Abstract
Loss of function of the Phosphatase and tensin homologue deleted on chromosome 10 (PTEN) tumor suppressor gene is associated with many human cancers. In the cytoplasm, PTEN antagonizes the Phosphatidylinositol 3′ kinase (PI3K) signaling pathway. PTEN also accumulates in the nucleus, where its function remains poorly understood. We demonstrate that SUMOylation (SUMO) of PTEN controls its nuclear localization. In cells exposed to genotoxic stress, SUMO-PTEN was rapidly excluded from the nucleus dependent on the protein kinase Ataxia telangiectasia mutated (ATM). Cells lacking nuclear PTEN were hypersensitive to DNA damage, while PTEN-deficient cells were susceptible to killing by a combination of genotoxic stress and a small molecule PI3K inhibitor both in vitro and in vivo. Our findings may have implications for individualized therapy for patients with PTEN-deficient tumors.
Main text
PTEN (Phosphatase and Tensin Homolog on chromosome ten) is encoded by one of the most commonly deleted tumor suppressor genes in human cancer. PTEN acts as a 3′-specific phosphatidylinositol phosphatase, and counters the activity of the phosphatidylinositol 3′ kinase (PI3K) signaling pathway in the cytoplasm (1). The PTEN signaling network is implicated in the control of cell metabolism, growth, proliferation, survival and migration; processes invariably aberrant in cancer (2–6). PTEN may have roles in maintaining genomic stability, mediated, at least in part, by PI3K-independent mechanisms (7–10). PTEN is also found in the nuclei of many normal and cancerous cells and tissues and various molecular mechanisms of PTEN nuclear localization have been described (11).
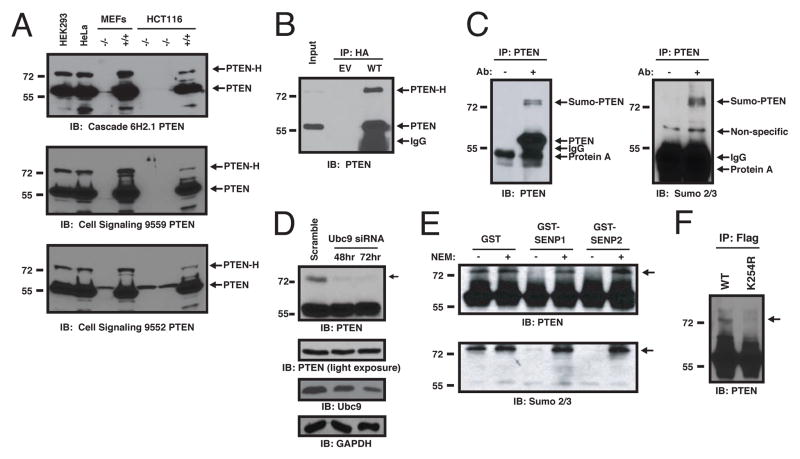
We noticed that, in addition to the expected PTEN protein of Mw ~55kDa, an ~75 kDa PTEN protein species could be detected with three PTEN antibodies in various mammalian cell lines (Fig. 1A). The ~75 kDa species, which we termed “PTEN-H,” represented < 10% of the total PTEN in the cell lines tested. PTEN-H was absent from mouse embryonic fibroblasts (MEFs) and human colon carcinoma HCT116 cells with a targeted disruption of the PTEN gene (Fig. 1A). Transfection of a vector expressing HA-epitope-tagged PTEN cDNA gave rise to PTEN and PTEN-H, establishing that PTEN-H arises as a consequence of post-translational modification, rather than alternative initiation or splicing (Fig. 1B). Use of mild detergents largely precluded detection of PTEN-H in cell lysates (Fig. S1A). Inclusion of N-ethylmaleamide (NEM), an inhibitor of cysteine-based enzymes, including deSUMOylases and deubiquitinases, during cell lysis, increased the detection of PTEN-H (Fig. S1B). Mono-SUMOylation could explain an apparent ~20 kD increase in molecular weight. Indeed, PTEN-H, but not “regular” PTEN reacted with the antibodies to SUMO 2 and SUMO 3 in PTEN immunoprecipitates from human embryonic kidney (HEK293) cells (Fig. 1C). Depletion of Ubc9, the sole SUMO-conjugating E2 protein (12), overexpression of the SENP1 or SENP2 deSUMOylases (Fig. 1D, S2), or treatment of PTEN immunoprecipitates with recombinant SENP1 or SENP2 decreased amounts of PTEN-H levels (Fig. 1E), further indicating that PTEN-H is a SUMOylated form of PTEN (SUMO-PTEN).
Fig. 1. SUMOylation of PTEN at lysine 254 in vivo.
(A) Multiple cell lines express a 75 kDa form of PTEN. Whole cell lysates were immunoblotted with 3 PTEN antibodies: 6H2.1 mouse mAb (top), rabbit mAb CST #9559 (middle), or a rabbit pAb (bottom) CST #9552. PTEN-H is indicated (arrow). (B) Transfection of minimal PTEN cDNA leads to PTEN-H formation. pcDNA3.1-HA-PTEN (WT) or pcDNA3.1-HA (EV) were transfected into HEK293 cells and HA-immunoprecipitates immunoblotted for PTEN. (C) SUMOylation of endogenous PTEN. PTEN was immunoprecipitated from HEK293 lysates with protein A beads alone or PTEN antibody (CST #9559) and immunoblotted with the PTEN 6H2.1 antibody (left) then stripped and re-probed with antibodies to Sumo2/3 (right). (D) Formation of SUMO-PTEN requires Ubc9. HEK293 cells were transfected with scrambled or Ubc9 siRNAs, and harvested at 48 (middle lane) or 72 (right lane) hrs post-transfection. Lysates were immunoblotted for PTEN, Ubc9 and GAPDH, as indicated. Arrow indicates SUMO-PTEN. (E) Sensitivity of PTEN-H to deSUMOylases. Immunoprecipitated Flag-PTEN was incubated with GST, GST-SENP1 or GST-SENP2, as indicated, in the presence or absence of 20 mM NEM. Reactions were immunoblotted with anti-PTEN 6H2.1 (top panel) or anti-Sumo2 (lower panel) antibodies. (F) PTEN is SUMOylated on lysine 254 in vivo. HEK293 cells were transfected with FLAG-PTEN-wt or FLAG-PTEN-K254R, together with His-SUMO2 and myc-His-Ubc9, as indicated. Flag immunoprecipitates were immunoblotted for PTEN.
In vitro SUMOylation of a series of PTEN deletion mutations (Fig. S3A) identified amino acids 238 to 320 as the minimal SUMOylated PTEN polypeptide (Fig. S3B, S3C). This PTEN portion contains a strong predicted SUMOylation site at position 254, a mutation of which precluded SUMOylation in vitro (Fig. S3C). Nanoflow liquid chromatography-tandem mass spectrometry (nLC-MS/MS) of SUMOylated PTEN polypeptides combined with SUMmOn pattern recognition software (13) independently identified K254 as a bona fide SUMOylation site (Fig. S4A, S4B). Consistently, wt but not Flag-PTEN K254R was readily SUMOylated in HEK293 cells (Fig. 1F) identifying K254 as the major PTEN SUMOylation site. The PTEN K254R mutant retained the ability to counter PI3K signaling as its expression in PTEN-deficient human U87MG glioblastoma cells (6) resulted in comparable decreases in phosphorylation of protein kinase B (PKB) (also called Akt) and of the PKB target GSK3β, as did expression of wt PTEN (Fig. S5A) and in vitro, displayed equal phosphatase activity towards PI(3,4,5)P3 (Fig. S5B).
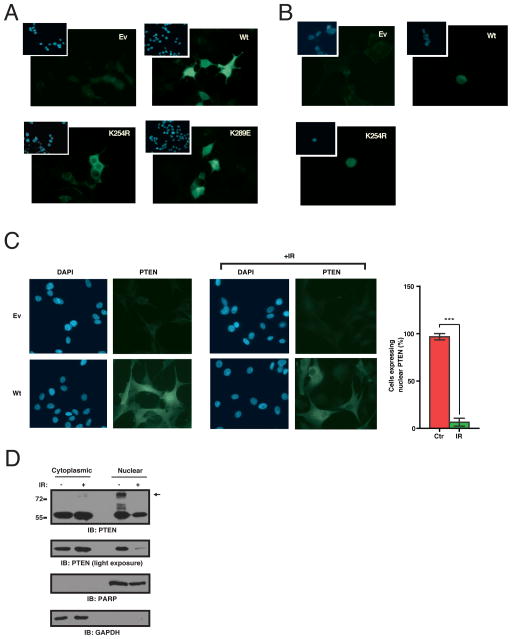
Unlike wt PTEN and an unrelated lysine PTEN K289E mutant, PTEN K254R expressed in HEK293 (Fig. 2A) or U87MG (Fig. S6) cells failed to localize to the nucleus, suggesting that SUMOylation might promote nuclear localization of PTEN. In cells treated with leptomycin B, an inhibitor of nuclear export, PTEN-K254R localized in the nucleus, indicating that this mutant can enter the nucleus but is not retained there (Fig. 2B). Treatment of cells with ionizing radiation (IR) led to loss of nuclear PTEN (Fig. 2C). Judging by fractionation of cellular lysates, SUMOylated PTEN was predominantly nuclear and reduced within hours of exposure to DNA damage (Fig. 2D).
Fig. 2. PTEN SUMOylation regulates nuclear retention, which is sensitive to genotoxic stress.
(A) Exclusion of the SUMO-deficient mutant PTEN K254R from the nucleus. Flag-FITC immunofluorescence images of HEK293 cells transfected as indicated. Insets show DAPI staining. (B) Nuclear retention of SUMOylated PTEN. Immunofluorescence as in (A). Cells were treated with 10 ng/ml of leptomycin B for 4 hours. (C) Decreased nuclear PTEN localization following genotoxic stress. Immunofluorescence as in (A) of PTEN-FITC U87MG cells transfected as indicated, 4 hours post-IR. Bar graph represents percentage of cells with nuclear PTEN in control or IR-treated cells (p<0.001, t test, n=5, bars represent SEM). (D) Decreased nuclear PTEN following genotoxic stress. HeLa cells were treated as indicated, harvested after 4 hours and separated into cytoplasmic and nuclear fractions followed by immunoblotting for PTEN. Fractionation was monitored by immunoblotting for PARP (nuclear protein) and GAPDH (cytoplasmic protein). Arrow indicates SUMO-PTEN.
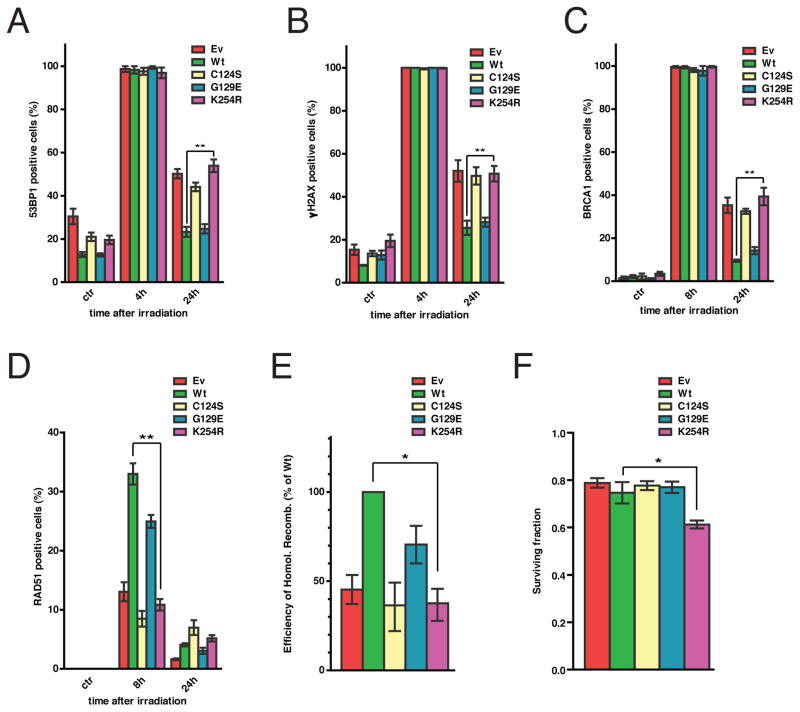
We investigated the DNA damage response (DDR) of U87MG cells, HCT116 cells, and their variants with the PTEN gene disrupted (HCT116 PTEN−/−) (14), engineered to express PTEN wt, K254R, a mutant lacking all phosphatase activity (C124S) or only lipid-phosphatase (G129E) activity, or empty vector control (Fig. S7A) by monitoring the formation of p53 binding protein 1 (53BP1) foci (15) (Fig. 3A, S7B, S8A). Four hours after IR exposure, 53BP1 foci were visible in all cell lines indicating the presence of double strand breaks (DSBs). However, by 24 hours, 53BP1 foci had largely resolved in wt PTEN- and G129E-reconstituted cells, but not in PTEN-deficient cells, cells lacking nuclear PTEN (K254R) or cells lacking all PTEN phosphatase activity (C124S) (Fig. 3A, S7B, S8A) indicative of deficient DNA repair. Consistently, PTEN-deficient cells or cells expressing PTEN K254R, or PTEN C124S, were also defective in resolving phosphorylated histone variant H2AX (γH2AX) and Breast cancer gene 1, early onset (BRCA1) foci (Fig. 3B & S7C, 3C & S8B). Indicative of deficiency in homologous recombination (HR)-based repair, PTEN-null cells, as well as their K254R- and C124S-expressing counterparts, failed to recruit RAD51 to the sites of DNA damage (Fig. 3D, S8C, S7D), without affecting RAD51 mRNA or protein abundance (Fig. S9A, S9B, S9C). Moreover, U87MG cells and cells derived from a mouse mammary tumor with a conditional PTEN gene disruption (WAP-Cre PTEN−/− MMTC) expressing PTEN-K254R or PTEN-C124S were deficient in repair of a stably integrated, HR-mediated DSB repair reporter (16) (Fig. 3E, S10). To distinguish the importance of nuclear localization versus SUMOylation for PTEN function in the response to DSBs, we fused a nuclear localization sequence from SV40 Large T antigen (17) to the non-SUMOylatable PTEN mutant (NLS-PTEN K254R). Despite constitutive localization to the nucleus (Fig. S11A), NLS-PTEN-K254R (Fig. S11B) did not restore the impaired response of U87MG cells to IR (Fig. S11C and D) indicating that SUMOylation is required for PTEN’s function in DDR. Reexpression of wt PTEN or various PTEN mutants did not yield changes in cell cycle distribution (Fig. S12A) or the engagement of cell cycle checkpoints following IR (Fig. S12B), indicating that DNA repair deficiency was not secondary to the potential effects of PTEN on the cell cycle. We monitored the effects of PTEN on radiosensitivity of cells by scoring the surviving fraction of U87MG cells and PTEN−/− MMTC expressing PTEN mutants 5 days after exposure to IR. While PTEN K254R-expressing cells exhibited decreased surviving fraction after IR exposure (Fig. 3F, S13), the survival of PTEN-null cells was indistinguishable from that of wt PTEN-expressing cells (Fig. 3F, S13), possibly reflecting the activation of PI3K-mediated cell survival signaling in cells lacking PTEN.
Fig. 3. Requirement of nuclear SUMO-PTEN for homologous recombination repair of DNA double-strand breaks.
U87MG cells (panels A to F) were reconstituted the indicated proteins. Cells treated with IR (5 Gy) were immunostained at the indicated times with antibodies to γH2AX, 53BP1, BRCA1 or RAD51. Cells containing >5 foci were scored as positive. Bars represent SEM. PTEN SUMOylation is required for the resolution of 53BP1 foci (p=0.0014, t test, n=3) (A), γH2AX foci (p=0.007, t test, n=3) (B) and BRCA1 foci (p=0.0136, t test, n=3) (C). (D) PTEN SUMOylation is required for RAD51 focus formation. (p=0.0016, t test, n=3). (E) Impaired homologous recombination-based DNA repair in PTEN- and SUMO-PTEN-deficient cells. U87MG cells stably expressing a DR-GFP reporter were reconstituted with the indicated PTEN proteins and their HR-mediated repair assessed as previously described (16). HR efficiency is expressed relative to that of wt. (p=0.012, t test, n=2). (F) Lack of SUMO-PTEN increases radiosensitivity. Surviving fraction was determined by Sulforhodamine B staining 6 days following exposure to 3 Gy of IR (p=0.02, t test, n=3).
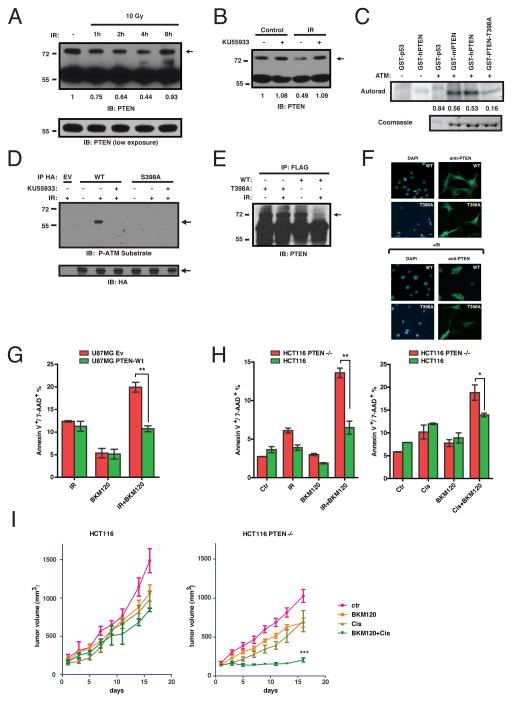
IR led to a gradual reduction in the amounts of SUMO-PTEN beginning 1 hour after IR exposure with the steady state amounts returning 8 hours later (Fig. 4A). Other forms of genotoxic stress, such as treatment with Cisplatin or Doxorubicin, also led to depletion of SUMO-PTEN, with the timing consistent with appearance of DNA damage elicited by these agents (Fig. S14). Protein kinases ATM and ATR phosphorylate multiple targets following DNA damage (15). Inhibition of ATM impaired SUMO-PTEN turnover in response to IR (Fig. 4B) while ATM immunoprecipitated from γ-irradiated cells phosphorylated both human and mouse GST-PTEN to a similar extent as it did p53, a known ATM substrate (Fig. 4C). PTEN contains a putative ATM phosphorylation site (18) at position 398. Mutation of this residue to alanine (T398A in human, S398A in mouse) decreased PTEN phosphorylation by ATM (Fig. 4C). PTEN was also phosphorylated at this site in vivo following IR in an ATM-dependent manner (Fig. 4D) establishing this residue as a likely ATM phosphorylation site within PTEN. Unlike wt SUMO-PTEN, SUMO-PTEN S/T398A was resistant to IR-induced turnover (Fig. 4E) and was not excluded from the nucleus in cells exposed to IR (Fig. 4F).
Fig. 4. PTEN is part of a ATM-regulated signaling cascade governing sensitivity to genotoxic agents.
(A) Decreased SUMO-PTEN after DNA damage. Lysates from HeLa cells treated as indicated were immunoblotted for PTEN. A ratio of SUMO-PTEN to PTEN immunoreactivity relative to the non-irradiated control is indicated. (B) ATM Regulation of SUMO-PTEN abundance by ATM. Lysates from HeLa cells treated as indicated were immunoblotted for PTEN and quantified as in (A). (C) ATM phosphorylates serine/threonine 398 of PTEN in vitro. The indicated GST-fusion proteins were incubated with active ATM in the presence of 32P-ATP. 32P incorporation was quantified by PhosphorImager and normalized to protein loading. (D) Phosphorylation of PTEN S398 in vivo by ATM. HA immunoprecipitates from HEK293 cells transfected and treated as indicated, were immunoblotted with antibody against phospho-ATM substrates (CST #9607). (E) SUMO-PTEN-T398A is not sensitive to IR. Flag immunoprecipitates from HEK293 cells transfected and treated as indicated were immunoblotted for PTEN. (F) PTEN-T398A remains in the nucleus after IR. Cells were treated and imaged as in Fig. 2C. (G-H) PTEN loss sensitizes cells to combination treatment with IR and a pan-PI3K inhibitor. Bars represent SEM. (G) U87MG cells reconstituted with empty vector or PTEN-wt were treated with either 2 Gy of IR, 500 nM BKM120 or both, and apoptosis was measured by annexin V+/7-AAD+ staining (p=0.0019, t test, n=3). (H) HCT116 PTEN-null and parental PTEN-WT cells were treated with either IR (2 Gy) or 1 mM cisplatin alone or in combination with 500 nM BKM120, as indicated. Apoptosis was measured as in (G) (IR+BKM120 p=0.0025, t test, n=3) (Cis+BKM120 p=0.0475, t test, n=3). (I) PTEN-deficient cells are sensitive to the Cisplatin/PI3K inhibitor combination in vivo. HCT116 parental or HCT116 PTEN−/− cells were injected subcutaneously into NOD/SCID mice (n=10). Mice were treated with cisplatin, BKM120, or both. Data points represent mean tumor volume ± SEM. Tumor growth difference was measured between day 1 and 16 and one way ANOVA was performed (p=0.0321 for HCT116 parental and p<0.0001 for HCT116 PTEN−/−) followed by Dunnet’s multiple comparison test (HCT116 PTEN−/− BKM120 + Cisplatin vs Ctr, p<0.0001).
We compared the sensitivity of U87MG cells reconstituted with either empty vector or wt PTEN, and the parental HCT116 and HCT116 PTEN−/− cells to BKM120, a small molecule pan-PI3K inhibitor, IR (2 Gy), or the combination thereof, at doses that produced minimal toxicity when administered alone (Fig. 4G, 4H, S15). PTEN-deficient U87MG cells displayed increased sensitivity to a combination of IR/BKM120, whereas a combination of BKM120 with the genotoxic agent cisplatin had an enhanced effect in HCT116 PTEN−/− cells (Fig. 4H). In vivo, immunocompromised mice carrying HCT116 and HCT116 PTEN−/− xenografts were treated with a single dose of cisplatin, daily BKM120 for 15 days, or the combination thereof (Fig. 4I). Although either drug alone had limited effect (<30% tumor growth inhibition (TGI) over the course of the treatment), the combination reduced the growth of HCT116 PTEN−/− xenografts (>90% TGI), but not their PTEN-proficient counterparts (Fig. 4I). Such synthetic sensitivity of PTEN-deficient cells to the combined action of DNA-damaging agents and PI3K pathway inhibitors might be useful in treating PTEN-deficient tumors (Fig S16). By contrast, our results suggest that administering genotoxic agents alone to such tumors could accelerate the acquisition of additional mutations (Fig S16). The ongoing clinical development of numerous agents countering activated PI3K signaling in cancer (19, 20) and next generation genotoxic agents (21) should facilitate testing of these concepts in the clinic.
Materials and methods
Antibodies
The following antibodies were purchased from Cell Signaling Technology: PTEN (138G6) (#9559), PTEN (#9552), SUMO-2/3 (18H8) (#4971), Ubc9 (#4918), PARP (#9542), Akt (#9272), phospho-Akt (Ser473) (193H12) (#4058), phospho-ATM substrate (#9607) and phospho-GSK-3β (Ser9) (#9336). Histone H3 (ab1971) and phospho-ATM (Ser1981) (ab2888) antibodies were from Abcam. FLAG M2-FITC (F4049), FLAG M2 (F3165) and FLAG M2 agarose (A2220) were from Sigma-Aldrich. GAPDH FL-335 (sc-25778), Rad51 H-92 (sc-8349) and BRCA1 I-20 (sc-646) antibodies were from Santa Cruz Biotechnology. PTEN antibody (6H2.1) (ABM-2052) was from Cascade Bioscience. Phospho-Histone H2A.X (Ser139) (JBW301) (05-636) was from Millipore. Antibody against 53BP1 (A300-272A) was from Bethyl Laboratories. ATM Ab-3 (PC116) was from Calbiochem. Anti-HA was produced in-house from the 12CA5 hybridoma.
Cell culture, transfections, viral infections and reagents
HEK293 cells, U87MG, HCT116 PTEN-null, parental PTEN wt cells and their derivatives were maintained in DMEM, supplemented with 10% FBS (Gibco) and Pen/Strep (100 mg/ml, Hyclone). Puromycin (1 ug/ml) was from Calbiochem. The mouse mammary tumor (MMTC) cell line was cultured in DMEM/F-12 (1:1), supplemented with 10% FBS (Gibco), Pen/Strep (100 mg/ml), insulin (5 ug/ml), hydrocortisone (1 ug/ml) (all from Sigma), and EGF (5 ng/ml, Peprotech).
Transient transfections of HEK293 cells were performed by using Lipofectamine (Invitrogen) or the CaCl2 and Hepes method, as described previously. High efficiency transfection (>90%) of U87MG cells for the homologous recombination assay was achieved using the Amaxa Cell Line Nucleofector Kit T (Lonza, VCA-1002), according to manufacturer’s instructions.
All plasmids used were constructed with the exception of plasmids encoding N-terminally His-tagged full-length SUMO1, SUMO2, and SUMO3, generously provided by Dr. Paul E. Fraser (University of Toronto), and pDR-GFP and pISceI (Addgene). For expression in mammalian cells, HA-PTEN was cloned into pcDNA3.1, and PTEN was cloned into p3XFLAG-CMV10 and pBabe-Flag. Ubc9 was cloned into pcDNA3.1-myc-His-A. Full-length SENP1 and SENP2 in pGEX4T3 were sub-cloned into pcDNA3.1-myc-His-A.
For expression of recombinant proteins in bacterial cells, full-length and truncation mutants of PTEN were cloned into pGEX4T3 and pET32a. Ubc9 and catalytic fragments of SENP1 and SENP2 were cloned into pGEX4T3. TP53 was cloned into pGEX2TK.
All point mutants were generated by using the QuickChange site-directed mutagenesis kit (Agilent), which also was used to insert the NLS derived from SV40 large T-antigen into p3XFLAG-CMV10-PTEN.
U87MG cells were engineered to express the ecotropic retroviral receptor by selecting a stable clone transfected with pWZL-Neo-EcoR. Viral transduction with retroviruses expressing FLAG-PTEN or mutants was performed in U87MG-EcoR and MMBC cells by following the procedures described previously (1). Cells were subsequently selected with puromycin (1 ug/ml) until negative control cells died (generally 3 days). Ubc9 siRNA was from Santa Cruz (sc-36773).
Cell lysis, immunoblotting and immunoprecipitations
Unless indicated otherwise, for immunoblotting cells were lysed in Laemmli sample buffer (60 mM Tris-Cl pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol), normalized for total protein content, resolved by SDS-PAGE, and transferred to PVDF membranes (Millipore). Membranes were blocked in 5% nonfat milk (Bioshop Canada) and probed with the indicated antibodies.
For immunoprecipitations, cells were lysed for 30 min on ice in RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1 mM EDTA), supplemented with fresh 20 mM N-ethylmaleimide (NEM), 1 mM dithiothreitol (DTT), 0.1 mM sodium orthovanadate and a protease inhibitor cocktail (Sigma). Insoluble material was removed by centrifugation at 15,000 × g for 15 min at 4 °C. Immunoprecipitations were performed by gentle rotation overnight at 4 °C, and then immune complexes were washed four times in cold RIPA buffer and resuspended in Laemmli buffer.
Immunoprecipitation of endogenous SUMO-PTEN was performed by lysing cells in Laemmli buffer, followed by sonication and centrifugation at 15,000 × g for 15 min at 4 °C. Clarified lysates were diluted 10-fold with PBS containing 1% Triton X-100 and the protease inhibitor cocktail, then anti-PTEN antibody was added and the mixture was allowed to rotate for 4 hours at 4 °C. Following the addition of protein A-Sepharose and rotation for 1 hr, immune complexes were washed 4 times with PBS containing 1% Triton x-100.
Immunofluorescence
Cells cultured on glass cover slips were rinsed in PBS, fixed with 3.7% formaldehyde in PBS for 10 min at room temperature, permeabilized with PBS plus 0.5% Triton X-100 for 5 min, blocked overnight at 4°C with PBS containing 1% bovine serum albumin (Fisher), and then incubated with primary antibodies (PTEN Cell signaling 9559, 1:200; RAD51 1:500; 53BP1 1:500; γH2AX 1:1000; BRCA1 1:100; FLAG-FITC 1:1000). After 3 washes with PBS, 5 min each, at room temperature, samples were incubated for 30 min with a 1:400 dilution (in PBS) of goat anti-rabbit IgG conjugated to the fluorescent Alexa 488 dye or with goat anti-mouse IgG conjugated to the fluorescent Alexa 546 dye (Invitrogen Molecular Probes), washed three times, stained with DAPI and mounted in Mowiol.
Cell proliferation assays
Cell number was assessed indirectly by using the Sulforhodamine B (SRB) assay (2).
Phosphorylation reactions
Cells were lysed in ATM buffer (20 mM HEPES, pH 7.4, 150 mM NaCl, 0.2% Tween 20, 1.5 mM MgCl2, 1 mM EGTA, 2 mM dithiothreitol, 50 mM NaF, 500 μM NaVO4, 1 mM phenylmethylsulfonyl fluoride, 0.1 μg/ml aprotinin, 0.1 μg/ml leupeptin), cleared by centrifugation, and subjected to immunoprecipitations with PC116 ATM antibody (Calbiochem). ATM immunoprecipitates were incubated with recombinant p53 or PTEN in kinase buffer (10 mM HEPES, pH 7.4; 10 mM MgCl2; 50 mM NaCl; 10 mM MnCl2), supplemented with 50 μM ATP and 10 μCi [γ-32P]ATP, for 15 min at 30°C (3).
Homologous recombination assays
To assess HR efficiency, U87MG cells were stably transduced with DR-GFP and various PTEN constructs, and subsequently transfected with pCMV3xnlsI-SceI, using Amaxa™ Nucleofector™ Technology (Lonza). At 2 days post-transfection, GFP signals were quantified by using a FACSCalibur flow cytometer (BD Biosciences). Recombination efficiency was calculated as the number of GFP-positive cells in the samples divided by the number of GFP-positive cells in WT-PTEN-reconstituted U87MG cells. For each experiment, 10,000 cells were scored per treatment group.
SUMOylation
Assays were performed with 150 ng of SAEI and SAEII (Boston Biochem), 1 μg of UBC9, 2 μl of 10× SUMOylation reaction buffer (200 mM HEPES, pH 7.5, 50 mM MgCl2 and 20 mM ATP), 1 μg of SUMO1 (Boston Biochem) and 300 ng of recombinant His-THX-PTEN. Reactions were incubated at 37°C for 2 hrs, quenched with SDS–PAGE sample buffer and analysed by SDS–PAGE and immunoblotting with anti-His antibody.
Lipid phosphatase assays
PTEN catalytic activity was determined by using the malachite green PTEN phosphatase assay kit, using PI(3,4,5)P3 diC8 as the substrate (Echelon Bioscence).
In vitro deSUMOylation
FLAG-PTEN immunoprecipitates were incubated in TBS with 1 ug of recombinant SENP catalytic fragments, expressed as His-fusion proteins, for 1 hr at 37 °C. Reactions were terminated by boiling in Laemmli loading buffer, and samples were analysed by SDS-PAGE and immunoblotting with anti-SUMO-2/3 antibodies.
RNA isolation, reverse transcription PCR, and real-time quantitative PCR
Total RNA was isolated by using RNeasy (QIAGEN) and treated with DNase I (Roche Diagnostics). Reverse transcription PCR (RT-PCR) was performed using the TaqMan Reverse Transcription Kit (Applied Biosystems). Quantitative real-time PCR analysis of RAD51 was performed using primers specific for human RAD51: hRAD51 F1 5′-CGTAAGCCAGGGGCGTTGGG-3′; hRAD51 R1 5′-TGCCATTACTCGGTCCGCAGC-3′.
Cell cycle analysis
Cell cycle analysis were carried out by flow cytometry. Briefly, U87MG cells were seeded into 6-well culture plates, treated as indicated, collected, fixed, stained with propidium iodide (100 μg/mL) and RNAse (20 μg/mL) in PBS for 1 hour, and analyzed on a FACScalibur flow cytometer instrument (BD Biosciences) and CellQuest software (BD Immunocytometry Systems, San Jose, CA). Similarly, indirect immunofluorescence on ethanol-fixed U87 cells (70% in PBS) was performed to quantify mitotic cells using an anti-phospho-H3 (Ser10) specific antibody (CellSignaling) detected by an Alexa488-labelled goat anti-rabbit secondary antibody (Invitrogen). Cells were analyzed using a FACScalibur instrument (BD Biosciences) and CellQuest software (BD Immunocytometry Systems, San Jose, CA).
Ten thousand events were analyzed for each sample.
Annexin V/7-AAD assay for apoptosis
For Annexin V/7-AAD assays, cells were stained with Annexin V–FITC and 7-AAD, and evaluated for apoptosis by flow cytometry according to the manufacturer’s protocol (BD PharMingen, San Diego, CA, USA). Briefly, 1 × 105 cells were washed twice with phosphate-buffered saline (PBS), and stained with 5 μl of Annexin V–FITC and 10 μl of &-AAD (5 μg/ml) in 1X binding buffer (10 mM HEPES, pH 7.4, 140 mM NaOH, 2.5 mM CaCl2) for 15 min at room temperature in the dark. The apoptotic cells were determined using a FACSCalibur flow cytometer (BD Biosciences).
In vivo studies
Compound preparation
NVP-BKM120 and AZD6244 were formulated in NMP/PEG300 (10/90, v/v). Solutions were freshly prepared for each day of dosing by dissolving the powder, first in N-Methyl-2-pyrrolidone (NMP) with sonication and then by adding the remaining volume of PEG300. The application volume was 10 mL/kg. Cisplatin treatment (6 mg/kg body weight) by intraperitoneal injections was given as a single dose.
Xenograft studies
Female nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice (6–10 weeks of age) were kept under sterile conditions (type III cage, in an Optimal Hygienic Conditions zone) with free access to food and water. Subcutaneous tumors were established by injection of 100 μL of PBS containing 2 × 106 tumor cells (HCT116 PTEN-null and isogenic PTEN-WT), in left flank of each animal. Treatments were initiated when the mean tumor volume in each randomized group (n = 8–10) reached 150 to 200 mm3. For BKM120, treatments were carried out orally, once per day, using an application volume of 10 mL/kg. Treatments were stopped and animals sacrificed when the tumor size in the vehicle control group reached 1,000 to 1,200 mm3. Tumor volumes were determined using calipers for measurement of longest (considered as length) and shortest (considered as diameter) dimensions of each tumor and calculated according to the modified ellipsoid formula (length × diameter2)/2. Data are presented as means ± one SEM. Comparisons between groups and vehicle control group were done using one-way ANOVA followed by the Dunnett tests. For all tests, the level of significance was set at P < 0.05. Calculations were carried out with GraphPad Prism. All experimental procedures strictly adhered to the Canadian Council on Animal Care guidelines.
Supplementary Material
Acknowledgments
We thank Drs. P. Fraser for reagents, D. Durocher for advice on DNA repair assays, R. Hakem for critical reading of the manuscript and Mr. A. Wakeham for advice on xenograft experiments, and Novartis for BKM120. Supported by the grant from the Canadian Cancer Society to VS (2011-700891), National Institutes of Health to BN (R37 CA49152) and partially supported by a grant from the Ontario Ministry of Health and Long Term Care and the Princess Margaret Hospital Foundation. BN and TWM hold Canada Research Chairs (Tier I) and BR holds Canada Research Chair (Tier II). CB was supported by the Excellence in Radiation Research for the 21st century (EIRR21st) fellowship and RJOD was supported by a Banting Postdoctoral Fellowship from the CIHR.
Footnotes
Contributions: CB and VS designed research, CB, JH, TS, RD, CG and SM performed research, CB, TS, BN, BR and VS analyzed data, TM, BN, BR and VS supervised research and CB, BN and VS wrote the paper.
References
- 1.Maehama T, Dixon JE. J Biol Chem. 1998;273:13375. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 2.Cairns P, et al. Cancer Res. 1997;57:4997. [PubMed] [Google Scholar]
- 3.Guldberg P, et al. Cancer Res. 1997;57:3660. [PubMed] [Google Scholar]
- 4.Ahmed NN, Grimes HL, Bellacosa A, Chan TO, Tsichlis PN. Proc Natl Acad Sci U S A. 1997;94:3627. doi: 10.1073/pnas.94.8.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasheed BK, et al. Cancer Res. 1997;57:4187. [PubMed] [Google Scholar]
- 6.Steck PA, et al. Nat Genet. 1997;15:356. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 7.Puc J, et al. Cancer Cell. 2005;7:193. doi: 10.1016/j.ccr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Fraser M, et al. Clin Cancer Res. 2012;18:1015. doi: 10.1158/1078-0432.CCR-11-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen WH, et al. Cell. 2007;128:157. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 10.Trotman LC, et al. Cell. 2007;128:141. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Planchon SM, Waite KA, Eng C. J Cell Sci. 2008;121:249. doi: 10.1242/jcs.022459. [DOI] [PubMed] [Google Scholar]
- 12.Anckar J, Sistonen L. Biochem Soc Trans. 2007;35:1409. doi: 10.1042/BST0351409. [DOI] [PubMed] [Google Scholar]
- 13.Pedrioli PG, et al. Nat Methods. 2006;3:533. doi: 10.1038/nmeth891. [DOI] [PubMed] [Google Scholar]
- 14.Samuels Y, et al. Cancer Cell. 2005;7:561. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 15.Thompson LH. Mutat Res. 2012 [Google Scholar]
- 16.Elliott B, Jasin M. Mol Cell Biol. 2001;21:2671. doi: 10.1128/MCB.21.8.2671-2682.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalderon D, Richardson WD, Markham AF, Smith AE. Nature. 1984;311:33. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 18.Matsuoka S, et al. Science. 2007;316:1160. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 19.Wong KK, Engelman JA, Cantley LC. Curr Opin Genet Dev. 2010;20:87. doi: 10.1016/j.gde.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu P, Cheng H, Roberts TM, Zhao JJ. Nat Rev Drug Discov. 2009;8:627. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lord CJ, Ashworth A. Nature. 2012;481:287. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.