ddm1 was smaller than the RDs of the heterozygous ddm1 plants (Fig. 7). However, some plants heterozygous for ddm1 were smaller than the average of the RD in F1 hybrid plants homozygous for ddm1 (Fig. 7). This result was also observed in the combinations between C24 (ddm1/ddm1) and Col (DDM1/ddm1), between Col (DDM1/ddm1) and C24 (ddm1/ddm1), between C24 (DDM1/ddm1) and Cvi (ddm1/ddm1), and between C24 (ddm1/ddm1) and Cvi (DDM1/ddm1) (SI Appendix, Fig. S9). These results suggest that the decrease in heterosis in the hybrid ddm1 homozygotes is associated with stochastic changes in the methylation state of loci either directly or indirectly contributing to the level of heterosis. The F1 hybrid between C24 ddm1 and Col met1-1 is heterozygous for DDM1 and MET1 and has a wild-type heterosis level (SI Appendix, Fig. S10).
Fig. 7.
Rosette diameter (RD) in the hybrid between C24 (DDM1/ddm1) x Col (ddm1/ddm1) at 14, 18, 22, and 26 DAS. (A) Frequency distribution of the RD for C24 (DDM1/ddm1) x Col (ddm1/ddm1) hybrids. (B) Mean of rosette diameter of heterozygous and homozygous ddm1 hybrids. Unknown indicates plants where the genotypes was not determined.
Discussion
It has been suggested that epigenetic systems may be involved in the alteration of gene expression in hybrids that, in turn, could contribute to the hybrid phenotype (12). One epigenetic system that has been studied is the small RNA system, particularly the 24nt-siRNAs, which are involved in the modification of DNA methylation of many genes of the genome (16). Some of the changes in DNA methylation in hybrids correlate with changes in transcription levels (11), but there is no consistent relationship among the changes in DNA methylation, the changes in transcription, and the generation of the heterotic phenotype.
We have considered the effects of three genes, POL IV, MET1, and DDM1, which are known to affect DNA methylation status, either directly or indirectly, of many loci in the genome. We have found that a loss of the RdDM system involving POL IV production of 24nt-siRNAs does not decrease the level of heterosis. The level of small siRNA expression in hybrids versus the levels of expression in the parents of the hybrids has been determined in rice, wheat, and maize, as well as in A. thaliana. In each of these species, a reduction in 24nt-siRNAs in the hybrid has been observed (13, 27, 28). In maize, mutants in the MOP1 (Mediator of Paramutation 1) gene, an ortholog of RDR2 of A. thaliana, reduced the level of siRNAs (29). Hybrids homozygous for the mop1 mutation retained their level of heterosis, indicating that siRNAs synthesized by MOP1 are not involved in generating the heterosis phenotype (27), consistent with our finding with the pol IV mutants in A. thaliana.
The second gene we studied was MET1, important in the maintenance of CG methylation (30). The null mutant of MET1 reduces the frequency of CG methylation but does not reduce the level of heterosis, indicating that MET1-dependent CG maintenance of DNA methylation is not involved in heterosis in these hybrids.
The third gene we examined was DDM1. This gene has a number of different effects through modification of chromatin structure, with resultant effects on DNA methylation levels (15, 31). We have examined seven loci in ddm1 hybrids and found DNA methylation to be reduced at these loci. These hybrids had a rosette diameter up to 25% smaller than the wild-type hybrid rosette at 14 DAS and up to 75% smaller than the wild-type hybrid rosette at 18 DAS. Hybrids heterozygous for the ddm1 mutation in general had a phenotype the same as wild-type hybrids, but up to 10% of the plants had a smaller rosette diameter. These DDM1/ddm1 hybrid plants with the smaller rosettes may result from the previous methylation state of the genome in the ddm1 parent. The ddm1 parent has progressive DNA demethylation over several generations with increasing levels of developmental abnormalities (26). In the DDM1/ddm1 heterozygous hybrid, the gene or segments important for heterosis coming from the ddm1 parent might already have an altered level of DNA methylation. It is possible that in the crosses involving first- and second-generation ddm1 mutants, not all prospective targets important for the heterotic phenotype have been subjected to DNA demethylation.
The difference in heterosis levels between DDM1/ddm1 and ddm1/ddm1 hybrids could be due to an unrelated effect of ddm1 on growth. We think this possibility is unlikely because the first-generation ddm1 mutants in the C24 and Col parents used in these crosses have rosette diameters similar to the rosette diameters of the wild-type parents (Table 1), consistent with the findings of Vongs et al. (31) who first described the ddm1 mutant. The level of heterosis in the rosette diameters in the ddm1/ddm1 hybrids is 10–25% lower than the rosette diameters of the wild-type hybrids when they are compared with their respective parents. Homozygous met1 plants can have reduced growth, but the level of heterosis is not altered (ref. 30 and Fig. 4).
The fact that the dmm1/ddm1 hybrid plants have reduced heterosis contrasting to the hybrids carrying met1 and pol IV mutation is likely to be a consequence of the different modes of involvement of these three loci in the methylation process. The POL IV locus is responsible for the production of 24nt-siRNAs, which act as guides for de novo methylation of sequences homologous to the sRNA molecules and for the maintenance of the CHH methylation. MET1 operates as a principal maintenance mechanism for the CG methylation context. DDM1 is a nucleosome remodeler that, through changes in chromatin structure, leads to hypomethylated loci in all three sequence contexts, mostly associated with heterochromatic transposable elements that, in turn, may result in changes in patterns of gene expression affecting the level of hybrid vigor.
The scale of the reduction in the level of heterosis in the ddm1/ddm1 hybrids could be due to the incomplete demethylation events caused by the ddm1 mutation in early generations of the homozygous mutant. The loci undergoing demethylation could have changes in their expression contributing to hybrid vigor. Another effect of the ddm1 allele is its propensity in successive generations to produce hypermethylation at some loci in the genome (32). The relative importance of hypermethylation and hypomethylation in affecting heterosis together with the identity of the critical loci is not clear. The observation that there is only a partial loss of heterosis in ddm1/ddm1 hybrids may be due to the levels of changes of methylation status of loci across the genome, but it may also point to the existence of other pathways that play a role in the generation of hybrid vigor.
Materials and Methods
Plant Materials.
Columbia-0 (Col), C24, and Cvi accessions were used as parental lines. Col and Cvi have “late” alleles of FLC and “inactive” alleles of FRI, C24 has an “early” allele of FLC and an “active” allele of FRI. The lines of ddm1-1 (Col) (31), ddm1-9 (C24) (33), met1-1 (Col) (30), nrpd1a-3 (Col) (SALK_128428), and sde4-2 (C24) (25) mutants were used. All reciprocal hybrids and backcross populations were generated by hand pollination. Plants were grown in growth chambers under a 16-h/8-h light/dark cycle at 22 °C. Seeds were sown on plastic dishes containing Murashige and Skoog agar medium supplemented with 1.0% sucrose (pH 5.7), and were transferred to soil at 14 DAS. RD was measured for evaluation of plant size or heterosis. RD equals the maximum diameter of the rosette as measured between the two largest leaves at a certain point in development. RD depends on leaf blade and petiole length. The ddm1-1 mutant in Col or Cvi was the first generation of the ddm1 homozygous mutation. The second generations of ddm1 mutants were developed by self-pollination. In ddm1-9 in C24, we crossed ddm1-9 to wild-type C24 and gained the first generation of ddm1-9 homozygous mutant from the F2 population, and first- and second-generation ddm1-9 mutants were used. Primer sequences used for genotyping of ddm1-1, ddm1-9, and met1-1 are shown in SI Appendix, Table S1.
RNA Extraction and Expression Analysis.
For RT-PCR or quantitative real-time (q) PCR, total RNA was isolated from leaves by using the SV Total RNA Isolation System (Promega). cDNA was synthesized from 500 ng of total RNA by ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo). For RT-PCR, absence of genomic DNA contamination was confirmed by the PCR of no RT control using EmeraldAmp MAX PCR Master Mix (Takara bio), and PCR conditions were 95 °C for 3 min followed by 30 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. RT-qPCR was performed by using a LightCycler Nano (Roche). The cDNA was amplified by using FastStart Essential DNA Green Master (Roche). PCR conditions were 95 °C for 10 min followed by 40 cycles of 95 °C for 10 s, 60 °C for 10 s, and 72 °C for 15 s, and Melting program (60–95 °C at 0.1 °C/s). After amplification cycles, each reaction was subjected to melt temperature analysis to confirm single amplified products. The relative expression level of each gene relative to ACTIN was calculated by using automatic CQ calling according to the manufacturer’s instructions (Roche). Data presented are the average and SE from three biological and experimental replications and statistically analyzed by using the Student’s t test, P < 0.05. Primer sequences used in this study are shown in SI Appendix, Table S2.
Small RNA Northern Blotting.
RNA was extracted from 250-ng flowering buds by using mirVana miRNA Isolation Kit (Ambion). RNA was separated by electrophoresis in 15% (vol/vol) denaturing polyacrylamide/urea gel and transferred to HybondN+ (GE Healthcare) by electroblotting using Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad). Probe sequences used in this study are shown in SI Appendix, Table S3. Membranes were hybridized with [γ-32P] ATP-labeled oligonucleotides by using Ultra Hyb oligo Buffer (Ambion) at 40 °C overnight. Membranes were washed twice at 40 °C in 2× SSC, 0.5% SDS for 15 min, and exposed to X-ray film for 3 d (siRNA02 and siRNA1003) or 5 h (miR171) at −80 °C.
Chop-PCR.
Chop-PCR experiment was performed as described by Sasaki et al. (34). Genomic DNAs were extracted from leaves by using DNeasy Plant Mini Kit (Qiagen). Fifty nanograms of genomic DNA was digested with Hae III in 20 μL of reaction mix at 37 °C overnight. After restriction digestion, 1 μL of digested DNA was used as a template for PCR in 10 μL of reaction mix. The PCR conditions were 95 °C for 2 min followed by 40 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. Primers used for chop-PCR are listed in SI Appendix, Table S4.
Bisulfite Sequencing.
Genomic DNAs from leaves was isolated by using DNeasy Plant Mini Kit (Qiagen). Five hundred nanograms of DNA was fragmented by sonication, and fragments were ∼300–800 bp in length. MethylCode Bisulfite Conversion Kit (Thermo Fisher) was used for chemical bisulfite reaction, and PCR was performed by using bisulfite-treated DNAs as templates. PCR conditions were 95 °C for 2 min followed by 40 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. Amplified PCR fragments were gel purified by using GENECLEAN III Kit (MP Biomedicals) and cloned into pGEM-T Easy vector (Promega). Ten independent clones were sequenced. Primers used for bisulfite-sequence are listed in SI Appendix, Table S4.
Methylated-DNA Immunoprecipitation-qPCR.
Methylated-DNA Immunoprecipitation (MeDIP) was performed as described (35). Genomic DNA isolation and fragmentation were the same as bisulfite sequencing described above, and anti-5-methylcytosine antibody (NA81, Merck Millipore) was used. MeDIP-qPCR was performed by the same methods as the RT-qPCR using the purified immunoprecipitated DNAs as templates. Primers used for MeDIP-qPCR are listed in SI Appendix, Table S4.
Generation of RNAi Construct and Plant Transformation.
The construct expressing double-strand RNA was created by using Gateway Technology according to the manufacturer’s specifications (Invitrogen). The DNA fragment corresponding to MET1 was amplified by PCR, and PCR products were cloned into the vector pDONR201 by BP recombinase reaction. DNA fragments were inserted into pDONR201 and transferred into the vector pHELLSGATE12 (36) by LR recombinase reaction. The MET1 RNAi construct was transformed into the C24 accession via Agrobacterium tumefaciens strain C58C1 by the floral dip procedure (37). Transgenic seedlings were selected through resistance to kanamycin on a selection medium. Primers used for constructing vector are listed in SI Appendix, Table S5.
Supplementary Material
Acknowledgments
We thank Dr. Tetsuji Kakutani, Dr. Craig S. Pikaard, and Dr. David C. Baulcombe for providing seeds. This work was supported in part by an Open Partnership Joint Projects of Japan Society for the Promotion of Science Bilateral Joint Research Projects (14544567) and by Precursory Research for Embryonic Science and Technology (12101066) (JST) (to R.F.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613372113/-/DCSupplemental.
References
- 1.Frascaroli E, et al. Classical genetic and quantitative trait loci analyses of heterosis in a maize hybrid between two elite inbred lines. Genetics. 2007;176(1):625–644. doi: 10.1534/genetics.106.064493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lippman ZB, Zamir D. Heterosis: Revisiting the magic. Trends Genet. 2007;23(2):60–66. doi: 10.1016/j.tig.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Kawamura K, et al. Genetic distance of inbred lines of Chinese cabbage and its relationship to heterosis. Plant Gene. 2016;5:1–7. [Google Scholar]
- 4.Birchler JA, Yao H, Chudalayandi S, Vaiman D, Veitia RA. Heterosis. Plant Cell. 2010;22(7):2105–2112. doi: 10.1105/tpc.110.076133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen ZJ. Genomic and epigenetic insights into the molecular bases of heterosis. Nat Rev Genet. 2013;14(7):471–482. doi: 10.1038/nrg3503. [DOI] [PubMed] [Google Scholar]
- 6.Groszmann M, Greaves IK, Fujimoto R, Peacock WJ, Dennis ES. The role of epigenetics in hybrid vigour. Trends Genet. 2013;29(12):684–690. doi: 10.1016/j.tig.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Schnable PS, Springer NM. Progress toward understanding heterosis in crop plants. Annu Rev Plant Biol. 2013;64:71–88. doi: 10.1146/annurev-arplant-042110-103827. [DOI] [PubMed] [Google Scholar]
- 8.He G, et al. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell. 2010;22(1):17–33. doi: 10.1105/tpc.109.072041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaughn MW, et al. Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 2007;5(7):e174. doi: 10.1371/journal.pbio.0050174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greaves IK, et al. Trans chromosomal methylation in Arabidopsis hybrids. Proc Natl Acad Sci USA. 2012;109(9):3570–3575. doi: 10.1073/pnas.1201043109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greaves IK, Groszmann M, Wang A, Peacock WJ, Dennis ES. Inheritance of trans chromosomal methylation patterns from Arabidopsis F1 hybrids. Proc Natl Acad Sci USA. 2014;111(5):2017–2022. doi: 10.1073/pnas.1323656111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greaves IK, et al. Epigenetic changes in hybrids. Plant Physiol. 2015;168(4):1197–1205. doi: 10.1104/pp.15.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen H, et al. Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell. 2012;24(3):875–892. doi: 10.1105/tpc.111.094870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng DW, et al. A role for CHH methylation in the parent-of-origin effect on altered circadian rhythms and biomass heterosis in Arabidopsis intraspecific hybrids. Plant Cell. 2014;26(6):2430–2440. doi: 10.1105/tpc.113.115980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zemach A, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153(1):193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pikaard CS, Mittelsten Scheid O. Epigenetic regulation in plants. Cold Spring Harb Perspect Biol. 2014;6(12):a019315. doi: 10.1101/cshperspect.a019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groszmann M, et al. Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc Natl Acad Sci USA. 2011;108(6):2617–2622. doi: 10.1073/pnas.1019217108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujimoto R, Taylor JM, Shirasawa S, Peacock WJ, Dennis ES. Heterosis of Arabidopsis hybrids between C24 and Col is associated with increased photosynthesis capacity. Proc Natl Acad Sci USA. 2012b;109(18):7109–7114. doi: 10.1073/pnas.1204464109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barth S, Busimi AK, Friedrich Utz H, Melchinger AE. Heterosis for biomass yield and related traits in five hybrids of Arabidopsis thaliana L. Heynh. Heredity (Edinb) 2003;91(1):36–42. doi: 10.1038/sj.hdy.6800276. [DOI] [PubMed] [Google Scholar]
- 20.Meyer RC, Törjék O, Becher M, Altmann T. Heterosis of biomass production in Arabidopsis. Establishment during early development. Plant Physiol. 2004;134(4):1813–1823. doi: 10.1104/pp.103.033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groszmann M, et al. Intraspecific Arabidopsis hybrids show different patterns of heterosis despite the close relatedness of the parental genomes. Plant Physiol. 2014;166(1):265–280. doi: 10.1104/pp.114.243998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer RC, et al. Heterosis manifestation during early Arabidopsis seedling development is characterized by intermediate gene expression and enhanced metabolic activity in the hybrids. Plant J. 2012;71(4):669–683. doi: 10.1111/j.1365-313X.2012.05021.x. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Varala K, Moose SP, Hudson ME. The inheritance pattern of 24 nt siRNA clusters in Arabidopsis hybrids is influenced by proximity to transposable elements. PLoS One. 2012;7(10):e47043. doi: 10.1371/journal.pone.0047043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye R, et al. A Dicer-independent route for biogenesis of siRNAs that direct DNA methylation in Arabidopsis. Mol Cell. 2016;61(2):222–235. doi: 10.1016/j.molcel.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308(5718):118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 26.Saze H, Kakutani T. Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J. 2007;26(15):3641–3652. doi: 10.1038/sj.emboj.7601788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barber WT, et al. Repeat associated small RNAs vary among parents and following hybridization in maize. Proc Natl Acad Sci USA. 2012;109(26):10444–10449. doi: 10.1073/pnas.1202073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, et al. Small RNAs as important regulators for the hybrid vigour of super-hybrid rice. J Exp Bot. 2014;65(20):5989–6002. doi: 10.1093/jxb/eru337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nobuta K, et al. Distinct size distribution of endogeneous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant. Proc Natl Acad Sci USA. 2008;105(39):14958–14963. doi: 10.1073/pnas.0808066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kankel MW, et al. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163(3):1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260(5116):1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- 32.Ito T, et al. Genome-wide negative feedback drives transgenerational DNA methylation dynamics in Arabidopsis. PLoS Genet. 2015;11(4):e1005154. doi: 10.1371/journal.pgen.1005154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang S, Pikaard CS. Transcript profiling in Arabidopsis reveals complex responses to global inhibition of DNA methylation and histone deacetylation. J Biol Chem. 2005;280(1):796–804. doi: 10.1074/jbc.M409053200. [DOI] [PubMed] [Google Scholar]
- 34.Sasaki T, Lorković ZJ, Liang SC, Matzke AJ, Matzke M. The ability to form homodimers is essential for RDM1 to function in RNA-directed DNA methylation. PLoS One. 2014;9(2):e88190. doi: 10.1371/journal.pone.0088190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawanabe T, Fujimoto R, Sasaki T, Taylor JM, Dennis ES. A comparison of transcriptome and epigenetic status between closely related species in the genus Arabidopsis. Gene. 2012;506(2):301–309. doi: 10.1016/j.gene.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Helliwell CA, Wesley SV, Wielopolska AJ, Waterhouse PM. High-throughput vectors for efficient gene silencing in plants. Funct Plant Biol. 2002;29(10):1217–1225. doi: 10.1071/FP02033. [DOI] [PubMed] [Google Scholar]
- 37.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.