Significance
Some cancers, termed carcinosarcomas (CSs), have mixed cell types, with either epithelial or mesenchymal features. Sequencing the genomes of uterine and ovarian CSs demonstrated that these different cell types derive from a common precursor cell that has many mutations typical of epithelial cancers. In addition, we find that these tumors have a significant burden of point mutations and amplification of histone genes, suggesting a potential role of these mutations in sarcomatous transformation. Consistent with this finding, expression of specific histone gene mutations in uterine carcinoma cells changed gene expression toward a mesenchymal state. These findings have potential implications for the treatment of these cancers.
Keywords: uterine carcinosarcoma, ovarian carcinosarcoma, exome sequencing
Abstract
Carcinosarcomas (CSs) of the uterus and ovary are highly aggressive neoplasms containing both carcinomatous and sarcomatous elements. We analyzed the mutational landscape of 68 uterine and ovarian CSs by whole-exome sequencing. We also performed multiregion whole-exome sequencing comprising two carcinoma and sarcoma samples from six tumors to resolve their evolutionary histories. The results demonstrated that carcinomatous and sarcomatous elements derive from a common precursor having mutations typical of carcinomas. In addition to mutations in cancer genes previously identified in uterine and ovarian carcinomas such as TP53, PIK3CA, PPP2R1A, KRAS, PTEN, CHD4, and BCOR, we found an excess of mutations in genes encoding histone H2A and H2B, as well as significant amplification of the segment of chromosome 6p harboring the histone gene cluster containing these genes. We also found frequent deletions of the genes TP53 and MBD3 (a member with CHD4 of the nucleosome remodeling deacetylase complex) and frequent amplification of chromosome segments containing the genes PIK3CA, TERT, and MYC. Stable transgenic expression of H2A and H2B in a uterine serous carcinoma cell line demonstrated that mutant, but not wild-type, histones increased expression of markers of epithelial–mesenchymal transition (EMT) as well as tumor migratory and invasive properties, suggesting a role in sarcomatous transformation. Comparison of the phylogenetic relationships of carcinomatous and sarcomatous elements of the same tumors demonstrated separate lineages leading to these two components. These findings define the genetic landscape of CSs and suggest therapeutic targets for these highly aggressive neoplasms.
Carcinosarcomas (CSs) of the female genital tract, also known as mixed malignant Müllerian tumors, are rare but highly aggressive tumors characterized by a biphasic histology. These cancers most commonly arise in the uterus, followed by the ovaries, fallopian tubes, and vagina (1–3). The diagnosis of CS requires the presence of both sarcomatous and carcinomatous components. Although the pathogenesis of CSs remains under debate, an increasing body of evidence supports the origin of both elements from a common epithelial cell that undergoes sarcomatous dedifferentiation, rather than two independent progenitors (2–5).
The overall 5-y survival is only 30 ± 9% for all stages, and the recurrence rate after surgery is extremely high (50–80%) (3–5). The uncertain origin and poor prognosis of uterine and ovarian CSs motivate determination of the molecular basis of CS aggressive behavior in the hope of developing novel and effective treatment modalities.
Results
The Genetic Landscape of CS.
A total of 68 patients with stage I–IV uterine (n = 44) and ovarian (n = 24) CSs were studied. Their clinical and histological features are presented in SI Appendix, Table S1. Upon surgical removal of tumors, primary cell lines were prepared (five tumors) or tumors were frozen (63 tumors). Among these tumors, 41 had matched normal tissues available and tumor-normal pairs were subjected to whole-exome sequencing, with 197 mean reads per targeted base for tumor samples and 101 for normal tissues; 94.5% of the targeted bases had at least 20 independent sequence reads (SI Appendix, Table S2). Tumor purity was estimated from the minor allele frequency of somatic mutations and segments showing loss of heterozygosity (LOH) or copy number variation (CNV); the mean tumor purity was 81%. Somatic point mutations were identified using MuTect (6) for normal-tumor pairs and were further filtered to reduce false-positive calls (Materials and Methods). Somatic indels were called by in-house Perl scripts (Materials and Methods). For unmatched tumors, we first called variants from the reference sequence and then excluded variants that have a frequency higher than 2 × 10−5 in the Exome Aggregation Consortium (ExAC) database (7). Driver mutations were confirmed by manual inspection of the reads plot, and selected mutations were confirmed by Sanger sequencing.
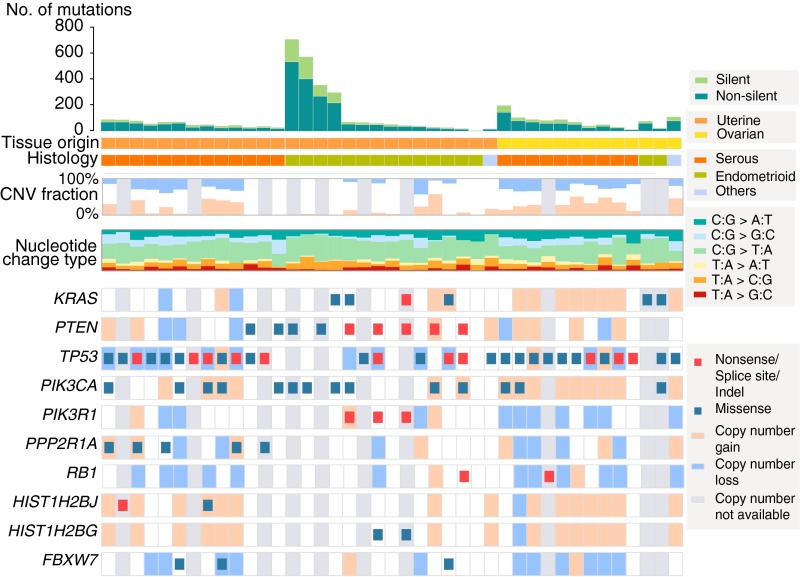
In total, 4,115 somatic point mutations (3,056 protein-altering mutations) and 49 indels from the 41 matched normal-tumor pairs were called. Based on tumor origin and histology of the carcinomatous elements, we grouped tumors into three categories: uterine serous, uterine endometrioid, and ovarian serous. Four samples from the uterine endometrioid group had markedly higher somatic mutation rates (mean = 16.8 per megabase pair) compared with the remainder (mean = 2.1 per megabase pair) (Fig. 1). Mutation spectrum analysis revealed a paucity of transversion mutations in the high-mutation group, a signature often observed in mismatch repair-deficient tumors. Mutation rates, mutation spectra, and the fraction of genome affected by CNV among the remaining 37 tumors were similar to those numbers previously reported for uterine and ovarian carcinomas (8–10).
Fig. 1.
Mutational landscape across 41 CS samples. Mutation spectra for each sample are shown. Samples are grouped based on origin and histology of the carcinomatous component. The CNV fraction is the fraction of the genome that is affected by copy number gain or loss. (Lower) Mutations found in genes with a burden q value less than 0.2 are shown.
Recurrent somatic mutations were identified in the 37 matched tumors and confirmed by Sanger sequencing; additional instances of the identical mutations were sought in the 27 unmatched tumors and were included in subsequent analysis. The results identified eight genes with recurrent somatic missense mutations (Table 1). These mutations included known activating mutations in the oncogenes PIK3CA (PI3-kinase subunit alpha) and KRAS (Kirsten rat sarcoma viral oncogene homolog), a recurrent mutation in PPP2R1A (phosphatase 2a regulatory subunit) that causes loss of normal regulation of serine-threonine phosphatase 2a (11), a known recurrent mutation (N1459S) in BCOR (BCL6 corepressor) that is likely gain of function (8), and a known recurrent mutation in CHD4 (DNA helicase) (9). There were also known recurrent mutations in the tumor suppressors TP53 (tumor protein P53) and PTEN (phosphatase and tensin homolog). All of these genes have been previously implicated in uterine and/or ovarian carcinomas.
Table 1.
Recurrent mutations identified in 64 CSs
| Gene name | No. recurrent | Status | Recurrent sites | Previously seen in gyn. carcinomas |
| TP53 | 19 | Missense, nonsense, and indel | I63F,R342P,R210R,S109Y,R141H,V84M/indel,R150W,G113D,R196stop | Yes |
| KRAS | 9 | Missense | G12D,G12V,G12C | Yes |
| PIK3CA | 8 | Missense | H1047R, E542K,R93W | Yes |
| PTEN | 5 | Missense and nonsense | R130G, R130stop | Yes |
| CHD4 | 3 | Missense | R1162Q | Yes |
| PPP2R1A | 3 | Missense | P179R | Yes |
| BCOR | 2 | Missense | N1459S | Yes |
| HIST1H2AB | 2 | Missense | E57Q | No |
The “Recurrent sites” column lists the original amino acid, amino acid change position, and mutated amino acid for all recurrent mutations identified in the gene. The “Previously seen in gyn. carcinomas” column indicates whether the mutated positions have ever been reported in gynecological (gyn.) carcinomas before. Data from COSMIC database (24).
In addition, we identified two tumors with a previously unidentified somatic mutation in HIST1H2AB [G > C at chromosome 6 (chr6): 26033628; amino acid change, E57Q]. HIST1H2AB encodes the histone H2A protein type 1-B/E. This gene has not previously been implicated in cancer, and this variant was absent from public and Yale University databases. The probability of having any recurrent G:C > C:G mutation in this dataset by chance, given the size of the exome, the number of tumors, and the observed substitution-specific mutation rate (0.5 per megabase pair), was low (P < 0.01).
To identify genes with an increased burden of somatic mutation, we used MutSigCV (12) to analyze all somatic damaging mutations (premature termination, splice site, and indel mutations) as well as missense mutations at phylogenetically conserved sites in the 37 matched tumors (Materials and Methods). With a genome-wide false discovery rate of 0.1, we identified eight significantly mutated genes (Fig. 1 and Table 2). In addition to genes identified above as having recurrent mutations, we found a significant burden of mutations in RB1 (a tumor suppressor gene significantly altered in high-grade ovarian serous carcinomas), PIK3R1 (inhibitor of the PI3K pathway), and HIST1H2BJ (core histone 2B type J). Close to the significance threshold, we found another core histone gene HIST1H2BG (core histone 2B type J; q value = 0.19) and FBXW7 [a known tumor suppressor gene previously identified in uterine serous carcinoma (USC); q value = 0.19].
Table 2.
Gene burden analysis results for 37 matched tumor-normal pairs by MutSigCV
| Gene | Cancer gene annotation | Coding size | No. nonsilent | No. silent | No. conserved missense | No. damaging | q |
| KRAS | Oncogene | 574 | 5 | 0 | 4 | 1 | 0 |
| PTEN | TSG | 1,221 | 10 | 0 | 4 | 6 | 0 |
| TP53 | TSG | 1,192 | 31 | 0 | 12 | 11 | 0 |
| PIK3CA | Oncogene | 3,227 | 11 | 0 | 11 | 0 | 0 |
| PIK3R1 | TSG | 2,190 | 4 | 0 | 0 | 4 | 0 |
| PPP2R1A | Oncogene | 1,785 | 5 | 0 | 5 | 0 | 7.81 × 10−3 |
| RB1 | TSG | 2,814 | 2 | 0 | 0 | 2 | 7.04 × 10−2 |
| HIST1H2BJ | Unknown | 382 | 2 | 0 | 2 | 0 | 9.38 × 10−2 |
| HIST1H2BG | Unknown | 382 | 2 | 0 | 1 | 1 | 1.89 × 10−1 |
| FBXW7 | TSG | 2,135 | 3 | 0 | 3 | 0 | 1.89 × 10−1 |
The “Cancer gene annotation” column indicates the role of the gene in carcinogenesis, either as an oncogene or tumor suppressor gene (TSG) (26) or unknown. The “q” column shows the q values returned by MutSigCV.
The four core histone proteins, H2A, H2B, H3, and H4, are small (98–136 aa in length), extremely highly conserved proteins that together form the nucleosome, which packages DNA in chromatin. Covalent modifications of these proteins play critical roles in regulating gene expression (13). Genes encoding these proteins are present in multiple copies in the genome in two clusters (chr1p and chr6p), permitting high expression in S-phase to allow assembly with newly replicated DNA. Further evaluation of histone genes demonstrated a total of 10 protein-altering mutations and one synonymous mutation (Table 3). Ten were confirmed as somatic mutations by sequencing normal DNA, whereas one was present in a tumor without an available matched normal sample. All of these mutations are absent in germline DNA databases. Although there are collectively 37 replication-dependent H2A and H2B genes at chr1p and chr6p, all 10 of the observed protein-altering mutations clustered among five of these genes, each with two mutations; analysis of individual sequence reads provided no evidence that mutations were mismapped (SI Appendix, Fig. S1). Lastly, these specific histone genes were expressed in CSs (SI Appendix, Fig. S2), and all observed mutations were mutually exclusive. Considering all 37 copies of core H2A and H2B genes as a single metagene yielded a P value for burden of mutation of 8 × 10−7. Moreover, the striking clustering of the 10 mutations among only five of these genes was highly unlikely to occur by chance (P < 6 × 10−4). In contrast to CSs, no excess burden of somatic mutation has been seen in these genes in USC analyzed at Yale University or by The Cancer Genome Atlas (TCGA), or in ovarian carcinoma, and no significant clustering of mutations in particular genes has been observed (SI Appendix, Table S3).
Table 3.
Somatic mutations in histone H2A or H2B in CS
| Gene | Mutations found in matched (n = 37) | Mutations found in unmatched (n = 27) | ||
| Status: no. | AA change | Status: no. | AA change | |
| HIST1H2AB | Missense: 1 | E57Q | Missense: 1 | E57Q* |
| HIST1H2AC | Missense: 1 | R4H | Missense: 1 | T102A* |
| HIST1H2BB | Missense: 1; silent: 1 | E36G, V49V | Missense: 1 | G27A |
| HIST1H2BG | Missense: 2 | A18G, M63K | ||
| HIST1H2BJ | Missense: 1; nonsense: 1 | T53S, K24stop | ||
Variants found in H2A/H2B genes with at least one nonsilent mutation in the matched cohort, excluding the four outliers, were added from the unmatched cohort. HIST1H2AB has another missense mutation (K16T) from one of the four outlier samples (353 mutations in total in that sample). AA, amino acid.
These two mutations were confirmed as somatic mutations by sequencing their matched normal tissues at a later stage.
We compared the mutation spectrum in CSs with carcinomas of the same tissue origin and histology; CSs included 26 uterine serous, 17 uterine endometrioid, and 19 ovarian serous tumors (SI Appendix, Fig. S3A). Comparator carcinomas were 92 USCs [46 from our previous study (9) and 46 from the TCGA database (8)], 104 uterine endometrioid carcinomas (8), and 316 ovarian serous carcinomas (10). We found similarities in the frequency of all driver mutations in CSs and these epithelial tumors. Mutations in PTEN, KRAS, ARID1A, and PIK3CA were predominant in uterine CSs with an endometrioid epithelial component, whereas TP53, PIK3CA, CHD4, PPP2R1A, and FBXW7 mutations were more frequently found in CSs with serous epithelial components. TP53 was mutated in most ovarian CSs, whereas the remaining genes were mutated only in a small fraction of tumors.
In contrast, mutations in histone H2A/H2B genes were significantly enriched in CSs compared with carcinomas (mutations in 21.2% of CSs and 5.2% of uterine and ovarian epithelial tumors; Fisher P = 0.002; SI Appendix, Fig. S3B). These findings implicate mutations in histone H2A/H2B genes in CSs, with further support from CNV (discussed below).
Analysis of CNV.
We identified CNV based on exome sequencing data for 29 CSs that passed CNV calling quality control and 25 USCs from a study by Zhao et al. (9) (Materials and Methods). We then used GISTIC2.0 (14) to identify chromosome segments showing significantly more CNV than expected by chance (Materials and Methods and SI Appendix, Fig. S4). Significantly amplified focal segments in CSs included chr3q27.1, which contains PIK3CA. Focal segments containing the known tumor suppressor genes TP53, PIK3R1, RB1, BRCA2, and MBD3 (the binding partner of CHD4) were significantly deleted (SI Appendix, Figs. S4 and S5). All of these CNVs are also frequently found in uterine and ovarian carcinomas. In addition to segments shared with uterine and ovarian carcinomas, we found significant broad CNV that was specifically enriched in CSs. This CNV included amplification of the segment of chr5p that includes TERT (telomerase reverse transcriptase) (15), which was amplified in 50% of CSs but only 17% of carcinomas (SI Appendix, Fig. S6 and Table S4). This amplification has been implicated in other cancers (16, 17). Similarly, chr6p was significantly amplified in CSs (SI Appendix, Fig. S6 and Table S4; eight of 19 uterine CSs and seven of 10 ovarian CSs). Interestingly, this segment contains the replication-dependent histone gene cluster (HIST1H) and includes all of the histone genes that are somatically mutated, as described above. We also noticed that 1q22, which was identified as a significantly amplified focal segment (found in 62% of CSs, SI Appendix, Figs. S5 and S6) contains the HIST2H gene cluster, the second largest cluster of core histone genes.
Impact of Histone H2A/H2B Mutations.
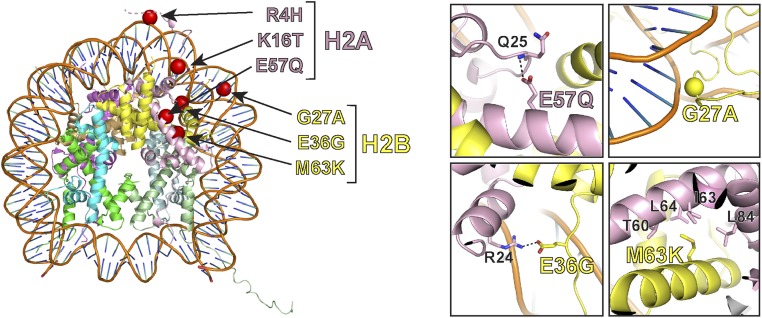
We mapped mutations in H2A and H2B onto the crystal structure of the core nucleosome particle [Protein Data Bank (PDB) ID code 1AOI] (18) (Fig. 2). Two mutations, H2A R4H and H2A K16T, occurred at sites of previously identified posttranslational modification. R4 in the N-terminal tail of H2A can be deaminated to citrulline (19), and is implicated in linear compaction of chromatin (20). K16 has been identified as a site of DNA damage-dependent ubiquitination and, when mutated, leads to decreased ubiquitination of H2A (21, 22). H2A residue E57, mutated to glutamine in two samples, is potentially involved in maintaining the structural integrity of the H2A fold. The side chain of E57 hydrogen bonds with the backbone nitrogen of Q25; the glutamine mutation is likely to disrupt this interaction.
Fig. 2.
Structural analysis of histone mutations. (Left) Identified mutations in H2A and H2B are mapped onto the crystal structures of the core nucleosome particle (PDB ID codes 1AOI and 1XK5). (Right) Zoomed-in view of four representative mutations predicted to have large impacts on normal histone functions.
Similarly, H2B G27 packs closely against the DNA double helix (23). The observed alanine substitution has the potential to disrupt this packing. Two other mutations in H2B, E36G and M63K, are located at the H2B–H2A heterodimerization interface. E36 makes a side chain-mediated salt bridge with the side chain of R24 in H2A, which would be lost with the glycine substitution. M63 has extensive hydrophobic interaction with a patch on H2A, which includes T60, I63, L64, and L84; this interaction would also be lost with the lysine substitution. These mutations are thus both likely to alter H2A–H2B interaction.
Somatic mutations in these H2A and H2B genes have not previously been implicated in cancer; however, some of these somatic mutations have been seen previously as sporadic somatic mutations in these or paralogous histone genes in diverse cancer types (24).
The association of these histone gene mutations with CS prompted exploration of the possibility that somatic histone H2A and H2B mutations play a role in epithelial–mesenchymal transition (EMT), contributing to evolution of carcinoma to sarcoma. We stably transfected a USC cell line (i.e., USC-ARK2) to express either the wild-type H2As (HIST1H2AB or HIST1H2AC) or the same genes with observed somatic mutations in CSs (four mutations were chosen) (Materials and Methods). All transgenes were tagged with Myc and DDK epitopes at the C terminus and were expressed under control of the CMV promoter. All six cell lines showed high expression of transgenes. These transgenic H2As were all incorporated in chromatin, as shown by imaging mitotic cells stained for anti-DDK antibodies (SI Appendix, Fig. S7).
EMT features the loss of epithelial markers, usually cell–cell junction molecules, permitting cell migration and invasion. We quantitated gene expression of several well-known epithelial markers (E-cadherin, claudin-3, and claudin-4) in each of the stable cell lines by quantitative RT-PCR (qRT-PCR). Compared with cell lines expressing transgenic wild-type histones, cell lines with mutant HIST1H2AC histones demonstrated significant reduction of all three epithelial markers (SI Appendix, Fig. S8). Increases in the expression of N-cadherin and transcription factor SNAI2/Slug are other common features seen in EMT and many cancers. The qRT-PCR results showed a two- to threefold increase of N-cadherin and SNAI2/Slug levels in cells expressing mutant HIST1H2AC or HIST1H2AB proteins compared with wild type. Finally, using transwell cell migration and invasion assays (SI Appendix, Fig. S9), we found mutated histone-core gene transfected USC cells, but not cell lines expressing transgenic wild-type histones, to acquire a significantly more migratory and more invasive phenotype. Taken together, these results suggest that histone mutations identified in our CS cohort may drive at least some of the molecular changes characteristics of EMT and the phenotypic divergence of CSs from their corresponding carcinomas.
Evolution of CSs Defined by Multiregion Sequencing.
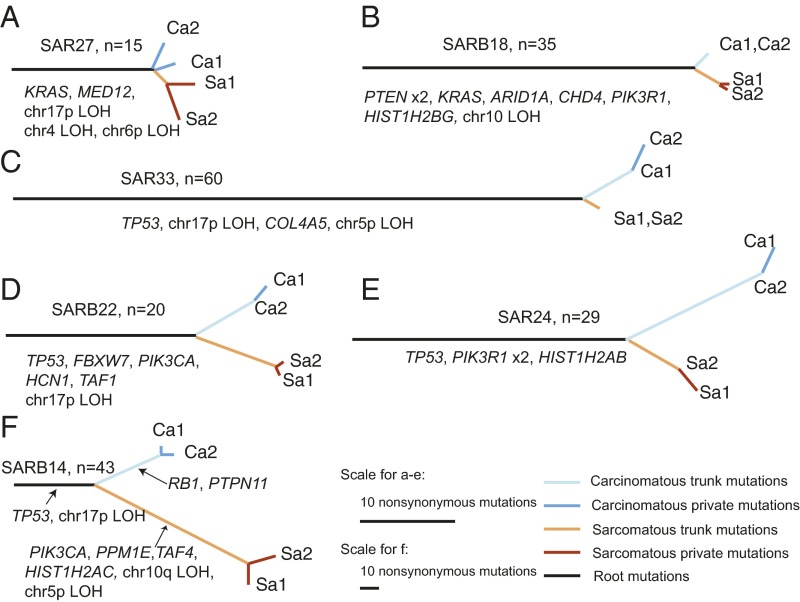
To evaluate the evolutionary histories of CSs, we dissected carcinomatous and sarcomatous elements from six CSs; we took two separate carcinomatous and two sarcomatous samples from each tumor and sequenced to a mean coverage depth of 261-fold (SI Appendix, Fig. S10 and Table S5). Each sample was taken from a site remote from other sites sampled in the same tumor.
A total of 473 different protein-altering somatic single-nucleotide mutations and indels were identified among these samples (Materials and Methods). A total of 202 of these mutations and indels were shared by all samples collected from the same tumors (root mutations). The number of root mutations varied from 15 to 59 per tumor, unequivocally establishing that the carcinomatous and sarcomatous elements arise from a common precursor. In addition, we found two to 53 nonsilent somatic mutations unique to carcinomatous elements and two to 109 mutations unique to sarcomatous elements. We next constructed phylogenetic trees for samples from each tumor (Fig. 3) by hierarchical clustering of allele frequencies for somatic mutations (Materials and Methods and SI Appendix, Fig. S11). For all six tumors, the multiple sarcomatous samples clustered with one another and separately from the multiple carcinomatous samples (which also clustered with one another). Of the 172 nonsilent mutations that were found in two of the four samples, 102 were shared by the two sarcomatous samples but not found in either of the carcinomatous components, 66 were found in the two carcinomatous components but not in either of the two sarcomatous components, and four were found present in both components. This distribution is not expected under the null hypothesis that the lineage of each sample is independent (P < 10−4). The divergence between carcinomatous and sarcomatous elements happened at different relative time points during the evolution of CSs. Three tumors (Fig. 3 A–C) demonstrated relatively late divergences, whereas the other three (Fig. 3 D–F) had earlier divergences.
Fig. 3.
Evolution paths of six CSs. (A–F) Each panel shows the phylogenetic tree for one CS tumor that has undergone multiregion sequencing. The tumor name and number of root mutations are indicated at the top of the phylogenetic tree. Ca1 and Ca2 are samples dissected from carcinoma areas, Sa1 and Sa2 are samples dissected from sarcoma areas, root mutations are mutations shared by all samples from the same tumor, sarcomatous (carcinomatous) trunk mutations are mutations shared by all sarcomatous (carcinomatous) samples but never in the carcinomatous (sarcomatous) samples, and sarcomatous (carcinomatous) are private mutations [mutations found in sarcomatous (carcinomatous) samples besides trunk mutations]. Branch length is proportional to the number of nonsynonymous mutations. Driver genes are significantly mutated histone genes, and LOH events are highlighted.
We next mapped mutations in cancer drivers on the phylogenetic trees. We defined potential driving mutations as nonsynonymous mutations in known ovarian and uterine cancer genes (8–10, 25, 26). CNV information was not available for most samples, because these samples were obtained from paraffin-embedded samples; however, we were able to identify LOH events affecting known driver genes in uterine and ovarian cancers (e.g., chr17p LOH containing TP53, chr4q LOH containing FBXW7, chr10q LOH containing PTEN, chr5p LOH, chr6p LOH). Among 34 such driving genetic events (point mutation and LOH), 29 were root mutations, whereas five were sarcomatous trunk-specific mutations (Fig. 3). Nonetheless, these mutations all occurred in the same tumor, indicating such events may not occur in most tumors. Four of the six tumors had TP53 root mutations and were frequently accompanied by LOH events, indicating the role of TP53 as an early driver. Among the rest of root driver mutations, we found one PIK3CA activating mutation (E542K), three PIK3R1 mutations (two indels and one missense mutation at a conserved site), and one CHD4 indel. These three genes are frequently mutated in uterine and ovarian carcinomas (8–10, 27) but have rarely been seen in sarcomas (28) (SI Appendix, Table S6). This finding suggests that CS likely begins as carcinoma, followed by sarcomatous transformation. Two histone H2A/H2B genes were mapped as root mutations, and another H2A gene was found having a sarcomatous component-specific mutation. The fact that histone mutations were acquired at different stages during tumor progression suggests that these mutations may not be sufficient for sarcomatous transformation of an epithelial tumor in CS. Other genetic or nongenetic factors may be required for this transformation.
Discussion
Our results demonstrate that CSs, as a whole, have mutation profiles that are similar to the mutation profiles found in epithelial cancers of the uterus and ovary. In addition, however, we found a significantly increased frequency of somatic mutations in histone genes and amplifications of the histone gene locus on chr6p, as well as significant amplification of TERT 5p in CSs, suggesting that these mutations play a role in development of the sarcomatous elements of these tumors. Separate sequencing of multiple isolates of carcinomatous and sarcomatous elements of CSs demonstrated that these separate elements share in common many somatic mutations, establishing unequivocally the common genetic origin of these tumors. The finding that these root mutations most often include mutations well-established to play a role in carcinomas of the uterus and ovary is consistent with an epithelial origin of these cancers. Because we have low sensitivity for detection of structural rearrangements, we cannot exclude the possibility that such events might also contribute to development of the sarcomatous elements of these tumors.
Previous studies suggested that the carcinomatous component represents the primary “driving force” of CSs and determines prognosis (29). The finding of frequent mutations characteristic of uterine and ovarian carcinomas, but not sarcomas, as well as carcinomatous mutations common to the root of all CSs, is consistent with this notion. These results also raise the possibility that treatments targeting genes and pathways within the carcinomatous elements, such as PIK3CA, CCNE1, and MYC (v-myc avian myelocytomatosis viral oncogene homolog), may prove efficacious in treating both carcinomatous and sarcomatous elements in these cancers.
Prior studies have found higher expression levels of specific mesenchymal markers and lower levels of epithelial markers in the sarcoma areas of CS compared with the carcinoma areas (30). Our results suggest that specific histone gene mutations contribute to this effect. Histone gene mutations have previously been implicated in several cancers. In follicular lymphoma, mutations in histone H1 genes have been found to affect binding affinities and residence times of affected H1 proteins (31). It was speculated that such mutations would lead to changes in chromatin conformation and altered gene regulation. In pediatric gliomas, recurrent mutations in histone H3.3 at lysine 27 inhibit PRC2 activity and drive an oncogenic expression pattern (32). Recently, a study pointed out that mutation of H2A R4 in our CS cohort, consistent with expectation, may lead to reduced chromatin compaction in cell culture experiments (20). These findings lend credence to the likely functional effects of histone gene mutations and amplifications identified in our cohort.
In conclusion, our results provide insight into the origins and development of CS and define the genetic landscape of this type of malignant tumor. These findings provide a useful starting point for further work to define the causes and best therapeutic approaches to this cancer.
Materials and Methods
Patients and Specimens.
The collection of the specimens and the study protocol were approved by the University of Arkansas for Medical Sciences, the University of Brescia, and the Yale University Human Investigation Committees. DNA was extracted from 68 tumors collected from newly diagnosed patients, and informed consent was obtained from all participants. The 1988 International Federation of Gynecology and Obstetrics staging system was used, and histology was further evaluated by board-certified pathologists to confirm the diagnosis (SI Appendix).
Whole-Exome Sequencing.
DNA was extracted from frozen samples by standard methods. For formalin-fixed, paraffin-embedded (FFPE) samples, DNA was extracted using a BiOstic FFPE Tissue DNA Isolation Kit (catalog no. 12250-50; MO BIO Laboratories) with a modified protocol. Genomic DNA was captured on the NimbleGen 2.1M human exome array and subjected to 74-bp end reads on an Illumina HiSeq 2000 instrument as described (33). Sequence reads were mapped to the reference genome (hg19) using the ELAND program.
Somatic Single-Nucleotide Mutation Calling and Related Analysis.
For matched normal-tumor pairs, somatic point mutations were called by MuTect. The output from MuTect was further filtered to remove false-positive calls. Somatic indels were called by an in-house pipeline, and all indels have been manually curated. For unmatched tumors, SAMtools was used to call variant bases appended with quality scores. Among these unmatched tumors, variants with a frequency greater than 2 × 10−5 in the ExAC database (7) were excluded. Details of all somatic mutation-related analysis are provided in SI Appendix.
Somatic Copy Number Mutation Calling.
The ratio of normalized coverage depth between tumor and normal samples was calculated for each exome capture probe. The genome was then de-noised and segmented by circular binary segmentation (34). Copy number was assigned to each segment based on both coverage depth ratio and deviation of the B-allele frequency using in-house scripts (SI Appendix).
Transgenic USC-ARK2 Cell Line.
pCMV6-Entry plasmids expressing HIST1H2AB (RC211585) or HIST1H2AC (RC210259) were purchased from OriGene, and desired mutations were introduced by site-directed mutagenesis (Stratagene QuickChangeII XL). The sequences of all plasmids were verified by direct Sanger sequencing. Constructs were transfected into USC-ARK2 cells and continuously selected using G418 (SI Appendix).
Immunofluorescence Microscopy, Western Blotting, and qRT-PCR.
Protocols of Western blotting and immunofluorescence microscopy (IF) followed previously described methods (35). An EpiSeeker Histone Extraction Kit (ab113476; Abcam) was used for histone extraction. Anti-myc antibody (Jackson Laboratory) was used for detection of exogenous expression of histones in Western blotting, and antibody against DDK tag (Sigma–Aldrich) was used in IF. RNA isolation and qRT-PCR were performed using standard protocols on an ABI 7500 Real-Time PCR instrument. Primer sequences are described in SI Appendix.
Cell Migration and Invasion Assays.
Migration and invasion assays were performed in 24-transwell chambers with polycarbonate membrane. Details are provided in SI Appendix.
Supplementary Material
Acknowledgments
R.P.L. is an Investigator of the Howard Hughes Medical Institute. This work was supported, in part, by NIH Grants R01 CA154460-01 and U01 CA176067-01A1, the Deborah Bunn Alley Foundation, the Tina Brozman Foundation, the Discovery to Cure Foundation, the Guido Berlucchi Foundation (A.D.S.), and Gilead Sciences. This investigation was also supported by NIH Research Grant CA-16359 from the National Cancer Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614120113/-/DCSupplemental.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983;15(1):10–17. doi: 10.1016/0090-8258(83)90111-7. [DOI] [PubMed] [Google Scholar]
- 3.Hendrickson M, Ross J, Eifel P, Martinez A, Kempson R. Uterine papillary serous carcinoma: A highly malignant form of endometrial adenocarcinoma. Am J Surg Pathol. 1982;6(2):93–108. doi: 10.1097/00000478-198203000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Lee KR, Tavassoli FA, Prat J. Classification of Tumours. Pathology and Genetics of Tumors of the Breast and Female Genital Organs. World Health Organization Classification of Tumours. IARC Press; Lyon, France: 2003. Surface epithelial stromal tumors of the ovary and peritoneum; pp. 117–161. [Google Scholar]
- 5.El-Sahwi KS, Schwartz PE, Santin AD. Development of targeted therapy in uterine serous carcinoma, a biologically aggressive variant of endometrial cancer. Expert Rev Anticancer Ther. 2012;12(1):41–49. doi: 10.1586/era.11.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kandoth C, et al. Cancer Genome Atlas Research Network Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao S, et al. Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proc Natl Acad Sci USA. 2013;110(8):2916–2921. doi: 10.1073/pnas.1222577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shih IeM, et al. Somatic mutations of PPP2R1A in ovarian and uterine carcinomas. Am J Pathol. 2011;178(4):1442–1447. doi: 10.1016/j.ajpath.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10(7):457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janknecht R. On the road to immortality: hTERT upregulation in cancer cells. FEBS Lett. 2004;564(1-2):9–13. doi: 10.1016/S0014-5793(04)00356-4. [DOI] [PubMed] [Google Scholar]
- 16.Cao Y, Bryan TM, Reddel RR. Increased copy number of the TERT and TERC telomerase subunit genes in cancer cells. Cancer Sci. 2008;99(6):1092–1099. doi: 10.1111/j.1349-7006.2008.00815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45(10):1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 19.Hagiwara T, Hidaka Y, Yamada M. Deimination of histone H2A and H4 at arginine 3 in HL-60 granulocytes. Biochemistry. 2005;44(15):5827–5834. doi: 10.1021/bi047505c. [DOI] [PubMed] [Google Scholar]
- 20.Macadangdang BR, et al. Evolution of histone 2A for chromatin compaction in eukaryotes. eLife. 2014;3:e02792. doi: 10.7554/eLife.02792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gatti M, et al. A novel ubiquitin mark at the N-terminal tail of histone H2As targeted by RNF168 ubiquitin ligase. Cell Cycle. 2012;11(13):2538–2544. doi: 10.4161/cc.20919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mattiroli F, et al. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012;150(6):1182–1195. doi: 10.1016/j.cell.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol. 2002;319(5):1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 24.Forbes SA, et al. COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue D1):D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinstein JN, et al. Cancer Genome Atlas Research Network The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dedes KJ, Wetterskog D, Ashworth A, Kaye SB, Reis-Filho JS. Emerging therapeutic targets in endometrial cancer. Nat Rev Clin Oncol. 2011;8(5):261–271. doi: 10.1038/nrclinonc.2010.216. [DOI] [PubMed] [Google Scholar]
- 28.Shern JF, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014;4(2):216–231. doi: 10.1158/2159-8290.CD-13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCluggage WG. Malignant biphasic uterine tumours: Carcinosarcomas or metaplastic carcinomas? J Clin Pathol. 2002;55(5):321–325. doi: 10.1136/jcp.55.5.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davidson N, Lehmann K-V, Kahles A, Perez A, Rätsch G. Integrative analysis of transcriptome variation in uterine carcinosarcoma and comparison to sarcoma and endometrial carcinoma. bioRxivorg. December 13, 2014 doi: 10.1101/012708. [DOI] [Google Scholar]
- 31.Okosun J, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46(2):176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis PW, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340(6134):857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi M, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci USA. 2009;106(45):19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5(4):557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 35.Shibata S, et al. Angiotensin II signaling via protein kinase C phosphorylates Kelch-like 3, preventing WNK4 degradation. Proc Natl Acad Sci USA. 2014;111(43):15556–15561. doi: 10.1073/pnas.1418342111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.