Significance
F-actin is assembled downstream of major actin nucleators like the actin-related proteins 2/3 (Arp2/3) complex and formins. Capping Protein (CP), conversely, halts actin assembly. The ankyrin-repeat protein V-1 has been shown to block CP activity in vitro. Here we show that V-1 overexpression and knockout in Dictyostelium amoeba phenocopy CP knockdown and overexpression, respectively. Moreover, V-1 must be capable of binding CP to exhibit overexpression phenotypes and rescue V-1–null cells. Finally, we present evidence that V-1’s ability to sequester CP is regulated by phosphorylation. Together, our data demonstrate that V-1 controls major actin-dependent cellular processes in vivo by virtue of its ability to inhibit CP and that its influence may be modulated in cells.
Keywords: actin, capping protein, V-1, myotrophin, CARMIL
Abstract
Capping Protein (CP) plays a central role in the creation of the Arp2/3-generated branched actin networks comprising lamellipodia and pseudopodia by virtue of its ability to cap the actin filament barbed end, which promotes Arp2/3-dependent filament nucleation and optimal branching. The highly conserved protein V-1/Myotrophin binds CP tightly in vitro to render it incapable of binding the barbed end. Here we addressed the physiological significance of this CP antagonist in Dictyostelium, which expresses a V-1 homolog that we show is very similar biochemically to mouse V-1. Consistent with previous studies of CP knockdown, overexpression of V-1 in Dictyostelium reduced the size of pseudopodia and the cortical content of Arp2/3 and induced the formation of filopodia. Importantly, these effects scaled positively with the degree of V-1 overexpression and were not seen with a V-1 mutant that cannot bind CP. V-1 is present in molar excess over CP, suggesting that it suppresses CP activity in the cytoplasm at steady state. Consistently, cells devoid of V-1, like cells overexpressing CP described previously, exhibited a significant decrease in cellular F-actin content. Moreover, V-1–null cells exhibited pronounced defects in macropinocytosis and chemotactic aggregation that were rescued by V-1, but not by the V-1 mutant. Together, these observations demonstrate that V-1 exerts significant influence in vivo on major actin-based processes via its ability to sequester CP. Finally, we present evidence that V-1’s ability to sequester CP is regulated by phosphorylation, suggesting that cells may manipulate the level of active CP to tune their “actin phenotype.”
The addition of Capping Protein (CP) to seed-initiated actin polymerization assays results in the rapid cessation of polymerization because CP binds with very high affinity to the fast-growing barbed end of the actin filament to block further monomer addition (1). Direct extrapolation of this simple, potent biochemical property would suggest that the cell’s content of F-actin should rise and fall as its content of CP is artificially forced to fall and rise, respectively. Indeed, this finding was reported many years ago in Dictyostelium amoeba (2). This simple view of CP’s role in regulating actin assembly in vivo falls short of the whole story, however. The additional complexity arises from the critical relationship between CP and the Arp2/3 complex, the major actin nucleating machine that generates the branched actin networks comprising lamellipodia and pseudopodia (3). At the heart of this relationship is the fact that CP increases the rate of Arp2/3-dependent filament nucleation and promotes optimal branching by rapidly capping filaments (4). As a result, CP promotes actin-related proteins 2 and 3 (Arp2/3)-driven actin assembly and motility (4, 5). This effect was evident from early solution experiments focused on defining the function of the Arp2/3 complex (6), verified by in vitro reconstitution of the Arp2/3-dependent motility of Listeria (5), and explained mechanistically by the elegant work of Akin and Mullins (4). Finally, evidence that CP promotes Arp2/3-dependent branched actin network assembly in vivo has been provided by several important studies where the cell’s content of CP was reduced by RNAi. Perhaps most dramatically, Mejillano et al. showed that the knockdown of CP in B16 melanoma cells resulted in a large-scale reduction in the size of their lamellipodia (7). Similarly, reduction in the level of CP in Drosophila S2 cells resulted in a large decrease in lamellipodial area (8).
Interestingly, a second consequence of CP knockdown is a dramatic increase in the number of filopodia, linear actin structures that extend away from the cell surface (2, 7). The extension of filopodia likely involves the action of formins and/or VASP (9–12), two actin polymerization machines that operate at the growing barbed end as processive polymerases to create the linear actin filaments that fill filopodia. Although both proteins are fairly effective at physically shielding the barbed end from CP (10, 13, 14), it is likely that their robustness as filopodia generators in vivo would be increased by a reduction in CP levels. Given the recent work demonstrating that formins and the Arp2/3 complex compete for G-actin in vivo (15–17), the increase in filopodia number seen upon CP knockdown may also be due in part to an increase in the amount of monomer available for formin/VASP after the reduction in Arp2/3-dependent nucleation caused by CP knockdown.
The studies discussed above suggest that cells could regulate their “actin phenotype” by regulating their level of active CP. Consistent with CP regulation in vivo, estimates of the half-life of CP on the barbed end near the plasma membrane in living cells are approximately three orders of magnitude shorter than CP’s half-life on the barbed in vitro (i.e., ∼2–15 s in cells vs. ∼30 min for pure proteins) (8, 18). To date, two direct regulators of CP activity have been identified. The first, CARMIL, is a ∼125-kDa protein that binds CP tightly via a small, highly conserved domain known as CAH3 or CPI (1, 19). CP bound to CPI binds the barbed end with an affinity of ∼50 nM, which, although still quite significant, is ∼100-fold weaker than free CP (20, 21). CPI also dramatically accelerates the dissociation of CP already present on the barbed end (i.e., it robustly uncaps CP-capped filaments) (22). Structural studies indicate that CPI reduces CP’s affinity for the barbed end in an allosteric fashion by restricting fluctuations within CP to a conformation that binds the barbed end weakly (23–25). Recent work suggests that CARMIL proteins function in vivo at the plasma membrane:cytoplasm interface of protruding edges, where they appear to be recruited, unfolded, and activated to promote actin assembly (26).
The second direct CP regulator, V-1 or myotrophin, is a ∼13-kDa ankyrin-repeat protein that binds CP 1:1 with an affinity of ∼20 nM to render CP incapable of binding the barbed end (i.e., V-1 sequesters CP) (27). Structural studies of mouse V-1 have shown that it abrogates CP’s barbed end capping activity in a steric fashion by occupying CP’s main barbed end interaction site (23, 28). Like CARMIL, V-1 is expressed throughout most of the Eukaryotic kingdom. Recent work in mouse embryo fibroblasts has indicated that V-1, like CP, is freely diffusing in the cytoplasm and that it is present in molar excess over CP (26). These observations argue that the majority of cellular CP may in fact be sequestered by V-1 at steady state, barring regulation. If true, this finding would represent a large departure from the traditional view that the cell’s entire complement of CP is active (3) and would force a very different view of how barbed end capping is regulated in cells.
Recent work by Fujiwara et al. seeking to define the functional interplay between CARMIL and V-1 has yielded a novel model for how barbed end capping may be regulated in cells (26). This model postulates that V-1 globally inhibits CP in the cytoplasm and that active, plasma membrane-associated CARMIL at protruding cell edges drives a complex exchange reaction converting sequestered CP (CP:V-1) into a version that binds the barbed end with ∼50 nM affinity (CP:CARMIL). Implicit in this model is the idea that the barbed end capping activity required to drive Arp2/3-dependent branched actin network assembly is only generated downstream of a CARMIL-driven complex exchange reaction that liberates CP from V-1. One prediction of this model is that cells expressing a version of CP that cannot see the CPI motif in CARMIL and related proteins (e.g., CD2AP and CKIP) should phenocopy Arp2/3-inhibited cells. This prediction has recently received direct support by the work of Edwards et al. (29).
Studies using gene ablation or RNAi have demonstrated that CARMIL proteins play important roles in the formation of cortical actin structures like pseudopodia and macropinocytic crowns in Dictyostelium and lamellipodia in tissue culture cells (1, 19). In contrast, relatively little is known regarding the importance of V-1 in vivo (30–32), especially with respect to effects on actin assembly and organization. Most relevantly, Takeda et al. showed that the overexpression of V-1 in PC12 cells results in higher levels of cellular F-actin, presumably due to enhanced CP sequestration (23). Here, we sought to determine the functional significance of V-1 with regard to actin organization and function using the cellular slime mold Dictyostelium discoideum and a combination of overexpression and gene knockout approaches. Special emphasis was placed on using as a control a version of V-1 containing a function blocking mutation (FBM) in its CP binding site to prove that the effects observed were because of V-1’s ability to sequester CP. In total, our data show that V-1 plays a major role in the regulation of CP activity in vivo, thereby influencing the organization, dynamics, and function of cortical actin structures that drive many fundamental cellular processes. Moreover, our results provide additional support for the novel model of CP regulation in vivo proposed recently by Fujiwara et al. (26).
Results
Biochemical Properties of Dictyostelium V-1 Mirror Those of Mouse V-1.
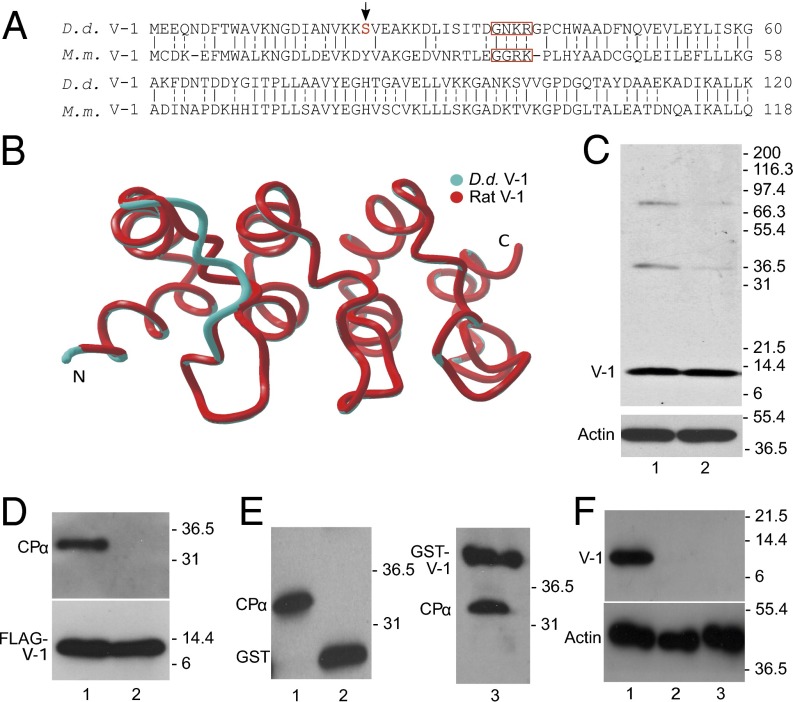
Dictyostelium discoideum (D.d.) contains a single gene encoding a protein with 49% identity and 82% overall similarity to mouse V-1 (Fig. 1A). Moreover, a Phyre2 homology model (33) of D.d. V-1 (blue) superposes almost perfectly with the experimentally determined backbone structure of rat V-1 [Protein Data Bank (PDB) ID code 2MYO; red) (Fig. 1B), with an overall pairwise rmsd of just 0.54 Å. A polyclonal antibody raised against full-length D.d. V-1 recognized almost exclusively a ∼13-kDa protein in whole cell extracts made from both vegetative cells and starved, developing cells (Fig. 1C, lanes 1 and 2, respectively). Consistent with the overall similarities in sequence and predicted structure between D.d. V-1 and mouse/rat V-1, pulldown of full-length, FLAG-tagged D.d. V-1 from cell extracts using anti-FLAG (M2) beads resulted in the coprecipitation of CP (Fig. 1D, lane 1). Similarly, Fig. 1E shows that application of Dictyostelium whole cell extracts to a resin loaded with GST-tagged, full-length D.d. V1 (lane 3), but not GST alone (lane 2), resulted in the retention of CP present in the extract (lane1) on the resin.
Fig. 1.
Dictyostelium possesses a V-1 homolog that interacts with CP. (A) Alignment of the amino acid sequences of Dictyostelium (D.d.) and mouse (M.m.) V-1 (identity, solid dash; similarly, split dash). When the boxed residues in M.m. V-1 are mutated to alanine, V-1’s affinity for CP is reduced by ∼200-fold (27). The red S and arrow indicate the phosphorylated serine (Ser-22). (B) Phyre2 homology model of D.d. V-1 (blue) superposed on the experimentally determined backbone structure of rat V-1 (red). (C, Upper) Western blot of whole cell extracts from vegetative (lane 1) and starved, developing cells (lane 2) probed with a polyclonal antibody to D.d. V-1. (C, Lower) Parallel Western blot probed with an antibody to G-actin as a loading control. (D, Upper) Western blot of the material eluted from anti-FLAG M2 beads that had been incubated with lysates of cells expressing FLAG-V-1 (lane 1) or FLAG-FBM V-1 (lane 2) and probed with a polyclonal antibody to D.d. CPα. (D, Lower) Parallel Western blot probed with an antibody to V-1 to demonstrate that approximately equal amounts of FLAG-V-1 (lane 1) and FLAG-FBM V-1 (lane 2) were precipitated. (E) Western blot of the material eluted from agarose beads coated with GST (lane 2) or GST-V-1 (lane 3) that had been incubated with lysates of D.d. amoeba and probed with the antibody to GST-D.d. CPα. The lysate itself is shown in lane 1. Note that the antibody to D.d. CPα also recognizes the GST and GST-V-1 eluted from the column because it was raised against D.d. CPα fused to GST. (F, Upper) Western blot of whole cell extracts from control cells (lane 1) and two independent V-1–null cell lines (lanes 2 and 3) probed with the polyclonal antibody to D.d. V-1. (F, Lower) Parallel Western blot probed with an antibody to G-actin as a loading control.
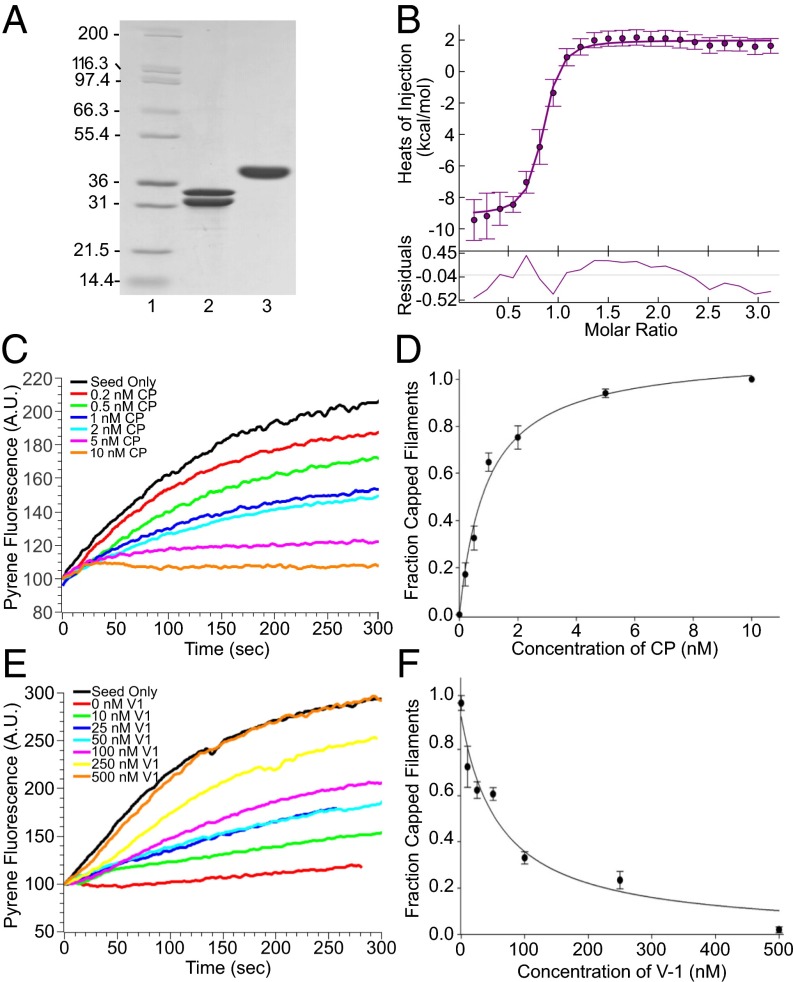
To confirm that D.d. V-1, like mouse V-1, binds CP with high affinity, we purified D.d. CP and GST-tagged D.d. V-1 from bacteria (Fig. 2A) and performed isothermal titration calorimetry (ITC). Fig. 2B shows the binding isotherm we obtained, which yielded an affinity of ∼85 nM for the 1:1 complex. This value is comparable to the values obtained for the mouse proteins, which ranged between ∼20 and ∼50 nM (23, 27, 28). To confirm that D.d. V-1, like mouse V-1, blocks CP’s barbed end capping activity, we performed standard, pyrene-based, seed-initiated actin polymerization assays (20) using rabbit skeletal muscle actin, D.d. CP, and D.d. V-1 tagged with GST. Control assays showed that, as expected, D.d. CP potently inhibited actin assembly, with ∼7 nM CP being sufficient to halt assembly in standard assays (Fig. 1C). This result yielded an approximate affinity for the barbed end of ∼1 nM (Fig. 1D), similar to estimates for mouse CP, which range between ∼0.1 and ∼1 nM (34–36). More importantly, the addition of increasing amounts of GST-tagged D.d. V-1 in the presence of 7 nM D.d. CP progressively restored polymerization, eventually yielding the seed-only rate (Fig. 1E). This latter observation indicates that the complex of D.d. CP and D.d. V-1, like the complex of mouse CP and mouse V-1 (27, 28, 37), has no affinity for the barbed end (i.e., that D.d. V-1 also sequesters CP). The half-maximal concentration of GST-tagged D.d. V-1 required to restore actin polymerization to the seed-only rate was 59.5 ± 14.8 nM (Fig. 1F), which is very similar to the value obtained for mouse V-1 in the presence of 5 nM mouse CP [40 ± 9 nM (27)] and indistinguishable from the value obtained for D.d. V-1 without the GST tag (62.8 ± 8.5 nM; Fig. S1). Finally, Fig. S2 shows that, like mouse V-1 (26, 27), D.d. V-1 is incapable of uncapping CP-capped filaments and readily dissociates from CP upon the addition of the CP-binding domain of D.d. CARMIL (CAH3).
Fig. 2.
Like mammalian V-1, Dictyostelium V-1 binds CP tightly to prevent it from capping the barbed end. (A) SDS/PAGE gel showing the purified D.d. CP (lane 2) and GST-tagged D.d. V-1 (lane 3) used for the ITC measurement (lane 1, molecular mass markers). (B, Upper) The calorimetric titration of V-1 into CP, where 1.75 µl aliquots of 150 µM V-1 were injected into 10 µM CP at 20 °C. The experimental data points were fit to a 1:1 binding isotherm (error bars show the integration errors for each titration peak). (B, Lower) The fit residuals. (C) Representative pyrene-based, seed-initiated, actin polymerization assays in which increasing amounts of CP were added before seed initiation (see the color-coded key). (D) Mean and SDs from three independent assays performed as in C and plotted as the fraction of capped filaments vs. CP concentration. (E) Representative pyrene-based, seed-initiated, actin polymerization assays performed in the presence of 7 nM CP and increasing amounts of V-1 added before seed initiation (see color-coded key). (F) Mean and SDs from three independent assays performed as in E and plotted as the fraction of capped filaments vs. V-1 concentration.
Fig. S1.
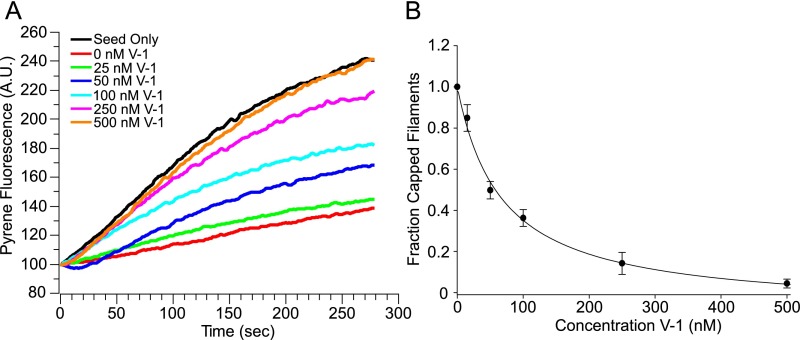
GST does not alter V-1’s ability to sequester CP. (A) Representative pyrene-based, seed-initiated, actin polymerization assays performed in the presence of 7 nM CP and increasing amounts of cleaved V-1 added before seed initiation (see color-coded key). (B) Mean and SDs from three independent assays performed as in A and plotted as the fraction of capped filaments vs. V-1 concentration. There is no statistical difference between cleaved V-1 and GST tagged V-1 (shown in Fig. 2 E and F) as regards their ability to sequester CP.
Fig. S2.
Like mouse V-1, D.d. V-1 cannot uncap CP-capped filaments and is readily displaced from CP by the CP binding domain of D.d. CARMIL. (A) Representative pyrene-based, seed-initiated, actin polymerization assays performed in the presence of 7 nM CP and increasing amounts of GST-V-1 added 20 s after seed initiation to detect the uncapping of CP-capped filaments (see color-coded key). No uncapping (i.e., restoration of polymerization) was observed even at the highest concentration of GST-V-1 tested (2 µM). (B) Shown is a Coomassie Blue-stained gel (inverted image) of a set of complex exchange reactions (26), in which resin coated with GST-V-1 was saturated with CP to create the CP:V-1 complex, washed, incubated for 10 min with the isolated CAH3 domain of D.d. CARMIL at the indicated molar ratios, and then fractionated into supernatant and pellet. The positions of GST-V-1, CPα, CPβ, and CAH3 are indicated. The CAH3 domain can be seen to drive in a dose dependent manner the dissociation of CP from GST-V-1, as evidenced by the progressive shift of CP from the pellet to the supernatant as more CAH3 is added.
Introduction of four consecutive point mutations into the first ankyrin loop of mouse V-1 (Fig. 1A) reduced its affinity for CP by ∼200-fold (27), effectively eliminating its ability to sequester CP. Similarly, pulldown of full-length, FLAG-tagged D.d. V-1 containing the analogous function blocking mutations [Fig. 1A; referred to hereafter as function blocking mutation V-1 (FBM V-1)] from Dictyostelium cell extracts using anti-FLAG M2 beads failed to coprecipitate CP (Fig. 1D, lane 2). This result suggests that D.d. FBM V-1 can serve as an effective negative control for overexpression and rescue experiments.
V-1’s potency as a CP regulator should scale positively with the degree to which its cellular concentration exceeds that of CP’s. To clarify this issue, we determined the amount of V-1 and CP in known numbers of vegetative Dictyostelium cells by Western blotting using antibodies to D.d. V-1 and D.d. CP, and standard curves created by using FLAG-tagged D.d. V-1 purified from cells and D.d. CP purified from bacteria (Fig. S3 A and B, respectively). Two independent determinations yielded values of 49 and 56 ng of V-1, and 122 and 141 ng of CP, per 106 cells. Using a value of 1 pL per cell (2) yielded mean cellular concentrations of 4.03 and 1.99 µM for V-1 and CP, respectively. Of note, these concentrations, and the ratio between them, are similar to the values obtained for V-1 and CP in mouse embryo fibroblasts [3.2 ± 0.3 and 1.0 ± 0.3 µM, respectively (26)]. Given these results, and the affinity of D.d. V-1 for D.d. CP, ∼98% of CP in Dictyostelium cells could be sequestered by V-1 at steady state, barring regulation.
Fig. S3.
Cellular concentrations of V-1 and CP, V-1 immunostaining, FCS, quantitation of V-1 overexpression, and CAR1 levels. (A, Upper) Western blot of a Dictyostelium whole cell extract (WCE; corresponding to 5 × 105 amoebas) and the indicated amounts of purified, FLAG-tagged D.d. V-1 (in ng) probed with the antibody to D.d. V-1. Molecular mass markers are indicated. (A, Lower) Intensities (in A.U.) of the purified V-1 bands, as determined by densitometry using Image J2, and plotted vs. the amount of V-1 loaded (in ng). The resulting standard curve was obtained by linear regression not anchored at 0. The content of V-1 in the WCE band was estimated from this standard curve as indicated. (B, Upper) Western blot of a Dictyostelium WCE (corresponding to 1 × 105 amoebas) and the indicated amounts of purified D.d. CP (in ng) that was probed with the antibody to D.d. CP. Molecular mass markers are indicated. (B, Lower) The intensities (in A.U.) of the purified CP bands, as determined by densitometry using Image J2, and plotted vs. the amount of CP loaded (in ng). The resulting standard curve was obtained by linear regression not anchored at 0. The content of CP in the WCE band was estimated from this standard curve as indicated. (C) A field of WT Dictyostelium amoeba fixed and stained for V-1. (Scale bar, 10 µm.) (D) A field of V-1–null cells fixed and stained for V-1 and imaged using the exact same parameters as in C. (Scale bar, 10 µm.) (E) A representative FCS plot for mRFP in the cytoplasm. (F) A representative FCS plot for mRFP-V-1 in the cytoplasm. (G) Western blot of WCEs made from cells that were expressing mRFP V-1 at L, M, and H levels (fractionated by a fluorescence activated cell sorter as described in Methods). The blot was probed with the antibody to D.d. V-1. For L, M, and H expressers, the ratio of mRFP V-1 to endogenous V-1 was 0.45 ± 0.07, 1.55 ± 0.07, and 3.00 ± 0.42, respectively. (H) As in G, except that the cells were expressing mRFP FBM-V-1. For L, M, and H expressers, the ratio of mRFP FBM-V-1 to endogenous V-1 was 0.65 ± 0.07, 1.95 ± 0.09, and 3.50 ± 0.28, respectively. Note that V-1 overexpression did not result in a lowering of the level of endogenous V-1. (I) Western blot of WCEs prepared from vegetative WT cells (lane 1), vegetative V-1–null cells (lane 2), ripple stage WT cells (lane 3), and ripple stage V-1–null cells (lane 4) and probed with a monoclonal antibody to the cAMP receptor CAR1.
Finally, staining of vegetative Dictyostelium cells for V-1 yielded a diffuse cytoplasmic signal (Fig. S3C) that was absent in V-1 knockout cells (Fig. S3D). This observation, together with the fact that the apparent diffusion constant for monomeric red fluorescent protein (mRFP)-tagged D.d. V-1 in the cytoplasm (112.1 ± 22.4 µm2/s; Fig. S3E), as determined by fluorescence correlation spectroscopy (FCS), is essentially identical to that of mRFP alone (113.9 ± 22.8 µm2/s; Fig. S3F) suggests that D.d. V-1, like mouse V-1 (26), is freely diffusing. In summary, the properties of D.d. V-1 mirror those of mouse V-1 in every way. Specifically, both bind CP tightly, block CP’s barbed end capping activity, do not uncap CP-capped filaments, are subject to CARMIL-driven complex exchange, are present in molar excess over CP, and are freely diffusing. In all subsequent figures, we refer to D.d. V-1 simply as V-1.
Overexpression of V-1, but Not FBM V-1, Shifts the Cell’s Actin Phenotype from Pseudopodial to Filopodial.
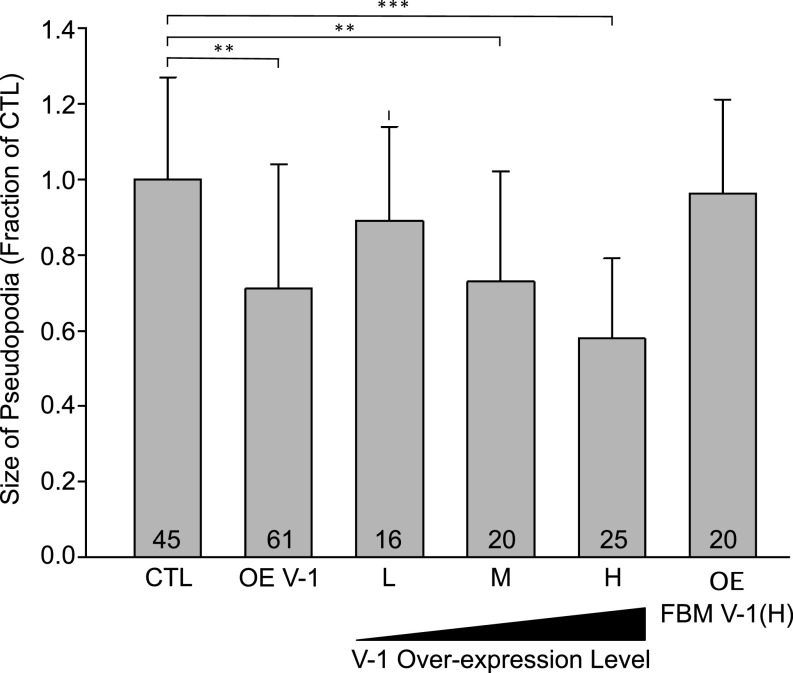
As discussed in the Introduction, CP knockdown in a variety of cell types has been shown to result in a decrease in both the size of lamellipodia and the cortical content of the Arp2/3 complex (7, 8). If V-1 functions to antagonize CP, then its overexpression in wild-type (WT) Dictyostelium amoeba should produce similar effects. To look for a decrease in pseudopodial area upon V-1 overexpression in vegetative amoeba, we fixed and stained control cells and cells overexpressing mRFP–V-1 with Alexa Fluor 488–phalloidin and measured the phalloidin signal within pseudopodia as a proxy for pseudopodial area (see Methods for details). In addition, we quantified on a per-cell basis total cellular mRFP fluorescence to correlate the degree of overexpression with the extent of change in pseudopodial area. Fig. 3 shows that Dictyostelium amoeba overexpressing V-1 do indeed exhibit on average a significant reduction (29%) in total pseudopodial area compared with control cells (compare OE V-1 to CTL) (note that in figures containing bar graphs, N values are noted at the bottom of each bar, mean and SD values are listed in the figure legend, and any experimental values that are different from control/WT in a statistically significant way are bracketed, with * denoting P < 0.05, ** denoting P < 0.01, and *** denoting P < 0.001). Moreover, the magnitude of the decrease in pseudopodial area scaled positively with the degree of V-1 overexpression [compare low (L), medium (M), and high (H) overexpressing cells to each other and to CTL), with high-expressing cells (which contain approximately four times more V-1 than control cells; Fig. S3 G and H) exhibiting a 42% decrease in pseudopodial area. Importantly, FBM V-1 had no effect on pseudopodial area, even at high levels of overexpression [compare OE FBM V-1 (H) to CTL], arguing that V-1‘s effect on the size of these Arp2/3-generated structures requires its ability to sequester CP.
Fig. 3.
V-1 overexpression reduces pseudopodial area in a dose-dependent fashion. Shown is the size of pseudopodia (as a fraction of control cells; CTL) for pooled overexpressing cells (OE-V-1), cells overexpressing L, M, and H levels of mRFP-V-1, and cells overexpressing H levels of mRFP-FBM V-1 [OE FBM V-1 (H)). Mean and SD values are as follows: CTL, 1.00 ± 0.27; OE V-1, 0.71 ± 0.33; L, 0.89 ± 0.25; M, 0.73 ± 0.29; H, 0.58 ± 0.21; OE FBM V-1 (H), 0.96 ± 0.25. Additional statistically different values are as follows: L vs. M, P < 0.05; L vs. H, P < 0.01; M vs. H, P < 0.05. Note that V-1–null cells exhibited a value of 0.94 ± 0.29, which was not significantly different from CTL. **P < 0.01; ***P < 0.001.
To look for a decrease in the cortical content of Arp2/3 upon V-1 overexpression, we stained control cells (Fig. 4A) and cells overexpressing high levels of mRFP-V-1 (Fig. 4B) with phalloidin (actin) and an antibody to D.d. Arp3 [also shown is the merge of these two signals (merge), the signal for mRFP–V-1, and the transmitted light image (differential interference contrast; DIC)). Comparison of these images suggested that overexpression of V-1 does indeed result in a significant reduction in the cortical content of the Arp2/3 complex (arrowheads in Fig. 4 A and B). Quantitation showed that this reduction was due to more than just the decrease in pseudopodial area described in Fig. 3, because measurement of the ratio of Arp3 fluorescence to F-actin fluorescence within 1 µm of the plasma membrane at protruding edges (excluding filopodial projections) revealed a ∼40% decrease in cells overexpressing high levels of V-1 compared with control cells (Fig. 4E, compare H to CTL). Importantly, this effect scaled positively with the degree of V-1 overexpression (Fig. 4 C and E) and was not due to a decrease in the cellular content of Arp3 in overexpressing cells (Fig. S4A). Moreover, the loss of cortical Arp3 signal in V-1–overexpressing cells could be accounted for by an increase in Arp3 signal in the central cytoplasm (Fig. S4B). As expected, cells expressing very high levels of FBM V-1 exhibited normal amounts of Arp3 signal at the edges of their actin-rich extensions (Fig. 4 D and E).
Fig. 4.
V-1 overexpression reduces the cortical content of the Arp2/3 complex. (A–D) Representative examples of a control cell (A; CTL), a cell overexpressing a high level of mRFP-V-1 [B; OE V-1 (H)), a cell overexpressing a low level of mRFP-V-1 [C; OE V-1 (L)), and a cell overexpressing a high level of mRFP-FBM V1 [D; OE FBM V-1 (H)) that had been stained for F-actin by using Alexa Fluor 647-labeled phalloidin (F-actin) and an antibody to D.d. Arp3 (Arp3). Also shown is the merge between these two signals (Merge), the signal for mRFP-V-1 or mRFP-FBM V-1, and the transmitted light image (DIC). All fluorescence images are maximum-intensity projections of complete optical sections. (Scale bar, 10 µm.) (E) The ratio of Arp3 fluorescence to F-actin fluorescence within 1 µm of the plasma membrane at protruding edges (excluding filopodia) for control cells (CTL), cells expressing L, M, and H levels of mRFP-V-1, and cells overexpressing high levels of mRFP-FBM V-1 (OE FBM V-1 (H)). Mean and SD values are as follows: CTL, 0.88 ± 0.18; L, 0.79 ± 0.16; M, 0.54 ± 0.17; H, 0.41 ± 0.12; OE FBM V-1 (H), 0.90 ± 0.20. Additional statistically different values are as follows: L vs. M, P < 0.05; L vs. H, P < 0.01; M vs. H, P < 0.05. Note that V-1–null cells exhibited a value of 0.82 ± 0.15, which was not significantly different from CTL. **P < 0.01; ***P < 0.001.
Fig. S4.
The cellular and cytoplasmic contents of Arp3 in V-1 overexpressing cells. (A) Western blot of whole cell extracts of control (lane 1) and mRFP-V-1 overexpressing cells (lane 2) probed for actin and Arp3, showing that the cellular content of Arp3 is unchanged in V-1 overexpressing cells. (B) The ratio of Arp3 fluorescence in pseudopodia to that in the cytoplasm for control cells (CTL) and cells expressing L, M, and H levels of mRFP-V-1, showing that the loss of cortical Arp3 signal seen in V-1 overexpressing cells reported in Fig. 4 can be accounted for by an increase in Arp3 signal in the central cytoplasm. Mean and SD values are: CTL, 3.29 ± 0.84; L, 2.81 ± 0.57; M, 1.83 ± 0.28; H, 1.32 ± 0.31.
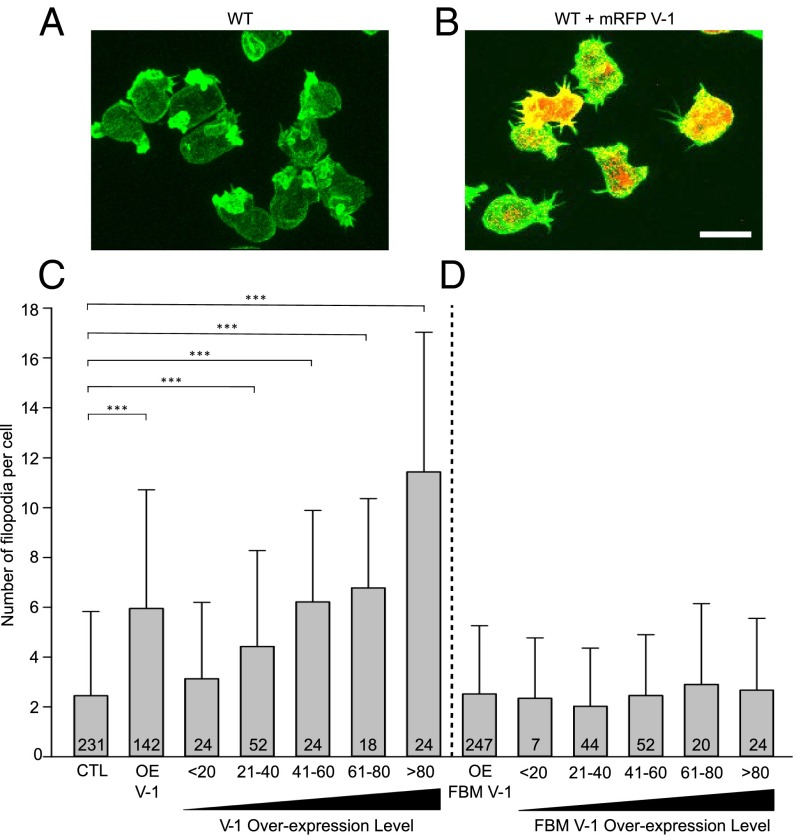
CP knockdown has also been shown in several cell types, including Dictyostelium, to result in an increase in the number of filopodia (2, 7). To look for an increase in the number of filopodia upon V-1 overexpression, we fixed control cells and cells overexpressing mRFP–V-1 with Alexa Fluor 488–phalloidin, counted the number of filopodia, and quantified on a per-cell basis total cellular mRFP fluorescence to correlate the degree of V-1 overexpression with the extent of filopodia induction. Comparison of typical control and V-1–overexpressing cells (Fig. 5 A and B, respectively) suggested that V-1 overexpression does indeed induce the formation of filopodia. Indeed, quantitation showed that, on average, overexpressing cells have ∼2.5 times as many filopodia as control cells (Fig. 5C, compare OE V-1 to CTL). Moreover, this effect scaled positively with the degree of V-1 overexpression (Fig. 5C). At the highest levels of V-1 overexpression, the mean number of filopodia per cell was more than fourfold higher than in control cells (Fig. 5C). Importantly, the overexpression of FBM V-1 had no effect on filopodial number per cell, even at the highest levels of expression (Fig. 5D). Therefore, the morphological consequence of V-1 overexpression with regard to filopodia formation requires V-1’s ability to sequester CP.
Fig. 5.
V-1 overexpression increases the number of filopodia. (A and B) Representative control cells (A) and cells overexpressing mRFP-V-1 (B) fixed and stained with Alexa Fluor 488-labeled phalloidin. Z-stack projections in the green (phalloidin) and red (mRFP-V-1) channels are superimposed. (Scale bar, 10 µm.) (C) Number of filopodia per cell in control cells (CTL), pooled mRFP-V-1 overexpressing cells (OE V-1), and mRFP-V-1 overexpressing cells binned for increasing V-1 expression level (<20, 21–40, 41–60, 61–80, and >80). Mean and SD values are as follows: CTL, 2.45 ± 3.38; OE V-1, 5.99 ± 4.32; <20, 3.13 ± 3.07; 21–40, 4.42 ± 3.86; 41–60, 6.21 ± 3.68; 61–80, 6.78 ± 3.59; >80, 11.43 ± 5.59. (D) The number of filopodia per cell in pooled mRFP-FBM V-1–overexpressing cells (OE FBM V-1), and mRFP-FBM V-1–overexpressing cells binned for increasing V-1 expression level. Mean and SD values are as follows: OE FBM V-1, 2.57 ± 2.92; <20, 2.31 ± 1.98; 21–40, 2.52 ± 2.34; 41–60, 2.55 ± 2.89; 61–80, 2.48 ± 3.24; >80, 2.67 ± 2.89. Note that V-1–null cells exhibited a value of 2.1 ± 2.59, which was not significantly different from CTL. ***P < 0.001.
To extend these results, we coexpressed the dynamic F-actin reporter GFP-LimEΔCC (38) with or without mRFP-V-1 and imaged the cells using lattice light sheet microscopy (39). Compared with control cells expressing only GFP-LimEΔCC (Movie S1, Left), cells overexpressing V-1 exhibited a dramatic increase in the number of filopodia on their surface (Movie S1, Right). Together, the data above indicate that V-1 overexpression phenocopies CP knockdown in terms of switching the actin cytoskeletal phenotype from lamellipodial/pseudopodial to filopodial and that V-1’s ability to drive this major switch in actin phenotype requires its ability to sequester CP.
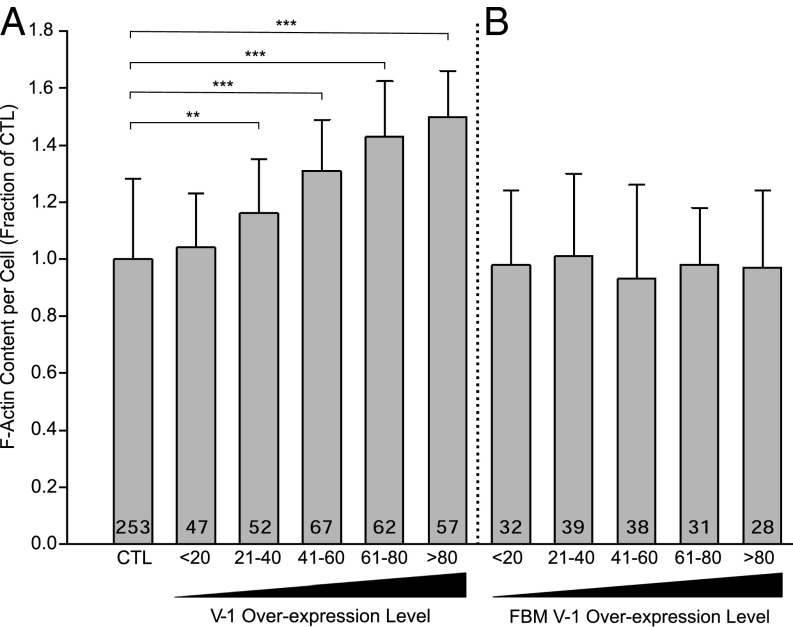
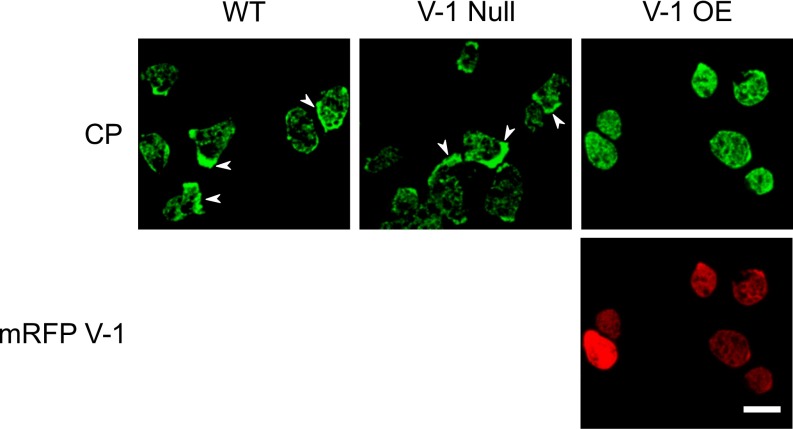
Finally, CP knockdown in Dictyostelium has been shown to result in an elevation in overall cellular F-actin content (2). To address the possibility that V-1 overexpression also increases cellular F-actin content, control cells and cell overexpressing mRFP–V-1 were fixed and stained with Alexa Fluor 488–phalloidin and optically sectioned, and the total fluorescence in all sections was summed in the 488-nm channel for F-actin and in the 543-nm channel for V-1. Fig. 6A shows that the overexpression of V-1 does indeed cause a rise in total cellular F-actin content, with the magnitude of the increase scaling positively with the degree of overexpression. At the highest levels of V-1 overexpression, cellular F-actin content is ∼1.5 times higher than in control cells (Fig. 6A). Importantly, the overexpression of mRFP–FBM V-1 had no effect on cellular F-actin content, even at high expression levels (Fig. 6B), which argues strongly that V-1 exerts its effect of F-actin content by sequestering CP. Finally, consistent with the observations above, the overexpression of mRFP V-1 causes a significant shift in the distribution of endogenous CP from the cortex to the cytoplasm (Fig. S5).
Fig. 6.
V-1 overexpression increases cellular F-actin content. (A) Cellular content of F-actin, expressed as a fraction of control cells, in control cells (CTL) and cells overexpressing mRFP-V-1 binned for increasing V-1 expression level (<20, 21–40, 41–60, 61–80, >80). Mean and SD values are as follows: CTL, 1.00 ± 0.28; <20, 1.04 ± 0.19; 21–40, 1.16 ± 0.19; 41–60, 1.31 ± 0.18; 61–80, 1.42 ± 0.19; >80, 1.50 ± 0.16. (B) Cellular content of F-actin, expressed as a fraction of control cells, in cells overexpressing mRFP-FBM V-1 binned for increasing FBM V-1 expression level. Mean and SD values are as follows: CTL, X ± X; <20, 0.99 ± 0.23; 21–40, 1.01 ± 0.19; 41–60, 0.93 ± 0.19; 61–80, 0.98 ± 0.20; >80, 1.01 ± 0.24. **P < 0.01; ***P < 0.001.
Fig. S5.
Localization of CP in WT, V-1–null, and mRFP-V-1–overexpressing cells. Shown are representative fields of WT, V-1–null, and mRFP-V-1–overexpressing cells stained with the antibody to CPα. Whereas WT and V-1–null cells show a preponderance of cortical localization for endogenous CP (arrowheads), cells overexpressing mRFP-V-1 show a preponderance of cytoplasmic localization for endogenous CP. (Scale bar, 10 µm.)
Dictyostelium Lacking V-1 Are Viable, but Grow Slowly and Have Less F-Actin.
The fact that V-1 is present within Dictyostelium in molar excess over CP, has a high affinity for CP, and has ready access to CP (both are freely diffusing) argues that V-1 may significantly suppress CP activity in the cytoplasm at steady state. If so, then the elimination of V-1 in Dictyostelium, like the overexpression of CP in these cells (2), should result in a decrease in total cellular F-actin content. To address this possibility, and to further gauge V-1’s physiologic significance, we used homologous recombination to create Dictyostelium cell lines that lack V-1. WT AX3 cells were transfected with a linear disruption fragment comprised of the selectable marker blasticidin flanked by portions of the Dictyostelium V-1 gene. Screening of ∼80 clones purified by serial dilution using Western blotting identified two clones that lacked V-1 (Fig. 1F, lanes 2 and 3). Disruption of the V-1 gene in these two cell lines via the intended double-crossover, gene-replacement event was confirmed by sequencing PCR products spanning the integration site. Because the behaviors of these two cell lines were indistinguishable in all subsequent assays, we refer to them below simply as V-1–null cells.
Our immediate impression upon passaging V-1–null cells was that they grew more slowly and reached a lower density at saturation. These impressions were borne out in representative growth curves (Fig. S6A) and by quantitative analyses. Specifically, V-1–null cells exhibited a 59% increase in doubling time (Fig. S6B) and a 43% decrease in cell density at saturation (Fig. S6C) relative to the parental cell line. Although the expression of FLAG-V-1 in null cells largely restored both doubling time and final cell density to WT values, expression of FLAG-FBM V-1 had no measureable effect on the growth of null cells (Fig. S6 A–C). Given that a slower growth rate could reflect a defect in cytokinesis, we also determined the number of nuclei per cell by attaching WT and V-1–null cells at midlog to coverslips and staining them with Hoechst 33342 to visualize nuclei and with the plasma membrane dye FM 4-64FX to unequivocally define individual cell boundaries. The histogram in Fig. S6D shows that V-1–null cells on average tend to be more multinucleate than WT cells, suggesting that V-1 contributes to the fidelity of cytokinesis, an actin-dependent process.
Fig. S6.
V-1–null cells grow slowly and may have a partial defect in cytokinesis. (A) Representative growth curves for WT cells (WT), V-1–null cells (V-1 Null), V-1–null cells expressing FLAG-V-1 (V-1 Null + V-1), and V-1–null cells expressing FLAG-FBM V-1 (V-1 Null + FBM V-1) (see key). The values are means of duplicate samples. (B) Doubling times for the same groups of cells in A. Mean and SD values are as follows: WT, 12.7 ± 1.1; V-1–null, 21.6 ± 1.0; V-1–null + V-1, 14.7 ± 1.7; V-1–null + FBM V-1, 20.8 ± 1.0. (C) Cell densities at saturation for the same group of cells in A. Mean and SD values are as follows: WT, 10.8 ± 0.9; V-1–null, 6.2 ± 0.7; V-1–null + V-1, 9.1 ± 0.8; V-1–null + FBM V-1, 6.0 ± 0.8. Note that V-1–null cells were not quite fully rescued by the reexpression of FLAG-V-1. (D) The distributions of WT and V-1–null cells (see key) with one, two, or three and greater nuclei. *P < 0.05; ***P < 0.001.
To address the possibility that the loss of V-1, like the overexpression of CP (2), decreases cellular F-actin content, we measured total cellular fluorescence in WT and V-1–null cells stained with FITC-labeled phalloidin. Fig. 7A shows that null cells contain on average 73.4% as much F-actin as WT cells. This difference was confirmed by measuring the F-actin content of Triton-insoluble cytoskeletons (Fig. 7B), where the value for V-1–null cells was on average 72.6% of WT, and by FACS of phalloidin-stained cells (Fig. 7C), where the value for V-1–null cells was on average 51.7% of WT. Importantly, reintroduction of FLAG-V-1 into null cells restored cellular F-actin content (Fig. 7A). Conversely, reintroduction of FLAG-FBM V-1 into null cells did not restore F-actin content (Fig. 7A), arguing that V-1 exerts its effects of F-actin content in vivo by sequestering CP.
Fig. 7.
V-1–null cells exhibit reduced F-actin content. (A) F-actin content (as a fraction of WT) in WT cells, V-1–null cells, V-1–null cells expressing FLAG-V-1, and V-1–null cells expressing FLAG-FBM V-1 determined by measuring total cellular fluorescence in cells stained with FITC-labeled phalloidin. Mean and SD values are as follows: WT, 1.00 ± 0.28; V-1–null, 0.73 ± 0.23; V-1–null + V-1, 0.92 ± 0.32; V-1–null + FBM V-1, 0.75 ± 0.21. (B) F-actin content (as a fraction of WT) in WT cells and V-1–null cells determined by measuring the signal for FITC-labeled phalloidin in Triton X-100–insoluble cytoskeletons. Mean and SD values are: WT, 1.00 ± 0.09; V-1–null, 0.73 ± 0.11. (C) Total cellular F-actin content of WT cells (green) and V-1–null cells (red) determined by flow cytometry of cells stained with FITC-labeled phalloidin. Each sample represents ∼30,000 cells (the black peak shows the background fluorescence). Mean and median values in arbitrary fluorescence units (A.U.) are as follows: WT, 6,996 and 6,301; V-1–null, 3,621 and 3,491. **P < 0.01.
Vegetative V-1–Null Cells Exhibit a Defect in Macropinocytosis That Is Rescued by V-1, but Not by FBM V-1.
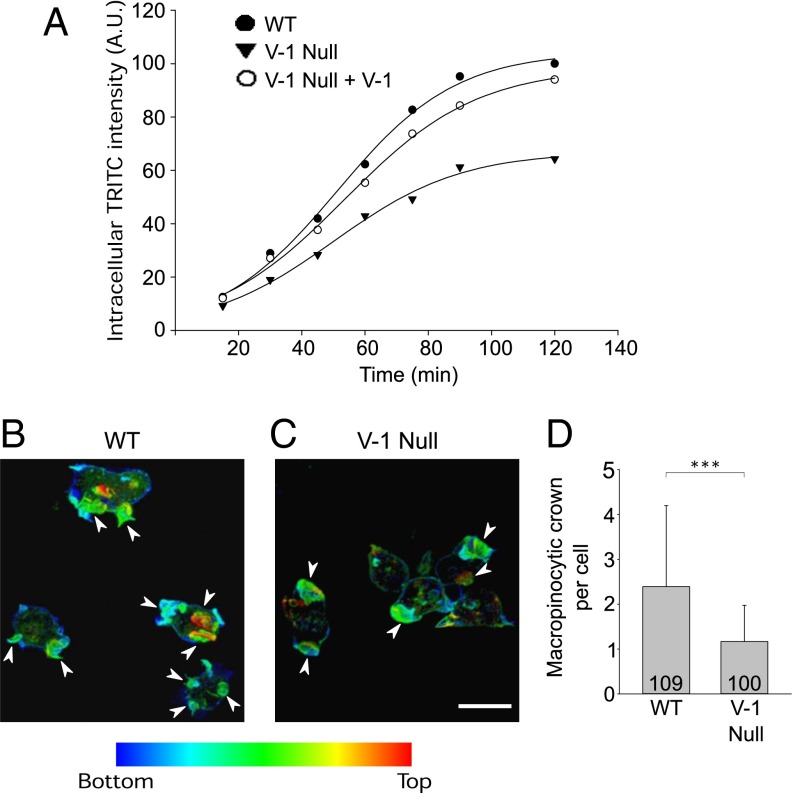
A major fraction of actin assembly in vegetative amoeba is devoted to the formation of dorsal, crown-like projections that drive macropinocytosis, the major route by which axenically grown cells obtain nutrients (40). To address V-1’s role in this actin-based process, we compared the rate of uptake of the nondigestible fluid phase marker TRITC–dextran in WT and V-1–null cells. Of note, the rate of accumulation of this marker within cells during the first ∼60 min after its addition to the medium accurately reports the rate of fluid phase endocytosis because the time interval between the uptake and exoctyosis of nondigestible markers in Dictyostelium, which lack a rapid recycling component, is ∼60 min (41). Fig. 8A shows that V-1–null cells exhibited a ∼30% reduction in the rate of fluid phase pinocytosis relative to WT cells based on the slopes between the 30- and 60-min values. Moreover, the amount of cell-associated dextran at steady state (∼90 min) was reduced by approximately one-third in the mutant. Of note, both values were largely restored to WT levels upon expression of FLAG-V-1 in V-1–null cells (Fig. 8A). Values that were very similar to those shown in Fig. 8 were obtained in a second experiment.
Fig. 8.
V-1-–null cells exhibit a defect in macropinocytosis. (A) Time course of intracellular accumulation of the nondigestible fluid phase pinocytic marker TRITC-dextran vs. time in WT cells (WT), V-1–null cells (V-1 Null), and V-1–null cells expressing FLAG-V-1 (V-1 Null + V-1) (see key). The values are the mean of duplicate samples. (B and C) Representative macropinocytic crowns in WT cells (B) and V-1–null cells (C) (arrowheads; the color coding indicates increasing distance from the coverslip). (D) The average number of macropincytic crowns per cell in WT and V-1–null cells. (Scale bar, 10 μm.) Mean and SD values are as follows: WT, 2.39 ± 1.80; V-1–null, 1.16 ± 0.80. ***P < 0.001.
To address the underlying cause of the reduced rate of macropinocytosis, we measured the number of macropinocytic crowns per cell by plating on coverslips, fixing, staining for actin, optically sectioning at 0.4 µm intervals on a Zeiss 780 LSM confocal microscope, and rendering the sections in 3D. Fig. 8 B and C show representative macropinocytic crowns in WT and V-1–null cells, respectively (arrowheads; the color coding indicates increasing distance from the coverslip). Quantitation revealed that V-1–null cells possess on average 48.8% as many crowns as WT cells (Fig. 8D). This difference was mirrored in time-lapse movies of WT and V-1–null cells expressing GFP-LimEΔCC and imaged by lattice light sheet microscopy (Movie S2, Left and Right, respectively). Together, these results show that a major actin-based process in vegetative amoeba is significantly impacted by the loss of V-1 and suggest that the decreased growth rate exhibited by V-1–null cells may be due, at least in part, to a reduction in the rate of fluid phase endocytosis.
Starved V-1–Null Cells Exhibit Defects in Chemotactic Aggregation, Motility, Polarity, and Adhesion That Are Rescued by V-1, but Not by FBM V-1.
When starved for nutrients, Dictyostelium amoeba initiate a developmental program that eventually culminates in the formation of a stalk with a spore-filled head (42). This structure is created from ∼100,000 cells that coalesce by crawling up a gradient of extracellular cAMP generated initially by small numbers of pioneer cells. Amoeba initially chemotax toward these pioneer cells as individual cells, but soon merge in head-to-tail fashion to create large streams of cells moving together toward what has now become an aggregation center. To gauge V-1’s contribution to this major actin-based process, we performed streaming assays in which vegetative cells were plated on plastic dishes at a density of 2 × 106 cells per cm2 in the presence of a simple salt solution and time-lapse imaged by using a stereo microscope. At this cell density, streams typically become prominent within 4–6 h, and aggregation is usually complete by 12 h. Fig. 9 A, Upper shows a representative example of WT cells undergoing streaming 6 h after plating, whereas Fig. 9 A, Lower shows the large aggregates that had formed by 12 h. In striking contrast, V-1–null cells made many tiny streams (Fig. 9 B, Upper) that culminated in the formation of many tiny aggregates (Fig. 9 B, Lower). Importantly, the streaming defects exhibited by null cells were largely rescued by the expression of FLAG-V-1 (Fig. 9C), but not by the expression of FLAG-FBM V-1 (Fig. 9D and Movie S3). Together, these results indicate that V-1’s ability to sequester CP is required for efficient cell migration.
Fig. 9.
V-1–null cells exhibit defects in chemotactic aggregation. (A–D) Representative examples of streaming assays performed using WT cells (A; WT), V-1–null cells (B; V-1 Null), V-1–null cells expressing FLAG-V-1 (C; V-1 Null + V-1), and V-1–null cells expressing FLAG-FBM V-1 (D; V-1 Null + FBM V-1) imaged at 6 h (Upper) and 12 h (Lower) with bright field. (Scale bar, 100 µm.) (E–G) Representative 15-min path plots (taken from phase images) for ripple-stage WT cells (E) and V-1–null cells (F), and the migration speeds of ripple-stage WT cells, V-1–null cells, V-1–null cells expressing FLAG-V-1, and V-1–null cells expressing FLAG-FBM V-1 (G). (Scale bar, 50 μm.) Mean and SD values (in µm/min) are as follows: WT, 11.58 ± 3.32; V-1–null, 5.52 ± 2.56; V-1–null + V-1, 10.2 ± 3.28; V-1–null + FBM V-1, 5.58 ± 1.94. ***P < 0.001.
To address the underlying cause of the defect in chemotactic aggregation, we starved WT and V-1–null cells at high density on black filters until “ripple stage,” which took ∼5 h for WT cells and ∼5.5 h for V-1–null cells. At this stage, Dictyostelium amoebae exhibit their highest rate of motility, which is approximately two to four times faster than the rate exhibited by vegetative cells (41, 43, 44). Ripple-stage cells were then harvested by trituration and allowed to attach at low density on chamber slides for 20 min. Cell motility rates were then measured by determining the centroid of every cell in the field of view every 15 s for 15 min. Representative path plots for WT and V-1–null cells (Fig. 9 E and F, respectively) suggested that V-1–null cells were significantly slower (see also Movie S4). Indeed, quantitation showed that the average rate of motility for V-1–null cells was 51% that of WT cells (Fig. 9G). Moreover, the motility of V-1–null cells was rescued by the expression of FLAG-V-1, but not by the expression of FLAG-FBM V-1 (Fig. 9G), indicating that V-1’s effect on motility requires its ability to sequester CP. Importantly, Fig. S3I shows that ripple-stage V-1–null cells exhibit the same degree of up-regulation of the cAMP receptor CAR1, a key reporter of development, as ripple-stage WT cells, arguing that the slower speed exhibited by V-1–null cells is not due to their being less well developed.
During chemotactic aggregation, the fast speed of amoeba is associated with an elongated, highly polarized shape that aligns with the direction of migration. Higher-magnification images of individual cells in the streaming assays performed in Fig. 9 showed that V-1–null cells appeared, on average, to be much less polarized than control cells (Fig. S7 A and B). To quantitate this result, we measured the ratio of cell length to cell width using still images from the movies used to determine the motility rates of ripple-stage cells. Fig. S7C shows that V-1–null cells are indeed significantly less polarized than WT cells. Again, expression of FLAG-V-1 in null cells rescued cell polarity, whereas expression of FLAG-FBM V-1 did not (Fig. S7C). Concurrent with the decrease in cell polarity associated with the lack of V-1 was an apparent decrease in cell adhesiveness. Specifically, the ratio of the area under each adherent cell that was black in an interference reflection (IRM) image (which defines that portion of the cell’s ventral plasma membrane that is very close to the glass) to the total ventral surface area in the DIC image was 38% lower in V-1–null cells than in WT cells (Fig. S7D).
Fig. S7.
V-1–null cells exhibit defects in cell polarity and adhesion. (A and B) Representative fields of WT (A) and V-1–null (B) cells 5 h after the initiation of streaming (arrows indicate the direction of migration) (DIC). (Scale bar, 20 µm.) (C) Degree of cell polarization exhibited by ripple stage WT cells, V-1–null cells, V-1–null cells expressing FLAG-V-1, and V-1–null cells expressing FLAG-FBM V-1. Mean and SD values are as follows: WT, 2.03 ± 0.52; V-1–null, 1.31 ± 0.35; V-1–null + V-1, 1.81 ± 0.57; V-1–null + FBM V-1, 1.38 ± 0.32. (D) Ratio of IRM surface area to total surface area in the DIC image of WT cells and V-1–null cells. The mean and SD values are as follows: WT, 0.56 ± 0.22; V-1–null, 0.37 ± 0.20. ***P < 0.001.
The V-1–Dependent Regulation of CP Activity May Be Controlled by the Phosphorylation of V-1.
The preceding data demonstrate that artificially manipulating V-1 levels, like artificially manipulating CP levels, has large consequences for actin-based processes and can alter the cell’s actin phenotype. The question then arises, do cells use some form of posttranslational modification, such as phosphorylation, to regulate V-1’s sequestering activity to modulate the level of active CP? Consistent with this possibility, blots of Dictyostelium whole cell extracts resolved by using urea-glycerol gels [used traditionally to separate phosphorylated from unphosphorylated myosin II RLCs (45)] and probed with the V-1 antibody yielded two bands (Fig. 10A, lane 1; lane 2 was a negative control performed using a V-1–null cell extract). Importantly, the upper band disappeared when the extract was pretreated with phosphatase (Fig. 10B, compare lane 2 to 1), suggesting that it corresponded to phosphorylated V-1. Identical results were obtained for FLAG-V-1 after its purification from cell extracts. Specifically, it ran as two bands in the absence of phosphatase treatment (Fig. 10C, lane 1) and one band after phosphatase treatment (Fig. 10C, lane 2). In complete agreement with these observations, mass-spectroscopic analysis of FLAG-V-1 purified from Dictyostelium extracts identified a single serine residue that is phosphorylated in vivo (Ser-22; Fig. 1A). Notably, Ser-22 resides immediately adjacent to the loop comprising the first ankryin repeat that, when mutated, abrogates V-1:CP interaction (27). Moreover, examination of the structure of V-1 suggests that the addition of a phosphate on Ser-22 would disrupt the conformation of this loop, likely reducing V-1’s affinity for CP (Fig. S8 A and B). Interestingly, mouse and other vertebrate V-1s contain a potentially phosphorylatable tyrosine at the position analogous to Ser-22 in D.d. V-1 (Fig. S8C) (note, however, that this implies a different kinase/signaling pathway than in Dictyostelium). Finally, precedents exist for the regulation of ankryin repeat proteins by the phosphorylation of residues adjacent to their ankyrin repeats (46, 47).
Fig. 10.
V-1 is phosphorylated on Ser-22 in vivo, and a phosphomimetic version of V-1 interacts weakly with CP in vitro. (A) Western blot of extracts of WT cells (lane 1) and V-1–null cells (lane 2) that were resolved on urea-glycerol gels and probed with the antibody to D.d. V-1. (B) Western blot of extracts of WT cells that were not (lane 1) or were (lane 2) treated with phosphatase before loading on urea-glycerol gels and probed with the antibody to D.d. V-1. (C) Western blot of the material eluted from anti-FLAG M2 beads that had been incubated with lysates of cells expressing FLAG-V-1 and that were not (lane 1) or were (lane 2) treated with phosphatase before loading on urea-glycerol gels and probed with the antibody to D.d. V-1. (D) Western blot of the material eluted from resins that had been loaded with equal amounts of GST-WT V-1 (lane 1) or GST-V-1 S22E (lane 2), incubated with cell extracts, and washed. The blot was probed with an antibody against D.d. CPβ. (E) Representative pyrene-based, seed-initiated, actin polymerization assays performed in the presence of 7 nM CP and increasing amounts of GST-V-1 S22E added before seed initiation (see color-coded key). (F) Mean and SDs from three independent assays performed as in E and plotted as the fraction of capped filaments vs. V-1 concentration.
Fig. S8.
Phosphorylation of Ser-22 may reduce D.d. V-1’s affinity for CP. (A and B) Two views of the structure of mouse V-1 (23) that provide insight into the possible effect Ser-22 phosphorylation might have on the function of D.d. V-1. (A) Loop 1 in mouse V-1 is critical for its interaction with CP, as mutation of the four residues marked (E33, G34, G35, and R36) results in a ∼200-fold decrease in V-1’s affinity for CP (27). Data presented here shows that loop 1 is also critical for the interaction of D.d. V-1 with D.d. CP. (A and B) In mouse V-1, Tyr-21 (Y21) sits in the position occupied by Ser-22 in D.d. V-1 and is likely hydrogen-bonded to Arg-30 (R30), thereby stabilizing the conformation of loop 1. Importantly, the overall folding of this region is probably conserved between mouse V-1 and D.d. V-1 because the hydrophobic residues involved in packing required to stabilize ankyrin repeat 1 (residues 5–23) and ankyrin repeat II (residues 39–57) are conserved. Given this, the introduction of a phosphate group on Ser-22 in D.d. V-1 would likely disrupt the conformation of loop 1, resulting in a decrease in the protein’s affinity for CP. (C) Alignment of the amino acid sequence of Dictyostelium V-1 with the amino acid sequences of human, mouse, rat, and chicken V-1 showing that these vertebrate V-1s contain a potentially phosphorylatable tyrosine (red box) at the position analogous to Ser-22 in D.d. V-1. Note that this implies that different kinases/signaling pathways are regulating V-1 in vertebrate cells and Dictyostelium. Interestingly, while Kitizawa et al. (50) presented indirect evidence that PKA is involved in the phosphorylation of mammalian V-1, precedents exist for the phosphorylation of tyrosines downstream of PKA (55).
To gain further support for the possibility that the phosphorylation of Ser-22 inhibits V-1’s ability to sequester CP, we generated a GST-tagged version of D.d. V-1 in which Ser-22 was changed to a glutamate residue (GST-V-1 S22E). Fig. 10D shows that this fusion protein pulled down far less CP from whole cell extracts (lane 2) than did an equal amount of WT GST-V-1 (lane 1) (the reduction was 80.5% and 74.9% in two separate experiments). Moreover, Fig. 10 E and F show that GST-V-1 S22E was far less capable of sequestering CP in solution assays than WT GST-V-1. Specifically, the highest concentration of GST-V-1 S22E tested (5 µM) restored polymerization to only ∼50% of the seed-only rate, yielding an approximate half-maximal concentration of GST-V-1 S22E required to restore actin polymerization of 493 ± 191 nM (vs. 59.5 ± 14.8 nM for WT GST-V-1; note that this decrease may be an underestimation, because the value for GST-V-1 S22E rises to ∼4 µm if one extrapolates the curve in Fig. 10F to saturation). Importantly, the negative impact on V-1’s CP sequestering activity of adding an actual phosphate group to Ser-22 is likely to exceed the negative impact of replacing Ser-22 with a glutamate. Together, these observations argue strongly that the ability of V-1 to sequester CP is regulated by phosphorylation, and they suggest that Dictyostelium may tune the activity of CP by controlling the phosphorylation state of V-1.
Discussion
We recently proposed a model for CP regulation in vivo in which V-1 globally inhibits CP in the cytoplasm, and active, plasma membrane-associated CARMIL at protruding cell edges drives a complex exchange reaction converting sequestered CP (CP:V-1) into a version that binds the barbed end with ∼50 nM affinity (CP:CARMIL) (26). In this model, therefore, the barbed end capping activity required to drive Arp2/3-dependent branched actin network assembly is only generated downstream of the CARMIL-driven complex exchange reaction that liberates CP from V-1. Importantly, two predictions of this model have already been confirmed. First, the half-life of CP on barbed ends near the plasma membrane in living cells is much closer to the half-life of the CP:CARMIL complex on the barbed end in vitro (∼8 s) (22) than to the half-life of CP on the barbed end in vitro (∼1,800 s) (8, 18). Second, cells engineered to express a version of CP that cannot see the CPI motif in CARMIL and related proteins (e.g., CD2AP or CKIP) phenocopy Arp2/3-inhibited cells (29).
A third prediction of our model—that cells containing elevated levels of V-1 should phenocopy cells containing reduced levels of CP—has now been confirmed in this study. Perhaps most telling, the overexpression of V-1, like the knockdown of CP, shifted the cell’s actin phenotype from pseudopodial to filopodial. Importantly, the extent of this phenotypic switch scaled positively with the degree of V-1 overexpression, as one would predict if V-1 is acting as a CP buffer. Moreover, this phenotypic switch did not occur in cells overexpressing FBM V-1, indicating that the switch was due to V-1’s CP-sequestering activity.
Finally, given CP’s central role in actin assembly—and the likelihood that V-1 sequesters a major fraction of CP at steady state—our data confirmed a fourth prediction of our model, which is that cells lacking V-1 should exhibit significant defects in cellular processes dependent on actin assembly. Specifically, we showed that cells lacking V-1, like cells overexpressing CP (2), exhibit reduced levels of F-actin. More importantly, we showed that V-1–null cells exhibit myriad defects in actin-dependent cellular processes, including macropinocytosis in vegetative amoeba and cell migration/chemotactic aggregation in starved amoeba. Critically, all of these defects were rescued by the expression of V-1, but not by the expression of FBM V-1, indicating that V-1 exerts its effects on the cell’s actin phenotype via its ability to sequester CP.
With regard to the underlying mechanism by which V-1 overexpression converts Dictyostelium amoeba from a pseudopodial to a filopodial phenotype, we think it revolves around the opposite relationship the two major nucleation machines have with CP. In the case of the Arp2/3 complex, CP promotes the quantity and quality of the branched actin arrays created by Arp2/3, such as lamellipodia and pseudopodia, by accelerating Arp2/3 complex-dependent nucleation and by promoting optimal branching within the resulting network (4, 5). Formins and VASP, conversely, consider CP a “pest” and use physical mechanisms to keep it at bay as they build linear actin structures such as filopodia (48, 49). Importantly, although formins are fairly effective at physically shielding the barbed end from CP, it is likely that their robustness as filopodia generators in vivo would be increased by a reduction in CP levels (13, 14). Given all this information, interventions that impair CP function, such as CP knockdown or V-1 overexpression, would be predicted to reduce the formation of cortical actin structures built by the Arp2/3 complex (lamellipodia and pseudopodia) and promote the formation of cortical actin structures built by formins and VASP (filopodia). Of note, the increase in filopodia number seen upon CP knockdown/V-1 overexpression is probably also due in significant part to an increase in the amount of monomer available for formin/VASP after the reduction in Arp2/3-dependent nucleation (15–17).
With regard to why Dictyostelium amoeba missing V-1 exhibit such widespread defects in actin-based processes, we think it is because the full function of such processes requires an optimum level of active CP. In the case of too little active CP, such as that seen in CP-knockdown cells or cells overexpressing V-1, Arp2/3-dependent actin assembly is compromised. In the case of too much active CP, such as that seen in cells overexpressing CP (2) or the V-1 knockout cells described here, there are also negative consequences, one of which is a reduction in the cell’s total content of F-actin. This consequence, as well as the many defects in actin-based processes exhibited by V-1–null cells, most likely reflects the negative consequences of excessive barbed end capping in vivo. Importantly, the in vitro reconstitution of the Arp2/3- and CP-dependent motility of Listeria also demonstrated an optimum for CP—that is, both too little and too much CP impaired the actin polymerization-driven motility of the bacterium (5). Together, these observations argue that cells must maintain the level of active CP within a certain range to promote robust actin-dependent processes.
Finally, although cells probably avoid extremes in both directions in the level of active CP, they might regulate the level of active CP over a limited range to modify their actin phenotype. One way to accomplish this goal would be through a posttranslational modification of V-1 that inhibits its ability to sequester CP. Relevant to this possibility, a previous study of vertebrate V-1 suggested that it might be phosphorylated and that phosphorylated V-1 might have a reduced affinity for CP (50). Moreover, other ankyrin repeat proteins have been shown to be inhibited by phosphorylation of residues adjacent to their ankyrin repeats (46, 47). Consistently, we demonstrated here that D.d. V-1 is phosphorylated in vivo at a single serine residue adjacent to its first ankryin repeat, which is critical for V-1:CP interaction. Moreover, a version of D.d. V-1 in which this serine was replaced with a glutamate to partially mimic serine phosphorylation exhibited a pronounced reduction it its ability to sequester CP, implying that the phosphorylation of this serine might fully abrogate V-1:CP interaction. Although much remains to be determined regarding the biological consequences of V-1 phosphorylation, these initial results argue that cells probably regulate the activity of this CP buffer to suit certain physiological purposes.
Methods
Cell Cultures, Clones, and Reagents.
Dictyostelium axenic strain AX3 (WT) was cultured in HL5 medium (41). Untagged, full-length versions of D.d. V-1 with or without the FBM (GNKRΔAAAA) or the S22E mutation were synthesized as Bam HI/Eco RI fragments by using the D.d. V-1 genomic sequence (DictyBase Gene ID No. DDB_G0268038). WT and S22E versions were cloned into pGST-Parallel 2 to create the GST fusion proteins used for pulldown experiments and raising the rabbit polyclonal antibody to D.d. V-1. For expression in Dictyostelium, WT and FBM versions were cloned into mRFP-mars (51). For FLAG-V-1 and FLAG-FBM V-1, full-length versions of D.d. V-1 with and without the FBM were synthesized with the 8-amino-acid FLAG sequence (DYKDDDDK) and a three-residue spacer (SGA) following the ATG and cloned into Exp4 (52). The dynamic F-actin reporter GFP-LimEΔCC (38) and the rabbit polyclonal antibody against D.d. Arp3 (19) have been described. The rabbit polyclonal antibody against D.d. CPα was a gift of John Cooper, Washington University, St. Louis. The rabbit polyclonal antibody against D.d. CPβ was generated by using gel-purified CPβ. The mouse monoclonal antibody against CAR1 was a gift of Carol Parent, National Cancer Institute, Bethesda. The mouse monoclonal antibody against β-actin was purchased from Santa Cruz Biotechnology (sc-47778). Labeled phalloidins and secondary antibodies were purchased from Thermo Fisher Scientific.
V-1 Knockout and Stable Cell Lines.
The generation of D.d. V-1 knockout cells by homologous recombination, and the generation of stable cell lines expressing tagged versions of D.d. V-1, are described in SI Methods.
Microscopy and Imaging-Based Quantitation.
For staining, cells were fixed with 15% (vol/vol) picric acid/2% (vol/vol) formaldehyde in HL5 medium at room temperature (RT) followed by 70% ethanol. Confocal imaging was performed on a Zeiss LSM 510 or LSM 780 microscope equipped with a 63× 1.4-NA objective (40×, 1.2 NA for the motility assays). To measure the effect of V-1 overexpression on pseudopodia size as a function of V-1 overexpression level, WT cells expressing mRFP-V-1 or mRFP-FBM V-1 were fixed and stained with Alexa Fluor 647–phalloidin. Actin-rich pseudopodia visible in 1.5-µm-thick ventral confocal sections were outlined, and the total fluorescence within these outlines was measured in the 647-nm (F-actin) and 543-nm (mRFP-V-1) channels. Note that for this and similar measurements, gain was minimized to avoid pixel saturation while still maintaining good dynamic range, and rapid scans with no frame averaging were used to avoid bleaching. In addition, note that for this and all overexpression experiments, results were expressed as a function of V-1 overexpression level by dividing the range of 543 nm (mRFP-V-1) channel values into thirds, yielding low (L), medium (M), and high (H) overexpression levels; or fifths, yielding <20, 21–40, 41–60, 61–80, and >80 overexpression levels. To measure the effect of V-1 overexpression on the cortical content of the Arp2/3 complex as a ratio of cortical F-actin content, WT cells overexpressing mRFP-V-1 or mRFP-FBM V-1 were fixed and stained with Alexa Fluor 647–phalloidin and an antibody to D.d. Arp3 (followed by an Alexa 488-conjugated goat anti-rabbit secondary antibody). Actin-rich pseudopodia visible in 1.5-µm-thick ventral confocal sections were identified, and the total fluorescence within 1 µm of the cell edge (with filopodia excluded) was measured in the 647-nm (F-actin), 543-nm (mRFP-V1), and 488-nm (Arp3) channels. To measure the effect of V-1 overexpression on filopodia number, WT cells overexpressing mRFP-V-1 or mRFP-FBM V-1 were fixed and stained with Alexa Fluor 488–phalloidin and optically sectioned in 0.4-µm slices, and the number of filopodia >1 µm in length were scored in projected images as a function of the strength of the mRFP-V-1 signal in the 543-nm channel. To measure the effect of V-1 overexpression on the total cellular content of F-actin, WT cells overexpressing mRFP-V-1 or mRFP-FBM V-1 were fixed and stained with Alexa Fluor 488–phalloidin and optically sectioned in 1-µm slices, and the total fluorescence in all sections summed in the 488-nm (F-actin) and 543-nm (mRFP-V-1) channels. To estimate the fold increase in cellular V-1 level in L, M, and H mRFP-V-1–overexpressing cells, the dimmest 20% (L), middle 60% (M), and brightest 20% (H) of cells were fractionated and collected by using an Aria II Flow Cytometer and subjected to Western blotting using the anti-V-1 antibody, and the untagged and mRFP-tagged V-1 bands were quantitated by densitometry. Imaging of macropinocytic crowns and filopodia by lattice light sheet microcopy (39) was performed on a custom-built instrument at Janelia Research Center, Howard Hughes Medical Institute.
FCS.
Measurements of the diffusion rates of mRFP and mRFP-V-1 in cells by FCS are described in SI Methods.
Cell Biological Assays.
Measurements of fluid-phase macropinocytosis, the preparation of ripple-stage cells, and measurements of rates of random cell motility were performed as described (41). Steaming assays were performed as described (41) and imaged every minute for 12 h on a Zeiss V12 stereo microscope equipped with a 1.0× objective. To estimate the number of nuclei per cell, cells were stained at RT with Hoechst 33342 to reveal nuclei, followed by the plasma membrane dye FM 4-64FX at 4 °C to reveal cell boundaries for cells in clumps. The growth rate of cells in suspension was determined as described (41), except that a Cellometer Auto T4 (Nexcelom Bioscience) cell counter was used. To measure cell adhesiveness, DIC and IRM images were captured on the Zeiss 510 LSM confocal microscope. The content of F-actin in Triton-insoluble cytoskeletons was determined as described (41). To determine the cellular content of F-actin by FACS, 1 × 106 cells suspended in 100 µL of PBS were fixed for 20 min at RT by the addition of paraformaldehyde to a final concentration of 4%, washed with PBS, permeabilized for 10 min using 0.4% Saponin in PBS, washed with PBS, labeled with 4 µM FITC–phalloidin for 1 h, washed with PBS, and resuspended in 1 mL of PBS. The total fluorescence intensity per cell for ∼30,000 individual cells was then determined by flow cytometry using a BD Biosciences model LSR II flow cytometer.
Biochemistry.
FLAG-V-1 and FLAG-FBM V-1 proteins were purified directly from Dictyostelium cells stably expressing them. Amoebas were broken by using Triton X-100 and spun at 10,000 × g for 15 min at 4 °C. The supernatant was then applied to anti-FLAG M2 affinity resin (Sigma-Aldrich, catalog no. A2220), and the FLAG-V-1 protein was eluted by using FLAG-peptide as described by the manufacturer (Sigma-Aldrich, catalog no. F3290). FLAG-V-1 was dialyzed into 1× PBS and concentrated by using a centrifugal filter (Amicon Ultra 4; 10-kDa cutoff). D.d. CP was purified from Escherichia coli strain BL21-RILP harboring the expression plasmid pET-28B containing both the α and β subunits of D.d. CP, essentially as described for mouse CP (22), but by using the CAH3 domain of D.d. CARMIL fused to GST as the affinity matrix. Eluted CP was dialyzed into 1× PBS and concentrated by using a centrifugal filter (Amicon Ultra 4; 10-kDa cutoff). To determine the affinity and stoichiometry of D.d. V-1 for D.d. CP, we performed ITC using 10 µM D.d. CP and 150 µM GST-tagged D.d. V-1 in 1× PBS and a iTC 200 ITC machine (Melvern Instruments). To determine the cellular concentrations of V-1 and CP, known numbers of vegetative AX3 cells and known amounts of purified D.d. FLAG-V-1 and D.d. CP were subjected to Western blotting using the antibodies to D.d. V-1 and D.d. CP and an IRDye 800 goat anti-rabbit IgG as a secondary antibody (Li-COR Biosciences; catalog no. 925-32211). Band intensities were measured by using an Odyssey infrared imager (LI-COR Biosciences). Lyophilized, pyrene-labeled rabbit skeletal muscle actin (Cytoskeleton, catalog no. AP05-A) was reconstituted in G-buffer (2 mM Tris⋅Cl, pH 8.0, 0.2 mM ATP, 0.5 mM DTT, 01 mM CaCl, and 1 mM NaN3) and determined to be ∼90% labeled. Standard, pyrene-based, seed-initiated actin polymerization assays were carried out as described (26). For the pulldown of CP using GST, GST-V-1, or GST-V-1 S22E, 50 µL of glutathione Sepharose resin containing 20 µg of fusion protein was incubated with cell extract (8 × 107 cells in 1.5 mL) for 2 h at 4 °C. After washing, CP was quantitatively eluted by using 5× TBS, the elute was resolved by SDS/PAGE, and the resulting Western blot was probed with an antibody to CP. The CAH3 domain-driven complex exchange reaction shown in Fig. S2B was performed as described by Fujiwara et al. (26). Protein concentrations were determined by Bradford assay (Bio-Rad).
Phosphorylation and Mass Spectroscopy.
The demonstration that D.d. V-1 is phosphorylated in vivo using urea-glycerol gels, and the identification of Ser-22 as the site of phosphorylation by mass spectroscopy, are described in SI Methods.
SI Methods
V-1 Knockout and Stable Cell Lines.
A linear gene-disruption fragment designed to promote a double-crossover gene replacement event was created by fusing nucleotides 1–283 and 346–672 of the D.d. V-1 genomic sequence to the 5′ and 3′ ends of the Blasticidin resistance cassette in plasmid Bsr2 (53), respectively, by using the described approach (54). The introduction of this fragment into WT cells by electroporation and the isolation of Blasticidin-resistant clones by serial dilution in 96-well plates were performed as described (54). V-1–null cells were identified by Western blotting using the D.d. V-1 antibody and confirmed by sequencing PCR products spanning the integration site. Stable cell lines expressing mRFP-V-1 or FLAG-V-1 with or without the FBM were created in the same fashion using the appropriate antibiotic. Note that Dictyostelium cell lines containing stably integrated DNA typically exhibit a range of expression levels despite being clonal (which can be leveraged to create dose–response curves as done here).
FCS.
FCS was performed on the cytoplasm of D.d. amoeba expressing either mRFP or mRFP-V-1 by using a Zeiss 780 ConfoCor3 equipped with 40× 1.2-NA water objective. After a 3-s photobleaching step to reduce the number of fluorescent molecules, 30 autocorrelation measurements of 1 s each were made per cell in the mRFP channel by using a 561-nm laser for excitation. The apparent diffusion constants presented in the text, which correspond to the apparent diffusion quantification provided by the Zeiss FCS default setting, represent the mean and SD for 10 cells in each condition (after omitting any obvious outliers in the 30-run measurements).
Phosphorylation and Mass Spectroscopy.
For Western blotting, WT and V-1–null cell extracts, or FLAG-V-1 precipitated from cell extracts using anti-FLAG M2 beads, were prepared in the presence of phosphatase inhibitor mixture 3 (Sigma-Aldrich, catalog no. P0044), resolved on urea–glycerol–acrylamide gels (45), and probed with the antibody to D.d. V-1. Some WT extracts and some precipitated FLAG-V-1 samples were pretreated with Lambda phosphatase (NEB, catalog no. P0753) before blotting. For mass spectroscopy, 60 µg of FLAG-V-1 was digested by using Glu-C enzyme (Promega, catalog no. V1651). The resulting peptides were enriched for phosphopeptides by using the Fe-NTA phosphopeptide enrichment kit (Thermo Fisher Scientific, catalog no. 88300). Liquid chromatography tandem mass spectrometry was performed by using an Eksigent nanoLC-Ultra 1D Plus system coupled to an LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific) using collision-induced dissociation fragmentation. Peptides were first loaded onto a Zorbax 300SB-C18 trap column (Agilent) and then separated on a reversed-phase Picofrit analytical column (New Objective). The LTQ-Orbitrap Elite was operated in a data-dependent mode (i.e., one MS1 high-resolution scan for precursor ions followed by six data-dependent MS2 scans for precursor ions above a threshold ion count of 500 with collision energy of 35%). The raw files that were generated from the LTQ Orbitrap Elite were analyzed by using Proteome Discoverer software (Version 1.4; Thermo Fisher Scientific). Data were submitted to Mascot (Version 2.5.1; Matrix Sciences) search engine with the following search criteria: phospho (serine and threonine), phospho (tyrosine); MS peptide tolerance 20 ppm; MS/MS tolerance 0.8 Da. The peptides were filtered for a false discovery rate at 1%.
Supplementary Material
Acknowledgments
We thank Dr. J. Philip McCoy and Ms. Leigh Samsel (National Heart, Lung, and Blood Institute Flow Cytometry Core), Dr. Marjan Gucek and Ms. Sajni Patel (National Heart, Lung, and Blood Institute Proteomics Core), and Drs. Nico Tjandra, Jim Gruschus, Paul Kriebel, Carol Parent, and Peter Devreotes for contributions to this study.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605350113/-/DCSupplemental.
References
- 1.Edwards M, et al. Capping protein regulators fine-tune actin assembly dynamics. Nat Rev Mol Cell Biol. 2014;15(10):677–689. doi: 10.1038/nrm3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hug C, et al. Capping protein levels influence actin assembly and cell motility in dictyostelium. Cell. 1995;81(4):591–600. doi: 10.1016/0092-8674(95)90080-2. [DOI] [PubMed] [Google Scholar]
- 3.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112(4):453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 4.Akin O, Mullins RD. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell. 2008;133(5):841–851. doi: 10.1016/j.cell.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401(6753):613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- 6.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95(11):6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mejillano MR, et al. Lamellipodial versus filopodial mode of the actin nanomachinery: Pivotal role of the filament barbed end. Cell. 2004;118(3):363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 8.Iwasa JH, Mullins RD. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr Biol. 2007;17(5):395–406. doi: 10.1016/j.cub.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bear JE, Gertler FB. Ena/VASP: Towards resolving a pointed controversy at the barbed end. J Cell Sci. 2009;122(Pt 12):1947–1953. doi: 10.1242/jcs.038125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breitsprecher D, et al. Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J. 2008;27(22):2943–2954. doi: 10.1038/emboj.2008.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kovar DR. Molecular details of formin-mediated actin assembly. Curr Opin Cell Biol. 2006;18(1):11–17. doi: 10.1016/j.ceb.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci USA. 2004;101(41):14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bombardier JP, et al. Single-molecule visualization of a formin-capping protein ‘decision complex’ at the actin filament barbed end. Nat Commun. 2015;6:8707. doi: 10.1038/ncomms9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shekhar S, et al. Formin and capping protein together embrace the actin filament in a ménage à trois. Nat Commun. 2015;6:8730. doi: 10.1038/ncomms9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burke TA, et al. Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. Curr Biol. 2014;24(5):579–585. doi: 10.1016/j.cub.2014.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lomakin AJ, et al. Competition for actin between two distinct F-actin networks defines a bistable switch for cell polarization. Nat Cell Biol. 2015;17(11):1435–1445. doi: 10.1038/ncb3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rotty JD, et al. Profilin-1 serves as a gatekeeper for actin assembly by Arp2/3-dependent and -independent pathways. Dev Cell. 2015;32(1):54–67. doi: 10.1016/j.devcel.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyoshi T, et al. Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: Evidence of frequent filament severing. J Cell Biol. 2006;175(6):947–955. doi: 10.1083/jcb.200604176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung G, Remmert K, Wu X, Volosky JM, Hammer JA., 3rd The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J Cell Biol. 2001;153(7):1479–1497. doi: 10.1083/jcb.153.7.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uruno T, Remmert K, Hammer JA., 3rd CARMIL is a potent capping protein antagonist: Identification of a conserved CARMIL domain that inhibits the activity of capping protein and uncaps capped actin filaments. J Biol Chem. 2006;281(15):10635–10650. doi: 10.1074/jbc.M513186200. [DOI] [PubMed] [Google Scholar]
- 21.Yang C, et al. Mammalian CARMIL inhibits actin filament capping by capping protein. Dev Cell. 2005;9(2):209–221. doi: 10.1016/j.devcel.2005.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujiwara I, Remmert K, Hammer JA., 3rd Direct observation of the uncapping of capping protein-capped actin filaments by CARMIL homology domain 3. J Biol Chem. 2010;285(4):2707–2720. doi: 10.1074/jbc.M109.031203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeda S, et al. Two distinct mechanisms for actin capping protein regulation--steric and allosteric inhibition. PLoS Biol. 2010;8(7):e1000416. doi: 10.1371/journal.pbio.1000416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zwolak A, Uruno T, Piszczek G, Hammer JA, 3rd, Tjandra N. Molecular basis for barbed end uncapping by CARMIL homology domain 3 of mouse CARMIL-1. J Biol Chem. 2010;285(37):29014–29026. doi: 10.1074/jbc.M110.134221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim T, Ravilious GE, Sept D, Cooper JA. Mechanism for CARMIL protein inhibition of heterodimeric actin-capping protein. J Biol Chem. 2012;287(19):15251–15262. doi: 10.1074/jbc.M112.345447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujiwara I, Remmert K, Piszczek G, Hammer JA. Capping protein regulatory cycle driven by CARMIL and V-1 may promote actin network assembly at protruding edges. Proc Natl Acad Sci USA. 2014;111(19):E1970–E1979. doi: 10.1073/pnas.1313738111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhattacharya N, Ghosh S, Sept D, Cooper JA. Binding of myotrophin/V-1 to actin-capping protein: Implications for how capping protein binds to the filament barbed end. J Biol Chem. 2006;281(41):31021–31030. doi: 10.1074/jbc.M606278200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zwolak A, Fujiwara I, Hammer JA, 3rd, Tjandra N. Structural basis for capping protein sequestration by myotrophin (V-1) J Biol Chem. 2010;285(33):25767–25781. doi: 10.1074/jbc.M110.135848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards M, McConnell P, Schafer DA, Cooper JA. CPI motif interaction is necessary for capping protein function in cells. Nat Commun. 2015;6:8415. doi: 10.1038/ncomms9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das B, et al. Influence of p53 in the transition of myotrophin-induced cardiac hypertrophy to heart failure. Cardiovasc Res. 2010;87(3):524–534. doi: 10.1093/cvr/cvq068. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Gupta S, Purcell NH, Lin A, Sen S. Activation of nuclear factor-kappaB is necessary for myotrophin-induced cardiac hypertrophy. J Cell Biol. 2002;159(6):1019–1028. doi: 10.1083/jcb.200207149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarkar S, et al. Cardiac overexpression of myotrophin triggers myocardial hypertrophy and heart failure in transgenic mice. J Biol Chem. 2004;279(19):20422–20434. doi: 10.1074/jbc.M308488200. [DOI] [PubMed] [Google Scholar]
- 33.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caldwell JE, Heiss SG, Mermall V, Cooper JA. Effects of CapZ, an actin capping protein of muscle, on the polymerization of actin. Biochemistry. 1989;28(21):8506–8514. doi: 10.1021/bi00447a036. [DOI] [PubMed] [Google Scholar]
- 35.Wanger M, Wegner A. Equilibrium constant for binding of an actin filament capping protein to the barbed end of actin filaments. Biochemistry. 1985;24(4):1035–1040. doi: 10.1021/bi00325a035. [DOI] [PubMed] [Google Scholar]
- 36.Yamashita A, Maeda K, Maéda Y. Crystal structure of CapZ: Structural basis for actin filament barbed end capping. EMBO J. 2003;22(7):1529–1538. doi: 10.1093/emboj/cdg167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taoka M, et al. V-1, a protein expressed transiently during murine cerebellar development, regulates actin polymerization via interaction with capping protein. J Biol Chem. 2003;278(8):5864–5870. doi: 10.1074/jbc.M211509200. [DOI] [PubMed] [Google Scholar]
- 38.Prassler J, et al. DdLIM is a cytoskeleton-associated protein involved in the protrusion of lamellipodia in Dictyostelium. Mol Biol Cell. 1998;9(3):545–559. doi: 10.1091/mbc.9.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen BC, et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346(6208):1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hacker U, Albrecht R, Maniak M. Fluid-phase uptake by macropinocytosis in Dictyostelium. J Cell Sci. 1997;110(Pt 2):105–112. doi: 10.1242/jcs.110.2.105. [DOI] [PubMed] [Google Scholar]
- 41.Jung G, Wu X, Hammer JA., 3rd Dictyostelium mutants lacking multiple classic myosin I isoforms reveal combinations of shared and distinct functions. J Cell Biol. 1996;133(2):305–323. doi: 10.1083/jcb.133.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kessin RH. 1999. The Evolution of the Cellular Slime Molds, Frontiers Science Series (Springer, Berlin), pp 3–14.
- 43.Hoeller O, Kay RR. Chemotaxis in the absence of PIP3 gradients. Curr Biol. 2007;17(9):813–817. doi: 10.1016/j.cub.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 44.Varnum B, Edwards KB, Soll DR. The developmental regulation of single-cell motility in Dictyostelium discoideum. Dev Biol. 1986;113(1):218–227. doi: 10.1016/0012-1606(86)90124-7. [DOI] [PubMed] [Google Scholar]
- 45.Persechini A, Kamm KE, Stull JT. Different phosphorylated forms of myosin in contracting tracheal smooth muscle. J Biol Chem. 1986;261(14):6293–6299. [PubMed] [Google Scholar]
- 46.Barrick D. Biological regulation via ankyrin repeat folding. ACS Chem Biol. 2009;4(1):19–22. doi: 10.1021/cb900003f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Löw C, Homeyer N, Weininger U, Sticht H, Balbach J. Conformational switch upon phosphorylation: Human CDK inhibitor p19INK4d between the native and partially folded state. ACS Chem Biol. 2009;4(1):53–63. doi: 10.1021/cb800219m. [DOI] [PubMed] [Google Scholar]
- 48.Hansen SD, Mullins RD. VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J Cell Biol. 2010;191(3):571–584. doi: 10.1083/jcb.201003014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Higgs HN. Formin proteins: A domain-based approach. Trends Biochem Sci. 2005;30(6):342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 50.Kitazawa M, et al. Intracellular cAMP controls a physical association of V-1 with CapZ in cultured mammalian endocrine cells. Biochem Biophys Res Commun. 2005;331(1):181–186. doi: 10.1016/j.bbrc.2005.03.127. [DOI] [PubMed] [Google Scholar]
- 51.Fischer M, Haase I, Simmeth E, Gerisch G, Müller-Taubenberger A. A brilliant monomeric red fluorescent protein to visualize cytoskeleton dynamics in Dictyostelium. FEBS Lett. 2004;577(1-2):227–232. doi: 10.1016/j.febslet.2004.09.084. [DOI] [PubMed] [Google Scholar]
- 52.Dynes JL, et al. LagC is required for cell-cell interactions that are essential for cell-type differentiation in Dictyostelium. Genes Dev. 1994;8(8):948–958. doi: 10.1101/gad.8.8.948. [DOI] [PubMed] [Google Scholar]
- 53.Sutoh K. A transformation vector for Dictyostelium discoideum with a new selectable marker bsr. Plasmid. 1993;30(2):150–154. doi: 10.1006/plas.1993.1042. [DOI] [PubMed] [Google Scholar]
- 54.Jung G, Titus MA, Hammer JA., 3rd The Dictyostelium type V myosin MyoJ is responsible for the cortical association and motility of contractile vacuole membranes. J Cell Biol. 2009;186(4):555–570. doi: 10.1083/jcb.200810147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Piiper A, et al. Protein kinase A mediates cAMP-induced tyrosine phosphorylation of the epidermal growth factor receptor. Biochem Biophys Res Commun. 2003;301(4):848–854. doi: 10.1016/s0006-291x(03)00055-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.