Abstract
Manipulating cytokine function with protein-based drugs has proven effective for treating a wide variety of autoimmune and auto-inflammatory disorders. However, the limited ability of protein-based drugs to modulate intracellular targets, including many implicated by studies of the genetics and physiology of these diseases, and to coordinately neutralize redundant inflammatory cytokines, suggest an important and complementary role for small molecules in immunomodulatory drug development. The recent clinical approval of Janus kinase and phosphodiesterase inhibitors, along with emerging evidence from other compound classes, firmly establish small molecules as effective tools for modulating therapeutically relevant proteins that give rise to aberrant cytokine signaling or mediate its downstream consequences.
Introduction
The incidence of autoimmune and auto-inflammatory disorders is rapidly increasing in developed countries [1]. Addressing this clinical need will require continued innovation in immunomodulatory drug development. Data from many sources, including analysis of how human genetic variation affects disease susceptibility, implicate aberrant cytokine production and signaling in the pathophysiology of these disorders (see Box 1 for background on the application of disease genetics to drug discovery). For example, mutations in the cellular machinery that processes the inflammatory cytokine interleukin-1β (IL-1β) to its mature form cause hereditary auto-inflammatory diseases known as cryopyrin disorders (Figure 1a) [2]. Protein therapies inhibiting IL-1β (canakinumab; rilonacept) or its receptor (anakinra) are used to treat cryopyrin disorders, as well as immune disorders with more complex etiologies, including gout, type-2 diabetes, rheumatoid arthritis (RA) and chronic granulomatous disease [3,4]. The clinical success of biopharmaceuticals targeting IL-1β or other cytokines (TNF-α, IL-6, IL-12/23) derives from their ability to disrupt protein–protein interactions with exquisite selectivity and predictable, long-lasting pharmacology [5].
Box 1. Using human disease genetics to guide drug development.
The study of human genetics can uncover factors that contribute to the initiation and maintenance of disease, and suggest new strategies for therapeutic intervention. Therapies directed at correcting defects associated with causative alleles in Mendelian disease – protein therapeutics targeting IL-1β in cyropyrin disoders (see main text) or ivacaftor to modulate the CFTR-G551D allele in cystic fibrosis – illustrate the potential of genetics to inform discovery of effective medicines. In contrast to Mendelian diseases, many autoimmune/auto-inflammatory diseases have a complex genetic architecture in which susceptibility is influenced by multiple alleles as well as environmental factors. For instance, a recent genome-wide association study of inflammatory bowel disease (IBD) identified single nucleotide polymorphisms (SNPs) in 163 genetic loci (i.e., chromosomal regions) associated with altered disease risk [20]. Leveraging these insights for drug discovery will require understanding how disease genes contribute to pathophysiology. For example, the ATG16L-T300A SNP that confers increased risk of Crohn’s disease (CD) is associated with defects in bacteria clearance and aberrant inflammatory cytokine production [54,55]. Small molecules that correct these defects may be useful for treating CD. While potentially less straightforward than monogenic diseases, the fact that several FDA-approved drugs have been shown retrospectively to modulate genes with risk-associated polymorphisms (e.g. thiazolidinediones targeting PPARγ for treatment of type 2 diabetes) and the early evidence of success for emerging targeting (e.g., PCSK9 in cardiovascular disease) suggests the approach may extend to complex inherited diseases (reviewed in [56]).
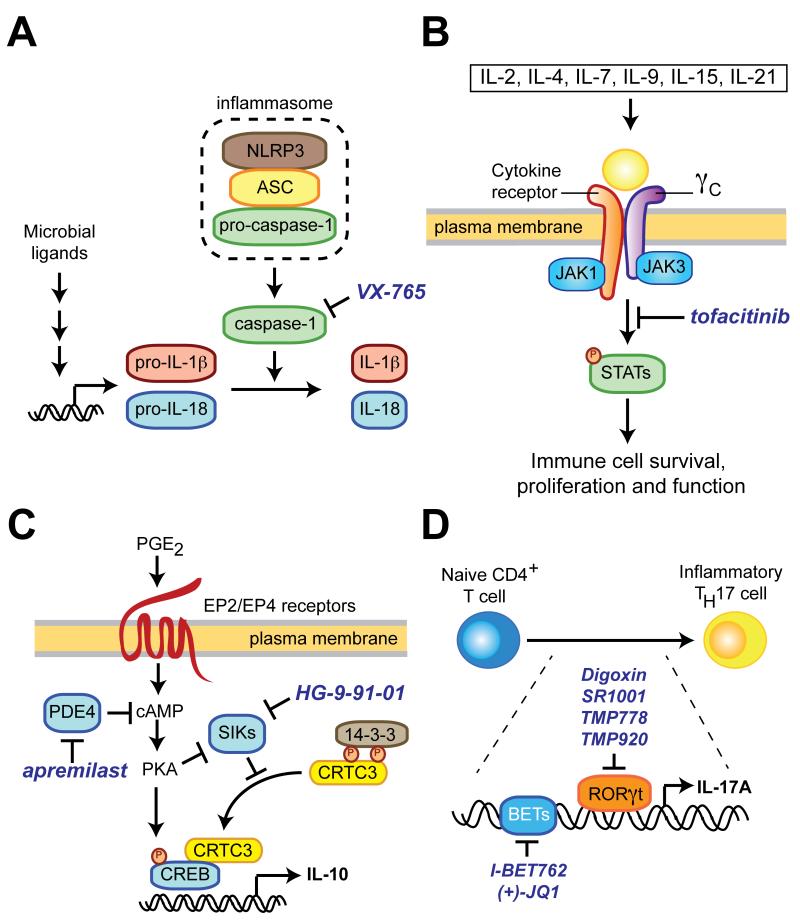
Figure 1.
Representative pathways regulating cytokine production and signaling that have been targeted by small molecules. (A) Microbial stimulation of innate immune cells induces transcription of full-length IL-1β and IL-18. Additional stress signals trigger inflammasome assembly leading to proteolytic processing of IL-1β and IL-18 to their mature forms by caspase-1. VX-765 and other caspase-1 inhibitors specifically block the final stage in this process. NLRP3, NOD-like receptor family, pyrin domain containing 3; ASC, apoptosis-associated speck-like protein containing a carboxy-terminal CARD. (B) The common gamma chain (γc) is a shared component of the receptor for IL-2 and many cytokines and preferentially associates with Janus kinase-3 (JAK3). Stimulation of γc triggers JAK3-dependent phosphorylation of STAT family transcription factors, which leads to pleiotropic effects on immune cell function. Suppression of γc signaling with the small-molecule JAK3 inhibitor tofacitinib is approved therapy for rheumatoid arthritis. STAT, signal transducer and activator of transcription. (C) Salt-inducible kinases (SIKs) restrain IL-10 production by phosphorylation of CRTC3 (CREB-regulated transcriptional co-activator 3), which results in its cytosolic sequestration by 14-3-3 proteins. Stimuli that elevate intracellular cAMP levels (e.g., prostaglandin E2 or PDE4 inhibitors) suppress SIK activity and robustly potentiate IL-10 production by macrophages and dendritic cells, a phenotype that can be mimicked by small molecules that directly inhibit SIKs such as HG-9-91-01. PKA, protein kinase A. (D) Differentiation of inflammatory TH17 cells requires the activity of the transcription factor RORγt and the chromatin-binding BET proteins.
Despite this success, several limitations of biopharmaceuticals hamper therapeutic manipulation of cytokine networks. Most notably, protein-based therapies are unable to regulate intracellular proteins, including many potential targets identified by disease genetics and recent studies of the mechanisms that regulate immune cell development and function [6-8]. Also, while systemic administration of blocking antibodies or decoy receptors can effectively neutralize individual cytokines in circulation, these effects can be undermined by functional redundancy among inflammatory cytokines or limited delivery of protein-based reagents to mucosal tissues [5,9]. Finally, biopharmaceuticals are expensive to produce and lack oral availability, often necessitating administration by specialists.
Small molecules constitute a complementary approach to immunomodulatory drug development by enabling modulation of intracellular proteins that give rise to aberrant cytokine signaling or mediate its downstream consequences. Endogenous small molecules such as eicosanoids have long been recognized to play a key role in controlling tissue-specific inflammation [10], and the impact of metabolites made by commensal microbes on cytokine-producing cells is increasingly clear [11-13]. Moreover, drugs modulating intracellular signaling proteins (rapamycin targeting mTOR [14]; FK506 and cyclosporine A targeting calcineurin [15]) were among the first immunomodulatory therapies approved by the FDA. More recently, small molecules have been discovered that modulate cytokine function through a range of mechanisms-of-action (Table 1). These successes establish small molecules as a complementary alternative to protein-based therapies for regulating cytokine networks. Here, we review recent findings that motivate discovery of small molecules targeting kinases, other classes of signaling proteins and transcriptional regulators implicated in aberrant cytokine signaling by the genetics and physiology of autoimmune/auto-inflammatory disorders.
Table 1.
Examples of small molecule modulators of cytokine function
| Target protein |
Small molecule |
Effects of target modulation on immune cell function |
Approval status |
|---|---|---|---|
| Kinases and phosphatases | |||
| mTOR | rapamycin | Inhibits T and B cell activation; promotes Treg differentiation [14] |
Approved for suppression of transplant rejection |
| Calcineurin | Cyclosporine A; FK506 |
Inhibits NFAT-mediated IL-2 production [15] |
Approved for suppression of transplant rejection |
| JAK3 | Tofacitinib [19] |
Inhibits cytokine via receptors using the common gamma chain (γc) [18] |
Approved for RA |
| SGK1 | GSK650394 [57] |
Inhibits IL-17A production and other phenotypes in salt-induced TH17 cells [29,30]. |
Pre-clinical |
| Lyp/PTPN22 | LTV-1 [58] | Potentiates T and B cell responses [58,59] | Pre-clinical |
| PIKfyve | Apilimod [32] | Inhibits TLR-induced IL-12p40 production [31,32] |
Phase II for Crohn’s Disease; Phase I for Psoriasis |
| SIK2 | HG-9-91-01 [21] |
Potentiates IL-10 production [21,22] | Pre-clinical |
| IRAK1 | IRAK1/4inh [60] |
Inhibits virus-stimulated interferon production [61] |
Pre-clinical |
| LRRK2 | GNE-7915 and others [62] |
Reducing LRRK2 levels suppresses inflammatory cytokine production [26,27] |
Pre-clinical |
| Chromatin modulators | |||
| HDACs | TSA, SAHA and others [63] |
Inhibits inflammatory cytokine production; increase Treg survival/function [64] |
Pre-clinical |
| JMJD3 and UTX |
GSK-J4 [44] | Inhibits inflammatory cytokine production [44] |
Pre-clinical |
| BET proteins | (+)-JQ1, I- BET762 and others [49] |
Inhibits inflammatory cytokine production [65] [50] [51] [52] |
Pre-clinical; phase II for type 2 diabetes (I- BET762) |
| RORγt | Digoxin, SR1001 and others [38,39] |
Inhibits TH17 cells differentiation [38,39] | Pre-clinical |
| AhR | TCDD, kynurenines and others [41,42] |
Stimulates IL-10 production; promotes Treg differentiation [41,42] |
Pre-clinical |
| RAR | ATRA and others |
Promotes Treg differentiation; inhibits TH17 cell differentiation [40] |
Pre-clinical |
| Proteases | |||
| Caspase-1 | VX-765 [66] | Inhibits proteolytic processing of IL-1β and iL-18 to secreted forms [67] |
Phase I for Muckle- Wells syndrome; Phase II for psoriasis |
| Immuno- proteasome |
ONX 0914 and others [68] |
Inhibits antigen processing and inflammatory cytokine production (IL-23; IFNγ; IL-2) [69]. |
Pre-clinical |
| Regulators of second messengers | |||
| PDE4 | Apremilast and others [24] |
Inhibits cAMP degradation [70] | Approved for psoriatic arthritis |
| Receptors | |||
| Toll-like receptors |
Rintatolimod (TLR3) [71]; imiquimod (TLR7) [72]; IMO-2125 (TLR9) [73] |
Stimulates inflammatory cytokine production (e.g. type I IFNs) [16]. |
Phase II for HIV infection (Rintatolimod); Approved for treatment of skin disorders (imiquimod); phase I for Hepatitis C (IMO-2125) |
| IL-2/IL-2R | SP-4206 and others [74] |
Inhibit IL-2 binding to IL-2R [75] | Pre-clinical |
| Prostanoid receptors |
EP4 selective agonists [10] |
Stimulates intracellular cAMP levels [22] | Phase II for ulcerative colitis |
Regulation of immune cell signaling with kinase inhibitors
Small molecules have been used successfully to manipulate immune cell signaling at several levels. Prostanoid receptor agonists are being explored as IBD therapies [10], whereas pathogen receptor agonists (imiquimod) are approved to treat skin disorders [16]. Phosphodiesterase-4 inhibitors such as apremilast, approved for treatment of psoriatic arthritis, demonstrate the utility of modulating intracellular targets within cytokine signaling networks. Given the central role of kinases in cellular networks that control cytokine production and signaling, it is likely that novel kinase inhibitors will be important for treating autoimmune/auto-inflammatory disorders going forward [17].
Although inhibitors of protein kinases have been developed largely for neoplastic disorders in recent years, the first drug of this class (rapamycin) initially obtained FDA approval for use as an immunosuppressant following organ transplantation. Rapamycin forms a ternary complex with FKBP12 and mTOR resulting in an immune cell state reminiscent of nutrient starvation [14]. A consequence is suppression of T and B cell responses normally elicited by activation of antigen receptor and/or IL-2 signaling. This seminal example illustrates the ability of kinase modulators to disrupt coordinately multiple signals needed for lymphocyte activation. The more recent approval of the JAK3 inhibitor tofacatinib for treatment of RA illustrates how small molecules can target redundancies within cytokine signaling networks. JAK3 preferentially associates with the common gamma chain (γc), which is a shared component of the receptor for IL-2 and many other cytokines (Figure 1b) [18]. Blocking γc/JAK3 signaling with tofacatinib affects several immune processes including reducing survival of activated T cells [19].
In addition to suppressing inflammatory cytokine function, kinase inhibitors may be exploited to stimulate production of anti-inflammatory cytokines such as IL-10. The importance of the IL-10 pathway in IBD is evidenced by disease-associated polymorphisms near IL10 and its receptor (IL10RA), as well as near genes that control its production, such as PTGER4 (which encodes the EP4 prostanoid receptor) and the transcriptional co-activator CRTC3 [20]. Salt-inducible kinase 2 (SIK2) normally suppresses IL-10 production by phosphorylation of CRTC3 (CREB-regulated transcriptional co-activator 3), which results in its cytosolic sequestration by 14-3-3 proteins (Figure 1c) [21,22]. Stimuli that enhance cAMP levels (e.g., prostaglandin E2 or PDE4 inhibitors) suppress SIK2 activity and robustly potentiate IL-10 production by macrophages and dendritic cells (DCs), a phenotype that can be mimicked by small molecules that directly inhibit SIK2 [21,22]. Whereas recombinant IL-10 supplementation is ineffective in Crohn’s disease (CD) patients [23], perhaps due to insufficient delivery to the gut mucosa [24], these data suggest that SIK2 inhibition may be effective at increasing IL-10 levels directly in this tissue. The additional ability of SIK2 inhibitors to suppress production of IL-12 and other inflammatory cytokines makes this kinase a promising target for further investigation in IBD.
Studies from genetics, physiology and chemical biology continue to implicate kinases as potential targets for restoring normal cytokine function in disease (Table 1). Novel polymorphisms in leucine-rich repeat kinase 2 (LRRK2, a gene previously linked to Parkinson’s disease) confer increased risk of IBD [25]. Functional studies suggest that LRRK2 regulates production of reactive oxygen species and inflammatory cytokines by macrophages [26,27]. In addition, SNPs near IRAK1, a kinase required for production of interferons (IFNs) following viral infection, confer increased risk of systemic lupus erythematosus [28]. The serum/glucocorticoid-regulated kinase 1 (SGK1) regulates differentiation of TH17 cells, a CD4+ T cell subset that produces IL-17A and other inflammatory cytokines, in response to environmental factors including NaCl; small-molecule inhibition of SGK1 suppresses high salt-induced TH17 development [29,30]. Mechanism-of-action studies have implicated the phoshatidylinositol kinase PIKfyve as the target of the clinical candidate apilimod, an inhibitor of IL-12/23 production discovered through phenotypic screening [31,32]. Targeting kinases implicated in cytokine regulation, with novel inhibitors or those repurposed from other indications, is a critical step for testing novel therapeutic hypotheses and may yield valuable starting points for drug development.
Controlling inflammation by targeted modulation of transcription
Signaling cascades downstream of immune receptors converge on transcription factors to regulate cytokine expression. The clinical success of calcineurin inhibitors, which suppress IL-2 production following T cell receptor stimulation by preventing dephosphorylation of NFAT [15], demonstrates the effectiveness of small molecules that target transcriptional regulation in immune cells. In addition to acute transcriptional responses, activation of immune cells leads to chromatin modifications that can promote acquisition of distinct effector states [6-8]. Genomic studies correlating transcription factor binding and histone modifications with gene expression have identified super-enhancers and other chromatin features that regulate immune cell function [33-35]. These insights, coupled with new tools for targeting transcription factors and chromatin-modifying proteins (Table 1), suggest that small-molecule modulators of transcription will be useful for therapeutic manipulation of cytokine networks.
RORγt (retinoid-related orphan receptor γt) is a nuclear hormone receptor (NHR) implicated in CD by human genetics that promotes differentiation of TH17 cells (Figure 1d) [20,36]. Although a monoclonal antibody targeting IL-17A (secukinumab) has demonstrated potential for treating psoriasis and ankylosing spondylitis, it is ineffective in CD patients [37]. The failure of IL-17A blockade in CD may suggest the need to suppress a wider set of cytokines produced by TH17 cells, possibly by interfering with TH17 differentiation. RORγt contains a deep binding pocket for endogenous small-molecule ligands, which has facilitated development of RORγt antagonists that suppress TH17 cell differentiation and display efficacy in murine models of graft-versus-host disease, demyelinating neurological disorders and cutaneous inflammation [38,39].
Their established roles in immune cell function, coupled with their ability to bind small molecules, make other NHRs intriguing drug targets. Activation of the retinoic acid receptor (RAR) by vitamin A metabolites enhances development of anti-inflammatory CD4+ regulatory T cells (Treg’s), an effect that contribute to the therapeutic activity of all-trans retinoic acid in murine models of autoimmune disease [40]. Binding of the aryl hydrocarbon receptor (AhR) by the tryptophan metabolite kynurenine stimulates IL-10 production by DCs and promotes differentiation of Treg’s [41,42]; two mechanisms that may underlie the finding that sub-lethal doses of bacteria enhance resistance to subsequent infections [43].
NHRs often work in concert with chromatin-modifying enzymes, several classes of which have been targeted with small molecules to modulate cytokine production. The novel small-molecule inhibitor of the jumonji family histone demethylases JMJD3 and UTX (GSK-J4) suppresses inflammatory cytokine production in macrophages [44]. Pan-histone deacetylase (HDAC) inhibitors suppress inflammatory cytokine production by macrophages, promote Treg differentiation and display efficacy in murine models of inflammation [45]. Of note, physiological concentrations of the microbial metabolite and pan-HDAC inhibitor butyrate specifically suppress IL-6, IL-12 and nitric oxide production in gut macrophages suggesting that HDAC inhibition may serve to limit auto-inflammatory responses to commensal microbes [11]. While Hdac3−/− macrophages display reduced inflammatory cytokine production [46], selective deletion of HDAC3 in intestinal epithelial cells alters intestinal architecture and increases sensitivity to experimentally induced colitis [47]. Defining the tissue- and cell-specific functions of individual HDACs, coupled with development of isoform-selective HDAC inhibitors [48], will be needed to discover optimal therapeutic strategies for targeting this class of chromatin modulators.
Proteins that recognize differentially modified histones and transcription factors to effect changes in cell state may themselves be promising points of intervention. For example, the BET (bromodomain and extra terminal) family member BRD4 associates with the master regulator of inflammatory cytokine production NF-κB following acetylation at Lys310 [49]. Disrupting this interaction with the small-molecule pan-BET inhibitor I-BET762 suppresses inflammatory cytokine production by macrophages and protects mice from bacteria-induced sepsis [50]. In addition, inhibiting BRD4 with I-BET762 or (+)-JQ1 is protective in murine models of demyelinating disease by suppressing development of TH1 and TH17 cells [51,52], which are responsible for production of IFNγ and IL-17A, respectively..
Future perspectives: emerging targets from other protein classes
The success of biopharmaceuticals has validated modulation of cytokine function as a therapeutic approach in autoimmune/auto-inflammatory disorders. However, there are clear examples (e.g., IL-10 supplementation; IL-17A blockade in CD) where manipulation of individual cytokines has been ineffective, and studies of the genetics and physiology of these disorders has identified many intracellular proteins that contribute to disease pathogenesis. A desire to overcome these challenges has renewed interest in using in the historically productive approach of regulating cytokine networks with small molecules.
To date, small-molecule regulation of cytokine function has primarily focused on established targets like kinases and transcriptional regulators. However, recent studies are pointing to other protein classes as targets for treating autoimmune/auto-inflammatory disorders. Components of the ubiquitin-proteasome system (e.g., TNFAIP3) are critical for cytokine and pathogen receptor signaling, and have been linked to IBD, SLE, RA and type 1 diabetes by genetics [17]. In addition, the discovery of risk and protective alleles for IBD in CARD9 suggests that scaffolding proteins may likewise be useful points of intervention [53]. Although traditional drug discovery has little experience with many emerging classes of targets, recent innovations in small-molecule science suggest that significant advances in this field will be forthcoming.
Acknowledgements
The Leona M. and Harry B. Helmsley Charitable Trust (R.J.X, A.F.S., T.B.S., S.L.S.) and Howard Hughes Medical Institute (S.L.S.) provided support for the authors of this manuscript. The authors declare that the funding sources did not play a role in the preparation of or decision to submit this manuscript.
REFERENCES
- 1.Belkaid Y, Hand TW. Role of the Microbiota in Immunity and Inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.So A, Ives A, Joosten LA, Busso N. Targeting inflammasomes in rheumatic diseases. Nat Rev Rheumatol. 2013;9:391–399. doi: 10.1038/nrrheum.2013.61. [DOI] [PubMed] [Google Scholar]
- 3.de Luca A, Smeekens SP, Casagrande A, Iannitti R, Conway KL, Gresnigt MS, Begun J, Plantinga TS, Joosten LAB, van der Meer JWM, et al. IL-1 receptor blockade restores autophagy and reduces inflammation in chronic granulomatous disease in mice and in humans. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:3526–3531. doi: 10.1073/pnas.1322831111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinarello CA, Simon A, van der Meer JWM. Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nature Reviews Drug Discovery. 2012;11:633–652. doi: 10.1038/nrd3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Averting inflammation by targeting the cytokine environment. Nature Reviews Drug Discovery. 2010;9:703–718. doi: 10.1038/nrd2805. • An informative overview of the major classes of inflammatory cytokines and strategies for their therapeutic manipulation.
- 6.Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, Wu C, Karwacz K, Xiao S, Jorgolli M, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496:461–468. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quintin J, Saeed S, Martens JH, Giamarellos-Bourboulis EJ, Ifrim DC, Logie C, Jacobs L, Jansen T, Kullberg BJ, Wijmenga C, et al. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe. 2012;12:223–232. doi: 10.1016/j.chom.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, De Nardo D, Gohel TD, Emde M, Schmidleithner L, et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Shahbazi M-A, Almeida PV, Santos HA. In: Mucus as a Barrier for Biopharmaceuticals and Drug Delivery Systems. Neves Jd, Sarmento B., editors. Springer; 2014. [Google Scholar]
- 10.Sugimoto Y, Narumiya S. Prostaglandin E receptors. J Biol Chem. 2007;282:11613–11617. doi: 10.1074/jbc.R600038200. [DOI] [PubMed] [Google Scholar]
- 11.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corbett AJ, Eckle SBG, Birkinshaw RW, Liu LG, Patel O, Mahony J, Chen ZJ, Reantragoon R, Meehan B, Cao HW, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014;509:361. doi: 10.1038/nature13160. + [DOI] [PubMed] [Google Scholar]
- 14.Thomson AW, Turnquist HR, Raimondi G. Immunoregulatory functions of mTOR inhibition. Nat Rev Immunol. 2009;9:324–337. doi: 10.1038/nri2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schreiber SL. Immunophilin-sensitive protein phosphatase action in cell signaling pathways. Cell. 1992;70:365–368. doi: 10.1016/0092-8674(92)90158-9. [DOI] [PubMed] [Google Scholar]
- 16.Hennessy EJ, Parker AE, O'Neill LA. Targeting Toll-like receptors: emerging therapeutics? Nat Rev Drug Discov. 2010;9:293–307. doi: 10.1038/nrd3203. [DOI] [PubMed] [Google Scholar]
- 17.Cohen P. Immune diseases caused by mutations in kinases and components of the ubiquitin system. Nat Immunol. 2014;15:521–529. doi: 10.1038/ni.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol. 2009;9:480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vijayakrishnan L, Venkataramanan R, Gulati P. Treating inflammation with the Janus kinase inhibitor CP-690,550. Trends Pharmacol Sci. 2011;32:25–34. doi: 10.1016/j.tips.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. • A comprehensive catalog of genetic variants that affect susceptibility to inflammatory bowel disease.
- 21.Clark K, MacKenzie KF, Petkevicius K, Kristariyanto Y, Zhang J, Choi HG, Peggie M, Plater L, Pedrioli PG, McIver E, et al. Phosphorylation of CRTC3 by the salt-inducible kinases controls the interconversion of classically activated and regulatory macrophages. Proc Natl Acad Sci U S A. 2012;109:16986–16991. doi: 10.1073/pnas.1215450109. •• Seminal study identifying inhibition of salt-inducible kinases as a strategy to enhance production of the anti-inflammatory cytokine IL-10 by innate immune cells.
- 22.MacKenzie KF, Clark K, Naqvi S, McGuire VA, Noehren G, Kristariyanto Y, van den Bosch M, Mudaliar M, McCarthy PC, Pattison MJ, et al. PGE(2) induces macrophage IL-10 production and a regulatory-like phenotype via a protein kinase A-SIK-CRTC3 pathway. J Immunol. 2013;190:565–577. doi: 10.4049/jimmunol.1202462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colombel JF, Rutgeerts P, Malchow H, Jacyna M, Nielsen OH, Rask-Madsen J, Van Deventer S, Ferguson A, Desreumaux P, Forbes A, et al. Interleukin 10 (Tenovil) in the prevention of postoperative recurrence of Crohn's disease. Gut. 2001;49:42–46. doi: 10.1136/gut.49.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marlow GJ, van Gent D, Ferguson LR. Why interleukin-10 supplementation does not work in Crohn's disease patients. World J Gastroenterol. 2013;19:3931–3941. doi: 10.3748/wjg.v19.i25.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardet A, Benita Y, Li C, Sands BE, Ballester I, Stevens C, Korzenik JR, Rioux JD, Daly MJ, Xavier RJ, et al. LRRK2 is involved in the IFN-gamma response and host response to pathogens. J Immunol. 2010;185:5577–5585. doi: 10.4049/jimmunol.1000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Z, Lee J, Krummey S, Lu W, Cai H, Lenardo MJ. The kinase LRRK2 is a regulator of the transcription factor NFAT that modulates the severity of inflammatory bowel disease. Nat Immunol. 2011;12:1063–1070. doi: 10.1038/ni.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacob CO, Zhu J, Armstrong DL, Yan M, Han J, Zhou XJ, Thomas JA, Reiff A, Myones BL, Ojwang JO, et al. Identification of IRAK1 as a risk gene with critical role in the pathogenesis of systemic lupus erythematosus. Proc Natl Acad Sci U S A. 2009;106:6256–6261. doi: 10.1073/pnas.0901181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleinewietfeld M, Manzel A, Titze J, Kvakan H, Yosef N, Linker RA, Muller DN, Hafler DA. Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature. 2013;496:518–522. doi: 10.1038/nature11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu C, Yosef N, Thalhamer T, Zhu C, Xiao S, Kishi Y, Regev A, Kuchroo VK. Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature. 2013;496:513–517. doi: 10.1038/nature11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai X, Xu Y, Cheung AK, Tomlinson RC, Alcazar-Roman A, Murphy L, Billich A, Zhang B, Feng Y, Klumpp M, et al. PIKfyve, a class III PI kinase, is the target of the small molecular IL-12/IL-23 inhibitor apilimod and a player in Toll-like receptor signaling. Chem Biol. 2013;20:912–921. doi: 10.1016/j.chembiol.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wada Y, Lu R, Zhou D, Chu J, Przewloka T, Zhang S, Li L, Wu Y, Qin J, Balasubramanyam V, et al. Selective abrogation of Th1 response by STA-5326, a potent IL-12/IL-23 inhibitor. Blood. 2007;109:1156–1164. doi: 10.1182/blood-2006-04-019398. • Along with ref 31, provides an illustrative example of using phenotypic screening followed by mechanism-of-action studies to identify a novel target (PIKfyve) for regulation of inflammatory cytokine production.
- 33.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Consortium EP. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anders L, Guenther MG, Qi J, Fan ZP, Marineau JJ, Rahl PB, Loven J, Sigova AA, Smith WB, Lee TI, et al. Genome-wide localization of small molecules. Nat Biotechnol. 2014;32:92–96. doi: 10.1038/nbt.2776. • Describes an innovative approach for elucidating how gene expression is affected by small molecules that target transcriptional machinery.
- 36.Littman DR, Rudensky AY. Th17 and regulatory T cells in mediating and restraining inflammation. Cell. 2010;140:845–858. doi: 10.1016/j.cell.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 37.Hueber W, Sands BE, Lewitzky S, Vandemeulebroecke M, Reinisch W, Higgins PD, Wehkamp J, Feagan BG, Yao MD, Karczewski M, et al. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn's disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut. 2012;61:1693–1700. doi: 10.1136/gutjnl-2011-301668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huh JR, Littman DR. Small molecule inhibitors of RORgammat: targeting Th17 cells and other applications. Eur J Immunol. 2012;42:2232–2237. doi: 10.1002/eji.201242740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao S, Yosef N, Yang J, Wang Y, Zhou L, Zhu C, Wu C, Baloglu E, Schmidt D, Ramesh R, et al. Small-molecule RORgammat antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity. 2014;40:477–489. doi: 10.1016/j.immuni.2014.04.004. • Recent example in which profiling of transcription factor binding identified functionally significant differences in the mechanisms-of-action of RORγt antagonists.
- 40.Xiao S, Jin H, Korn T, Liu SM, Oukka M, Lim B, Kuchroo VK. Retinoic acid increases Foxp3+ regulatory T cells and inhibits development of Th17 cells by enhancing TGF-beta-driven Smad3 signaling and inhibiting IL-6 and IL-23 receptor expression. J Immunol. 2008;181:2277–2284. doi: 10.4049/jimmunol.181.4.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mezrich JD, Fechner JH, Zhang XJ, Johnson BP, Burlingham WJ, Bradfield CA. An Interaction between Kynurenine and the Aryl Hydrocarbon Receptor Can Generate Regulatory T Cells. Journal of Immunology. 2010;185:3190–3198. doi: 10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen NT, Kimura A, Nakahama T, Chinen I, Masuda K, Nohara K, Fujii-Kuriyama Y, Kishimoto T. Aryl hydrocarbon receptor negatively regulates dendritic cell immunogenicity via a kynurenine-dependent mechanism. Proc Natl Acad Sci U S A. 2010;107:19961–19966. doi: 10.1073/pnas.1014465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bessede A, Gargaro M, Pallotta MT, Matino D, Servillo G, Brunacci C, Bicciato S, Mazza EM, Macchiarulo A, Vacca C, et al. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature. 2014 doi: 10.1038/nature13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, Bantscheff M, Bountra C, Bridges A, Diallo H, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488:404–408. doi: 10.1038/nature11262. •• Illustrative example of using small-molecule discovery to define novel roles for demethylases in regulation of inflammatory cytokine production.
- 45.Sweet MJ, Shakespear MR, Kamal NA, Fairlie DP. HDAC inhibitors: modulating leukocyte differentiation, survival, proliferation and inflammation. Immunol Cell Biol. 2012;90:14–22. doi: 10.1038/icb.2011.88. [DOI] [PubMed] [Google Scholar]
- 46.Chen X, Barozzi I, Termanini A, Prosperini E, Recchiuti A, Dalli J, Mietton F, Matteoli G, Hiebert S, Natoli G. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc Natl Acad Sci U S A. 2012;109:E2865–2874. doi: 10.1073/pnas.1121131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alenghat T, Osborne LC, Saenz SA, Kobuley D, Ziegler CG, Mullican SE, Choi I, Grunberg S, Sinha R, Wynosky-Dolfi M, et al. Histone deacetylase 3 coordinates commensal-bacteria-dependent intestinal homeostasis. Nature. 2013;504:153–157. doi: 10.1038/nature12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta P, Reid RC, Iyer A, Sweet MJ, Fairlie DP. Towards isozyme-selective HDAC inhibitors for interrogating disease. Curr Top Med Chem. 2012;12:1479–1499. doi: 10.2174/156802612802652420. [DOI] [PubMed] [Google Scholar]
- 49.Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov. 2014;13:337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 50.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mele DA, Salmeron A, Ghosh S, Huang HR, Bryant BM, Lora JM. BET bromodomain inhibition suppresses TH17-mediated pathology. J Exp Med. 2013;210:2181–2190. doi: 10.1084/jem.20130376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bandukwala HS, Gagnon J, Togher S, Greenbaum JA, Lamperti ED, Parr NJ, Molesworth AM, Smithers N, Lee K, Witherington J, et al. Selective inhibition of CD4+ T-cell cytokine production and autoimmunity by BET protein and c-Myc inhibitors. Proc Natl Acad Sci U S A. 2012;109:14532–14537. doi: 10.1073/pnas.1212264109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beaudoin M, Goyette P, Boucher G, Lo KS, Rivas MA, Stevens C, Alikashani A, Ladouceur M, Ellinghaus D, Torkvist L, et al. Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet. 2013;9:e1003723. doi: 10.1371/journal.pgen.1003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lassen KG, Kuballa P, Conway KL, Patel KK, Becker CE, Peloquin JM, Villablanca EJ, Norman JM, Liu TC, Heath RJ, et al. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc Natl Acad Sci U S A. 2014;111:7741–7746. doi: 10.1073/pnas.1407001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murthy A, Li Y, Peng I, Reichelt M, Katakam AK, Noubade R, Roose-Girma M, DeVoss J, Diehl L, Graham RR, et al. A Crohn's disease variant in Atg16l1 enhances its degradation by caspase 3. Nature. 2014;506:456–462. doi: 10.1038/nature13044. [DOI] [PubMed] [Google Scholar]
- 56.Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat Rev Drug Discov. 2013;12:581–594. doi: 10.1038/nrd4051. •• Informative introduction to the application of disease genetics to drug discovery.
- 57.Sherk AB, Frigo DE, Schnackenberg CG, Bray JD, Laping NJ, Trizna W, Hammond M, Patterson JR, Thompson SK, Kazmin D, et al. Development of a small-molecule serum- and glucocorticoid-regulated kinase-1 antagonist and its evaluation as a prostate cancer therapeutic. Cancer Research. 2008;68:7475–7483. doi: 10.1158/0008-5472.CAN-08-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vang T, Liu WH, Delacroix L, Wu S, Vasile S, Dahl R, Yang L, Musumeci L, Francis D, Landskron J, et al. LYP inhibits T-cell activation when dissociated from CSK. Nat Chem Biol. 2012;8:437–446. doi: 10.1038/nchembio.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rhee I, Veillette A. Protein tyrosine phosphatases in lymphocyte activation and autoimmunity. Nat Immunol. 2012;13:439–447. doi: 10.1038/ni.2246. [DOI] [PubMed] [Google Scholar]
- 60.Powers JP, Li S, Jaen JC, Liu J, Walker NP, Wang Z, Wesche H. Discovery and initial SAR of inhibitors of interleukin-1 receptor-associated kinase-4. Bioorg Med Chem Lett. 2006;16:2842–2845. doi: 10.1016/j.bmcl.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 61.Uematsu S, Sato S, Yamamoto M, Hirotani T, Kato H, Takeshita F, Matsuda M, Coban C, Ishii KJ, Kawai T, et al. Interleukin-1 receptor-associated kinase-1 plays an essential role for Toll-like receptor (TLR)7- and TLR9-mediated interferon-{alpha} induction. J Exp Med. 2005;201:915–923. doi: 10.1084/jem.20042372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kavanagh ME, Doddareddy MR, Kassiou M. The development of CNS-active LRRK2 inhibitors using property-directed optimisation. Bioorg Med Chem Lett. 2013;23:3690–3696. doi: 10.1016/j.bmcl.2013.04.086. [DOI] [PubMed] [Google Scholar]
- 63.Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, Mazitschek R. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Glauben R, Siegmund B. Inhibition of histone deacetylases in inflammatory bowel diseases. Mol Med. 2011;17:426–433. doi: 10.2119/molmed.2011.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Belkina AC, Nikolajczyk BS, Denis GV. BET Protein Function Is Required for Inflammation: Brd2 Genetic Disruption and BET Inhibitor JQ1 Impair Mouse Macrophage Inflammatory Responses. Journal of Immunology. 2013;190:3670–3678. doi: 10.4049/jimmunol.1202838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.MacKenzie SH, Schipper JL, Clark AC. The potential for caspases in drug discovery. Curr Opin Drug Discov Devel. 2010;13:568–576. [PMC free article] [PubMed] [Google Scholar]
- 67.Wannamaker W, Davies R, Namchuk M, Pollard J, Ford P, Ku G, Decker C, Charifson P, Weber P, Germann UA, et al. (S)-1-((S)-2-{[1-(4-amino-3-chloro-phenyl)-methanoyl]-amino}-3,3-dimethyl-butanoy l)-pyrrolidine-2-carboxylic acid ((2R,3S)-2-ethoxy-5-oxo-tetrahydro-furan-3-yl)-amide (VX-765), an orally available selective interleukin (IL)-converting enzyme/caspase-1 inhibitor, exhibits potent anti-inflammatory activities by inhibiting the release of IL-1beta and IL-18. J Pharmacol Exp Ther. 2007;321:509–516. doi: 10.1124/jpet.106.111344. [DOI] [PubMed] [Google Scholar]
- 68.Miller Z, Ao L, Kim KB, Lee W. Inhibitors of the immunoproteasome: current status and future directions. Curr Pharm Des. 2013;19:4140–4151. doi: 10.2174/1381612811319220018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Muchamuel T, Basler M, Aujay MA, Suzuki E, Kalim KW, Lauer C, Sylvain C, Ring ER, Shields J, Jiang J, et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat Med. 2009;15:781–787. doi: 10.1038/nm.1978. [DOI] [PubMed] [Google Scholar]
- 70.Schafer P. Apremilast mechanism of action and application to psoriasis and psoriatic arthritis. Biochem Pharmacol. 2012;83:1583–1590. doi: 10.1016/j.bcp.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 71.Jasani B, Navabi H, Adams M. Ampligen: a potential toll-like 3 receptor adjuvant for immunotherapy of cancer. Vaccine. 2009;27:3401–3404. doi: 10.1016/j.vaccine.2009.01.071. [DOI] [PubMed] [Google Scholar]
- 72.Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, Horiuchi T, Tomizawa H, Takeda K, Akira S. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 73.Agrawal S, Kandimalla ER. Synthetic agonists of Toll-like receptors 7, 8 and 9. Biochem Soc Trans. 2007;35:1461–1467. doi: 10.1042/BST0351461. [DOI] [PubMed] [Google Scholar]
- 74.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 75.Braisted AC, Oslob JD, Delano WL, Hyde J, McDowell RS, Waal N, Yu C, Arkin MR, Raimundo BC. Discovery of a potent small molecule IL-2 inhibitor through fragment assembly. Journal of the American Chemical Society. 2003;125:3714–3715. doi: 10.1021/ja034247i. [DOI] [PubMed] [Google Scholar]