Significance
NMDA receptors are ionotropic glutamate receptors and crucial for excitatory synaptic transmissions in the brain. We studied the regulation of the exocytosis of NMDA receptors by directly visualizing their exocytic events in rat hippocampal neurons by using total internal reflection florescence microscopy (TIRFM). We characterized the constitutive exocytic events of NMDA receptors and identified the Rab proteins involved in the exocytosis. Moreover, combining the TIRFM and immunocytochemistry methods, we found that the SNAP25–VAMP1–syntaxin4 complex mediated the constitutive exocytosis of NMDA receptor and maintained the expression of NMDA receptors at both extrasynaptic and synaptic surfaces. These results suggest that the SNAP25–VAMP1–syntaxin4 complex is important for neural functions by mediating the delivery of NMDA receptors to the plasma membrane.
Keywords: NMDA receptor, constitutive exocytosis, TIRFM, SNARE, Rab
Abstract
In the central nervous system, NMDA receptors mediate excitatory neurotransmissions and play important roles in synaptic plasticity. The regulation of NMDA receptor trafficking is critical for neural functions in the brain. Here, we directly visualized individual exocytic events of NMDA receptors in rat hippocampal neurons by total internal reflection fluorescence microscopy (TIRFM). We found that the constitutive exocytosis of NMDA receptors included both de novo exocytic and recycling events, which were regulated by different Rab proteins. We also identified the SNAP25–VAMP1–syntaxin4 complex mediating the constitutive exocytosis of NMDA receptors. Transient knockdown of each component of the SNARE complex interfered with surface delivery of NMDA receptors to both extrasynaptic and synaptic membranes. Our study uncovers the postsynaptic function of the SNAP25–VAMP1–syntaxin4 complex in mediating the constitutive exocytosis of NMDA receptors, suggesting that this SNARE complex is involved in excitatory synaptic transmission.
NMDA (N-methyl-d-aspartate) receptors are ionotropic glutamate receptors, mediating excitatory neurotransmission in the brain and playing crucial roles in synaptogenesis, synaptic plasticity, and excitotoxicity. NMDA receptors consist of tetrameric combinations of the homologs subunits NR1, NR2A-D, and NR3A-B. In hippocampal neurons during synaptogenesis, most NMDA receptors are NR1-NR2B heteromers. As the neurons mature, the abundance of NR1-NR2A and NR1-NR2A-NR2B heteromers increased (1). Electrophysiological and immunocytochemical evidences showed that in mature neurons, NMDA receptors primarily clustered at excitatory synapses, which were localized on dendritic spines (2–8). To achieve this specific distribution, NMDA receptors undergo regulations in various trafficking steps, including de novo exocytosis, endocytosis, recycling, lateral movement between synaptic and extrasynaptic membranes, and stabilization by synaptic scaffolding proteins (1, 9).
Recent evidences suggest that SNARE (soluble N-ethylmaleimide–sensitive factor attachment receptor) proteins are involved in NMDA receptor trafficking (10–16). SNARE proteins are a large family of proteins mediating fusion events between target membranes and vesicles. SNARE family members are categorized as three subfamilies based on their localizations. tSNAREs (target SNAREs) are localized on target membranes and consist of SNAPs (synaptosome-associated proteins) and syntaxins. vSNAREs (vesicle SNAREs) are VAMPs (vesicle associated membrane proteins or synaptobrevin) and localized on the vesicles. The four-helix SNARE complex, formed by SNAP, syntaxin and VAMP, brings the target membrane and the vesicle to close proximity and allows their fusion (17). This process can be inhibited by clostridial neurotoxins that specifically cleave different SNARE proteins (18). In the central nervous system, the presynaptic function of SNARE complexes in mediating neurotransmitter release is extensively studied (19); however, the postsynaptic function of these complexes remain to be investigated.
In the current study, we specifically visualized the constitutive exocytic events of NMDA receptors using total internal reflection florescence microscopy (TIRFM), which was previously used to image individual exocytic events of AMPA and GABAA receptors in hippocampal neurons (20–23). This imaging technique in combination with other molecular and cellular methods enabled us to characterize the constitutive exocytosis of NMDA receptors and further identify the SNAP25–VAMP1–syntaxin4 complex mediating the NMDA receptor exocytosis. Our study reveals the postsynaptic function of the SNAP25–VAMP1– syntaxin4 complex in regulating NMDA receptor trafficking.
Results
Dynamic TIRF Events of NMDA Receptors.
To study the exocytosis of NMDA receptors, we chose NR1 subunit because it is common in most NMDA receptor complexes (1). Two approaches were combined to ensure the direct observation of the exocytic events of NR1. First, we took advantage of TIRFM (24), which allows selective visualization of the trafficking of fluorescently tagged proteins near the plasma membrane (within 100 nm) of cells attaching to the glass coverslip. Second, we tagged the extracellular N terminus of NR1 with superecliptic pHluorin (pH), which is a variant of EGFP with bright fluorescence at pH 7.4 and quenched in endocytic organelles with a pH < 6 (25). Following the exocytosis of pH-NR1, the pHluorin tag will be exposed to the extracellular solution with pH 7.4 and its fluorescence will dramatically increase. Previous studies showed that neither pHluorin nor EGFP fluorescent tag at the N terminus of NR1 affected its trafficking in neurons (26–28).
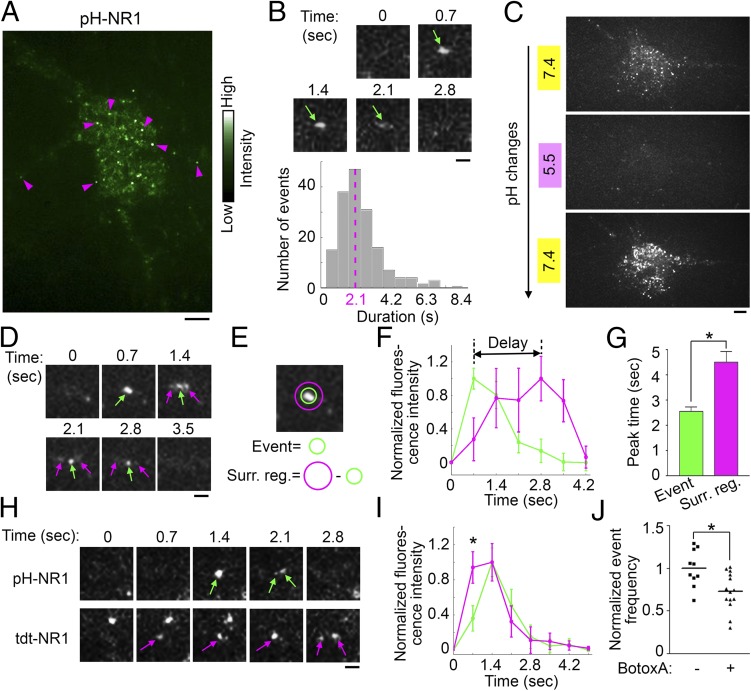
To visualize the exocytosis of NR1, dissociated hippocampal neurons grown on glass coverslips were cotransfected with pH-NR1 and untagged NR2A subunit to form NMDA receptor complexes, and imaged 24–48 h after transfection. Following the photobleaching of preexisting fluorescence on the plasma membrane (21, 23), new fluorescent events of pH-NR1 were directly visualized under the TIRFM (500-ms exposure and 1.4-Hz frequency). We observed robust dynamic events of pH-NR1 as 25 ± 2 events per s per 100 µm2 throughout the plasma membrane, primarily on extrasynaptic surface of soma and dendrites. Synaptic events of pH-NR1 were rarely observed (Movie S1 and Fig. 1A). These dynamic events were transient: 87.5% events lasted less than 4 s and the average event duration was approximately 2.1 s (Fig. 1B). To further confirm that the dynamic events are pH-NR1 insertions to the plasma membrane, rather than fluorescent signals in intracellular organelles with neutral pH (29), we tested the pH sensitivity of the events by rapidly switching the extracellular imaging solutions with different pH values (23, 30) (Fig. 1C). The dynamic pH-NR1 events, which were clearly detected in the imaging solution with pH 7.4, were rapidly quenched after the perfusion of the pH 5.5 solution and soon recovered following the perfusion with the pH 7.4 solution. This phenomenon indicates that the dynamic events of pH-NR1 are on the plasma membrane.
Fig. 1.
Exocytic events of pH-NR1 under TIRFM. (A) TIRF events of pH-NR1 are highlighted based on their intensity (magenta arrowheads). (B) Dynamics of pH-NR1 events. (B, Upper) Time series of an event (green arrow). (B, Lower) The distribution of event durations with the main duration of 2.1 s (magenta dotted line). n = 168. (C) Acidification-neutralization test of the pH-NR1 events. Cells in the extracellular solutions at pH 7.4 (Top), 5.5 (Middle), and 7.4 (Bottom). (D) Lateral diffusion (magenta arrows) of pH-NR1 following the exocytosis (green arrows). (E) Diagram showing the event location and its surrounding region. (F) Averaged fluorescence time courses of pH-NR1 events on the event location (green) and surrounding region (magenta). n = 16. (G) The peak fluorescence of pH-NR1 on the exocytic location appeared earlier than that in the surrounding region. n = 16. (H) Dynamics of a coinsertion event containing tdt- and pH-NR1. (I) Averaged fluorescence time courses of tdt- (magenta) and pH-NR1 (green). Each time course was normalized so that the maximal intensity (1 at .4 s for both curves) was 1. The fluorescence of tdt-NR1 at 0.7 s was higher than that of pH-NR1. n = 18. (J) BotoxA inhibited the TIRF events of pH-NR1. Each data point is one cell. The line shows mean frequency of the dataset. Event frequencies were normalized by the mean of the control (BotoxA −). (Scale bars: A and C, 5 µm; B, D, and H, 1 µm.) * indicates statistical significances: *P < 0.05; **P < 0.01; ***P < 0.001. n.s., no statistical significance.
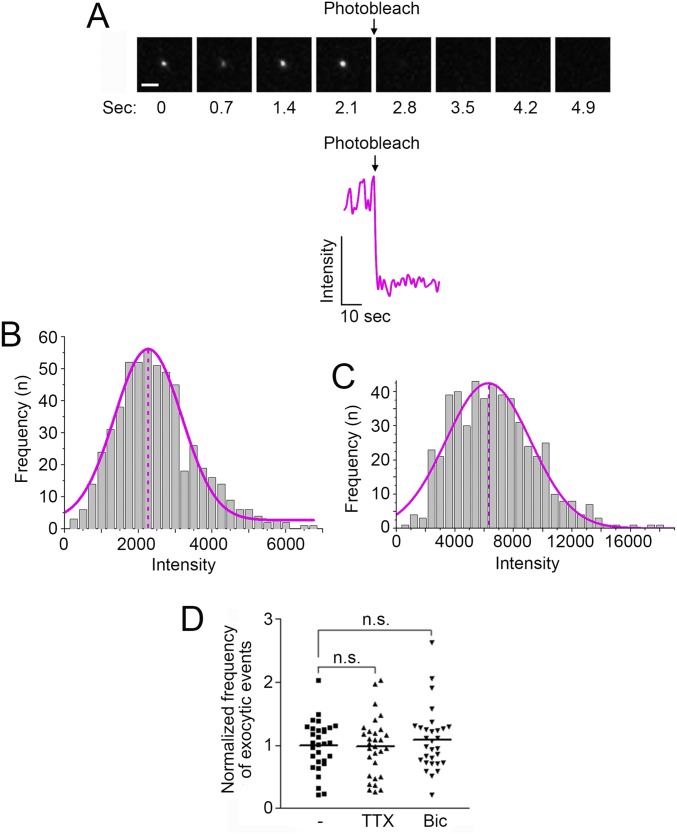
We further characterized the TIRF events of pH-NR1 by determining the number of fluorescently tagged NR1 subunits in each vesicle (21, 23). Based on the fact that the fluorescent intensity of the EGFP monomer is close to the intensity of pHluorin under the environment of pH 7.4 (25, 30, 31), we compared the fluorescent intensity of EGFP monomers to the intensity of single events of pH-NR1 under TIRFM. EGFP monomers were confirmed by their single-step photobleaching property and blinking dynamics (21, 23, 32, 33) (Fig. S1A). The distributions of fluorescent intensity of EGFP monomers and pH-NR1 followed the Gaussian distribution (Fig. S1 B and C). According to the peak intensities of fitted Gaussian curves for EGFP monomers and pH-NR1 events, we estimated that each event contained an average of three pH-NR1 subunits (2.8 ± 0.1 subunits per event).
Fig. S1.
Properties of dynamic TIRF events of pH-NR1. (A, Upper) Sequential images showing the kinetics of surface-absorbed EGFP monomer under single-step photobleaching under the TIRFM. Image interval: 0.7 s. (Scale bar: 1 µm.) (A, Bottom) An example of fluorescence time course of EGFP monomer under single-step photobleaching. This dataset is the same as used in the previous study (23). (B) Binned fluorescence intensity distribution of individual EGFP monomers (gray histogram) and fitted Gaussian curve based on the distribution (magenta curve). The magenta dashed line shows the peak intensity of the Gaussian curve. n = 548. This dataset is the same as used in the previous study (23). (C) Binned fluorescence intensity distribution of individual pH-NR1–containing vesicles (gray histogram) and fitted Gaussian curve based on the distribution (magenta curve). The magenta dashed line shows the peak intensity of the Gaussian curve. n = 508. (D) Exocytic frequency of NR1 was not affected by the changes of neuronal activity. One micromolar TTX or 20 µM Bic were applied to hippocampal neurons in the media for 2 h, and the cells were then imaged under TIRFM in ACSF containing the same concentrations of TTX or Bic, respectively. n.s., no statistical significance.
Dynamic TIRF Events of NR1 Are Constitutive Exocytic Events.
To investigate whether the dynamic TIRF events of NR1 are exocytic events, we examined their three properties. First, it has been shown that receptors disperse away from the insertion spot to the surrounding regions after exocytosis (20, 23). Indeed, we observed that pH-NR1 events exhibited increased fluorescence in the surrounding region following the insertion (Fig. 1D). The fluorescence in the surrounding region showed a significantly delayed increase in comparison to that in the insertion center (Fig. 1 E–G). Moreover, many events split into two to three small events during this dispersion process (Fig. 1 B, D, and H). These results together suggest that the dynamic events are exocytic events carrying a small number of receptor subunits, which independently disperse to the surrounding region after the exocytosis.
Second, as we demonstrated previously, the exocytic events exhibit stereotypic dynamics under TIRFM (23). A coinsertion event, which contains two subunits of the same receptor differentially tagged with pHluorin and a pH-insensitive red fluorescent protein, exhibits immediate increase of red fluorescence when it enters the TIRF field. However, its pHluorin signal is only elevated when the pHluorin is exposed to the extracellular space with pH 7.4 (Fig. S2). To investigate whether the pH-NR1 events show this stereotypic dynamics, a pH-insensitive red fluorescent protein, tdTomato, was used to tag an NR1 subunit at its N-terminal (tdt-NR1) (34). We observed that tdt-NR1 was highly colocalized with EGFP-NR1 throughout the cell body, dendrites, and spines (Fig. S3), indicating that in hippocampal neurons, tdt-NR1 was trafficked as well as EGFP-NR1.
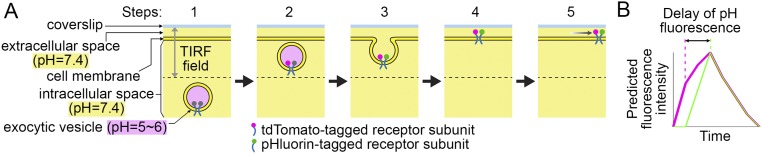
Fig. S2.
Dynamics of coinsertion events under TIRFM. (A) Dynamics of an exocytic vesicle containing two receptor subunits differentially tagged with pHluorin and tdTomato. (B) Predicted dynamics of tdTomato and pHluorin receptors in the same exocytic vesicle under TIRFM. These two figures are cited from the previous study (reproduced from ref. 23).
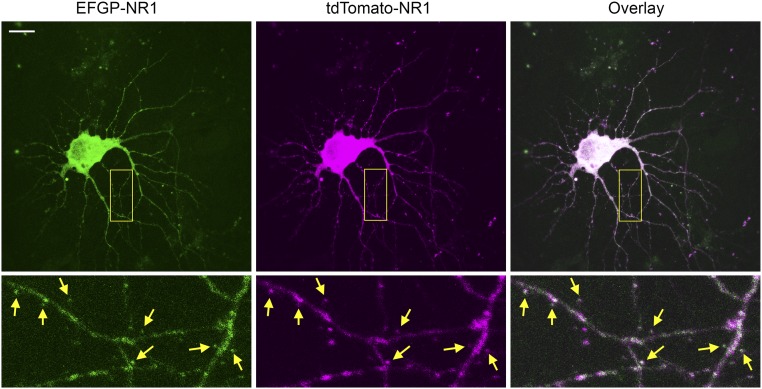
Fig. S3.
Characterization of tdTomato-NR1. Coexpression of EGFP-NR1 (Top Left) and tdTomato-NR1 (Top Middle) and their overlaid signal (Top Right) in live hippocampal neurons. (Scale bar: 20 µm.) High-magnification images from the boxed region are shown at Bottom. Yellow arrows at corresponding locations indicate the expressions of EGFP-NR1 and tdTomato-NR1 at the same spines.
pH-NR1 and tdt-NR1 were coexpressed in hippocampal neurons, and their dynamic events were simultaneously visualized under dual-color TIRFM with 488-nm and 568-nm lasers, which excited pHluorin and tdTomato, respectively. We analyzed the dynamics of the coinsertion events of pH-NR1 and tdt-NR1 and found that the fluorescence of pH-NR1 exhibited delayed increase comparing to that of tdt-NR1 (Fig. 1 H and I). This observation strongly suggests that the dynamic events of NR1 are exocytic events.
The last evidence of exocytosis is the effect of botulinum toxin (Botox) on the frequency of the pH-NR1 events. Botoxs specifically cleave different components of the SNARE complex, which mediates the fusion of exocytic vesicles to the plasma membrane (18). The treatment of BotoxA, which cleaves SNAP25, significantly reduced the frequency of the pH-NR1 events (Fig. 1J). This result supports the fact that the dynamic TIRF events of pH-NR1 are exocytic events mediated by the SNARE complex.
We further asked whether these exocytic events of pH-NR1 are regulated by neuronal activity. Acute application of tetrodotoxin (TTX) or bicuculline (Bic), which suppresses or elevates neuronal activity, respectively, did not alter the exocytic frequency of pH-NR1 (Fig. S1D), suggesting that these exocytic events mostly contribute to constitutive delivery of NMDA receptors to the plasma membrane.
In conclusion, the dynamic TIRF events of pH-NR1 are constitutive exocytic events.
Constitutive Exocytosis of NR1 Is Regulated by Rabs.
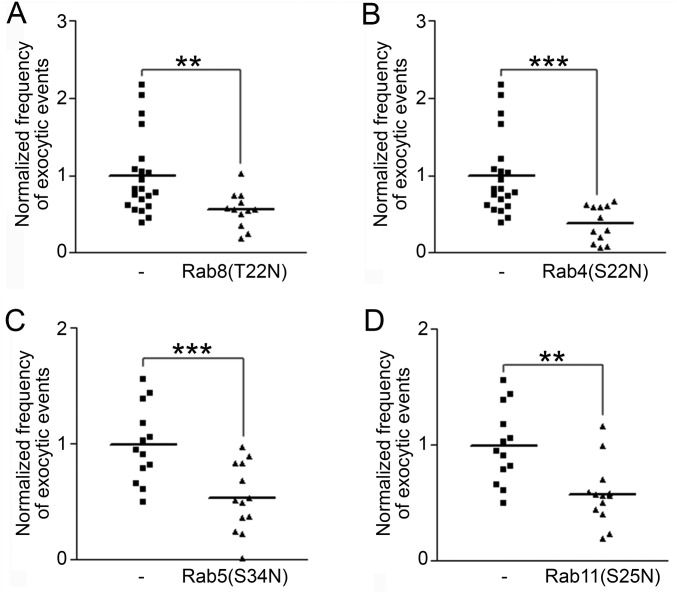
To investigate the source of the pH-NR1–containing exocytic events, we visualized them while coexpressing dominant negative mutants of different Rab proteins, which regulate various steps of intracellular vesicle trafficking (35). Dominant negative mutant of Rab8 [Rab8(T22N)] specifically interferes with de novo exocytosis from the Golgi apparatus to the plasma membrane. Rab5(S34N), Rab4(S22N), and Rab11(S25N) block different steps of recycling, including endocytosis, recycling from the early endosomes to the plasma membrane, and recycling from the recycling endosomes to the plasma membrane, respectively (36). As a result, we observed significant reductions of pH-NR1 exocytic frequencies with the coexpressions of all of the Rab dominant-negative mutants mentioned above (Fig. S4). These results indicate that the exocytic vesicles of pH-NR1 originate from both de novo exocytic and recycling pathways.
Fig. S4.
Exocytosis of NR1 is regulated by Rab proteins. Empty vector or dominant negative mutant [Rab8(T22N) (A), Rab4(S22N) (B), Rab5(S34N) (C), or Rab11(S25N) (D)] was coexpressed with pH-NR1 in dissociated rat hippocampal neurons for 24 h. The cells were then imaged under TIRFM. The same control dataset was used in A and B. Another control dataset was used in both C and D. * indicates statistical significances: **P < 0.01; ***P < 0.001.
Constitutive Exocytosis of NR1 Is Mediated by SNAREs.
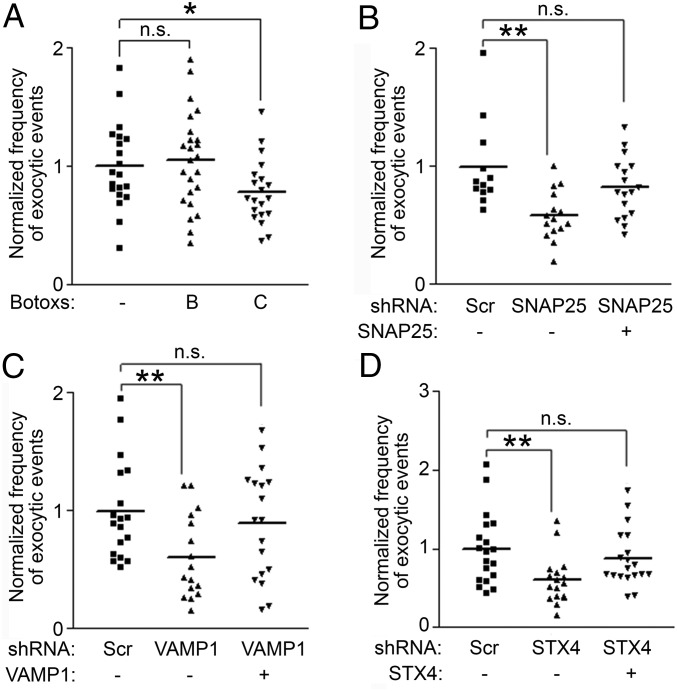
As we showed previously, BotoxA treatment significantly reduced exocytic frequency of pH-NR1 (Fig. 1J), suggesting that SNAP25 is involved in the exocytosis of NMDA receptors. To investigate the role of other SNARE components in regulating exocytosis of NMDA receptors, we examined the effects of two other types of Botoxs: Botoxs B and C (Fig. 2A). As expected, the treatment of BotoxC, which cleaves SNAP25 and syntaxin 1, 2, and 3, reduced the exocytic frequency of pH-NR1. However, the treatment of BotoxB, which cleaves most VAMPs, had no effect on pH-NR1 exocytosis, suggesting that the fusion of pH-NR1–containing vesicles to the plasma membrane requires a BotoxB-insensitive VAMP.
Fig. 2.
Exocytosis of NR1 is mediated by SNARE complexes. (A) Exocytosis of pH-NR1 was inhibited by the treatment of Botox C, but not BotoxB. (B–D) SNARE proteins mediate the exocytosis of pH-NR1. Scramble shRNA (Scr), the shRNA of each SNARE protein [SNAP25 (B), VAMP1 (C), syntaxin4 (STX4) (D)], or the shRNA-resistant SNARE protein was coexpressed with pH-NR1 in dissociated hippocampal neurons. *P < 0.05; **P < 0.01. n.s., no statistical significance.
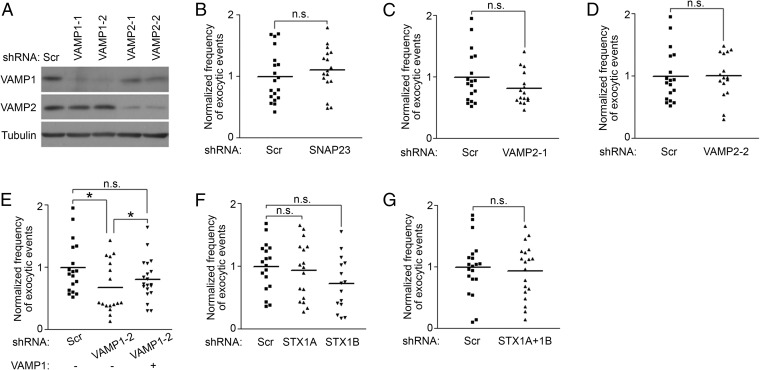
To further confirm the results of the Botox treatments and identify SNARE proteins mediating the exocytosis of NMDA receptors, we next used a shRNA-based approach to specifically knock down individual SNARE proteins and examined the effect of the knockdown on the exocytic frequency of NR1 (Fig. S5A) (23). Consistently, the knockdown of SNAP25 significantly reduced the exocytic frequency of pH-NR1. This effect was rescued by a shRNA-resistant SNAP25 (Fig. 2B). However, SNAP23, which is a homolog of SNAP25 and insensitive to the cleavages of Botoxs A and C, is also expressed in hippocampal neurons (37). SNAP23 was previously reported to regulate the trafficking of NMDA receptors (10, 11). However, we found that the knockdown of SNAP23 did not affect the exocytic frequency of pH-NR1 (Fig. S5B). Overall, these results confirmed the result of BotoxA treatment that SNAP25, rather than SNAP23, mediates the exocytosis of NMDA receptors.
Fig. S5.
Specific SNARE proteins plays different roles in regulating the exocytosis of pH-NR1. (A) Knockdown of VAMP1 or VAMP2 by specific shRNAs. Three micrograms of scramble shRNA and shRNAs specifically targeting VAMP1 or VAMP2 was electroporated into dissociated rat cortical neurons at DIV0 immediately before plating. The cells were lysed 4 d after electroporation, and expression of endogenous VAMP1 and VAMP2 proteins were analyzed. Tubulin was used as an internal control in all experiments. There were two shRNAs targeting VAMP1 gene (VAMP1-1, -2) and two shRNAs targeting VAMP2 gene (VAMP2-1, -2). The shRNA VAMP1-1 was used in Figs. 2C, 3C, and 4. The shRNAs VAMP1-2, VAMP2-1, and VAMP2-2 were used in Fig. S5 E, C, and D, respectively. (B) SNAP23 was not involved in the regulation of the exocytosis of NR1. In dissociated hippocampal neurons, scramble shRNA (Scr) or SNAP23 shRNA were coexpressed with pH-NR1 in dissociated hippocampal neurons. Twenty-four hours after transfection, exocytosis of pH-NR1 was imaged under TIRFM. (C) VAMP2 was not required for exocytosis of NR1. Scramble shRNA (Scr) or shRNA VAMP2-1 were coexpressed with pH-NR1 in dissociated hippocampal neurons. Forty-eight hours after transfection, exocytosis of pH-NR1 was imaged under TIRFM. (D) Knockdown of VAMP2 using another shRNA did not affect exocytic frequency of pH-NR1. Scramble shRNA (Scr) or shRNA VAMP2-2 were coexpressed with pH-NR1 in dissociated hippocampal neurons. Forty-eight hours after transfection, exocytosis of pH-NR1 was imaged under TIRFM. (E) Knockdown of VAMP1 using another shRNA reduced exocytic frequency of pH-NR1. Scramble shRNA (Scr), shRNA VAMP1-2, or shRNA-resistant VAMP1 were coexpressed with pH-NR1 in dissociated hippocampal neurons. Forty-eight hours after transfection, exocytosis of pH-NR1 was imaged under TIRFM. (F) Knockdown of two syntaxin isoforms individually did not affect the exocytosis of pH-NR1. Scramble shRNA (Scr), syntaxin1A shRNA (STX1A), or syntaxin1B shRNA (STX1B) were coexpressed with pH-NR1 in dissociated hippocampal neurons. Thirty-six hours after transfections, exocytosis of pH-NR1 was imaged under TIRFM. (G) Knockdown of two syntaxin isoforms together did not affect the exocytosis of pH-NR1. Scramble shRNA (Scr) or syntaxin1A shRNA (STX1A) plus syntaxin1B shRNA (STX1B) were coexpressed with pH-NR1 in dissociated hippocampal neurons. Thirty-six hours after transfections, exocytosis of pH-NR1 was imaged under TIRFM. *P < 0.05. n.s., no statistical significance.
The results of BotoxB treatment suggest that a BotoxB-insensitive VAMP is involved in exocytosis of NMDA receptors. Among seven mammalian VAMPs (VAMP1, 2, 3, 4, 5, 7, and 8), VAMP1 and VAMP7 are the only two isoforms insensitive to BotoxB. In addition, VAMP1 and VAMP2 are specifically expressed in the brain and mediate presynaptic vesicle fusion events (38–40). Consistent with the result of BotoxB treatment, the knockdowns of VAMP2 by two shRNAs, which specifically targeted different regions of VAMP2, did not alter the exocytic frequency of pH-NR1 (Fig. S5 C and D). However, knockdown of VAMP1 by two independent shRNAs significantly reduced the exocytic frequency of pH-NR1, and these effects were rescued by shRNA-resistant VAMP1s (Fig. 2C and Fig. S5E). These results suggest that VAMP1 is involved in the exocytosis of NMDA receptors.
We then asked which syntaxin is important for the exocytosis of NR1. In rat hippocampal neurons, there are five syntaxins expressed on the plasma membrane: syntaxin1A, 1B, 2, 3, and 4 (41). Most syntaxins, except syntaxin4, are sensitive to the proteolytic activity of BotoxC (42). We specifically investigated roles of syntaxin1A, 1B, and 4, which have higher affinity to SNAP25 than other syntaxins (43, 44). The knockdown of syntaxin4 reduced the exocytic frequency of pH-NR1, and this effect was rescued by the overexpression of shRNA-resistant syntaxin4 (Fig. 2D). However, the knockdown of syntaxin1A and 1B individually or together did not affect the exocytic frequency of NR1 (Fig. S5 F and G). These results indicate that the exocytosis of NR1 is mediated by syntaxin4.
In conclusion, these results show that the SNAP25–VAMP1–syntaxin4 complex potentially mediates the exocytosis of NMDA receptors.
SNAREs Regulates Surface Expression of NMDA Receptors.
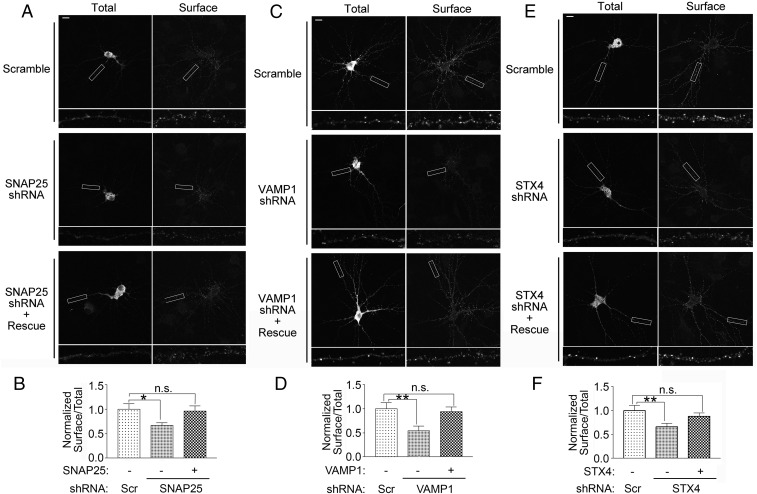
Given the role of the SNAP25–VAMP1–syntaxin4 SNARE complex in mediating the exocytosis of NMDA receptors, we investigated whether the disruption of the SNARE complex affects surface expression of NMDA receptors. We found that individual knockdown of SNAP25, VAMP1, or syntaxin4 reduced the surface expression of pH-NR1, and these effects were rescued by shRNA-resistant mutant of each SNARE protein (Fig. 3). These data further indicate that the SNAP25–VAMP1–syntaxin4 complex is important for maintaining the surface level of NMDA receptors.
Fig. 3.
SNARE proteins are important for surface expression of NR1. (A, C, and E) Knockdowns of SNARE proteins reduced surface expression of pH-NR1. Scramble shRNA, shRNA of each SNARE protein [SNAP25 (A), VAMP1 (C), syntaxin4 (STX4) (E)], or shRNA-resistant SNARE protein (rescue) were coexpressed with pH-NR1, as indicated in the figure. Lower show higher-magnification images of the processes boxed at Top. (Scale bars: 20 µm.) (B, D, and F) Quantification of surface pH-NR1 in A, C, and E, respectively. The knockdown of SNAP25 (B), VAMP1 (D), or syntaxin4 (F) reduced the surface level of pH-NR1, which was the ratio of surface and total pH-NR1 in dendrites. The surface pH-NR1 of each cell was further normalized by the average of the control group transfected with the scramble shRNA. Scr, scramble shRNA. Five dendrites were used from each neuron. n = 12, 29, and 26 neurons for B, D, and F, respectively. *P < 0.05; **P < 0.01. n.s., no statistical significance.
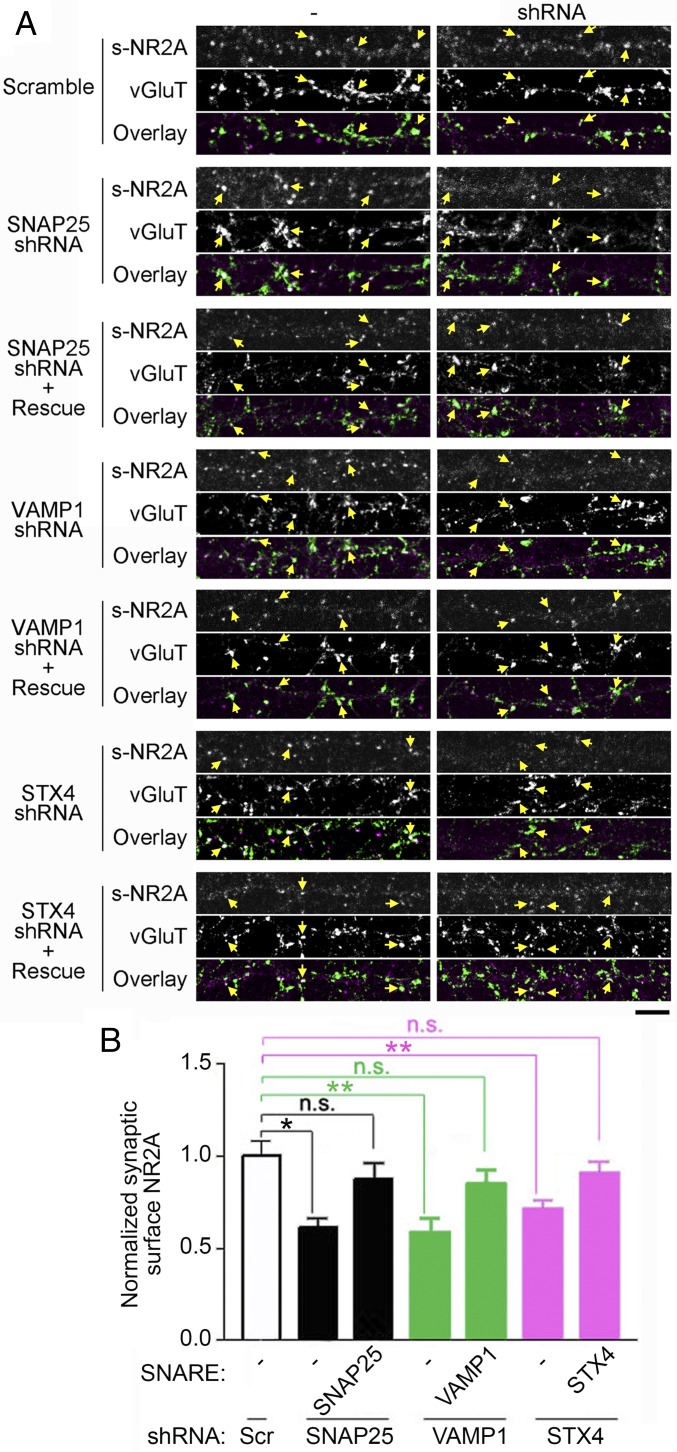
We next examined whether the SNAP25–VAMP1–syntaxin4 complex specifically regulates surface expression of NMDA receptors at excitatory synapses. We found that the knockdown of SNAP25, VAMP1, or syntaxin4 significantly reduced synaptic surface expressions of endogenous NR2A, which was colocalized with the excitatory presynaptic marker VGluT (vesicular glutamate transporter), and these effects were rescued by the overexpression of shRNA-resistant mutants of individual SNARE proteins (Fig. 4).
Fig. 4.
SNARE proteins are important for synaptic surface expression of NR2A. (A) The knockdown of SNAP25, VAMP1, or syntaxin4 reduced the expression of endogenous NR2A at the excitatory postsynaptic membrane. In dissociated hippocampal neurons, scramble shRNA, shRNA of each SNARE protein, or shRNA-resistant SNARE protein (rescue) were coexpressed with EGFP to label transfected cells. For each dataset: Top, surface NR2A; Middle, VGluT; Bottom, overlay of Top and Middle. White puncta show the colocalization of surface NR2A (magenta) and VGluT (green). The synaptic surface NR2A in shRNA-transfected cells (shRNA) was compared with that of nontransfected cells (-) on the same coverslip. Yellow arrows at corresponding locations indicate the synaptic surface NR2A, VGlut, and overlaid signal at the same synapse. (Scale bar: 10 µm.) (B) Quantification of the synaptic surface NR2A in A. The knockdown of SNAP25, VAMP1, or syntaxin4 reduced the synaptic surface level of NR2A, which was determined by the surface NR2A colocalized with VGluT. The synaptic surface NR2A level in each shRNA-transfected cell was compared with nontransfected cells by taking a ratio of the synaptic surface NR2A of the two populations. The ratio in each sample was further normalized by the average of the control transfected with the scramble shRNA. n = 10–15 neurons in each group. Ten synapses were used from each neuron. *P < 0.05; **P < 0.01. n.s., no statistical significance.
Overall, these results suggest that the SNAP25–VAMP1–syntaxin4 SNARE complex regulates surface expression of NMDA receptors at both extrasynaptic and synaptic membranes. Since most NR1-containing vesicles insert into the extrasynaptic membranes, the effects of SNARE proteins on synaptic surface NMDA receptors could result from the depletion of the extrasynaptic pool that replenishes synaptic surface receptors.
Discussion
In the present study, we characterized the constitutive exocytic events of NMDA receptors and identified the SNARE complex required for the exocytosis by combining the TIRFM and immunocytochemical methods. The constitutive exocytosis of NMDA receptors were transient and predominantly occurred on extrasynaptic surface of the soma and dendrites. The frequency of these exocytic events, which originated from both de novo exocytic and recycling pathways, was mediated by the SNAP25–VAMP1–syntaxin4 SNARE complex, and the interruption of this SNARE complex reduced the expression of NMDA receptors on both extrasynaptic and synaptic membranes. These results indicate that the SNAP25–VAMP1–syntaxin4 complex mediates the fusion of NMDA receptor-containing vesicles to the plasma membrane and, therefore, is important for the excitatory synaptic transmission.
The constitutive NMDA receptor exocytic events had fast kinetics and occurred at high frequency on the plasma membrane (Table S1). These features are reminiscent to those of the constitutive exocytic events of AMPA and GABAA receptors (23). In contrast, the activity-dependent exocytic events of GluA1 and GluA2 had slower kinetics and lower frequencies (20–22). Moreover, the constitutive exocytic vesicles contained a small number of receptors compared with the activity-dependent exocytic vesicles (21, 23). These observations suggest two types of exocytic events of neurotransmitter receptors: (i) the constitutive exocytosis occurs through the vesicles with low numbers of the receptors and quickly delivers them to the plasma membrane at high frequency, maintaining the surface receptor level at basal state. (ii) The activity-dependent exocytosis occurs slowly and less frequently through vesicles with a large number of the receptors, supplying receptors in response to the fast and robust changes of neuronal activity. Furthermore, we and others observed that the exocytosis primarily occurred at extrasynaptic sites (Table S1) (20–23, 45). The interruptions of specific SNARE complexes, which mediate the exocytosis, led to the reduction of surface receptors on both extrasynaptic and synaptic membranes (23). These results suggest that neurotransmitter receptors are generally delivered to the extrasynaptic plasma membrane. The synaptic distribution of these receptors is mostly achieved by lateral diffusion of the receptors from extrasynaptic zones to synaptic zones and further stabilization of receptors on the synaptic membrane by scaffold proteins.
Table S1.
Summary of properties of exocytic events of NMDA, AMPA, and GABAA receptors
| Features of exocytic events | Constitutive exocytosis | Activity-dependent exocytosis | |||
| NMDA receptor | AMPA receptor (23) | GABAA receptor (23) | AMPA receptor subunit GluA1 (20-21) | AMPA receptor subunit GluA2 (22) | |
| Duration, s | 2.1 | 2.8 | 2.1 | 10–12 | 10–12 |
| Frequency, events/sec per 100 µm2 | 25 ± 2 | 15 ± 1 | 22 ± 2 | 0.11 (cell body); | 0.04 (estimated based on no. of events per s |
| 0.05 (dendrites) | |||||
| No. of receptor subunits per vesicle | 2.8 ± 0.1 | 2.2 ± 0.1 | 3.9 ± 0.2 | 50 | 30–50 (estimated based on the brightness of events) |
| Exocytic location | Extrasynaptic | Extrasynaptic | Extrasynaptic | Extrasynaptic | Extrasynaptic |
The roles of SNARE proteins, especially SNAP23 and SNAP25, on NMDA receptor exocytosis were shown in many studies. The results of BotoxA treatment, lentiviral-mediated shRNA, or dominant-negative peptide applications showed that SNAP25 was required for activity-dependent NMDA receptor exocytosis including PKC-dependent exocytosis and mGluR1α-induced delivery to the plasma membrane (12–14). SNAP25 RNAi only induced little change of NMDA current at basal state (12). However, the knockdown of SNAP25 expression with lentiviral-mediated shRNA revealed no postsynaptic role of SNAP25 in NMDA receptors trafficking (10). Similarly, no postsynaptic defects were detected after the application of glutamate agonists in SNAP25-deficient neurons (46). On the other hand, SNAP23 regulated the constitutive recycling of NMDA receptors at CA1 synapses, and surface levels of NMDA receptor were reduced in SNAP23+/− mice (10). SNAP23 was also required for exocytosis of NMDA receptor in an early developmental stage (11). All of the studies above, including our current study, show significant inconsistencies, which could be caused by several factors. First, instead of specifically detecting exocytic events, some biochemical and immunocytochemical approaches examine surface receptor levels, which could be the summation of many trafficking steps. Second, the disruption of SNARE complex using genetic ablation and lentiviral approaches could induce compensatory overexpression of other related proteins and the alteration of global neuronal activity as some SNARE proteins are required for neurotransmitter release (39). All of these effects could affect the trafficking of NMDA receptors. To avoid these caveats and specifically study the role of postsynaptic SNARE complex on NMDA receptor excytosis, we took the advantage of the unique features of pHluorin tag and TIRM to isolate exocytic events, as well as combined the acute Botox treatments and shRNA-mediated gene silencing at single cell level.
The function of VAMPs on NMDA receptor exocytosis was indirectly shown in several studies based on the results of neurotoxin treatments. BotoxB had no effect on PKC-induced potentiation of NMDA-elicited currents (13). However, BotoxB blocked the potentiation of NMDA currents induced by PKM, the constitutive active form of PKC (12). In addition, early studies showed that BotoxB treatment did not alter the NMDA receptor-mediated EPSCs (47) and NMDA receptor functions during LTP (48). Tetanus toxin (TeTx), which also cleaves BotoxB-sensitive VAMPs (18), did not prevent the enhancement of NMDA receptor-mEPSCs during glycine-induced LTP (49). Overall, most studies indicate that the exocytosis of NMDA receptor could be mediated by a BotoxB-insensitive VAMP, which is consistent with our current results that the BotoxB-insensitive VAMP1 mediates the exocytosis of NMDA receptor. Additionally, it was shown that syntaxin1 was colocalized with NR2B in spines (16). However, this correlated protein distributions did not provide direct evidence for the role of synatxin1 in the exocytosis of NMDA receptor. The inhibition of syntaxin4 led to the elimination of the APDC-dependent enhancement of NMDAR currents (15). This result is consistent with our conclusion that syntaxin4 mediates the exocytosis of NMDA receptor.
In conclusion, we directly visualized the constitutive exocytic events of NMDA receptors and identified the SNARE complex required for NMDA receptor exocytosis. Our study suggests the function of the SNAP25–VAMP1–syntaxin4 complex in NMDA receptor trafficking and the role of this SNARE complex in NMDA receptor-mediated neuronal functions, especially synaptic transmission and plasticity.
Materials and Methods
Animal Use.
The use and care of animals followed the guideline of the Institutional Animal Care and Use Committee at the Johns Hopkins University.
Fusion Constructs.
pHluorin-NR1 and tdTomato-NR1 were made by replacing GFP in pCI-GFP-NR1 with pHluorin or tdTomato so that they were inserted after amino acid 21 in rat NR1 (28).
shRNAs.
pSuper was used as a vector to generate shRNAs targeting individual rat VAMPs, SNAPs, and syntaxins. The oligos were annealed for direct subcloning into pSuper between BglII and HindIII sites or between BglII and XhoI sites. The sequences of shRNAs are described in SI Materials and Methods.
TIRFM Imaging.
An Olympus IX71 microscope with a plan-Apo objective (100×, N.A. 1.45, oil; Olympus) was used for TIRFM imaging with a 488-nm excitation laser.
Detailed materials and methods are described in SI Materials and Methods.
SI Materials and Methods
Primary Neuron Culture.
Primary hippocampal neurons were dissociated from rat hippocampi of day 18 embryos and seeded on poly-l-lysine precoated 25-mm (for TIRFM imaging) or 18-mm (for immunocytochemistry) coverslips in Neurobasal medium (Invitrogen) supplemented with 50 U/mL penicillin, 50 µg/mL streptomycin, 2 mM glutamax, 2% (vol/vol) B27, and 5% (vol/vol) FBS (plating medium). Media were replaced 24 h after plating with feeding medium (plating medium without serum), and neurons were fed twice a week thereafter. Primary cortical neurons were prepared similarly from rat cortex and seeded on poly-l-lysine–precoated 12-well plates for electroporation. The neurons were grown at 37 °C and 5% (vol/vol) CO2/95% (vol/vol) air.
Transfection and Electroporation.
Dissociated hippocampal neurons were transfected with Lipofectamine 2000 (Invitrogen) at day in vitro (DIV) 11–DIV14 and incubated in Neurobasal medium with supplements [50 U/mL penicillin, 50 µg/mL streptomycin, 2 mM glutamax, 2% (vol/vol) B27], 1 µM MK801 (Tocris), and 100 µM DL-APV (Tocris) for 24–72 h. To test the knockdown efficiency of shRNAs, Amaxa Nucleofector kit (Lonza; catalog no. VPG-1003) and Nucleofector device (Lonza) were used for electroporation of cortical neurons. At DIV0, 3 × 106 neurons were electroporated with 3-µg pSuper-shRNAs, seeded, and harvested 4 d after the electroporation.
shRNAs.
pSuper was used as a vector to generate shRNAs targeting individual rat VAMPs, SNAPs, and syntaxins. The oligos were annealed for direct subcloning into pSuper between BglII and HindIII sites (for scramble and SNAP23 shRNAs) or between BglII and XhoI sites (for VAMP1, VAMP2, SNAP25, syntaxin1A, syntaxin1B, and syntaxin4 shRNAs). The scramble shRNA sequence and shRNA sequences against SNAP23, SNAP25, syntaxin1A, syntaxin1B, syntaxin4, VAMP1-shRNA-1 and VAMP2-shRNA-2 were reported (23). The sequences of VAMP1-shRNA-2 were obtained from ON-TARGET plus siRNAs of Dharmacon (Lafayette, CO) (catalog no. J-091077-10): 5′ GAT CCC CGG ACA TCA TGC GCG TGA ATT TCA AGA GAA TTC ACG CGC ATG ATG TCC TTT TTC 3′ (sense) and 5′ TCG AGA AAA AGG ACA TCA TGC GCG TGA ATT CTC TTG AAA TTC ACG CGC ATG ATG TCC GGG 3′ (antisense). The sequences of VAMP2-shRNA-1 were designed based on the published sequence (50): 5′ GAT CCC CAA CAA GTG CAG CCA AGC TCA ATT CAA GAG ATT GAG CTT GGC TGC ACT TGT TTT TTT C 3′ (sense) and 5′ TCG AGA AAA AAA CAA GTG CAG CCA AGC TCA ATC TCT TGA ATT GAG CTT GGC TGC ACT TGT TGG G-3′.
Chemicals, Pharmacology, and Molecular Biology.
All chemicals were purchased from Sigma-Aldrich unless otherwise specified.
For TIRF experiments, 260 nM BotoxA, activated by 5 mM DTT, was applied to hippocampal neurons in media for 1 h, and the cells were then imaged under TIRFM. Cells treated with the same amount of DTT for 1 h were imaged as controls.
Botulinum type B and C toxins were purchased from METAbiologics. For TIRF experiments, 100 nM BotoxB or C (dissolved in PBS) was applied to hippocampal neurons in media for 6 h and the cells were then imaged under TIRFM. Cells treated with the same amount of PBS for 6 h were imaged as controls.
TTX and bicuculline were purchased from Tocris Bioscience. For molecular biology, all restriction enzymes were purchased from New England Biolabs. DNA sequencing was performed at the Johns Hopkins University Bloomberg School of Public Health Sequencing Facility and Genewiz Sequencing.
TIRFM Imaging and TIRF Data Analysis.
TIRFM imaging was performed on an Olympus IX71 microscope with a plan-Apo objective (100×, N.A. 1.45, oil; Olympus). The excitation laser was a 1W Kr/Ar TIRF laser with launch (model no. SP_2018; Prairie Technologies). pHluorin was excited with a 488-nm laser (1.7 mW to the back aperture of the objective) and collected through a 525- to 550-nm emission filter. Fluorescent signals were captured by a 512 × 512 back-illuminated CCD camera with on-chip multiplication gain and 16 × 16 µm pixels (Cascade 512B; Photometrics) and MetaMorph acquisition software (Molecular Device) at a rate of 1.4 Hz and 500-ms exposure. Neurons between DIV 13 and 16 were used for imaging. All imaging experiments were carried out in artificial cerebrospinal fluid [ACSF; 120 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 25 mM Hepes (pH 7.4), 30 mM glucose] containing 1 µM MK801 (Tocris) and 100 µM DL-APV (Tocris). Temperature controller (model no. TC-202A; Harvard Apparatus) was used to maintain the imaging temperature at 35–37 °C. To increase the signal-to-noise ratio, we photobleached the fluorescent signals for 20 s with maximal laser power under TIRF mode before data acquisition. For the dual-TIRFM imaging, the fluorescent signal of pHlourin was imaged as described above, whereas the fluorescent signal of tdTomato was imaged with a 568-nm laser (2.8 mW to the back aperture of the objective) and a 605- to 655-nm emission filter.
As described previously (23), exocytic frequencies of pHluorin-NR1 were analyzed by combining DiaTrack 3.03 software (Vallotton Semasopht) and ImageJ (US National Institutes of Health). Exocytic events were generated by subtraction of the sequential images 2–11 (acquired in 0.7–7.7 s) by the series of images from 1 to 10 (acquired in 0–7 s) (ImageJ). The number of events of each subtracted image from each channel was picked up and counted by DiaTrack. To get normalized exocytic frequency of a particular cell, numbers of exocytic events from each subtracted images were averaged and further normalized by the surface area of the cell including somatic membrane and proximal dendritic membrane (ImageJ).
Single-Molecule Analysis.
The number of pH-NR1 in individual exocytic vesicles was estimated as described (21, 23). TIRF events of pH-NR1 in transfected hippocampal neurons were imaged under TIRFM. Under the same condition, fluorescent signals of purified EGFP monomer absorbed to poly-d-lysine–coated coverslips were also imaged under TIRFM. Single molecules of EGFP were identified by the blinking and single-step photobleaching characters. The fluorescent intensities of pH-NR1 events and EGFP monomers were background-subtracted, plotted, and fitted by Gaussian curve (OriginPro 8; OriginLab). The intensity corresponding to the peak of the curve was used to estimate number of pH-NR1 in each event.
Immunocytochemistry and Image Data Analysis.
Surface expression of pH-NR1 in dissociated hippocampal neurons was labeled by incubating live neurons in ACSF containing 10% (wt/vol) BSA and anti-GFP antibody (JH4030, rabbit) (51) for 20 min at 10 °C. The neurons were then washed twice with ice-cold ACSF containing 10% (wt/vol) BSA for washout of the excess antibodies and then fixed with 4% (vol/vol) paraformaldehyde, 4% (wt/vol) sucrose in PBS at room temperature for 15 min. Alexa568-conjugated secondary antibodies (Invitrogen) were applied following fixation to visualize the surface staining of GFP antibody [in PBS containing 10% (wt/vol) BSA, room temperature, 60 min]. The neurons were then permeabilized with PBS containing 0.25% Triton X-100 at room temperature for 20 min. The neurons were blocked in 10% (wt/vol) BSA in PBS at room temperature for 1 h. Total expression pH-GluR2 was stained with a different anti-GFP antibody (mouse 3E6; Invitrogen) in PBS containing 10% (wt/vol) BSA at room temperature for 2 h. Alexa488-conjugated secondary antibodies (Invitrogen) were applied to visualize the total staining of GFP antibody (in PBS containing 10% (wt/vol) BSA, room temperature, 60 min). Surface expression of endogenous NR2A was stained with NR2A antibody (JH6097) (51) by following the similar protocol of surface pH-NR1 staining. Excitatory presynaptic marker VGluT was stained with anti-VGluT antibody (Millipore). Alexa647-conjugated secondary antibodies (Invitrogen) were applied to visualize the staining of VGluT. Fluorescence images were acquired on a Zeiss LSM510 by using a 63× objective (N.A. 1.40). Fluorescent intensities were quantified by using ImageJ.
Statistics.
Unpaired t test was used for statistical analysis and is performed by using Prism software (GraphPad Software). The statistically significant differences were determined when the two-tailed P value < 0.05. Values were expressed as mean ± SEM (standard error of the mean).
Supplementary Material
Acknowledgments
We thank members of the R.L.H. laboratory for their constructive comments during the execution of this study, Dr. John Goutsias for advice on TIRFM imaging analysis, Dr. José Esteban for providing cDNAs of Rab dominant negative mutants, and Dr. A. Kimberley McAllister for providing pCI-GFP-NR1 construct. TIRF microscope and relevant technical assistance were provided by the Microscope Facility & Imaging for Institute for Basic Biomedical Sciences at the Johns Hopkins School of Medicine. This research was supported by NIH Grants R01 NS036715 and R01 MH64856. R.L.H. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614042113/-/DCSupplemental.
References
- 1.Lau CG, Zukin RS. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat Rev Neurosci. 2007;8(6):413–426. doi: 10.1038/nrn2153. [DOI] [PubMed] [Google Scholar]
- 2.Bekkers JM, Stevens CF. NMDA and non-NMDA receptors are co-localized at individual excitatory synapses in cultured rat hippocampus. Nature. 1989;341(6239):230–233. doi: 10.1038/341230a0. [DOI] [PubMed] [Google Scholar]
- 3.Jones KA, Baughman RW. Both NMDA and non-NMDA subtypes of glutamate receptors are concentrated at synapses on cerebral cortical neurons in culture. Neuron. 1991;7(4):593–603. doi: 10.1016/0896-6273(91)90372-7. [DOI] [PubMed] [Google Scholar]
- 4.Petralia RS, Yokotani N, Wenthold RJ. Light and electron microscope distribution of the NMDA receptor subunit NMDAR1 in the rat nervous system using a selective anti-peptide antibody. J Neurosci. 1994;14(2):667–696. doi: 10.1523/JNEUROSCI.14-02-00667.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel SJ, et al. Regional, cellular, and ultrastructural distribution of N-methyl-D-aspartate receptor subunit 1 in monkey hippocampus. Proc Natl Acad Sci USA. 1994;91(2):564–568. doi: 10.1073/pnas.91.2.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson RR, Jiang X, Burkhalter A. Regional and laminar differences in synaptic localization of NMDA receptor subunit NR1 splice variants in rat visual cortex and hippocampus. J Comp Neurol. 1996;368(3):335–355. doi: 10.1002/(SICI)1096-9861(19960506)368:3<335::AID-CNE2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 7.Siegel SJ, et al. Distribution of the excitatory amino acid receptor subunits GluR2(4) in monkey hippocampus and colocalization with subunits GluR5-7 and NMDAR1. J Neurosci. 1995;15(4):2707–2719. doi: 10.1523/JNEUROSCI.15-04-02707.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kharazia VN, Phend KD, Rustioni A, Weinberg RJ. EM colocalization of AMPA and NMDA receptor subunits at synapses in rat cerebral cortex. Neurosci Lett. 1996;210(1):37–40. doi: 10.1016/0304-3940(96)12658-6. [DOI] [PubMed] [Google Scholar]
- 9.Groc L, Bard L, Choquet D. Surface trafficking of N-methyl-D-aspartate receptors: Physiological and pathological perspectives. Neuroscience. 2009;158(1):4–18. doi: 10.1016/j.neuroscience.2008.05.029. [DOI] [PubMed] [Google Scholar]
- 10.Suh YH, et al. A neuronal role for SNAP-23 in postsynaptic glutamate receptor trafficking. Nat Neurosci. 2010;13(3):338–343. doi: 10.1038/nn.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Washbourne P, Liu XB, Jones EG, McAllister AK. Cycling of NMDA receptors during trafficking in neurons before synapse formation. J Neurosci. 2004;24(38):8253–8264. doi: 10.1523/JNEUROSCI.2555-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau CG, et al. SNAP-25 is a target of protein kinase C phosphorylation critical to NMDA receptor trafficking. J Neurosci. 2010;30(1):242–254. doi: 10.1523/JNEUROSCI.4933-08.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lan JY, et al. Protein kinase C modulates NMDA receptor trafficking and gating. Nat Neurosci. 2001;4(4):382–390. doi: 10.1038/86028. [DOI] [PubMed] [Google Scholar]
- 14.Lan JY, et al. Activation of metabotropic glutamate receptor 1 accelerates NMDA receptor trafficking. J Neurosci. 2001;21(16):6058–6068. doi: 10.1523/JNEUROSCI.21-16-06058.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng J, Liu W, Duffney LJ, Yan Z. SNARE proteins are essential in the potentiation of NMDA receptors by group II metabotropic glutamate receptors. J Physiol. 2013;591(16):3935–3947. doi: 10.1113/jphysiol.2013.255075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hussain S, Ringsevjen H, Egbenya DL, Skjervold TL, Davanger S. SNARE protein Syntaxin-1 colocalizes closely with NMDA receptor subunit NR2B in postsynaptic spines in the hippocampus. Front Mol Neurosci. 2016;9:10. doi: 10.3389/fnmol.2016.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Südhof TC, Rothman JE. Membrane fusion: Grappling with SNARE and SM proteins. Science. 2009;323(5913):474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schiavo G, Matteoli M, Montecucco C. Neurotoxins affecting neuroexocytosis. Physiol Rev. 2000;80(2):717–766. doi: 10.1152/physrev.2000.80.2.717. [DOI] [PubMed] [Google Scholar]
- 19.Sudhof TC. Neurotransmitter release. Handb Exp Pharmacol. 2008;(184):1–21. doi: 10.1007/978-3-540-74805-2_1. [DOI] [PubMed] [Google Scholar]
- 20.Lin DT, et al. Regulation of AMPA receptor extrasynaptic insertion by 4.1N, phosphorylation and palmitoylation. Nat Neurosci. 2009;12(7):879–887. doi: 10.1038/nn.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yudowski GA, et al. Real-time imaging of discrete exocytic events mediating surface delivery of AMPA receptors. J Neurosci. 2007;27(41):11112–11121. doi: 10.1523/JNEUROSCI.2465-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Araki Y, Lin DT, Huganir RL. Plasma membrane insertion of the AMPA receptor GluA2 subunit is regulated by NSF binding and Q/R editing of the ion pore. Proc Natl Acad Sci USA. 2010;107(24):11080–11085. doi: 10.1073/pnas.1006584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu Y, et al. Differential vesicular sorting of AMPA and GABAA receptors. Proc Natl Acad Sci USA. 2016;113(7):E922–E931. doi: 10.1073/pnas.1525726113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Axelrod D. Total internal reflection fluorescence microscopy in cell biology. Traffic. 2001;2(11):764–774. doi: 10.1034/j.1600-0854.2001.21104.x. [DOI] [PubMed] [Google Scholar]
- 25.Miesenböck G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394(6689):192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 26.Bard L, et al. Dynamic and specific interaction between synaptic NR2-NMDA receptor and PDZ proteins. Proc Natl Acad Sci USA. 2010;107(45):19561–19566. doi: 10.1073/pnas.1002690107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Washbourne P, Bennett JE, McAllister AK. Rapid recruitment of NMDA receptor transport packets to nascent synapses. Nat Neurosci. 2002;5(8):751–759. doi: 10.1038/nn883. [DOI] [PubMed] [Google Scholar]
- 28.Barria A, Malinow R. Subunit-specific NMDA receptor trafficking to synapses. Neuron. 2002;35(2):345–353. doi: 10.1016/s0896-6273(02)00776-6. [DOI] [PubMed] [Google Scholar]
- 29.Khiroug SS, et al. Dynamic visualization of membrane-inserted fraction of pHluorin-tagged channels using repetitive acidification technique. BMC Neurosci. 2009;10:141. doi: 10.1186/1471-2202-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sankaranarayanan S, De Angelis D, Rothman JE, Ryan TA. The use of pHluorins for optical measurements of presynaptic activity. Biophys J. 2000;79(4):2199–2208. doi: 10.1016/S0006-3495(00)76468-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patterson GH, Knobel SM, Sharif WD, Kain SR, Piston DW. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys J. 1997;73(5):2782–2790. doi: 10.1016/S0006-3495(97)78307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haggie PM, Verkman AS. Monomeric CFTR in plasma membranes in live cells revealed by single molecule fluorescence imaging. J Biol Chem. 2008;283(35):23510–23513. doi: 10.1074/jbc.C800100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moerner WE, Peterman EJ, Brasselet S, Kummer S, Dickson RM. Optical methods for exploring dynamics of single copies of green fluorescent protein. Cytometry. 1999;36(3):232–238. doi: 10.1002/(sici)1097-0320(19990701)36:3<232::aid-cyto13>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 34.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22(12):1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 35.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10(8):513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 36.Gerges NZ, Backos DS, Esteban JA. Local control of AMPA receptor trafficking at the postsynaptic terminal by a small GTPase of the Rab family. J Biol Chem. 2004;279(42):43870–43878. doi: 10.1074/jbc.M404982200. [DOI] [PubMed] [Google Scholar]
- 37.Ravichandran V, Chawla A, Roche PA. Identification of a novel syntaxin- and synaptobrevin/VAMP-binding protein, SNAP-23, expressed in non-neuronal tissues. J Biol Chem. 1996;271(23):13300–13303. doi: 10.1074/jbc.271.23.13300. [DOI] [PubMed] [Google Scholar]
- 38.Bennett MK, et al. The syntaxin family of vesicular transport receptors. Cell. 1993;74(5):863–873. doi: 10.1016/0092-8674(93)90466-4. [DOI] [PubMed] [Google Scholar]
- 39.Lin RC, Scheller RH. Mechanisms of synaptic vesicle exocytosis. Annu Rev Cell Dev Biol. 2000;16:19–49. doi: 10.1146/annurev.cellbio.16.1.19. [DOI] [PubMed] [Google Scholar]
- 40.Hong W. SNAREs and traffic. Biochim Biophys Acta. 2005;1744(2):120–144. doi: 10.1016/j.bbamcr.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 41.Teng FY, Wang Y, Tang BL. The syntaxins. Genome Biol. 2001;2(11):REVIEWS3012. doi: 10.1186/gb-2001-2-11-reviews3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schiavo G, Shone CC, Bennett MK, Scheller RH, Montecucco C. Botulinum neurotoxin type C cleaves a single Lys-Ala bond within the carboxyl-terminal region of syntaxins. J Biol Chem. 1995;270(18):10566–10570. doi: 10.1074/jbc.270.18.10566. [DOI] [PubMed] [Google Scholar]
- 43.Chen D, Minger SL, Honer WG, Whiteheart SW. Organization of the secretory machinery in the rodent brain: Distribution of the t-SNAREs, SNAP-25 and SNAP-23. Brain Res. 1999;831(1-2):11–24. doi: 10.1016/s0006-8993(99)01371-2. [DOI] [PubMed] [Google Scholar]
- 44.Pevsner J, et al. Specificity and regulation of a synaptic vesicle docking complex. Neuron. 1994;13(2):353–361. doi: 10.1016/0896-6273(94)90352-2. [DOI] [PubMed] [Google Scholar]
- 45.Makino H, Malinow R. AMPA receptor incorporation into synapses during LTP: The role of lateral movement and exocytosis. Neuron. 2009;64(3):381–390. doi: 10.1016/j.neuron.2009.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Washbourne P, et al. Genetic ablation of the t-SNARE SNAP-25 distinguishes mechanisms of neuroexocytosis. Nat Neurosci. 2002;5(1):19–26. doi: 10.1038/nn783. [DOI] [PubMed] [Google Scholar]
- 47.Lüscher C, et al. Role of AMPA receptor cycling in synaptic transmission and plasticity. Neuron. 1999;24(3):649–658. doi: 10.1016/s0896-6273(00)81119-8. [DOI] [PubMed] [Google Scholar]
- 48.Lledo PM, Zhang X, Südhof TC, Malenka RC, Nicoll RA. Postsynaptic membrane fusion and long-term potentiation. Science. 1998;279(5349):399–403. doi: 10.1126/science.279.5349.399. [DOI] [PubMed] [Google Scholar]
- 49.Lu W, et al. Activation of synaptic NMDA receptors induces membrane insertion of new AMPA receptors and LTP in cultured hippocampal neurons. Neuron. 2001;29(1):243–254. doi: 10.1016/s0896-6273(01)00194-5. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki J, Jin ZG, Meoli DF, Matoba T, Berk BC. Cyclophilin A is secreted by a vesicular pathway in vascular smooth muscle cells. Circ Res. 2006;98(6):811–817. doi: 10.1161/01.RES.0000216405.85080.a6. [DOI] [PubMed] [Google Scholar]
- 51.Hayashi T, Thomas GM, Huganir RL. Dual palmitoylation of NR2 subunits regulates NMDA receptor trafficking. Neuron. 2009;64(2):213–226. doi: 10.1016/j.neuron.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.