Significance
The onset of type 1 diabetes autoimmunity is indicated by the development of multiple islet autoantibodies, produced by B cells with the help of T follicular helper (TFH) cells. MicroRNAs (miRNAs) are small noncoding RNAs that regulate cellular states, as immune activation, making them suitable targets for disease intervention. Here, we show an enrichment of insulin-specific C-X-C chemokine receptor type 5 (CXCR5)+CD4+ TFH precursors correlating with high miRNA92a abundance during onset of autoimmunity and identify Krueppel-like factor 2 (KLF2) as a target for miRNA92a. We demonstrate that miRNA92a inhibition blocks TFH induction and reduces murine islet autoimmunity in vivo. Therefore, we propose miRNA92a and the phosphatase and tension homolog-phosphoinositol-3-kinase-KLF2 signaling network as possible innovative precision medicine targets to interfere with aberrant immune activation in islet autoimmunity.
Keywords: miRNA92a, KLF2, PTEN-PI3K signaling, T follicular helper cells, type 1 diabetes
Abstract
Aberrant immune activation mediated by T effector cell populations is pivotal in the onset of autoimmunity in type 1 diabetes (T1D). T follicular helper (TFH) cells are essential in the induction of high-affinity antibodies, and their precursor memory compartment circulates in the blood. The role of TFH precursors in the onset of islet autoimmunity and signaling pathways regulating their differentiation is incompletely understood. Here, we provide direct evidence that during onset of islet autoimmunity, the insulin-specific target T-cell population is enriched with a C-X-C chemokine receptor type 5 (CXCR5)+CD4+ TFH precursor phenotype. During onset of islet autoimmunity, the frequency of TFH precursors was controlled by high expression of microRNA92a (miRNA92a). miRNA92a-mediated TFH precursor induction was regulated by phosphatase and tension homolog (PTEN) - phosphoinositol-3-kinase (PI3K) signaling involving PTEN and forkhead box protein O1 (Foxo1), supporting autoantibody generation and triggering the onset of islet autoimmunity. Moreover, we identify Krueppel-like factor 2 (KLF2) as a target of miRNA92a in regulating human TFH precursor induction. Importantly, a miRNA92a antagomir completely blocked induction of human TFH precursors in vitro. More importantly, in vivo application of a miRNA92a antagomir to nonobese diabetic (NOD) mice with ongoing islet autoimmunity resulted in a significant reduction of TFH precursors in peripheral blood and pancreatic lymph nodes. Moreover, miRNA92a antagomir application reduced immune infiltration and activation in pancreata of NOD mice as well as humanized NOD Scid IL2 receptor gamma chain knockout (NSG) human leucocyte antigen (HLA)-DQ8 transgenic animals. We therefore propose that miRNA92a and the PTEN-PI3K-KLF2 signaling network could function as targets for innovative precision medicines to reduce T1D islet autoimmunity.
Autoimmune type 1 diabetes (T1D) is presumed to result from T-cell–mediated destruction of the pancreatic insulin-secreting islet β cells (1). In children, the development of multiple autoantibodies reacting with the well-established autoantigens (insulin, glutamic decarboxylase, insulinoma antigen, and islet zinc transporter) indicates the onset of islet autoimmunity (pre-T1D) (2, 3). Autoantibodies against insulin are often the first to appear, indicating an essential impact of insulin in the onset of islet autoimmunity (2, 4). In young children, clinically overt T1D can occur within months of the appearance of autoantibodies, but may take more than a decade to occur in some children (5), referring to the so-called slowly progressing phenotype. Moreover, children with slowly progressing phenotypes can lose some of their earliest islet autoantibodies, especially insulin autoantibodies. Despite these insights, the cellular and molecular mechanisms involved in triggering the onset, as well as the progression, of human islet autoimmunity remain incompletely understood.
T follicular helper (TFH) cells support antibody responses by the induction of B-cell activation. Murine data suggested that islet autoantibodies can enhance the survival of proliferating autoreactive CD4+ T cells, whereas blocking Fcγ receptor delayed and reduced the incidence of autoimmune diabetes (6). TFH cells are characterized by a memory phenotype, and thereby retain their capacity to recall their TFH-specific effector functions upon reactivation to provide help for B-cell responses (7). After interactions with dendritic cells in the T-cell zones of secondary lymphoid organs, a fraction of activated CD4+ T cells migrate toward B-cell follicles by up-regulating C-X-C chemokine receptor type 5 (CXCR5) (8–10). The transcription factor B-cell lymphoma 6 (Bcl6) plays an essential role in initiating TFH-cell differentiation (11). Inducible T-cell costimulator (ICOS) signaling transiently inactivates forkhead box protein O1 (Foxo1), which, in turn, relieves Foxo1-dependent inhibition of Bcl6 expression and promotes TFH differentiation (12). Reduced Foxo1 abundance, either resulting from increased expression of ICOS induced by loss of Foxp1 or due to degradation by the E3 ubiquitin ligase ITCH, may enhance TFH-cell differentiation (13, 14). Moreover, induced deficiency of the zinc finger transcription factor Krueppel-like factor 2 (KLF2) in activated CD4+ T cells leads to increased TFH-cell generation and B-cell priming, whereas KLF2 overexpression prevented TFH-cell production (15).
Recent studies in man have provided new insights into the ontogeny of circulating CXCR5+CD4+ T cells (16–20). In particular, the frequency of circulating TFH precursor C-C chemokine receptor type 7 (CCR7)low programmed cell death protein 1 (PD1)highCXCR5+CD4+ T cells is associated with active TFH-cell differentiation in secondary lymphoid organs (21).
However, involved signaling pathways that regulate the differentiation of human TFH precursors, as well as their contribution in the initiation and progression of human islet autoimmunity, are incompletely understood (22).
MicroRNAs (miRNAs) function as critical regulators in the mammalian immune system (23–26), and thereby affect complex cellular states, including immune activation and regulation (27, 28). The miRNA17∼92 transcript, which is encoded by mouse chromosome 14 and human chromosome 13, is the precursor for six mature miRNAs (miRNA17, miRNA18a, miRNA19a, miRNA20a, miRNA19b, and miRNA92a). Transgenic (Tg) mice overexpressing miRNA17∼92 in lymphocytes developed lymphoproliferative disorders and autoimmunity (29), and miRNA92a promoted the generation of TFH cells (30).
In man, the impact of the miRNA17∼92 family and its critical targets to the posttranscriptional regulation of human TFH precursor cell induction in the development of human islet autoimmunity are currently unclear. We therefore studied insulin-specific TFH precursors and their signaling pathways during the onset of human islet autoimmunity. We provide direct evidence that during the onset of human islet autoimmunity, the insulin-specific target T-cell population is enriched with a CXCR5+CD4+ TFH precursor phenotype. The frequency of CCR7lowPD1highCXCR5+ TFH precursor cells was controlled by high abundance of miRNA92a. miRNA92a-mediated TFH-cell induction was regulated by phosphatase and tension homolog (PTEN) - phosphoinositol-3-kinase (PI3K) signaling involving down-regulation of PTEN, PH domain and leucine-rich repeat protein phosphatase 2 (PHLPP2), and Foxo1. Moreover, we identify KLF2 as a target of miRNA92a and show that upon blockade of KLF2 signaling, miRNA92a-mediated induction of human TFH precursors was severely abolished. Importantly, a miRNA92a antagomir completely blocked human TFH precursor induction in vitro. Of note, in vivo application of a miRNA92a antagomir to nonobese diabetic (NOD) mice significantly lowered TFH precursors. More importantly, application of a miRNA92a antagomir critically reduced pancreatic immune activation. miRNA92a and the PTEN-PI3K-KLF2 signaling network might therefore function as targets for innovative precision intervention that can limit the activation of T1D islet autoimmunity.
Results
Enhancement of Insulin-Specific CXCR5+ TFH Precursors During the Onset of Human Islet Autoimmunity.
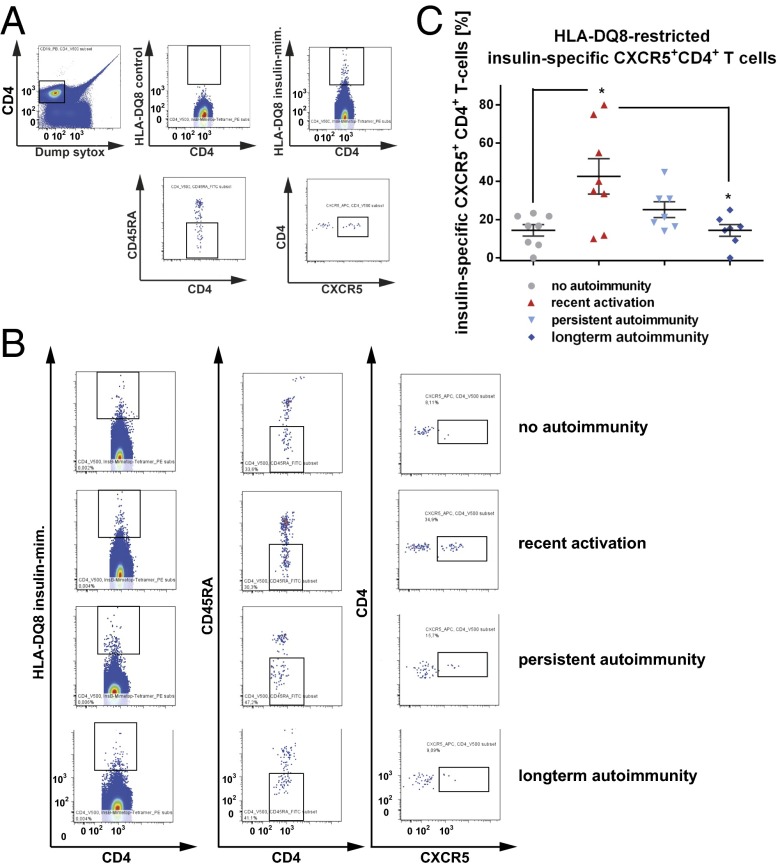
Given the critical role of the autoantigen insulin in initiating islet autoimmunity, we studied the TFH precursor characteristics of the insulin-specific target T-cell population. We used CD4+ T cells from nondiabetic children without autoimmunity (islet autoantibody-negative children), with recent onset of autoimmunity (recent activation, multiple autoantibodies for <5 y), persistent autoimmunity (multiple autoantibodies for >5 to <10 y), and long-term autoimmunity (multiple autoantibodies for >10 y without overt T1D). We applied recently developed fluorescent insulin-specific HLA-DQ8 tetramers (14E-22E and 14E-21G 22E tetramers) based on a set of two insulin B-chain 10–23 mimetopes to identify human HLA-DQ8–restricted insulin-specific CD4+ T cells ex vivo directly (31). We detected no tet+CD4+ T cells using the control tetramers, whereas a population of insulin-specific CD4+ T cells was readily identified ex vivo with the insulin-specific tetramers (Fig. 1 A and B). Frequencies of tet+CD4+ T cells were correlated with CD3 expression. Importantly, we provide direct evidence that during onset of islet autoimmunity and in the absence of clinically overt T1D, the insulin-specific target T-cell population is enriched with a CXCR5+CD4+ TFH precursor phenotype (no autoimmunity vs. recent onset of autoimmunity: 14.4 ± 2.9% vs. 42.5 ± 9.2% of tet+CD4+ T cells; P < 0.05; Fig. 1C).
Fig. 1.
Ex vivo identification of HLA-DQ8–restricted insulin-specific CXCR5+ TFH precursor cells from children with or without ongoing islet autoimmunity. (A, Upper Left) Human CD4+ T cells were analyzed by flow cytometry, first gating on live, CD19−, CD14−, CD8a−, CD11b−, CD4+, and CD3+ cells and then examining the tetramer binding. (A, Upper Right) Representative set of FACS plots for the identification of HLA-DQ8–restricted insulin-specific CD4+ T cells. mim, mimic. (A, Upper Center) Control staining was used to assess the quality and specificity of the tetramer staining using a combination of two control tetramers fused to irrelevant peptides. (A, Lower) Insulin-specific CD4+ memory TFH precursor cells were then identified by gating on CD45RA− and CXCR5+. (B) FACS plots for insulin-specific memory CXCR5+ TFH precursor cells purified from children without autoimmunity (islet autoantibody-negative), with recent onset of autoimmunity (recent activation = multiple autoantibodies for ≤5 y), with persistent autoimmunity (multiple autoantibodies for >5 to ≤10 y), and with long-term autoimmunity (multiple autoantibodies for >10 y without T1D). (C) Summary of identified HLA-DQ8–restricted insulin-specific tet+CD4+CXCR5+ TFH precursor cells purified from children without autoimmunity (no autoimmunity, n = 8), with recent onset of autoimmunity (recent activation, n = 8), with persistent autoimmunity (n = 7), and with long-term autoimmunity (n = 7). Data represent the mean ± SEM. *P < 0.05.
Of note, the frequency of these insulin-specific TFH precursors in nondiabetic children with long-term autoimmunity was 14.3 ± 3.0% of tet+CD4+ T cells, which was significantly lower than in children with recent onset of islet autoimmunity (P < 0.05; Fig. 1C). These findings are in accordance with the fact that nondiabetic children with latency in progression to clinically overt disease can lose some of their earliest islet autoantibodies, especially insulin autoantibodies.
Increase of Circulating CCR7lowPD1highCXCR5+ TFH Precursor Cells During the Onset of Human Islet Autoimmunity.
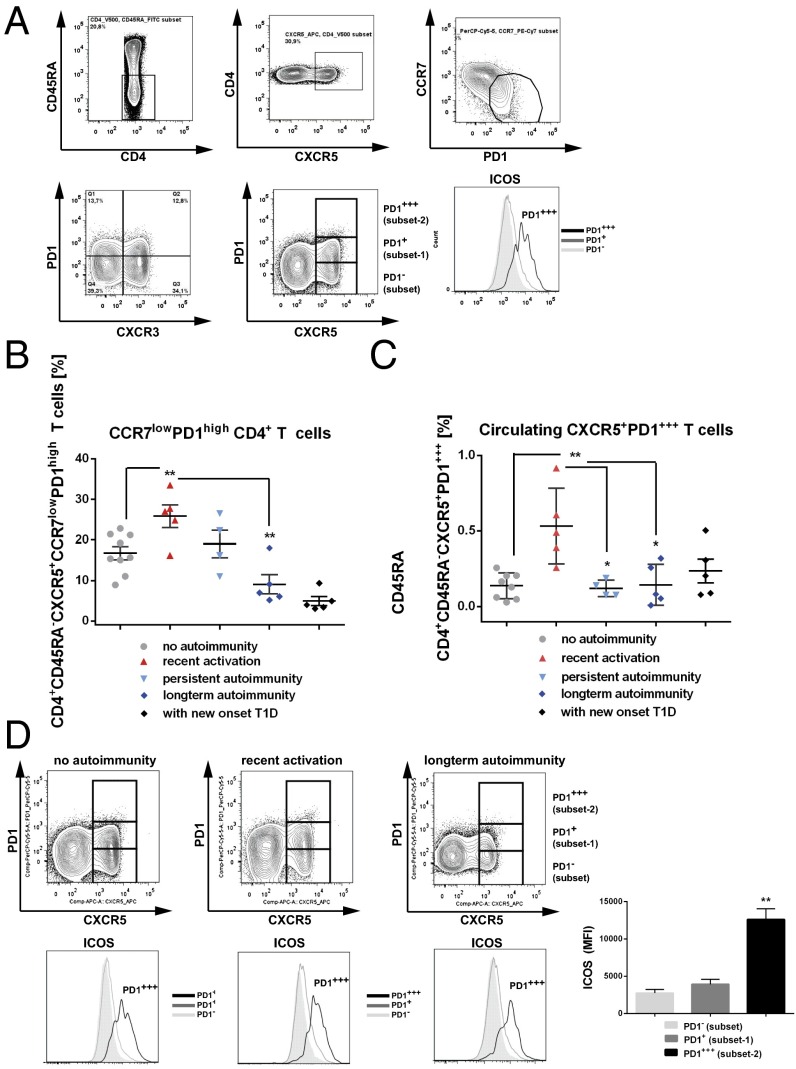
During islet autoimmunity onset, we also identified up-regulated PD1 and ICOS expression levels within circulating CXCR5+ T cells, together with increased frequencies of TFH precursors at the polyclonal level (Fig. 2). Specifically, the frequency of CCR7lowPD1highCXCR5+CD4+ T cells (gating example in Fig. 2A, Upper) was significantly greater in children with onset of islet autoimmunity than in children without islet autoimmunity (no autoimmunity vs. recent onset of autoimmunity: 16.8 ± 1.6% vs. 25.8 ± 2.8% of CD4+CD45RA− T cells; P < 0.01; Fig. 2B), and was significantly reduced in nondiabetic children with long-term autoimmunity (9.0 ± 2.3% of CD4+CD45RA− T cells; P < 0.01; Fig. 2B) compared with the other two groups of children. The frequency of CXCR5+CD4+ T cells harboring the highest level of PD1 expression (PD1+++; gating example in Fig. 2A, Lower) was significantly greater in children with recent onset of islet autoimmunity than in children without autoimmunity (no autoimmunity vs. recent onset of autoimmunity: 0.14 ± 0.03% vs. 0.53 ± 0.11% of CD4+ T cells; P < 0.01; Fig. 2 C and D). The abundance of CXCR5+PD1+++CD4+ T cells in nondiabetic children with long-term autoimmunity was 0.16 ± 0.06% of CD4+ T cells, which was significantly lower than in children with recent onset of autoimmunity (P < 0.05) and similar to the abundance in children without autoimmunity (P > 0.05; Fig. 2 C and D). CXCR5+PD1+++CD4+ T cells also harbored the highest ICOS expression level (ICOS mean fluorescence intensity (MFI) within PD1+++ subset = 12,600 ± 1,457 vs. ICOS MFI within PD1+ subset = 3,919 ± 666; P < 0.01; Fig. 2D, Lower Right). These findings therefore support the hypothesis that during the onset of autoimmunity, these circulating CXCR5+PD1+++CD4+ T cells with the highest ICOS expression level can boost immune activation and trigger autoantibody production.
Fig. 2.
Identification and characterization of blood-residing CXCR5+CCR7lowPD1high and CXCR5+PD1+++ cells in CD4+ T cells from children with or without ongoing islet autoimmunity or with new onset T1D. (A) Identification of CD4+CD45RA−CXR5+CCR7lowPD1high T cells (Upper) and CXCR5+PD1−, CXCR5+PD1+, and CXCR5+PD1+++ T cells (Lower). (Lower Right) Histogram indicates ICOS expression levels in the PD1− subset (light gray line, plot filled), PD1+ subset 1 (gray line, plot unfilled), and PD1+++ subset 2 (black line). (B) Summary graphs for the frequencies of circulating CD4+CD45RA−CXR5+CCR7lowPD1high T cells (no autoimmunity, n = 9; recent onset of autoimmunity, n = 5; persistent autoimmunity, n = 4; long-term autoimmunity, n = 5; new onset T1D, n = 5). Data represent the mean ± SEM. **P < 0.01. (C) Summary graphs for the frequencies of circulating CD4+CD45RA−CXR5+PD1+++ T cells (no autoimmunity, n = 8; recent onset of autoimmunity, n = 5; persistent autoimmunity, n = 4; long-term autoimmunity, n = 5; new onset T1D, n = 5). Data represent the mean ± SEM. *P < 0.05; **P < 0.01. (D, Upper) Identification of CD4+CD45RA−CXCR5+PD1+++ T cells from children with or without pre-T1D (no autoimmunity, recent activation of autoimmunity, and long-term autoimmunity). (D, Lower) Histograms indicate ICOS expression levels in CXCR5+PD1−, CXCR5+PD1+, and CXCR5+PD1+++ subsets. (D, Lower Right) Summary graph shows ICOS MFIs, dependent on the PD1 expression levels. Data represent the mean ± SEM. **P < 0.01.
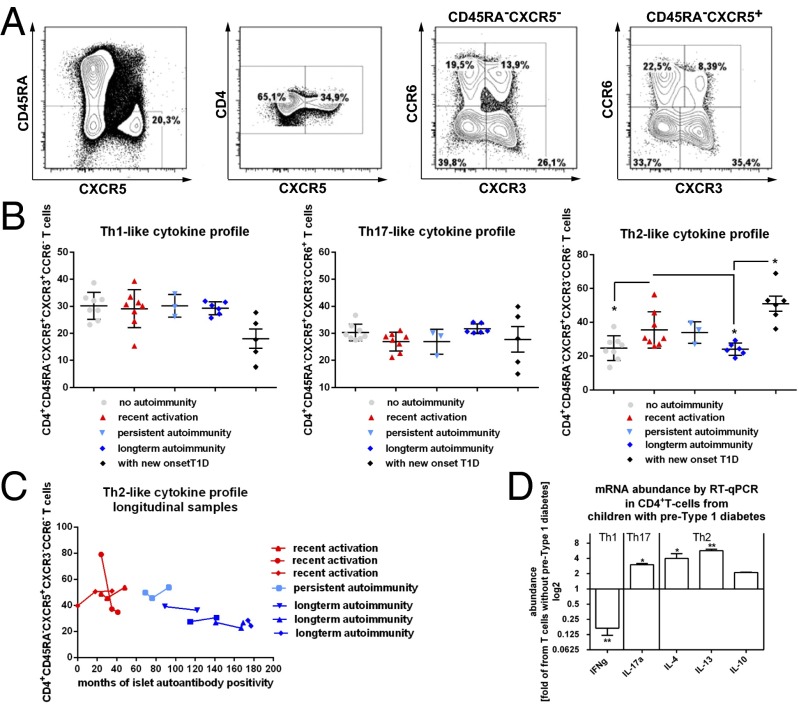
To investigate the frequencies of CCR7lowPD1high and CXCR5+PD1+++CD4+ T cells in children or adolescents near to the diagnosis of clinically overt T1D (newly manifest T1D), we studied T cells from the German new onset diabetes in the young incident cohort study (32, 33). We found the abundance of CCR7lowPD1high and CXCR5+PD1+++CD4+ T cells in young individuals with newly manifest T1D to be unaltered compared with nondiabetic children with long-term autoimmunity (Fig. 2 B and C). Next, we analyzed T helper (Th)-like subtypes within the CXCR5+CD4+ T-cell population based on the coexpression of CXCR3 and CCR6 (gating example in Fig. 3A). CXCR3+CCR6− cells mainly secrete IFN-γ (Th1-like profile) but not Th2 or Th17 cytokines; CXCR3−CCR6− cells exclusively secrete Th2 cytokines [interleukin (IL)-4, IL-5, and IL-13]; and CXCR3−CCR6+ cells secrete Th17 cytokines (IL-17A and IL-22) (17). CXCR5+ Th2 cells and CXCR5+ Th17 cells were found to induce naive B cells to secrete immunoglobulins via IL-21 but to modulate isotype switching differentially, whereas human CXCR5+ Th1 cells were unable to provide B-cell help (17). We found no differences in the frequencies of Th1 cells and Th17 cells within the CXCR5+CD4+ T-cell compartment between disease groups, including young individuals with new onset T1D (Fig. 3B). However, we observed a significant increase in the TFH-Th2 subset in children with recent onset of islet autoimmunity compared with T cells from children without ongoing autoimmunity or with long-term autoimmunity (no autoimmunity vs. recent onset of autoimmunity vs. long-term autoimmunity: 24.6 ± 2.5% vs. 35.4 ± 3.8%; P < 0.05 vs. 23.9 ± 1.4% of CD4+CD45RA− T cells; P < 0.05; Fig. 3B). In parallel to the increase in the TFH-Th2 subset during onset of islet autoimmunity, we found a significant enhancement in this subset of TFH cells in young individuals with new onset T1D. This increase in TFH-Th2 cells was significant compared with T cells from nondiabetic children with long-term autoimmunity (long-term autoimmunity: 23.9 ± 1.4% vs. with new onset T1D: 50.9 ± 4.4; P < 0.05; Fig. 3B).
Fig. 3.
Identification of circulating CXCR5+CD4+ T cells and Th1, Th2, or Th17 cytokine expression profiles in CD4+ T cells from children with or without ongoing islet autoimmunity or with new onset T1D. (A) Identification of CD4+CD45RA−CXCR5+ T cells and respective CXCR3+CCR6− (Th1), CXCR3−CCR6− (Th2), or CXCR3−CCR6+ (Th17) subsets. (B) Summary graphs for the frequencies of circulating CXCR5+CD4+ T cells with Th1, Th2, or Th17 characteristics (no autoimmunity, n = 8; recent onset of autoimmunity, n = 8; long-term autoimmunity, n = 6; new onset T1D, n = 5). Data represent the mean ± SEM. *P < 0.05. (C) Frequencies of the TFH-Th2 subset in longitudinal samples from children with recent activation, persistent islet autoimmunity, or long-term islet autoimmunity. (D) Summary graphs for the mRNA abundance of cytokine expression levels (IFN-γ = Th1; IL-17a = Th17, IL-4, IL-13; and IL-10 = Th2) within respective CD4+ T-cell subsets by RT-qPCR from children with islet autoimmunity (pre-T1D). Results are shown in abundance as the fold of T cells from children without ongoing islet autoimmunity (no pre-T1D), with no autoimmunity (n = 6), or with ongoing islet autoimmunity (n = 7). Data represent the mean ± SEM. *P < 0.05; **P < 0.01.
When we analyzed individual longitudinal samples from nondiabetic children with ongoing islet autoimmunity, we again observed the highest frequencies of Th2-like TFH cells within recent activation of islet autoimmunity (<5 y of islet autoimmunity), whereas frequencies were lower during persistent islet autoimmunity (>5 to < 10 y of islet autoimmunity) and further decreased in children with long-term autoimmunity (>10 y of islet autoimmunity; Fig. 3C).
In accordance, the respective T-cell subset presented with an increased abundance of Th2 cytokines, such as IL-4 and IL-13, with no change in IL-10 (Fig. 3D).
Increase of miRNA17∼92 Family Abundance in CD4+ T Cells from Children with Recent Onset of Islet Autoimmunity.
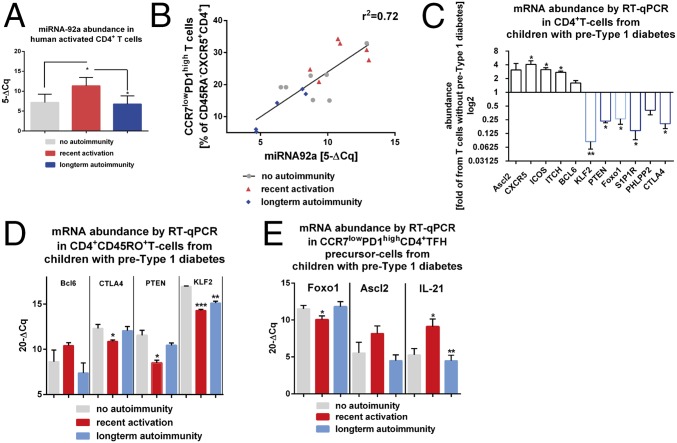
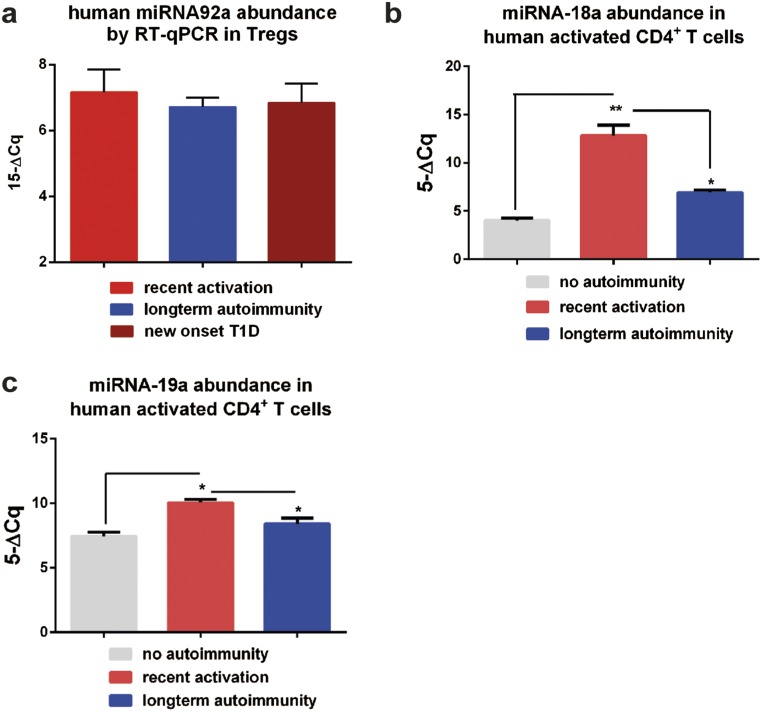
To define involved signaling pathways, we next examined miRNA92a abundance of CD4+ T cells from children with or without islet autoimmunity. Importantly, miRNA92a expression levels were significantly increased in CD4+ T cells from children with recent onset of islet autoimmunity (no autoimmunity vs. recent onset of autoimmunity: 7.1 ± 0.6 vs. 11.3 ± 0.7; P < 0.05; Fig. 4A). However, its expression was down-regulated in nondiabetic children with long-term autoimmunity (6.7 ± 0.7), in accordance with reduced TFH precursor frequencies. These changes in miRNA92a expression levels in relation to duration of islet autoimmunity were not observed within the CD127lowCD25high regulatory T (Treg)-cell population (Fig. S1A).
Fig. 4.
Enhanced abundance of miRNA92a promotes increased frequencies of circulating CXCR5+CD4+ T cells during onset of human islet autoimmunity. (A) miRNA92a abundance in human CD4+ T cells purified from children with or without pre-T1D (no autoimmunity, n = 10; recent onset of autoimmunity, n = 6; long-term autoimmunity, n = 6) by RT-qPCR analyses. Data represent the mean ± SEM. *P < 0.05. (B) Correlation of CXCR5+CCR7lowPD1highCD4+ T cells with miRNA92a abundance. (C) Abundance of predicted signaling pathways regulated by miRNA92a in human islet autoimmunity. Down-regulation of the PTEN-Foxo1-KLF2 signaling network to promote human TFH precursor cells and onset of islet autoimmunity is shown. mRNA abundance of Ascl2, CXCR5, ICOS, ITCH, Bcl6, KLF2, PTEN, Foxo1, PHLPP2, CTLA4, and S1P1R from CD4+ T cells of individual children with islet autoimmunity (pre-T1D) is shown. Results are shown in abundance as the fold of T cells from children without ongoing islet autoimmunity (no pre-T1D), with no autoimmunity (n = 6), and with ongoing islet autoimmunity, n = 7). *P < 0.05; **P < 0.01. (D) mRNA abundance from a selection of genes predictively targeted by miRNA92a in CD4+CD45RO+ T cells from individual children with or without ongoing pre-T1D as assessed by RT-qPCR analyses (no autoimmunity, n = 7; recent onset of autoimmunity, n = 6; long-term autoimmunity, n = 5). Data represent the mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. (E) mRNA abundance of Foxo1, Ascl2, and IL-21 in CCR7lowPD1highCD4+ TFH precursor cells in accordance with the duration of islet autoimmunity (no autoimmunity, n = 7; recent onset of autoimmunity, n = 6; long-term autoimmunity, n = 5). Data represent the mean ± SEM. *P < 0.05; **P < 0.01.
Fig. S1.
(A) miRNA92a abundance in CD4+CD3+CD127lowCD25high Treg cells. miRNA18a (B) and miRNA19a (C) abundance in human CD4+ T cells purified from children with or without pre-T1D (no autoimmunity, n = 10; recent onset of autoimmunity, n = 6; long-term autoimmunity, n = 6) as assessed by qRT-PCR analyses. Data represent the mean ± SEM. *P < 0.05; **P < 0.01.
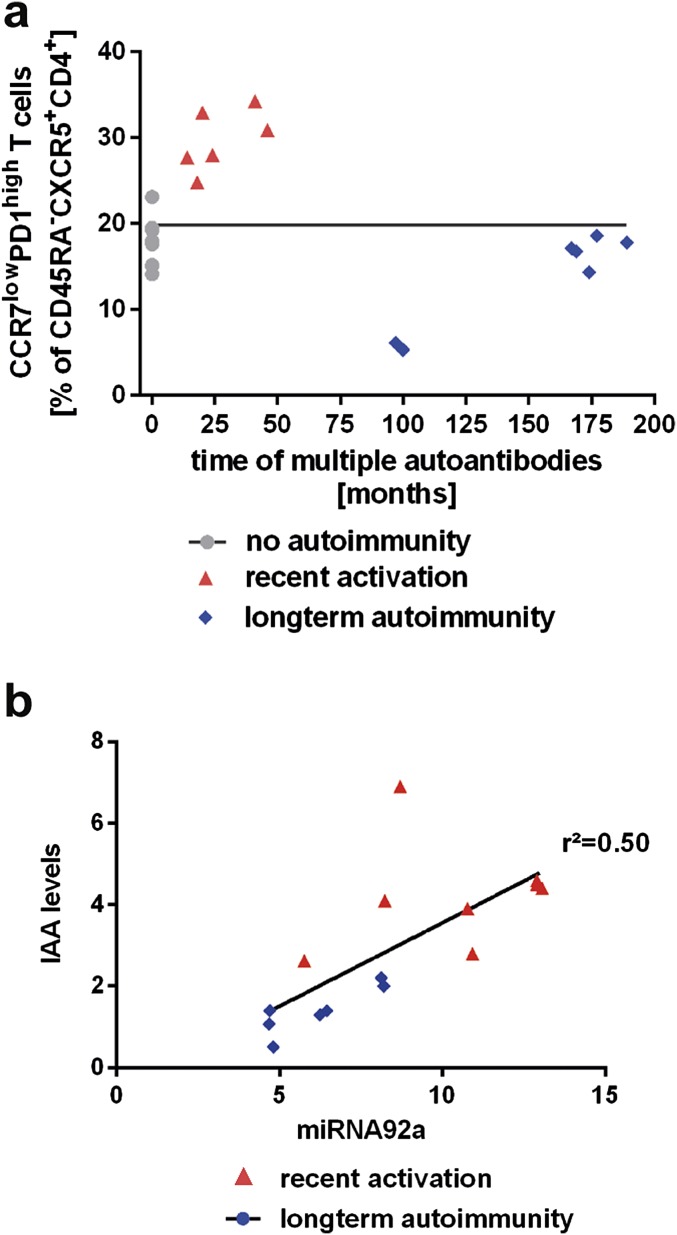
The expression profiles of two other members of the miRNA17∼92 cluster, miRNA-18a and miRNA-19a, were altered in a similar fashion to the abundance of miRNA92a (Fig. S1 B and C). Of note, we identified a strong correlation between the abundance of miRNA92a and the frequency of CXCR5+ TFH precursors. CD4+ T cells from children without islet autoimmunity and from nondiabetic children with a slow-progressor phenotype presented with the lowest frequencies of CXCR5+ TFH precursors and the lowest abundance of miRNA92a. In contrast, during onset of islet autoimmunity, in states of strong immune activation, more CD4+ T cells harbored a CXCR5+ TFH precursor phenotype accompanied with increased abundance of miRNA92a (r2 = 0.72; Fig. 4B). In contrast, no correlation was observed between the duration of islet autoimmunity (months of multiple autoantibodies) and the frequencies of CCR7lowPD1highCD4+ T cells, supporting concepts that point to a relapsing/remitting autoimmunity phenotype (Fig. S2A). However, we did find a correlation between miRNA92a abundance in CD4+ T cells and levels of insulin autoantibodies (IAA) within disease groups (Fig. S2B).
Fig. S2.
(A) Correlation of duration of islet autoimmunity (months of multiple islet autoantibody positivity) with CXCR5+CCR7lowPD1highCD4+ T cells. (B) Correlation of miRNA92a abundance in CD4+ T cells with IAA levels in serum.
Reduced Abundance of Predicted Signaling Pathways Regulated by miRNA92a in Human Islet Autoimmunity: PTEN, PHLPP2, and Cytotoxic T-Lymphocyte–Associated Protein 4.
We next determined mRNA abundances of predicted signaling pathways regulated by miRNA92a. As bioinformatics tools, we used miRDB [MirTarget (34)]. Additionally, TargetScan (Human 6.2) was used to predict biological targets for miRNA92a (35). In accordance with the observed enhanced abundance of miRNA92a in CD4+ T cells from islet autoantibody-positive children, we found repression of predicted target genes of miRNA92a (Fig. 4C). Specifically, in CD4+ T cells from children with ongoing autoimmunity, we observed repression of the phosphatase PTEN, which was outlined as a direct target of the miRNA92a family in mice (29). In our human T-cell setting, we likewise saw reduced abundance of the miRNA92 target PHLPP2 (36), which encodes an Akt phosphatase (37) and functions as a critical negative regulator of the PI3K-Akt signaling pathway as identified in murine studies (Fig. 4C). We also observed reduced mRNA abundance of predicted targets of miRNA92a that negatively regulate T-cell activation, such as cytotoxic T-lymphocyte–associated protein 4 (CTLA4) (Fig. 4C).
miRNA92a Targets the Zinc-Finger Transcription Factor KLF2 During Human Islet Autoimmunity.
Next, using TargetScan analyses (35, 38), we identified an evolutionarily conserved binding site for human miRNA92a in the 3′ UTR of KLF2. Importantly, abundance of KLF2 was found to be critically repressed in CD4+ T cells from children with ongoing islet autoimmunity (Fig. 4C). These findings support a potential role of KLF2 as a target of miRNA92a in human CD4+ T cells.
Down-Regulation of PTEN-Foxo1-KLF2 Signaling Network to Promote Human TFH Precursor Cells and Onset of Islet Autoimmunity.
To dissect involved signaling pathways relevant for TFH differentiation further, we next identified a significantly increased abundance of ICOS in such CD4+ T cells from children with ongoing autoimmunity (pre-T1D), whereas expression levels of Foxo1, which represses Bcl6 transcription, as well as PTEN and KLF2, were significantly down-regulated. In accordance with a low abundance of KLF2, we found a high abundance of CXCR5 and low expression of S1PR1 (Fig. 4C) in such CD4+ T cells from children with islet autoimmunity compared with children without ongoing islet autoimmunity (no pre-T1D).
When we determined the mRNA abundance of genes involved in T-cell activation and TFH-cell differentiation and function according to the duration of islet autoimmunity, we confirmed that KLF2 abundance was significantly down-regulated, particularly during the onset of islet autoimmunity (P < 0.001; Fig. 4D). Moreover, during onset of islet autoimmunity, we likewise observed negative regulators of T-cell activation (e.g., CTLA4, PTEN) to be repressed (Fig. 4D).
Foxo1 expression was significantly reduced in CCR7lowPD1high TFH precursors from children with recent onset of islet autoimmunity (Fig. 4E). Additionally, we observed increased levels of Ascl2 and IL-21 in CCR7lowPD1high TFH precursors from children with recent onset of islet autoimmunity in contrast to cells from nondiabetic children with long-term autoimmunity (Fig. 4E).
Induction of Human TFH Precursor Cells by miRNA92a in Vitro.
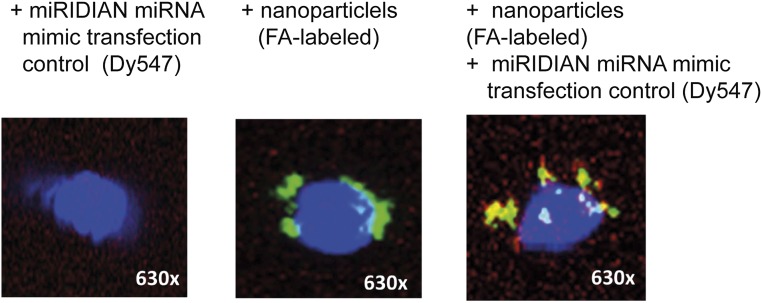
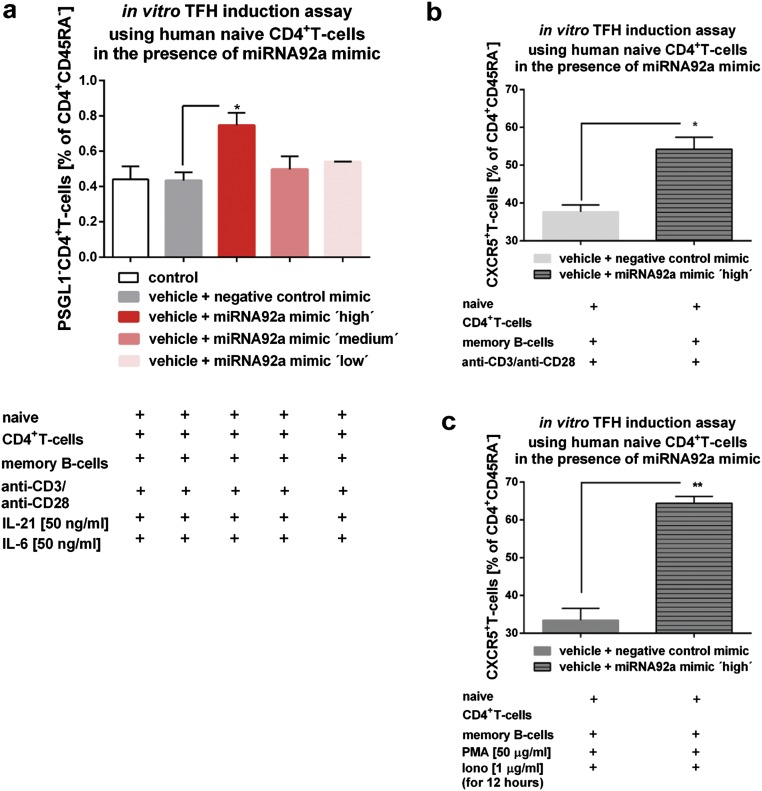
We next investigated whether activation of the miRNA92a pathway can induce human TFH precursor cells in vitro. Chitosan-coated poly(d,l-lactide-coglycolide) (PLGA) nanoparticles (39) were used to deliver mature miRNA92a mimics with a triple-RNA strand design (40). The uptake, intracellular localization of labeled nanoparticles, and delivery of fluorescently labeled control miRNAs were verified by confocal microscopy (Fig. S3). Next, we performed in vitro TFH precursor induction experiments (16, 21) with human naive CD4+ T cells and autologous memory B cells, sorted as CD20+CD27+, in the presence or absence of a miRNA92a mimic (Fig. S4A). Importantly, the miRNA92a mimic significantly increased the frequencies of CXCR5+CD4+ TFH precursors in vitro (vehicle + negative control mimic vs. vehicle + miRNA92a mimic: 37.6 ± 1.8 vs. 54.2 ± 3.1 CD45RA−CXCR5+CD4+ T cells; P < 0.05; Fig. S4 B and C).
Fig. S3.
Assessment of uptake, intracellular colocalization of FA-labeled nanoparticles, and Dy457-labeled transfection control miRNA mimic in CD4+ T cells upon stimulation with anti-CD3/anti-CD28 by confocal microscopy.
Fig. S4.
(A) TFH precursor induction using human naive CD4+ T cells in the presence of memory B cells and IL-6, IL-21, and a titration of a miRNA92a mimic (low = 0.0375 μg in 200 μL per 100,000 cells, medium = 0.075 μg in 200 μL per 100,000 cells, and high = 0.15 μg in 200 μL per 100,000 cells) or a negative miRNA mimic control. Summary graph for frequencies of PSGL1−CD4+ T cells is shown as percentages of CD4+CD45RA− T cells (n = 4). Data represent the mean ± SEM from duplicate wells per individual. *P < 0.05. (B) TFH precursor induction using human naive CD4+ T cells in the presence of memory B cells and a miRNA92a mimic or a negative miRNA mimic control. Summary graph for frequencies of CXCR5+CD4+ T cells is shown as percentages of CD4+CD45RA− T cells (n = 4). Data represent the mean ± SEM from duplicate wells per individual. *P < 0.05. (C) TFH precursor induction using human naive CD4+ T cells in the presence of memory B cells and a miRNA92a mimic or a negative miRNA mimic control. In such assays, PMA (50 ng/mL) and ionomycin (1 μg/mL) were added for the last 12 h of the experiments, and frequencies of CXCR5+CD4+ T cells shown as percentages of CD4+CD45RA− T cells were analyzed (n = 4). Data represent the mean ± SEM from duplicate wells per individual. **P < 0.01.
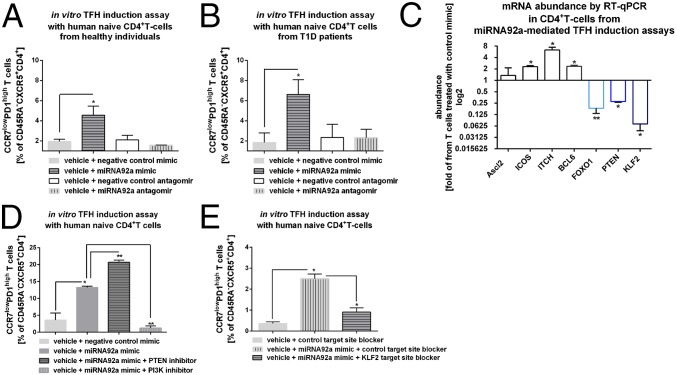
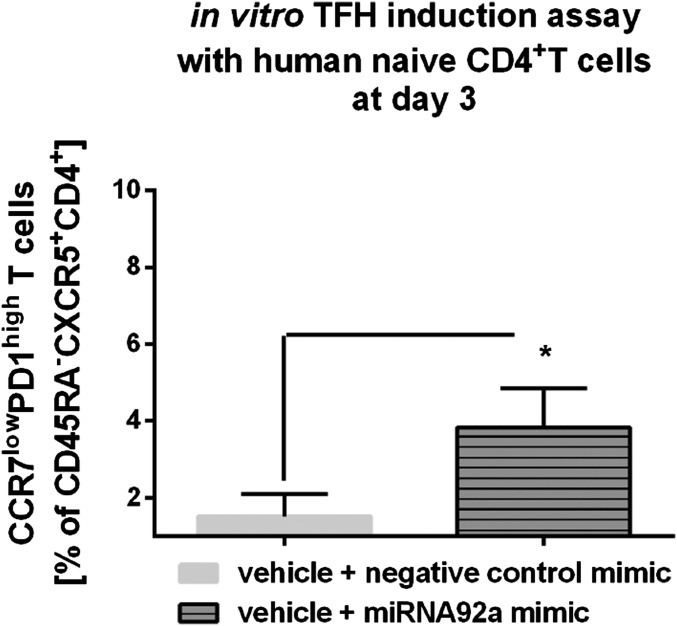
The miRNA92a mimic also significantly enhanced the frequency of CCR7lowPD1highCD4+ TFH precursor cells using T cells from healthy individuals (vehicle + negative control mimic vs. vehicle + miRNA92a mimic: 1.8 ± 0.2 vs. 4.6 ± 0.8 CCR7lowPD1highCD4+ T cells; P < 0.05; Fig. 5A) and from T1D patients (Fig. 5B). An induction of CCR7lowPD1highCD4+ TFH precursor cells by miRNA92a mimic was observed already after 3 d of incubation (Fig. S5).
Fig. 5.
miRNA92a regulates human TFH precursor induction in vitro. (A) TFH precursor induction using human naive CD4+ T cells from healthy individuals in the presence of memory B cells with or without a miRNA92a mimic, a miRNA92a antagomir, or respective negative control mimics or antagomirs. Summary graphs are shown for frequencies of CCR7lowPD1high cells presented as percentages of CXCR5+CD45RA−CD4+ T cells. Data represent the mean ± SEM. *P < 0.05. (B) TFH precursor induction as outlined in A using human naive CD4+ T cells from T1D individuals. Summary graphs are shown for frequencies of CCR7lowPD1high cells presented as percentages of CXCR5+CD45RA−CD4+ T cells. Data represent the mean ± SEM. *P < 0.05. (C) mRNA abundance of predicted signaling pathways controlled by miRNA92a from CD4+ T cells of TFH precursor induction assays in the presence of miRNA92a mimic quantified by RT-qPCR analyses. Results are shown in abundance as the fold of T cells treated with negative miRNA mimic controls. Data represent the mean ± SEM from duplicate wells of four independent experiments. *P < 0.05; **P < 0.01. (D) TFH induction using human naive CD4+ T cells in the presence of memory B cells with or without a miRNA92a mimic or negative miRNA mimic controls. In such assays, PMA (50 ng/mL) and ionomycin (1 μg/mL) were added for the last 12 h of the experiments, and frequencies of CCR7lowPD1highCD4+ T cells were analyzed. Experiments were performed in the presence or absence of a PTEN or PI3K inhibitor, respectively. Summary graphs for frequencies of CCR7lowPD1high TFH precursor cells (percentage of CD45RA−CXCR5+CD4+ T cells) (n = 4). Data represent the mean ± SEM from duplicate wells of four independent experiments. *P < 0.05; **P < 0.01. (E) TFH precursor induction using human naive CD4+ T cells in the presence of memory B cells with or without a miRNA92a mimic, a control TSB, a miRNA92a KLF2 TSB, or a combination. Summary graphs are shown for frequencies of CCR7lowPD1high cells presented as percentages of CXCR5+CD45RA−CD4+ T cells (n = 4). Data represent the mean ± SEM from duplicate wells of four independent experiments. *P < 0.05.
Fig. S5.
TFH precursor induction using human naive CD4+ T cells in the presence of memory B cells with or without a miRNA92a mimic or respective negative control mimics. Summary graphs are shown for frequencies of CCR7lowPD1high cells presented as percentages of CXCR5+CD45RA−CD4+ T cells after a 3-d culture. Data represent the mean ± SEM. *P < 0.05.
mRNA Abundance of Predicted Signaling Pathways Controlled by miRNA92a During miRNA92a-Mediated TFH Precursor Induction in Vitro.
We next studied whether activation of the miRNA92a pathway in vitro can modulate mRNA abundance of signaling pathways predictively targeted by miRNA92a. Upon miRNA92a mimic application, we observed significantly enhanced abundance of ITCH (miRNA92-mimic: ITCH: 2.6 ± 0.3, fold of T cells treated with control mimic; P < 0.05; Fig. 5C). Bcl6 expression was moderately but significantly up-regulated in line with findings indicating that upon in vitro differentiation, such TFH precursors present with intermediate levels of Bcl6 (41), in contrast to bona fide germinal center TFH cells that harbor high Bcl6 abundance (41). Moreover, expression levels of Foxo1 (miRNA92a mimic: Foxo1: 0.18 ± 0.04, fold of T cells treated with control mimic; P < 0.01) as well as PTEN (miRNA92a mimic: PTEN: 0.27 ± 0.01, fold of T cells treated with control mimic; P < 0.05) were significantly reduced compared with T cells that had been incubated with control mimics (Fig. 5C). In addition, KLF2 mRNA abundance of CD4+ T cells from miRNA92a-mediated TFH induction assays was significantly down-regulated compared with T cells treated with control mimics (miRNA92a mimic: KLF2: 0.07 ± 0.02, fold of T cells treated with control mimic; P < 0.05; Fig. 5C). These data further support the concept that KLF2 functions as a target of miRNA92a.
miRNA92a-Mediated Human in Vitro TFH Precursor Induction Is Regulated by PTEN-PI3K Signaling.
TFH precursor induction by miRNA92a was further enhanced upon stimulation with phorbol 12-myristate 13-acetate (PMA)/ionomycin in terms of CCR7lowPD1highCD4+ T cells (Fig. 5D). To analyze the involvement of the PTEN-PI3K signaling network in the regulation of TFH precursor induction, we performed such assays with a miRNA92a mimic in the presence or absence of a PI3K or PTEN inhibitor, respectively. Coapplication of a PI3K inhibitor to the miRNA92a mimic completely abolished TFH precursor induction (CCR7lowPD1high T cells: miRNA92a mimic: 13.3 ± 0.3 vs. miRNA92a mimic + PI3K inhibitor: 1.2 ± 0.6; P < 0.01). In accordance, miRNA92a mimic treatment in the presence of a PTEN inhibitor resulted in a significant boost of TFH precursor cell induction (CCR7lowPD1high T cells: miRNA92a mimic: 12.6 ± 0.7 v. miRNA92a mimic + PTEN-inhibitor: 20.6 ± 0.6; P < 0.01; Fig. 5D).
miRNA92a Targets KLF2 to Promote Human TFH Precursor Induction in Vitro.
To underline KLF2 as a target of miRNA92a, we used a specific miRNA92a KFL2 target site blocker (TSB). miRNA TSBs are antisense oligonucleotides that bind to the miRNA target site of an mRNA, thereby preventing miRNAs from gaining access to that site. Application of a miRNA92a mimic in the presence of a control TSB resulted in a significant increase of CCR7lowPD1highCD4+ TFH precursor cells (vehicle + control TSB: 0.4 ± 0.07 vs. vehicle + miRNA92a mimic + control TSB: 2.5 ± 0.2; P < 0.05; Fig. 5E) in vitro. In contrast, coapplication of a specific KLF2 TSB significantly reduced the miRNA92a-mediated induction of TFH precursors (vehicle + miRNA92a mimic + control TSB: 2.5 ± 0.2 vs. vehicle + miRNA92a mimic + KLF2 TSB: 0.9 ± 0.2; P < 0.05; Fig. 5E), further indicating that KLF2 is a target of miRNA92a.
Blockade of Human TFH Precursor Cell Induction by a miRNA92a Antagomir in Vitro.
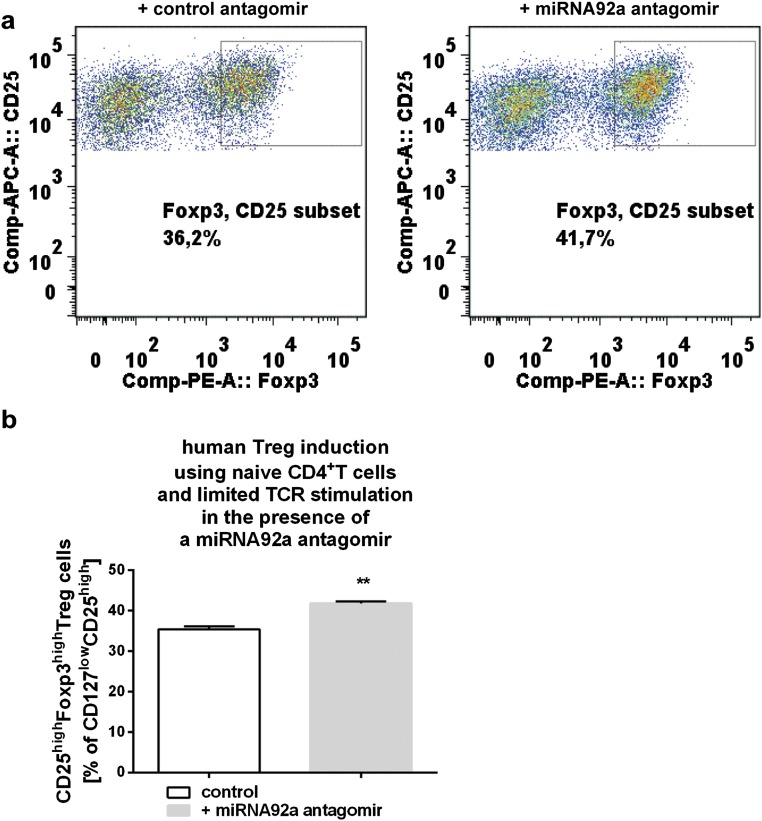
To assess a potential relevance of blocking miRNA92a activity to limit islet autoimmunity, we first performed in vitro TFH induction assays in the presence of a miRNA92a antagomir. Importantly, application of a miRNA92a antagomir completely blocked the induction of human CCR7lowPD1highCD4+ TFH precursor cells (vehicle + negative control antagomir: 2.1 ± 0.4 vs. vehicle + miRNA92a antagomir: 1.6 ± 0.05; Fig. 5 A and B) in vitro. The inhibition of human TFH differentiation was seen with CD4+ T cells from healthy donors (Fig. 5A), as well as from individuals with T1D (Fig. 5B). Accordingly, in human Treg-cell induction assays using established protocols with naive CD4+ T cells and limited T-cell receptor stimulation (31), a miRNA92a antagomir resulted in an increase in induced human CD127lowCD25highFoxp3high Treg cells (+ control antagomir: 35.4 ± 0.8 vs. + miRNA92a antagomir: 41.8 ± 0.5; P < 0.01; Fig. S6).
Fig. S6.
(A) Human Treg-cell induction using human naive CD4+ T cells in the presence of a miRNA92a antagomir (0.075 μg in 200 μL per 100,000 cells) or respective negative control antagomir and limited T-cell receptor stimulation. (B) Summary graph for frequencies of induced human CD127lowCD25highFoxp3high Treg cells (n = 4). Data represent the mean ± SEM from triplicate wells. **P < 0.01.
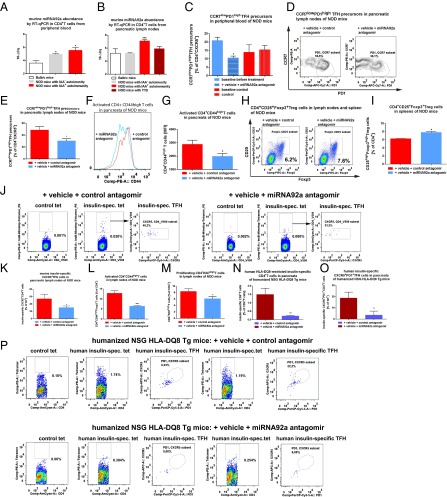
Inhibition of Murine TFH Precursors by a miRNA92a Antagomir in NOD Mice in Vivo.
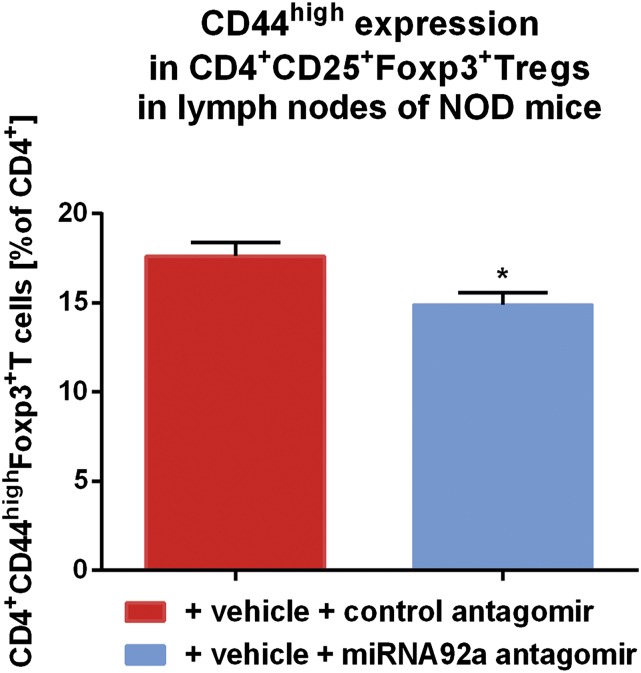
As a critical model of human autoimmune T1D, NOD mice share many similarities with human T1D, including the presence of specific autoantibodies and autoreactive T cells. In addition, in humans and mice, the T-cell response to insulin is dominated by a major histocompatibility complex II (MHCII) (IAg7)-restricted segment of the insulin B chain comprising residues 9–23. In contrast to CD4+ T cells from nonautoimmune-prone BALB/c and NOD mice without insulin-specific autoantibodies, CD4+ T cells from NOD mice with IAA+ autoimmunity presented with significantly increased expression levels of miRNA92a, especially in pancreatic lymph nodes (Fig. 6 A and B). Next, to test a possible therapeutic value of blocking miRNA92a activity in halting immune activation in vivo, we used NOD mice with IAA+ autoimmunity and a custom-designed locked nucleic acid (LNA) miRNA92a antagomir (Exiqon). Of note, short-term in vivo application of a miRNA92a antagomir (10 mg/kg i.p. on day 0, day 2, and day 6) to NOD mice significantly lowered TFH precursors in peripheral blood compared both with baseline levels assessed before beginning of the treatment and with mice treated with control antagomirs [baseline before miRNA92a antagomir application: CCR7lowPD1highCD4+ T cells (percentage of CXCR5+CD4+ T cells): 20.9 ± 1.9 vs. after miRNA92a antagomir application: 10.6 ± 1.0; P < 0.05; Fig. 6C]. Application of miRNA92a antagomir also resulted in significantly reduced frequencies of CCR7lowPD1high TFH precursors in pancreatic lymph nodes [CCR7lowPD1highCD4+ T cells (percentage of CXCR5+CD4+ T cells): control antagomir: 48.4 ± 5.9 vs. miRNA92a antogomir: 30.4 ± 3.6; P < 0.05; Fig. 6 D and E]. More importantly, the miRNA92a antagomir application reduced immune activation directly in the pancreas, because frequencies of CD4+CD44high T cells were significantly lowered in such treated mice compared with control animals [CD4+CD44high T cells (MFI) control antagomir: 2,877 ± 316 vs. miRNA92a antagomir: 1,976 ± 224; P < 0.05; Fig. 6 F and G]. Additionally, upon treatment with a miRNA92a antagomir, frequencies of CD4+CD25+Foxp3+ Treg cells were increased in lymph nodes (Fig. 6 H and I).
Fig. 6.
In vivo miRNA92a antagomir application reduces immune activation in NOD and humanized mice. The miRNA92a abundance in murine CD4+ T cells from BALB/c mice and NOD mice with or without ongoing islet autoimmunity, as well as with T1D in peripheral blood (A) or in pancreatic lymph nodes (B), as assessed by RT-qPCR analyses, is shown. Data represent the mean ± SEM. *P < 0.05; ***P < 0.001. (C) Summary graphs for CCR7lowPD1high T cells in peripheral blood of NOD mice before and after treatment with either control antagomirs or a specific miRNA92a antagomir. Data represent the mean ± SEM from two independent experiments (n = 4). *P < 0.05. (D) FACS plots for the identification of CCR7lowPD1high TFH precursors in pancreatic lymph nodes of NOD mice after treatment with control antagomir or miRNA92a antagomir. (E) Summary graphs for CCR7lowPD1high T cells in pancreatic lymph nodes of NOD mice given a control antagomir or a specific miRNA92a antagomir. Data represent the mean ± SEM from two independent experiments (n = 4). *P < 0.05. (F) FACS histograms for the identification of CD4+CD44high T cells of pancreas-infiltrating T cells of NOD mice treated with either control antagomirs or a miRNA92a antagomir. (G) Summary graphs for CD4+CD44high T cells of pancreas-infiltrating T cells of NOD mice as indicated in F. Data represent the mean ± SEM from two independent experiments (n = 4). *P < 0.05. (H) Identification of CD4+CD25+Foxp3+ T cells of NOD mice given a control antagomir or a miRNA92a antagomir. (I) Summary graphs for CD4+CD25highFoxp3high T cells of NOD mice as indicated in H. Data represent the mean ± SEM from two independent experiments (n = 4). *P < 0.05. (J) Identification of IAg7-restricted, insulin-specific CD4+ T cells and CXCR5+ TFH cells in pancreatic lymph nodes of NOD mice given a control antagomir or miRNA92a antagomir. (K) Frequencies of insulin-specific TFH cells as in J (mean ± SEM from two independent experiments; n = 4). *P < 0.05. (L) Frequencies of activated CD44high T cells upon treatment with a control or a miRNA92a antagomir. Data represent the mean ± SEM from two independent experiments (n = 4). ***P < 0.01. (M) Proliferating CD4+Ki67high T cells upon treatment with a control or a miRNA92a antagomir. Data represent the mean ± SEM from two independent experiments (n = 4). *P < 0.05. (N) Frequencies of human insulin-specific CD4+ T cells in pancreata of humanized mice as in P (mean ± SEM; n = 5). **P < 0.01. (O) Frequencies of human insulin-specific CXCR5+PD1+CD4+ T cells in pancreata of humanized mice as in P (mean ± SEM; n = 5). *P < 0.05. (P) Identification of human HLA-DQ8–restricted, insulin-specific CD4+ T cells and CXCR5+PD1+ TFH cells in pancreata of humanized NSG HLA-DQ8 Tg mice given a control antagomir or a miRNA92a antagomir. APC, allophycocyanin; PerCP, peridinin chlorophyll protein; spec., specific.
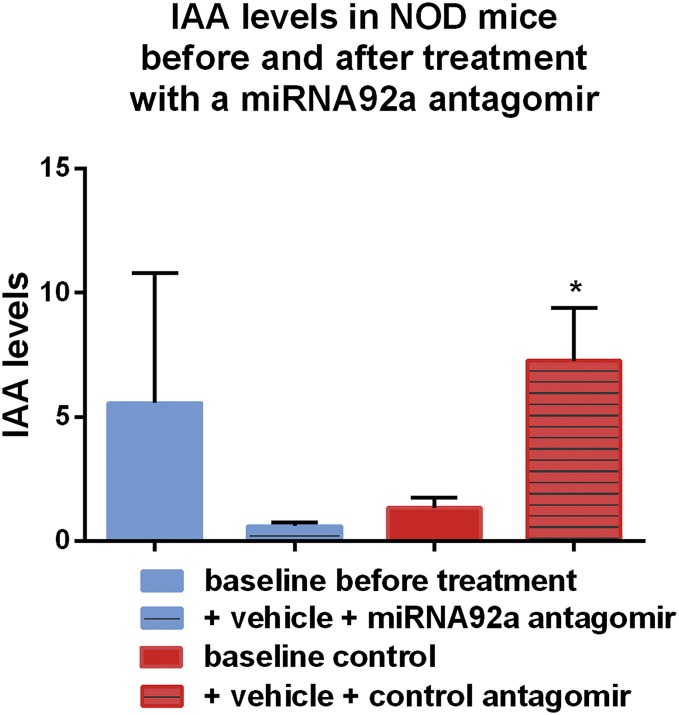
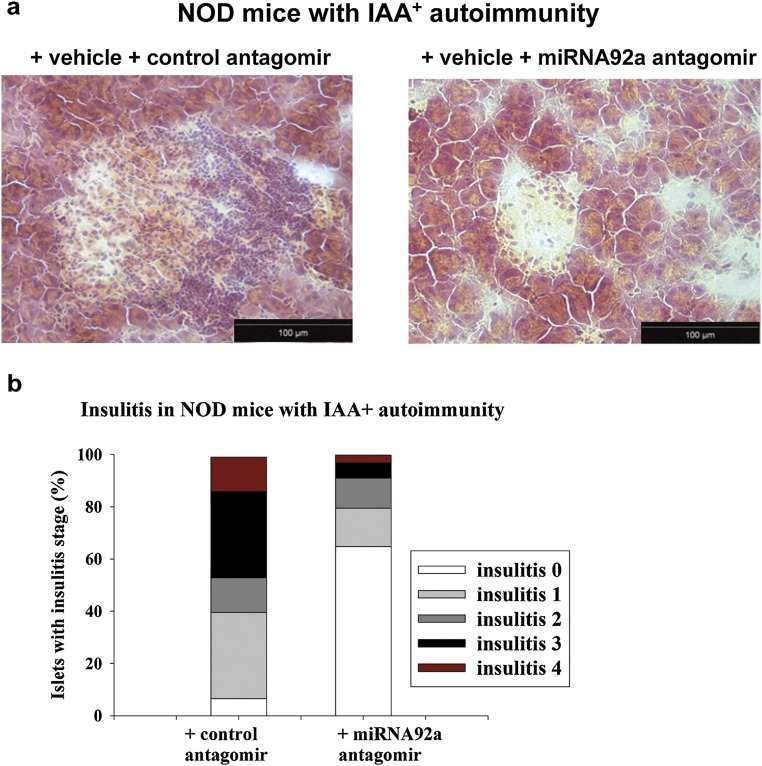
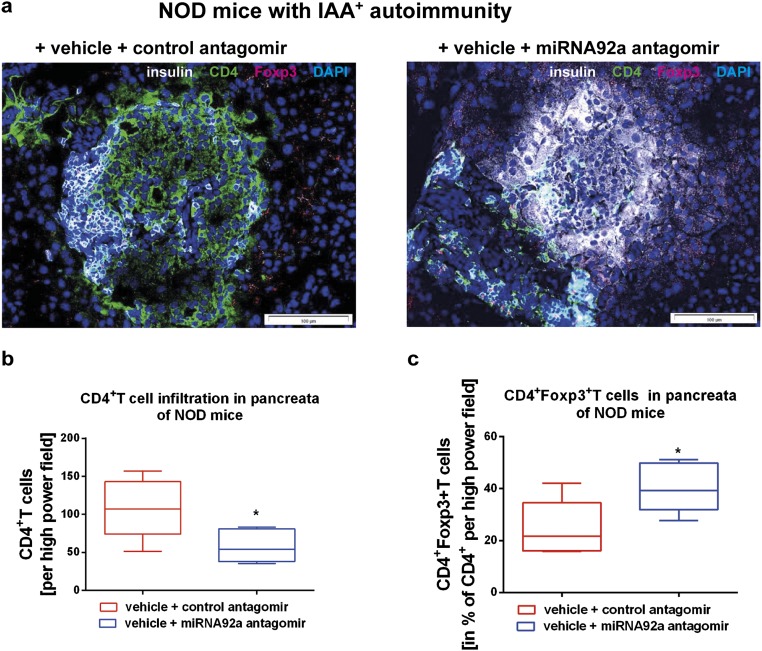
Furthermore, when we treated NOD mice for a longer period of 14 d (with injections four times per week at 5 mg/kg), the miRNA92a antagomir significantly reduced the frequencies of insulin-specific CD4+CXCR5+ TFH cells in pancreatic lymph nodes (Fig. 6 J and K). Additionally, the longer treatment period resulted in a more pronounced reduction of CD4+CD44high T cells (Fig. 6L). In contrast, the CD44high T cells within the CD4+CD25highFoxp3+ Treg-cell population was affected to a lesser extent (Fig. S7). Consistent with a reduced autoimmune activation upon miRNA92a antagomir application, we observed lower levels of proliferating Ki67highCD4+ T cells in these NOD mice (Fig. 6M). Although IAA levels had increased from baseline in control animals, IAA levels were found to be decreased in mice that were given the miRNA92a antagomir (Fig. S8). Accordingly, histopathological evaluation of pancreata from these NOD mice revealed that miRNA92a antagomir application had reduced pancreatic infiltration, resulting in more morphologically intact β cells (Fig. S9 A and B). These results were confirmed using analyses by immunofluorescence in pancreatic cryosections of such NOD mice (Fig. S10 A–C). NOD pancreata of mice that had received a miRNA92a antagomir showed distinctly reduced CD4+ T-cell infiltration accompanied by an enhanced abundance of local CD4+Foxp3+ Treg cells (Fig. S10 A–C).
Fig. S7.
Summary graph for CD44high expression in CD4+CD25+Foxp3+ Treg cells purified from lymph nodes of NOD mice given a control antagomir or a miRNA92a antagomir (14 d of treatment, with injections four times per week at 5 mg/kg) (n = 5 per group). Data represent the mean ± SEM. *P < 0.05.
Fig. S8.
Summary graphs for IAA levels from NOD mice before and after treatment with either control antagomirs or a specific miRNA92a antagomir (14 d of treatment, with injections four times per week at 5 mg/kg) (n = 5 per group). Data represent the mean ± SEM. *P < 0.05.
Fig. S9.
(A) Histopathological evaluation of pancreas sections from NOD mice with IAA+ autoimmunity that were given a control antagomir or a miRNA92a antagomir (14 d of treatment, with injections four times per week at 5 mg/kg). Representative hematoxylin and eosin-stained pancreas cryosections are shown. (B) Grading of insulitis from NOD mice as in A (n = 5 per group).
Fig. S10.
(A) Immunofluorescence for insulin (white), CD4 (green), and Foxp3 (red) in pancreatic cryosections of NOD mice given a control inhibitor or a miRNA92a inhibitor (14 d of treatment, with injections four times per week at 5 mg/kg). (B) CD4+ T cells infiltrating the pancreas as in A. Shown are box and whiskers plots of CD4+ T cells per high-power field. (C) CD4+Foxp3+ T cells in the percentage of infiltrating CD4+T cells per high-power field of mice as in A (n = 5 per group).
Next, to assess a potential human in vivo relevance of miRNA92a antagomir application, we investigated human HLA-DQ8–restricted insulin-specific TFH cells in humanized NOD Scid IL2 receptor gamma chain knockout (NSG) human leucocyte antigen (HLA)-DQ8 Tg mice in accordance with previously established procedures (31). Specifically, we focused on human pancreas-infiltrating CD4+ T cells upon treatment with a miRNA92a antagomir (1 wk of treatment, with injections three times per week at 10 mg/kg) or a control antagomir. In contrast to humanized mice that were given the control antagomir, mice that received the miRNA92a antagomir showed distinctly reduced frequencies of HLA-DQ8–restricted insulin-specific CD4+ T cells in pancreata of humanized NSG HLA-DQ8 mice (Fig. 6 N and O). More importantly, miRNA92a antagomir application significantly lowered the frequencies of insulin-specific CD4+CXCR5+PD1+ TFH cells in pancreata of humanized mice (Fig. 6 N and P). Overall, these findings further support a role of blocking miRNA92a activity in limiting autoimmune activation.
Discussion
Aberrant immune activation mediated by effector T-cell populations is pivotal in promoting the onset of autoimmunity, as in autoimmune T1D (2, 42). In children with ongoing islet autoimmunity, the time to progression to clinically overt T1D is highly plastic, ranging from a few months to more than two decades (2, 5), which highlights the critical contribution of differences in immune activation and pathways of regulation in disease progression. Therefore, studies of T cells from nondiabetic children according to the duration of islet autoimmunity can provide important insights into the regulation of aberrant immune activation, as well as into the mechanisms involved in triggering the onset of islet autoimmunity.
Currently, the role of TFH cells in promoting the onset of human islet autoimmunity remains unclear, especially concerning their potential function in accelerating progression to clinically overt T1D. Furthermore, our understanding of relevant signaling pathways regulating human TFH induction is limited. This knowledge gap resulted from the fact that previous studies had focused on analyses of TFH-cell characteristics in patients with clinically overt T1D (43, 44), a situation where the main target of the autoimmune attack, the insulin-producing β cells, have been destroyed already. Until now, the role of human TFH precursors during the onset of autoimmunity before clinically manifest T1D and relevant signaling pathways involved in their induction has remained elusive.
To tame aberrant TFH-cell responses specifically (45), the dissection of molecular mechanisms that regulate TFH differentiation and function during the onset of autoimmunity is required. Here, we provide direct evidence that during the onset of human islet autoimmunity, the insulin-specific target T-cell population is enriched with a CXCR5+CD4+ TFH precursor phenotype.
Mechanistically, we show that miRNA92a is critically involved in the induction of human TFH precursors and links the frequency of CXCR5+CD4+ T cells and the abundance of miRNA92a with the level of autoimmune activation. Specifically, our findings in CD4+ T cells from nondiabetic children with persistent and long-term autoimmunity demonstrate that a reduced frequency of peripheral CXCR5+ TFH precursors can be indicative of reduced immune activation in accordance with a slow progression to clinically overt T1D. In line with these results in human CD4+ T cells from nondiabetic children with ongoing islet autoimmunity, we identify miRNA92a to be significantly up-regulated in CD4+ T cells purified from pancreatic lymph nodes of NOD mice upon initiation of islet autoimmunity.
These findings highlight the importance to study miRNAs in the context of human autoimmunity, particularly in disease-relevant lymphocyte populations, to advance our understanding of gene regulation in autoimmunity, as well as for the identification of novel biomarkers indicative of pathogenic processes (46). Moreover, such miRNAs could function as future drug targets, because sequence-specific miRNA inhibitors were proven to work in patients (47). The delivery of such molecules to immune cells has been challenging; however, there are encouraging results for local or systemic application of miRNA inhibitors in autoimmunity and allergy (48, 49). Of note, in vivo application of a miRNA92a antagomir to NOD mice with ongoing islet autoimmunity reduced TFH precursors in peripheral blood and pancreatic lymph nodes. More importantly, application of a miRNA92a antagomir significantly lowered immune activation directly in the pancreas.
The identification of specific miRNA targets and defining their functional relevance remain challenging, because a single miRNA can directly regulate hundreds of genes and the amplitude of control is generally rather modest (50). Here, we provide evidence for repression of PTEN and PHLPP2, thereby permitting a boost of PI3K-Akt kinase signaling to trigger immune activation and TFH differentiation (36). This finding is consistent with an important role of PI3K in the germinal center response (51, 52). Down-regulation of PHLPP2 was shown to be critical for signaling via ICOS-PI3K, which drives the migration of T cells into B-cell follicles (53), a critical part of the TFH differentiation program (51).
We likewise found CTLA4 abundance to be significantly down-regulated during the onset of islet autoimmunity. Accordingly, T cells in CTLA4-deficient mice differentiate spontaneously into TFH cells in vivo, supporting a model in which CTLA4 mediates regulation of TFH-cell differentiation by graded control of CD28 engagement (54).
Moreover, during the onset of islet autoimmunity, Foxo1 abundance was significantly reduced. In accordance, murine data showed that Foxo1 negatively regulates Bcl6 and that its enforced nuclear localization prevents TFH-cell differentiation (12). In line with the fact that ICOS coreceptor signaling inactivates Foxo1 (12), we saw here significantly enhanced abundance of ICOS in CD4+ T cells from children with recent onset of islet autoimmunity.
In mice, KLF2 functions as a key modulator of homing receptors involved in lymphocyte migration (55, 56). KLF2 serves to restrain murine TFH-cell generation (15), and induced KLF2 deficiency in activated CD4+ T cells promotes TFH-cell generation and B-cell priming. Additionally, KLF2 promotes expression of the trafficking receptor S1PR1, and S1PR1 down-regulation was essential for efficient TFH-cell production (15).
Importantly, based on the observed significant down-regulation of KLF2 in CD4+ T cells from children with ongoing islet autoimmunity, we demonstrate that reduced KLF2 abundance can promote human TFH precursor induction during the onset of islet autoimmunity. In accordance with a low abundance of KLF2, we found a high abundance of CXCR5 and low expression of S1PR1 in T cells from children with ongoing islet autoimmunity. In addition, ICOS was highlighted to maintain the TFH phenotype by repressing KLF2 (57).
Of note, we highlight KLF2 as a potential target of miRNA92a in the regulation of islet autoimmunity. In accordance, during TFH induction assays, a miRNA92a mimic promoted a significant down-regulation of KLF2 mRNA abundance in CD4+ T cells. Moreover, miRNA92a-mediated induction of TFH precursor cells was significantly blunted in the presence of a specific miRNA92a-KLF2 TSB.
In addition, in T cells from such miRNA92a-mediated TFH precursor induction assays, we observed an enhanced abundance of the E3 ubiquitin ligase ITCH, which was found to be essential for the differentiation of TFH cells (13), and a reduced expression of Foxo1. Importantly, KLF2 functions as a direct downstream transcriptional target of Foxo1 (58, 59). Therefore, the reduced abundance of KLF2 could be aligned with the fact that during onset of islet autoimmunity, we observed Foxo1 abundance to be significantly down-regulated, whereas ICOS and miRNA92a abundance was found to be up-regulated.
With respect to a potential future targeting of TFH cells within the autoimmune disease process, we found CXCR5+CD4+ TFH cells at higher frequencies in at-risk children with islet autoantibody conversions within the past 5 y (recent onset of islet autoimmunity). Those children with multiple autoantibodies defined as having persistent (5–10 y) and long-term (>10 y) autoimmunity usually harbored frequencies similar to autoantibody-negative children. These observations are in line with concepts that children with a slow-progressor phenotype might have regulated the disease process or might at least be in a transient state of immune regulation. Furthermore, these findings also support the hypothesis that autoimmune T1D might have a relapsing/remitting disease phenotype. The fact that we observed frequencies of TFH cells to be particularly increased in early stages of the disease process is in accordance with a critical role of TFH cells in promoting B-cell activation and autoantibody production, and would therefore suggest that targeting of this population may be important during the very early disease stages. Consistent with this concept, in a limited set of longitudinal samples of children with various durations of islet autoimmunity, we again found the highest frequencies of TFH cells, and particularly Th2-like TFH cells, in early stages of islet autoimmunity. In the future, a more detailed analysis of longitudinal samples from islet autoantibody-positive children will help to clarify further the role of TFH cells in promoting progression to clinical disease.
In conclusion, the findings of this work provide evidence that during the onset of human islet autoimmunity, the insulin-specific target T-cell population is enriched in CXCR5+ TFH precursor cells. Moreover, we present a model in which, during the onset of human islet autoimmunity, miRNA92a-mediated TFH precursor cell induction is regulated by PTEN-PI3K signaling involving up-regulation of ICOS and down-regulation of PTEN, PHLPP2, and Foxo1. In addition, we show that KLF2 can function as a target of miRNA92a in promoting induction of TFH precursors. Importantly, miRNA92a antagomir application blocks TFH induction in vitro and significantly reduces islet autoimmunity in NOD mice with ongoing islet autoimmunity in vivo. Of note, in humanized NSG HLA-DQ8 Tg mice, administration of a miRNA92a antagomir significantly lowered frequencies of pancreas-residing, insulin-specific TFH cells. We therefore propose that manipulating miRNA92a or the PTEN-PI3K-KLF2 signaling network may facilitate the development of innovative regimens for the blockade of human T1D islet autoimmunity.
Materials and Methods
Human Subjects and Blood Samples.
Blood samples were collected from first-degree relatives of patients with T1D (children or adults). All subjects have already been enrolled into longitudinal studies with prospective follow-up from birth (60–62). Written informed consent was received from participants before inclusion in the Munich Bioresource Project (approval no. 5049/11; Technische Universität München). The age of islet autoantibody seroconversion (onset of islet autoimmunity) was documented. Blood samples from children with newly manifest T1D were collected from the German new onset diabetes in the young incident cohort study (approval no. 08043; Ethikkommission der Bayerischen Landesärztekammer) (32, 33). Blood samples consisted of venous blood collected in sodium heparin tubes, and blood volumes collected were based on European Union guidelines, with a maximal blood volume of 2.4 mL/kg body weight. Subjects have been stratified based on the presence or absence of multiple islet autoantibodies (with or without pre-T1D) and based on the duration of islet autoantibody positivity as follows: no autoimmunity: first-degree relatives of patients with T1D who are islet autoantibody-negative [n = 11, median age = 8 y, interquartile range (IQR) = 6–15 y, six males and five females]; recent onset of islet autoimmunity: subjects with multiple islet autoantibodies for less than 5 y (n = 8, median age = 7 y, IQR = 4–14 y, six males and two females); persistent autoimmunity: subjects with multiple islet autoantibodies for more than 5 but less than 10 y (n = 9, median age = 12 y, IQR = 9.25–17.5 y, seven males and two females); and long-term autoimmunity: subjects with multiple islet autoantibodies for more than a decade who had not yet developed T1D (n = 7, median age = 15 y, IQR = 14–25 y, three males and four females). For the blood samples, the investigators were not blinded to the presence and duration of assessed islet autoimmunity of children as described above during analyses of ex vivo T-cell phenotypes.
Human Cell Isolation.
Peripheral blood mononuclear cells (PBMCs) were isolated by density centrifugation over Ficoll–Paque PLUS (GE Healthcare). For TFH-cell induction assays, human CD4+ T cells were isolated from fresh PBMCs via positive magnetic bead enrichment (CD4 microbeads; Miltenyi Biotec) and B cells were isolated from the flow-through using CD19 microbeads (Miltenyi Biotec) following the manufacturer’s protocol. For tetramer stainings, human CD4+ T cells were isolated by negative enrichment from fresh PBMCs (EasySep Human CD4+ T-Cell Isolation Kit; Stemcell).
Cell Staining, Flow Cytometry, and Cell Sorting.
A description of monoclonal antibodies used for FACS stainings can be found in SI Materials and Methods.
Cells were acquired on a BD FACS Aria III cell sorting system flow cytometer using FACS Diva software (both from BD Biosciences) with optimal compensation and gain settings determined for each experiment based on unstained and single-color–stained samples. Doublets were excluded based on side scatter area (SSC-A) vs. side scatter width (SSC-W) plots. Live-cell populations were gated on the basis of cell side and forward scatter, as well as the exclusion of cells positive for Sytox Blue (Life Technologies). At least 50,000 gated events were acquired for each sample and analyzed using FlowJo Software Version 7.6.1 (TreeStar, Inc.).
Insulin-Specific HLA-DQ8–Restricted Tetramer Stainings.
Fluorescent HLA-DQ8 tetramers based on insulin B-chain 10–23 mimetopes were developed in collaboration with the NIH tetramer facility. Specifically, a 14E-22E tetramer and a 14E-21G-22E-HLA-DQ8-phycoerythrin (PE)–labeled tetramer were combined in stainings. Untouched CD4+ T cells (as described above) were incubated with tetramers for 1 h at 37 °C in humidified 10% (vol/vol) CO2, with gentle agitation every 20 min. Surface marker staining was performed directly afterward for 20 min at 4 °C. A set of exclusion markers [CD8, CD11b, CD19, CD14, and a dead cell exclusion marker (Sytox)] was used to increase specificity of the staining. As negative controls, we used a combination of two HLA-DQ8 tetramers fused to irrelevant peptides (PVSKMRMATPLLMQA and QDLELSWNLNGLQADL) and labeled with PE. Virtually no CD4+ T cells were detected with the control tetramers.
Insulin-Specific IAg7-Restricted Tetramer Stainings.
Fluorescent IAg7-tetramers based on insulin B-chain 10–23 mimetopes were received from the NIH tetramer core facility. Specifically a 22E tetramer and a 21G-22E-IAg7-PE–labeled tetramer were combined in stainings. A set of exclusion markers [CD8a, CD11b, CD11c, CD14, B220, F4/80, and a dead cell exclusion marker (Sytox)] was used to increase specificity of the staining. As negative controls, we used one IAg7 tetramer fused to an irrelevant peptide (AMKRHGLDNYRGYSL) and labeled with PE (31).
Isolation and Processing of Small RNAs/miRNAs.
Small RNAs/miRNAs were isolated using the miRNeasy Micro Kit (Qiagen). RNA concentration and purity were determined using a NanoDrop (Epoch; Biotech) and/or RNA Nano Chip and Agilent 2100 Bioanalyzer (Agilent Technologies). For cDNA synthesis, the Universal cDNA Synthesis Kit II (Exiqon) was used according to the instructions. Quantitative PCR (qPCR) was performed using ExiLENT SYBR Green PCR Master Mix (Exiqon) in combination with miRCURY LNA primers for miRNA92a-3p, miRNA19a-3p, and miRNA18a-5p. For normalization, miRCURY LNA primers for the housekeeper 5s rRNA were used (Exiqon). The reaction was run on a CFX96 real-time system (BioRad).
Isolation and Processing of mRNAs.
mRNAs were isolated using the miRNeasy Micro Kit (Qiagen). cDNA synthesis was performed with either the SMARTer Ultra Low Input RNA Kit for Sequencing Version 3 (Takara Clontech) when cell material was limited or the iScript cDNA Synthesis Kit (BioRad) when cell material was not limited. For the SMARTer kit, cycle numbers for amplification were adjusted for the lower RNA content of T cells. For qPCR, SsoFast Evagreen Supermix (BioRad) was used (primer information can be found in SI Materials and Methods). For normalization, the primers for the housekeeping genes 18s (Quantitect Primer Assay; Qiagen) and Histone (forward: ACT GGC TAC AAA AGC CGC TC, reverse: ACT TGC CTC CTG CAA AGC AC) were used. The reactions were run on a CFX96 Real-Time System.
In Vitro TFH Precursor Cell Induction Assay.
Naive T cells were sort-purified as CD4+, CD3+, CD45RA+, CD45RO−, and CXCR5−, and memory B cells were sorted as CD20+ and CD27+. Both cell types were cocultured at a ratio of 1:1. The cells were stimulated with 5 μg/mL anti-CD3 (Okt3) and anti-CD28 (CD28.2), both from Biolegend; in some experiments, IL-6 and IL-21, both at 50 ng/mL (Peprotech), were added in accordance with previously established procedures (16, 21). On day 5, CD4+ T cells were analyzed by flow cytometry on a BD FACS Aria III for expression of CXCR5, PSGL1, PD-1, and CCR7. In some experiments, PMA/ionomycin stimulation (PMA at 50 ng/mL and ionomycin at 1 μg/mL) was added to the assay 12 h before analysis. TFH induction assays have likewise been performed in presence of a PI3K inhibitor (LY294002 at 10 μM) or a PTEN inhibitor (SF1670 at 325 nM), respectively.
Application of miRNA92a Mimic, Antagomir, and KLF2 TSB to Human CD4+ T Cells.
PLGA-coated nanoparticles, loaded with miRNA92a mimic (miRCURY LNA microRNA Mimic; Exiqon) were added to the wells of the TFH differentiation assay at a ratio of 1:50 mimic/nanoparticles. The final concentration of mimic was 0.75 ng/μL per 100,000 cells (referred to as “high”). Titration was performed, with “medium” referring to 0.375 ng/μL per 100,000 cells and “low” referring to 0.1875 ng/μL per 100,000 cells. For the miRNA92a antagomir, the final concentration was 2 ng/μL per 100,000 cells. Control experiments were performed with nanoparticles and negative miRCURY LNA microRNA Mimic controls. The guide strands were found to have no homology to any known miRNA or mRNA sequences in mouse, rat, or human (63). For investigation of KLF2 as a possible target of miRNA92a, a KLF2 TSB or control TSB (custom-designed by Exiqon) was added to the assay at 2 ng/μL per 100,000 cells.
Mice.
For ex vivo analysis of miRNA92a abundance in CD4+ T cells, pancreatic lymph nodes from NOD/ShiLtJ mice (The Jackson Laboratory) (according to their insulin autoantibody status; the insulin autoantibody assay protocol is described in SI Materials and Methods) or age-matched BALB/c controls were ground through 70-μm cell strainers, stained with antibodies (Materials and Methods, Cell Staining, Flow Cytometry, and Cell Sorting), and magnetic activated cell sorting (MACS)-purified using CD4-Biotin and Streptavidin Microbeads (Miltenyi Biotec). Total CD4+ T cells were sort-purified, and miRNA abundance was determined as described above.
For the in vivo inhibition of miRNA92a, age-matched, insulin autoantibody-positive female NOD/ShiLtJ mice (The Jackson Laboratory) were randomized into two groups. For short-term treatment, the mice were injected i.p. with 10 mg/kg custom-designed in vivo LNA miRNA92a antagomir (Exiqon) or control antagomir on day 0, day 2, and day 6. Analysis of insulin-specific or polyclonal TFH frequencies was performed from peripheral blood, pancreatic lymph nodes, and all remaining lymph nodes on day 7. In addition, pancreata of the treated mice and control mice were analyzed for the activation status of infiltrating T cells on day 7. Therefore, pancreata were digested in 1 mg/mL Collagenase V for 5 min at 37 °C and passed through a 100 μM cell strainer. For long-term treatment, the mice were injected i.p. with 5 mg/kg in vivo miRNA92a antagomir or control antagomir every second day for 14 d. Insulin-specific or polyclonal TFH frequencies were analyzed in peripheral blood and pancreatic lymph nodes on day 14. To analyze pancreas pathology, pancreata were embedded in Cryomold and frozen on dry ice for cryosections. For analysis of the in vivo effect of miRNA92a on the human immune cells, NOD.Cg-Prkdcscid H2-Ab1tm1Gru Il2rgtm1Wjl Tg (HLA-DQA1, HLA-DQB1)1Dv//Sz (NSG HLA-DQ8 Tg) mice, developed by Leonard Shultz, The Jackson Laboratory, Bar Harbor, ME, were used. These mice lack mouse MHC class II, although they express the human HLA-DQ8 transgenically. NSG HLA-DQ8 mice are immunodeficient and develop a human immune system after reconstitution with human hematopoietic cells. Donors for reconstitution were sex-matched. The treatment of humanized mice with the in vivo miRNA92a antagomir was performed with three i.p. injections of 10 mg/kg over 7 d. No animals were excluded due to illness or outlier results; therefore, no exclusion determination was required.
Ethical approval for all mouse experimentation has been received by the District Government of Upper Bavaria, Munich, Germany (approval nos. 5.2-1-54-2532-81-12 and 55.2-1-54-2532-84-12). The investigators were not blinded to group allocation during the in vivo experiments or to the assessment of experimental end points.
Histopathology of NOD Pancreata.
Pancreata were embedded with TissueTek O.C.T. Compound and frozen on dry ice, and serial sections were stained with hematoxylin and eosin. Insulitis scoring was performed as previously described (64, 65). The following scores were assigned: 0, intact islets/no lesions; 1, periislet infiltrates; 2, <25% islet destruction; 3, >25% islet destruction; and 4, complete islet destruction.
Immunofluorescence.
Immunofluorescence staining was done using rabbit–anti-mouse insulin antibodies (Cell Signaling) and donkey–anti-rabbit Alexa647 antibodies (Dianova). For CD4 staining, rat–anti-mouse (Becton, Dickinson and Company) antibodies were used, followed by goat–anti-rat AlexaFluor488 (Dianova). For FOXP3 staining, cells were incubated with rat–anti-mouse antibodies (eBioscience) and goat–anti-ratbio (Becton, Dickinson and Company) combined with TSA Cyanine3 amplification (PerkinElmer). Nuclei were counterstained with HOECHST 33342 dye (Invitrogen). Negative control slides were incubated with secondary antibodies. Cells were analyzed by confocal microscopy (Olympus).
Statistics.
Results are presented as the mean and SEM or as percentages, where appropriate. For normally distributed data, the Student’s t test for unpaired values was used to compare means between independent groups and the Student’s t test for paired values was used to compare values for the same sample or subject tested under different conditions. The nonparametric Wilcoxon signed-ranks test was applied when data did not show a Gaussian distribution. Group size estimations were based upon a power calculation to yield minimally an 80% chance to detect a significant difference in the respective parameter of P < 0.05 between the relevant groups. For all tests, a two-tailed P value of <0.05 was considered to be significant. Statistical significance is shown as *P < 0.05, **P < 0.01, ***P < 0.001, or not significant (P > 0.05). Analyses were performed using the programs GraphPad Prism 6 and the Statistical Package for the Social Sciences (SPSS 19.0; SPSS, Inc.).
SI Materials and Methods
Antibodies for Flow Cytometry.
The following monoclonal antibodies were used for human FACS staining: anti-CD45RO allophycocyanin (APC)-H7 (UCHL1), anti-CD4 V500 (RPA-T4), anti-PD1 peridinin chlorophyll protein (PerCP)-Cy5.5 (EH12.1), and anti-CD45 APC-H7 (2D1) (all from BD Biosciences) and anti-CD45RA FITC (HI100), anti-CD8a Pacific Blue (RPA-T8), anti-CD11b Pacific Blue (ICRF44), anti-CD14 Pacific Blue (HCD14), anti-CD19 Pacific Blue (HIB19), anti-CCR7 PE-Cy7 (G043H7), anti-CXCR5 APC (J252D4), anti-CD3 Alexa Fluor 700 (HIT3a), anti-CXCR3 APC-Cy7 (G025H7), anti-CXCR3 PerCP-Cy5.5 (G025H7), anti-CCR6 PE-Cy7 (G034E3), anti-ICOS PE (C398.4A), anti-PSGL1 PE (KPL-1), anti-CD20 FITC (2H7), and anti-CD27 APC (M-T271) (all from Biolegend). For murine FACS staining, the following monoclonal antibodies were used: anti-CD4 Biotin (GK1.5), anti-CD14 BD Horizon 450 (rmC5-3), and anti-Bcl6 Alexa Fluor 647 (K112-91) (all from BD Biosciences); anti-CD44 PE (IM7), anti-CD25 PerCP-Cy5.5 (PC61), anti-ICOS FITC (C398.4A), anti-CXCR5 PerCP-Cy5.5 (L138D1), anti–PD-1 Alexa Fluor 780 (J43), anti-CD4 Alexa Fluor 700 (RM4-5), anti-CD8a Pacific Blue (53-6.7), anti-CD11b Pacific Blue (M1/70), anti-B220 Pacific Blue (RA3-6B2), anti-F4/80 Pacific Blue (BM8), and anti-CD11c Brilliant Violet 421 (N418) (all from Biolegend); and anti-CCR7 PE (4B12) and anti-Foxp3 FITC (FJK-16s) (both from eBiosciences).
Primers.
For RT-qPCR analysis of gene expression, the following primers were used: QuantiTect Primer Assays (Qiagen) for IFN-γ, IL-13, IL-10, CXCR5, ICOS, ITCH, Bcl6, KLF2, PTEN, Foxo1, S1P1R, PHLPP2, and CTLA4 and PrimePCR PreAmp for SYBR Green Assay (BioRad) for Ascl2, IL-17a, and IL-4. The sequence for the primers for ICOS is as follows: forward (Fwd): GCA CGA CCC TAA CGG TGA AT and reverse (Rev): GAA AAC TGG CCA ACG TGC TT, and the sequence for the primers for IL-21 is as follows: Fwd: CTC CCA AGG TCA AGA TCG CC and Rev: TGG CAG AAA TTC AGG GAC CA.
Chitosan-Coated PLGA Nanoparticle Preparation and Characterization.
The nanoparticles were produced following the method described by Ravi Kumar et al. (39) using PLGA Resomer RG 752H (Evonik) and Protasan UP CL 113 Chitosan (NovaMatrix). For confocal imaging, the nanoparticles were produced with a 5-fluorescineamine (FA)-PLGA conjugate. The measured hydrodynamic diameter was 146.7 ± 0.8 nm (152.8 ± 1.2 nm), the polydispersity index was 0.068 ± 0.009 (0.056 ± 0.007), and the zeta potential was +29.6 ± 0.3 mV (+29.6 ± 0.7 mV) for the chitosan-coated PLGA nanoparticles and FA-labeled nanoparticles.
Analysis of miRNA/Nanoparticles Uptake by Confocal Imaging of CD4+ T-Cell Cytospins.
Chitosan-coated PLGA nanoparticles (∼130 nm in size) with or without fluorescent label (FA) were obtained from Claus-Michael Lehr at the Department of Drug Delivery, Helmholtz Institute for Pharmaceutical Research Saarland, Saarland University, Germany. Successful uptake and delivery of miRNA to human and murine CD4+ T cells was determined: Respective naive CD4+ T cells were stimulated with anti-CD3/anti-CD28 for 18 h in the presence of FA-labeled nanoparticles complexed with miRIDIAN miRNA transfection control (Dy547). The uptake was initially confirmed by identification of a clearly distinguishable FA+ population using FACS analysis. Additionally the uptake was confirmed by confocal imaging: Human naive CD4+ T cells were stimulated as described above for 18 h in the presence or absence of nanoparticles and labeled miRIDIAN miRNA mimic transfection control (Dy547). After the stimulation, CD4+ T cells were washed and fixed. Upon generation of cytospins, intracellular localization of nanoparticles and delivered miRNA were assessed by confocal microscopy.
Insulin Autoantibody Assay.
Insulin autoantibodies were determined using an ELISA system as described previously. In brief, high-binding, 96-well plates (Costar) were coated with human recombinant insulin (10 μg/mL; Sigma–Aldrich) overnight at 4 °C. Unspecific blocking was done using PBS containing 2% BSA for 2 h at room temperature. Preincubated serum (diluted 1:10) with or without insulin competition was added and incubated for 2 h at room temperature. After four wash steps, biotinylated anti-mouse IgG1 (Abcam) diluted 1:10,000 in PBS/BSA was added for 30 min at room temperature. After washing the plate, horseradish peroxidase-labeled streptavidin was added for 15 min. The plate was washed five times, and tetramethylbenzidine (TMB) substrate solution was added (OptEIA reagent set; Becton, Dickinson and Company). The reaction was stopped using sulfuric acid, and fluorescent intensity was determined in the Epoch plate reader (Biotech). Each sample was run in duplicate with and without competition using human insulin. For each sample, an index was calculated based on the mean of the results.
To determine levels of IAA in NOD mice, a Protein A/G radiobinding assay based on 125I-labeled recombinant human insulin was applied as previously described (62). Serum from nonautoimmune prone BALB/c mice was used as negative control.
Acknowledgments
We thank Ruth Chmiel and Susanne Hummel for blood sample collection and patient follow-up; Melanie Spornraft and the European Molecular Biology Laboratory Genecore facility, especially Vladimir Benes, for providing reagents and analysis tools; Peter Achenbach and Claudia Matzke for supporting IAA measurements in NOD mice; and M. H. Tschöp for critical reading of the manuscript. B.W. is supported by Grant WE 4656/2 and Deutsche Forschungsgemeinschaft (DFG) CRC1811 (B02). C.D. is supported by the Young Investigator Group (Helmholtz Zentrum München) and received support through an associated membership in the CRC1054 of the DFG. The work was supported by grants from the Juvenile Diabetes Research Foundation (JDRF 2-SRA-2014-161-Q-R to C.D. and A.-G.Z.), JDRF 17-2012-16 (to A.G.Z.), and JDRF 6-2012-20 (to A.G.Z.); the Kompetenznetz Diabetes Mellitus (Competence Network for Diabetes Mellitus), funded by the Federal Ministry of Education and Research Grants (FKZ 01GI0805-07 and FKZ 01GI0805); and the German Center for Diabetes Research. A.-G.Z. is Principal Investigator of human birth cohort studies.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606646113/-/DCSupplemental.
References
- 1.Bluestone JA, Herold K, Eisenbarth G. Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature. 2010;464(7293):1293–1300. doi: 10.1038/nature08933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ziegler AG, Nepom GT. Prediction and pathogenesis in type 1 diabetes. Immunity. 2010;32(4):468–478. doi: 10.1016/j.immuni.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Insel RA, et al. Staging presymptomatic type 1 diabetes: A scientific statement of JDRF, the Endocrine Society, and the American Diabetes Association. Diabetes Care. 2015;38(10):1964–1974. doi: 10.2337/dc15-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Unanue ER. Antigen presentation in the autoimmune diabetes of the NOD mouse. Annu Rev Immunol. 2014;32:579–608. doi: 10.1146/annurev-immunol-032712-095941. [DOI] [PubMed] [Google Scholar]
- 5.Ziegler AG, et al. Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA. 2013;309(23):2473–2479. doi: 10.1001/jama.2013.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silva DG, et al. Anti-islet autoantibodies trigger autoimmune diabetes in the presence of an increased frequency of islet-reactive CD4 T cells. Diabetes. 2011;60(8):2102–2111. doi: 10.2337/db10-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hale JS, Ahmed R. Memory T follicular helper CD4 T cells. Front Immunol. 2015;6:16. doi: 10.3389/fimmu.2015.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ansel KM, McHeyzer-Williams LJ, Ngo VN, McHeyzer-Williams MG, Cyster JG. In vivo-activated CD4 T cells upregulate CXC chemokine receptor 5 and reprogram their response to lymphoid chemokines. J Exp Med. 1999;190(8):1123–1134. doi: 10.1084/jem.190.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flynn S, Toellner KM, Raykundalia C, Goodall M, Lane P. CD4 T cell cytokine differentiation: The B cell activation molecule, OX40 ligand, instructs CD4 T cells to express interleukin 4 and upregulates expression of the chemokine receptor, Blr-1. J Exp Med. 1998;188(2):297–304. doi: 10.1084/jem.188.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumjohann D, et al. Persistent antigen and germinal center B cells sustain T follicular helper cell responses and phenotype. Immunity. 2013;38(3):596–605. doi: 10.1016/j.immuni.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 11.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325(5943):1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stone EL, et al. ICOS coreceptor signaling inactivates the transcription factor FOXO1 to promote Tfh cell differentiation. Immunity. 2015;42(2):239–251. doi: 10.1016/j.immuni.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao N, et al. The E3 ubiquitin ligase Itch is required for the differentiation of follicular helper T cells. Nat Immunol. 2014;15(7):657–666. doi: 10.1038/ni.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, et al. The transcription factor Foxp1 is a critical negative regulator of the differentiation of follicular helper T cells. Nat Immunol. 2014;15(7):667–675. doi: 10.1038/ni.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JY, et al. The transcription factor KLF2 restrains CD4⁺ T follicular helper cell differentiation. Immunity. 2015;42(2):252–264. doi: 10.1016/j.immuni.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Locci M, et al. International AIDS Vaccine Initiative Protocol C Principal Investigators Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39(4):758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morita R, et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34(1):108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bentebibel SE, et al. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Sci Transl Med. 2013;5(176):176ra32. doi: 10.1126/scitranslmed.3005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmitt N, Bentebibel SE, Ueno H. Phenotype and functions of memory Tfh cells in human blood. Trends Immunol. 2014;35(9):436–442. doi: 10.1016/j.it.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ueno H, Banchereau J, Vinuesa CG. Pathophysiology of T follicular helper cells in humans and mice. Nat Immunol. 2015;16(2):142–152. doi: 10.1038/ni.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He J, et al. Circulating precursor CCR7(lo)PD-1(hi) CXCR5⁺ CD4⁺ T cells indicate Tfh cell activity and promote antibody responses upon antigen reexposure. Immunity. 2013;39(4):770–781. doi: 10.1016/j.immuni.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 22.Scherm MG, Ott VB, Daniel C. Follicular helper T cells in autoimmunity. Curr Diab Rep. 2016;16(8):75. doi: 10.1007/s11892-016-0770-2. [DOI] [PubMed] [Google Scholar]
- 23.Xiao S, et al. Retinoic acid increases Foxp3+ regulatory T cells and inhibits development of Th17 cells by enhancing TGF-beta-driven Smad3 signaling and inhibiting IL-6 and IL-23 receptor expression. J Immunol. 2008;181(4):2277–2284. doi: 10.4049/jimmunol.181.4.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuipers H, Schnorfeil FM, Brocker T. Differentially expressed microRNAs regulate plasmacytoid vs. conventional dendritic cell development. Mol Immunol. 2010;48(1-3):333–340. doi: 10.1016/j.molimm.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 25.Kuipers H, Schnorfeil FM, Fehling HJ, Bartels H, Brocker T. Dicer-dependent microRNAs control maturation, function, and maintenance of Langerhans cells in vivo. J Immunol. 2010;185(1):400–409. doi: 10.4049/jimmunol.0903912. [DOI] [PubMed] [Google Scholar]
- 26.Turner ML, Schnorfeil FM, Brocker T. MicroRNAs regulate dendritic cell differentiation and function. J Immunol. 2011;187(8):3911–3917. doi: 10.4049/jimmunol.1101137. [DOI] [PubMed] [Google Scholar]
- 27.Cobb BS, et al. A role for Dicer in immune regulation. J Exp Med. 2006;203(11):2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cobb BS, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201(9):1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9(4):405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baumjohann D, et al. The microRNA cluster miR-17∼92 promotes TFH cell differentiation and represses subset-inappropriate gene expression. Nat Immunol. 2013;14(8):840–848. doi: 10.1038/ni.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serr I, et al. Type 1 diabetes vaccine candidates promote human Foxp3(+)Treg induction in humanized mice. Nat Commun. 2016;7:10991. doi: 10.1038/ncomms10991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thümer L, et al. German new onset diabetes in the young incident cohort study: DiMelli study design and first-year results. Rev Diabet Stud. 2010;7(3):202–208. doi: 10.1900/RDS.2010.7.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warncke K, et al. Does diabetes appear in distinct phenotypes in young people? Results of the diabetes mellitus incidence Cohort Registry (DiMelli) PLoS One. 2013;8(9):e74339. doi: 10.1371/journal.pone.0074339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong N, Wang X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43(Database issue):D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 36.Kang SG, et al. MicroRNAs of the miR-17∼92 family are critical regulators of T(FH) differentiation. Nat Immunol. 2013;14(8):849–857. doi: 10.1038/ni.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang P, Rao EY, Meng N, Zhao Y, Wang JJ. MicroRNA-17-92 significantly enhances radioresistance in human mantle cell lymphoma cells. Radiat Oncol. 2010;5:100. doi: 10.1186/1748-717X-5-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ravi Kumar MN, Bakowsky U, Lehr CM. Preparation and characterization of cationic PLGA nanospheres as DNA carriers. Biomaterials. 2004;25(10):1771–1777. doi: 10.1016/j.biomaterials.2003.08.069. [DOI] [PubMed] [Google Scholar]
- 40.Bramsen JB, et al. Improved silencing properties using small internally segmented interfering RNAs. Nucleic Acids Res. 2007;35(17):5886–5897. doi: 10.1093/nar/gkm548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma CS, Deenick EK, Batten M, Tangye SG. The origins, function, and regulation of T follicular helper cells. J Exp Med. 2012;209(7):1241–1253. doi: 10.1084/jem.20120994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Boehmer H, Daniel C. Therapeutic opportunities for manipulating T(Reg) cells in autoimmunity and cancer. Nat Rev Drug Discov. 2013;12(1):51–63. doi: 10.1038/nrd3683. [DOI] [PubMed] [Google Scholar]
- 43.Kenefeck R, et al. Follicular helper T cell signature in type 1 diabetes. J Clin Invest. 2015;125(1):292–303. doi: 10.1172/JCI76238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferreira RC, et al. IL-21 production by CD4+ effector T cells and frequency of circulating follicular helper T cells are increased in type 1 diabetes patients. Diabetologia. 2015;58(4):781–790. doi: 10.1007/s00125-015-3509-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rekers NV, von Herrath MG, Wesley JD. Immunotherapies and immune biomarkers in Type 1 diabetes: A partnership for success. Clin Immunol. 2015;161(1):37–43. doi: 10.1016/j.clim.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 46.Simpson LJ, Ansel KM. MicroRNA regulation of lymphocyte tolerance and autoimmunity. J Clin Invest. 2015;125(6):2242–2249. doi: 10.1172/JCI78090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Janssen HL, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368(18):1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 48.Murugaiyan G, et al. MicroRNA-21 promotes Th17 differentiation and mediates experimental autoimmune encephalomyelitis. J Clin Invest. 2015;125(3):1069–1080. doi: 10.1172/JCI74347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc Natl Acad Sci USA. 2009;106(44):18704–18709. doi: 10.1073/pnas.0905063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gigoux M, et al. Inducible costimulator promotes helper T-cell differentiation through phosphoinositide 3-kinase. Proc Natl Acad Sci USA. 2009;106(48):20371–20376. doi: 10.1073/pnas.0911573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rolf J, et al. Phosphoinositide 3-kinase activity in T cells regulates the magnitude of the germinal center reaction. J Immunol. 2010;185(7):4042–4052. doi: 10.4049/jimmunol.1001730. [DOI] [PubMed] [Google Scholar]
- 53.Xu H, et al. Follicular T-helper cell recruitment governed by bystander B cells and ICOS-driven motility. Nature. 2013;496(7446):523–527. doi: 10.1038/nature12058. [DOI] [PubMed] [Google Scholar]
- 54.Du P, Ma X, Wang C. Associations of CTLA4 gene polymorphisms with Graves’ ophthalmopathy: A meta-analysis. Int J Genomics. 2014;2014:537969. doi: 10.1155/2014/537969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sebzda E, Zou Z, Lee JS, Wang T, Kahn ML. Transcription factor KLF2 regulates the migration of naive T cells by restricting chemokine receptor expression patterns. Nat Immunol. 2008;9(3):292–300. doi: 10.1038/ni1565. [DOI] [PubMed] [Google Scholar]
- 56.Carlson CM, et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442(7100):299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 57.Weber JP, et al. ICOS maintains the T follicular helper cell phenotype by down-regulating Krüppel-like factor 2. J Exp Med. 2015;212(2):217–233. doi: 10.1084/jem.20141432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kerdiles YM, et al. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol. 2009;10(2):176–184. doi: 10.1038/ni.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fabre S, et al. FOXO1 regulates L-Selectin and a network of human T cell homing molecules downstream of phosphatidylinositol 3-kinase. J Immunol. 2008;181(5):2980–2989. doi: 10.4049/jimmunol.181.5.2980. [DOI] [PubMed] [Google Scholar]
- 60.Ziegler AG, Bonifacio E. BABYDIAB-BABYDIET Study Group Age-related islet autoantibody incidence in offspring of patients with type 1 diabetes. Diabetologia. 2012;55(7):1937–1943. doi: 10.1007/s00125-012-2472-x. [DOI] [PubMed] [Google Scholar]
- 61.Ziegler AG, Hummel M, Schenker M, Bonifacio E. Autoantibody appearance and risk for development of childhood diabetes in offspring of parents with type 1 diabetes: The 2-year analysis of the German BABYDIAB Study. Diabetes. 1999;48(3):460–468. doi: 10.2337/diabetes.48.3.460. [DOI] [PubMed] [Google Scholar]
- 62.Achenbach P, et al. Autoantibodies to zinc transporter 8 and SLC30A8 genotype stratify type 1 diabetes risk. Diabetologia. 2009;52(9):1881–1888. doi: 10.1007/s00125-009-1438-0. [DOI] [PubMed] [Google Scholar]
- 63.Kozomara A, Griffiths-Jones S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jaeckel E, Lipes MA, von Boehmer H. Recessive tolerance to preproinsulin 2 reduces but does not abolish type 1 diabetes. Nat Immunol. 2004;5(10):1028–1035. doi: 10.1038/ni1120. [DOI] [PubMed] [Google Scholar]
- 65.Krishnamurthy B, et al. Responses against islet antigens in NOD mice are prevented by tolerance to proinsulin but not IGRP. J Clin Invest. 2006;116(12):3258–3265. doi: 10.1172/JCI29602. [DOI] [PMC free article] [PubMed] [Google Scholar]