Significance
The ability of pathogenic microorganisms to acquire iron from heme has been shown to be a valid antimicrobial target. However, all of the known mechanisms for heme catabolism that lead to iron release in bacteria and higher eukaryotes are dependent on molecular oxygen as a cosubstrate. The human gut is dominated by strictly anaerobic bacteria, and in this work, we provide evidence for anaerobic heme degradation in enteric pathogens. The anaerobic mechanism is significantly different from what has been observed in other bacteria and may provide a unique opportunity for a new class of antimicrobial compounds.
Keywords: heme, pathogen, anaerobic, intestinal microbiome, radical SAM
Abstract
All of the heme-degrading enzymes that have been characterized to date require molecular oxygen as a cosubstrate. Escherichia coli O157:H7 has been shown to express heme uptake and transport proteins, as well as use heme as an iron source. This enteric pathogen colonizes the anaerobic space of the lower intestine in mammals, yet no mechanism for anaerobic heme degradation has been reported. Herein we provide evidence for an oxygen-independent heme-degradation pathway. Specifically, we demonstrate that ChuW is a radical S-adenosylmethionine methyltransferase that catalyzes a radical-mediated mechanism facilitating iron liberation and the production of the tetrapyrrole product we termed “anaerobilin.” We further demonstrate that anaerobilin can be used as a substrate by ChuY, an enzyme that is coexpressed with ChuW in vivo along with the heme uptake machinery. Our findings are discussed in terms of the competitive advantage this system provides for enteric bacteria, particularly those that inhabit an anaerobic niche in the intestines.
Iron acquisition is essential to the survival of all cellular organisms and is a major barrier for a microorganism during pathogenesis of a mammalian host (1). For example, an enteric pathogen entering through the digestive tract must compete with the host, as well as other intestinal microflora for iron. Interestingly, the most abundant source of dietary iron is found in heme (2), a molecule that is also responsible for essential cellular processes by serving as a cofactor in a variety of enzymes (3). The degradation of heme also plays an important role in iron homeostasis and cell signaling in cyanobacteria, plants, and mammals (1, 4). Considering the bioavailability of dietary heme and the importance of iron as a nutrient, it is not surprising that pathways have evolved in pathogenic bacteria to transport and degrade heme for the sole purpose of iron acquisition (5). Furthermore, the ability to accomplish the liberation of iron from heme under strictly anaerobic conditions would be advantageous to the enteric bacteria that can inhabit certain niches of the intestines.
Enzymes that catalyze the opening of the porphyrin ring have been well characterized and are collectively referred to as heme oxygenases (6). This classification is based on the common mechanistic property of activating molecular oxygen by a “P450-like” mechanism to catalyze the oxidative degradation of the heme cofactor (7–9). Recently, two heme oxygenases have been characterized in the organisms Staphylococcus aureus and Mycobacterium tuberculosis that degrade heme to the unique chromophores staphylobilin (10) and mycobilin (11), respectively. Although molecular oxygen is still required, neither of these degradation mechanisms result in carbon monoxide production, presumably due to the pathophysiology of these organisms (12). These reports significantly advance the hypothesis that organisms evolved a variety of heme degradation mechanisms to satisfy their own physiological needs (13), thus raising the following question: how do hemolytic organisms use heme as an iron source during infection or colonization in an anaerobic environment?
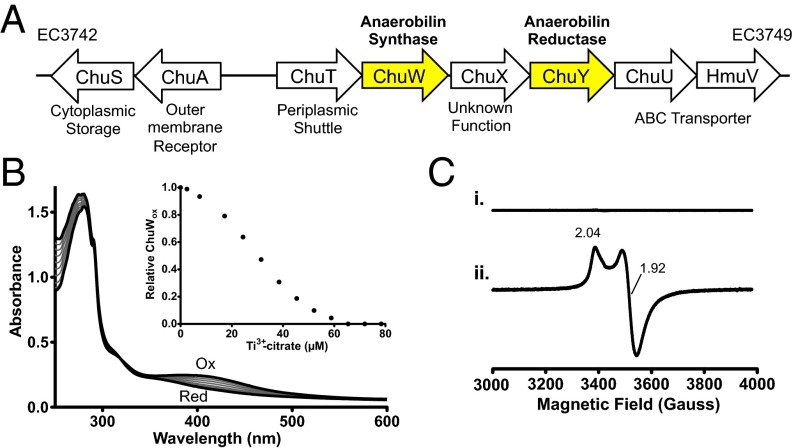
Vibrio cholerae and Escherichia coli O157:H7 are hemolytic enteric pathogens that colonize the nonsterile region of the lower intestines. Despite their ability to use heme as an iron source, a bona fide heme oxygenase has not been characterized for either organism. However, both organisms contain a heme uptake operon that is up-regulated during iron duress (14, 15). The operon encodes for the heme uptake and transport machinery, as well as three additional genes, chuW, chuX, and chuY. (Fig. 1A) (16). ChuW is annotated as a member of the radical S-adenosylmethionine (SAM) superfamily, a large class of enzymes that perform a wide array of difficult chemical reactions (17, 18). Enzymes belonging to the radical SAM superfamily share common cofactor requirements and mechanistic features, such as the coordination of a redox active [4Fe-4S] cluster ligated by three protein-derived cysteine residues, often located within a CX3CX2C motif. This coordination environment facilitates the interaction of SAM at the unique iron site of the cluster. In the reduced state (formally 1+), the [4Fe-4S] cluster reductively cleaves SAM, forming a highly oxidative 5′-deoxyadenosyl-5′-radical (5′-dA•) that is subsequently used for catalysis. ChuW was originally annotated as the radical SAM enzyme HemN, an anaerobic coproporphyrinogen oxidase. HemN is involved in the anaerobic biosynthesis of heme, performing two subsequent decarboxylation reactions to produce protoporphyrinogen IX (19). However, previous work has shown that ChuW homologs do not retain the essential functional motifs, nor do they rescue a HemN KO in Salmonella enterica, and thus likely catalyze a different reaction (14, 20). A role in heme biosynthesis is also inconsistent with the location of chuW in an iron-regulated operon adjacent to genes that are required for heme uptake (Fig. 1).
Fig. 1.
Genetic organization of the heme utilization operon in E. coli O157:H7 and characterization of the Fe-S cluster in purified ChuW. (A) ChuW is genetically adjacent to the heme uptake machinery that is expressed during iron starvation. (B) UV-visible spectra of isolated ChuW and reduction of the Fe-S cluster. Scans were recorded after injecting with equivalents of Ti3+citrate. (Inset) Percentage of oxidized ChuW based on the relative change in the adsorption at 400 nm plotted with respect to the concentration of Ti3+citrate added. (C) EPR spectra of ChuW in the absence and presence of 1 mM sodium dithionite (top and bottom trace, respectively). Spectra are a result of three scans recorded at 10 K with 1-MW microwave power, a modulation frequency of 100 kHz, and modulation amplitude of 6.477 gauss (G).
Sequence alignments indicate that ChuW has homology to the radical SAM methyltransferases (RSMTs). RSMTs are a subfamily of radical SAM enzymes that function in the transfer of methyl groups to unreactive carbon atoms and, in some cases, promote significant chemical rearrangements (21, 22). However, they are the least understood of the four distinguished classes (23), making it difficult to determine function or mechanism using only a bioinformatics approach. To address the precise function of ChuW from the enterohemorrhagic serotype of E. coli O157:H7, we isolated and characterized the recombinant enzyme. Our data provide in vitro evidence that ChuW functions as an anaerobic heme-degrading enzyme that uses a mechanism similar to what has been reported for other RSMTs.
Results
Isolation of ChuW with an Intact [4Fe-4S] Cluster.
Similar to what has been reported for other radical SAM enzymes, the most stable form of ChuW was obtained when we coexpressed the Azotobacter vinelandii iron sulfur cluster (isc) biosynthesis genes (pDB1282 vector, a generous gift from Dennis Dean, Department of Biochemistry, Virginia Tech University, Blacksburg, VA) and chuW in E. coli using minimal media that was supplemented with exogenous iron and cysteine during expression. Isolation of ChuW using this approach yielded a protein with 3.8 ± 0.2 irons per monomer of purified enzyme, following the reconstitution and purification procedure described in SI Materials and Methods. We investigated the spectroscopic characteristics of the reconstituted enzyme using UV-visible spectroscopy. The purified enzyme (20 μM) exhibited a broad absorption feature with a maximum near 400 nm, indicative of the coordination of a [4Fe-4S] cluster (Fig. 1B). Injecting incremental aliquots of Ti3+-citrate into the sample resulted in a bleaching of color and decreased the absorption feature around 400 nm. The relative percentage of oxidized ChuW was plotted with respect to the concentration of Ti3+-citrate injected (Fig. 1B, Inset) and suggests a single electron reduction of the cluster. The oxidation state of the [4Fe-4S] cluster was further investigated by electron paramagnetic resonance (EPR) (Fig. 1C). In the presence of sodium dithionite, we observed an axial EPR spectrum (g|| = 2.04, g | = 1.92; Fig. 1C, spectra ii) that is typical for radical SAM enzymes that contain only the catalytic [4Fe-4S] cluster (24, 25). As expected, the [4Fe-4S] cluster of ChuW was extremely oxygen sensitive, and enzyme activity was lost if exposed to any level of molecular oxygen.
Flavodoxin Is the Electron Source for the Radical SAM Enzyme ChuW.
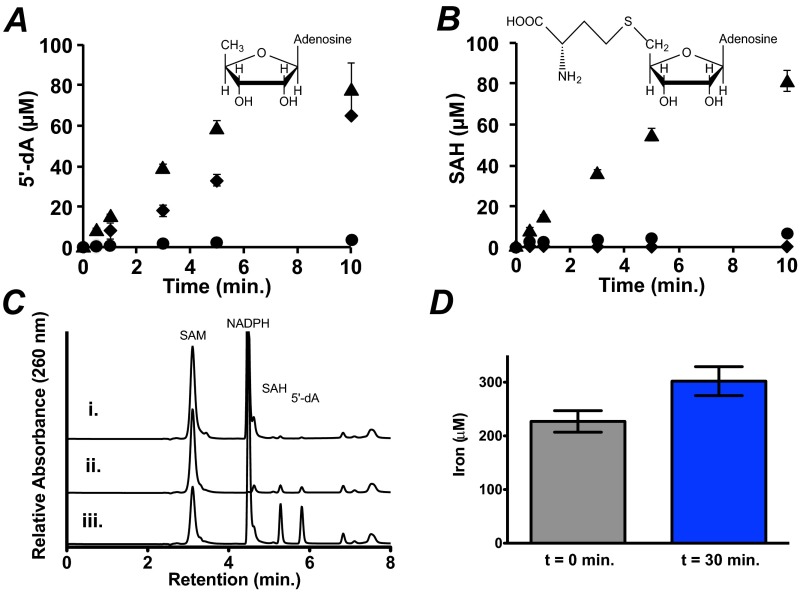
In addition to being a radical SAM enzyme, sequence analysis indicates that ChuW is distinct from HemN and more similar to the class C RSMTs (Fig. S1). Therefore, we looked for both 5′-deoxyadenosine (5′-dA) and S-adenosylhomocysteine (SAH) production during ChuW turnover by HPLC. In our assays, we used the heme analog deuteroheme as the substrate due to the insolubility of hemin. Consistent with what others have reported, we observed “abortive cleavage” of SAM to 5′-dA in the absence of deuteroheme when using the chemical reductant sodium dithionite (Fig. S2A). Minimal SAH production was observed when deuteroheme was absent and the chemical reductant was used (Fig. S2B). However, when using the E. coli flavodoxin (EcFldA)/ferredoxin (flavodoxin):NADP+ oxidoreductase (EcFpr)/NADPH system (26), abortive cleavage was not observed, and both 5′-dA and SAH were produced at 1:1 stoichiometries only when substrate was present (Fig. S2C, trace ii). These observations further underscore the importance of using a physiological electron source for the in vitro characterization of radical SAM enzymes (26–28). Therefore, we used the physiological electron donor system in place of a chemical reductant. Quantitation of the total iron that was subject to chelation (labile iron) was also performed. Measurements of the labile iron were performed at the start of the reaction and after 30 min. It should be recognized that the catalytic cluster is fragile and also subject to chelation in contrast to iron bound to the porphyrin. Notably, we observe an increase in the labile iron during ChuW turnover; specifically, we measured 227 ± 20 μM labile iron at the start of the assay and 302 ± 27 μM after a 30-min reaction (Fig. S2D).
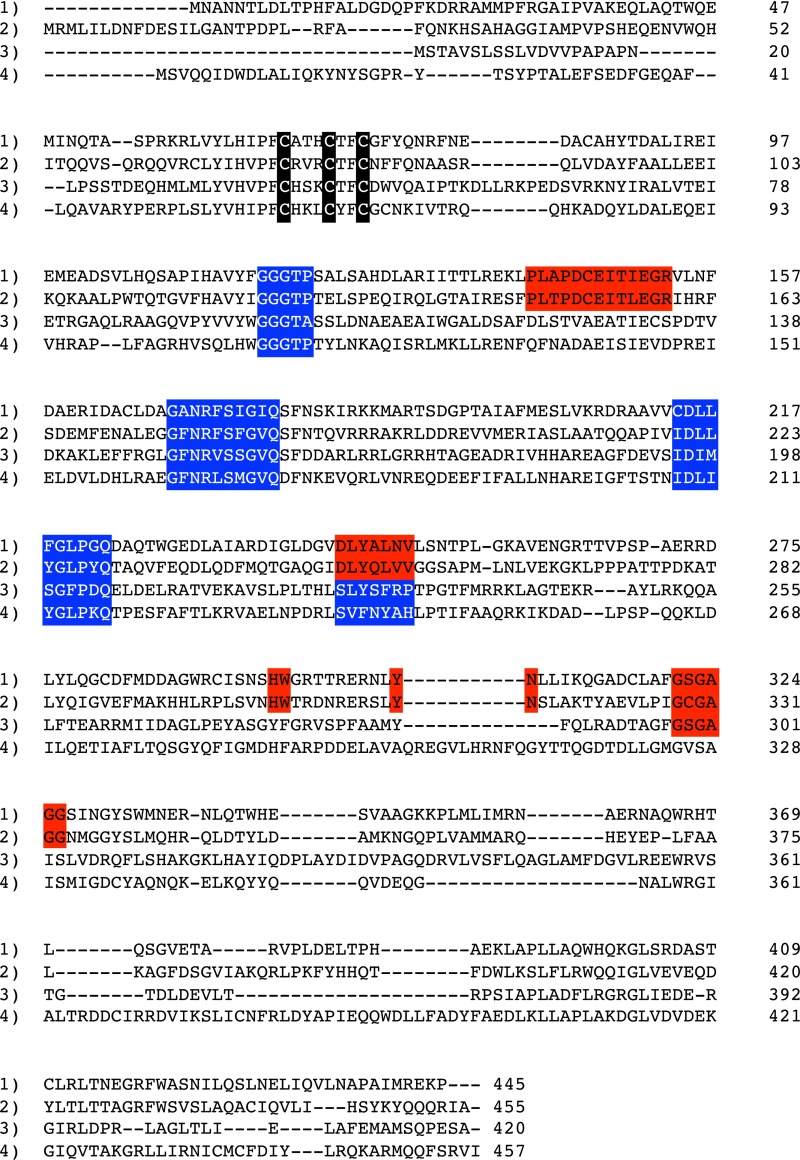
Fig. S1.
Protein sequence alignment of ChuW from E. coli O157:H7 (1) with V. cholera HutW (2), S. actuosus NosN (3), and E. coli HemN (4). Sequences highlighted in red represent motifs specific to ChuW/HutW homologs, whereas sequences in blue are homologous in all proteins presented. Residues highlighted in black correspond to the cysteines in the radical SAM CxxxCxxC motif.
Fig. S2.
Quantification of 5′-dA, SAH, and iron produced during ChuW reaction(s). Separation and quantification of 5′-dA and SAH using HPLC was performed as described in SI Materials and Methods. In the absence of sodium dithionite (circles), no SAH or 5′-dA was produced (cf. A and B); however, in the absence of only deuteroheme (diamonds) 5′-dA was still produced in a linear manner, whereas SAH was not detected (cf. A and B). In the presence of deuteroheme (triangles), both SAH and 5′-dA were produced in a 1:1 ratio (cf. A and B). Products were quantified using a standard curve, and assays were performed in triplicate. (C) HPLC traces showing the results of a 20-min assay using the physiological electron donor system flavodoxin, NADPH/flavodoxin reductase in the absence of NADPH (trace i), the absence of heme (trace ii), or with the complete assay mix (trace iii). (D) Total labile iron present in the ChuW assay at the start of the reaction and after 30 min.
Heme, Mesoheme, and Deuteroheme Are Substrates for ChuW.
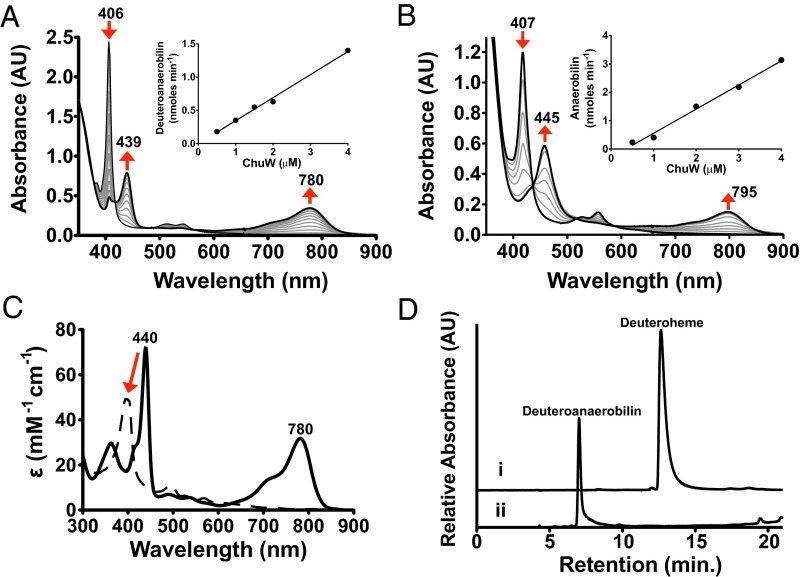
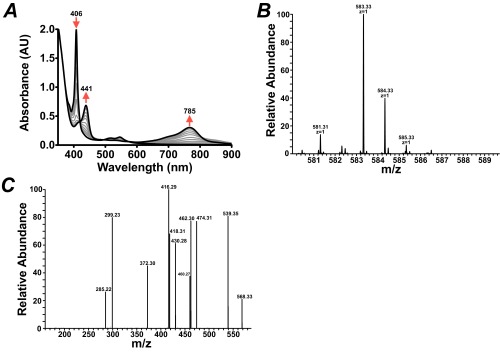
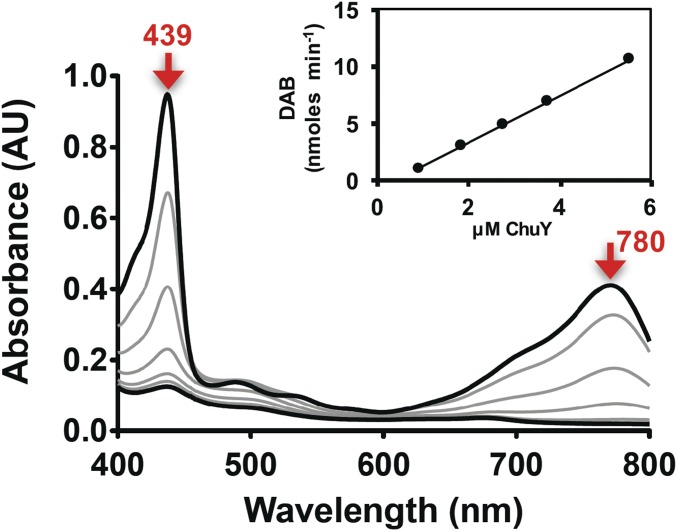
No notable changes in the UV-visible spectrum of heme could be directly associated with heme-iron coordination by ChuW. However, when all of the components required for RSMT activity were present, distinct absorption changes for heme or deuteroheme (between 350 and 900 nm) were observed during a 30-min reaction (Fig. 2 A and B, respectively). Turnover of mesoheme was also monitored by UV-visible spectroscopy, and the products were analyzed by MS (Fig. S3A). Under our assay conditions, deuteroheme was soluble and preferentially used as a substrate, although similar results were obtained with mesoheme and hemin. For deuteroheme, turnover resulted in a decrease in its major Soret band at 407 nm, with a corresponding increase in absorbance at 439 and 780 nm (Fig. 2A). Based on the isosbestic point observed at 418 nm and the initial concentration of deuteroheme, we calculated an extinction coefficient for the product(s) absorbing at 780 nm to be 31.5 mM−1⋅cm−1. Similar results were obtained when the physiological substrate, heme, is used in the assay, albeit with red-shifted spectral features (Fig. 2B). The calculated extinction coefficients for the product were used to determine the specific activities by plotting the initial rate of product formation with respect to the concentration of ChuW (Fig. 2, A and B, Inset). From this analysis the specific activities were calculated to be 5.5 and 8.4 nmol⋅min−1⋅mg−1 for deuteroheme and heme, respectively. We have not established an accurate Vmax for any of the ChuW catalyzed reactions and therefore cannot report a turnover number. In addition, we are not using a coupled assay and cannot account for potential feedback inhibition of product, as has been observed for the canonical heme oxygenase. Finally, we cannot account for any photodegradation that may be occurring. Given this precedent, it is possible that we are underestimating the true rate and the development of a coupled assay is underway. However, to make a catalytic comparison, it is useful to look at the rate relative to the molar amount of ChuW in the assay. In this case, the enzyme is catalyzing the conversion of 0.289 and 0.442 nmol⋅min−1⋅(nmol ChuW)−1 for deuteroheme and heme, respectively. By comparison, using a coupled assay, hHO-1 has been shown to consume heme at a rate of ∼3.3 nmol⋅min−1⋅(nmol of hHO-1)−1 (29). Consistent with the heme oxygenase literature, we have named products of ChuW-catalyzed heme, deuteroheme, and mesoheme degradation “anaerobilin,” “deuteroanaerobilin,” and “mesoanaerobilin,” respectively.
Fig. 2.
Characterization of anaerobic ChuW-dependent heme degradation by UV-visible spectroscopy. UV-visible spectroscopic assays were performed by adding ChuW (2 μM) to a solution containing 10 μM deuteroheme (A) or heme (B), respectively, as well as the EcFld/EcFpr electron donor system (5 and 2 μM, respectively) with 200 μM NADPH. Assays were initiated by the addition of SAM (250 μM), and spectra were recorded every 2 min over the course of 30 min. The red arrows indicate the direction of spectral changes during the course of the experiment. (Insets) Plot of the product formation rate at various ChuW concentrations. (C) The UV-visible spectrum of isolated DAB in methanol resulted in similar spectroscopic features observed in the enzyme assay (solid spectra). Exposure to sunlight resulted in significant changes to the spectra (dashed line). (D) HPLC chromatogram (detection at 397 nm) following the isolation of the deuteroheme substrate and the product of ChuW turnover, herein termed DAB.
Fig. S3.
(A) Spectroscopic assay using mesoheme as a substrate, (B) MS spectra of the isolated mesoanaerobilin product acquired by direct infusion on an LTQ-Orbitrap mass spectrometer, and (C) MS/MS of the 583.33 peak acquired in B.
To assess the properties of the tetrapyrrole that was produced, we isolated the deuteroheme-derived product, deuteroanaerobilin (DAB). Exposure of DAB to sunlight for 5 min resulted in a significant change in the UV-visible spectrum (Fig. 2C, dashed spectrum). In contrast, if the isolation procedure was performed while avoiding light exposure of the sample, the isolated tetrapyrrole had a spectra that was identical to the absorption spectrum observed at the end of the assay (Fig. 2C, solid spectrum). These data demonstrate that, similar to biliverdin, DAB is photolabile and must be isolated in the absence of light. HPLC analysis of DAB was performed to ensure ChuW catalyzes the formation of a single product. Although deuteroheme elutes at 12.6 min, DAB eluted at 7 min and appeared as a single peak (Fig. 2 D, traces i and ii, respectively).
ChuW Is a Radical SAM Methyltransferase That Catalyzes the Anaerobic Liberation of Iron and Formation of a Tetrapyrrole.
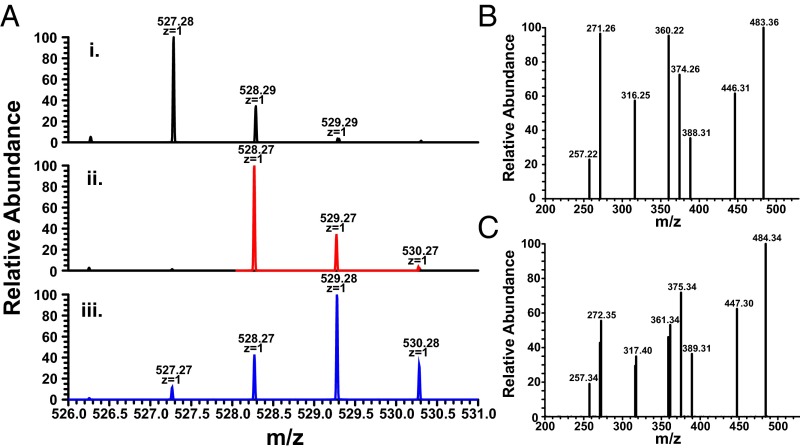
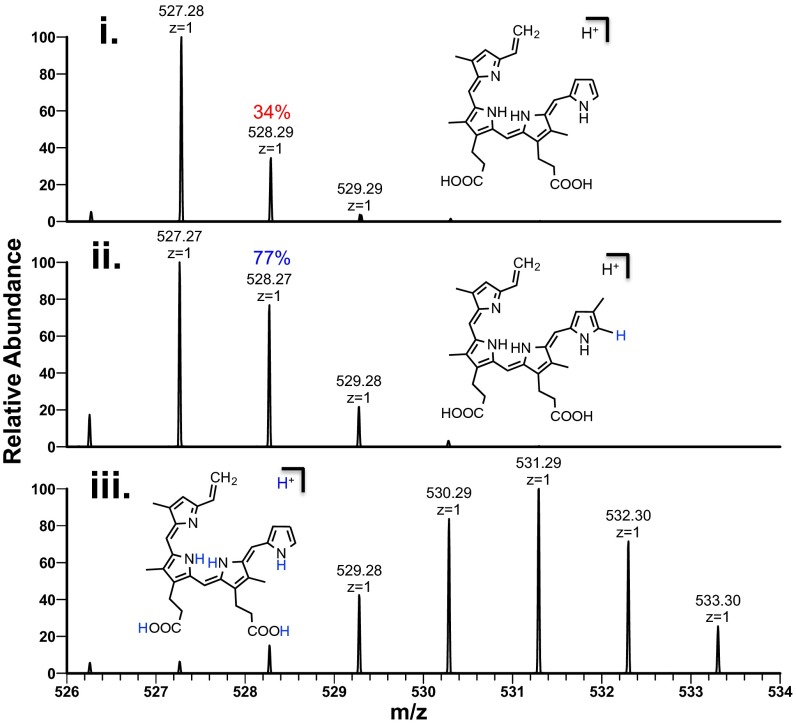
The light sensitivity of anaerobilin and DAB has prohibited the accumulation of product that is sufficient for NMR studies at this time. Therefore, we used nanospray ionization MS (NSI-MS) to address the structure of DAB by direct infusion of the isolated product. When unlabeled SAM was used in the ChuW assay, we observed a dominant peak of 527.28 m/z [M+H]+ (Fig. 3, spectra i). The natural abundance of heavy isotopes is represented by the [A+1] peak in the mass spectrum. In this case, the [A+1] is predominately determined by the number of carbon atoms in the molecule. For DAB, the 528 m/z [A+1] peak is 34% of the parent ion, suggesting a molecule containing 31 carbon atoms. This observation was further substantiated by analysis of the MS for the product of mesoheme turnover, mesoanaerobilin. Mesoanaerobilin has a [M+H]+ of 583 m/z (Fig. S3B), which corresponds to the difference in ethyl groups on the 3 and 8 positions of the porphyrin ring compared with deuteroheme. The [A+1] peak for mesoanaerobilin was ∼38% of the parent ion and is in agreement with a 35-carbon molecule.
Fig. 3.
MS of isolated DAB. (A) NSI-MS of DAB was performed as described in SI Materials and Methods using deuteroheme and (trace i) (12C-methyl) SAM, (trace ii) (13C-methyl) SAM, or (trace iii) (d3-methyl) SAM as substrates. (B) The MS/MS of DAB or (C) 13C-methylated DAB revealed multiple fragments when using CID at 40%. Predicted fragment structures are shown in Fig. S4.
To further address the methyl transfer reaction, [13C-methyl]-SAM was used in the assay, and the isolated product exhibited an enrichment of 13C with a dominant [M+H]+ of 528 m/z (Fig. 3, spectra ii). This observation provides direct evidence that ChuW catalyzes the transfer of a methyl group from the SAM moiety to the product. To assess the mechanism of methyl transfer, we used [d3-methyl]-SAM in the ChuW assay. The use of the deuterium-labeled SAM molecule resulted in a DAB mass shifted by two units yielding a primary peak of 529.28 m/z (Fig. 3, spectra iii), thereby confirming the transfer of only two deuterium atoms from the SAM-derived methyl group. The use of [d3-methyl]-SAM in the assay resulted in a kinetic isotope effect (KIE) of 2.3 based on specific activities.
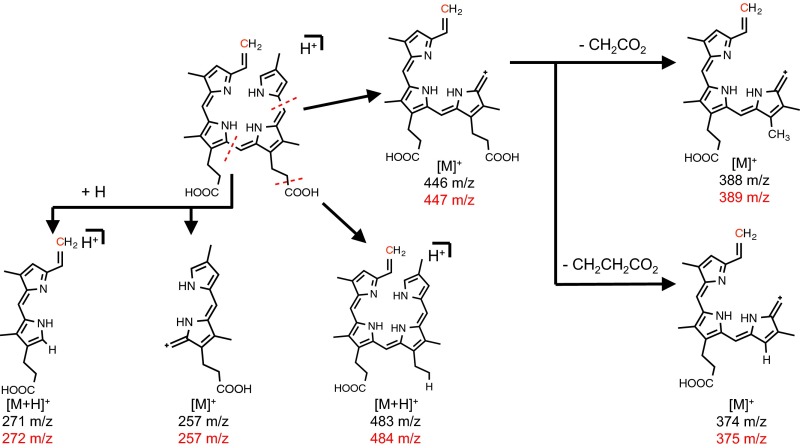
To investigate the state of the pyrrolic ring system post turnover, we acquired the fragmentation pattern of DAB (Fig. 3B). The MS/MS revealed multiple fragments consistent with the cleavage of the macrocycle. Upon fragmentation of 13C-methylated DAB, some of these fragments also shifted by one mass unit, indicating the retention of the isotopic label (Fig. 3C). A similar fragmentation profile was observed when MS analysis was performed on the product mesoanaerobilin (Fig. S3C). These data are in stark contrast to the fragmentation typically observed for closed porphyrin ring systems, which only results in the cleavage of substituents surrounding the ring. This observation suggests that ChuW ultimately catalyzes the opening of the porphyrin ring. This conclusion is also supported by the increase in labile iron (Fig. S2). A proposed fragmentation pathway was constructed based on the predicted structure of DAB (Fig. S4).
Fig. S4.
Potential structures for the MS/MS peaks observed for DAB.
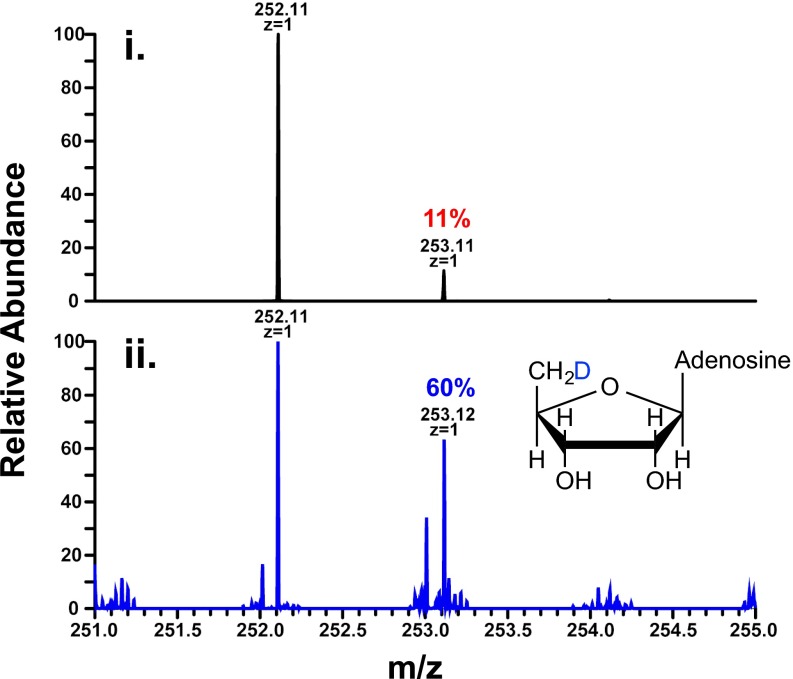
We further probed the methyl transfer mechanism through NSI-MS analysis of HPLC-isolated 5′-dA when [d3-methyl]-SAM was used in the assay (Fig. S5). The [A+1] peak for 5′-dA increased from 11% to 60% (Fig. S6, compare traces i and ii). These data are consistent with what has been reported for other RSMTs and indicate that the 5′-dA•, generated on one SAM molecule, abstracts a hydrogen atom from the methyl group of another SAM molecule. In addition to sequence similarities between ChuW, HemN, and several other class C RSMTs, these data would further support the presence of two distinct SAM binding sites. In fact, two SAM binding sites would be consistent with the proposed HemN mechanism and what has already been confirmed for some RSMTs.
Fig. S5.
NSI-MS of 5′-dA obtained after ChuW turnover. (i) 5′-dA standard was analyzed by NSI-MS with a [M+H]+ of 252.11 m/z. (ii) 5′-dA was isolated by HPLC post ChuW turnover using (d3-methyl) SAM and resulted in a 49% increase in the A+1 peak at 253.12 m/z.
Fig. S6.
NADPH-dependent turnover of DAB by purified ChuY monitored with UV-visible spectroscopy. The arrows indicate the direction of spectroscopic changes during the time course of the assay. (Inset) Rate of DAB consumption at various ChuY concentration.
Because lyase reactions may require proton transfer or incorporation, we performed the assay in buffered 2H2O (70%) using deuteroheme as the substrate to determine if a solvent-derived proton is incorporated into DAB. Upon purification in methanol, NSI-MS of DAB resulted in a 43% increase in the [A+1] peak to the parent ion (Fig. 4, compare traces i and ii). This signal was retained after incubation with methanol for 24 h, suggesting that a deuteron is incorporated during the reaction and originates from solvent. Similarly, the number of exchangeable hydrogens that are present on the product was determined by analyzing isolated DAB in d4-methanol. NSI-MS analysis resulted in a Gaussian distributed signal centered at 531 m/z (Fig. 4, trace iii).
Fig. 4.
MS following solvent-derived deuterium incorporation into DAB. ChuW was assayed in buffered H2O (trace i) and 70% buffered D2O (trace ii). DAB was isolated as described in SI Materials and Methods and analyzed by NSI-MS. When assayed in buffered D2O, we observed a 43% enrichment of the A+1 peak at 528.27 m/z for the product (trace ii). NSI-MS of isolated DAB in d4-methanol (trace iii).
DAB Is a Substrate for ChuY.
To further confirm the role of ChuW in an anaerobic pathway, we isolated and characterized ChuY, the protein that is expressed downstream from ChuW in the heme uptake operon. The current function for ChuY is annotated as an NAD(P)H oxidoreductase, so we performed activity assays using ChuY in the presence of NADPH and isolated DAB. On addition of enzyme into the assay mixture, we observed a decrease in the UV-visible features of DAB with respect to time (Fig. S6). Increasing the amount of ChuY present in the assay resulted in a linear increase in that activity, indicative of an enzyme-catalyzed reaction. No biliverdin reductase activity was observed for ChuY, although the function appears to be similar, specifically to reduce a potentially toxic product of heme degradation.
SI Materials and Methods
Chemicals.
Porphyrins were purchased from Frontier Scientific with the exception of hemin chloride, which was purchased from Sigma Chemical Company. SAM HCl, 5′-dA, and SAH were also purchased from Sigma. Buffer components were purchased from Fischer Scientific.
Overexpression and Isolation of ChuW.
ChuW was expressed in E. coli strain BL21 DE3 using a commercially available pET expression plasmid exhibiting kanamycin resistance with an isopropyl β-d-1-thiogalactopyranoside (IPTG)-inducible promoter. Consistent with what others have observed, we also coexpress the pDB1282 plasmid (ampicillin resistant) containing the Isc operon during cell growth at low temperature as has been previously established (18). In short, a 20-mL starter culture was added to 2 L M9 minimal media (4-L culture flasks) containing 0.02% casamino acids and grown at 37 °C, shaking at 200 rpm. At an OD of 0.3, arabinose was added to 0.2% (wt/vol). Once the OD600 reached 0.6, IPTG was added to a final concentration of 200 μM, and cultures were incubated at 17 °C shaking at 180 rpm for 17 h. Cells were harvested by centrifugation (10,000 × g) and stored at −80 °C until further use.
The cells were resuspended in degassed buffer (20 mM Tris buffer at pH 8.1 with 300 mM KCl, and 10% (vol/vol) glycerol containing DNase, Lysozyme, and PMSF) in an anaerobic gloveblox, sealed, and then brought out of the glovebox to further degas with argon while stirring. The solubilized cells were then lysed anaerobically by equilibrating a closed-system French press with anaerobic buffer while maintaining a stream of argon in the drawing and collection flasks. The lysate was centrifuged at 60,000 × g for 1.5 h. The supernatant was collected, and ChuW was purified anaerobically by a gravity flow cobalt column equilibrated with buffer. The column was then washed with three column volumes of buffer containing 0 and 10 mM imidazole. ChuW was eluted in buffer containing 250 mM imidazole, and fractions were analyzed by SDS/PAGE. The protein was then diluted to 1 mg/mL and reconstituted following previous established methods (18). In short, the protein was diluted in 20 mM Tris, pH 8.1, 300 mM KCl, 10% glycerol, and 10 mM DTT. After incubation for 5 min, 100 mM ferric chloride was added to 4× the protein concentration while swirling. After a 30-min incubation, 15 μL 100 mM sodium sulfide was added every 10 min until reaching 1 equivalent (eq) of the iron added. The mixture was capped and incubated in the glovebox for 12 h. The precipitate was removed by centrifugation, and the supernatant was run over a DEAE Sepharose anion exchange column by gravity flow in the glovebox. The column was washed with 20 mM Tris, pH 8.1, and 10% glycerol (buffer A), and the protein was eluted using stepwise gradient of 1 M KCl in buffer A. Protein fractions were pooled, concentrated anaerobically, and stored in liquid nitrogen until further use.
Iron Analysis of Purified ChuW.
Iron quantification of ChuW was performed as previously described (38). Iron standards were prepared in acid-washed glassware at a concentration of 0.5 mM ferrous ammonium sulfate heptahydrate and diluted to various concentrations between 0.012 and 0.2 mM to a final volume of 250 μL with identical buffers (20 mM Tris, pH 8.0, 300 mM KCl, and 10% glycerol). After acid precipitation and heat incubation at 80 °C, 750 μL dH2O was added, and then precipitates were pelleted by centrifugation; 750 μL of each solution was transferred to a new microcentrifuge tube, where 50 μL 10% hydroxylamine and 250 μL 0.1% bathophenanthroline were added with vortexing between additions. Samples were incubated at room temperature for 1 h and measured at 535 nm following the incubation period. ChuW samples were prepared in the same manner with triplicate measurements per concentration tested, whereas independent experiments were performed in triplicate as well. The same protocol was used to detect the total amount of iron available to the chelator in the ChuW assay before and after turnover. Any protein-bound iron is released during the acid hydrolysis and is subject to chelation by bathophenanthroline, whereas heme-, deuteroheme-, or mesoheme-bound iron is not.
EPR Spectroscopy of Purified ChuW.
Samples were prepared by diluting ChuW to a final concentration of 400 μM either in the presence or absence of 2 mM sodium dithionite. Spectra are a result of three scans recorded at 10 K with 1-MW microwave power, modulation frequency of 100 kHz, and modulation amplitude of 6.477 G.
HPLC of SAM Cleavage by ChuW.
Single turnover assays were performed in 2-mL reaction volumes with a final concentration of 1 mM SAM, 100 μM deuteroheme, 100 μM ChuW, and 2 mM sodium dithionite in a 20 mM Tris, pH 8.1, 125 mM KCl, and 10% glycerol buffer. Controls were run in side-by-side reactions in the absence of either deuteroheme or sodium dithionite, and 90-μL aliquots were subjected to acid precipitation with 50 μL 1% trifluoroacetic acid (TFA) at 0, 0.5, 1, 3, 5, and 10 min. Samples were centrifuged, and the supernatant was applied to a Denali C18 HPLC column (15 cm × 4.6 mm; 5 μm). Cleavage products were separated by a gradient of acetonitrile 0–80% in 0.1% TFA at a flow rate of 1 mL/min. When using NADPH, flavodoxin, and flavodoxin reductase, the flow rate was lowered to 0.8 mL/min. All HPLC assays were monitored at 260 nm, and quantitation of 5′-dA and SAH was performed by comparing the peak areas to a standard curve. To confirm the deuteron transfer from (d3-methyl) SAM, 5′-dA was collected by manual fractionation and subjected to vacuum centrifugation. MS was performed on the collected fractions by direct infusion. Details of the instrument used and the conditions are described below.
Monitoring ChuW Activity by UV-Visible Spectroscopy.
All UV-visible spectra were recorded on an Agilent 8A53 diode array spectrophotometer using a Peltier temperature controller set to 25 °C. Typical enzyme assays were conducted anaerobically with degassed buffer containing 5% DMSO, 1 μM ChuW, 5 μM 3EcFldA, 2 μM EcFpr, 250 μM NADPH, and 10 μM of either hemin or deuteroheme. Reactions were initiated by the addition of SAM to a final concentration of 250 μM. Spectra were taken from 350 to 900 nm every 2 min for 30 min. Extinction coefficients of DAB and anaerobilin were determined by the isosbestic point at 418 nm using the following relationship:
MS of Reaction Products.
DAB and mesoanaerobilin was purified by separation of small molecules on a C1 sep pack attained from Silicycle. In short, a 5-mL reaction mixture containing 100 μM deuteroheme or mesoheme, 20 μM ChuW, 400 μM SAM, 500 μM NADPH, 20 μM EcFld, and 5 μM EcFpr was incubated anaerobically for 4 h at room temperature and in the dark. The assay mixture was applied to the sep pack preequilibrated with 1 eq of methanol, 1 eq of dH2O, and 1 eq of assay buffer. Once the assay mixture ran through the column, it was washed with 1 eq of buffer, 2 eq of water, 1 eq of 1% TFA, and 3 eq of water, before elution in 100% methanol. To study product Y, the same assay was conducted but with 10 μM ChuY. The eluent was diluted in methanol and analyzed using NSI-MS by direct infusion into a linear ion trap mass spectrometer (LTQ-Orbitrap Discovery; Thermo Fisher Scientific) using a nanoelectrospray source at a syringe flow rate of 0.40 μL/min and capillary temperature set to 210 °C. Fragmentation by collision-induced dissociation (CID) in MS/MS was used with normalized collision energy of 35–40%. Automated acquisition of MS2 fragmentation was achieved using the total ion mapping (TIM) functionality of the Xcalibur software package (version 2.0; Thermo Fisher Scientific). Identical assays were performed with labeled SAM substrate prepared as previously described (39).
HPLC of Isolated DAB.
Detection of isolated DAB was performed using a SAS-Hypersil C1 column (150 × 4.6 mm) from Thermo Scientific equipped with a 1-mL precolumn and monitoring at 400 nm. Isolated DAB solubilized in 100% methanol was manually injected (20 μL). Separation was carried out using 100% methanol (solvent A) and 1 M ammonium acetate-acetic acid buff (pH 4.6) (solvent B) using a flow rate of 1.0 mL/min. Elution was achieved by applying a linear gradient starting with 62% solvent A and increasing to 80% by 20 min. The column was washed for 10 min with 80% solvent A before starting the next sample. The elution of DAB was compared with deuteroheme based on retention times.
Expression, Purification, and Enzymatic Assay of ChuY.
ChuY was expressed by growing the E. coli strain BL21 DE3 strain containing an IPTG-inducible expression plasmid that carried the gene coding for ChuY with a 6xHis tag (pET15a). In short, a 20-mL starter culture was grown for 6 h before inoculating six 1-L flasks containing TB media. Cultures were grown at 37 °C shaking at 200 rpm until the OD at 600 nm reached 0.8. IPTG was added to a final concentration of 500 μM. Cultures were incubated at 18 °C overnight shaking at 200 rpm and harvested by centrifugation (10,000 × g) before being stored at −80 °C. The frozen cell pellets were solubilized in buffer containing 20 mM Tris, pH 8.1, 300 mM KCL, and 10% glycerol with trace amounts of PMSF, lysozyme, and DNase. Cells were lysed by French pressure cell, and lyase was ultracentrifuged at 65,000 × g for 1.5 h. The supernatant was applied to Talon resin, washed with 2 column eq of water, and 8 column eq of solubilization buffer. After washing the column with 3 column eq of 10 mM imidazole, ChuY was eluted with buffer containing 250 mM imidazole. Fractions containing ChuY were pooled, buffer exchanged to remove imidazole, and then concentrated. Protein content was analyzed by SDS/PAGE and Biuret analyses before freezing aliquots at −80 °C.
Monitoring Activity of ChuY by UV-Visible Spectroscopy.
All UV-visible spectra were recorded on an Agilent 8A53 diode array spectrophotometer using a Peltier temperature controller set to 25 °C. Typical enzyme assays were conducted anaerobically with degassed buffer containing 250 μM NADPH and 20 μM of isolated DAB. Reactions were initiated by the addition of ChuY to 1 μM. Spectra were taken from 350 to 900 nm every 2 min for 10 min. The activity was monitored by the disappearance of the 780-nm feature of DAB and was converted to concentration by the extinction coefficient for DAB.
Discussion
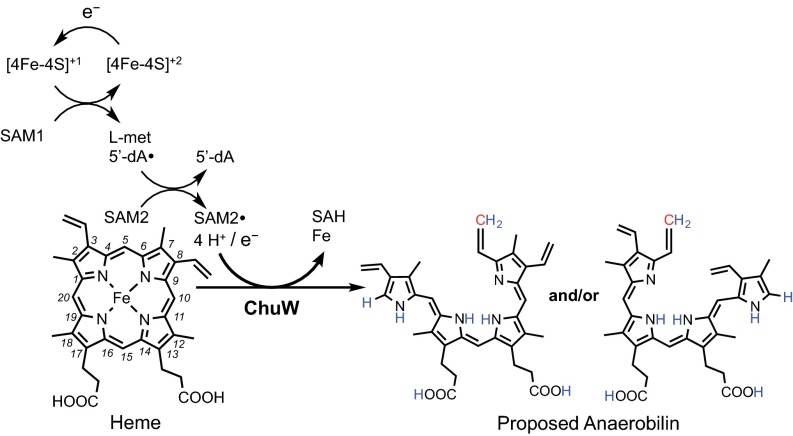
In this investigation, we present evidence for enzyme-catalyzed oxygen-independent heme degradation using isotope labeling, activity assays, and MS to gain insight into the reaction catalyzed by ChuW from enterohemorrhagic E. coli (Fig. 5). Our data suggest that ChuW is a radical SAM enzyme and specifically a member of the class C RSMT subgroup based on the domain structure, sequence analysis, and similar mechanisms for methylation that have been proposed for these enzymes (23). The current mechanistic paradigm for all heme oxygenases reported to date involves activation of molecular oxygen. In stark contrast, the data presented herein suggests that ChuW uses the oxidizing power of a primary-carbon radical to catalyze methyl transfer and rearrangement of the porphyrin ring, resulting in the liberation of iron. Similar to other radical SAM enzymes, any exposure to molecular oxygen inactivates ChuW, likely due to the degradation of the catalytic [4Fe-4S] cluster. Without question, the ability to acquire iron from a porphyrin macrocycle under low oxygen tension would provide a microorganism with a distinct competitive advantage. Such a competitive advantage is especially important for an enteric organism that is competing for survival in an anaerobic environment, such as the human intestine, where heme is readily available from dietary sources.
Fig. 5.
Proposed reaction scheme and products for ChuW. Predicted placement of hydrogens observed during deuterium incorporation are labeled in blue, whereas the SAM-derived methyl group is shown in red.
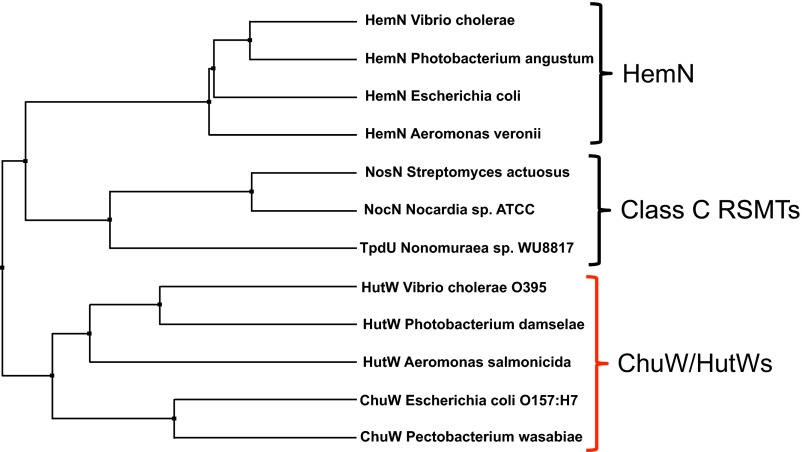
Radical SAM enzymes are known to perform difficult chemical rearrangements and, in some cases, oxygen-using enzymes have taken over a particular metabolic step, presumably after the appearance of oxygen-evolving photosynthesis. The current broad classification of genes as a “HemN” is ambiguous due to the limited scope of the sequence motif used for the classification (20). However, distinction between genes is coming to light. Bioinformatic analyses have identified more than 300 similar sequences across 36 genera within the Structure-Function Linkage Database (SFLD) (30), all from organisms that do not encode a heme oxygenase (HO). Evolutionarily speaking, it is interesting that two distinct enzymes, HemN and ChuW (HutW in V. cholerae), contain a similar functional domain, yet one enzyme is important for the anaerobic formation of heme (HemN), whereas the other enzyme can catalyze the anaerobic degradation of heme (ChuW).
A radical-based mechanism, instead of the activation of molecular oxygen, makes mechanistic sense; however, it is unclear how such a mechanism proceeds. Clues to the ChuW mechanism come from what has been recently observed for other class C RSMTs. All known class C RSMTs catalyze methylations of sp2-hybridized carbon atoms that, in some cases, lead to considerable structural rearrangement (22, 23, 31, 32). Although ChuW/HutW’s appear to be evolutionarily distinct (Fig. S7) compared with other RSMTs or HemN, all of these enzymes have been shown to use the reductive cleavage of SAM as the starting point in their catalytic cycle. In addition, activation of the substrate or the methyl group of one SAM moiety by reductive cleavage of another SAM molecule has already been presented for several RSMTs (23). Building on our observations and these postulated mechanisms, Fig. 5 depicts the proposed reaction scheme for the ChuW-dependent decyclization of the porphyrin ring. Although the complete mechanism is incomplete, our data indicate that after generation of the 5′-dA•, this radical abstracts a hydrogen atom from the methyl group of a second SAM molecule. The newly formed methylene radical attacks the substrate directly before causing a rearrangement of the porphyrin ring that results in the liberation of iron. At the present time, there are other possible product structures that would satisfy this goal and our MS data (Fig. S8), and this is under further investigation. Regardless, the most likely mechanism for ring opening would involve the formation of an intermediate that spontaneously results in a distortion of the porphyrin structure.
Fig. S7.
Phylogenetic comparison of HemN, class C RSMTs, and ChuW/HutW. A phylogenetic tree was generated by Jalview (40) from a sequence alignment based on % identity.
Fig. S8.
Alternative structures that are also consistent with the MS data obtained for anaerobilin.
Several intermediates can be considered based on our observations and the observations for other class C RSMTs. Of particular interest are the products of YtkT and Jaw5 that have been shown to contain other “high energy” C1-derived moieties, such as cyclopropane rings (22, 31). Such a moiety may be unstable if placed across the 4–5 or 5–6 carbon–carbon bond of deuteroheme (Fig. 5) or heme and would provide structural reasoning for any of the products presented depending on resolution of the cyclopropane ring cleavage (Fig. 5 and Fig. S8). Similar to what has been described for the class C RSMTs YtkT and Jaw5 (22), whether or not such a mechanism involves radical addition or nucleophilic attack by the methylene radical or ylide, respectively, is also not clear. Either mechanism for methyl transfer and ring opening requires an additional electron, a problem discussed in a recent review (23) but that is still unresolved for these class C RSMTs.
The fragmentation pattern that we observe for DAB is particularly useful in that cleavage at the central bridging methine carbon produces a pyrrolium ion [M′+H]+ at 271 m/z and a pyrrolic ion [M+] at 257 m/z (Fig. S4), which is similar to what has been observed in the mass spectral analysis of biliverdin (33). To this end, we compared structures that coincided with ring cleavage at either the α or γ methine bridge while accounting for the addition of a methyl group at or near the cleavage site. The resulting even mass fragments suggest a loss of one nitrogen atom and are best explained by fragmentation at either the β or δ methine bridges. It is unclear if there is a preference for which methine bridging carbon–carbon bond is ultimately broken (C4-C5 or C5-C6) during catalysis, but both outcomes are possible (Fig. 5). Although our observations for MS/MS analysis of DAB are explained by the proposed structure (Figs. 4 and 5), other potential isomers are described in Fig. S8 that cannot be ruled out at this time. Due to the photolability and oxygen sensitivity of the reaction products, efforts to isolate DAB, anaerobilin, and mesoanaerobilin to a purity and quantity sufficient for NMR analysis have proven challenging.
Heme has limited solubility in aqueous solution; therefore, the standard protocol for assaying HOs is to load the enzyme with 1:1 equivalents of heme:protein followed by initiating turnover with cytochrome P450 reductase and near stoichiometric amounts of NADPH. These experiments are essentially a single turnover reaction that is often slow and that can be the result of nonenzymatic coupled oxidation when catalase is not present in the assay (13). Therefore, the identification of genuine HOs can be difficult. ChuS from E. coli O157:H7 was originally identified as a HO (34), and although the authors reported activity in the presence of catalase, a later report revealed significantly decreased activity in the presence of catalase for a homologous protein (35). Similarly, the heme utilization protein HutZ from V. cholerae was described as a HO based on spectroscopic assays (36); however, there was no indication that catalase or superoxide dismutase was included in the assay reported for the initial characterization, and the products were never identified, nor was this activity observed by others (14). Consequently, the benefit of studying an anaerobic heme-degrading enzyme in vitro is the minimal risk of nonenzymatic coupled oxidation artifacts. However, if ChuS and HutZ were bona fide heme degraders, this would allow enterohemorrhagic E. coli and V. cholerae the ability to acquire iron from heme in varying oxygenic environments.
The existence of an anaerobic mechanism for heme degradation is consistent with (i) the recent revelations in the heme biosynthetic pathways (20, 37), (ii) the characterization of radical SAM enzymes involved in catalyzing important steps in the anaerobic biosynthesis of protoheme (19, 25), and (iii) recent observations that heme degradation has evolved to suit the physiological needs of the organism (12). Our in vitro data suggest that ChuW functions as a RSMT that methylates heme as part of a decyclization mechanism, resulting in an open tetrapyrrole and the release of the essential nutrient iron (Fig. 5). Many of our observations overlap with what has been published for the class C RSMTs. We further demonstrated that the product of the ChuW reaction is a photosensitive substrate that is reduced by ChuY in a NADPH-dependent reaction. We propose that ChuX may facilitate the transfer of anaerobilin or the removal of iron from a ChuW-generated intermediate. In regard to the latter, a zinc ion has been observed bound to a conserved histidine residue in the structure of a ChuX homolog (Protein Data Bank ID code 3FM2). However, despite a reported dissociation constant (KD) of 1.99 ± 0.02 μM, a structure for ChuX with heme bound is not available at this time. We propose that both the iron atom and anaerobilin may be toxic to the cell, so a logical role for ChuX could be to chelate the exiting iron atom and/or transport anaerobilin to ChuY.
In addition to iron acquisition, the products of anaerobic heme or porphyrin degradation may have more diverse functions. When our findings are considered in light of the data for HemN, and the fact that the products of other class C RSMTs have antitumor and antibiotic properties, further characterization of these radical mechanisms will have profound implications. In particular, the presence of an enzyme that catalyzes a radical-mediated porphyrin degradation reaction in a select group of enteric pathogens will open the door to research and discovery of a new class of antimicrobial compounds.
Materials and Methods
A typical assay for E. coli O157:H7 ChuW, expressed and purified as described SI Materials and Methods, consisted of 1 μM enzyme, 5 μM EcFld, 2 μM EcFpr 250 μM NADPH, and 10 μM of hemin, mesoheme, or deuteroheme. Assays were performed anaerobically and initiated by the injection of SAM to a final concentration of 250 μM. Deuteroanaerobilin and mesoanaerobilin were isolated anaerobically in the dark using a disposable C1 column. NSI-MS was performed using a linear ion trap mass spectrometer (LTQ-Orbitrap Discovery; Thermo Fischer Scientific). Unless stated otherwise, all experimental procedures were performed in an anaerobic chamber under positive nitrogen:hydrogen (95:5) pressure with continuous scrubbing to remove any molecular oxygen. To ensure that there was no molecular oxygen present in the glovebox during experiments, we continuously monitored the atmosphere of the chamber using an electronic detector, as well as a chemical mixture of reduced methyl viologen. All additional experimental procedures are described in SI Materials and Methods.
Acknowledgments
We thank Clark Lagarias and members of the H. A. Dailey and Z. Wood laboratory for numerous helpful discussions; Tamara A. Dailey for editing of this manuscript; the Squire J. Booker laboratory for their generous donation of the E. coli strain DM22pK8; and Dennis Dean for the donation of the pDB1282 plasmid. We thank the Lance Wells and Michael Tiemeyer laboratories for help with MS. We acknowledge the support and access to instrumentation provided through a grant from the National Institute of General Medical Sciences (P41GM103490) of the NIH. This research was also supported by National Science Foundation Grant MCB 0835432 (to W.N.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. I.H. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603209113/-/DCSupplemental.
References
- 1.Wilks A, Burkhard KA. Heme and virulence: How bacterial pathogens regulate, transport and utilize heme. Nat Prod Rep. 2007;24(3):511–522. doi: 10.1039/b604193k. [DOI] [PubMed] [Google Scholar]
- 2.Carpenter CE, Mahoney AW. Contributions of heme and nonheme iron to human nutrition. Crit Rev Food Sci Nutr. 1992;31(4):333–367. doi: 10.1080/10408399209527576. [DOI] [PubMed] [Google Scholar]
- 3.Hooda J, Shah A, Zhang L. Heme, an essential nutrient from dietary proteins, critically impacts diverse physiological and pathological processes. Nutrients. 2014;6(3):1080–1102. doi: 10.3390/nu6031080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frankenberg-Dinkel N. Bacterial heme oxygenases. Antioxid Redox Signal. 2004;6(5):825–834. doi: 10.1089/ars.2004.6.825. [DOI] [PubMed] [Google Scholar]
- 5.Wandersman C, Delepelaire P. Bacterial iron sources: From siderophores to hemophores. Annu Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 6.Wilks A. Heme oxygenase: Evolution, structure, and mechanism. Antioxid Redox Signal. 2002;4(4):603–614. doi: 10.1089/15230860260220102. [DOI] [PubMed] [Google Scholar]
- 7.Friedman J, Lad L, Li H, Wilks A, Poulos TL. Structural basis for novel delta-regioselective heme oxygenation in the opportunistic pathogen Pseudomonas aeruginosa. Biochemistry. 2004;43(18):5239–5245. doi: 10.1021/bi049687g. [DOI] [PubMed] [Google Scholar]
- 8.Wilks A, Schmitt MP. Expression and characterization of a heme oxygenase (Hmu O) from Corynebacterium diphtheriae. Iron acquisition requires oxidative cleavage of the heme macrocycle. J Biol Chem. 1998;273(2):837–841. doi: 10.1074/jbc.273.2.837. [DOI] [PubMed] [Google Scholar]
- 9.Zhu W, Wilks A, Stojiljkovic I. Degradation of heme in gram-negative bacteria: The product of the hemO gene of Neisseriae is a heme oxygenase. J Bacteriol. 2000;182(23):6783–6790. doi: 10.1128/jb.182.23.6783-6790.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reniere ML, et al. The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Mol Microbiol. 2010;75(6):1529–1538. doi: 10.1111/j.1365-2958.2010.07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nambu S, Matsui T, Goulding CW, Takahashi S, Ikeda-Saito M. A new way to degrade heme: The Mycobacterium tuberculosis enzyme MhuD catalyzes heme degradation without generating CO. J Biol Chem. 2013;288(14):10101–10109. doi: 10.1074/jbc.M112.448399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsui T, et al. Heme degradation by Staphylococcus aureus IsdG and IsdI liberates formaldehyde rather than carbon monoxide. Biochemistry. 2013;52(18):3025–3027. doi: 10.1021/bi400382p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilks A, Ikeda-Saito M. Heme utilization by pathogenic bacteria: Not all pathways lead to biliverdin. Acc Chem Res. 2014;47(8):2291–2298. doi: 10.1021/ar500028n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wyckoff EE, Schmitt M, Wilks A, Payne SM. HutZ is required for efficient heme utilization in Vibrio cholerae. J Bacteriol. 2004;186(13):4142–4151. doi: 10.1128/JB.186.13.4142-4151.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Septer AN, Wang Y, Ruby EG, Stabb EV, Dunn AK. The haem-uptake gene cluster in Vibrio fischeri is regulated by Fur and contributes to symbiotic colonization. Environ Microbiol. 2011;13(11):2855–2864. doi: 10.1111/j.1462-2920.2011.02558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torres AG, Payne SM. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 1997;23(4):825–833. doi: 10.1046/j.1365-2958.1997.2641628.x. [DOI] [PubMed] [Google Scholar]
- 17.Broderick JB, Duffus BR, Duschene KS, Shepard EM. Radical S-adenosylmethionine enzymes. Chem Rev. 2014;114(8):4229–4317. doi: 10.1021/cr4004709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanz ND, et al. RlmN and AtsB as models for the overproduction and characterization of radical SAM proteins. Methods Enzymol. 2012;516:125–152. doi: 10.1016/B978-0-12-394291-3.00030-7. [DOI] [PubMed] [Google Scholar]
- 19.Layer G, Verfürth K, Mahlitz E, Jahn D. Oxygen-independent coproporphyrinogen-III oxidase HemN from Escherichia coli. J Biol Chem. 2002;277(37):34136–34142. doi: 10.1074/jbc.M205247200. [DOI] [PubMed] [Google Scholar]
- 20.Dailey HA, Gerdes S, Dailey TA, Burch JS, Phillips JD. Noncanonical coproporphyrin-dependent bacterial heme biosynthesis pathway that does not use protoporphyrin. Proc Natl Acad Sci USA. 2015;112(7):2210–2215. doi: 10.1073/pnas.1416285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q, van der Donk WA, Liu W. Radical-mediated enzymatic methylation: A tale of two SAMS. Acc Chem Res. 2012;45(4):555–564. doi: 10.1021/ar200202c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang W, et al. Characterization of yatakemycin gene cluster revealing a radical S-adenosylmethionine dependent methyltransferase and highlighting spirocyclopropane biosynthesis. J Am Chem Soc. 2012;134(21):8831–8840. doi: 10.1021/ja211098r. [DOI] [PubMed] [Google Scholar]
- 23.Bauerle MR, Schwalm EL, Booker SJ. Mechanistic diversity of radical S-adenosylmethionine (SAM)-dependent methylation. J Biol Chem. 2015;290(7):3995–4002. doi: 10.1074/jbc.R114.607044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Layer G, et al. Radical S-adenosylmethionine enzyme coproporphyrinogen III oxidase HemN: Functional features of the [4Fe-4S] cluster and the two bound S-adenosyl-L-methionines. J Biol Chem. 2005;280(32):29038–29046. doi: 10.1074/jbc.M501275200. [DOI] [PubMed] [Google Scholar]
- 25.Lobo SA, et al. Characterisation of Desulfovibrio vulgaris haem b synthase, a radical SAM family member. Biochim Biophys Acta. 2014;1844(7):1238–1247. doi: 10.1016/j.bbapap.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 26.Grove TL, Lee KH, St Clair J, Krebs C, Booker SJ. In vitro characterization of AtsB, a radical SAM formylglycine-generating enzyme that contains three [4Fe-4S] clusters. Biochemistry. 2008;47(28):7523–7538. doi: 10.1021/bi8004297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grove TL, et al. Further characterization of Cys-type and Ser-type anaerobic sulfatase maturating enzymes suggests a commonality in the mechanism of catalysis. Biochemistry. 2013;52(17):2874–2887. doi: 10.1021/bi400136u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bruender NA, Young AP, Bandarian V. Chemical and biological reduction of the radical SAM enzyme 7-carboxy-7-deazaguanine [corrected] synthase. Biochemistry. 2015;54(18):2903–2910. doi: 10.1021/acs.biochem.5b00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilks A. 2003. Purification and characterization of heme oxygenase. Curr Protoc Toxicol Chap 9:Unit 9.9.
- 30.Pegg SC, et al. Leveraging enzyme structure-function relationships for functional inference and experimental design: The structure-function linkage database. Biochemistry. 2006;45(8):2545–2555. doi: 10.1021/bi052101l. [DOI] [PubMed] [Google Scholar]
- 31.Hiratsuka T, et al. Biosynthesis of the structurally unique polycyclopropanated polyketide-nucleoside hybrid jawsamycin (FR-900848) Angew Chem Int Ed Engl. 2014;53(21):5423–5426. doi: 10.1002/anie.201402623. [DOI] [PubMed] [Google Scholar]
- 32.Yu Y, et al. Nosiheptide biosynthesis featuring a unique indole side ring formation on the characteristic thiopeptide framework. ACS Chem Biol. 2009;4(10):855–864. doi: 10.1021/cb900133x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorchein A, Lim CK, Cassey P. Extraction and analysis of colourful eggshell pigments using HPLC and HPLC/electrospray ionization tandem mass spectrometry. Biomed Chromatogr. 2009;23(6):602–606. doi: 10.1002/bmc.1158. [DOI] [PubMed] [Google Scholar]
- 34.Suits MD, et al. Identification of an Escherichia coli O157:H7 heme oxygenase with tandem functional repeats. Proc Natl Acad Sci USA. 2005;102(47):16955–16960. doi: 10.1073/pnas.0504289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee MJ, Schep D, McLaughlin B, Kaufmann M, Jia Z. Structural analysis and identification of PhuS as a heme-degrading enzyme from Pseudomonas aeruginosa. J Mol Biol. 2014;426(9):1936–1946. doi: 10.1016/j.jmb.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 36.Uchida T, Sekine Y, Matsui T, Ikeda-Saito M, Ishimori K. A heme degradation enzyme, HutZ, from Vibrio cholerae. Chem Commun (Camb) 2012;48(53):6741–6743. doi: 10.1039/c2cc31147j. [DOI] [PubMed] [Google Scholar]
- 37.Dailey HA, Gerdes S. HemQ: An iron-coproporphyrin oxidative decarboxylase for protoheme synthesis in Firmicutes and Actinobacteria. Arch Biochem Biophys. 2015;574:27–35. doi: 10.1016/j.abb.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lovenberg W, Buchanan BB, Rabinowitz JC. Studies on the chemical nature of clostridial ferredoxin. J Biol Chem. 1963;238:3899–3913. [PubMed] [Google Scholar]
- 39.Grove TL, et al. A radically different mechanism for S-adenosylmethionine-dependent methyltransferases. Science. 2011;332(6029):604–607. doi: 10.1126/science.1200877. [DOI] [PubMed] [Google Scholar]
- 40.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]