Significance
The transcription factor T-bet (Tbox protein expressed in T cells), a master regulator of T-cell lineage commitment, is a member of the Tbox family but coordinately regulates many more genes than other Tbox proteins. How T-bet simultaneously recognizes distant elements that may be thousands of base pairs apart is unknown. We have determined the crystal structure of the Tbox DNA binding domain of T-bet complexed with a 24-bp palindromic DNA. The structure shows a dimer where each monomer binds simultaneously to two independent DNA molecules. Fluorescence-based assays show T-bet can synapse two DNA molecules in solution. Chromosome conformation capture assays confirm that T-bet can directly mediate the formation of chromatin loops at the IFN-γ gene locus in the absence of other transcription-related proteins.
Keywords: T-bet, transcriptional regulation, DNA looping, crystal structure, master regulator
Abstract
The transcription factor T-bet (Tbox protein expressed in T cells) is one of the master regulators of both the innate and adaptive immune responses. It plays a central role in T-cell lineage commitment, where it controls the TH1 response, and in gene regulation in plasma B-cells and dendritic cells. T-bet is a member of the Tbox family of transcription factors; however, T-bet coordinately regulates the expression of many more genes than other Tbox proteins. A central unresolved question is how T-bet is able to simultaneously recognize distant Tbox binding sites, which may be located thousands of base pairs away. We have determined the crystal structure of the Tbox DNA binding domain (DBD) of T-bet in complex with a palindromic DNA. The structure shows a quaternary structure in which the T-bet dimer has its DNA binding regions splayed far apart, making it impossible for a single dimer to bind both sites of the DNA palindrome. In contrast to most other Tbox proteins, a single T-bet DBD dimer binds simultaneously to identical half-sites on two independent DNA. A fluorescence-based assay confirms that T-bet dimers are able to bring two independent DNA molecules into close juxtaposition. Furthermore, chromosome conformation capture assays confirm that T-bet functions in the direct formation of chromatin loops in vitro and in vivo. The data are consistent with a looping/synapsing model for transcriptional regulation by T-bet in which a single dimer of the transcription factor can recognize and coalesce distinct genetic elements, either a promoter plus a distant regulatory element, or promoters on two different genes.
Transcription factors that control the expression of large families of genes involved in cell lineage commitment are often termed master regulators of cell-fate determination (1). In the development of both innate and adaptive immunity, one such master regulator is the transcription factor T-bet (Tbox protein expressed in T cells) (2). In T-cell–mediated adaptive immunity, T-bet controls the differentiation of naive CD4+ T helper lymphocytes into TH1 cells by activating transcription of TH1-specific genes, such as IFN-γ, while repressing transcription of TH2-specific genes, such as IL-4 (3). CD4+ T cells lacking T-bet are severely impaired in their ability to produce IFN-γ, susceptible to Leishmania major infection, and have a marked in vivo shift of the TH1/TH2 balance toward the TH2 pathway (4–6). Ectopic expression of T-bet is sufficient to “reprogram” fully polarized TH2 cells into the TH1 pathway, as demonstrated by induction of IFN-γ transcription and shutdown of the expression of genes encoding TH2 cytokines (7). T-bet–deficient and T-bet–overexpressing strains of mice have provided confirming evidence that T-bet is required for the generation of TH1 cells in vivo and that it simultaneously represses IL-4 production and shuts down the TH2 gene program (2). T-bet is required for the effective handling of pathogens (6, 8) and cancer cells (9–11), is pathogenic in the setting of autoimmunity (12), and is protective in the asthmatic response (4). In addition to its roles in CD4+ cells and the adaptive arm of the immune system, T-bet controls the development and effector function of CD8+ cells (13–15) and natural killer cells (16), is required for isotype switching to Ig G2a in B cells (12), and also plays essential roles in dendritic and natural killer cell function in innate immunity (16, 17).
T-bet belongs to the Tbox family of transcription regulators, whose members are characterized by the presence of the Tbox DNA binding domain (DBD). Tbox family proteins are required both for early cell-fate decisions, such as those for formation of the basic vertebrate body plan, and for differentiation and organogenesis (18). Tbox proteins are typically around 60 kDa in molecular mass and comprise at least two structural and functional domains: the Tbox sequence-specific DBD and a second domain that is thought to be involved in interactions with other proteins. The relative position of the domains varies between different members of the family, but the order is conserved for any one member and its orthologs. Despite sequence variations within the Tbox DBD between family members, all members of the family appear to bind to the same DNA consensus sequence, TCACACCT. In several in vitro binding-site selection studies, members of the Tbox family were found to bind preferentially sequences containing two or more of these core motifs arranged in various orientations; however, the significance of such double sites in vivo is uncertain, as most Tbox target gene sites have been found to contain only a single consensus motif (18).
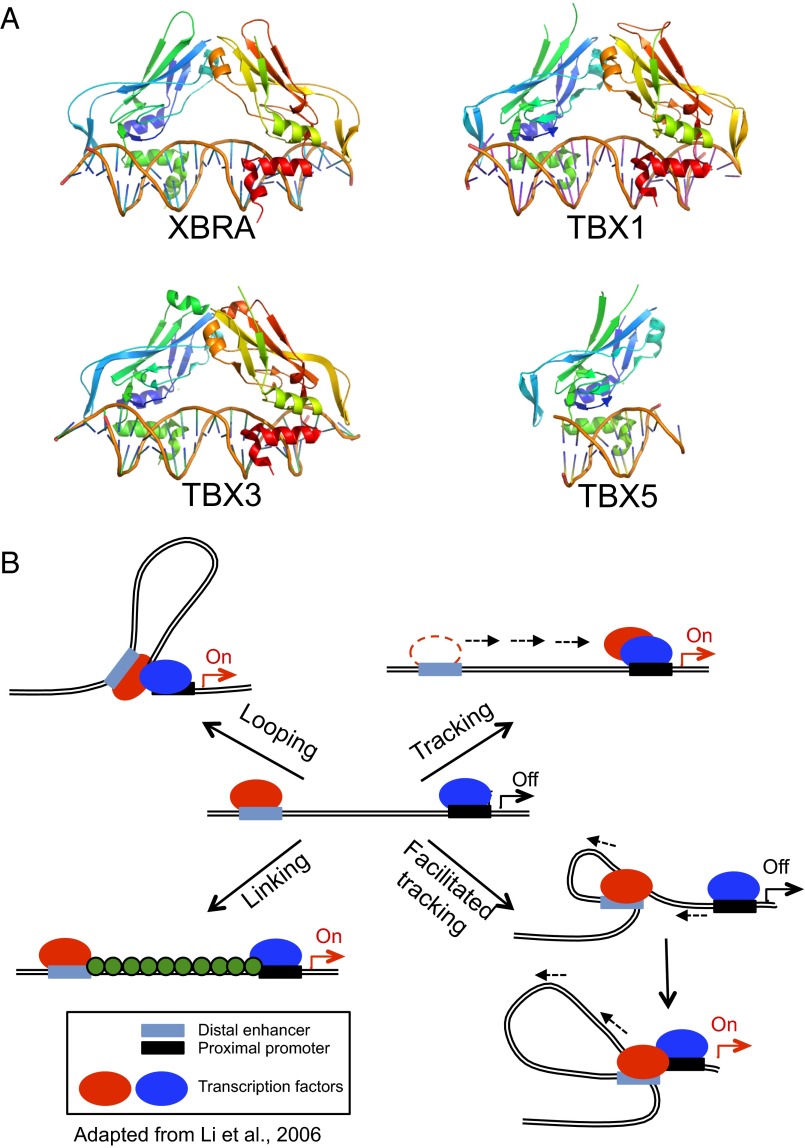
To date, crystallographic analysis of Tbox proteins has been reported only for the Tbox DBDs of Xbra, the Xenopus homolog of the mammalian protein Brachyury (19), and the Tbox protein TBX3 (20), both in complex with a palindromic consensus DNA oligomer; and, more recently, Tbox protein TBX5 in both its DNA-bound and unbound states (21) and TBX1 in its DNA-bound state (22). The structures of all four Tbox domains in the bound complexes are very similar (Fig. 1A), consisting of a seven-stranded β-barrel domain that is closed by a smaller β-pleated sheet, with two projecting helices that are involved in DNA recognition. Although the monomers of Xbra, TBX3, and TBX1 differ little from one another, they do differ in the way they come together on their two-site DNA duplexes. In the crystal structure, Xbra binds as a dimer, and binding to each site appears to be stabilized by monomer–monomer interactions. The dimer interface, however, is not extensive, and it is likely that dimerization is an artifact of crystallization coupled with the presence of a palindromic DNA molecule. Consistent with this possibility, TBX3 and TBX1 have no dimer interface and recognize each site independently, although the arrangement of each monomer on the DNA is virtually identical to that of the Xbra dimer and the overall structures of the two protein–DNA complexes strongly resemble each other (Fig. 1A). Furthermore, dimers seen in the published Tbox crystal structures require a palindromic DNA sequence to form, but only nonpalindromic half sites have been found in the promoters of target genes (23), which further call into question the physiological relevance of these weak dimers.
Fig. 1.
The Tbox transcription factors bind contiguous DNA elements. (A) Structures of the Tbox DBD–DNA complexes of Xbra (PDB ID code 1XBR), TBX1 (PDB ID code 4A04), TBX3 (PDB ID code 1H6F), and TBX5 (PDB ID code 2X6V). Note that the overall folds of the four Tbox domains are essentially identical, as is their mode of recognition of the B-form DNA double helix. Xbra, TBX1, and TBX3 crystallized as homodimers and bind two consensus target sequences, respectively, but the dimers are not tightly associated and probably bind their two sites independently; TBX5 binds as a monomer to a single site. (B) Models for action at a distance by a dimeric transcription factor. The two subunits of the dimer are shown in red and blue and may be either different (heterodimer) or identical (homodimer). In a looping model, each subunit binds independently to a promoter recognition element and a distant element, which may be either another promoter or, as shown, an enhancer sequence. Formation of the dimer juxtaposes the distant sites. Tracking postulates that the different elements serve to recruit the protein subunits to the DNA, but that one of them then scans along the chromosome, leaving its site behind, until it encounters its cognate partner. Facilitated tracking combines elements of tracking and looping, in that the distant site is brought along with its binding protein. In a linking model, the two subunits and their sites remain separate and other proteins (shown in green) connect them. The previously known Tbox protein structures shown in A are either monomeric or bind contiguous DNA elements and consequently would not be expected to act at a distance as depicted in these models.
The TBX5 structure is the first to be reported of a Tbox domain in both the bound and unbound states. Both structures are similar, but a 310-helix at the C terminus of the TBX5 Tbox domain only forms upon DNA binding and is unstructured in the DNA-unbound form (21). TBX5 is a monomer in solution and binds as a monomer to the single consensus sequence present in its DNA complex; how it would bind if presented with a palindromic sequence is unknown.
All four Tbox proteins recognize DNA in the same way, by means of two contiguous C-terminal helices: an α-helix and the 310-helix (Fig. 1A). Only a small number of amino acids make direct contact with the bases in the minor groove of the DNA, and these are conserved among Tbox family members (18), explaining the consensus recognition sequence.
Tbox proteins can function as transcriptional activators and as repressors. Which activity occurs for any given target gene has been shown to depend on sequences located in the carboxyl-terminal portion of the protein, outside the Tbox DNA binding domain.
Enhancer sequences for some of the Tbox proteins have been identified; these typically lie hundreds to thousands of base pairs from the promoter site. For example, an enhancer sequence for T-bet activation of the IFN-γ promoter is located ∼22 kb upstream of the promoter site and contains a consensus Tbox binding sequence (24). Tbox enhancer sequences are very similar in sequence to the promoter recognition sequences (25), suggesting that the Tbox DBD binds directly to them; this has been demonstrated directly in some cases. Although thus far only single consensus sequence sites have been identified in natural promoters of Tbox target genes, further calling into question the relevance of the palindromic sites used in some of the crystallographic studies and in many in vitro binding experiments, a few of the enhancer sites do contain imperfect tandem recognition elements; the significance of this is unclear.
The great separation between enhancer and promoter sites bound by the same DBD raises the question of action at a distance, a problem that is particularly relevant for master regulators of gene expression, which coordinately control the expression of multiple genes. To date, four different—but not mutually exclusive—hypotheses have been put forward for how an enhancer activates its cognate genes across distances up to 800 kb (Fig. 1B) (26): (i) Chromatin looping between the enhancer and promoters, in which two protein molecules, which may be different or identical, bind to the separate elements and then form either a hetero- or homodimer, bringing the distant sites into juxtaposition (27). (ii) A tracking or scanning model, which hypothesizes that the transcription-activating protein recruited by an enhancer linearly tracks along chromatin until it encounters a competent promoter with its own bound transcription factor and a dimer is formed (28); in this model, the enhancer element is left behind and the scanning process does not alter the proximity between the enhancer and the promoter. Early evidence for tracking was first presented at T4 late promoters by Herendeen et al. (29). (iii) Facilitated tracking, which incorporates aspects of both the looping and tracking models (30), where both the enhancer element and its bound protein migrate along the chromatin fiber until they encounter the cognate promoter complex, and the intervening chromatin between the enhancer and the promoter “reels out” through the enhancer complex and forms a loop, which is progressively enlarged during tracking. (iv) A linking model, which postulates that the binding of facilitator proteins between an enhancer–protein complex and its cognate promoter–protein complex mediates transcription-enhancing activity, without bringing the two sites into juxtaposition (31).
Although studies of several multigene clusters, such as the β-globin gene cluster, have shown that gene activation by, for example, a remote enhancer can be associated with chromatin looping (32, 33), the molecular mechanism of loop formation and its involvement in gene regulation are not fully understood. Recent evidence for the coordinate regulation of multiple genes, sometimes even located on distinct chromosomes, in “transcription factories” has pointed out the importance of understanding how action at a distance occurs (34): for example, the IL-4 gene locus was shown to associate with the IFN-γ gene locus on a different chromosome in naive T cells that are committed to differentiate into cells expressing only one of the two cytokines in a monoallelic manner (35).
To gain insight into the nature of transcription control by the master regulator T-bet, we crystallized its Tbox domain in complex with a 24-bp DNA oligomer containing two consensus binding sequences arranged palindromically. The crystal structure shows two Tbox monomers forming a tight dimer that is completely different from the quaternary structure observed for other Tbox proteins. Each monomer of the T-bet dimer binds a single recognition site on two independent DNA double helical oligomers; the second site of each palindrome is unoccupied. The structure immediately suggests that the dimeric T-bet DBD is capable of looping DNA by recognizing two distant sites simultaneously. Size-exclusion chromatography confirms that T-bet DBD dimers exist, and a FRET experiment demonstrates that T-bet is able to synapse two independent pieces of DNA in solution. Furthermore, in vitro and in vivo chromosome conformation capture (3C) experiments establish that T-bet functions in the direct formation of chromatin loops or tethers at the IFN-γ gene locus. Taken together, our data provide multiple independent lines of evidence that T-bet is able to recognize remote distal regulatory sequences and coordinate the expression of multiple genes simultaneously, at least in part by using its unique dimeric structure to bring distant recognition elements into juxtaposition.
Results
Structure Determination of the T-bet DBD–DNA Complex.
Expression of Mus musculus full-length T-bet in Escherichia coli resulted in inclusion bodies. Better results were obtained with the T-bet Tbox DNA binding domain (amino acids 135–326; T-bet DBD). Although the domain on its own failed to crystallize, trials including various length oligonucleotides representing consensus or specific T-bet DNA binding sequences produced diffraction-quality crystals. The best results were obtained with the full Tbox domain and a 24-bp, palindromic, consensus T-bet recognition sequence (the two core recognition elements are underlined):
AATTTCACACCTAGGTGTGAAATT
TTAAAGTGTGGATCCACACTTTAA
However, these crystals initially diffracted only to about 4-Å resolution, even with synchrotron radiation. After optimizing the buffer conditions for purification and the crystallization conditions, crystals were finally obtained that diffracted to around 3-Å resolution. The crystals had the symmetry of the hexagonal space group P61 with unit cell dimensions a = b = 70.45 Å, c = 438.4 Å. Packing considerations suggested that the asymmetric unit probably contained two Tbox domains and two DNA duplexes, but a protein:palindromic-DNA stoichiometry of 2:1 was also possible, and would be expected for a Tbox:DNA complex such as that observed (Fig. 1A) in the crystal structures of the transcription factors Xbra (19), TBX3 (20), and TBX1 (22).
The T-bet DBD structure (PDB ID code 5T1J) was solved by molecular replacement using the known structure of B-form DNA and a homology model of the T-bet DBD monomer generated with SWISS MODEL (36) based on the Tbox DNA binding domain of the Brachyury/Xbra transcription factor (PDB ID code 1XBR), which is 50% identical in sequence to the T-bet DBD. Following model adjustment, the structure was refined by conventional means. The current Rcryst and Rfree are 26% and 30%, respectively, using all data to 2.95-Å resolution (although data beyond 3.2 Å are weak), and the rmsd from ideal bond lengths and angles are 0.004 Å and 0.9°, respectively. Details of the structure determination and statistics are given in SI Appendix, Supplemental Methods and Materials and SI Appendix, Table S1.
The Structure of the T-bet DBD Monomer and Its Interactions with DNA Are Typical of Other Tbox Domains.
The asymmetric unit of the crystal structure of the T-bet DBD comprises two monomers in tight association (Fig. 2A). The polypeptide chain folds of the two subunits of this T-bet DBD dimer are virtually identical to one another, and are extremely similar to the canonical Tbox fold first seen in the Xbra and TBX3 structures (19, 20). Each subunit uses its two projecting helices to recognize the DNA consensus sequence in the same way observed in all four previously determined Tbox protein structures, in agreement with the known ability of T-bet to bind to the same sequences as TBX1, TBX3, TBX5, and Xbra. The C-terminal α-helix of T-bet inserts into the minor groove of the DNA, parallel to the axis of the double helix, and the 310 helix, which is approximately perpendicular to the α-helix, lies across the minor groove (Fig. 2A). Recognition of the consensus sequence involves very few specific contacts with the base pairs or sugars: the side-chain of Arg164 forms a hydrogen bond to the guanine base at position 17 in the complementary strand of the consensus sequence, and phenylalanines 320 and 324 partially wedge themselves between the strands of the oligonucleotide, where they make nonbonded contacts primarily with the ribose groups of guanine 15 and thymine 16, respectively. Nearly all other contacts are with the phosphate groups of the double helices: the side-chains of Arg198, Ser262, Asn246, and Glu326, and the backbone NH of Gly323 make hydrogen bonds to phosphate groups on the consensus strand, whereas the side-chains of Thr311, Tyr305, Arg165, Asn318, and Lys243 make hydrogen bonds with phosphates on the complementary strand.
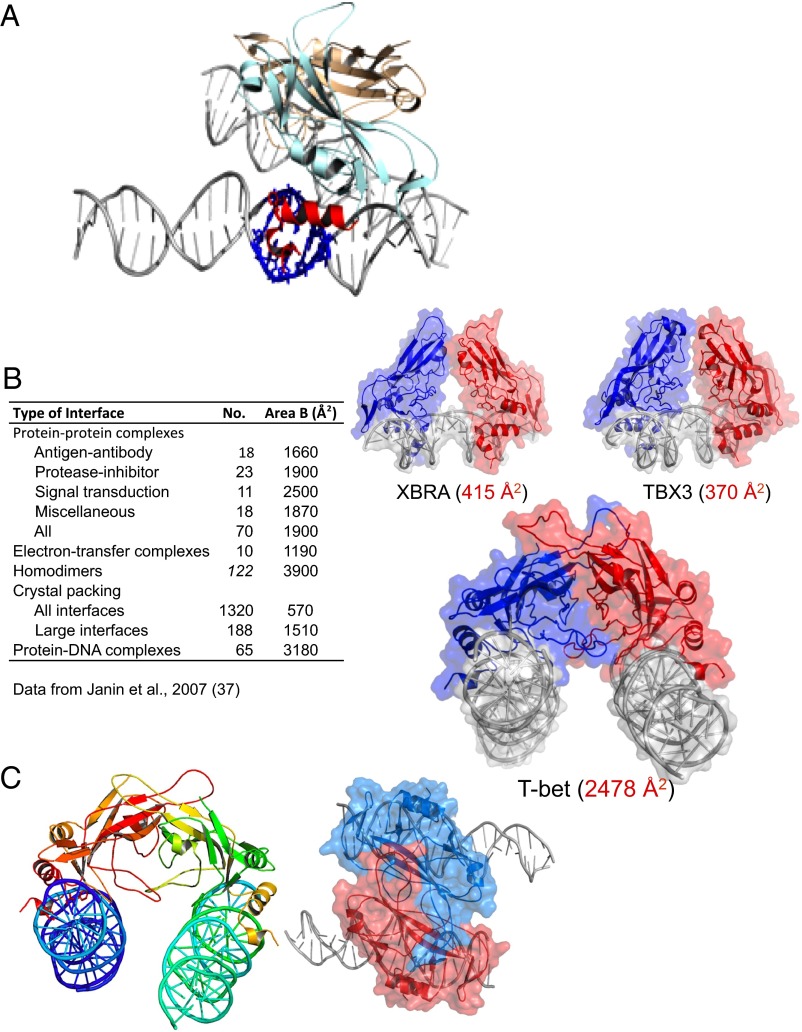
Fig. 2.
The DNA binding domain of T-bet forms a dimer that cross-links two independent DNA strands. (A) The two T-bet DBD monomers (light blue and brown) form a tight dimer. Each monomer binds a single T-bet recognition site on a separate strand of DNA. The protruding helices that make contact with the minor groove of the DNA are shown in red for the light blue monomer; the residues on the double helix that interact with this part of the Tbox domain are shown in navy blue. (B) The T-bet DBD–DNA complex with the molecular surface overlaid, and a similar surface model of the Tbox DBD–DNA complexes of Xbra and TBX3. The Xbra dimer interface is much smaller, and TBX3 binds essentially as two monomers. (C) Orthogonal views of the T-bet Tbox–DNA complex showing binding of two independent DNA strands to each subunit of the dimer. The view on the right has a surface representation overlaid, indicating the tightness of the complex.
The DNA Double Helix Is Slightly Distorted by T-bet.
The best fit of the electron density in the region of DNA where the T-bet DBD makes contact with the double helix could not be obtained with a classic B-DNA conformation. At our resolution the nature of the distortion cannot be characterized with precision, but it appears similar to that in the TBX5–DNA complex (21). In that structure, the minor groove in the TBX5–DNA-binding site is wider at base pairs 4–6 of the consensus sequence compared with B-DNA, because of the insertion of the Tbox 310-helix into the groove. Specifically, two aromatic residues, Phe232 and Phe236 in the TBX5 sequence, distort the DNA by nonpolar, steric interactions. As noted above, the corresponding residues in T-bet (Phe320 and Phe324) appear to do the same thing to the same region of the consensus sequence in our structure. The TBX5–DNA structure also shows a slight bending of the DNA at base pairs 7 and 8. Our electron density in this region is consistent with a similar distortion.
The T-bet DBD Forms a Dimer That Recognizes Two Independent Pieces of DNA.
The striking difference between the T-bet DBD–DNA structure and the structures of DNA complexes of the Tbox DBDs from TBX1, TBX3, TBX5, and Brachyury/Xbra, is that the T-bet DBD forms a unique, very tight dimer that binds to single half-sites of two independent double-stranded palindromic molecules of DNA, rather than forming a weaker dimer bound to two adjacent half sites on a single palindromic DNA strand as in the Xbra structure, or simply binding as nonassociating monomers to adjacent half-sites, as observed in the TBX3–DNA, TBX5–DNA, and TBX1–DNA complexes (Figs. 1A and 2).
The dimer interface in the T-bet DBD is very different from that of the Xbra Tbox dimer in terms of both size and location of the contact regions. The total surface area buried in the T-bet dimer is about 2,478 Å2, compared with only 415 Å2 in the Xbra dimer (Fig. 2B). This much greater contact area for the T-bet dimer is characteristic of specific, physiologically relevant interactions (37). We conclude, therefore, that in contrast to other Tbox transcription factors, which appear to be monomeric in solution, T-bet should be able to exist as a homodimer, and will be unlikely to form heterodimers with other Tbox protein DBDs.
The Xbra Tbox dimer is primarily held together by interactions at the “fingertips” of the extended domain structure, as shown in Figs. 1A and 2B. This minimal contact keeps the rest of the protein structure in each subunit well separated, and makes the binding regions of the two subunits colinear, allowing both monomers to bind to the same face of the two-turn palindromic DNA sequence. In contrast, the T-bet dimer has extensive contact between the sides of the fingers, orienting the two subunits such that it is impossible for them to bind simultaneously to both sites on a palindromic sequence (Fig. 2C). Instead, the second subunit is splayed away from the DNA binding site of the first, and their binding sites are parallel, allowing the second subunit to bind to a different, remote recognition sequence on the same (or possibly even a different) chromosome (Fig. 2C). The actual interactions between the DNA and the recognition helices in the subunit are, as indicated above, the same as those of the other Tbox proteins (not surprisingly, because all those residues are conserved); it is the different manner of oligomerization that causes one dimeric molecule of T-bet to bind to two half sites on two different recognition sequences, unlike the other Tbox domains.
The T-bet DBD is able to form this unique dimer because T-bet has two loops (loop 1, residues 248–256 in the full-length T-bet sequence; loop 2, residues 276–293) that are different in length and in sequence than the analogous loops in the other Tbox proteins (SI Appendix, Fig. S3). These two loops largely make up the new dimer interface, where they extend out like two arms reaching to the adjacent monomer to form an interlocking “hug.” Although these loops and their flanking residues are not conserved in other Tbox proteins (and which is also why heterodimer formation of T-bet with those proteins is unlikely), they are conserved in all T-bet protein sequences determined to date. Loop 2 in particular has several negatively charged residues, which interact with positively charged ones on its own monomer and on the twofold-related monomer. The result is a conserved network of salt-bridges and long-range electrostatic interactions that stabilize the unique T-bet dimer (SI Appendix, Fig. S3C).
The Crystal Structure Suggests That T-bet Can Loop and Synapse DNA.
The structure provides direct evidence that a single dimeric molecule of a eukaryotic transcriptional factor can bind simultaneously to two independent double-stranded DNA molecules. A similar synapsing of independent DNA molecules by a dimeric protein involved in recombination was observed in the crystal structure of the complex of the RAG1 nonamer binding domain with its nonamer DNA recognition site (38). Coincidentally the functional counterpart of T-bet, GATA3, which is a master transcriptional regulator that coordinately regulates the expression of TH2 cytokines and represses the expression of TH1 cytokines, has also been suggested to mediate long-range DNA interactions (39). The implication of this mode of DNA binding is that one T-bet dimer has the potential to juxtapose distant DNA sequences on the same chromosome and, in the process, loop out the intervening DNA (SI Appendix, Fig. S4), or that it could bind simultaneously to sequences on two different chromosomes, thereby synapsing them, as has been proposed for transcription factories (34, 35).
An extensively studied transcription factor that has been shown to involve both DNA looping and interchromosomal tethering is the CCCTC-binding factor, although its direct mechanical involvement in the process is uncertain (40). Coincidentally, similar to T-bet, CTCF can act either as an activator or a suppressor of various genes (40). CTCF physically binds to itself to form homodimers (41), which causes the bound DNA to form loops (42). Using ChIP followed by ChIP-seq, it was found that CTCF localizes with cohesin genome-wide and affects gene regulatory mechanisms and higher-order chromatin structure (43).
We cannot distinguish between a looping model and a facilitated tracking model (Fig. 1B), which are variations of the same theme, because both would result in a dimeric species bound to two separate recognition sequences with a loop of DNA in between. For convenience, we will refer to both of these models as “looping.”
T-bet Homodimerizes in Solution.
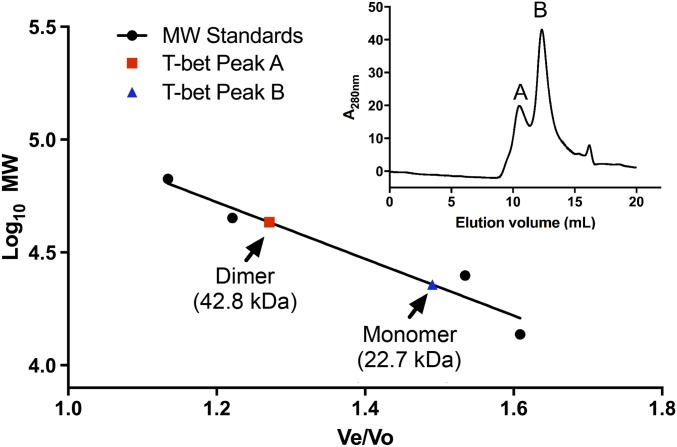
If the function of T-bet involves binding of a single dimeric T-bet molecule to two independent DNA sites, T-bet must be able to homodimerize in solution, in contrast to the other Tbox DNA binding proteins, which are solution monomers. To assess the oligomerization state of T-bet, we performed size-exclusion chromatography on the purified T-bet Tbox domain in the absence of DNA. The results (Fig. 3) clearly show a mixture of monomer and homodimer in solution with a molecular mass of 22,707 kDa and 42,838 kDa respectively. This matched well with the expected molecular mass for the T-bet DNA binding domain of the monomer and homodimer of 22,015 kDa and 44,030 kDa respectively.
Fig. 3.
The DNA binding domain of T-bet forms a dimer in solution. Size-exclusion chromatography data show that T-bet DBD exists as a mixture of monomeric and dimeric species in solution.
T-bet Can Synapse DNA in Solution.
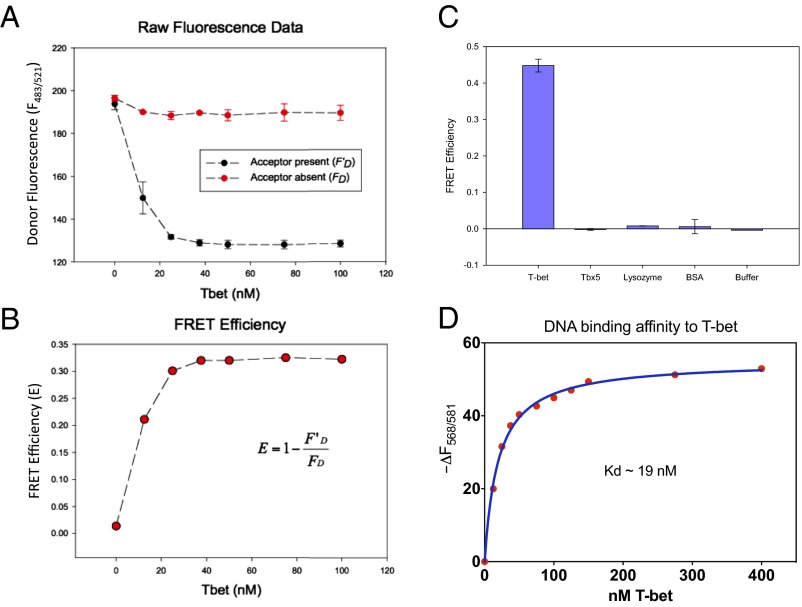
We next devised an assay to determine whether a single molecule of T-bet could bind simultaneously to two different DNA double helices in solution. Following Yin et al. (38), we prepared two DNA oligomers, each labeled at both ends with a donor fluorophore or an acceptor fluorophore, where the donor emission spectrum overlaps with the acceptor excitation spectrum (SI Appendix, Fig. S5). In the absence of T-bet, these independent constructs should display no FRET between their fluorophores; however, if each binds to one T-bet subunit in the manner observed in our structure, the donor-to-acceptor distance would become less than 40 Å and a significant FRET signal should be observable.
The two fluorescently labeled DNA oligomers gave no FRET signal on their own or when mixed together. Addition of BSA or the LexA DNA binding protein, which binds as a homodimer to two adjacent target sequences on the same DNA strand (44), also failed to produce a signal. However, when a purified T-bet Tbox domain was added to the mixture of oligonucleotides, a strong T-bet concentration-dependent FRET signal was observed (Fig. 4).
Fig. 4.
FRET experiments confirm T-bet can synapse DNA in vitro. (A) Titration of FAM- and TAMRA-labeled DNA with T-bet. A strong FRET signal is observed when T-bet is present together with both donor- and acceptor-labeled DNA, but not in the absence of T-bet. (B) Relative FRET efficiency vs. T-bet concentration. (C) Fluorescently labeled DNA FRET efficiency induced by T-bet, TBX5, lysozyme, BSA, and buffer using 10 nM FAM-labeled DNA and 10 nM TAMRA-labeled DNA. Only T-bet produced a FRET signal. Even the monomeric T-bet homolog TBX5, which binds to the same consensus sequence, fails to do so. The concentrations of T-bet, TBX5, lysozyme, and BSA used in this experiment were 1.16 mM, 3.75 mM, 5.35 mM, and 2.5 mM, respectively. (D) The binding affinity of DNA to T-bet was determined by titrating T-bet to 1.25 nM TAMRA-DNA and observing the non-FRET decrease in the fluorescence of the T-bet bound TAMRA-DNA (SI Appendix, Supplementary Methods and Materials). The dissociation constant determined by this method was ∼19 nM.
As a further test, we carried out the same assay with different control proteins, including the T-bet homolog TBX5 [a generous gift of Christoph Muller and Christian Stirnimann, Structural and Computational Biology Unit, European Molecular Biology Laboratory (EMBL) Heidelberg, Heidelberg], which binds to the same consensus oligonucleotide sequence as T-bet. TBX5 has been shown to be a monomer both in solution and when bound to DNA (21), and so would not be expected to produce a FRET signal in this assay. Again, we only observed energy transfer when T-bet was present in the oligonucleotide mixture; neither other control proteins nor TBX5 gave any signal (Fig. 4C).
We observed that the fluorescence of the acceptor-DNA used in the FRET assay [carboxytetramethylrhodamine (TAMRA)-DNA] was quenched upon binding to T-bet independent of the donor-DNA [6-carboxyfluorescein (FAM)-DNA]. We used this non-FRET effect as a binding assay to determine the Kd for the T-bet consensus sequence complex by titrating T-bet Tbox protein to TAMRA-DNA and monitoring the non-FRET quenching of TAMRA-DNA fluorescence. The Tbox domain of T-bet has a relatively high affinity for its recognition site; the measured Kd is ∼19 nM (Fig. 4D).
Dissociation Kinetics of the T-bet–DNA Complex.
We used the loss of fluorescence quenching on dissociation of the T-bet/FAM-DNA/TAMRA-DNA complex and the T-bet/TAMRA-DNA complex upon addition of excess unlabeled DNA and dilution to determine the kinetic dissociation parameters (SI Appendix, Fig. S6). Two different fluorescence signals were used to capture the dissociation process. The loss of TAMRA-DNA fluorescence quenching captures the dissociation of total bound DNA to T-bet, whereas the loss of FAM-DNA fluorescence quenching in the presence of TAMRA-DNA captures the initial dissociation step that corresponds to either the dissociation of the first double helix or the dissociation into monomers of DNA-bound T-bet dimer (i.e., the loss of FRET step). The dissociation kinetics were biphasic as detected by the loss of TAMRA-DNA fluorescence quenching, with a fast dissociation rate constant koff(fast) of 5.0 × 10−3 s−1 and a slow dissociation rate constant [koff(slow)] of 0.28 × 10−3 s−1, but monophasic when detected by the loss of FAM-DNA fluorescence quenching (loss of the FRET signal), with a dissociation rate constant of 2.5 × 10−3 s−1. Taken together, the dissociation kinetics data suggest that the dissociation of the T-bet–DNA quaternary complex must occur via a minimum of two steps and that the fast dissociation rate constant corresponds to the initial step that leads to the loss of FRET signal.
Two simple schemes consistent with these observations are shown in SI Appendix, Fig. S7. In model 1, the T-bet dimer dissociates first, followed by dissociation of the DNA from the monomers. Model 2 starts with the dissociation of one DNA oligomer from the dimer, followed by dissociation of the other oligomer. Model 1 may be preferred because it does not require different dissociating rate constants for the two oligonucleotides to the protein; in this model the fast step is dimer dissociation and both monomers lose their DNA at the same rate. The two models can be used to predict the concentration dependence of the FRET signal observed when T-bet binds to DNA (SI Appendix, Fig. S8A). Comparison with our FRET measurements suggests that model 1 is a better fit to the observed data (SI Appendix, Fig. S8 B and C), although a mixture of both models may be possible. Assuming primarily model 1, T-bet appears able to bind to DNA first as a pair of independent monomers, followed by protein dimerization to bring the two DNA sites together (49).
T-bet Directly Mediates Long-Range DNA Interactions at the IFN-γ Gene Locus.
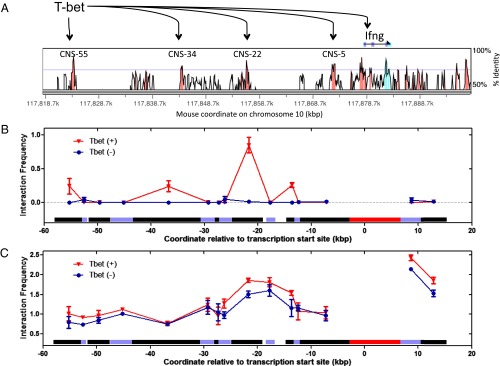
To assess if T-bet mediates long-range DNA interactions by direct juxtaposition of distant DNA regions in cis to form DNA loops, we measured long-range DNA interactions within one of T-bet’s physiological substrates, the IFN-γ gene locus, by using an in vitro 3C assay on samples containing purified T-bet DBD and a BAC plasmid containing the IFN-γ gene locus (45). The IFN-γ gene is a hallmark TH1 cytokine gene activated by T-bet, and conserved noncoding sequences (CNS) far upstream of the IFN-γ gene are known to physically interact with T-bet and to enhance transcription of IFN-γ in a T-bet–dependent manner (24, 46) (Fig. 5A). We therefore sought to determine if the proximal promoter of IFN-γ physically interacts with these distal CNS enhancer sites in a T-bet–dependent manner, forming DNA loops (data in SI Appendix, Fig. S9). As shown in Fig. 5B, the in vitro 3C profiles showed that the IFN-γ promoter region interacts with CNS-22, CNS-34, or CNS-55 fragments (the numbers refer to their locations upstream from the promoter in kilobases) only in the presence of the T-bet DBD. These interactions were absent when T-bet was omitted and replaced with a nonphysiologically relevant protein BSA. Interaction of the promoter fragment with the CNS-22 fragment was the strongest, which coincides with the finding that this CNS is the strongest known T-bet–dependent enhancer of IFN-γ transcription (24). Taken together, our in vitro 3C experiment clearly shows that the T-bet DBD, in the absence any other protein or protein complexes, is sufficient for the direct mediation of long-range DNA interactions in the IFN-γ gene locus.
Fig. 5.
The 3C experiments show T-bet can synapse IFN-γ (Ifn-γ) enhancer and promoter sites. (A) VISTA plot comparing human and mouse sequences at the Ifng gene locus. Locations of CNS are highlighted in pink. These CNS sites are also conserved in rhesus, dog, horse, and rat. CNS-55, CNS-34, CNS-22, CNS-5, and the Ifng promoter are known to bind to T-bet as determined by ChIP (24). (B) In vitro 3C profiles of samples with T-bet (T-bet DBD) or without T-bet (BSA). (C) In vivo 3C profiles of samples with T-bet (WT mouse CD4+ T-cells) or without T-bet (T-bet−/− mouse CD4+ T-cells). In B and C, horizontal bars directly above the x axis depict the length and location of each 3C fragments analyzed. The promoter fragment, shown as a red bar, was used as the 3C anchor for both the in vitro and in vivo 3C experiments.
To further assess T-bet mediation of direct loop formation, we also performed standard in vivo 3C on mouse CD4+ T-cells harvested and expanded in vitro from either T-bet+/+ wild-type or T-bet−/− knockout mice. As shown in Fig. 5C, a strong DNA looping interaction was observed between the promoter fragment and the CNS-22 fragment in both T-bet+/+ and T-bet−/− samples, although the interaction frequency was higher in the T-bet+/+ sample. The fact that nearly identical DNA interaction profiles at the IFN-γ locus were observed in both T-bet+/+ and T-bet−/− mouse T-cells implies that these 3C fragment interactions were at least partially formed independent of T-bet, presumably by other unidentified protein complexes that “prime” the TH1 gene cluster.
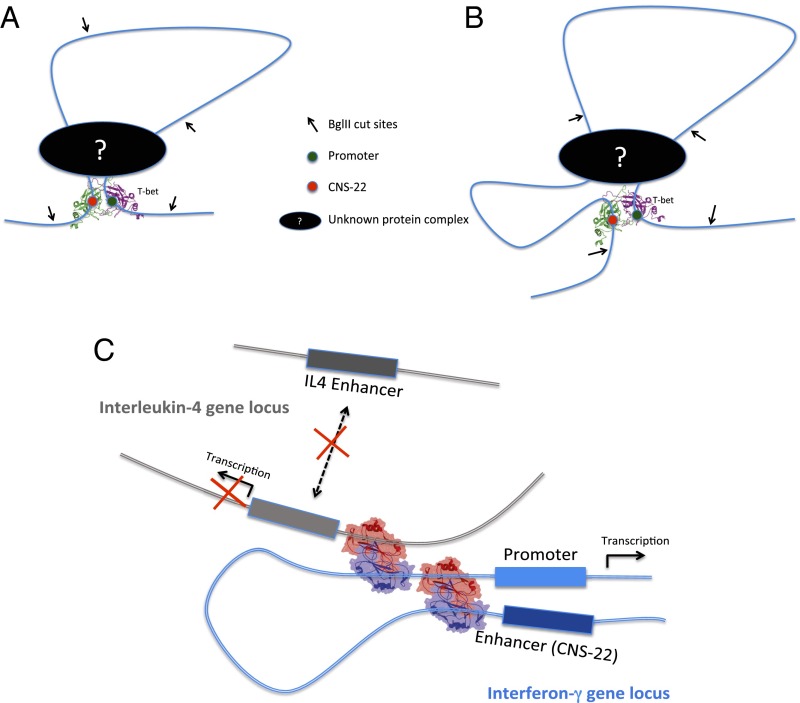
This result suggests a model for the physiological role of T-bet DNA looping activity consistent with the observed in vivo and in vitro 3C data (Fig. 6). As shown in Fig. 6A, a plausible physiological role of T-bet may be to stabilize and maintain existing loops formed between distant transcription regulatory elements by other protein complexes; in this model, no new long-range DNA interactions need be formed by T-bet, but efficient transcription requires stabilization of previously synapsed or “primed” DNA regions by T-bet. This model is consistent with the finding that transcription of β-globin genes requires long contact times between the β-globin–locus control region and β-globin gene, in the range of 45–80 min (47): based on our dissociation kinetic studies of the T-bet–DNA complex (SI Appendix, Fig. S6), the overall mean lifetime of dimeric T-bet bound to DNA is ∼60 min, indicating that T-bet is capable of stabilizing long-range DNA interactions in the time range of 45–80 min in vivo. We note that Splinter et al. (48) have found that CTCF binds sites around the mouse β-globin locus that spatially cluster in the erythroid cell nucleus, and showed that both conditional deletion of CTCF and targeted disruption of its DNA-binding site destabilize these long-range interactions and cause local loss of histone acetylation and gain of histone methylation, but without affecting transcription at the locus. These data show that CTCF is directly involved in chromatin architecture and regulates local balance between active and repressive chromatin marks, and Splinter et al. further postulate that throughout the genome, relative position and stability of CTCF-mediated loops determine their effect on enhancer–promoter interactions, with gene insulation as one possible outcome. CTCF or a similar protein could thus preform the appropriate loops without activating transcription until T-bet binds and the loops are stabilized and the rest of the transcriptional machinery is brought to bear.
Fig. 6.
Proposed models for the physiological role of T-bet DNA looping activity based on the in vivo 3C data. (A) T-bet functions to stabilize and maintain existing DNA loops. No new long-range DNA interactions are created. (B) T-bet creates new long-range DNA interactions, but these new interactions were not detected uniquely as T-bet–dependent interactions because of the existence of prior T-bet–independent interactions between the same 3C fragments. (C) Schematic of T-bet molecules binding simultaneously to promoter sites on genes located on different chromosomes, activating one while repressing the other.
A second plausible model consistent with the in vivo 3C data suggests that the physiological role of T-bet is indeed to initiate de novo, new long-range DNA interactions that bring distal enhancers into close proximity to the promoter, but these T-bet–dependent loops nearly coincide with T-bet–independent loops formed within the same 3C DNA fragments (Fig. 6B). Again, a protein like CTCF could establish these less-specific loops. Because of the limiting resolution of the 3C assay, where some of the digested DNA fragments were in the multiple thousands of base pairs long, such T-bet–mediated loops may not have been uniquely resolved from T-bet–independent loops. Both models would be consistent with an additional role for T-bet as a master regulator of cell fate, that of bringing previously looped enhancer–promoter “hubs” from different genes together (49).
Discussion
T-bet Has a Unique Quaternary Structure Enabling It to Bind Two Distinct DNA Sites.
T-bet (also called TBX21) is a member of the Tbox family of eukaryotic transcription factors, whose proteins regulate key processes in development (2). T-bet expression during T-cell activation is strongly dependent on IFN-γ signaling and STAT1 activation (50). A genomic map of T-bet binding in primary human T cells showed that, in TH1 cells, T-bet associates with genes of diverse function, including those with roles in transcriptional regulation, chemotaxis, and adhesion, whereas the TH2-specific transcription factor GATA-3 shares a large proportion of targets with T-bet (51). The choice between TH1 and TH2 lineage commitment appears to be the result of the opposing action of these two master regulators, T-bet and GATA-3, at a shared set of target genes.
In contrast to other Tbox proteins, T-bet expression is sufficient to reprogram a fully differentiated TH2 cell into a TH1 cell (7). Nevertheless, the DNA binding domain of T-bet has been assumed to be similar in structure to other Tbox domains, and to recognize the Tbox family consensus DNA sequence in the same way.
Although the crystal structure of the Xbra–DNA complex showed a Tbox homodimer binding to both sites on a single palindromic recognition element (19), the interface between the monomers is so small that it is unlikely to persist in solution. Consistent with this hypothesis, the structure of the related TBX3–DNA and TBX1–DNA complexes showed almost no association between the monomers—although they each bound to a half-site of the palindrome in essentially the same manner as Xbra (20)—and TBX5, another Tbox family transcriptional regulator, was shown to be monomeric both when bound to DNA in the crystal structure of the complex, and in solution (21). It seems likely, therefore, that most members of the Tbox family are monomers and act by binding as monomers to single recognition sequences (half-sites of the palindromic constructs). This generalization is borne out by the observation that most Tbox binding sites identified to date, whether in promoter regions or other distal regulatory elements, are not palindromic and consist only of a single recognition sequence (18).
The structure of the T-bet Tbox domain in complex with DNA presented here shows that this particular Tbox family member is a striking exception. Because of sequence differences with all other known Tbox proteins in two loop regions distal to the DNA binding site, T-bet forms a homodimer with an extensive interface. This quaternary structure makes it impossible for a single dimeric molecule of T-bet to bind a palindromic double site, but instead enables the dimer to bind to two separate single recognition elements, which could be located thousands of base pairs apart or even on different chromosomes. Size-exclusion chromatography confirmed that the Tbox domain of T-bet forms dimers in vitro. The subunit interface region of the T-bet dimer may represent a target for small molecules aimed at either inhibiting T-bet function by disrupting the dimer or increasing the steady-state level of the active form of the protein by stabilizing the association between the monomers.
To test the hypothesis from the crystal structure that a single dimer of T-bet is able to juxtapose distant or disconnected DNA sites, we devised a FRET assay and showed that T-bet could bring two independent recognition sequences together, whereas the monomeric TBX5 protein could not. We were also able to measure the dissociation constant of the T-bet–DNA complex, which was found to be about 19 nM. Furthermore, we also devised 3C assays to explore the architectural role of T-bet on the IFN-γ gene locus and showed in vitro that T-bet alone is sufficient for the direct formation of chromatin loops or tethers that bring about the juxtaposition of IFN-γ promoter and distal enhancer DNA fragments, which is consistent with our crystal structure. Similar 3C assays performed in vivo showed that similar loops existed in cells in the absence of T-bet but were stabilized in its presence (see below).
A Model for Simultaneous Regulation of Multiple Genetic Loci by T-bet.
As a master regulator of TH1 lineage commitment, T-bet binds to consensus recognition sites in the promoter region of numerous genes, where it activates the expression of TH1-specific genes while simultaneously repressing transcription of TH2-specific genes. Its regulatory properties also involve enhancer elements with similar sequences to the promoter sites, suggesting that the Tbox domain of T-bet binds to those loci as well.
A DNA looping model has been proposed to account for the transcription activation properties of noncoding enhancer DNA regions that are >3 kb away from the promoter region of a gene (26, 31). In such a model, a gene is activated when a distal enhancer region is brought into close proximity to the promoter region by DNA looping or facilitated tracking, because each site is bound to one subunit of an oligomeric transcription factor (Fig. 1B). Other protein and protein–DNA complexes important for transcription that are located far away from a gene can also be brought to the promoter by a DNA-synapsing transcription factor if they bind at or near sequences recognized by that DNA binding protein (34, 35).
Our structure of the T-bet DBD suggests that it has DNA looping activity, and the FRET and in vitro 3C experiments confirm that this activity is present for the isolated Tbox domain in solution. The data presented in this paper are consistent with a model in which a single T-bet dimer is able to bind simultaneously to recognition sites on two different chromosomal regions (Fig. 6C). One site, for example, could be a promoter recognition sequence, whereas the other could be either a distal enhancer site or a promoter recognition sequence on a completely different gene. In principle, that gene need not be located on the same chromosome as the first one, allowing T-bet to initiate the formation or stabilization of so-called transcriptional factories that coordinately regulate different genes regardless of their location (49). Depending on the function of the N- and C-terminal domains of T-bet and the various other proteins that are known to bind to them, including other transcription factors, such as GATA-3 (51), factories involving a number of different genes could coalesce around multiple transcription factors, all nucleated by, for example, one or more T-bet dimers to form an active chromatin hub. It is also not necessary that transcription of all genes in such a factory be activated, depending again, on what else is bound to its other domains, a single T-bet molecule could both activate a gene and repress another simultaneously (Fig. 6C).
This model fits well with biochemical data on the transcriptional regulation by T-bet of the IFN-γ locus. Recent studies have identified multiple cis-regulatory elements within ∼110 kb surrounding the IFN-γ locus, including enhancers and boundary elements, many of which bind the Th1-specific transcription factors T-bet and STAT4; moreover, CNSs greater than 5 kb upstream and downstream of the IFN-γ gene were found to enhance the transcription of IFN-γ in a T-bet–dependent manner (3, 24, 25, 52–58). DNA colocalization assays, such as 3C, have found that these CNS sites can colocalize with the promoter region of the IFN-γ gene in vivo (59, 60, and present work). It has also been found that the CNS sites, as well as the IFN-γ promoter region, contain conserved Tbox binding half sites, but not full palindromic sites, just as would be expected from our model. It will be important to determine how the N- and C-terminal domains of T-bet modulate its transcriptional activity, and to assess its interactions with other protein complexes known to facilitate enhancer–promoter association, such as the general transcription factor mediator (61).
The crystal structure and the various in vitro biochemical and biophysical data taken together strongly suggest that the DNA binding domain of T-bet dimerizes to bring distal elements together to modulate transcription. As discussed, in light of the small T-bet dependent “looping” effects observed in the in vivo 3C experiment in Th1 cells, a plausible physiological role of T-bet consistent with all of the data is that T-bet may function to stabilize and solidify existing loops performed by other “looping” transcription factors, such as CTCF (or other components of the transcriptional machinery) during T-cell differentiation. T-bet could also be forming new loops within larger T-bet–independent loops that we are not able to resolve with the 3C experiment. The resolution of these ideas will be the basis of future experiments.
In summary, our T-bet DBD–DNA crystal structure, fluorescence binding assays, and 3C assays all indicate that T-bet functions as a homodimer that activates the transcription of loci, such as the IFN-γ gene via a DNA looping mechanism, and differs from other Tbox transcription factors in its ability to bind to more than one recognition sequence at the same time. Such activity may underlie the particular potency of T-bet as a master regulator of gene expression in the immune system (2) and may be a more general characteristic of other transcription factors that are also master regulators of cell-fate determination, such as FOXP3 (62) and GATA3 (39), for which there are also crystallographic evidence for synapsing of DNA.
Materials and Methods
Construction, expression, and purification of the T-bet DBD were done using conventional cloning, expression, and purification methods. Purified T-bet DBD in complex with a 24-bp palindromic DNA were crystallized by the hanging-drop method. Crystallographic data were collected at the Advanced Photon Source of Argonne National Laboratory, and the final structure was solved by Molecular Replacement using a homology model generated from the crystal structure of the Xenopus laevis Brachyury T-domain (PDB ID code 1XBR). Size-exclusion chromatography of T-bet DBD was carried out using an YMC-Pack Diol-120 column on an Agilent HPLC system. Fluorescence DNA binding and kinetic experiments were carried out using fluorescently tagged DNA molecules. A traditional 3C assay was carried out as previously described (45, 63). Long-range DNA interactions induced by the T-bet DBD within the IFN-γ gene locus were also probed using a novel in vitro 3C assay. In this modified 3C assay, purified T-bet DBD and purified naked DNA containing the IFN-γ gene locus (BAC RP24-348011) were incubated together, and DNA interactions induced by the addition of T-bet DBD were detected as previously described in the original in vivo 3C assay (45, 63). Details of all experimental procedures can be found in SI Appendix, Supplemental Methods and Materials.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant P01-AI056296 (to L.H.G. and G.A.P.). G.S.B. was supported in part by National Research Service Award Individual Training Grant Fellowship 5F32GM069057. This work is based on research conducted at the GM/CA CAT (GM/CA@APS) and the Structural Biology Center of the Advanced Photon Source. GM/CA@APS has been funded in whole or in part with Federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 5T1J).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613914113/-/DCSupplemental.
References
- 1.Sundrud MS, Nolan MA. Synergistic and combinatorial control of T cell activation and differentiation by transcription factors. Curr Opin Immunol. 2010;22(3):286–292. doi: 10.1016/j.coi.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Glimcher LH. Trawling for treasure: Tales of T-bet. Nat Immunol. 2007;8(5):448–450. doi: 10.1038/ni0507-448. [DOI] [PubMed] [Google Scholar]
- 3.Avni O, et al. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002;3(7):643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 4.Finotto S, et al. Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science. 2002;295(5553):336–338. doi: 10.1126/science.1065544. [DOI] [PubMed] [Google Scholar]
- 5.Mullen AC, et al. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science. 2001;292(5523):1907–1910. doi: 10.1126/science.1059835. [DOI] [PubMed] [Google Scholar]
- 6.Szabo SJ, et al. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295(5553):338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 7.Szabo SJ, et al. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100(6):655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 8.Ravindran R, Foley J, Stoklasek T, Glimcher LH, McSorley SJ. Expression of T-bet by CD4 T cells is essential for resistance to Salmonella infection. J Immunol. 2005;175(7):4603–4610. doi: 10.4049/jimmunol.175.7.4603. [DOI] [PubMed] [Google Scholar]
- 9.Garrett WS, et al. Colitis-associated colorectal cancer driven by T-bet deficiency in dendritic cells. Cancer Cell. 2009;16(3):208–219. doi: 10.1016/j.ccr.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng SL, Townsend MJ, Hecht JL, White IA, Glimcher LH. T-bet regulates metastasis rate in a murine model of primary prostate cancer. Cancer Res. 2004;64(2):452–455. doi: 10.1158/0008-5472.can-03-3401. [DOI] [PubMed] [Google Scholar]
- 11.Werneck MB, Lugo-Villarino G, Hwang ES, Cantor H, Glimcher LH. T-bet plays a key role in NK-mediated control of melanoma metastatic disease. J Immunol. 2008;180(12):8004–8010. doi: 10.4049/jimmunol.180.12.8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng SL, Szabo SJ, Glimcher LH. T-bet regulates IgG class switching and pathogenic autoantibody production. Proc Natl Acad Sci USA. 2002;99(8):5545–5550. doi: 10.1073/pnas.082114899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joshi NS, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27(2):281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Intlekofer AM, et al. Requirement for T-bet in the aberrant differentiation of unhelped memory CD8+ T cells. J Exp Med. 2007;204(9):2015–2021. doi: 10.1084/jem.20070841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Intlekofer AM, et al. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science. 2008;321(5887):408–411. doi: 10.1126/science.1159806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Townsend MJ, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20(4):477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 17.Lugo-Villarino G, Ito S, Klinman DM, Glimcher LH. The adjuvant activity of CpG DNA requires T-bet expression in dendritic cells. Proc Natl Acad Sci USA. 2005;102(37):13248–13253. doi: 10.1073/pnas.0506638102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson V, Conlon FL. The Tbox family. Genome Biol. 2002;3:3008.3001–3008.3007. doi: 10.1186/gb-2002-3-6-reviews3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Müller CW, Herrmann BG. Crystallographic structure of the T domain-DNA complex of the Brachyury transcription factor. Nature. 1997;389(6653):884–888. doi: 10.1038/39929. [DOI] [PubMed] [Google Scholar]
- 20.Coll M, Seidman JG, Müller CW. Structure of the DNA-bound T-box domain of human TBX3, a transcription factor responsible for ulnar-mammary syndrome. Structure. 2002;10(3):343–356. doi: 10.1016/s0969-2126(02)00722-0. [DOI] [PubMed] [Google Scholar]
- 21.Stirnimann CU, Ptchelkine D, Grimm C, Müller CW. Structural basis of TBX5-DNA recognition: The T-box domain in its DNA-bound and -unbound form. J Mol Biol. 2010;400(1):71–81. doi: 10.1016/j.jmb.2010.04.052. [DOI] [PubMed] [Google Scholar]
- 22.El Omari K, et al. Structure of the DNA-bound T-box domain of human TBX1, a transcription factor associated with the DiGeorge syndrome. Proteins. 2012;80(2):655–660. doi: 10.1002/prot.23208. [DOI] [PubMed] [Google Scholar]
- 23.Papaioannou VE. The T-box gene family: Emerging roles in development, stem cells and cancer. Development. 2014;141(20):3819–3833. doi: 10.1242/dev.104471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatton RD, et al. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity. 2006;25(5):717–729. doi: 10.1016/j.immuni.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Cho JY, Grigura V, Murphy TL, Murphy K. Identification of cooperative monomeric Brachyury sites conferring T-bet responsiveness to the proximal IFN-gamma promoter. Int Immunol. 2003;15(10):1149–1160. doi: 10.1093/intimm/dxg113. [DOI] [PubMed] [Google Scholar]
- 26.Li Q, Barkess G, Qian H. Chromatin looping and the probability of transcription. Trends Genet. 2006;22(4):197–202. doi: 10.1016/j.tig.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Wang JC, Giaever GN. Action at a distance along a DNA. Science. 1988;240(4850):300–304. doi: 10.1126/science.3281259. [DOI] [PubMed] [Google Scholar]
- 28.Tuan D, Kong S, Hu K. Transcription of the hypersensitive site HS2 enhancer in erythroid cells. Proc Natl Acad Sci USA. 1992;89(23):11219–11223. doi: 10.1073/pnas.89.23.11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herendeen DR, Kassavetis GA, Geiduschek EP. A transcriptional enhancer whose function imposes a requirement that proteins track along DNA. Science. 1992;256(5061):1298–1303. doi: 10.1126/science.1598572. [DOI] [PubMed] [Google Scholar]
- 30.Blackwood EM, Kadonaga JT. Going the distance: A current view of enhancer action. Science. 1998;281(5373):60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 31.Bulger M, Groudine M. Looping versus linking: Toward a model for long-distance gene activation. Genes Dev. 1999;13(19):2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 32.de Laat W, Grosveld F. Spatial organization of gene expression: The active chromatin hub. Chromosome Res. 2003;11(5):447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- 33.Palstra RJ, de Laat W, Grosveld F. β-Globin regulation and long-range interactions. Adv Genet. 2008;61:107–142. doi: 10.1016/S0065-2660(07)00004-1. [DOI] [PubMed] [Google Scholar]
- 34.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416(6880):499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 35.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435(7042):637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 36.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31(13):3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janin J, Rodier F, Chakrabarti P, Bahadur RP. Macromolecular recognition in the Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 1):1–8. doi: 10.1107/S090744490603575X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin FF, et al. Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis. Nat Struct Mol Biol. 2009;16(5):499–508. doi: 10.1038/nsmb.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y, et al. DNA binding by GATA transcription factor suggests mechanisms of DNA looping and long-range gene regulation. Cell Reports. 2012;2(5):1197–1206. doi: 10.1016/j.celrep.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohlsson R, Bartkuhn M, Renkawitz R. CTCF shapes chromatin by multiple mechanisms: The impact of 20 years of CTCF research on understanding the workings of chromatin. Chromosoma. 2010;119(4):351–360. doi: 10.1007/s00412-010-0262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13(2):291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 42.Hou C, Zhao H, Tanimoto K, Dean A. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc Natl Acad Sci USA. 2008;105(51):20398–20403. doi: 10.1073/pnas.0808506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee BK, Iyer VR. Genome-wide studies of CCCTC-binding factor (CTCF) and cohesin provide insight into chromatin structure and regulation. J Biol Chem. 2012;287(37):30906–30913. doi: 10.1074/jbc.R111.324962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang AP, Pigli YZ, Rice PA. Structure of the LexA-DNA complex and implications for SOS box measurement. Nature. 2010;466(7308):883–886. doi: 10.1038/nature09200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295(5558):1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 46.Wilson CB, Schoenborn J. BACing up the interferon-gamma locus. Immunity. 2006;25(5):691–693. doi: 10.1016/j.immuni.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wijgerde M, Grosveld F, Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature. 1995;377(6546):209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]
- 48.Splinter E, et al. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes Dev. 2006;20(17):2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chakalova L, Debrand E, Mitchell JA, Osborne CS, Fraser P. Replication and transcription: Shaping the landscape of the genome. Nat Rev Genet. 2005;6(9):669–677. doi: 10.1038/nrg1673. [DOI] [PubMed] [Google Scholar]
- 50.Afkarian M, et al. T-bet is a STAT1-induced regulator of IL-12R expression in naïve CD4+ T cells. Nat Immunol. 2002;3(6):549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- 51.Jenner RG, et al. The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proc Natl Acad Sci USA. 2009;106(42):17876–17881. doi: 10.1073/pnas.0909357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balasubramani A, et al. Modular utilization of distal cis-regulatory elements controls Ifng gene expression in T cells activated by distinct stimuli. Immunity. 2010;33(1):35–47. doi: 10.1016/j.immuni.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaneko T, et al. Chromatin remodeling at the Th2 cytokine gene loci in human type 2 helper T cells. Mol Immunol. 2007;44(9):2249–2256. doi: 10.1016/j.molimm.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 54.Lee DU, Avni O, Chen L, Rao A. A distal enhancer in the interferon-gamma (IFN-gamma) locus revealed by genome sequence comparison. J Biol Chem. 2004;279(6):4802–4810. doi: 10.1074/jbc.M307904200. [DOI] [PubMed] [Google Scholar]
- 55.Schoenborn JR, et al. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8(7):732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shnyreva M, et al. Evolutionarily conserved sequence elements that positively regulate IFN-gamma expression in T cells. Proc Natl Acad Sci USA. 2004;101(34):12622–12627. doi: 10.1073/pnas.0400849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tong Y, Aune T, Boothby M. T-bet antagonizes mSin3a recruitment and transactivates a fully methylated IFN-gamma promoter via a conserved T-box half-site. Proc Natl Acad Sci USA. 2005;102(6):2034–2039. doi: 10.1073/pnas.0409510102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang Y, Ochando JC, Bromberg JS, Ding Y. Identification of a distant T-bet enhancer responsive to IL-12/Stat4 and IFNgamma/Stat1 signals. Blood. 2007;110(7):2494–2500. doi: 10.1182/blood-2006-11-058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rowell E, Merkenschlager M, Wilson CB. Long-range regulation of cytokine gene expression. Curr Opin Immunol. 2008;20(3):272–280. doi: 10.1016/j.coi.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5(10):1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 61.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30(5):235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 62.Bandukwala HS, et al. Structure of a domain-swapped FOXP3 dimer on DNA and its function in regulatory T cells. Immunity. 2011;34(4):479–491. doi: 10.1016/j.immuni.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miele A, Dekker J. Mapping cis- and trans- chromatin interaction networks using chromosome conformation capture (3C) Methods Mol Biol. 2009;464:105–121. doi: 10.1007/978-1-60327-461-6_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.