Abstract
Class I-like CD1 molecules are in a family of antigen-presenting molecules that bind lipids and lipopeptides, rather than peptides for immune surveillance by T cells. Since CD1 lacks the high degree of polymorphism found in their major histocompatibility complex (MHC) class I molecules, different species express different numbers of CD1 isotypes, likely to be able to present structurally diverse classes of lipid antigens. In this review, we will present a historical overview of the structures of the different human CD1 isotypes and also discuss species-specific adaptations of the lipid-binding groove. We will discuss how single amino acid changes alter the shape and volume of the CD1 binding groove, how these minor changes can give rise to different numbers of binding pockets, and how these pockets affect the lipid repertoire that can be presented by any given CD1 protein. We will compare the structures of various lipid antigens and finally, we will discuss recognition of CD1-presented lipid antigens by antigen receptors on T cells (TCRs).
Keywords: CD1 isoforms, TCR recognition, Lipid antigens, Lipid-binding pockets
Introduction
Genes of the human CD1 family (CD1A-E) were first cloned by Calabi and Milstein in 1986 (Calabi et al. 1989; Calabi and Milstein 1986). They encode a family of antigen-presenting molecules that are structurally related to the peptide-presenting major histocompatibility complex (MHC) class I molecules. Yet, rather than presenting peptides, CD1 proteins present an array of different classes of lipids and glycolipids to T cells for immune surveillance (Adams and Luoma 2013; Bendelac et al. 2007; Girardi and Zajonc 2012; Mori et al. 2016; Rossjohn et al. 2015). Most CD1-restricted T cells express an antigen receptor (TCR) composed of an αβ heterodimer (Brigl and Brenner 2004), but T cells expressing an γδ (Luoma et al. 2013; Uldrich et al. 2013) or δ/αβ TCR (Pellicci et al. 2014) have also been reported. Since the discovery of the human CD1 locus, CD1 has been found in many more species and each species expresses different numbers of CD1 genes (see also Reinink and Van Rhijn 2016).
Human CD1 proteins are categorized into group 1 (CD1a–c), group 2 (CD1d), and group 3 (CD1e). CD1e is the only isotype that does not directly present lipid antigens to T cells but instead participates in lipid processing and subsequent loading onto other CD1 family members (Angenieux et al. 2005; Angenieux et al. 2000; de la Salle et al. 2005; Facciotti et al. 2011). Mice express two highly conserved pseudoalleles of CD1d (CD1d1 and CD1d2), suggesting a recent gene duplication event, and while mouse CD1 transcripts are expressed equally well in the thymus, CD1d1 is generally expressed at tenfold higher levels in other organs (Bradbury et al. 1988).
In addition to mammals, CD1 has also been identified in birds and reptiles, including lizards but not in amphibians (Maruoka et al. 2005; Miller et al. 2005; Salomonsen et al. 2005; Yang et al. 2015). Guinea pigs and cows express multiple isoforms of CD1b (Dascher et al. 1999; Van Rhijn et al. 2006). The functional consequence of expressing multiple isoforms is not always known and whether these isoforms can compensate for the lack of other isoforms, or whether they were shaped by the lipid ligand repertoire of these organisms, is not clear. While functional CD1d is found in cattle, guinea pigs, for example, express CD1a, CD1b, CD1c, and CD1e orthologs but not CD1d (Dascher et al. 1999; Nguyen et al. 2012). Mammalian CD1 molecules are widely expressed as glycosylated proteins on many immune cells, including myeloid dendritic cells, thymocytes, B cells, and Langerhans cells (Brigl and Brenner 2004).
CD1 protein structure and trafficking
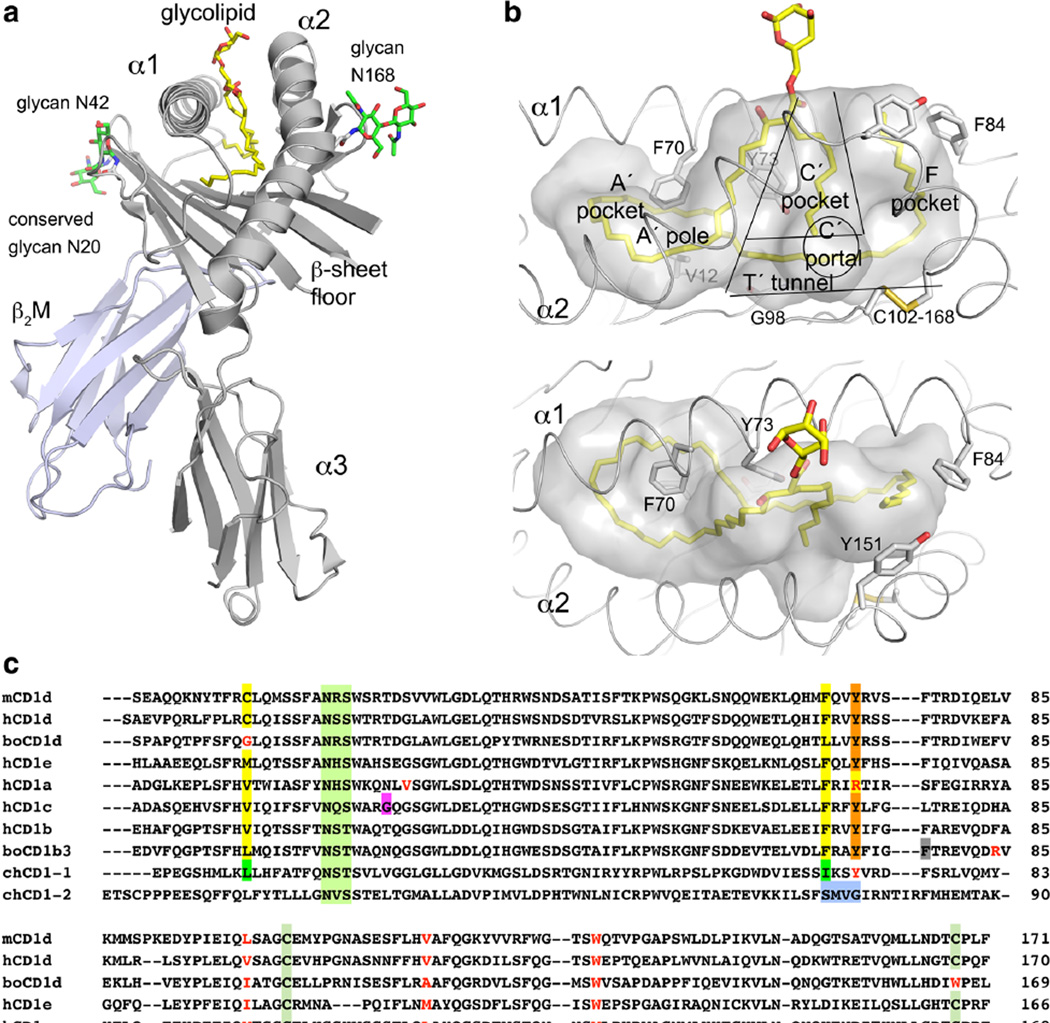
Similar to MHC I, the CD1 heavy chain non-covalently associates with β2-microbglobulin (β2M) to form a heterodimer of about 45–50 kDa molecular weight, including N-linked carbohydrates (Fig. 1). The CD1 heavy chain is organized into three domains, namely α1, α2, and α3, and is anchored to the cell membrane by a transmembrane domain. While the α1 and α2 domains combine to form the central binding groove of CD1 and are more divergent among the various isotypes, the α3 domain is more conserved as it associates with β2M (Fig. 1a). Like classical class I molecules, the central binding groove of CD1 is formed by two antiparallel α-helices (α1, α2), which sit on top of a six-stranded β-sheet platform. The binding groove of all mammalian CD1 isotypes contains at least the two major binding pockets, A′ and F′, while human CD1b has the most elaborate pocket architecture with an additional T′ tunnel that connects the A′ and F′ pockets just above the β-sheet floor, as well as a C′ portal that opens underneath the α2 helix for lipid tail egress into the solvent (Fig. 1b) (Moody et al. 2005). As a consequence, the CD1b binding groove had originally been coined “a maze for alkyl chains” (Gadola et al. 2002). A unique feature of the CD1c binding groove is the D′/E′ portal, which is analogous to the C′ portal of CD1b and provides an exit portal underneath the α1 helix at the terminus of the A′ pocket for possible egress of the antigen into the solvent (Scharf et al. 2010). All mammalian CD1 isotypes have a conserved A′ pole (Val/Cys/Met12 and Phe70) that allows the alkyl chain to circle within the A′ pocket either fully (CD1b, c, d, and e) or partially (CD1a). An excellent depiction of all the human CD1 binding grooves is found in Garcia-Alles et al. (2011b). A sequence alignment of the crystallized CD1 proteins illustrates the CD1 isoform and species-specific adaptations of CD1 binding grooves that will be discussed later in more detail (Fig. 1c).
Fig. 1.
CD1 structure overview and binding groove architecture. a Cartoon representation of the mouse CD1d-BbGl-2c structure. CD1d heavy chain in gray, b2M in blue gray. N-linked glycans in green and lipid in yellow. b Lipid-binding groove of human CD1b in gray space filling view with bound GMM lipid in yellow in a side view (top) and top view, looking into the groove (bottom). Individual pockets (A′-T′) and residues that delineate pockets are labeled. c Sequence alignment of all crystallized CD1 proteins. Mammalian CD1 shares many features, while avian CD1 is distinct in amino acid sequence and structure. Conserved features are highlighted yellow (A′ pole, green in chCD1-1), light green (conserved N-linked glycan), green (T′ tunnel in CD1b), dark green (disulfide bond in A′ pocket), cyan (disulfide bond at C′ portal), blue (A′ loop), and orange (acyl chain guide into A′ pocket), while residues in red block a particular feature
Although the overall three-dimensional structure among the CD1 family is very similar, amino acid substitutions in the α1-α2 superdomain are responsible for shaping the individual grooves and for the formation of isotype-specific pockets. These pockets or tunnels are very hydrophobic and reach deep inside the protein. As a result, the lipid backbone of all CD1 antigens is usually almost completely buried inside the CD1 protein and shielded from the surrounding solvent (Fig. 1a). The different carbohydrate or peptide headgroups are exposed at the CD1 surface and can either directly serve as the major T cell epitopes or affect the structure of the TCR binding site by an induced fit mechanism to modulate TCR recognition without direct antigen contact (Birkinshaw et al. 2015; Girardi and Zajonc 2012; Rossjohn et al. 2015).
A tyrosine-based sorting motif is encoded on the short cytoplasmic tail of CD1b, CD1c, and CD1d to which various adaptor proteins (AP1-3) bind but is absent in CD1a (Sugita et al. 2004). As a result, CD1b–d are sorted differentially into late endosomal and lysosomal compartments (Jackman et al. 1998; Moody and Porcelli 2003; Sugita et al. 2004; Sugita et al. 1999), while CD1a mainly recycles through early endosomes (Salamero et al. 2001). While CD1a is S-palmitoylated on its short intracellular tail (RKRCFC), possibly impacting its intracellular trafficking and association with detergent resistant membrane microdomains (lipid rafts), no effect of palmitoylation on CD1a trafficking had been observed (Barral et al. 2008). The ability of the CD1 family members to traverse different endosomal compartments enables each type of CD1 molecule access to unique lipids, since based on their particular structure, lipids associate and partition into distinct cellular compartments (Maxfield and Hao 2013).
This differential sampling of the various intracellular compartments allows CD1 to effectively monitor the lipid content of antigen presenting cells. In addition to the isotype-specific adaptation of the individual CD1 binding grooves, the colocalization of certain lipids with particular CD1 isotypes is a key factor in determining which antigens can be presented by each of the CD1 molecules in vivo.
Lipid antigens and CD1 loading
The CD1 family can bind and display an array of structurally diverse lipids, ranging from monoacylated lipids or lipopeptides to tetra-acylated lipids. In addition to the number of acyl chains contained in an antigen, the chain length as well as natural alkyl chain substitutions (methylation, hydroxylation, cyclization) can differ greatly. While several unique mycobacterial antigens, including mycolates (Moody et al. 1997), lipoglycans (Ernst et al. 1998; Fischer et al. 2004; Sieling et al. 1995), diacylated sulfoglycolipids (Gilleron et al. 2004), lipopeptides (Moody et al. 2004; Van Rhijn et al. 2005), and phosphomycoketides (Matsunaga et al. 2004; Moody et al. 2000), can only be presented by certain CD1 isotypes, such as CD1b and CD1c (phosphomycoketide), other more common lipids, such as self and foreign glycosphingolipids (Goff et al. 2004; Jahng et al. 2004; Kain et al. 2014; Kawano et al. 1997; Kinjo et al. 2005; Mattner et al. 2005; Miyamoto et al. 2001; Schmieg et al. 2003; Shamshiev et al. 1999; Shamshiev et al. 2002; Wu et al. 2005; Wu et al. 2003; Yu et al. 2005; Zhou et al. 2004b) and phosphoglycerolipids (Agea et al. 2005; Gumperz et al. 2000; Joyce et al. 1998; Rauch et al. 2003), can be presented by most CD1 isoforms, and specificity of recognition is determined by the CD1-restricted T cell. An overview of CD1-presented glycolipids is shown in Fig. 2. It is important to note that while many lipids can be ligands for CD1, they are not necessarily T cell antigens. Many self-antigens associate with CD1 during de novo expression to stabilize the CD1 protein on its way to the cell surface. CD1-bound lipids can be replaced either at the cell surface or during CD1 trafficking through endosomal compartments. While the focus of this article is not on the details of lipid loading, many lipid transfer proteins are involved in this process, including the GM2 activator protein (Zhou et al. 2004a), saposins (Kang and Cresswell 2004; Winau et al. 2004; Zhou et al. 2004a), apolipoprotein E (van den Elzen et al. 2005), microsomal triglyceride tranfer protein (MTP) (Brozovic et al. 2004; Dougan et al. 2005), and fatty acid amide hydrolase (FAAH) (Freigang et al. 2010).
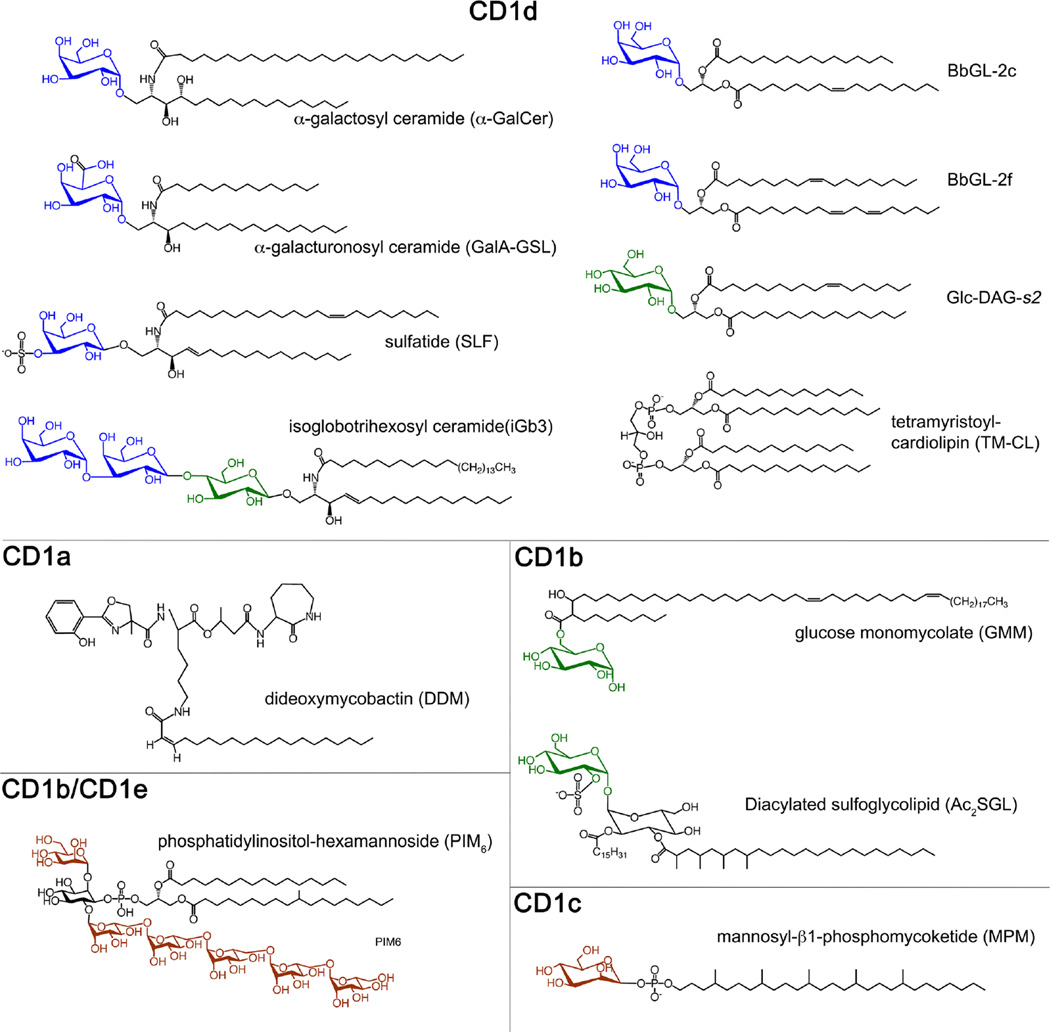
Fig. 2.
Structures of certain crystallized CD1 lipid antigens. For glycolipids, sugars are colored as follows: galactose in blue, glucose in green, and mannose in brown. Only representative lipids are shown. While CD1d-presented glycolipids are mostly based on either a ceramide or a diacylglycerol backbone, CD1a, b, and c presents structurally more unique lipids that, except for PIM6, cannot be presented by CD1d, while many of the CD1d antigens can be presented by the other CD1 proteins.
T cell diversity and recognition
CD1-restricted T cells comprise roughly 10 % of all αβ T lymphocytes in human peripheral blood, similar to MR1-restricted T cells, while the remaining 80 % are peptide classical class I reactive. The majority of lipid-reactive T cells are specific for CD1c (~7 %), while CD1a (~2 %), CD1b (~1%), and CD1d (0.1 %) are less abundant (Young and Gapin 2011). However, most of our understanding of lipid-reactive T cells stems from the work on CD1d-restricted T cells, termed Natural Killer T (NKT) cells, which are activated within hours after antigen challenge and express both NK cell receptors and a TCR (Bendelac et al. 2007). A large subset of NKT cells express a semi-invariant TCR formed by a conserved Vα24Jα18 chain (Vα14Jα18 in mice) that pairs with Vβ11 (Vβ8.2/Vβ7, and Vβ2 in mice). These semi-invariant or type I NKT cells have broad substrate specificity and recognize different carbohydrate epitopes (mostly galactose and glucose) that are predominantly linked to either a diacylglycerolipid backbone or ceramide backbone using an α-anomeric linkage (Girardi and Zajonc 2012). Many microbial antigens have been discovered that follow these structural principles, and type I NKT cells have been demonstrated to protect against microbial infection (Kinjo et al. 2011; Kinjo et al. 2006; Kinjo et al. 2005; Mattner et al. 2005; Sriramet al. 2005; Zajonc and Girardi 2015). Several β-anomeric glycolipids can also be recognized by type I NKT cells, including the self-antigen iGb3 (Zhou et al. 2004b). However, β-anomeric glycolipids are much weaker antigens for type I NKT cells than their α-anomeric counterparts. Sulfatide, a β-anomeric myelin-derived glycosphingolipid is a potent antigen for a subset of type II NKT cells (Blomqvist et al. 2009; Jahng et al. 2004; Jahng et al. 2001; Rhost et al. 2012). Type II NKT cells do not have a conserved TCR rearrangement and can be formed by many different TCR α and β genes in mice, most commonly Vα3/Vα1-Jα7/Jα9 and Vβ8.1/Vβ3.1-Jβ2.7 (Arrenberg et al. 2010).
While type I NKT cells are immunomodulatory, due to their potent production of both pro- and antiinflammatory cytokines, type II NKT cells are considered immunosuppressive. Activation of type II NKT cells can in turn lead to inhibition of type I NKT, reducing their IFN-y production and thus prevent liver damage (Arrenberg et al. 2011).
Interestingly CD1b-restricted T cells contain a subset called germline-encoded mycolyl lipid-reactive (GEM) T cells that is characterized by higher binding affinity to CD1d-glucose monomycolate (GMM) (Van Rhijn et al. 2013). Similar to type I NKT cells, GEM T cells also utilize a limited TCR repertoire using TRAV1-2/TRAJ9 with few N additions. CD1b-GMM-reactive T cells that bind with lower affinity revealed a more diverse TCR repertoire. GMM-specific T cells were also shown to be antimicrobial (Van Rhijn et al. 2013).
CD1a-restricted T cells are autoreactive and recognize antigens that lack a polar headgroup, suggesting an indirect recognition of the antigen (de Jong et al. 2014). Interestingly, lipids that contained a polar headgroup blocked T cell activation (de Jong et al. 2014). Since many glycolipids share common structural features, such as number and length of alkyl chains or the same carbohydrate moiety, recognition of any given lipid is generally achieved by TCR-binding specificity toward both the CD1 protein and the glycolipid, while CD1a autoreactive T cells follow different rules for activation. TCR recognition of permissive CD1a-presented antigens versus direct recognition of CD1d-presented antigen will be discussed in detail later.
Human/mouse CD1 isotype-specific adaptations
Mouse/human CD1d (1997/2005)
In 1997, the crystal structure of mouse (m) CD1d was first reported, followed by human CD1d in 2005 (Koch et al. 2005; Zeng et al. 1997). CD1d is the most studied isotype, since it is the only CD1 molecule expressed in mice. The CD1d structures represent the benchmark for comparing the CD1 isotype-specific adaptations of the lipid-binding groove. CD1d has a medium size binding groove (~1650 Å3 volume) and the two main pockets A′ and F′ (Fig. 3). The A′ pocket is deeply buried and doughnut shaped, while the F′ pocket descents straight down from the groove opening toward the β-sheet floor. The A′ pocket is larger in length and can accommodate the alkyl chains of up to 29 carbons, while the F′ pocket is shorter and limits alkyl chain length to roughly 18 carbons. This size distribution favors sphingolipid binding to CD1d, which generally contains a 26-carbon fatty acid that is N-amide-linked to a sphingoid base of 18 carbons. Lipids that are considerably shorter are usually found in conjunction with spacer lipid molecules that fill and presumably stabilize the remainder of the groove. These can range from short C8 to C16 fatty acid, although the exact nature has not always been precisely determined. The spectrum of lipid antigens that bind CD1d ranges from sphingolipid to glycerolipids, to cholesterol derivates, to small hydrophobic molecules, to even amphipathic α-helical peptides and lipopeptides (Fig. 2). Lipid antigens that have been crystallized bound to mouse or human CD1d include phosphatidylcholine (PC), acquired during expression of CD1d in insect cells (Giabbai et al. 2005); C8-α-galactosylceramide (PBS-25) together with a C16 spacer lipid (Zajonc et al. 2005a); α-galactosylceramide (α-GalCer) (Koch et al. 2005) and α-galacturonosylceramide (GalAGsl) (Wu et al. 2006); sulfatide (Luoma et al. 2013; Zajonc et al. 2005c); phosphatidylinositol-dimannoside (PIM-2) (Zajonc et al. 2006); isoglobotrihexosyl ceramide (iGb3) (Zajonc et al. 2008a); phenyl group-containing α-GalCer analogs C6Ph, C8Ph, C10Ph, and C8PhF (Schiefner et al. 2009); short chain α-GalCer analog OCH (Sullivan et al. 2010); Borrelia burgdorferi glycolipid 2c and 2f (BbGL-2c, -2f) (Wang et al. 2010); tetra-myristoyl cardiolipin (Dieude et al. 2011); Streptococcus pneumonia glucosyl-diacylglycerol (Glc-DAG-s2) (Kinjo et al. 2011); ganglioside GD3 (Mallevaey et al. 2011); lyso PC (Lopez-Sagaseta et al. 2012); and even a synthetic peptide p99, as well as the related lipopeptide p99p (Girardi et al. 2016).
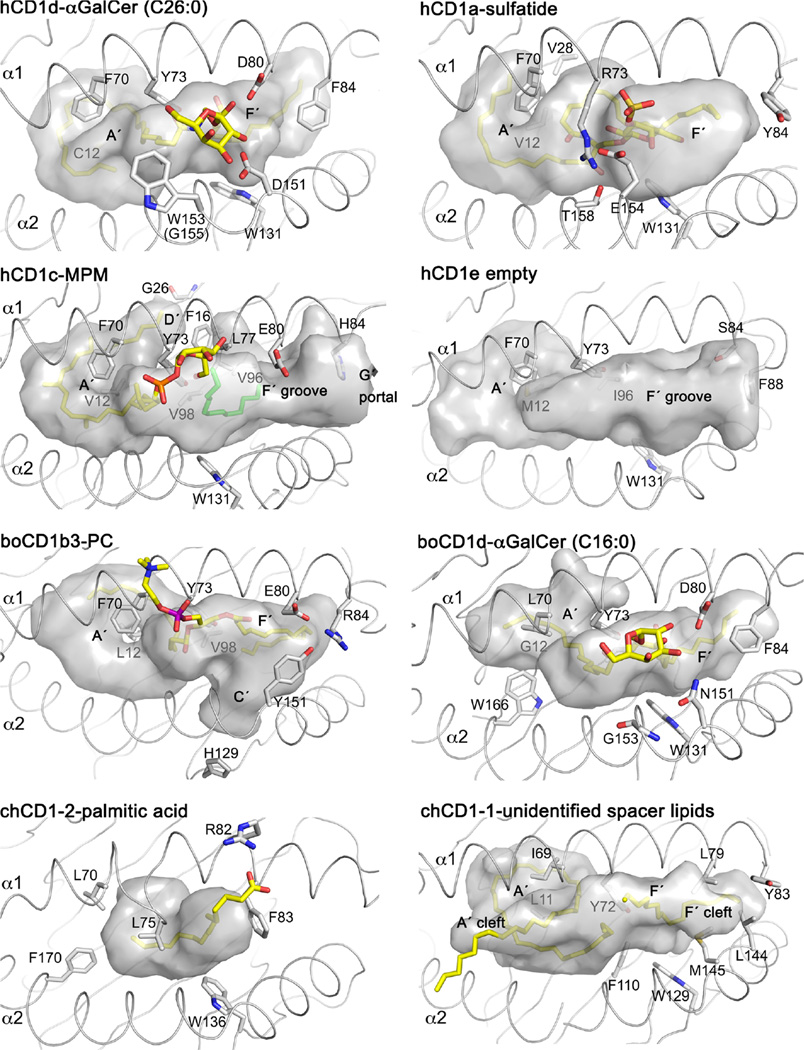
Fig. 3.
CD1-binding pockets and structural features. All CD1 proteins, expect for chCD1-2 have an A′ pole and at least the two pockets A′ and F′. Structural differences are the closed A′ roof of CD1a; the D′ portal, open F′ pocket, and G′ portal of CD1c; the open groove of CD1e; the T′ tunnel of CD1b; the partially closed A′ pocket of bovine CD1d; the lipid-binding pore of chCD1-2; and the A′ and F′ cleft of chCD1-1. CD1 residues beneath the binding groove are labeled in gray
A common feature of antigen presentation by CD1d is the intimate interaction of the core CD1d residues Asp80, Asp151 (Asp153 in mice), and Thr154 (Thr156 in mice) with the polar regions of the lipid antigen. For glycosphingolipids, an intricate hydrogen bond network orients the glycolipid at the entrance of the binding groove for the carbohydrate epitope to be presented to the corresponding TCR for subsequent TCR binding and T cell activation (Borg et al. 2007; Pellicci et al. 2009). The shape complementarity of the CD1d binding groove portal together with the precise interaction of the CD1d core residues with the antigen, rather than the size of the lipid alkyl chains or the antigen binding pockets, are the key factors that determine the binding orientation of all glycosphingolipids. Since most common glycosphingolipids consist of a fatty acid with a length of 24–26 carbons, which can only be bound within the A′ pocket of CD1d, the typically C18 sphingoid base becomes inserted into the F′ pocket. However, even if the acyl chain is truncated to 8–16 carbons, the sphingolipid binds in the same orientation, rather than inserting the longer sphingoid base into the larger A′ pocket and the shorter acyl chain into the smaller F′ pocket (Zajonc et al. 2005a). This “headgroup anchoring” for glycolipids is manifested with generally very well-defined electron density for glycosphingolipids. Much of our understanding of the binding of diacylglycerolipids (DAG lipids) has been through studies of bacterial antigens (Girardi et al. 2011; Kinjo et al. 2011; Kinjo et al. 2006; Wang et al. 2010). DAG lipids are more flexible in structure, since they lack the planar N-amide linkage. As a consequence opposite binding orientations have been observed (Wang et al. 2010). In these cases, it appeared as if the nature of the fatty acid (length and level of unsaturations) dictated the binding orientation of the DAG lipid, which directly affected the presentation of the carbohydrate headgroup and its T cell antigenicity (Kinjo et al. 2011; Kinjo et al. 2006; Wang et al. 2010). Since DAG lipids can bind in opposite orientation, meaning that either the acyl chain that is connected to sn-1 or sn-2 position of the glycerol can bind inside either the A′ or F′ pocket, antigenicity for DAG-based glycolipids is difficult to predict. For Borrelia glycolipids, the A′ pocket of mCD1d favors binding of oleic acid. In the glycolipid BbGL-2c, the oleic acid was at the sn-1 position and a palmitic acid at sn-2, presenting a viable antigen for iNKT cell recognition (Kinjo et al. 2006). The glycolipid BbGL-2f, however, which has an sn-2-linked oleic acid but an sn-1-linked linoleic acid bound in the reversed orientation, was not recognized by murine iNKT cells. Surprisingly, however, human CD1d can present BbGL-2f as an antigen to human iNKT cells, suggesting that subtle differences in antigen presentation exist between human and mouse CD1d that govern differences in antigenicity. Antigenicity of a DAG lipid can also be achieved by the combination of a unique fatty acid with an otherwise low antigenic carbohydrate epitope, such as glucose (Girardi et al. 2011; Kinjo et al. 2011). The S. pneumonia antigen GlcDAG-s2 contains an sn-1-linked palmitic acid and an sn-2-linked vaccenic acid (C18:1, n-7). Since the sn-2-linked vaccenic acid, similar to the oleic acid of the Borrelia glycolipids, binds in the A′ pocket and as such mimicks the binding orientation of BbGl-2f, we would expect the lipid to be presented in a non-antigenic orientation to the T cell. However, the combination of the unusual fatty acid, which, compared to oleic acid, has the unsaturation moved by 1 carbon with the glucose, leads to a novel interaction of the glucose with CD1d, which would not be seen using an oleic acid in combination with glucose or a vaccenic acid with galactose (Girardi et al. 2011). As a result, Glc-DAG-s2 stimulates both mouse and human iNKT cells.
CD1b (2002)
CD1b has the largest binding groove of all CD1 isotypes (~2200 Å3) and the most elaborate pocket network. Not surprisingly, CD1b can bind the largest of the CD1 antigens, namely mycolates of up to C80 in length. The first crystal structures of CD1b in complex with either phosphatidylinositol (PI) or gangliosides (GM2) (Gadola et al. 2002) revealed that the binding groove was composed of four interconnected pockets, termed A′, C′, F′, and T′ (Fig. 3). The T′ tunnel was essentially created by small, CD1b-specific glycine residues 98 and 116 on the β-sheet floor. In other CD1 isotypes, valine or leucine residues blocked the T′ tunnel. The structure of glucose monomycolate bound to CD1b revealed the connection of the pockets in such a way that the longer β-hydroxy chain was inserted into the A′ pocket, traversed through the T′ tunnel, and up through the F′ pocket with the tail end sticking out into the solvent, while the shorter α-alkyl side chain descended down into the C′ pocket (between the A′ and F′ pocket) and out into the solvent through the C′ portal (Batuwangala et al. 2004). This portal, which is kept open by a disulfide bridge (Cys131-Cys145) is instead blocked in CD1d and other isotypes by bulky residues, such as Trp133 (Fig. 3). Other CD1 isotypes are more restricted in size and cannot bind lipids that would exceed the size of the groove. Since the first human CD1b-lipid structures, phosphatidylcholine (Garcia-Alles et al. 2006) as well as synthetic diacylsulfoglycolipid SGL12 (Garcia-Alles et al. 2011a), have been crystallized bound to CD1b. Interestingly, except for SGL12, which has a di-saccharide headgroup, little hydrogen bond interaction between the ligands and CD1b is formed. Since CD1b has the largest binding groove, CD1b recruits the most spacer lipids when average size glycolipids (~40 carbons) are presented.
CD1a (2003)
The first structure of CD1a in complex with the glycosphingolipid sulfatide revealed that CD1a has the smallest and most restricted of the human CD1 binding grooves (~1350 Å3 volume) (Zajonc et al. 2003). In contrast to CD1b and d, the A′ pocket is not doughnut-shaped as Val28 (glycine in all human CD1 isotypes and serine in mCD1d) blocks full encircling of the A′ pole (Fig. 3). As a consequence, the A′ pocket is shaped like a hook which terminates underneath the α1 helix and allows a total of approximately 36 carbon atoms to fit into the groove (Zajonc et al. 2003). CD1a has an atypical A′ and F′ pocket groove organization. The A′ pocket is not directly connected to the CD1 surface, since it is closed at the top (A′ roof). Instead, the A′ pocket gradually merges with the F′ pocket, which runs less deeply compared to CD1b and d. The A′ roof is formed by CD1a-specific Arg73 that points toward the α2 helix, where it forms a hydrogen bond with Thr158 and a salt bridge with Glu154. In CD1b and CD1d, the corresponding Tyr73 points down into the binding groove, where it guides the descending alkyl chain into the A′ pocket. The crystal structure of CD1a in complex with sulfatide revealed a different glycolipid presentation compared to CD1b and d. Sulfatide sits much deeper inside the CD1a binding pocket than it does when bound to mCD1d (Zajonc et al. 2003; Zajonc et al. 2005c). Also, the fatty acid, rather than the sphingoid base binds inside the A′ pocket and the fatty acid tail rather than reaching down into the F′ pocket, ascends from the bottom of the F′ pocket up to the groove opening. This deeply buried antigen binding places only the 3′-sulfate group of the galactose moiety at the CD1 surface for T cell recognition.
However, since the fatty acid of sphingolipids can vary in size, this deeply buried sphingolipid headgroup presentation is greatly affected by the fatty acid chain length. A recent structure of CD1a with sphingomyelin, which had a longer C24:1 fatty acid (instead of the C16), demonstrated that the PC headgroup is much more exposed compared to sulfatide, since the longer fatty acid elevates the headgroup presentation (Birkinshaw et al. 2015). In this case, the sphingoid base descends down into the F′ pocket, similar to how CD1b and d bind glycosphingolipids.
The structure of a synthetic lipopeptide (Zajonc et al. 2005b), which is similar in structure to the mycobacterial didehydroxymycobactin (Moody et al. 2004; Van Rhijn et al. 2005), revealed a binding mode in which the alkyl chain was inserted into the end of the A′ pocket, with the peptidic moiety folding up inside the F′ pocket for presentation to T cells.
CD1c (2010)
CD1c is best known for its ability to bind phosphomycoketide antigens, which are mycobacterial single alkyl chain antigens with methyl substitution (Matsunaga et al. 2004; Moody 2001). CD1c structures are available with bound phosphomycoketide and mannosyl-b1-phosphomycoketide, as well as with fatty acids that were captured during protein expression and serve as natural spacer lipids to stabilize the groove (Mansour et al. 2016; Roy et al. 2014; Scharf et al. 2010). CD1c has the second largest human CD1 binding groove (~1780 Å3 volume). The binding groove contains an A′ pocket that wraps almost fully around the A′ pole (Val12 and Phe70) but connects to the solvent via the D′ portal (Fig. 3). The F′ pocket is rather broad, contains a small portal, and is open to the solvent, as it does not form a F′ roof. The authors, therefore, refer to the F′ pocket as the F′ groove (Scharf et al. 2010). CD1c has neither a T′ tunnel nor a C′ portal.
The single chain phosphomycoketide antigen binds deep inside the A′ pocket, with the terminal phosphate or phosphoryl-mannose protruding from the groove opening into the solvent. Phosphomycoketide presentation does not require the F′ pocket, and instead, the F′ pocket is occupied by a fatty acid spacer lipid. The structural data also provide a model of how dual alkyl chain antigens, such as sulfatide, would be able to bind within both pockets, as sulfatide is common lipid that can bind to CD1a–d (Shamshiev et al. 2002; Zajonc et al. 2005c). A unique feature of CD1c is the D′ and E′ portals. The E′ portal is located in the F′ pocket, where it is formed by the unique CD1c residues Phe16, Leu77, and Val96, with contribution of residues that are shared with other CD1 isoforms (Phe18, Thr78, Ile81). However, the E′ portal is not formed in all CD1c structures and appears too small to serve as a functional exit portal unless ligand-induced structural changes occur to open it further. In contrast, the D′ portal appears to form a true exit portal within the lateral wall of the A′ pocket next to the E′ portal. A combination of an elevated α1 helix that in contrast to CD1b increases the distance to the β-sheet floor of the A′ pocket, as well as substitution of Thr26 with Gly26, which allows the antigen to traverse underneath the helix and into the solvent form the D′ portal (Fig. 3). Another unique feature of the F′ pocket is its open nature, which gave it the name “F′ groove” (Scharf et al. 2010). Such an open pocket architecture had not been observed to this time for any other human CD1 isoform. A recent study demonstrated that the open F′ groove can form an F′ roof, similar to other CD1 isoforms, especially CD1d. While the F′ roof will close the F′ groove at the top, it does not close the end of the F′ groove, which is open to the solvent and referred to by the authors as G′ portal (Fig. 3). In contrast to CD1a and CD1d, which have a defined A′ pocket size and a closed F′ pocket, CD1c can bind lipids that exceed the size of the lipid-binding groove by using both the D′ and G′ portals. As a result, while CD1a- and CD1d-presented antigens have a maximum length in both alkyl chains, there is no length restriction for CD1b and CD1c, since both alkyl chains of a dual alkyl chain antigen can extend from the A′ and F′ pocket into the solvent.
More recently, a study reported flexibility of the CD1c binding groove (Mansour et al. 2016). The structure of refolded CD1c identified two short spacer lipids in the F′ groove, which are bound on top of each other. This binding mode suggests that larger lipid moieties can fit into the F′ groove of CD1c. Indeed, using thermodynamic simulations and functional T cell activation assays, the authors suggested that cholesterol-based lipids can be presented by CD1c and even stabilize the CD1c molecule for recognition by CD1c-self-reactive T cells (Mansour et al. 2016).
CD1e (2011)
CD1e is the only human isotype that does not directly present lipids to T cells. It is predominantly found in the lysosome of antigen presenting cells and not on the cell surface (Angenieux et al. 2005). The function of CD1e is to assist with the processing of complex phosphatidylinositol mannosides (PIMs) and their loading to CD1b for subsequent presentation to CD1b-restricted T cells (de la Salle et al. 2005). The only available CD1e structure identified a few unique characteristics, including an A′ and F′ pocket architecture similar to CD1d (Garcia-Alles et al. 2011b). The doughnut-shaped A′ pocket wraps fully around the A′ pole formed by Met12 and Phe70 (Phe70 is conserved across all human and mouse CD1 isoforms), while the F′ pocket descends toward the β-sheet floor and is closed at the end by Phe88 (conserved in CD1b) (Fig. 3). In contrast to CD1b, however, the F′ portal is wider, since it is lined by Ser84. In CD1a, CD1b, and CD1d, bulky hydrophobic residues (Tyr, Phe, Leu) form the lid or neck of the F′ pocket (Zajonc and Wilson 2007). Unique to CD1e, however, is the open nature of the binding groove, which is very accessible to solvent and rather large (2000 Å3 volume). This suggests that in contrast to other CD1 isoforms, CD1e binds lipids more transiently for fast transfer to CD1b. Therefore, one would assume that CD1e interacts less intimately with the lipid. Likely, a reflection of the transient lipid-binding properties of CD1e is the lack of a well-defined electron density for any self-antigen or spacer lipid in the crystal structure. This also suggests that CD1e can remain temporarily empty without collapse of the binding groove. Another feature of CD1e is its slightly positive charge around the binding groove portal, while all other CD1 isoforms are rather negatively charged. This opposite charge may help to transiently associate with CD1b for lipid exchange.
CD1 in other species
While human and mouse CD1 are the best studied members of this family, structural data has also been obtained on CD1 from other species, including ruminants (Bos taurus) and birds (Gallus gallus), which provide insight into species-specific adaptations of the lipid-binding grooves and the lipid antigens.
Bovine CD1b3 (2010)
Cattle have three potentially expressed CD1b proteins, namely CD1b1, CD1b3, and CD1b5 (Van Rhijn et al. 2006). The structure of bovine CD1b3 (boCD1b3) with endogenously bound lipids PC and phosphatidylethanolamine (PE) identified a binding groove that is identical in size to that of human CD1b (Girardi et al. 2010). A doughnut-shaped A′ pocket that circles around the conserved A′ pole (Leu12 and Phe70) directly connects with the solvent accessible F′ pocket (Fig. 3). Interestingly, the T′ tunnel found in human CD1b is closed by Val98. BoCD1b3 also has a centrally located C′ pocket ending in a C′ portal that leads into the solvent. However, His129 (Ala129 in hCD1b) blocks the terminal C′ portal found in hCD1b and diverts the portal opening to both lateral sides. In addition, the binding groove is narrower compared to hCD1b, due to closure of the F′ roof through a triad of residues including Glu80, Arg84, and Tyr151. Mass spectrometric analysis of identified PC and PE as endogenously acquired antigens during the course of recombinant protein expression in insect cells. PC has before been identified as an endogenously acquired antigen during mCD1d expression in insect cells; however, boCD1b presents PC and PE differently in the crystal structure. Since the F′ pocket is closed at the top, one acyl chain binds inside the A′ pocket and one inserts into the C′ pocket, from which it extends into the F′ pocket, rather than entering the C′ portal (Fig. 3). In case of PE, the acyl chain follows from the C′ pocket down into the F′ pocket and turns upward, while the opposite orientation is observed for PC. Here, the acyl chain traverses from the C′ pocket directly over to the F′ pocket and down to the β-sheet floor, where it turns upward to end close to the C′ portal. The difference in lipid binding compared to hCD1b is likely driven by the lack of the T′ tunnel, which now restricts the length of acyl chains accommodated inside the A′ pocket. In this binding orientation, longer acyl chains in the F′ pocket could potentially exit through the C′ portal. The lack of the T′ tunnel suggests that boCD1b3 samples are different lipids, compared to hCD1b. The presented lipid repertoire found in different organisms is likely the reason for the species-specific adaptations (evolution) of the CD1 binding grooves.
Bovine CD1d (2012)
While originally believed that cattle contain a CD1d pseudogene that is not expressed due to the lack of a start codon (Van Rhijn et al. 2006), cell surface expression of bovine CD1d (boCD1d) was later confirmed, revealing the usage of an alternate start codon (Nguyen et al. 2012). In addition to boCD1d, cattle express the TCR genes necessary to produce functional NKT cells, which are characterized by their reactivity to CD1d-presented α-GalCer (Reinink and Van Rhijn 2009). However, α-GalCer treatment of cattle did not show any immune response against this glycolipid, which raised the question as to whether NKT cells are generated in cows.
Lipid-binding assays first demonstrated that boCD1d can bind glycosphingolipids with an acyl chain length of C18 (GT1B) but not C24 (sulfatide) (Wang et al. 2012). This led to the investigation whether the structure of boCD1d differed greatly from that of hCD1d, resulting in the inability of long chain glycosphingolipids to be presented. Two crystal structures of boCD1d loaded with either disulfatide (C12 acyl chain) or medium length α-GalCer (C16 instead of C26 acyl chain) were determined (Wang et al. 2012). These structures revealed that similar to human and mouse CD1d, boCD1d has both A′ and F′ pockets and presents glycolipid with similar binding chemistries using many conserved CD1d residues, such as Asp80, Tyr73, Phe77, and Thr154 (Thr156 in mice). However, quite strikingly, the A′ pocket has a different architecture. The conserved A′ pole is absent (Gly12 and Leu70, instead of Cys/Val12 and Phe70) and instead of the typical doughnut shape, the A′ pocket is straight and approximately 300 Å3 smaller in volume than that of mouse or human CD1d (Wang et al. 2012). While a disulfide bond (Cys102-Cys166) in hCD1d widens the A′ pocket below the α2 helix, the bulky residue Trp166 in boCD1d blocks that part of the pocket and closes the side of the A′ pocket. As a result, acyl chains of only up the C18 can bind. Also, the A′ pocket of boCD1d seems to be more flexible and can slightly change its shape upon binding to different size lipids. Most notably, Trp40 can “swing in” when shorter acyl chains (C12) are bound, and slight rotation of Trp166 and Leu161 has also been observed that can broaden or restrict the volume but not necessarily the length of the A′ pocket. However, despite the ability of α-GalCer of medium-chain length to bind to boCD1d, as mentioned, no immune reactivity has been observed when injected in cattle (Nguyen et al. 2012), suggesting that the self-antigens that would be responsible for positive selection of this T cell subset might either not bind to boCD1 or, alternatively, that boCD1d presents a different lipid repertoire to different T cell subsets.
Avian CD1
While reptiles are the evolutionary oldest group of animals that is known to express CD1, we lack any structural information for reptile CD1 (Yang et al. 2015). Birds, however, express two CD1 genes that have likely evolved from a common ancestral CD1 gene ~310 mya (Dascher 2007; Miller et al. 2005; Salomonsen et al. 2005). Since MHC molecules are found even earlier in evolution (>450 mya), as early as in cartilaginous fish (e.g., shark), it is tempting to speculate that a primordial CD1 can be found that is derived from an evolutionary early MHC gene. The crystal structures of both chicken CD1-1 (chCD1-1) and chCD1-2 had been determined to address this question (Dvir et al. 2010; Zajonc et al. 2008b).
Chicken CD1-2 (2008)
ChCD1-2 has the most primitive and smallest of all CD1 binding grooves (470 Å3 volume) (Zajonc et al. 2008b). Instead of a binding groove that contains two (A′ and F′) or more (C′, T′) pockets, chCD1-2 has a simple pore, rather than a broader pocket (Fig. 3). This already suggested that chCD1-2 is not capable of binding common dual alkyl chain lipid antigens, such as diacylglycerols or sphingolipids. Instead, the structure revealed a linear electron density that could be best described using a fatty acid, such as palmitic acid, acquired during protein expression in insect cells (Zajonc et al. 2008b). While resembling the three-dimensional structures of other CD1 or MHC molecules, the α1 helix is intersected in the center by the A′ loop, a stretch of residues (Ser73, Met74, Val75, and Gly76) that bridge over to the α2 helix and restrict the groove opening dramatically. The fatty acid found inside the chCD1-2 groove binds with the alkyl chain deep inserted into the single pocket with the carboxylate extending out toward the solvent. Here, the positively charged residue Arg82 binds to the carboxylate using an electrostatic interaction. This binding is reminiscent of how Arg79 of human or mouse CD1d can bind glycolipids and is in line with a possible direct antigen presentation to T cells. While this structure revealed how primitive a lipid-binding pocket of CD1 can look, it did not identify an evolutionary and potentially hybrid structure: a peptide-binding, MHC-like binding pocket with the ability to bind lipids. Interestingly, however, the structure of the classical chicken MHC YF1*7.1 was determined, in which binding of an alkyl chain was observed (Hee et al. 2010). However, whether this structure represents a molecule related to the common ancestor of classical class I and CD1 is not known.
Chicken CD1-1 (2010)
Since the structure of chCD1-2 revealed a primitive binding pocket, the structure of chCD1-1 had been determined in an attempt to structurally characterize all CD1 proteins of a single species (Dvir et al. 2010). The structure of chCD1-1 identified an unexpectedly complex lipid-binding groove. The size of the binding groove is that of between human CD1a and CD1d (1440 Å3 volume) (Dvir et al. 2010). It consists of a rather large A′ pocket and a more restricted and narrow F′ pocket (Fig. 3). Both pockets are separated by Tyr72, by which most other CD1 proteins guides the alkyl chain into the F′ pocket. While the F′ pocket can accommodate alkyl chains of up to 16 carbons in length, the A′ pocket is doughnut-shaped and tilted 90° compared to mammalian A′ pockets. The A′ pole is not vertical but horizontally formed between the α1 and α2 helices, however, using equivalent residues (Leu11 and Ile 69). Interestingly, both A′ and F′ pockets open into a hydrophobic cleft at the protein surface, which also participate in antigen binding. This lipid presentation is on top of the binding pocket, effectively increasing the size of lipids that can be presented by chCD1-1 (Dvir et al. 2010). The crystal structure also revealed the presence of endogenous ligands in both the A′ and F′ pockets. A C45 alkyl chain was found in the A′ pocket and extended into the A′ cleft, while a C16 alkyl chain was found in the F′ pocket. Moreover, electron density for an unidentified molecule was observed in the F′ cleft. The A′ pocket binds lipids differently compared to mammalian A′ pockets. Since the pocket is tilted by 90°, the terminal end of the acyl chain that encircles the A′ pole ends up at the protein surface in the A′ cleft from where it can extend into the solvent. Therefore, the alkyl chain of lipid antigens that are bound within the A′ pocket is not restricted and can exceed the length of 45 carbons, while the size of the F′ pocket restricts lipid tails to 16 carbons. Mycolic acids contain features that are compatible with their presentation by chCD1-1, and lipid-binding studies demonstrated that both glycosphingolipids and mycolic acid can indeed bind (Dvir et al. 2010). Since Mycobacterium avium is a known bird pathogen, its lipid repertoire could have shaped the evolution of the chCD1-1 lipid-binding pocket. Another surprising feature of the A′ pocket was the presence of three short sidepockets that could potentially bind branched alkyl chains, such as those found in either mycolic acids or phosphomycoketides, also a common lipid class found in mycobacteria.
TCR interaction with CD1-lipid (since 2007)
TCR recognition was first structurally characterized for CD1d-restricted type I NKT cells presenting α-GalCer (Borg et al. 2007; Pellicci et al. 2009). In contrast to the typical diagonal binding orientation of the TCR above the antigen binding groove of the MHC molecule, the semi-invariant TCR of type I NKT cells bound parallel to the α helices of CD1d and centered above the F′ pocket, rather than spanning over both A′ and F′ pockets (Fig. 4). This places the invariant TCRα chain directly over the carbohydrate headgroups of CD1d-presented glycolipids. Complementarity determining region (CDR) loops 1α (Asn30α in mouse, Ser31α in human) and 3α (Gly96α) exclusively contact the antigen, while CDR3 residue Arg95α (encoded by Jα18) also contacts the 3′-hydroxyl of the ceramide backbone. Leu99α forms critical contacts with CD1d residues forming the F′ roof (L84, Val149) and contributes greatly to the stability of the complex. Before crystal structures were available, alanine-scanning mutagenesis of CDR3α residues demonstrated that every amino acid of this loop was required for activation of type I NKT cell hybridomas, regardless of the presented antigen (Scott-Browne et al. 2007). Since the structural characterization of the recognition of α-GalCer and structural analogs of α-GalCer, many microbial antigens, as well as self-antigens such as iGb3, have been structurally characterized (Aspeslagh et al. 2011; Aspeslagh et al. 2013; Girardi et al. 2011; Kerzerho et al. 2012; Li et al. 2010; Lopez-Sagaseta et al. 2012; Mallevaey et al. 2011; Patel et al. 2011; Pellicci et al. 2011; Wun et al. 2008; Wun et al. 2011; Wun et al. 2012; Yu et al. 2011). Surprisingly, despite being structurally diverse (Fig. 2), the antigens are engaged by the TCR using nearly identical binding chemistries. While α-GalCer is recognized by the TCR using a lock-and-key mechanism (explaining both the high-affinity TCR binding and potency of the antigen), other lipids have to be molded into the position that α-GalCer already adopted when it is presented by CD1d (Li et al. 2010; Pellicci et al. 2011; Yu et al. 2011). As a consequence, selfantigens and microbial antigens activate type I NKT cells less potently compared to α-GalCer. In case of iGb3, the TCR squashes the triglycosyl headgroup over the α2 helix of CD1d, where CD1d now interacts and binds the terminal α-linked galactose to allow the TCR to bind with relatively high affinity (Pellicci et al. 2011; Yu et al. 2011) (Fig. 4). This TCR molding forces the proximal β-anomeric glucose into a position where it mimics an α-anomeric sugar, which is the main structural signature of potent type I NKT cell antigens.
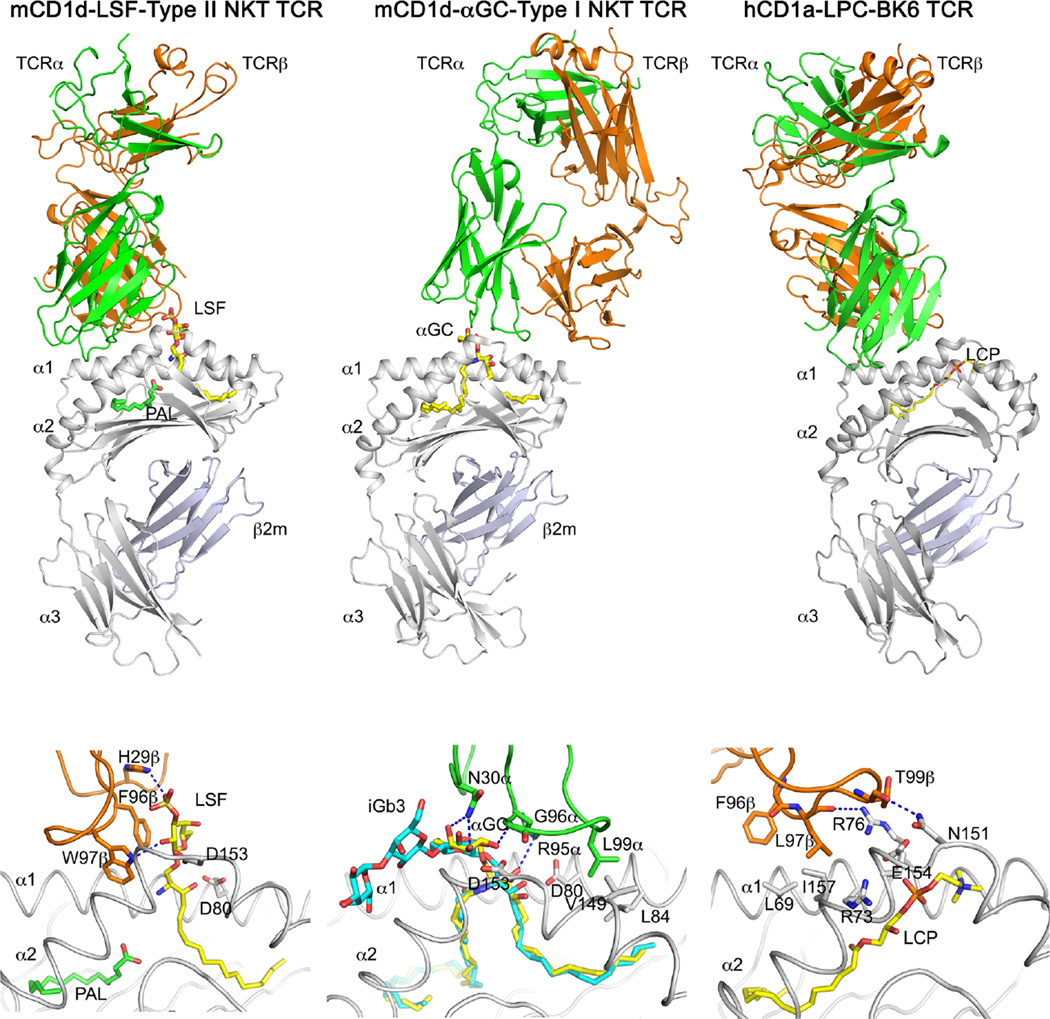
Fig. 4.
TCR recognition of CD1-presented antigens. Structures of the XV19.3 type II NKT TCR in complex with CD1d-lysosulfatide (LSF), the type I NKT TCR bound to mCD1d-presented αGalCer (αGC), and the BK6 TCR bound to CD1a presenting the permissive ligand lysophosphatidyl choline (LPC) are shown at the top, with detailed interactions shown below. Note that the BK6 TCR does not directly contact the lipid antigen, while both types I and II NKT TCRs require contacts with the antigen for binding. Hydrogen bonds in blue dashed lines; lipids in yellow and cyan; TCRα chain in green; TCRβ chain in orange; CD1 ingray. Note that the β-anomeric glucose of iGb3 is molded upon TCR binding into the approximate position of α-GalCer to allow for conserved TCR interactions (bottom middle panel)
Interestingly, type II NKT cells that recognize the β-anomeric self-antigen sulfatide bind their antigen with strikingly different chemistries (Fig. 4). Here, the TCR sits in an almost perpendicular orientation over the A′ pocket of CD1d and interacts with the ligand only through the TCRβ chain, while the TCRα chain only contacts CD1d (Girardi et al. 2012; Patel et al. 2012). While CDR1β residue His29β forms a single H bond with the sulfate moiety of sulfatide, Phe96β of CDR3β packs against the β-anomeric galactose for hydrophobic interactions. As a result, the type II NKT TCR is optimized for binding to extended ligands, especially β-anomeric sugars. Also, since this TCR can recognize also the uncharged lipid β-GlcCer and the phospholipid LPC, it appears that the contact with H29β is dispensable for T cell activation (Maricic et al. 2014; Rhost et al. 2012), while Phe96β and Trp97β are crucial for TCR binding (Girardi et al. 2012). Trp97β forms crucial interactions with the A′ roof of CD1d, similar to what L99α does in the type I NKT TCR does with the F′ roof of CD1d. In addition, CDR3α residues Asn96α, Asn97α, andTyr98α are also indispensable for TCR binding, as demonstrated by single alanine scanning mutagenesis, since these residues interact with CD1d (Girardi et al. 2012). In summary, both types I and II NKT TCRs (from hybridoma XV19; Cardell et al. 1995) recognize the antigen with both CDR1 and 3 of only a single TCR chain, while peptide/MHC-restricted T cells generally discriminate the peptide antigens with both CDR3α and 3β (Rossjohn et al. 2015).
For CD1a, two modes of T cell recognition of antigens exist. Both are antigen-dependent, but in the case of sulfatide, the TCR is expected to directly contact the glycolipid (Shamshiev et al. 2002), while permissive antigens that are mostly buried within CD1a allow the BK6 TCR to contact the CD1a molecule, without directly contacting the antigen (Birkinshaw et al. 2015; de Jong et al. 2014) (Fig. 4). Interestingly, similar to the type II NKT TCR, the CD1a-restricted BK6 TCR also binds in a perpendicular orientation above the A′ pocket (Birkinshaw et al. 2015). However, since the A′ pocket is closed at the top compared to CD1d, the TCR does not engage the antigen directly. Instead, the TCR uses its β chain to form H bond interactions with CD1a residues that form the neck of the A′ pocket (Arg76 and Asn151) (Fig. 4). Also, in analogy to both types I and II NKT cell TCRs, the BK6 TCR also uses a hydrophobic finger (L97β) to bind to the hydrophobic A′ roof (Leu69 and Ile157 among others) of CD1a. TCR interaction with Leu69 appears crucial for T cell activation (Birkinshaw et al. 2015). It is proposed that glycolipids, such as sulfatide, disrupt a second salt bridge formed between Arg76 and Glu154 (in addition to Arg73/Glu 154) and that the Arg76 would than collide with the BK6 TCR to prevent binding. However, ligands that bind CD1a and do not disrupt this interaction (permissive ligands, e.g., oleic acid) would allow the autoreactive BK6 TCR to bind to CD1a (Birkinshaw et al. 2015). Since the normal human T cell repertoire contains a number of CD1a-autoreactive T cells (Bourgeois et al. 2015; de Jong et al. 2014; de Lalla et al. 2011; Jarrett et al. 2016), this mode of recognizing permissive CD1a-presented ligands could be a common theme for CD1a.
Recently, γδ T cell recognition of CD1d-presented lipids has also been reported, and the binding orientation roughly corresponds with that of the type II NKT TCR (Luoma et al. 2013; Uldrich et al. 2013). In contrast to αβ TCR recognition, however, the structurally characterized γδ T cells are highly autoreactive and often bind the antigen-presenting molecule also in the absence of a defined added antigen, drawing some parallels to autoreactive CD1a-restricted T cells. It appears that addition of certain antigens can increase the TCR binding affinity by providing additional TCR interactions. Since recognition of glycolipids by γδ T cells is not the focus of this article, more information can be found here (Adams et al. 2015; Luoma et al. 2013; Uldrich et al. 2013).
Conclusion
T cells are able to sense three broad classes of antigens. Peptides, presented by classical MHC I and MHC II are recognized by CD8 and CD4 T cells, respectively; lipids and lipopeptides are recognized by CD1-restricted T cells; and microbial vitamin b metabolites are presented by MR1 to MAIT cells (Mori et al. 2016; Rossjohn et al. 2015). While peptides represent the largest number of antigens and are also presented by the largest family of polymorphic antigen presenting molecules, MR1-restricted T cells are seemingly limited in recognizing a particular biosynthetic pathway that does not exist in mammals. CD1-restricted T cells recognize an intermediate number of different antigens that are found across most living organisms. Lack of CD1 polymorphism is, in part, compensated for by the expression of different CD1 isotypes within a given species. In addition, certain CD1-restricted T cells, such a type I NKT cells, are multi-specific and share properties of innate pattern recognition receptors, as they seem to have evolved to optimally recognize the α-anomeric linkage (pattern) of glycosphingolipids. However, quite strikingly, phospholipids, lysolipids, ether bonded lipids, and even cholesterol-derived antigens are recognized by this T cell population (Chang et al. 2011; Ito et al. 2013). Our understanding of CD1 group 1-restricted T cell still falls short of that of NKT cells, but recent technological advances, such as group 1 and group 2 CD1 tetramers, are now paving the way for studying the entire family of lipid-reactive T cells in health and disease (Birkinshaw et al. 2015; Kasmar et al. 2011; Ly et al. 2013; Matsuda et al. 2000).
Acknowledgments
This work was supported by grants from the National Institute of Health, R01 AI074952 and R21 AI107318 (to DMZ). We thank Enrico Girardi and Martin Flajnik for critically reading this manuscript.
References
- Adams EJ, Gu S, Luoma AM. Human gamma delta T cells: evolution and ligand recognition. Cell Immunol. 2015 doi: 10.1016/j.cellimm.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams EJ, Luoma AM. The adaptable major histocompatibility complex (MHC) fold: structure and function of nonclassical and MHC class I-like molecules. Annu Rev Immunol. 2013;31:529–561. doi: 10.1146/annurev-immunol-032712-095912. [DOI] [PubMed] [Google Scholar]
- Agea E, et al. Human CD1-restricted T cell recognition of lipids from pollens. J Exp Med. 2005;202:295–308. doi: 10.1084/jem.20050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenieux C, et al. The cellular pathway of CD1e in immature and maturing dendritic cells. Traffic. 2005;6:286–302. doi: 10.1111/j.1600-0854.2005.00272.x. [DOI] [PubMed] [Google Scholar]
- Angenieux C, Salamero J, Fricker D, Cazenave JP, Goud B, Hanau D, de La Salle H. Characterization of CD1e, a third type of CD1 molecule expressed in dendritic cells. J Biol Chem. 2000;275:37757–37764. doi: 10.1074/jbc.M007082200. [DOI] [PubMed] [Google Scholar]
- Arrenberg P, Halder R, Dai Y, Maricic I, Kumar V. Oligoclonality and innate-like features in the TCR repertoire of type II NKT cells reactive to a beta-linked self-glycolipid. Proc Natl Acad Sci U S A. 2010;107:10984–10989. doi: 10.1073/pnas.1000576107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrenberg P, Maricic I, Kumar V. Sulfatide-mediated activation of type II natural killer T cells prevents hepatic ischemic reperfusion injury in mice. Gastroenterology. 2011;140:646–655. doi: 10.1053/j.gastro.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspeslagh S, et al. Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis. EMBO J. 2011;30:2294–2305. doi: 10.1038/emboj.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspeslagh S, et al. Enhanced TCR footprint by a novel glycolipid increases NKT-dependent tumor protection. J Immunol. 2013;191:2916–2925. doi: 10.4049/jimmunol.1203134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral DC, et al. CD1a and MHC class I follow a similar endocytic recycling pathway. Traffic. 2008;9:1446–1457. doi: 10.1111/j.1600-0854.2008.00781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batuwangala T, et al. The crystal structure of human CD1b with a bound bacterial glycolipid. J Immunol. 2004;172:2382–2388. doi: 10.4049/jimmunol.172.4.2382. [DOI] [PubMed] [Google Scholar]
- Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- Birkinshaw RW, et al. Alphabeta T cell antigen receptor recognition of CD1a presenting self lipid ligands. Nat Immunol. 2015;16:258–266. doi: 10.1038/ni.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomqvist M, et al. Multiple tissue-specific isoforms of sulfatide activate CD1d-restricted type II NKT cells. Eur J Immunol. 2009;39:1726–1735. doi: 10.1002/eji.200839001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg NA, et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- Bourgeois EA, et al. Bee venom processes human skin lipids for presentation by CD1a. J Exp Med. 2015;212:149–163. doi: 10.1084/jem.20141505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury A, Belt KT, Neri TM, Milstein C, Calabi F. Mouse CD1 is distinct from and co-exists with TL in the same thymus. EMBO J. 1988;7:3081–3086. doi: 10.1002/j.1460-2075.1988.tb03173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- Brozovic S, et al. CD1d function is regulated by microsomal triglyceride transfer protein. Nat Med. 2004;10:535–539. doi: 10.1038/nm1043. [DOI] [PubMed] [Google Scholar]
- Calabi F, Jarvis JM, Martin L, Milstein C. Two classes of CD1 genes. Eur J Immunol. 1989;19:285–292. doi: 10.1002/eji.1830190211. [DOI] [PubMed] [Google Scholar]
- Calabi F, Milstein C. A novel family of human major histocompatibility complex-related genes not mapping to chromosome 6. Nature. 1986;323:540–543. doi: 10.1038/323540a0. [DOI] [PubMed] [Google Scholar]
- Cardell S, Tangri S, Chan S, Kronenberg M, Benoist C, Mathis D. CD1-restricted CD4+ T cells in major histocompatibility complex class II-deficient mice. J Exp Med. 1995;182:993–1004. doi: 10.1084/jem.182.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YJ, et al. Influenza infection in suckling mice expands an NKT cell subset that protects against airway hyperreactivity. J Clin Invest. 2011;121:57–69. doi: 10.1172/JCI44845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dascher CC. Evolutionary biology of CD1. Curr Top Microbiol Immunol. 2007;314:3–26. doi: 10.1007/978-3-540-69511-0_1. [DOI] [PubMed] [Google Scholar]
- Dascher CC, et al. Conservation of a CD1 multigene family in the guinea pig. J Immunol. 1999;163:5478–5488. [PubMed] [Google Scholar]
- de Jong A, et al. CD1a-autoreactive T cells recognize natural skin oils that function as headless antigens. Nat Immunol. 2014;15:177–185. doi: 10.1038/ni.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Salle H, et al. Assistance of microbial glycolipid antigen processing by CD1e. Science. 2005;310:1321–1324. doi: 10.1126/science.1115301. [DOI] [PubMed] [Google Scholar]
- de Lalla C, et al. High-frequency and adaptive-like dynamics of human CD1 self-reactive T cells. Eur J Immunol. 2011;41:602–610. doi: 10.1002/eji.201041211. [DOI] [PubMed] [Google Scholar]
- Dieude M, et al. Cardiolipin binds to CD1d and stimulates CD1d-restricted gammadelta T cells in the normal murine repertoire. J Immunol. 2011;186:4771–4781. doi: 10.4049/jimmunol.1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan SK, et al. Microsomal triglyceride transfer protein lipidation and control of CD1d on antigen-presenting cells. J Exp Med. 2005;202:529–539. doi: 10.1084/jem.20050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvir H, Wang J, Ly N, Dascher CC, Zajonc DM. Structural basis for lipid-antigen recognition in avian immunity. J Immunol. 2010;184:2504–2511. doi: 10.4049/jimmunol.0903509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst WA, et al. Molecular interaction of CD1b with lipoglycan antigens. Immunity. 1998;8:331–340. doi: 10.1016/s1074-7613(00)80538-5. [DOI] [PubMed] [Google Scholar]
- Facciotti F, et al. Fine tuning by human CD1e of lipid-specific immune responses. Proc Natl Acad Sci U S A. 2011;108:14228–14233. doi: 10.1073/pnas.1108809108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K, et al. Mycobacterial phosphatidylinositol mannoside is a natural antigen for CD1d-restricted T cells. Proc Natl Acad Sci U S A. 2004;101:10685–10690. doi: 10.1073/pnas.0403787101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freigang S, et al. Fatty acid amide hydrolase shapes NKT cell responses by influencing the serum transport of lipid antigen in mice. J Clin Invest. 2010;120:1873–1884. doi: 10.1172/JCI40451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadola SD, et al. Structure of human CD1b with bound ligands at 2.3 Å, a maze for alkyl chains. Nat Immunol. 2002;3:721–726. doi: 10.1038/ni821. [DOI] [PubMed] [Google Scholar]
- Garcia-Alles LF, et al. Structural reorganization of the antigen-binding groove of human CD1b for presentation of mycobacterial sulfoglycolipids. Proc Natl Acad Sci U S A. 2011a;108:17755–17760. doi: 10.1073/pnas.1110118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alles LF, et al. Crystal structure of human CD1e reveals a groove suited for lipid-exchange processes. Proc Natl Acad Sci U S A. 2011b;108:13230–13235. doi: 10.1073/pnas.1105627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alles LF, et al. Endogenous phosphatidylcholine and a long spacer ligand stabilize the lipid-binding groove of CD1b. EMBO J. 2006;25:3684–3692. doi: 10.1038/sj.emboj.7601244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giabbai B, et al. Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: a molecular basis for NKT cell activation. J Immunol. 2005;175:977–984. doi: 10.4049/jimmunol.175.2.977. [DOI] [PubMed] [Google Scholar]
- Gilleron M, et al. Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating CD1-restricted T cells during infection with Mycobacterium tuberculosis. J Exp Med. 2004;199:649–659. doi: 10.1084/jem.20031097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi E, Maricic I, Wang J, Mac TT, Iyer P, Kumar V, Zajonc DM. Type II natural killer T cells use features of both innate-like and conventional T cells to recognize sulfatide self antigens. Nat Immunol. 2012;13:851–856. doi: 10.1038/ni.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi E, et al. Crystal structure of bovine CD1b3 with endogenously bound ligands. J Immunol. 2010;185:376–386. doi: 10.4049/jimmunol.1000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi E, Wang J, Zajonc DM. Structure of an alpha-helical peptide and lipopeptide bound to the non-classical MHC class I molecule CD1d. J Biol Chem. 2016 doi: 10.1074/jbc.M115.702118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi E, et al. Unique interplay between sugar and lipid in determining the antigenic potency of bacterial antigens for NKT cells. PLoS Biol. 2011;9:e1001189. doi: 10.1371/journal.pbio.1001189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi E, Zajonc DM. Molecular basis of lipid antigen presentation by CD1d and recognition by natural killer T cells. Immunol Rev. 2012;250:167–179. doi: 10.1111/j.1600-065X.2012.01166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff RD, et al. Effects of lipid chain lengths in α-galactosylceramides on cytokine release by natural killer T cells. J Am Chem Soc. 2004;126:13602–13603. doi: 10.1021/ja045385q. [DOI] [PubMed] [Google Scholar]
- Gumperz JE, et al. Murine CD1d-restricted T cell recognition of cellular lipids. Immunity. 2000;12:211–221. doi: 10.1016/s1074-7613(00)80174-0. [DOI] [PubMed] [Google Scholar]
- Hee CS, Gao S, Loll B, Miller MM, Uchanska-Ziegler B, Daumke O, Ziegler A. Structure of a classical MHC class I molecule that binds “non-classical” ligands. PLoS Biol. 2010;8:e1000557. doi: 10.1371/journal.pbio.1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, et al. Helicobacter pylori cholesteryl alpha-glucosides contribute to its pathogenicity and immune response by natural killer T cells. PLoS One. 2013;8:e78191. doi: 10.1371/journal.pone.0078191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman RM, et al. The tyrosine-containing cytoplasmic tail of CD1b is essential for its efficient presentation of bacterial lipid antigens. Immunity. 1998;8:341–351. doi: 10.1016/s1074-7613(00)80539-7. [DOI] [PubMed] [Google Scholar]
- Jahng A, Maricic I, Aguilera C, Cardell S, Halder RC, Kumar V. Prevention of autoimmunity by targeting a distinct, noninvariant CD1d-reactive T cell population reactive to sulfatide. J Exp Med. 2004;199:947–957. doi: 10.1084/jem.20031389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahng AW, et al. Activation of natural killer T cells potentiates or prevents experimental autoimmune encephalomyelitis. J Exp Med. 2001;194:1789–1799. doi: 10.1084/jem.194.12.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett R, et al. Filaggrin inhibits generation of CD1a neolipid antigens by house dust mite-derived phospholipase. Sci Transl Med. 2016;8:325ra318. doi: 10.1126/scitranslmed.aad6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce S, et al. Natural ligand of mouse CD1d1: cellular glycosylphosphatidylinositol. Science. 1998;279:1541–1544. doi: 10.1126/science.279.5356.1541. [DOI] [PubMed] [Google Scholar]
- Kain L, et al. The identification of the endogenous ligands of natural killer T cells reveals the presence of mammalian alpha-linked glycosylceramides. Immunity. 2014;41:543–554. doi: 10.1016/j.immuni.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SJ, Cresswell P. Saposins facilitate CD1d-restricted presentation of an exogenous lipid antigen to T cells. Nat Immunol. 2004;5:175–181. doi: 10.1038/ni1034. [DOI] [PubMed] [Google Scholar]
- Kasmar AG, et al. CD1b tetramers bind alphabeta T cell receptors to identify a mycobacterial glycolipid-reactive T cell repertoire in humans. J Exp Med. 2011;208:1741–1747. doi: 10.1084/jem.20110665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano T, et al. CD1d-restricted and TCR-mediated activation of Vα14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- Kerzerho J, et al. Structural and functional characterization of a novel nonglycosidic type I NKT agonist with immunomodulatory properties. J Immunol. 2012;188:2254–2265. doi: 10.4049/jimmunol.1103049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjo Y, et al. Invariant natural killer T cells recognize glycolipids from pathogenic gram-positive bacteria. Nat Immunol. 2011;12:966–974. doi: 10.1038/ni.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjo Y, et al. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7:978–986. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- Kinjo Y, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- Koch M, et al. The crystal structure of human CD1d with and without α-galactosylceramide. Nat Immunol. 2005;8:819–826. doi: 10.1038/ni1225. [DOI] [PubMed] [Google Scholar]
- Li Y, Girardi E, Wang J, Yu ED, Painter GF, Kronenberg M, Zajonc DM. The Vα14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode. J Exp Med. 2010;207:2383–2393. doi: 10.1084/jem.20101335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Sagaseta J, Sibener LV, Kung JE, Gumperz J, Adams EJ. Lysophospholipid presentation by CD1d and recognition by a human natural killer T-cell receptor. EMBO J. 2012;31:2047–2059. doi: 10.1038/emboj.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoma AM, et al. Crystal structure of Vdelta1 T cell receptor in complex with CD1d-sulfatide shows MHC-like recognition of a self-lipid by human gammadelta T cells. Immunity. 2013;39:1032–1042. doi: 10.1016/j.immuni.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly D, et al. CD1c tetramers detect ex vivo T cell responses to processed phosphomycoketide antigens. J Exp Med. 2013;210:729–741. doi: 10.1084/jem.20120624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallevaey T, et al. A molecular basis for NKT cell recognition of CD1d-self-antigen. Immunity. 2011;34:315–326. doi: 10.1016/j.immuni.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour S, et al. Cholesteryl esters stabilize human CD1c conformations for recognition by self-reactive T cells. Proc Natl Acad Sci U S A. 2016;113:E1266–E1275. doi: 10.1073/pnas.1519246113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricic I, Girardi E, Zajonc DM, Kumar V. Recognition of lysophosphatidylcholine by type II NKT cells and protection from an inflammatory liver disease. J Immunol. 2014;193:4580–4589. doi: 10.4049/jimmunol.1400699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruoka T, Tanabe H, Chiba M, Kasahara M. Chicken CD1 genes are located in the MHC: CD1 and endothelial protein C receptor genes constitute a distinct subfamily of class-I-like genes that predates the emergence of mammals. Immunogenetics. 2005;57:590–600. doi: 10.1007/s00251-005-0016-y. [DOI] [PubMed] [Google Scholar]
- Matsuda JL, et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga I, et al. Mycobacterium tuberculosis pks12 produces a novel polyketide presented by CD1c to T cells. J Exp Med. 2004;200:1559–1569. doi: 10.1084/jem.20041429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattner J, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- Maxfield FR, Hao M. Lipid trafficking in cells. In: Roberts GCK, editor. Encyclopedia of biophysics. Berlin Heidelberg: Springer; 2013. pp. 1289–1296. [Google Scholar]
- Miller MM, et al. Characterization of two avian MHC-like genes reveals an ancient origin of the CD1 family. Proc Natl Acad Sci U S A. 2005;102:8674–8679. doi: 10.1073/pnas.0500105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Miyake S, Yamamura T. A synthetic glycolipid prevents autoimmune encephalomyelitis by inducing TH2 bias of natural killer T cells. Nature. 2001;413:531–534. doi: 10.1038/35097097. [DOI] [PubMed] [Google Scholar]
- Moody DB. Polyisoprenyl glycolipids as targets of CD1-mediated T cell responses. Cell Mol Life Sci. 2001;58:1461–1474. doi: 10.1007/PL00000789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody DB, Porcelli SA. Intracellular pathways of CD1 antigen presentation. Nat Rev Immunol. 2003;3:11–22. doi: 10.1038/nri979. [DOI] [PubMed] [Google Scholar]
- Moody DB, et al. Structural requirements for glycolipid antigen recognition by CD1b- restricted T cells. Science. 1997;278:283–286. doi: 10.1126/science.278.5336.283. [DOI] [PubMed] [Google Scholar]
- Moody DB, et al. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature. 2000;404:884–888. doi: 10.1038/35009119. [DOI] [PubMed] [Google Scholar]
- Moody DB, et al. T cell activation by lipopeptide antigens. Science. 2004;303:527–531. doi: 10.1126/science.1089353. [DOI] [PubMed] [Google Scholar]
- Moody DB, Zajonc DM, Wilson IA. Anatomy of CD1-lipid antigen complexes. Nat Rev Immunol. 2005;5:387–399. doi: 10.1038/nri1605. [DOI] [PubMed] [Google Scholar]
- Mori L, Lepore M, De Libero G. The immunology of CD1- and MR1-restricted T cells. Annu Rev Immunol. 2016;34:479–510. doi: 10.1146/annurev-immunol-032414-112008. [DOI] [PubMed] [Google Scholar]
- Nguyen TK, et al. The bovine CD1D gene has an unusual gene structure and is expressed but cannot present alpha-galactosylceramide with a C26 fatty acid. Int Immunol. 2012 doi: 10.1093/intimm/dxs092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel O, et al. NKT TCR recognition of CD1d-alpha-C-galactosylceramide. J Immunol. 2011;187:4705–4713. doi: 10.4049/jimmunol.1100794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel O, et al. Recognition of CD1d-sulfatide mediated by a type II natural killer T cell antigen receptor. Nat Immunol. 2012;13:857–863. doi: 10.1038/ni.2372. [DOI] [PubMed] [Google Scholar]
- Pellicci DG, et al. Recognition of beta-linked self glycolipids mediated by natural killer T cell antigen receptors. Nat Immunol. 2011;12:827–833. doi: 10.1038/ni.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicci DG, et al. Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity. 2009;31:47–59. doi: 10.1016/j.immuni.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicci DG, et al. The molecular bases of delta/alphabeta T cell-mediated antigen recognition. J Exp Med. 2014;211:2599–2615. doi: 10.1084/jem.20141764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch J, et al. Structural features of the acyl chain determine self-phospholipid antigen recognition by a CD1d-restricted invariant NKT (iNKT) cell. J Biol Chem. 2003;278:47508–47515. doi: 10.1074/jbc.M308089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinink P, Van Rhijn I. The bovine T cell receptor alpha/delta locus contains over 400 V genes and encodes V genes without CDR2. Immunogenetics. 2009;61:541–549. doi: 10.1007/s00251-009-0384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinink P, Van Rhijn I. Mammalian CD1 and MR1 genes. 2016 doi: 10.1007/s00251-016-0926-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhost S, et al. Identification of novel glycolipid ligands activating a sulfatide-reactive, CD1d-restricted, type II natural killer T lymphocyte. Eur J Immunol. 2012;42:2851–2860. doi: 10.1002/eji.201142350. [DOI] [PubMed] [Google Scholar]
- Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, McCluskey J. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol. 2015;33:169–200. doi: 10.1146/annurev-immunol-032414-112334. [DOI] [PubMed] [Google Scholar]
- Roy S, Ly D, Li NS, Altman JD, Piccirilli JA, Moody DB, Adams EJ. Molecular basis of mycobacterial lipid antigen presentation byCD1c and its recognition by alphabeta T cells. Proc Natl Acad Sci U S A. 2014;111:E4648–E4657. doi: 10.1073/pnas.1408549111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamero J, et al. CD1a molecules traffic through the early recycling endosomal pathway in human Langerhans cells. J Investig Dermatol. 2001;116:401–408. doi: 10.1046/j.1523-1747.2001.01264.x. [DOI] [PubMed] [Google Scholar]
- Salomonsen J, et al. Two CD1 genes map to the chicken MHC, indicating that CD1 genes are ancient and likely to have been present in the primordial MHC. Proc Natl Acad Sci U S A. 2005;102:8668–8673. doi: 10.1073/pnas.0409213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf L, et al. The 2.5 a structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity. 2010;33:853–862. doi: 10.1016/j.immuni.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefner A, Fujio M, Wu D, Wong CH, Wilson IA. Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands. J Mol Biol. 2009;394:71–82. doi: 10.1016/j.jmb.2009.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieg J, Yang G, Franck RW, Tsuji M. Superior protection against malaria and melanoma metastases by a C-glycoside analogue of the natural killer T cell ligand α-galactosylceramide. J Exp Med. 2003;198:1631–1641. doi: 10.1084/jem.20031192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Browne JP, et al. Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat Immunol. 2007;8:1105–1113. doi: 10.1038/ni1510. [DOI] [PubMed] [Google Scholar]
- Shamshiev A, Donda A, Carena I, Mori L, Kappos L, De Libero G. Self glycolipids as T-cell autoantigens. Eur J Immunol. 1999;29:1667–1675. doi: 10.1002/(SICI)1521-4141(199905)29:05<1667::AID-IMMU1667>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Shamshiev A, Gober HJ, Donda A, Mazorra Z, Mori L, De Libero G. Presentation of the same glycolipid by different CD1 molecules. J Exp Med. 2002;195:1013–1021. doi: 10.1084/jem.20011963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieling PA, et al. CD1-restricted T cell recognition of microbial lipoglycan antigens. Science. 1995;269:227–230. doi: 10.1126/science.7542404. [DOI] [PubMed] [Google Scholar]
- Sriram V, Du W, Gervay-Hague J, Brutkiewicz RR. Cell wall glycosphingolipids of Sphingomonas paucimobilis are CD1d-specific ligands for NKT cells. Eur J Immunol. 2005;35:1692–1701. doi: 10.1002/eji.200526157. [DOI] [PubMed] [Google Scholar]
- Sugita M, Cernadas M, Brenner MB. New insights into pathways for CD1-mediated antigen presentation. Curr Opin Immunol. 2004;16:90–95. doi: 10.1016/j.coi.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Sugita M, Grant EP, van Donselaar E, Hsu VW, Rogers RA, Peters PJ, Brenner MB. Separate pathways for antigen presentation by CD1 molecules. Immunity. 1999;11:743–752. doi: 10.1016/s1074-7613(00)80148-x. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, et al. Mechanisms for glycolipid antigen-driven cytokine polarization by Vα14i NKT cells. J Immunol. 2010;184:141–153. doi: 10.4049/jimmunol.0902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uldrich AP, et al. CD1d-lipid antigen recognition by the γδ TCR. Nat Immunol. 2013 doi: 10.1038/ni.2713. [DOI] [PubMed] [Google Scholar]
- van den Elzen P, et al. Apolipoprotein-mediated pathways of lipid antigen presentation. Nature. 2005;437:906–910. doi: 10.1038/nature04001. [DOI] [PubMed] [Google Scholar]
- Van Rhijn I, et al. A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat Immunol. 2013;14:706–713. doi: 10.1038/ni.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rhijn I, et al. The bovine CD1 family contains group 1 CD1 proteins, but no functional CD1d. J Immunol. 2006;176:4888–4893. doi: 10.4049/jimmunol.176.8.4888. [DOI] [PubMed] [Google Scholar]
- Van Rhijn I, Zajonc DM, Wilson IA, Moody DB. T-cell activation by lipopeptide antigens. Curr Opin Immunol. 2005;17:222–229. doi: 10.1016/j.coi.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Wang J, Guillaume J, Pauwels N, Van Calenbergh S, Van Rhijn I, Zajonc DM. Crystal structures of bovine CD1d reveal altered alphaGalCer presentation and a restricted A′ pocket unable to bind long-chain glycolipids. PLoS One. 2012;7:e47989. doi: 10.1371/journal.pone.0047989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. Lipid binding orientation within CD1d affects recognition of Borrelia burgdorferi antigens by NKT cells. Proc Natl Acad Sci U S A. 2010;107:1535–1540. doi: 10.1073/pnas.0909479107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winau F, et al. Saposin C is required for lipid presentation by human CD1b. Nat Immunol. 2004;5:169–174. doi: 10.1038/ni1035. [DOI] [PubMed] [Google Scholar]
- Wu D, et al. Bacterial glycolipids and analogs as antigens for CD1d-restricted NKT cells. Proc Natl Acad Sci U S A. 2005;102:1351–1356. doi: 10.1073/pnas.0408696102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, et al. Design of natural killer T cell activators: structure and function of a microbial glycosphingolipid bound to mouse CD1d. Proc Natl Acad Sci U S A. 2006;103:3972–3977. doi: 10.1073/pnas.0600285103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DY, Segal NH, Sidobre S, Kronenberg M, Chapman PB. Cross-presentation of disialoganglioside GD3 to natural killer T cells. J Exp Med. 2003;198:173–181. doi: 10.1084/jem.20030446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wun KS, et al. A minimal binding footprint on CD1d-glycolipid is a basis for selection of the unique human NKT TCR. J Exp Med. 2008;205:939–949. doi: 10.1084/jem.20072141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wun KS, et al. A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells. Immunity. 2011;34:327–339. doi: 10.1016/j.immuni.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wun KS, et al. Human and mouse type I natural killer T cell antigen receptors exhibit different fine specificities for CD1d-antigen complex. J Biol Chem. 2012;287:39139–39148. doi: 10.1074/jbc.M112.412320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, et al. Analysis of the reptile CD1 genes: evolutionary implications. Immunogenetics. 2015;67:337–346. doi: 10.1007/s00251-015-0837-2. [DOI] [PubMed] [Google Scholar]
- Young MH, Gapin L. Group 1 CD1-restricted T cells take center stage. Eur J Immunol. 2011 doi: 10.1002/eji.201141408. [DOI] [PubMed] [Google Scholar]
- Yu ED, Girardi E, Wang J, Zajonc DM. Cutting edge: structural basis for the recognition of {beta}-linked glycolipid antigens by invariant NKT cells. J Immunol. 2011;187:2079–2083. doi: 10.4049/jimmunol.1101636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu KO, et al. Modulation of CD1d-restricted NKT cell responses by using N-acyl variants of α-galactosylceramides. Proc Natl Acad Sci U S A. 2005;102:3383–3388. doi: 10.1073/pnas.0407488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajonc DM, Ainge GD, Painter GF, Severn WB, Wilson IA. Structural characterization of mycobacterial phosphatidylinositol mannoside binding to mouse CD1d. J Immunol. 2006;177:4577–4583. doi: 10.4049/jimmunol.177.7.4577. [DOI] [PubMed] [Google Scholar]
- Zajonc DM, et al. Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor. Nat Immunol. 2005a;8:810–818. doi: 10.1038/ni1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajonc DM, et al. Molecular mechanism of lipopeptide presentation by CD1a. Immunity. 2005b;22:209–219. doi: 10.1016/j.immuni.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Zajonc DM, Elsliger MA, Teyton L, Wilson IA. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 Å. Nat Immunol. 2003;4:808–815. doi: 10.1038/ni948. [DOI] [PubMed] [Google Scholar]
- Zajonc DM, Girardi E. Recognition of microbial glycolipids by natural killer T cells. Front Immunol. 2015;6:400. doi: 10.3389/fimmu.2015.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajonc DM, et al. Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity. J Exp Med. 2005c;202:1517–1526. doi: 10.1084/jem.20051625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajonc DM, Savage PB, Bendelac A, Wilson IA, Teyton L. Crystal structures of mouse CD1d-iGb3 complex and its cognate Vα14 T cell receptor suggest a model for dual recognition of foreign and self glycolipids. J Mol Biol. 2008a;377:1104–1116. doi: 10.1016/j.jmb.2008.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajonc DM, Striegl H, Dascher CC, Wilson IA. The crystal structure of avian CD1 reveals a smaller, more primordial antigen-binding pocket compared to mammalian CD1. Proc Natl Acad Sci U S A. 2008b;105:17925–17930. doi: 10.1073/pnas.0809814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajonc DM, Wilson IA. Architecture of CD1 proteins. Curr Top Microbiol Immunol. 2007;314:27–50. doi: 10.1007/978-3-540-69511-0_2. [DOI] [PubMed] [Google Scholar]
- Zeng Z, Castano AR, Segelke BW, Stura EA, Peterson PA, Wilson IA. Crystal structure of mouse CD1: an MHC-like fold with a large hydrophobic binding groove. Science. 1997;277:339–345. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- Zhou D, et al. Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science. 2004a;303:523–527. doi: 10.1126/science.1092009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004b;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]