Abstract
Due to recent advances in high-resolution detection technology, multiple primary lung cancer (MPLC) is becoming an increasingly common diagnosis. However, the genotype-phenotype correlations in MPLC patients have not yet been assessed. In this study, we analyzed the clinical and pathological data for 129 consecutive MPLC patients who received curative surgery at the Tongji University Shanghai Pulmonary Hospital, China. We have screened 129 patients in the present study and found mutations in EGFR, BRAF, ROS1 and KRAS genes, as well as the rearrangement of the EML4-ALK gene in 113 patients. The mean patient age was 59.9 (25–78) years old and 41 patients were males (31.8%). Among the total patients, 123 (95.4%) had two primary lesions, 5 (3.9%) had three primary lesions, and 1 (0.8%) had four primary lesions. In 38.8% of the patients, all lesions were located on only one side of the body. Most of the detected mutations (98 patients) were in the EGFR gene. The patients exhibited significant differences in the EGFR mutation, age at diagnosis, and foci location.
Multiple primary lung cancer (MPLC) refers to the synchronous or metachronous appearance of more than one primary lung cancer in a single patient. MPLC was first reported in 19241. In recent years, the rate of diagnosis worldwide has rapidly increased due to the improvement in radiological diagnostic procedures such as high-resolution multi-slice spiral computed tomography (CT) and positron emission tomography (PET)2,3,4,5. However, preoperative diagnosis can be problematic because it is often difficult to differentiate MPLC from lung cancer metastasis based on radiological and histopathological criteria alone.
Identification of oncologic biomarkers by molecular genetic analysis may aid in the accurate diagnosis of MPLC. Potential oncologic biomarkers for analysis include allelic loss of heterozygosity (LOH) markers and specific gene mutations5,6,7,8,9,10,11,12. Recent advances in tumor molecular biology have resulted in the identification of several candidate markers such as P5313 and the EGFR gene14 that can be used for MPLC diagnosis. In addition, several genes have been confirmed to be driver genes in non-small cell lung cancer (NSCLC), including anaplastic lymphoma receptor tyrosine kinase (ALK), v-akt murine thymoma viral oncogene homolog 1 (AKT1), B-Raf proto-oncogene, serine/threonine kinase (BRAF), v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 (ERBB2), Kirsten rat sarcoma viral oncogene homolog (KRAS) and phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha (PIK3CA)15,16,17,18,19,20. However, current knowledge of the molecular characteristics of MPLC is insufficient to allow for validation of the accuracy and sensitivity of molecular diagnosis. In this study, we screened for mutations in several oncogenic driver genes in a cohort of Chinese MPLC patients and analyzed the data to correlate our genetic findings with the clinical phenotypes.
Materials and Methods
Ethical approval
This study was conducted in accordance with the amended Declaration of Helsinki, and it was approved by the Institutional Review Board (IRB) of Tongji University Shanghai Pulmonary Hospital, China. Written informed consent was obtained from all participants in this study. The methods were carried out in accordance with the approved guidelines.
Patients and specimen collection
From May 2011 to January 2015, 129 consecutive NSCLC patients with MLPC received surgical resection of their lung lesions at the Shanghai Pulmonary Hospital. Post-operative histological analyses confirmed the presence of primary lung cancer in all patients. Fresh primary tumor tissues containing more than 50% tumor cells were collected from all patients during surgery and were used for subsequent gene mutation analyses.
Demographic and clinical data were collected from the patient notes and computer records, including age, gender, the tumor size and location, the TNM stage and the histological type.
Candidate gene mutation analysis
Genomic DNA and the total RNA were immediately extracted from fresh tissues using a commercial QIAamp DNA Tissue Kit and RNeasy Kit (Qiagen, Germany), respectively. Mutations in the EGFR, BRAF, ROS1 and KRAS genes as well as the rearrangement of EML4-ALK were detected with Amoy Diagnostics kits (Xiamen, China) utilizing proprietary real-time PCR technology to detect mutations in the target genes.
Statistical analysis
Statistical analysis was performed using SPSS software (Version 18.0, Chicago, IL). The Chi-square tests and one-way ANOVA were used to detect associations between the clinical characteristics and studied genes. A P value of <0.05 was regarded as being significant.
Results
Demographic and Clinical characteristics
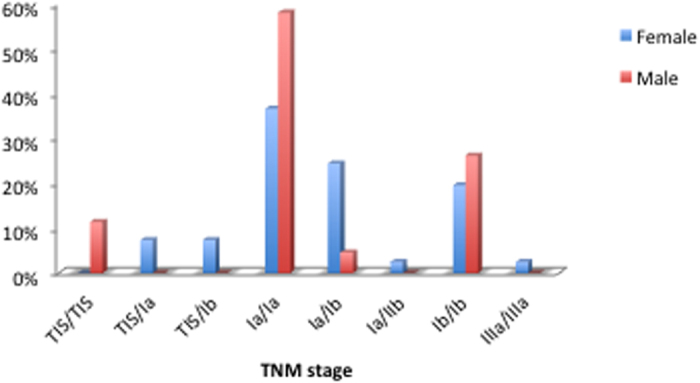
This cohort study included 41 male and 88 female patients,. The mean age was 59.9 (25–83) years old (Fig. 1). All the 5 youngest patients (<40 years) were females. A total of 299 foci were removed during the surgery. All lesions were detected simultaneously in each patient (Fig. 2). In total, 123 patients (95.35%) had two lesions, 5 patients (3.88%) had three lesions and 1 patient (0.78%) had four lesions. In 50 patients (38.8%), all the tumors were located on the same side. Most of the tumors were detected at an early stage, including 26 foci (8.70%) at the Tis stage (Fig. 3). Multiple lesions with an identical histological type were recorded in 88 patients, whereas multiple lesions with different histological types were detected in the other 41 patients.
Figure 1. The age distribution of the 129 MPLC patients analyzed.
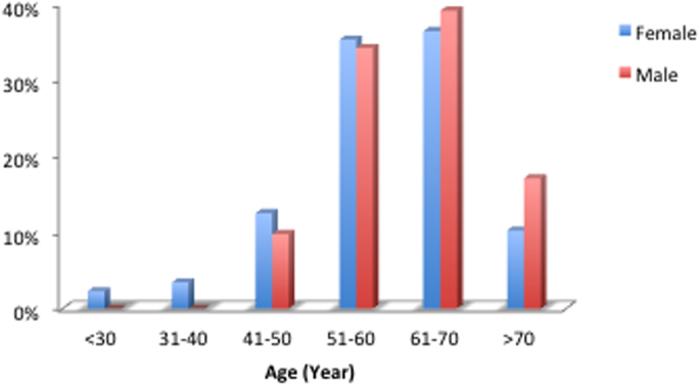
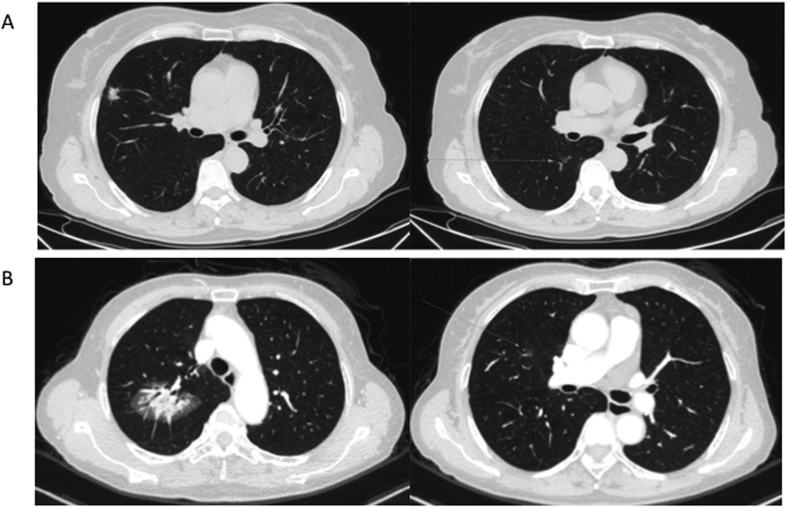
Figure 2. CT scan images for two MPLC patients.
(A) Images of Patient I. Left, CT scan of upper right lobi pulmonis; and right, CT scan of lower right lobi pulmonis. (B) Images of Patient II. Left, CT scan of upper right lobi pulmonis; and right, CT scan of middle right lobi pulmonis.
Figure 3. The TNM stages profile of these MPLC patients.
Mutation spectrum
Mutations in the EGFR, KRAS, BRAF and ROS1 genes and the rearrangement of EML4-ALK were detected in 113 patients (87.6%). Most of the patients (98) had EGFR mutations. No mutations were detected in 8 (19.5%) male and 8 (9.1%) female patients, and mutations in two genes were identified in 10 patients (Tables 1, 2, 3 and 4). A total of 12 subtypes of EGFR gene mutations were identified, including 6 types of complicated mutations. Complicated mutations (L858R/S768I, L858R/20ins, G719X/T790M, 19del/T790M, 19del/L858R and 19del/20ins) were only found in female patients, whereas the mutation spectrum of the male patients was relatively simple. Only exon 19del and L858R mutations were detected in the male patients (Tables 5 and 6).
Table 1. The distribution of gene mutations among 129 patients.
| Male | Female | |||
|---|---|---|---|---|
| EGFR | 24 | 58.54% | 64 | 72.73% |
| KRAS | 7 | 17.07% | 3 | 3.41% |
| BRAF | 0 | 0.00% | 3 | 3.41% |
| ROS1 | 0 | 0.00% | 1 | 1.14% |
| EML4-ALK | 1 | 2.44% | 0 | 0.00% |
| EGFR + KRAS | 1 | 2.44% | 2 | 2.27% |
| EGFR + EML4-ALK | 0 | 0.00% | 6 | 6.82% |
| EGFR + ROS1 | 0 | 0.00% | 1 | 1.14% |
| No mutation in the above genes | 8 | 19.51% | 8 | 9.09% |
Table 2. Details of somatic mutations in EGFR, BRAF and KRAS genes analyzed in this study.
| Gene | Mutation | Exon | Base Change |
|---|---|---|---|
| EGFR | |||
| G719A | 18 | 2156G > C | |
| G719S | 18 | 2155G > A | |
| G719C | 18 | 2155G > T | |
| E746_A750del (1) | 19 | 2235_2249del15 | |
| E746_A750del (2) | 19 | 2236_2250del15 | |
| L747_P753 > S | 19 | 2240_2257del18 | |
| E746_T751 > I | 19 | 2235_2252 > AAT(complex) | |
| E746_T751del | 19 | 2236_2253del18 | |
| E746_T751 > A | 19 | 2237_2251del15 | |
| E746_S752 > A | 19 | 2237_2254del18 | |
| E746_S752 > V | 19 | 2237_2250 > T(complex) | |
| E746_S752 > D | 19 | 2238_2255del18 | |
| L747_A750 > P | 19 | 2238_2248 > GC(complex) | |
| L747_T751 > Q | 19 | 2238_2252 > GCA(complex) | |
| L747_E749del | 19 | 2239_2247del9 | |
| L747_T751del | 19 | 2239_2253del15 | |
| L747_S752del | 19 | 2239_2256del18 | |
| L747_A750 > P | 19 | 2239_2248TTAAGAGAAG > C (complex) | |
| L747_P753 > Q | 19 | 2239_2258 > CA (complex) | |
| L747_T751 > S | 19 | 2240_2251del12 | |
| L747_T751del | 19 | 2240_2254del15 | |
| L747_T751 > P | 19 | 2239_2251 > C (complex) | |
| T790M | 20 | 2369C > T | |
| S768I | 20 | 2303G > T | |
| H773_V774insH | 20 | 2319_2320insCAC | |
| D770_N771insG | 20 | 2310_2311insGGT | |
| V769_D770insASV | 20 | 2307_2308insgccagcgtg | |
| L858R | 21 | 2573T > G | |
| L861Q | 21 | 2582T > A | |
| KRAS | |||
| Gly12Asp | GGT > GA T | ||
| Gly12Ala | GGT > GCT | ||
| Gly12Val | GGT > GTT | ||
| Gly12Ser | GGT > AGT | ||
| Gly12Arg | GGT > CGT | ||
| Gly12Cys | GGT > TGT | ||
| Gly13Asp | GGC > GAC | ||
| BRAF | |||
| V600E | 15 | 1799T > A | |
Table 3. EML4-ALK fusions detected in this study.
| Alternate name | EML4 spliced exon | Breakpoint variety (bp)* | ALK spliced exon |
|---|---|---|---|
| EML4-ALK variant 1 | 13 | −/−/ins69/−/−/− | 20 |
| EML4-ALK variant 3a/b | 6 | −/−/ins33/− | 20 |
| EML4-ALK variant 2 | 20 | −/−/ins18/− | 20 |
| EML4-ALK variant 4 | 15 | del71 | 20 |
| EML4-ALK variant 4′ | 14 | ins11del49/del12/del36 | 20 |
| EML4-ALK variant 5′ | 18 | — | 20 |
| EML4-ALK variant 5a/b | 2 | −/ins117 | 20 |
| / | 17 | ins68 | 20 |
*Differing breakpoints result in different cDNA isoforms: ins indicates insertion, and del indicates deletion.
Table 4. ROS1 gene fusions detected in this study.
| Alternate name | Fused gene | ROS1 spliced sites |
|---|---|---|
| ROS1-1 | SLC34A2 exon 4 | ROS1 exon 32 |
| SLC34A2 exon 4 | ROS1 exon 34 | |
| ROS1-2 | SLC34A2 exon 13 | ROS1 exon 32 |
| SLC34A2 exon 13 | ROS1 exon 34 | |
| ROS1-3 | CD74 exon 6 | ROS1 exon 32 |
| CD74 exon 6 | ROS1 exon 34 | |
| ROS1-4 | SDC exon 2 | ROS1 exon 32 |
| SDC exon 2 | ROS1 exon 34 | |
| ROS1-5 | SDC exon 4 | ROS1 exon 32 |
| SDC exon 4 | ROS1 exon 34 | |
| EZR exon 10 | ROS1 exon 34 | |
| ROS1-6 | TPM3 exon 8 | ROS1 exon 35 |
| LRIG3 exon 16 | ROS1 exon 35 | |
| ROS17 | FIG exon 8 | ROS1 exon 35 |
| FIG exon 4 | ROS1 exon 36 |
Table 5. The clinicopathological characteristics of the male MPLC patients.
| 19del + L858R | 19del + WT | L858R | L858R + WT | WT | |
|---|---|---|---|---|---|
| Age | |||||
| <30 | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% |
| <40 | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% |
| <50 | 20.00% | 33.33% | 0.00% | 0.00% | 12.50% |
| <60 | 20.00% | 33.33% | 40.00% | 57.14% | 25.00% |
| <70 | 40.00% | 33.33% | 40.00% | 42.86% | 37.50% |
| <80 | 0.00% | 0.00% | 10.00% | 0.00% | 25.00% |
| >80 | 20.00% | 0.00% | 10.00% | 0.00% | 0.00% |
| Site | |||||
| Left | 60.00% | 33.33% | 35.00% | 42.86% | 50.00% |
| Right | 40.00% | 66.67% | 65.00% | 57.14% | 50.00% |
| Pathology | |||||
| Lung adenocarcinoma | 90.00% | 66.67% | 95.00% | 85.71% | 81.25% |
| Squamous carcinoma | 10.00% | 0.00% | 0.00% | 0.00% | 15.63% |
| Large cell carcinoma | 0.00% | 0.00% | 0.00% | 7.14% | 3.13% |
| Atypical hyperplasia of adenoma | 0.00% | 33.33% | 5.00% | 7.14% | 0.00% |
| TNM stage | |||||
| Tis | 0.00% | 0.00% | 5.00% | 14.29% | 9.38% |
| Ia | 70.00% | 16.67% | 60.00% | 50.00% | 53.13% |
| Ib | 30.00% | 83.33% | 25.00% | 35.71% | 34.38% |
| IIb | 0.00% | 0.00% | 0.00% | 0.00% | 3.13% |
| IIIa | 0.00% | 0.00% | 10.00% | 0.00% | 0.00% |
Table 6. The clinicopathological characteristics of the female MPLC patients.
| 19del | 19del + L858R | 19del + rare mutation | 19del + WT | L858R + rare mutation | L858R | L858R + WT | G719X | 20ins + WT | WT | |
|---|---|---|---|---|---|---|---|---|---|---|
| Age | ||||||||||
| <30 | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 8.33% | 0.00% | 0.00% | 0.00% |
| <40 | 0.00% | 0.00% | 33.33% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 10.53% |
| <50 | 0.00% | 0.00% | 33.33% | 9.09% | 0.00% | 0.00% | 16.67% | 0.00% | 0.00% | 31.58% |
| <60 | 50.00% | 25.00% | 0.00% | 36.36% | 50.00% | 23.08% | 25.00% | 100.00% | 100.00% | 42.11% |
| <70 | 50.00% | 50.00% | 0.00% | 45.45% | 33.33% | 53.85% | 41.67% | 0.00% | 0.00% | 15.79% |
| <80 | 0.00% | 25.00% | 33.33% | 9.09% | 16.67% | 23.08% | 8.33% | 0.00% | 0.00% | 0.00% |
| Site | ||||||||||
| Left | 33.33% | 15.79% | 50.00% | 43.48% | 43.75% | 38.46% | 42.31% | 0.00% | 0.00% | 48.72% |
| Right | 66.67% | 84.21% | 50.00% | 56.52% | 56.25% | 61.54% | 57.69% | 100.00% | 100.00% | 51.28% |
| Pathology | ||||||||||
| Adenocarcinoma | 100.00% | 100.00% | 83.33% | 78.26% | 100.00% | 100.00% | 100.00% | 100.00% | 100.00% | 89.74% |
| Squamous carcinoma | 0.00% | 0.00% | 16.67% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% |
| Atypical hyperplasia of adenoma | 0.00% | 0.00% | 0.00% | 21.74% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 10.26% |
| TNM stage | ||||||||||
| Tis | 0.00% | 0.00% | 0.00% | 9.09% | 0.00% | 7.69% | 0.00% | 0.00% | 0.00% | 15.79% |
| Ia | 66.67% | 75.00% | 66.67% | 54.55% | 83.33% | 23.08% | 91.67% | 100.00% | 100.00% | 68.42% |
| Ib | 33.33% | 25.00% | 33.33% | 27.27% | 0.00% | 46.15% | 8.33% | 0.00% | 0.00% | 15.79% |
| Ia + Ib | 0.00% | 0.00% | 0.00% | 9.09% | 16.67% | 23.08% | 0.00% | 0.00% | 0.00% | 0.00% |
Phenotype-genotype correlation in MPLC patients
After determining the MPLC patient genotypes, we analyzed the phenotype-genotype correlations among these individuals. Significant differences were detected between the presence of EGFR mutations and the age at diagnosis and foci location (Table 7). Rare EGFR mutations (G719X, S768I, T790M and L861Q) were detected in 18 female patients. These rare mutations were accompanied by common mutations (L858R, exon 19del and exon 20ins) in multiple lesions. Among the lesions with L858R and a rare mutation, 6.25% were mucinous adenocarcinomas, whereas 62.5% were invasive adenocarcinomas. Further, 31.25% of these lesions were located in left upper lobe. Notably, none of the young patients had these rare mutations.
Table 7. Correlation between EGFR mutations and clinicopathological features.
|
EGFR mutation |
P value | ||
|---|---|---|---|
| Positive | Negative | ||
| Gender | 0.2035 | ||
| Male | 25 | 16 | |
| Female | 69 | 19 | |
| Age | 0.0236 | ||
| <60 | 46 | 22 | |
| ≥60 | 48 | 13 | |
| Location | 0.0456 | ||
| Left | 73 | 35 | |
| Right | 109 | 36 | |
| Pathology | 0.2781 | ||
| Adenocarcinoma | 170 | 66 | |
| Squamous carcinoma | 2 | 0 | |
| Large cell carcinoma | 9 | 4 | |
| Atypical hyperplasia of adenoma | 1 | 1 | |
| TNM stage | 0.0869 | ||
| <Ib | 112 | 52 | |
| ≥Ib | 70 | 18 | |
Discussion
The etiology of MPLC remains unclear. At present, the field cancerization theory is the most common explanation12. The accurate preoperative diagnosis of MPLC can be problematic. Hitherto, differentiating between MPLC and lung metastases in patient with multi-focal lung cancer has mainly depended on morphological methods. The diagnostic criteria for MPLC recommended by the American College of Chest Physicians (ACCP) are as follows: (1) the presence of anatomically separated tumors with the same histology, with the cancers in different lobes, no N2 or N3 involvement, and no systemic metastases; (2) the presence of temporally separated tumors with the same histology, with a 4-yr interval between cancers and no systemic metastases from any of the cancers; and (3) the presence of tumors with different histologies, including tumors with different histologic types and different molecular genetic characteristics or tumors arising separately from in situ carcinoma foci21,22,23.
In spite of these difficulties, it remains critical to distinguish between primary and metastatic lung cancer because the long-term survival of these two types of cancer is significantly different. Metastatic lung cancers, especially in patients with multiple metastases, should not be resected as the prognosis is extremely poor. On the other hand, most MPLC patients have separate lesions that are each individually at an early stage. The 5-year survival rate for synchronous MPLC after curative surgery could be as high as 75.8%24. Therefore, the proper diagnsis of MPLC is key to the appropriate treatment of patients.
Molecular genetic diagnosis was introduced into the diagnostic criteria for ACCP in 2013; however, the lack of effective molecular markers has limited its clinical application in practice. Hence, there is a pressing need for comprehensive phenotypic and genotypic analyses of MPLC patients.
Multiple factors can contribute to the development of MPLC, including genetic factors, smoking and exposure to environmental pollutants. Previous studies3 have shown that MPLC is more common in 50- to 70-year-old male smokers; most tumors in these patients have been reported to be squamous carcinoma-squamous carcinoma, squamous carcinoma-adenocarcinoma or adenocarcinoma-adenocarcinoma. In contrast with these data, our findings suggests that MPLC is more common in females, although they confirm that 50- to 70-year-old patients represent the highest-risk age group.
In our study, we screened for mutations in the EGFR, BRAF, ROS1 and KRAS genes, as well as the rearrangement of the EML4-ALK gene.
Since the direct sequencing of nucleotides is a time-consuming and labor-intensive process25,26, we used a real-time PCR-based method to analyze these mutations. Both previous studies and our experience suggested that a real-time PCR-based method would detect the mutations of interest with the same efficiency as direct sequencing27,28,29,30. Only 13.95% of the patients screened were found to be free from any mutations in the genes analyzed in the present study. The frequency of EGFR gene mutations (76.1% in the female patients and 61.0% in the male patients) was higher in our study than that reported in Chinese lung cancer patients with a single focus31,32. Furthermore, only 30 (32.6%) of the MPLC patients had identical gene mutations. These data suggest that more than half of second primary lung cancers result from different mechanisms compared with primary cancers. In addition, a previous study indicated that the risk of second primary lung cancers in patients treated with surgical resection for stage I NSCLC is 1.99 per 100 patient-years33. Based on these results, we conclude that each MPLC lesion should be independently diagnosed and treated by surgical resection.
Our results suggested that rare mutations in the EGFR gene contributed significantly to the diversity of mutations detected in female patients. Although these data provide evidence of genotype-phenotype correlations in MPLC patients, further study involving more patients will be needed to confirm these conclusions.
Additional Information
How to cite this article: Yang, Y. et al. Phenotype-genotype correlation in multiple primary lung cancer patients in China. Sci. Rep. 6, 36177; doi: 10.1038/srep36177 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
This work was supported by the National Natural Scientific Foundation of China (81370107; 81501750; 81602412) and Scientific research project of Shanghai Municipal Commission of Health and Family Planning (2016Y0121). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions Y.Y., W.C., W.H., W.Y., K.F. and G.J. designed the study. Y.Y., W.C., W.H., W.Y., C.J., X.Z., X.S. and J.Z. performed the study and analyzed the data. Y.Y., W.Y., W.C., K.F. and G.J. wrote the main manuscript. All authors reviewed the manuscript. W.C. and G.J. approved the manuscript.
References
- Beyreuther H. Multipicate von carcinomen bei einem fall von sog. Schenecberger Lungenkrebs mit tuberkulose. Virchows Arc. 250, 230–243 (1924). [Google Scholar]
- Okada M. et al. Operative approach for multiple primary lung carcinomas. J Thorac Cardiovasc Surg. 115, 836–840 (1998). [DOI] [PubMed] [Google Scholar]
- Fujita S. et al. Multiple Primary Malignancies in Patients with Non-Small Cell Lung Cancer. Intern Med. 54, 325–331 (2015). [DOI] [PubMed] [Google Scholar]
- Gogakos A. S. et al. Double primary non-small cell lung cancer with synchronous small cell lung cancer N2 nodes: a case report. Ann Transl Med. 3, 157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C. et al. “Different trend” in multiple primary lung cancer and intrapulmonary metastasis. Eur J Med Res. 20, 17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau D. H. et al. Clonal origin of multiple lung cancers: K-ras and p53 mutations determined by nonradioisotopic single-strand conformation polymorphism analysis. Diagn Mol Pathol. 6, 179–184 (1997). [DOI] [PubMed] [Google Scholar]
- Wang X. et al. Carcinogen exposure, p53 alteration, and K-ras mutation in synchronous multiple primary lung carcinoma. Cancer. 85, 1734–1739 (1999). [PubMed] [Google Scholar]
- Califano J. et al. Second esophageal tumors in patients with head and neck squamous cell carcinoma: an assessment of clonal relationships. Clin Cancer Res. 5, 1862–1867 (1999). [PubMed] [Google Scholar]
- Van Oijen M. G., Leppers V., Straat F. G., Tilanus M. G. & Slootweg P. J. The origins of multiple squamous cell carcinoma in the aerodigestive tract. Cancer. 88, 884–893 (2000). [DOI] [PubMed] [Google Scholar]
- Shimizu S. et al. High frequency of clonally related tumors in cases of multiple synchronous lung cancers as revealed by molecular diagnosis. Clin Cancer Res. 6, 3994–3999 (2000). [PubMed] [Google Scholar]
- van der Sijp J. R. et al. Determination of the molecular relationship between multiple tumors within one patient is of clinical importance. J Clin Oncol. 20, 1105–1114 (2002). [DOI] [PubMed] [Google Scholar]
- Pasic A. et al. Multiple suspicious lesions detected by autofluorescence bronchoscopy predict malignant development in the bronchial mucosa in high risk patients. Lung Cancer. 41, 295–301 (2003). [DOI] [PubMed] [Google Scholar]
- van Rens M. T. et al. Survival in synchronous vs single lung cancer: upstaging better reflects prognosis. Chest. 118, 952–958 (2000). [DOI] [PubMed] [Google Scholar]
- Hanahan D. & Weinberg R. A. The hallmarks of cancer. Cell. 100, 57–70 (2000). [DOI] [PubMed] [Google Scholar]
- Hammerman P. S. et al. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discov. 1, 78–89 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard D. A. et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 23, 5900–5909 (2005). [DOI] [PubMed] [Google Scholar]
- Jin G. et al. EML4-ALK fusion gene in Korean non-small cell lung cancer. J Korean Med Sci. 27, 228–230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K. et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 18, 378–381 (2012). [DOI] [PubMed] [Google Scholar]
- Fukui T. et al. Clinicoradiologic characteristics of patients with lung adenocarcinoma harboring EML4-ALK fusion oncogene. Lung Cancer. 77, 319–325 (2012). [DOI] [PubMed] [Google Scholar]
- Wang L. et al. PIK3CA Mutations Frequently Coexist with EGFR/KRAS Mutations in Non-Small Cell Lung Cancer and Suggest Poor Prognosis in EGFR/KRAS Wildtype Subgroup. PLoS ONE. 9, e88291(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozower B. D. et al. Special treatment issues in non-small cell lung cancer: diagnosis and management of lung cancer,3rd ed: American college of chest physicians evidence-based clinical practice guidelines. Chest 143, 369–399 (2013). [DOI] [PubMed] [Google Scholar]
- Shen K. R. et al. Special treatment issues in lung cancer: ACCP evidence-based clinical practice guidelines (2nd edition) Chest. 132, 290–305 (2007). [DOI] [PubMed] [Google Scholar]
- Detterbeck F. C. et al. Lung cancer special treatment issues. Chest. 123, 244–258 (2013). [DOI] [PubMed] [Google Scholar]
- Mun M. & Kohno T. Single-stage surgical treatment of synchronous bilateral multiple lung cancers. Ann Thorac Surg. 83, 1146–1151 (2007). [DOI] [PubMed] [Google Scholar]
- Kim H. J. et al. Detection and comparison of peptide nucleic acid-mediated real-time polymerase chain reaction clamping and direct gene sequencing for epidermal growth factor receptor mutations in patients with non-small cell lung cancer. Lung Cancer. 75, 321–325 (2012). [DOI] [PubMed] [Google Scholar]
- Pao W. & Ladanyi M. Epidermal growth factor receptor mutation testing in lung cancer: searching for the ideal method. Clin Cancer Res. 13, 4954–5495 (2007). [DOI] [PubMed] [Google Scholar]
- Wang R. et al. The use of quantitative real-time reverse transcriptase PCR for 5′ and 3′ portions of ALK transcripts to detect ALK rearrangements in lung cancers. Clin Cancer Res. 18, 4725–4732 (2012). [DOI] [PubMed] [Google Scholar]
- Tuononen K. et al. Comparison of targeted next-generation sequencing (NGS) and real-time PCR in the detection of EGFR, KRAS, and BRAF mutations on formalin-fixed, paraffin-embedded tumor material of non-small cell lung carcinoma-superiority of NGS. Genes Chromosomes Cancer. 52, 503–511 (2013). [DOI] [PubMed] [Google Scholar]
- Er T. K. et al. Increase EGFR Mutations Detection Rate in Lung Adenocarcinoma by Real-Time PCR Screening Followed by Direct Sequencing. Appl Immunohistochem Mol Morphol. 23, 343–348 (2015). [DOI] [PubMed] [Google Scholar]
- van Eijk R. et al. Rapid KRAS, EGFR, BRAF and PIK3CA mutation analysis of fine needle aspirates from non-small-cell lung cancer using allele-specific qPCR. PLoS One. 6, e17791 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. et al. Elderly male smokers with right lung tumors are viable candidates for KRAS mutation screening. Scientific reports. 6, 18566 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W. et al. Integration of molecular profiling into the lung cancer clinic. Clin Cancer Res. 15, 5317–5322 (2009). [DOI] [PubMed] [Google Scholar]
- Rice D. et al. The risk of second primary tumors after resection of stage I nonsmall cell lung cancer. Ann Thorac Surg. 76, 1001–1007 (2003). [DOI] [PubMed] [Google Scholar]