Significance
Many living organisms use terpenes for ecological interactions. Terpenes are biosynthesized by terpene synthases (TPSs), but classic TPS genes are known to exist only in plants and fungi among the eukaryotes. In this study, TPS genes were identified in six species of amoebae with five of them being multicellular social amoebae. Amoebal TPSs showed closer relatedness to fungal TPSs than bacterial TPSs. In the social amoeba Dictyostelium discoideum, all nine TPS genes encoded active enzymes and most of their terpene products were released as volatiles in a development-specific manner. This study highlights a wider distribution of TPS genes in eukaryotes than previously thought and opens a door to studying the function and evolution of TPS genes and their products.
Keywords: terpene synthases, amoebae, volatiles, evolution, chemical ecology
Abstract
Terpenes are structurally diverse natural products involved in many ecological interactions. The pivotal enzymes for terpene biosynthesis, terpene synthases (TPSs), had been described only in plants and fungi in the eukaryotic domain. In this report, we systematically analyzed the genome sequences of a broad range of nonplant/nonfungus eukaryotes and identified putative TPS genes in six species of amoebae, five of which are multicellular social amoebae from the order of Dictyosteliida. A phylogenetic analysis revealed that amoebal TPSs are evolutionarily more closely related to fungal TPSs than to bacterial TPSs. The social amoeba Dictyostelium discoideum was selected for functional study of the identified TPSs. D. discoideum grows as a unicellular organism when food is abundant and switches from vegetative growth to multicellular development upon starvation. We found that expression of most D. discoideum TPS genes was induced during development. Upon heterologous expression, all nine TPSs from D. discoideum showed sesquiterpene synthase activities. Some also exhibited monoterpene and/or diterpene synthase activities. Direct measurement of volatile terpenes in cultures of D. discoideum revealed essentially no emission at an early stage of development. In contrast, a bouquet of terpenes, dominated by sesquiterpenes including β-barbatene and (E,E)-α-farnesene, was detected at the middle and late stages of development, suggesting a development-specific function of volatile terpenes in D. discoideum. The patchy distribution of TPS genes in the eukaryotic domain and the evidence for TPS function in D. discoideum indicate that the TPS genes mediate lineage-specific adaptations.
Terpenes constitute a structurally diverse class of natural products. They are synthesized from two universal precursors: isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), which are supplied by the mevalonate pathway and/or the methylerythritol phosphate pathway (1). From IPP and DMAPP, isoprenyl diphosphates of various chain lengths are produced by the action of isoprenyl diphosphate synthases (IDSs) (2). Among the many metabolic fates of isoprenyl diphosphates (3), they serve as substrates for terpene synthases, which convert isoprenyl diphosphates to different subclasses of terpenes of fascinating structural diversity, such as monoterpenes, sesquiterpenes, and diterpenes (4). The ability of an organism to produce terpenes depends on whether the organism contains terpene synthase genes.
Unlike IDS genes, which are ubiquitous in living organisms, the occurrence of terpene synthase genes and, thus, the production of terpenes appear to be lineage-specific. Presently, two general types of terpene synthases are recognized: classic terpene synthases (abbreviated as TPSs) and IDS-type terpene synthases. The majority of terpene synthases that have been characterized so far belongs to the classic TPSs. In prokaryotes, classic TPS genes are widely distributed in bacteria (5, 6), whereas none has been observed in archaea. In eukaryotes, classic TPS genes had been found only in land plants (7, 8) and fungi (9, 10), whereas the IDS-type terpene synthases have been identified recently in two species of insects (11, 12). Sequence analysis of these insect genes suggests that they have evolved recently from insect IDSs (12), whereas classic TPSs probably also evolved from IDSs, but anciently (13). TPS genes are major contributors to the chemical diversity exhibited by living organisms, so it is important to understand their distribution and evolution.
In the current global tree of eukaryotes, a domain that is composed of diverse organisms, the five supergroups Opisthokonta, Amoebozoa, Excavata, Archaeplastida, and SAR (stramenopiles + alveolates + Rhizaria) are recognized (14, 15). Only the supergroup Archaeplastida, which contains land plants, and Opisthokonta, which contains fungi, are known to contain classic TPS genes. It has been accepted that classic TPS genes are absent in insects (12), which are in the supergroup of Opisthokonta. The presence/absence of TPS genes in other eukaryotes has not been systematically investigated. Terpenes serve diverse functions in the organisms that produce them, including defense against predators and attraction of beneficial organisms (16), which implies that TPS genes play a role in evolutionary adaptations. The goals of this study were to systematically search for classic TPS genes in nonplant/nonfungus eukaryotes, infer their evolutionary relationship to known TPSs, and understand their biochemical and biological functions.
Results
Identification of Terpene Synthase Genes in Nonplant/Nonfungus Eukaryotes.
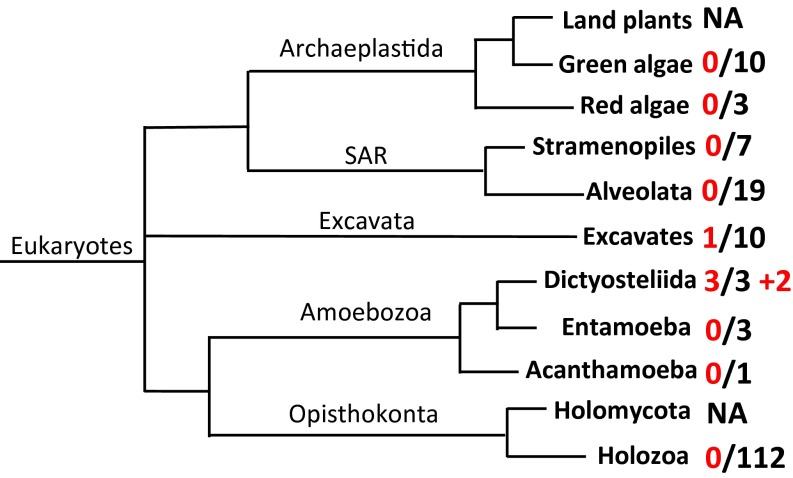
To determine the occurrence of TPS genes in eukaryotes other than plants and fungi, a HMMER search (17) was conducted by using a set of 168 well-annotated genomes (Table S1) of nonplant/nonfungus eukaryotes. Whereas the general absence of TPS genes in nonplant/nonfungus eukaryotes was confirmed, TPS genes were detected in the two supergroups of Amoebozoa and Excavata (Fig. 1). Among the seven species of Amoebozoa analyzed (Table S1), all three species from the genus Dictyostelium, Dictyostelium discoideum, Dictyostelium fasciculatum, and Dictyostelium purpureum, were found to contain TPS genes, whereas no TPS genes were found in the other four species Entamoeba histolytica, Entamoeba dispar, Entamoeba invadens, and Acanthamoeba castellanii. Among the 10 species of Excavata with sequenced genomes (Table S1), TPS genes were found only in Naegleria gruberi (Fig. 1). A search against the nonredundant (nr) database of National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov) revealed that TPS genes are also present in two additional species of eukaryotes, Polysphondylium pallidum and Acytostelium subglobosum, both of which are classified as Amoebozoa (Fig. 1). Both genera Polysphondylium and Dictyostelium belong to the family Dictyosteliidae, whereas A. subglobosum is a species in the Actyosteliidae family. Both Dictyosteliidae and Actyosteliidae belong to the same order of Dictyosteliida (18). It should be noted that all of the six eukaryotic species that were found to contain TPS genes in this study are amoebae (19), with the five species from Dictyosteliida belonging to the unique group of multicellular social amoebae (18).
Table S1.
Eukaryotes with well-annotated genomes analyzed in this study
| Category | Species |
| Holozoa | Homo sapiens |
| Holozoa | Pan troglodytes |
| Holozoa | Pan paniscus |
| Holozoa | Gorilla gorilla gorilla |
| Holozoa | Pongo abelii |
| Holozoa | Nomascus leucogenys |
| Holozoa | Macaca mulatta |
| Holozoa | Macaca fascicularis |
| Holozoa | Rhinopithecus roxellana |
| Holozoa | Callithrix jacchus |
| Holozoa | Mus musculus |
| Holozoa | Rattus norvegicus |
| Holozoa | Cricetulus griseus |
| Holozoa | Nannospalax galili |
| Holozoa | Heterocephalus glaber |
| Holozoa | Oryctolagus cuniculus |
| Holozoa | Tupaia chinensis |
| Holozoa | Canis familiaris |
| Holozoa | Ailuropoda melanoleuca |
| Holozoa | Ursus maritimus |
| Holozoa | Felis catus |
| Holozoa | Panthera tigris altaica |
| Holozoa | Bos taurus |
| Holozoa | Bos mutus |
| Holozoa | Pantholops hodgsonii |
| Holozoa | Capra hircus |
| Holozoa | Ovis aries |
| Holozoa | Sus scrofa |
| Holozoa | Camelus ferus |
| Holozoa | Balaenoptera acutorostrata scammoni |
| Holozoa | Lipotes vexillifer |
| Holozoa | Equus caballus |
| Holozoa | Myotis brandtii |
| Holozoa | Myotis davidii |
| Holozoa | Pteropus alecto |
| Holozoa | Monodelphis domestica |
| Holozoa | Sarcophilus harrisii |
| Holozoa | Ornithorhynchus anatinus |
| Holozoa | Gallus gallus |
| Holozoa | Meleagris gallopavo |
| Holozoa | Anas platyrhynchos |
| Holozoa | Taeniopygia guttata |
| Holozoa | Geospiza fortis |
| Holozoa | Ficedula albicollis |
| Holozoa | Pseudopodoces humilis |
| Holozoa | Corvus cornix |
| Holozoa | Falco peregrinus |
| Holozoa | Falco cherrug |
| Holozoa | Columba livia |
| Holozoa | Alligator sinensis |
| Holozoa | Alligator mississippiensis |
| Holozoa | Pelodiscus sinensis |
| Holozoa | Chelonia mydas |
| Holozoa | Anolis carolinensis |
| Holozoa | Python bivittatus |
| Holozoa | Xenopus laevis |
| Holozoa | Xenopus tropicalis |
| Holozoa | Danio rerio |
| Holozoa | Takifugu rubripes |
| Holozoa | Maylandia zebra |
| Holozoa | Oryzias latipes |
| Holozoa | Xiphophorus maculatus |
| Holozoa | Latimeria chalumnae |
| Holozoa | Callorhinchus milii |
| Holozoa | Branchiostoma floridae |
| Holozoa | Ciona intestinalis |
| Holozoa | Strongylocentrotus purpuratus |
| Holozoa | Saccoglossus kowalevskii |
| Holozoa | Drosophila melanogaster |
| Holozoa | Drosophila pseudoobscura |
| Holozoa | Drosophila ananassae |
| Holozoa | Drosophila erecta |
| Holozoa | Drosophila persimilis |
| Holozoa | Drosophila sechellia |
| Holozoa | Drosophila simulans |
| Holozoa | Drosophila willistoni |
| Holozoa | Drosophila yakuba |
| Holozoa | Drosophila grimshawi |
| Holozoa | Drosophila mojavensis |
| Holozoa | Drosophila virilis |
| Holozoa | Musca domestica |
| Holozoa | Anopheles gambiae |
| Holozoa | Aedes aegypti |
| Holozoa | Culex quinquefasciatus |
| Holozoa | Apis mellifera |
| Holozoa | Solenopsis invicta |
| Holozoa | Acromyrmex echinatior |
| Holozoa | Harpegnathos saltator |
| Holozoa | Camponotus floridanus |
| Holozoa | Nasonia vitripennis |
| Holozoa | Tribolium castaneum |
| Holozoa | Bombyx mori |
| Holozoa | Plutella xylostella |
| Holozoa | Acyrthosiphon pisum |
| Holozoa | Pediculus humanus corporis |
| Holozoa | Ixodes scapularis |
| Holozoa | Caenorhabditis elegans |
| Holozoa | Caenorhabditis briggsae |
| Holozoa | Brugia malayi |
| Holozoa | Loa |
| Holozoa | Trichinella spiralis |
| Holozoa | Helobdella robusta |
| Holozoa | Lottia gigantea |
| Holozoa | Crassostrea gigas |
| Holozoa | Octopus bimaculoides |
| Holozoa | Schistosoma mansoni |
| Holozoa | Nematostella vectensis |
| Holozoa | Hydra vulgaris |
| Holozoa | Trichoplax adhaerens |
| Holozoa | Amphimedon queenslandica |
| Holozoa | Monosiga brevicollis |
| Holozoa | Salpingoeca rosetta |
| Green algae | Chlamydomonas reinhardtii |
| Green algae | Volvox carteri f. nagariensis |
| Green algae | Ostreococcus lucimarinus |
| Green algae | Ostreococcus tauri |
| Green algae | Bathycoccus prasinos |
| Green algae | Micromonas sp. RCC299 |
| Green algae | Micromonas pusilla |
| Green algae | Coccomyxa subellipsoidea |
| Green algae | Chlorella variabilis |
| Green algae | Auxenochlorella protothecoides |
| Red algae | Cyanidioschyzon merolae |
| Red algae | Galdieria sulphuraria |
| Red algae | Chondrus crispus |
| Dictyosteliida | Dictyostelium discoideum |
| Dictyosteliida | Dictyostelium purpureum |
| Dictyosteliida | Dictyostelium fasciculatum |
| Entamoeba | Entamoeba histolytica |
| Entamoeba | Entamoeba dispar |
| Entamoeba | Entamoeba invadens |
| Acanthamoeba | Acanthamoeba castellanii |
| Alveolata | Plasmodium falciparum 3D7 |
| Alveolata | Plasmodium falciparum Dd2 |
| Alveolata | Plasmodium falciparum HB3 |
| Alveolata | Plasmodium yoelii |
| Alveolata | Plasmodium chabaudi |
| Alveolata | Plasmodium berghei |
| Alveolata | Plasmodium knowlesi |
| Alveolata | Plasmodium vivax |
| Alveolata | Plasmodium cynomolgi |
| Alveolata | Theileria annulata |
| Alveolata | Theileria parva |
| Alveolata | Theileria orientalis |
| Alveolata | Theileria equi |
| Alveolata | Babesia bovis |
| Alveolata | Cryptosporidium parvum |
| Alveolata | Cryptosporidium hominis |
| Alveolata | Toxoplasma gondii |
| Alveolata | Tetrahymena thermophila |
| Alveolata | Paramecium tetraurelia |
| Stramenopiles | Phaeodactylum tricornutum |
| Stramenopiles | Thalassiosira pseudonana |
| Stramenopiles | Aureococcus anophagefferens |
| Stramenopiles | Nannochloropsis gaditana |
| Stramenopiles | Phytophthora infestans |
| Stramenopiles | Phytophthora sojae |
| Stramenopiles | Saprolegnia parasitica |
| Excavata | Trypanosoma brucei |
| Excavata | Trypanosoma cruzi |
| Excavata | Leishmania major |
| Excavata | Leishmania infantum |
| Excavata | Leishmania donovani |
| Excavata | Leishmania mexicana |
| Excavata | Leishmania braziliensis |
| Excavata | Naegleria gruberi |
| Excavata | Trichomonas vaginalis |
| Excavata | Giardia lamblia |
Fig. 1.
Distribution of terpene synthase (TPS) genes among the major lineages of eukaryotes with sequenced genomes. A total of 168 species (Table S1), which did not include any species from land plants and fungi (Holomycota), were analyzed. The phylogeny of eukaryotes was adapted from Adl et al. (14) and Burki (15) with five supergroups recognized: Opisthokonta, Amoebozoa, Excavata, Archaeplastida, and SAR (stramenopiles + alveolates + Rhizaria). The first number (before the slash) indicates the number of species in certain lineages that were determined to contain TPS genes. The second number (after the slash) indicates the total number of species in that lineage that were analyzed. NA, not analyzed. The “+2” indicates that two additional species from Amoebozoa were identified to contain TPS genes in the nonredundant database at NCBI.
There are significant variations in the number of TPS genes found in the genome of each of the five Dictyosteliida species. Whereas A. subglobosum contains a single TPS gene, the four species from Dictyosteliidae possess small gene families ranging from three TPS genes in D. fasciculatum to 21 TPS genes in P. pallidum (Table S2). The amoeba N. gruberi contains seven TPS genes. The number of introns for the TPS genes from Dictyosteliida ranges between 0 and 3, whereas in contrast, all of the TPS genes from N. gruberi are intronless (Table S2).
Table S2.
Summary of the identified eukaryotic terpene synthase genes
| Gene name | Gene ID | Protein size (amino acids) | Intron | Note |
| DdTPS1 | DDB_G0269428 | 345 | 0 | — |
| DdTPS2 | DDB_G0272510 | 349 | 0 | — |
| DdTPS3 | DDB_G0272706 | 362 | 0 | — |
| DdTPS4 | DDB_G0277385 | 335 | 0 | — |
| DdTPS5 | DDB_G0281467 | 347 | 3 | — |
| DdTPS6 | DDB_G0284707 | 361 | 0 | — |
| DdTPS7 | DDB_G0278647 | 339 | 2 | — |
| DdTPS8 | DDB_G0293666 | 312 | 0 | — |
| DdTPS9 | DDB_G0278279 | 339 | 2 | — |
| DdTPS10 | DDB_G0270514 | 297 | 1 | Pseudogene |
| DdTPS11 | DDB_G0272508 | 104 | 0 | Partial |
| DpTPS1 | DPU_G0053652 | 377 | 1 | — |
| DpTPS2 | DPU_G0053138 | 333 | 1 | — |
| DpTPS3 | DPU_G0056038 | 317 | 0 | — |
| DpTPS4 | DPU_G0067512 | 366 | 0 | — |
| DpTPS5 | DPU_G0067516 | 360 | 0 | — |
| DpTPS6 | DPU_G0067956 | 310 | 0 | — |
| DpTPS7 | DPU_G0070306 | 329 | 1 | — |
| DpTPS8 | DPU_G0074508 | 331 | 1 | — |
| DpTPS9 | DPU_G0052218 | 341 | 2 | — |
| DpTPS10 | — | 344 | 1 | — |
| DpTPS11 | — | 325 | 0 | — |
| DpTPS12 | — | 358 | 1 | — |
| DpTPS13 | DPU_G0074482 | 217 | 0 | Partial |
| PpTPS1 | PPA_G1297554 | 332 | 1 | — |
| PpTPS2 | PPA_G1299870 | 365 | 0 | — |
| PpTPS3 | PPA_G1324710 | 298 | 1 | — |
| PpTPS4 | PPA_G1325482 | 244 | 1 | — |
| PpTPS5 | PPA_G1335516 | 328 | 1 | — |
| PpTPS6 | PPA_G1340734 | 330 | 1 | — |
| PpTPS7 | PPA_G1352704 | 258 | 2 | — |
| PpTPS8 | PPA_G1297554 | 332 | 1 | — |
| PpTPS9 | PPA_G1369888 | 298 | 1 | — |
| PpTPS10 | PPA_G1369898 | 311 | 1 | — |
| PpTPS11 | PPA_G1369924 | 298 | 1 | — |
| PpTPS12 | PPA_G1370524 | 298 | 1 | — |
| PpTPS13 | PPA_G1378996 | 323 | 3 | — |
| PpTPS14 | PPA_G1386590 | 357 | 0 | — |
| PpTPS15 | PPA_G1394454 | 310 | 3 | — |
| PpTPS16 | PPA_G1402282 | 285 | 3 | — |
| PpTPS17 | — | 298 | 0 | — |
| PpTPS18 | PPA_G1419016 | 324 | 2 | — |
| PpTPS19 | PPA_G1420290 | 332 | 0 | — |
| PpTPS20 | PPA_G1425380 | 121 | 0 | Partial |
| PpTPS21 | PPA_G1425388 | 159 | 3 | Partial |
| DfaTPS1 | DFA_G1558546 | 338 | 1 | — |
| DfaTPS2 | DFA_G1589356 | 354 | 2 | — |
| DfaTPS3 | DFA_G1589330 | 123 | 1 | — |
| AsTPS1 | ADB0003110 | 332 | 1 | — |
| NgTPS1 | 8861934 | 341 | 0 | — |
| NgTPS2 | 8858387 | 418 | 0 | — |
| NgTPS3 | 8855007 | 374 | 0 | — |
| NgTPS4 | 8855311 | 322 | 0 | — |
| NgTPS5 | 8859416 | 369 | 0 | — |
| NgTPS6 | — | 390 | 0 | — |
| NgTPS7 | 8849513 | 390 | 0 | — |
As, Actyostelium subglobosum; Dd, Dictyostelium discoideum; Dfa, Dictyostelium fasciculatum; Dp, Dictyostelium purpureum; Ng, Naegleria gruberi; Pp, Polysphondylium pallidum.
Identified Eukaryotic Terpene Synthases: Evolutionary Relatedness and Motifs.
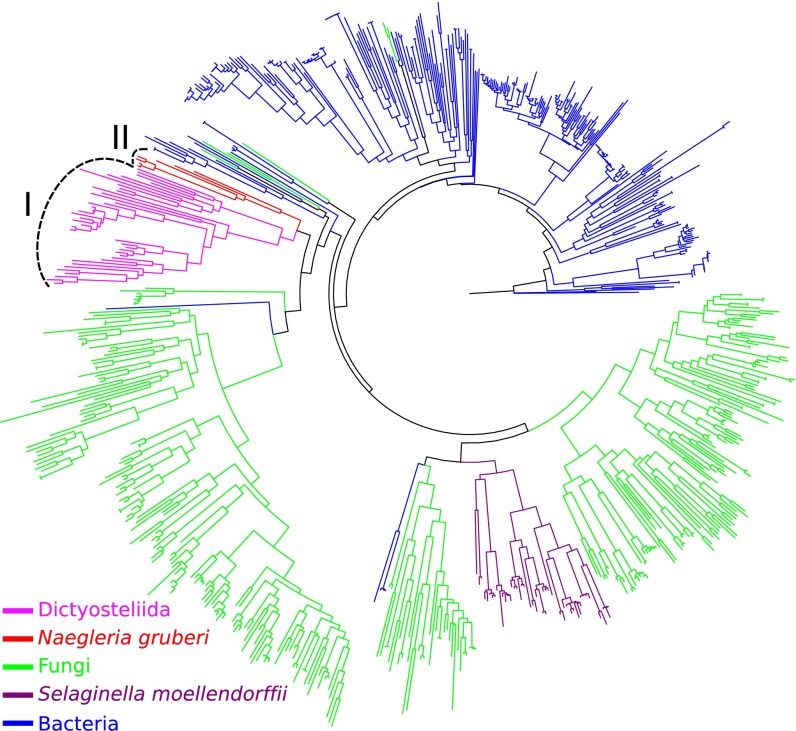
To understand the evolutionary relatedness of the identified eukaryotic TPSs with known TPSs, a phylogenetic tree was constructed that includes, besides the eukaryotic TPSs described here, representative bacterial and fungal TPSs, and microbial type TPSs from the lycophyte Selaginella moellendorffii (8). Notably, the TPSs from the five species of Dictyosteliida clustered together (clade I), whereas the seven TPSs from N. gruberi clustered in a separate, but closely related clade (clade II) (Fig. 2). Together, the amoebal TPSs showed closer relatedness to fungal TPSs than to bacterial TPSs (Fig. 2).
Fig. 2.
Phylogenetic reconstruction of newly identified eukaryotic TPSs with known TPSs. The set of known TPSs includes representative TPSs from fungi and bacteria. Also included were the microbial terpene synthase-like proteins identified from the plant Selaginella moellendorffii. The newly identified eukaryotic TPSs include a total of 50 putative full-length TPSs identified from six species (five species from Dictyosteliida and N. gruberi) (Table S2). TPSs are color-coded based on their source. The TPSs from Dictyosteliida form clade I and the TPSs from N. gruberi form clade II.
TPSs contain several highly conserved motifs that are important for catalytic activity including the aspartate-rich “DDxx(x)D/E” motif and the “NDxxSxxxD/E” motif, both of which are involved in complexing metal ions to coordinate the binding of the isoprenyl diphosphate substrate in the active site (20, 21). Both motifs are also highly conserved among all newly identified eukaryotic TPSs (Table S3). In addition, the diphosphate sensor that is involved in substrate recognition and critical for catalytic activity (Arginine) (6, 22) was also highly conserved (Table S3).
Table S3.
Conserved domains of the identified eukaryotic terpene synthases
| Genes | Domains | ||
| DDxx(x)D/E | R | NDxxSxxD/E | |
| AsTPS1 | FDDGLDA | LRK | INDLVSYEKEV |
| DdTPS1 | IDDFLED | IRK | VNDCASYDKEY |
| DdTPS2 | FDDLFDG | HRK | VNDCVSYEKEI |
| DdTPS3 | LDDFLER | IRT | FNDCVSYAKEL |
| DdTPS4 | LDDYIFE | KRS | INDLYSFNREI |
| DdTPS5 | FDDPLDT | IRK | VNDAASFCKEI |
| DdTPS6 | FDDVLDA | LRK | INDLVSYEKEV |
| DdTPS7 | IDDFLES | FKK | FNDCASYAKEV |
| DdTPS8 | LDDYIYE | SRH | INDCGSFKMEI |
| DdTPS9 | VDDFLES | FKR | MNDCASYAKEI |
| DfaTPS1 | FDDALDS | LRK | INDLVSYEKEV |
| DfaTPS2 | YDEMHDY | LRT | LNDIVSWEVEK |
| DpTPS1 | FDDALDA | LRK | INDLVSYEKEV |
| DpTPS2 | FDDDLDT | IRK | YNDCVSYAKEV |
| DpTPS3 | VDDFYFE | YRC | INDMVSFERER |
| DpTPS4 | LDDFLER | IRT | VNDCVSYAKEI |
| DpTPS5 | LDDFLER | IRT | FNDCVSYAKEI |
| DpTPS6 | IDDFYFE | SRY | INDCYSFNKEK |
| DpTPS7 | IDDLYLE | YRG | INDIHSFNKEI |
| DpTPS8 | VDDFYLE | SRG | INDIYSFNKET |
| DpTPS9 | LDDILDS | LRK | VNDMASYCKEV |
| DpTPS10 | IDDYLDS | IRT | VNDAVSYAKEI |
| DpTPS11 | YDDEYLE | TRS | VNDIYSFVKES |
| DpTPS12 | TDDFFDG | IRR | INDLHSFEKEF |
| NgTPS1 | VDDACDC | WRR | INDLASFPKEL |
| NgTPS2 | VDDACDS | LRE | INDICSLPKDL |
| NgTPS3 | VDDRIDS | RRI | VNDLLSYHKDV |
| NgTPS4 | VDDACDE | ERR | LNDVASIAKDT |
| NgTPS5 | VDDACDE | DRR | LNDVASIAKDT |
| NgTPS6 | VDDACDE | ERR | FNDVASIAKDA |
| NgTPS7 | VDDACDE | ERR | FNDVASIAKDA |
| PpTPS1 | FDDLIDT | MRR | YNDIFSYEKEL |
| PpTPS2 | FDEKMDA | IRI | INDICSYEKEF |
| PpTPS3 | FDDLIDS | IRR | YNDIFSYEKEL |
| PpTPS5 | FDDNIDT | LRR | YNDVLSYEKEM |
| PpTPS6 | FDDLIDS | MRR | FNDLFSYEKEL |
| PpTPS8 | FDDLIDT | MRR | YNDVFSYEKEL |
| PpTPS9 | FDDFIDS | LRI | YNDVLSYEKEV |
| PpTPS10 | FDDLIDS | LRR | YNDVLSYEKEV |
| PpTPS11 | FDDLIDT | MRN | YNDVLSYEKEV |
| PpTPS12 | FDDFIDC | LRI | YNDVFSYEKEV |
| PpTPS13 | FDDLIDT | MRI | YNDVFSYEKEL |
| PpTPS14 | IDDIVIE | FRS | VNDLYSFVSEF |
| PpTPS15 | FDECIES | LRR | YNDLFSYEKEV |
| PpTPS16 | FDDFIET | LRG | FNDITSYEKEL |
| PpTPS17 | FDDYIEK | IRL | VNDVYSFRKEM |
| PpTPS18 | FDDGLDA | LRK | INDLVSYEKEV |
| PpTPS19 | TDDQLLE | FRE | VNDLYSFNKEI |
Bold characters denote conserved amino acids in respective domains.
Expression Patterns of Individual Terpene Synthase Genes in Dictyostelium discoideum.
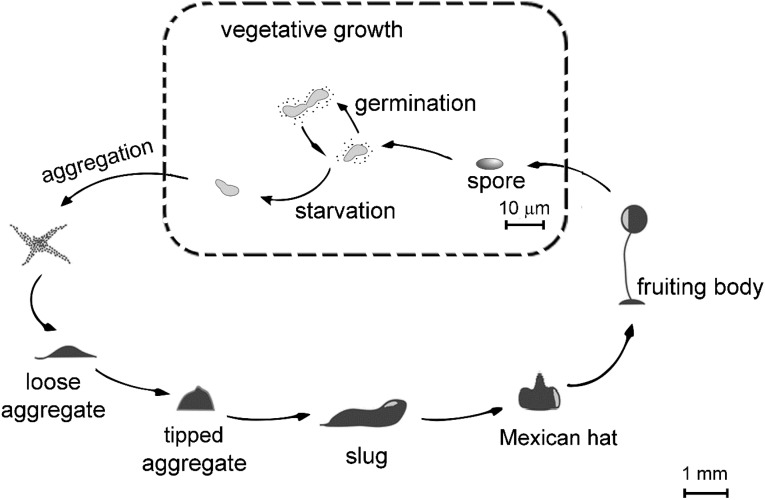
D. discoideum was selected as a model system to explore the function of the newly identified eukaryotic TPSs. As a social amoeba, D. discoideum has a distinctive life cycle (Fig. S1). It propagates vegetatively as a unicellular organism when food (bacteria in the natural environment) is abundant. Upon starvation, D. discoideum transitions into multicellular development in a highly coordinated process that causes individual cells to aggregate and differentiate with formation of a multicellular slug that migrates and finally turns into a fruiting body. This process lasts approximately 24 h (23).
Fig. S1.
Life cycle of the social amoeba D. discoideum. D. discoideum amoebae propagate vegetatively as a unicellular organism when food (naturally bacteria) is abundant. Upon starvation, social amoebae transition into multicellular development. During development, individual cells aggregate and differentiate, forming a multicellular slug that migrates and then forms a fruiting body in a highly coordinated process. The cartoons at the bottom half of the figure depict various stages during multicellular development: streaming, loose aggregate, tipped aggregate, slug, Mexican hat, and fruiting bodies.
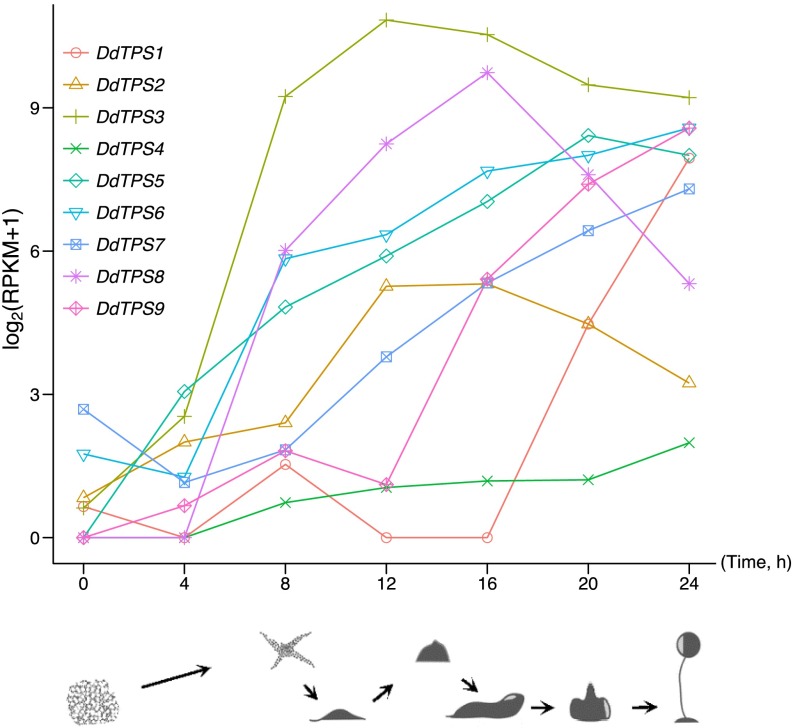
The D. discoideum genome contains 11 putative TPS genes, 9 of which show a full-length sequence and were designated DdTPS1 to DdTPS9. Analysis of the published gene expression dataset of D. discoideum (24) revealed that each TPS gene of D. discoideum was expressed at different times during multicellular development (Fig. 3). Specifically, as D. discoideum began to starve (0 h), mRNAs of DdTPS1, DdTPS2, DdTPS3, DdTPS6, and DdTPS7 were present at detectable but low levels, whereas mRNAs of DdTPS4, DdTPS5, DdTPS8, and DdTPS9 were almost undetectable, but in all cases, the mRNA abundance increased during development. During mound formation (approximately 8–12 h), DdTPS3 attained the highest level of expression among all nine genes, whereas DdTPS1 levels were still near the limit of detection. At the time of slug formation (approximately 16 h), DdTPS2 and DdTPS8 mRNAs reached their highest abundance, whereas the abundance of DdTPS3 started to decrease. Finally, during culmination (from 18 to 24 h), DdTPS2, DdTPS3, DdTPS5, and DdTPS8 levels decreased, whereas DdTPS1, DdTPS4, DdTPS6, and DdTPS9 mRNAs accumulated to higher levels, reaching their peaks at the ultimate stage of mature fruiting body (24 h).
Fig. 3.
Expression patterns of nine terpene synthase genes in D. discoideum (DdTPS1-9). This analysis was based on published RNAseq data (24), which were obtained at seven time points during a complete developmental program in which individual D. discoideum cells aggregated and differentiated, forming a multicellular slug that migrated and then formed a fruiting body in a highly coordinated process that lasted approximately 24 h. The expression levels of nine DdTPS genes were measured by RPKM (reads per kilobase per million sequenced reads) and then displayed on a log2(RPKM+1) scale in this line plot. The line plot shows the transcript abundance (y axis; log-scale) of nine DdTPS genes. The cartoons depict various stages during multicellular development: vegetative, individual cells (0 h), streaming (8 h), loose aggregate (10 h), tipped aggregate (14 h), slug (16 h), Mexican hat (20 h), and fruiting bodies (24 h).
DdTPS Genes in Dictyostelium discoideum Encode Active Terpene-Producing Enzymes.
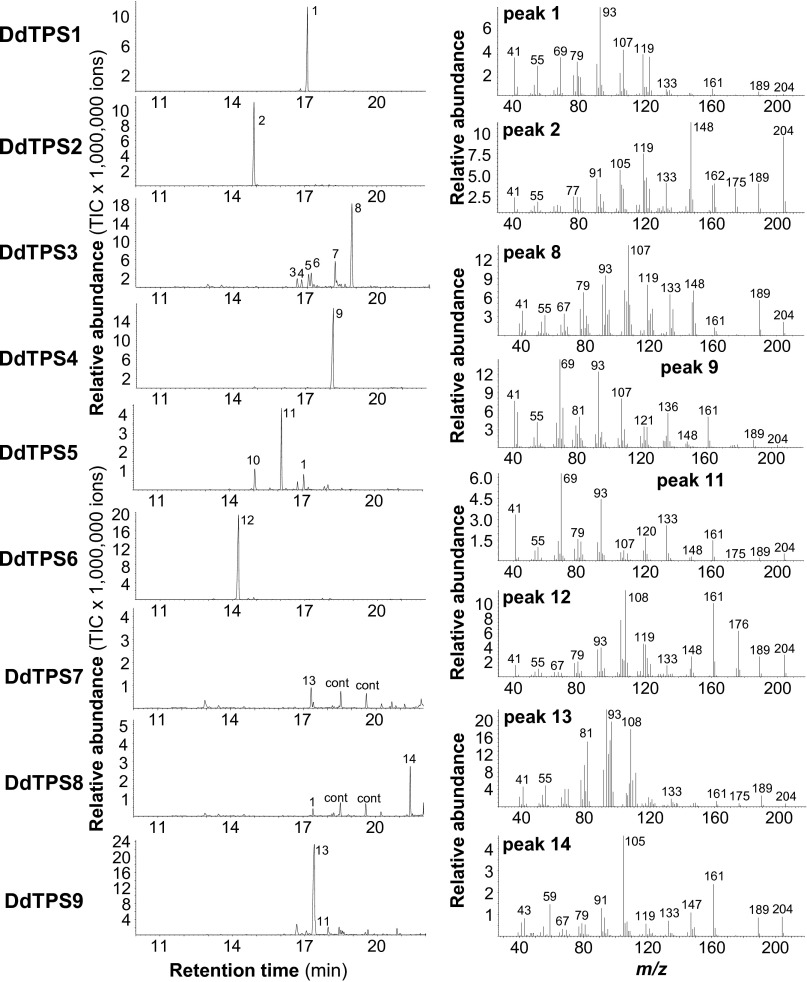
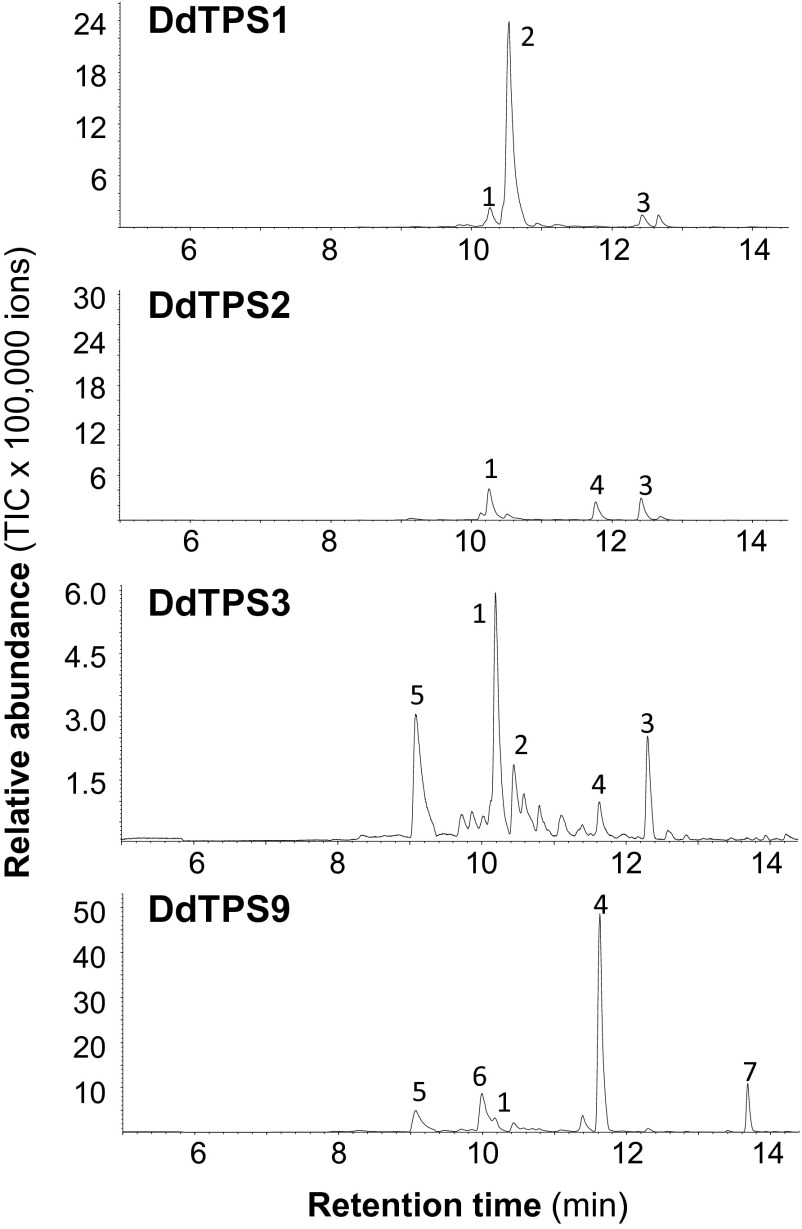
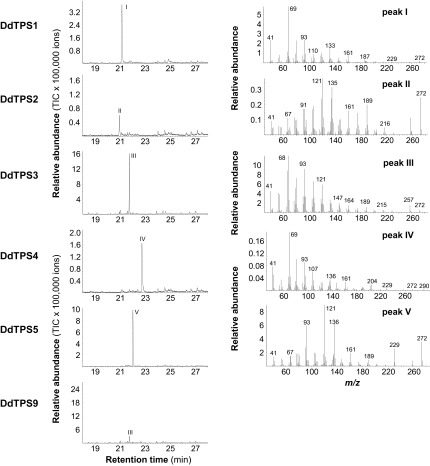
To further understand the function of D. discoideum terpene synthase genes, we characterized the biochemical activities of the enzymes they encode. Full-length cDNAs of DdTPS1–DdTPS9 were cloned and heterologously expressed in Escherichia coli. Individual recombinant DdTPS proteins were tested for terpene synthase activity. All nine enzymes were able to accept farnesyl diphosphate (FPP) as a substrate to produce either a single sesquiterpene or a mixture of compounds (Fig. 4). The major products of DdTPS1, DdTPS4, DdTPS5, and DdTPS7/9 were identified as (E,E)-α-farnesene, (E)-nerolidol, (E)-β-farnesene, and β-barbatene, respectively, whereas DdTPS2, DdTPS3, DdTPS6, and DdTPS8 produced unidentified sesquiterpenes. DdTPS1, DdTPS2, DdTPS3, and DdTPS9 were also able to convert geranyl diphosphate (GPP) into different mixtures of monoterpenes (Fig. S2). In addition, diterpene products from geranylgeranyl diphosphate (GGPP) could be observed for DdTPS1, DdTPS2, DdTPS3, DdTPS4, DdTPS5, and DdTPS9 (Fig. S3).
Fig. 4.
Sesquiterpene synthase activity of D. discoideum terpene synthases. Genes were heterologously expressed in E. coli, and crude protein extracts were incubated with the substrate FPP. Enzyme products were collected by using solid-phase microextraction and analyzed by GC/MS. GC traces (Left) and mass spectra of major products (Right) are shown. 1, (E,E)-α-farnesene*; 2, unidentified sesquiterpene hydrocarbon; 3, β-maaliene; 4, aristolene; 5, calarene; 6, unidentified sesquiterpene hydrocarbon; 7, unidentified sesquiterpene hydrocarbon; 8, unidentified sesquiterpene hydrocarbon; 9, (E)-nerolidol*; 10, β-elemene*; 11, (E)-β-farnesene*; 12, unidentified sesquiterpene hydrocarbon; 13, β-barbatene*; 14, unidentified sesquiterpene; cont, contamination. Compounds marked with asterisks (*) were identified by using authentic standards. Each assay was repeated at least three times, and a representative GC chromatogram is shown.
Fig. S2.
Monoterpene synthase activity of D. discoideum terpene synthases. Genes were heterologously expressed in E. coli, and crude protein extracts were incubated with the substrate GPP. Enzyme products were collected by using solid-phase microextraction and analyzed by GC/MS. 1, (Z)-β-ocimene; 2, (E)-β-ocimene; 3, allo-ocimene; 4, linalool; 5, β-myrcene; 6, limonene; 7, α-terpineol.
Fig. S3.
Diterpene synthase activity of D. discoideum terpene synthases. Genes were heterologously expressed in E. coli, and crude protein extracts were incubated with the substrate GGPP. Enzyme products were extracted with hexane and analyzed by using GC/MS. GC traces (Left) and mass spectra of major products (Right) are shown. I, unidentified diterpene; II, unidentified diterpene; III, unidentified diterpene; IV, unidentified oxygenated diterpene; V, unidentified diterpene.
Dictyostelium discoideum Emits Terpene-Dominated Volatiles During Multicellular Development.
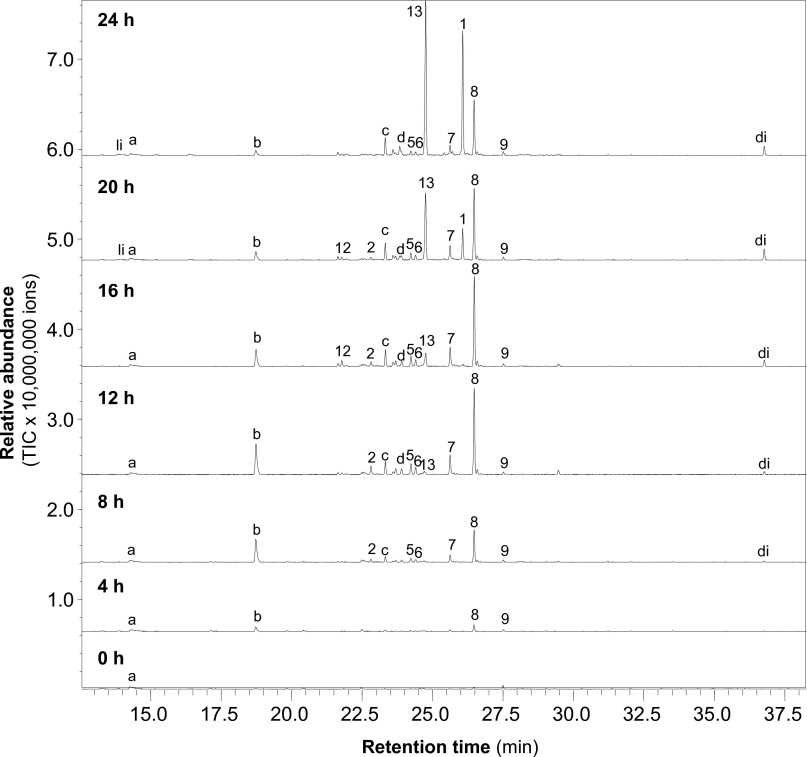
The elaborate temporal regulation of mRNA abundance during different life stages suggests that the DdTPS genes may play a role in development. Monoterpenes and sesquiterpenes are generally volatile compounds that can be trapped by use of, e.g., a closed-loop stripping apparatus or solid-phase microextraction (SPME) and analyzed by gas chromatography/mass spectrometry (GC/MS) (25). Based on the expression patterns of individual DdTPS genes (Fig. 3) and the in vitro biochemical activities of the respective proteins (Fig. 4 and Figs. S2 and S3), we hypothesized that DdTPSs might be involved in producing volatile compounds during development. To test this hypothesis, we performed volatile profiling of D. discoideum cultures at 4-h intervals during the 24 h of development (Fig. 5).
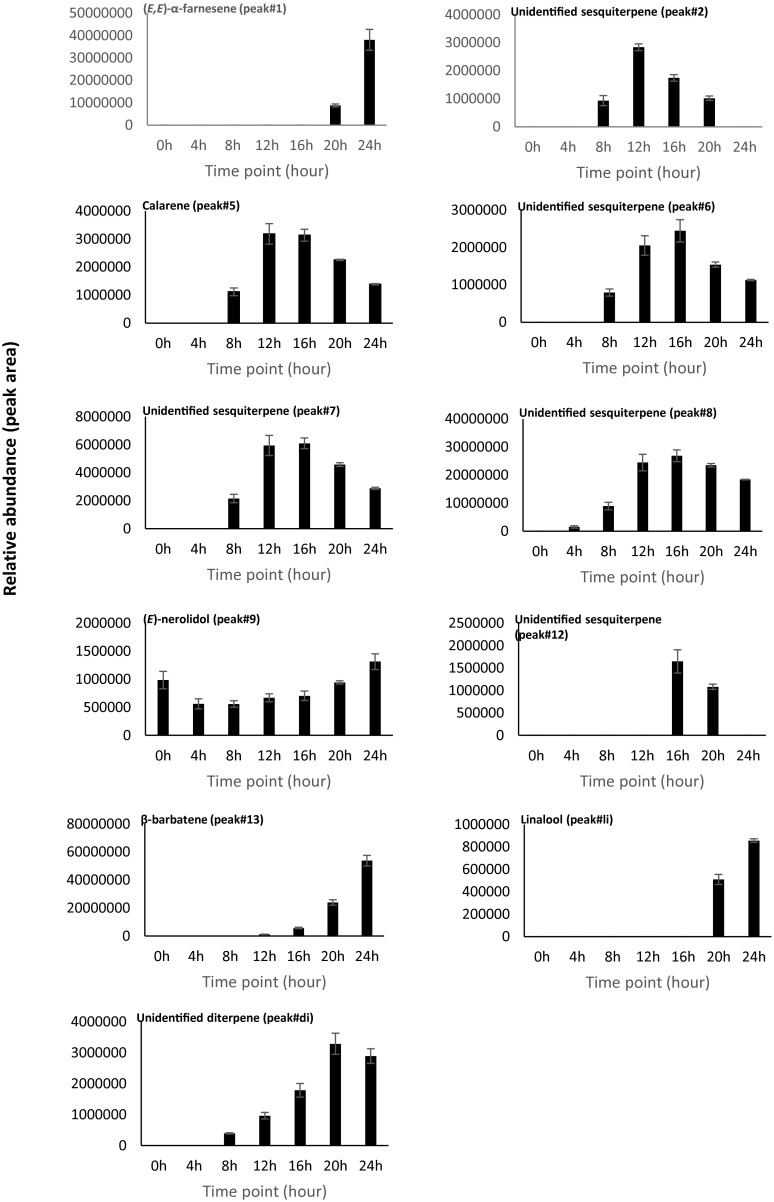
Fig. 5.
D. discoideum emits diverse volatile terpenes during development. The beginning of multicellular development was set as 0 h. Volatiles were collected once every 4 h. Shown are GC/MS chromatograms from one representative replicate. All of the peaks labeled with a number are sesquiterpenes, and the number corresponds to the number of the peak in Fig. 4. 1, (E,E)-α-farnesene*; 2, unidentified sesquiterpene hydrocarbon; 5, calarene; 6, unidentified sesquiterpene hydrocarbon; 7, unidentified sesquiterpene hydrocarbon; 8, unidentified sesquiterpene hydrocarbon; 9, (E)-nerolidol*; 12, unidentified sesquiterpene hydrocarbon; 13, β-barbatene*. Compounds marked with asterisks (*) were identified by using authentic standard. “li” represents the monoterpene linalool, and “di” represents an unidentified diterpene. The letters indicate nonterpene volatiles. “a” represents 2-phenylethanol; “b”, “c”, and “d” represent unidentified compounds.
Altogether, a total of 15 volatile compounds were detected from developing D. discoideum cultures (Fig. 5), including 11 terpenes and four nonterpene volatiles, one of which was identified as 2-phenylethanol. The terpene portion of the volatiles was dominated by nine detectable sesquiterpenes, of which four were identified as (E,E)-α-farnesene, calarene, (E)-nerolidol, and β-barbatene (Fig. 5). Comparison of the mass spectra of the unidentified sesquiterpenes in the headspace extracts to those obtained enzymatically with the expressed DdTPSs allowed assignment of each of the compounds emitted by D. discoideum with confidence to a specific DdTPS. As such, each sesquiterpene in Fig. 5 was labeled with the same peak number as used in Fig. 4. In addition, the monoterpene linalool and one diterpene, which was identical to in vitro diterpene product of DdTPS5 (Fig. S3), were detected in the culture extracts.
The relative abundance of individual volatile terpenes during the 24 h of development was calculated based on three biological replicates (Fig. 5 and Fig. S4). At the beginning of development (0 h), essentially no volatile terpenes were detected, whereas after 4 h of development, the emission of traces of terpenes including (E)-nerolidol was detected. The production of terpenes by D. discoideum gradually increased during the next hours of development, but some compounds showed an early maximum production, e.g., calarene peaked at 12 h and 16 h, whereas the production of other terpenes such as (E,E)-α-farnesene and β-barbatene exhibited a later maximum of production.
Fig. S4.
Relative abundance of individual volatile terpenes at each time point during the 24 h of multicellular development of D. discoideum. The emission rates were calculated based on the peak area of individual compound in GC chromatograms from three biological replicates with Fig. 5 as one of the replicates.
Discussion
This first report of the occurrence of canonical terpene synthase genes in the social amoebae raises questions about the functions of the terpene products in these organisms. The fact that TPS gene expression and terpene volatile emission in D. discoideum are restricted to specific periods during multicellular development suggests possible roles for these compounds if the unique biology of D. discoideum is considered in light of the known functions of volatile terpenes in other organisms.
One possible function of volatile terpenes emitted from D. discoideum is to attract other organisms to facilitate spore dispersal, resembling the function of volatile terpenes from the fruiting bodies of fungi (26). Forming fruiting bodies by social amoebae is considered to be an adaptation for spore dispersal (27). This hypothesis was directly supported by experimental studies in which fruiting bodies were shown to increase the rate at which spores are acquired by a model invertebrate Drosophila melanogaster (27). Although the primary vectors for D. discoideum spore dispersal are unknown (27), it will be an interesting future subject to identify such vectors in nature and then determine whether volatile terpenes have a role in attracting such vectors to facilitate spore dispersal. Consistent with this hypothesis, β-barbatene (Fig. 5) emitted from the fruiting bodies of the bracket fungus Fomitopsis pinicola has been implicated in attracting insects for spore dispersal (26).
Another possible function of D. discoideum volatile terpenes is defense. Social amoebae are preyed on by nematodes (28). They have evolved multiple defense mechanisms, which include the synthesis of a protective extracellular matrix called the slime sheath and the formation of protective coats at the surface of spores (28). In addition, individual amoebae protect themselves by secreting compounds that repel nematodes (28). It will be interesting to see whether any of the volatile terpenes emitted from D. discoideum serve such a function as well. Consistent with this hypothesis, (E,E)-α-farnesene (Fig. 5) emitted from the leaves of the model plant Arabidopsis thaliana has been implicated in defense against insects (29).
The third possibility is that D. discoideum terpenes may function as signals to coordinate multicellular development. The roles of terpenes in signaling have been relatively well-studied in plants (30). As volatile compounds, terpenes can signal over a distance in either multicellular organisms or multicellular aggregates. Previous studies showed that ammonia, a volatile by-product of gluconeogenesis, is involved in regulating several stages of D. discoideum development, including aggregation (31), slug migration (32), and culmination (33). It is certainly intriguing to ask whether volatile terpenes could have similar functions, with the diversity of chemical structures helping to provide functional specificity.
It is interesting that all of the five species of social amoebae (i.e., all from Dictyosteliida) with sequenced genomes contain TPS genes, whereas the three species from the genus Entamoeba (Table S1 and Fig. 1), which also belong to the supergroup Amoebozoa, do not contain any TPS gene. The genus Entamoeba are amoebae but not social amoebae, suggesting that TPS genes may provide adaptive functions for social amoeba, which share a unique lifestyle. However, N. gruberi of the supergroup Excavata, which is also an amoeba having no multicellular development (34), contains TPS genes (Fig. 1). Sharing similarity in lifestyle, N. gruberi and Entamoeba are evolutionarily distantly related. It will be interesting to investigate the biochemical and biological function of N. gruberi TPS genes and to understand how they may confer a fitness advantage.
To briefly summarize, we have found classic terpene synthase genes in six species of amoebae among a broad range of nonplant/nonfungus eukaryotes. Amoebal TPSs are more closely related to fungal TPSs and the microbial type TPSs from plants than bacterial TPSs (Fig. 2). The social amoeba D. discoideum is an organism other than plants, fungi, and bacteria from which classic terpene synthase genes have been functionally characterized (Fig. 4). This study provides insights into the occurrence, function and evolution of TPS genes, particularly in eukaryotes, and it is expected to stimulate important future research.
Materials and Methods
Sequence Retrieval and Analysis.
A total of 168 species of nonplant/nonfungus eukaryotes with well-annotated genome sequences (Table S1) archived at the KEGG genome database (www.genome.jp/kegg/catalog/org_list.html) were downloaded as the genome dataset. Another dataset was the nonredundant (nr) protein database from NCBI, which was downloaded on April 19, 2016. Both databases were searched against the Pfam-A database locally by using HMMER 3.0 with an e-value of 1e−2. Sequences with best hits from the following three HMM profiles were identified as putative terpene synthases: Terpene_synth_C (PF03936) and Terpene Synthase N-terminal domain (PF01397), and TRI5 (PF06330). For the search of the nr database, the terpene synthase hits identified from plants, fungi, archaea, and bacteria were removed. For phylogenetic reconstruction, known bacterial and fungal terpene synthases were retrieved from Pfam database (version 27). MAFFT (l-INS-i) was used to build the multiple sequence alignment with 1,000 iterations of improvement. The maximum-likelihood phylogenetic tree was built with RAxML through the CIPRES Science Gateway (https://www.phylo.org) by using the LG+G+F amino acid substitution model with 1,000 bootstrap replicates and then rendered by using FigTree (version 1.4.2).
Cloning of Full-Length cDNA of DdTPS Genes of Dictyostelium discoideum via RT-PCR.
D. discoideum (strain AX4) was obtained from the Dictybase Stock Center (www.dictybase.org). D. discoideum was cocultured with live Klebsiella pneumoniae bacteria on SM agar plates (35) by following the protocol described in the Dictybase Stock Center. When slugs formed, D. discoideum cells were collected and used for total RNA isolation following the protocol described by ref. 36. Full-length cDNA of each DdTPS gene was amplified by RT-PCR using the gene specific primers listed in Table S4. PCR products were cloned into the pEXP-5-CT/TOPO vector (ThermoFisher Scientific) and confirmed by sequencing.
Table S4.
Primers used for cloning of full-length cDNAs of DdTPS genes
| Gene name | Primers | Sequences |
| Forward | 5′-ATGAATATTTTATTAAAAGATTTTAAATAC-3′ | |
| DdTPS1 | Reverse | 5′-TTAAATGTATTTTGATTTTGTATTTTC-3′ |
| Forward | 5′-ATGGATATTAAAAATCTATCATTAAAAG-3′ | |
| DdTPS2 | Reverse | 5′-TTATTTATTTTTTAAATCATAAATTTTTG-3′ |
| Forward | 5′-ATGGAAACTATTAATCTTGAAAAAC-3′ | |
| DdTPS3 | Reverse | 5′-TTAATAAAGATTTGGATCTAAAATAG-3′ |
| Forward | 5′-ATGGAAGAAAAAATAAAATTTTATTTTG-3′ | |
| DdTPS4 | Reverse | 5′-TTAATTATATCTTTTACTTTTTTTATG-3′ |
| Forward | 5′-ATGTTATGCTTTAATGATTTTAATTTTC-3′ | |
| DdTPS5 | Reverse | 5′-TTAAATATAGTTTTTATTTGTTAATG-3′ |
| Forward | 5-ATGTCTTTATCTTTGGGTGATATC-3′ | |
| DdTPS6 | Reverse | 5′-TTAATTATTTAAATCAATTGGTGTTG-3′ |
| Forward | 5′-ATGTATTCAATTTATGATTTTAAATAC-3′ | |
| DdTPS7 | Reverse | 5′-TTATAATTTAATAATTTTATCAACAATATTTTC-3 |
| Forward | 5′-ATGGATTATGATATAAAATTTACTTG-3′ | |
| DdTPS8 | Reverse | 5′-CTAATTATAACGATTTGCATGAC-3′ |
| Forward | 5′-ATGTATTCTCTTCATGATTTCAAATTC-3′ | |
| DdTPS9 | Reverse | 5′-TTATTTTAATAAAATTTTATCAACAATTTTTTC-3′ |
Terpene Synthase Enzyme Assays.
Heterologous expression of DdTPS genes in E. coli, recombinant protein preparation, terpene synthase enzyme assays, and terpene product identification using a Hewlett-Packard 6890 gas chromatograph coupled to a Hewlett-Packard 5973 mass spectrometer were performed as described (8). Each expressed protein was assayed at least three times.
DdTPS Gene Expression Analysis.
DdTPS gene expression was analyzed by using published RNA-seq data from developing D. discoideum (24). Transcript abundance was quantified as described in ref. 37. The data are available on dictyExpress (38). Briefly, D. discoideum was grown on a thick lawn of K. aerogenes before the food source was removed to initiate the developmental program. The samples were obtained in 4-h intervals throughout the 24-h developmental program.
Headspace Collection and GC/MS Analysis.
A mixture of D. discoideum spores and freshly grown K. pneumoniae was spread onto SM agar plates to initiate D. discoideum culture. Under our experimental conditions, D. discoideum progressed from spore germination to vegetative growth to the completion of multicellular development in ∼48 h. At 24 h, the appearance of the culture plate changed from opaque (from the bacterial lawn) to translucent, indicating the clearing of bacteria. This time point was defined as the start of multicellular development, after which D. discoideum progressed through the various described developmental stages in the next 24 h with the eventual formation of fruiting bodies (Fig. S1). SPME combined with GC/MS was used for volatile profiling of the D. discoideum cultures. During the 24 h of development, volatiles were collected once every 4 h (Fig. 5). Before each collection, the lid of the culture plate was removed and the plate was left in the hood for 1 min to dispose of accumulated volatiles. Then the lid was put back on and a SPME fiber coated with 100-µm polydimethylsiloxane was inserted into the headspace of the plate to start volatile collection. After 1 h, the SPME fiber was retracted and inserted into the injector port (a splitless injection and injector temperature of 250 °C) of a Shimadzu 17A gas chromatograph coupled to a Shimadzu QP5050A quadrupole mass selective detector for chemical identification. Separation was performed on a Restek Rxi-5Sil MS column (30 m × 0.25 mm i.d. × 0.25 µm thickness; Restek) with helium as the carrier gas and a temperature program from 60 °C to 300 °C at 5 °C⋅min−1. The experiment was performed with three biological replicates.
Acknowledgments
We thank Wangdan Xiong for her assistance with the preparation of Fig. 3 and Fig. S1 and Natascha Rauch (Max Planck Institute for Chemical Ecology) for her technical assistance. This project was supported by a University of Tennessee AgResearch Innovation Grant (to F.C.) and the Max Planck Society (T.G.K. and J.G.). G.S. and B.S. acknowledge support from National Institutes of Health Grant P01 HD039691.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences for the biochemically characterized terpene synthases reported in this paper have been deposited in the GenBank database (accession nos. KX364374–KX364382).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610379113/-/DCSupplemental.
References
- 1.Eisenreich W, Bacher A, Arigoni D, Rohdich F. Biosynthesis of isoprenoids via the non-mevalonate pathway. Cell Mol Life Sci. 2004;61(12):1401–1426. doi: 10.1007/s00018-004-3381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jia Q, Chen F. Catalytic functions of the isoprenyl diphosphate synthase superfamily in plants: A growing repertoire. Mol Plant. 2016;9(2):189–191. doi: 10.1016/j.molp.2015.12.020. [DOI] [PubMed] [Google Scholar]
- 3.Wang KC, Ohnuma S. Isoprenyl diphosphate synthases. Biochim Biophys Acta. 2000;1529(1-3):33–48. doi: 10.1016/s1388-1981(00)00136-0. [DOI] [PubMed] [Google Scholar]
- 4.Degenhardt J, Köllner TG, Gershenzon J. Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry. 2009;70(15-16):1621–1637. doi: 10.1016/j.phytochem.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 5.Yamada Y, et al. Terpene synthases are widely distributed in bacteria. Proc Natl Acad Sci USA. 2015;112(3):857–862. doi: 10.1073/pnas.1422108112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dickschat JS. Bacterial terpene cyclases. Nat Prod Rep. 2016;33(1):87–110. doi: 10.1039/c5np00102a. [DOI] [PubMed] [Google Scholar]
- 7.Chen F, Tholl D, Bohlmann J, Pichersky E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011;66(1):212–229. doi: 10.1111/j.1365-313X.2011.04520.x. [DOI] [PubMed] [Google Scholar]
- 8.Li G, et al. Nonseed plant Selaginella moellendorffi [corrected] has both seed plant and microbial types of terpene synthases. Proc Natl Acad Sci USA. 2012;109(36):14711–14715. doi: 10.1073/pnas.1204300109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt-Dannert C. Biosynthesis of terpenoid natural products in fungi. Adv Biochem Eng Biotechnol. 2015;148:19–61. doi: 10.1007/10_2014_283. [DOI] [PubMed] [Google Scholar]
- 10.Keller NP, Turner G, Bennett JW. Fungal secondary metabolism - from biochemistry to genomics. Nat Rev Microbiol. 2005;3(12):937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 11.Gilg AB, Tittiger C, Blomquist GJ. Unique animal prenyltransferase with monoterpene synthase activity. Naturwissenschaften. 2009;96(6):731–735. doi: 10.1007/s00114-009-0521-1. [DOI] [PubMed] [Google Scholar]
- 12.Beran F, et al. Novel family of terpene synthases evolved from trans-isoprenyl diphosphate synthases in a flea beetle. Proc Natl Acad Sci USA. 2016;113(11):2922–2927. doi: 10.1073/pnas.1523468113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao R, et al. Diterpene cyclases and the nature of the isoprene fold. Proteins. 2010;78(11):2417–2432. doi: 10.1002/prot.22751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adl SM, et al. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59(5):429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burki F. The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb Perspect Biol. 2014;6(5):a016147. doi: 10.1101/cshperspect.a016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gershenzon J, Dudareva N. The function of terpene natural products in the natural world. Nat Chem Biol. 2007;3(7):408–414. doi: 10.1038/nchembio.2007.5. [DOI] [PubMed] [Google Scholar]
- 17.Finn RD, Clements J, Eddy SR. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Web Server issue):W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schilde C, Schaap P. The Amoebozoa. Methods Mol Biol. 2013;983:1–15. doi: 10.1007/978-1-62703-302-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pawlowski J, Burki F. Untangling the phylogeny of amoeboid protists. J Eukaryot Microbiol. 2009;56(1):16–25. doi: 10.1111/j.1550-7408.2008.00379.x. [DOI] [PubMed] [Google Scholar]
- 20.Starks CM, Back K, Chappell J, Noel JP. Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science. 1997;277(5333):1815–1820. doi: 10.1126/science.277.5333.1815. [DOI] [PubMed] [Google Scholar]
- 21.Christianson DW. Unearthing the roots of the terpenome. Curr Opin Chem Biol. 2008;12(2):141–150. doi: 10.1016/j.cbpa.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baer P, et al. Induced-fit mechanism in class I terpene cyclases. Angew Chem Int Ed Engl. 2014;53(29):7652–7656. doi: 10.1002/anie.201403648. [DOI] [PubMed] [Google Scholar]
- 23.Kessin RH. Dictyostelium: Evolution, Cell Biology, and the Development of Multicellularity. Cambridge Univ Press; Cambridge, UK: 2001. [Google Scholar]
- 24.Parikh A, et al. Conserved developmental transcriptomes in evolutionarily divergent species. Genome Biol. 2010;11(3):R35. doi: 10.1186/gb-2010-11-3-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dickschat JS. Capturing volatile natural products by mass spectrometry. Nat Prod Rep. 2014;31(6):838–861. doi: 10.1039/c3np70080a. [DOI] [PubMed] [Google Scholar]
- 26.Fäldt J, Jonsell M, Nordlander G, Borg-Karlson A-K. Volatiles of bracket fungi Fomitopsis pinicola and Fomes fomentarius and their functions as insect attractants. J Chem Ecol. 1999;25(3):567–590. [Google Scholar]
- 27.Smith J, Queller DC, Strassmann JE. Fruiting bodies of the social amoeba Dictyostelium discoideum increase spore transport by Drosophila. BMC Evol Biol. 2014;14:105. doi: 10.1186/1471-2148-14-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kessin RH, Gundersen GG, Zaydfudim V, Grimson M. How cellular slime molds evade nematodes. Proc Natl Acad Sci USA. 1996;93(10):4857–4861. doi: 10.1073/pnas.93.10.4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang M, et al. Variation of herbivore-induced volatile terpenes among Arabidopsis ecotypes depends on allelic differences and subcellular targeting of two terpene synthases, TPS02 and TPS03. Plant Physiol. 2010;153(3):1293–1310. doi: 10.1104/pp.110.154864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maag D, Erb M, Köllner TG, Gershenzon J. Defensive weapons and defense signals in plants: Some metabolites serve both roles. BioEssays. 2015;37(2):167–174. doi: 10.1002/bies.201400124. [DOI] [PubMed] [Google Scholar]
- 31.Bonner JT, Suthers HB, Odell GM. Ammonia orients cell masses and speeds up aggregating cells of slime molds. Nature. 1986;323(6089):630–632. [Google Scholar]
- 32.Bonner JT, Chiang A, Lee J, Suthers HB. The possible role of ammonia in phototaxis of migrating slugs of Dictyostelium discoideum. Proc Natl Acad Sci USA. 1988;85(11):3885–3887. doi: 10.1073/pnas.85.11.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feit IN, Sollitto RB. Ammonia is the gas used for the spacing of fruiting bodies in the cellular slime mold, Dictyostelium discoideum. Differentiation. 1987;33(3):193–196. [Google Scholar]
- 34.Fritz-Laylin LK, et al. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140(5):631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 35.Sussman M. Biochemical and genetic methods in the study of cellular slime mold development. Methods Cell Biol. 1966;2:397–410. [Google Scholar]
- 36.Pilcher KE, Gaudet P, Fey P, Kowal AS, Chisholm RL. A general purpose method for extracting RNA from Dictyostelium cells. Nat Protoc. 2007;2(6):1329–1332. doi: 10.1038/nprot.2007.191. [DOI] [PubMed] [Google Scholar]
- 37.Miranda ER, et al. Transcriptional profiling of Dictyostelium with RNA sequencing. Methods Mol Biol. 2013;983:139–171. doi: 10.1007/978-1-62703-302-2_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rot G, et al. dictyExpress: A Dictyostelium discoideum gene expression database with an explorative data analysis web-based interface. BMC Bioinformatics. 2009;10:265. doi: 10.1186/1471-2105-10-265. [DOI] [PMC free article] [PubMed] [Google Scholar]