Abstract
Complex interactions between the host and the gut microbiota govern intestinal homeostasis but remain poorly understood. Here we reveal a relationship between gut microbiota and caspase recruitment domain family member 9 (CARD9), a susceptibility gene for inflammatory bowel disease (IBD) that functions in the immune response against microorganisms. CARD9 promotes recovery from colitis by promoting interleukin (IL)-22 production, and Card9−/− mice are more susceptible to colitis. The microbiota is altered in Card9−/− mice, and transfer of the microbiota from Card9−/− to wild-type, germ-free recipients increases their susceptibility to colitis. The microbiota from Card9−/− mice fails to metabolize tryptophan into metabolites that act as aryl hydrocarbon receptor (AHR) ligands. Intestinal inflammation is attenuated after inoculation of mice with three Lactobacillus strains capable of metabolizing tryptophan or by treatment with an AHR agonist. Reduced production of AHR ligands is also observed in the microbiota from individuals with IBD, particularly in those with CARD9 risk alleles associated with IBD. Our findings reveal that host genes affect the composition and function of the gut microbiota, altering the production of microbial metabolites and intestinal inflammation.
The microbial community in the human gastrointestinal tract is fundamental to the health and nutrition of the host1. Loss of the fragile equilibrium within this complex ecosystem, termed dysbiosis, is involved in numerous pathologies, including IBD. The incidence of IBD increased during the 20th century and continues to rise2. IBD develops as a result of a combination of genetic predisposition, dysbiosis of the gut microbiota, and environmental influences3. CARD9, one of the numerous IBD susceptibility genes, encodes an adaptor protein that integrates signals downstream of pattern recognition receptors. CARD9 is particularly involved in the immune response to fungi via C-type lectin sensing4, but it also has a role in response to bacteria by mediating nucleotide-binding oligomerization domain containing 2 (NOD2)-dependent mitogen-activated protein kinase 14 (MAPK14; also known as p38)]–Janus kinase (JNK) signaling5 and modulating Toll-like receptor signaling6,7. CARD9 promotes recovery from colitis through activation of the IL-22 pathway, and Card9−/− mice are more susceptible to colitis and have an increased load of gut-resident fungi8. Dysbiosis is often viewed as a contributor to intestinal inflammation owing to increased numbers of pro-inflammatory microorganisms9. However, a lack of microorganisms that have regulatory functions may also enhance inflammation10,11.
Recent data have suggested that tryptophan catabolites generated by metabolism by the microbiota have a role in mucosal immune responses via AHR12 by modulating production of IL-22, a cytokine with well-known effects on intestinal homeostasis12,13. Here we assessed the interaction between CARD9, the microbiota and tryptophan in intestinal homeostasis.
RESULTS
Card9−/− mice are more susceptible to colitis
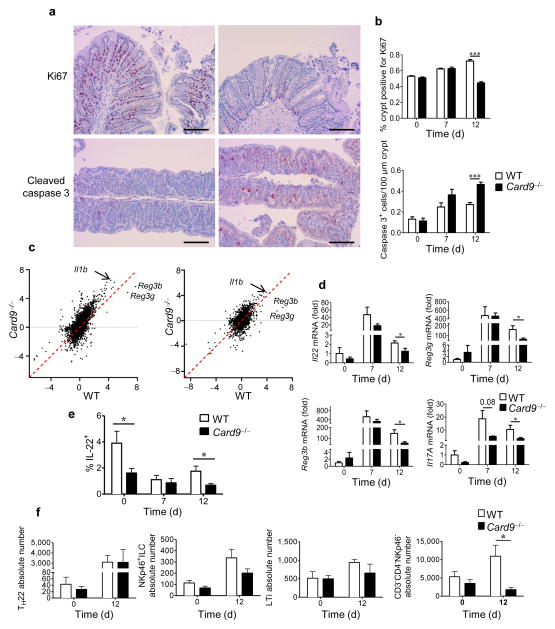
Recovery is impaired in Card9−/− mice after dextran sulfate sodium (DSS)-induced colitis, as compared to that in WT mice, with delayed weight gain and greater histopathologic alterations (Supplementary Fig. 1)8. Epithelial cell proliferation is reduced and apoptosis is increased, as shown by decreased staining for the proliferation marker Ki67 and increased staining for cleaved caspase 3, respectively, confirming impaired intestinal healing (Fig. 1a,b). To examine the mechanisms responsible for this defect, we compared the colon transcriptomes of wild-type (WT) and Card9−/− mice before and during DSS-induced colitis. The mouse transcriptomes clustered according to genotype (Supplementary Fig. 2a). The number of upregulated genes on day 7 was higher in Card9−/− mice than in WT mice. Pathway analyses of the induced transcripts showed a dominance of immune-related pathways, corresponding to a stronger signal in Card9−/− mice (Supplementary Fig. 2b). During the recovery period (day 12), the pathways involved in cell proliferation and replication were more activated in WT mice than in Card9−/− mice, confirming the healing defect in Card9−/− mice (Supplementary Fig. 2c). The most induced and differentially expressed genes between Card9−/− and WT mice on days 7 and 12 were regenerating islet-derived 3 gamma (Reg3g; encoding REGIII-γ), Reg3b (encoding REGIII-β) and interleukin 1beta (Il1b) (Fig. 1c). The expression of antimicrobial proteins, such as REGIII-γ and REGIII-β, by intestinal epithelial cells is induced by IL-22 (ref. 14,15). Moreover, IL-17A has a protective role in concert with IL-22 (ref. 16,17). As assessed by real-time qPCR, colonic expression of Il22, Reg3g, Reg3b, and Il17a was decreased on day 12 in Card9−/− mice (Fig. 1d). In the colon lamina propria, the percentage of IL-22+ cells was decreased in Card9−/− mice on days 0 and 12 (Fig. 1e). Among these cells, the CD3−CD4−NKp46− subset was reduced on day 12 in Card9−/− mice (Fig. 1f and Supplementary Fig. 3). These results highlight the role of CARD9 and its effector IL-22 in mediating recovery from DSS-induced colitis. The major role of IL-22 and its target genes Reg3g and Reg3b in the response to bacterial and fungal infections4,5,8,18 raises questions about the specific role of the microbiota in the hyper-susceptibility to colitis that is observed in Card9−/− mice.
Figure 1.
CARD9 is involved in recovery from colitis. (a) Representative immunohistochemistry images for cross-sections of the proximal colon from WT (left) and Card9−/− (right) mice stained for Ki67 (top) or cleaved caspase-3 (bottom) at day 12. Scale bars, 200 μm. (b) Quantification of Ki67 (top) and cleaved caspase-3 (bottom) staining in the proximal colon of WT and Card9−/− mice before and at days 7 and 12 after induction of colitis. Data are mean ± s.e.m. (c) Comparative expression of genes in the colon by microarray data analysis (log2-transformed (fold change, as compared to that at day 0)) on day 7 (left) and day 12 (right) after initiation of DSS treatment. (d) Il22, Reg3g, Reg3b, and Il17A transcript expression in the colon before (day 0, n = 3 mice per group) and after (day 7, n = 5 mice per group; day 12, n = 10 mice per group) initiation of DSS treatment. (e) Proportion of IL-22+ cells in the lamina propria of the colon in WT and Card9−/− mice. (f) Quantification of subpopulations of IL-22+ cells isolated from the lamina propria of the colon of WT and Card9−/− mice before (day 0) and after (day 12) DSS treatment. Cells were gated on CD3+CD4+ (for TH22), CD3−CD4−NKp46+ (for NKp46+ ILCs), CD3−CD4+NKp46− (for LTi), and CD3−CD4−NKp46−. In a–c, n = 3 (day 0) and n = 5 (days 7 and 12) mice per group; in e,f, n = 5 mice per group. Throughout, data are mean ± s.e.m. *P < 0.05; ***P < 0.001; by two-tailed Student’s t-test.
The gut microbiota is altered in Card9−/− mice
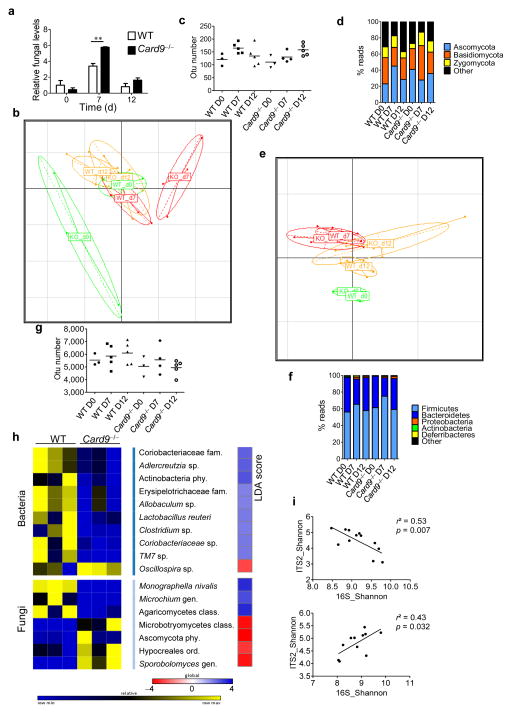
We next explored the composition of the microbiota at the fungal and bacterial level at baseline and during colitis (Fig. 2). In Card9−/− and WT mice, the colonic fungal load reached a peak at day 7, but it was higher in Card9−/− mice (Fig. 2a). Little is known regarding the diversity of the gut fungal microbiota and how fungi contribute to colitis in mice19,20. We therefore further analyzed the fungal microbiota in feces via high-throughput internal transcribed spacer 2 (ITS2) sequencing. Principal component analysis (PCA) on the basis of genus composition revealed major differences between WT and Card9−/− mice at days 0 and 7. The fungal composition in Card9−/− mice was altered at days 0, 7, and 12, whereas it changed only slightly with colitis in WT mice, showing more resilience to DSS-induced perturbation (Fig. 2b and Supplementary Fig. 4a). Diversity measurements also suggested increased resilience of the fungal microbiota in WT mice than in Card9−/− mice (Fig. 2c). The fungal microbiota of WT and Card9−/− mice was dominated by members of the phyla Ascomycota, Basidiomycota, and Zygomycota (Fig. 2d and Supplementary Fig. 4b). Using the linear discriminant analysis (LDA) effect size (LEfSe) pipeline21, we observed several differences in the baseline fecal fungal microbiota composition in Card9−/− mice, as compared to that in WT mice (Fig. 2h and Supplementary Fig. 4b). In line with the PCA results, the composition of the fungal microbiota was more altered at days 7 and 12 in Card9−/− mice than in WT mice (Supplementary Fig. 5). In parallel, we explored the composition of the fecal bacterial microbiota by using 16S rDNA sequencing. Although the alterations were less marked than for the fungal microbiota, PCA revealed that the baseline bacterial microbiota was different in WT and Card9−/− mice (Supplementary Fig. 4c). Moreover, the shift in the composition of the bacterial microbiota during colitis followed a similar pattern in WT and Card9−/− mice, but with decreased stability in Card9−/− mice (Fig. 2e,f and Supplementary Fig. 4c,d). No differences were observed regarding biodiversity between WT and Card9−/− mice at any of the time points (Fig. 2g). The LEfSe analysis revealed differences at baseline, including decreases in Adlercreutzia (genus), Actinobacteria (phylum), and Lactobacillus reuteri in the Card9−/− mouse microbiota (Fig. 2h). In WT mice, there was a negative correlation between bacterial and fungal biodiversity (Fig. 2i), suggesting an inter-kingdom relationship and possibly competition. In contrast, a positive correlation was observed in Card9−/− mice, suggesting abnormal interactions within the gut ecosystem between bacteria and fungi, with possible consequences on gut homeostasis (Fig. 2i). Overall, these data demonstrate that CARD9 has a role in shaping the bacterial and fungal gut microbiota and that it is required to control fungal microbiota expansion in colitis.
Figure 2.
The fungal and bacterial microbiota are altered in Card9−/− mice. (a) Fungal levels in the fecal microbiota were quantified using 18S rRNA qRT–PCR and were normalized to those of the bacterial population. Data are mean ± s.e.m. **P < 0.01 by two-tailed Student’s t-test. (b) Principal component analysis (PCA) based on fungal ITS2 rDNA gene sequence abundance in the feces. Axes correspond to principal components 1 (x axis) and 2 (y axis). d, day; KO, Card9−/− mice. (c) Fungal diversity on the basis of the operational taxonomic unit (OTU) number in the fecal samples from WT and Card9−/− mice. Horizontal line depicts the mean. (d) Fungal-taxon-based analysis at the phylum level in the fecal microbiota. (e) PCA plot based on bacterial 16S rDNA gene sequence abundance in fecal content. Axes correspond to principal components 1 (x axis) and 2 (y axis). (f) Bacterial-taxon-based analysis at the phylum level in the feces. (g) Bacterial diversity based on the OTU number in the fecal samples. Horizontal line depicts the mean. (h) Bacterial and fungal taxa differentially enriched in WT and Card9−/− mice (generated using LeFSE analysis). The heat map on the left shows the relative abundance of taxa, and the heat map on the right shows the linear differential analysis (LDA) scores. (i) Correlation between ITS2 and 16S rDNA Shannon diversity index in the fecal samples from DSS-treated WT (top) and Card9−/− (bottom) mice. Significance determined using linear regression. **P < 0.01, two-tailed Student’s t-test (a) and one-way analysis of variance (ANOVA) and post hoc Tukey test (c,g). Throughout, n = 3 mice per group for day 0, and n = 5 mice per group for days 7 and 12.
The Card9−/− microbiota has pro-inflammatory effects
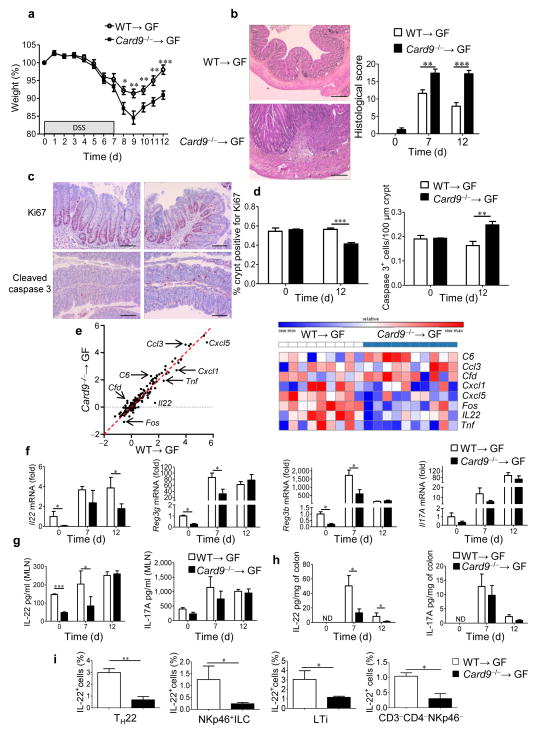
To isolate the effect of the gut microbiota, we colonized WT germ-free (GF) mice with the microbiota of WT (WT→GF) or Card9−/− (Card9−/−→GF) mice and exposed them to DSS. The microbiota transfer was sufficient to recapitulate the phenotype observed in Card9−/− mice, as evidenced by increased susceptibility to colitis and impaired recovery, with decreased epithelial cell proliferation and increased apoptosis (Fig. 3a–d and Supplementary Fig. 6a). However, the fungal component of the microbiota did not seem to be involved in this effect, as fungal levels were similar in WT→GF and Card9−/−→GF mice, and antifungal treatment did not modify the phenotype (Supplementary Fig. 6b–d). To decipher the mechanisms leading to the defective recovery from DSS-induced injury in Card9−/−→GF mice, we assessed the colonic expression levels of 179 inflammation-associated genes using NanoString technology (Supplementary Fig. 6e). Il22 was one of the most highly downregulated genes in Card9−/−→GF mice, as compared to those in WT→GF mice (Fig. 3e), in addition to the chemokine-encoding genes Cxcl1 and Cxcl5 and the transcription-factor-encoding gene Fos, all of which are known IL-22 target genes in epithelial cells22–24. IL-22 has been implicated in intestinal homeostasis13, and it mediates innate antimicrobial resistance in mice12,16. Furthermore, similar to Card9−/− and Card9−/−→GF mice, Il22−/− mice are more susceptible to DSS-induced colitis and show impaired healing during the recovery period25. Therefore, we postulated that a deficient IL-22 response may underlie impaired recovery of Card9−/−→GF mice during DSS-induced epithelial injury. In agreement with the NanoString results, Il22 expression in the colon was decreased in Card9−/−→GF mice on days 0 and 12 (Fig. 3f). Moreover, the expression levels of Reg3g and Reg3b were reduced in Card9−/−→GF mice, as compared to those in WT→GF mice at days 0 and 7 (Fig. 3f). In contrast, no differences were detected in the expression of Il17a (Fig. 3f) or defensin beta 1 (Defb1), which is a target of IL-17A17 (Supplementary Fig. 7a). We next confirmed the IL-22 defect in Card9−/−→GF mice at the protein level in the colon and mesenteric lymph nodes (MLNs) at baseline, day 7 and day 12 (Fig. 3g,h). We found no differences in the amounts of IL-17A, IL-6, IFN-γ, or IL-10 in the MLN or colon (Fig. 3g,h and Supplementary Fig. 7b,c), suggesting that the IL-22 axis is specifically impaired in Card9−/−→GF mice. No differences in IL-22 amounts were observed in splenocytes, suggesting a gut-limited defect in Card9−/−→GF mice (Supplementary Fig. 7d). Several sources of IL-22 have been identified in the gut, including innate lymphoid cells (ILCs), natural killer cells, T helper 17 (TH17) and TH22 cells, γδ T cells, and lymphoid tissue inducer (LTi) cells14,26. Following isolation of intraepithelial and lamina propria cells, we observed that IL-22 production by TH22, NKp46+ ILCs, LTi cells, and CD3−CD4−NKp46− cells was decreased in the colon lamina propria of Card9−/−→GF mice, as compared to that in the colon lamina propria of WT→GF mice on day 12 (Fig. 3i and Supplementary Fig. 8). In contrast, IL-17 production was not altered, and no difference was observed in IL-22 or IL-17 production by intraepithelial γδ T cells (Supplementary Figs. 8c and 9). Intestinal dendritic cells and macrophages, which are involved in the stimulation of IL-22-producing cells via IL-23 production14, were also explored, but no differences were observed between Card9−/−→GF and WT→GF mice (Supplementary Fig. 10). These data indicate that the microbiota of Card9−/− mice is defective in inducing IL-22 production by T cells and ILCs in the colon, leading to impaired recovery from DSS-induced colitis.
Figure 3.
Transfer of the microbiota from Card9−/− mice is sufficient to increase susceptibility to colitis and reduce IL-22 production. (a) Weight of DSS-exposed GF WT mice colonized with microbiota from WT (WT→GF) or Card9−/− (Card9−/−→GF) mice (n = 23 per condition). Data are mean ± s.e.m. of four experiments. (b) Representative H&E-stained images (of three experiments) of mouse proximal colon samples (left) and mouse histological scores at day 12 (right). Data are representative of three experiments. Scale bars, 200 μm. (c,d) Representative immunohistochemistry images (of three experiments) for Ki67 (top) and cleaved caspase-3 (bottom) staining in proximal colon samples from WT→GF (left) and Card9−/−→GF (right) mice (c) and quantification of Ki67 and cleaved caspase-3 staining in the proximal colon at day 12. Data are representative of three experiments. Scale bars, 200 μm. (e) Comparative analysis (left) and heat map (right) of the gene expression in the colons of WT→GF or Card9−/−→GF mice on day 12, using NanoString. (f) Transcript expression in the colon of WT→GF or Card9−/−→GF mice. (g) Secreted IL-22 (left) and IL-17A (right) amounts by MLN cells from WT→GF or Card9−/−→GF mice. (h) Secreted IL-22 (left) and IL-17A (right) amounts by colon explants from WT→GF or Card9−/−→GF mice. ND, not detected. (i) Quantification of IL-22+ cells isolated from the lamina propria of the colon of WT→GF and Card9−/−→GF mice on day 12. Cells were gated for TH22 cells, NKp46+ ILCs, LTi cells, and CD3−CD4−NKp46− cells (n = 5 mice per group). Throughout, data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001; by two-tailed Student’s t-test (a,b,d,g,h,i) and Mann–Whitney U-test (f). In b–h, n = 5 mice per group (days 0 and 7) and n = 10 mice per group (day 12).
Impaired tryptophan metabolism in Card9−/−→GF mice
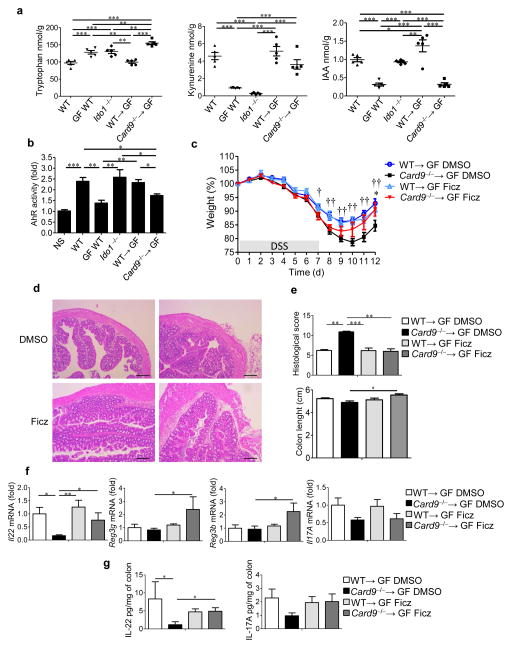
Our results suggest that the gut microbiota of Card9−/− mice contributes to susceptibility to DSS-induced colitis by altering IL-22 production. One mechanism that could link these findings is the modulation of AHR activation by the microbiota within the gastrointestinal tract. Tryptophan can be metabolized either by the gut bacteria into indole derivatives, such as indole-3-acetic acid (IAA) or by host cells into kynurenine (Kyn) via indoleamine 2,3-dioxygenase 1 (IDO1; Supplementary Fig. 11a)12,27,28. Indole derivatives are AHR ligands (Supplementary Fig. 11b) known to promote local IL-22 production29. Therefore, by examining the levels of AHR ligands in the lumen of the colons of germ-free, Ido1−/−, Card9−/−, Card9−/−→GF, WT, and WT→GF mice, we were able to analyze tryptophan metabolism by the host and the gut bacteria. As expected, production of Kyn was impaired in Ido1−/− mice, whereas normal IAA levels were observed. Production of IAA was impaired in GF mice, and Kyn levels were also low, probably because of the underdevelopment of the gut immune system (Fig. 4a and Supplementary Fig. 11c,d). Most notably, the levels of IAA in Card9−/−→GF and Card9−/− mice were decreased, whereas the bacterial concentrations within the colons were not modified (Fig. 4a and Supplementary Fig. 11c–e), indicating that the change in the microbiota in Card9−/− mice was responsible for the low level of IAA. In line with these data, culture supernatants of L. reuteri and Allobaculum sp., two bacteria with decreased abundance in the Card9−/− mouse microbiota (Fig. 2h), strongly activate AHR (Supplementary Fig. 12a). These results suggest that impaired tryptophan metabolism by the microbiota of the Card9−/− mice could be associated with, or even be responsible for, the hypersusceptibility of Card9−/− mice to colitis.
Figure 4.
Tryptophan metabolism is impaired in the gut microbiota of Card9−/− mice, leading to defective AHR activation and colitis recovery. (a) Tryptophan (left), kynurenine (middle), and IAA (right) concentrations in the feces of WT, GF WT, Ido1−/−, WT→GF or Card9−/−→GF mice (n = 5 mice per group). (b) Quantification of AHR activity from fecal samples of the indicated mice (n = 5 for GF WT and Ido−/− mice; n = 12 mice for all other groups). NS, not stimulated. (c) Weight of DSS-exposed mice that were treated with DMSO or Ficz. For statistical comparisons, dagger (†) indicates WT→GF DMSO versus Card9−/−→GF DMSO, and asterisk (*) indicates Card9−/−→GF DMSO versus Card9−/−→GF Ficz. (d) Representative H&E-stained images of proximal colon cross-sections from WT→GF (left) and Card9−/−→GF (right) mice at day 12 that were treated with DMSO (top) or Ficz (bottom). Scale bars, 200 μm. (e) Histological scores (top) and length (bottom) of the colons of the indicated mice at day 12. (f) Transcript expression in the colons of the indicated mice at day 12. (g) Secreted IL-22 (left) and IL-17A (right) amounts by colon explants from the indicated mice at day 12. Throughout, data are mean ± s.e.m. *P < 0.05; ** P < 0.01; ††P < 0.01; *** P < 0.001; by one-way ANOVA and post hoc Tukey test (a–c) or Kruskal–Wallis test followed by a post hoc Dunn’s test (e–g). In c–g, n = 11 DMSO-treated WT→GF mice per experiment; n = 12 DMSO-treated Card9−/−→GF mice per experiment; n = 9 Ficz-treated WT→GF mice per experiment; n = 6 Ficz-treated Card9−/−→GF mice per experiment.
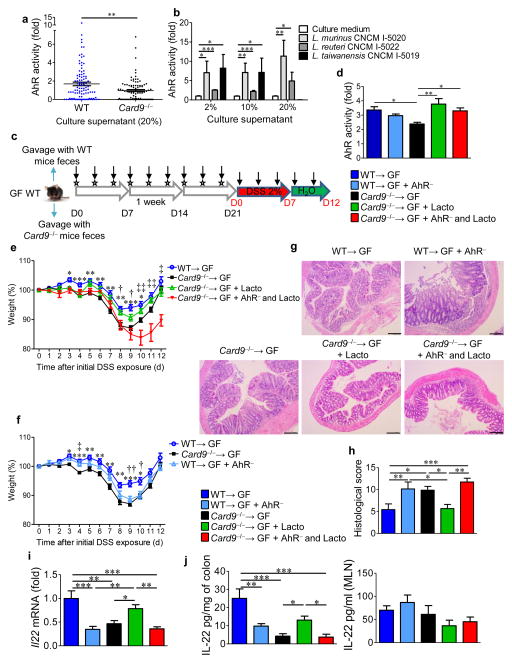
Using an AHR reporter system, we found that feces from Card9−/− and Card9−/−→GF mice were defective in their ability to activate AHR, to a similar extent as that for feces from GF mice (Fig. 4b and Supplementary Fig. 12b). To assess the in vivo importance of this finding, we administered 6-formylindolo(3,2-b)carbazole (Ficz), an AHR agonist, to DSS-exposed mice. The severity of colitis (body weight loss, histology score, and colon shortening) was reduced in Card9−/− and Card9−/−→GF mice that were treated with Ficz, as compared to their untreated counterparts during recovery, reaching the level of severity seen in WT or WT→GF mice (Fig. 4c–e and Supplementary Fig. 12c–e). Accordingly, defects in the colonic expression of Il22, Reg3g, and Reg3b in Card9−/− and Card9−/−→GF mice were rescued by Ficz administration (Fig. 4f and Supplementary Fig. 12f). This effect was also observed at the protein level for IL-22 in the colon (Fig. 4g and Supplementary Fig. 12g). Moreover, supplementation of Card9−/−→GF mice with three Lactobacillus strains, isolated from WT mice for their ability to activate AHR (Fig. 5a,b), rescued susceptibility of Card9−/−→GF mice to colitis, IL-22 expression, and AHR ligand production (Fig. 5c–j). These effects were mediated by AHR, as they were abrogated in the presence of an AHR antagonist (Fig. 5e–j).
Figure 5.
Inoculation with lactobacilli that metabolize tryptophan and produce AHR ligands reduces colitis in an AHR-dependent manner. (a) AHR activation by culture supernatants from strains isolated from feces of WT and Card9−/− mice, relative to that culture media (n = 3 replicates for each strain). (b) AHR activation by culture supernatants from L. murinus CNCM I-5020, L. reuteri CNCM I-5022, and L. taiwanensis CNCM I-5019 isolated from feces of WT mice, relative to that by culture medium alone (n = 3 replicates for each strain). (c) Experimental design of AHR antagonist treatment and Lactobacilli inoculation. Vertical arrows indicate injections of vehicle or AHR antagonist. Stars indicate intragastric gavage with PBS or bacterial suspension. D, day. (d) AHR activation by day 21 feces from WT→GF, Card9−/−→GF, or WT→GF mice treated with an AHR antagonist (WT→GF + AHR−), and Card9−/−→GF mice gavaged with L. murinus CNCM I-5020, L. reuteri CNCM I-5022, and L. taiwanensis CNCM I-5019 isolated from feces of WT mice and treated with either vehicle (Card9−/−→GF + Lacto) or an AHR antagonist (Card9−/−→GF + AHR− and Lacto). (e) Weight of DSS-exposed mice. For statistical comparisons, asterisk (*) indicates WT→GF versus Card9−/−→GF; dagger (†) indicates Card9−/−→GF versus Card9−/−→GF + Lacto; double dagger (‡) indicates Card9−/−→GF + Lacto versus Card9−/−→GF + AHR− and Lacto. (f) Weight of DSS-exposed mice. For statistical comparisons, asterisk (*) indicates WT→GF versus Card9−/−→GF; dagger (†) indicates WT→GF versus WT→GF + AHR−; double dagger (‡) indicates Card9−/−→GF versus WT→GF + AHR−. (g) Representative H&E-stained images of proximal colon cross-sections at day 12 after initial DSS exposure. Scale bars, 200 μm. (h) Histological scores at day 21. (i) Il22 expression in the colon. (j) Secreted IL-22 amounts by colon explants and MLN cells. Throughout, data are mean ± s.e.m. *P < 0.05; †P < 0.05; ‡P < 0.05; **P < 0.01; ††P < 0.01; ‡‡P < 0.01; ***P < 0.001, by Mann–Whitney U-test (a), two-tailed Student’s t-test (b), or one-way ANOVA with post hoc Tukey test (d–f, h–j). In d–j, n = 5 mice per group per experiment.
One study showed that the microbiota of Il22−/− mice is altered, and its transfer increases susceptibility of WT mice to colitis30. Therefore, we postulated that a deficient IL-22 response may also be involved in the decreased production of AHR ligands by the microbiota. Indeed, microbiota from Il22−/− mice had impaired AHR activity and decreased levels of IAA (Supplementary Fig. 13a,b). Moreover, administration of exogenous IL-22 was sufficient to normalize AHR ligand production and colitis susceptibility in Card9−/− mice (Supplementary Fig. 13c–h). These results show that the gut microbiota of Card9−/− mice contributes to the susceptibility of the mice to colitis by altering the IL-22 signaling pathway via impaired tryptophan metabolism, leading to defective AHR activation. In addition, IL-22 is required for the production of AHR ligands by the microbiota (Supplementary Fig. 14). An AHR agonist can effectively counter these defects.
Reduced AHR activity and tryptophan metabolites in subjects with IBD
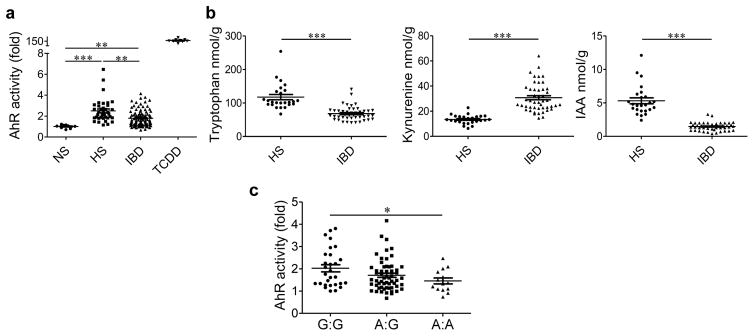
We explored whether these findings were relevant to human disease and analyzed fecal samples from individuals with IBD (Supplementary Table 1) and healthy subjects for their ability to activate AHR. The fecal samples from healthy subjects induced greater activation of AHR than those from patients with IBD (Fig. 6a). This defect was observed in both Crohn’s disease and ulcerative colitis (Supplementary Fig. 15a). This finding was associated with decreased levels of tryptophan and IAA, and increased levels of Kyn, in the fecal samples from patients with IBD (Fig. 6b and Supplementary Fig. 15b–d). Therefore, activated immune cells in the gut of individuals with IBD may use tryptophan to produce Kyn via IDO1, whereas the metabolism of tryptophan by the gut microbiota is impaired, leading to defective AHR activation. We next searched for a connection between CARD9 and the ability of the microbiota to produce AHR ligands in humans. We genotyped patients with IBD for an IBD-associated single-nucleotide polymorphism (SNP) within CARD9 (rs10781499)31 and found that the risk allele was associated with reduced AHR activation by microbiota-derived metabolites extracted from fecal samples in our in vitro assay (Fig. 6c and Supplementary Fig. 15c,d). No association was observed among other major IBD-associated SNP-containing alleles of genes, including NOD2, autophagy-related 16-like 1 (ATG16L1), and leucine-rich repeat kinase 2 (LRRK2) (Supplementary Fig. 15e). These results should be confirmed in an independent cohort but suggest a connection between IBD, CARD9, and the ability of the microbiota to produce AHR agonists in humans.
Figure 6.
Reduced tryptophan metabolism and AHR activation in the gut microbiota of individuals with IBD, and its association with the CARD9 genotype. (a) Quantification of AHR activation from the feces of healthy subjects (HS; n = 37) and individuals with IBD (n = 102) in remission. NS, not stimulated (basal AHR activity of cells) (n = 9); TCDD, feces treated with AHR agonist (n = 9). (b) Concentrations of tryptophan, kynurenine, and IAA in the feces of HS (n = 32) and patients with IBD in remission (n = 54). (c) Quantification of AHR activation by the feces of HS and individuals with IBD in remission, as classified on the basis of SNP rs10781499, with ‘A’ denoting the risk allele (n = 101). Throughout, data are mean ± s.e.m. *P < 0.05; **P < 0.001; ***P < 0.0001; by the Kruskal–Wallis test followed by a post hoc Dunn’s test (a), Mann–Whitney U-test (b) or one-way ANOVA and post hoc Tukey test (c).
DISCUSSION
The gut microbiota is a key player in mammalian physiology, and its composition is influenced by genetics, environment, and diet1–3. Any change in these factors can predispose the host to metabolic or inflammatory disorders, including obesity and IBD1–3. However, the mechanisms by which the microbiota influences the host remain unknown. It is also unclear whether dysbiosis is a cause or a consequence of these diseases. Our results showed that Card9 deletion has an effect on the gut microbiota in mice and that transfer of microbiota from Card9−/− mice to WT GF recipient mice is sufficient to recapitulate the defective IL-22 activation and increased sensitivity to colitis observed in Card9−/− mice. These alterations were due to an impaired ability of the microbiota of Card9−/− mice to catabolize tryptophan into AHR ligands.
Metabolomics studies have revealed large effects of the gut microbiota on host metabolism32. Indole derivatives, which are tryptophan catabolites generated by the microbiota, were recently identified as activators of AHR, which consequently regulates local IL-22 production12,13,29. Any modification in AHR ligand production impacts IL-22 levels and, therefore, acts on the fragile equilibrium between the microbiota and the host cells12,30,33. In accordance with this model, the Card9−/− mouse microbiota has decreased levels of bacteria that have tryptophan-catabolizing functions, such as L. reuteri, and members of the genus Allobaculum and the phylum Actinobacteria12,34. Moreover, the susceptibility of GF mice that received microbiota from Card9−/− mice (Card9−/−→GF mice) to colitis and the decreased AHR ligand production were rescued by supplementation with AHR-ligand-producing Lactobacillus strains. Thus, our results provide evidence that defects in expression of factors involved in innate immunity, such as CARD9, can shape an altered microbiota, which can then modify the host immune response. Our results are relevant to humans, as impaired microbial production of AHR ligands is observed in patients with IBD and correlates with an IBD-associated SNP within CARD9 (rs10781499). Consequently, the tryptophan catabolites derived from gut microbiota could be used as biomarkers for dysbiosis and may be targeted for the development of new therapeutic drugs for individuals with IBD. For instance, indole derivatives, or the probiotic organisms that produce them12, could be used as a supportive therapy in individuals with intestinal dysbiosis. Our findings support a more general concept that suggests that, owing to their tight relationship, the respective roles of the host factors and the gut microbiota in IBD pathogenesis cannot be completely distinguished. Thus, dysbiosis should neither be considered a cause nor a consequence of IBD, but both simultaneously. We hypothesize that the altered immune response in Card9−/− mice has an effect on the microbiota. In turn, the modified microbiota alters tryptophan catabolite production, affecting the host’s immune response and amplifying dysbiosis in a vicious cycle that leads to the loss of intestinal homeostasis.
ONLINE METHODS
Mice
Card9-deficient mice (Card9−/−) on the C57BL/6J background have been described previously35. After rederivation at Charles River Laboratories, the animals were housed under specific pathogen-free conditions at the Saint-Antoine Research Center. Heterozygous mice (Card9+/−) were used as breeders. At weaning, mice were separated according to genotype. Germ-free C57BL/6J mice were bred in germ-free isolators at the CDTA (Transgenese et Archivage Animaux Modèles, CNRS, UPS44, Orléans, France). Conventional mice were fed a standard chow diet (R03, SAFE, Augy, France) and germ-free mice were fed a diet without yeast (R04, SAFE, Augy, France). All conventional WT and Card9−/− mice used in this study were 8-week-old mice. Animal experiments were performed according to the institutional guidelines approved by the local ethics committee of the French authorities, the ‘Comité d’Ethique en Experimentation Animale’ (COMETHEA) and registered with the following national number C2EA-24.
Gut microbiota transfer and isolation of bacterial strains from feces
Fresh stool samples from WT or Card9−/− mice (8-week-old males) were immediately transferred to an anaerobic chamber, in which the stool samples were suspended and diluted in LYHBHI (Brain-heart infusion) medium (BD Difco, Le Pont De Claix, France) supplemented with cellobiose (1 mg/ml; Sigma-Aldrich, St. Louis, MO, USA), maltose (1 mg/ml; Sigma-Aldrich), and cysteine (0.5 mg/ml; Sigma-Aldrich). These fecal suspensions were used either to inoculate mice or to isolate bacterial strains. WT germ-free mice (4- to 5-week-old females) were randomly assigned to two groups and inoculated via oral gavage with 400 μl of fecal suspension (1:100) from the conventional wild-type (WT→GF) or Card9−/− (Card9−/−→GF) mice and maintained in separate isolators. One aliquot of each fecal suspension was stored at –80 °C. All experiments in WT→GF and Card9−/−→GF mice were performed three weeks after inoculation. For the isolation of bacterial strains, the highest countable dilution of fecal suspensions was isolated in an anaerobic chamber in YHBHI agar medium (BD Difco) supplemented with cellobiose (1 mg/ml; Sigma-Aldrich), maltose (1 mg/ml; Sigma-Aldrich), and cysteine (0.5 mg/ml; Sigma-Aldrich) or in MRS agar medium (BD Difco) supplemented with cysteine (0.5 mg/ml; Sigma-Aldrich). Bacterial strains from fecal suspensions were also isolated in aerobic conditions in bile esculin azide agar medium (Biokar Diagnostics, Beauvais, France) or in desoxycholate agar medium (BD Difco). All isolates were routinely maintained by growing for 18–20 h at 37 °C in agar and liquid MRS, YBHI, and Luria–Bertani (BD Difco) medium. Cultures were examined by phase-contrast microscopy for morphology and purity, and stored at –80 °C.
Induction of DSS colitis and treatments
To induce colitis, mice were administered drinking water supplemented with 2% (wt./vol.) dextran sulfate sodium (DSS; MP Biomedicals, LLC, Aurora, OH, USA) for 7 d and were then allowed to recover by drinking unsupplemented water for the next 5 d (Supplementary Fig. 1a). The 6-formylindolo(3,2-b)carbazole (Ficz; Enzo Life Sciences, Lausen, Switzerland) and the AHR antagonist CH223191 (AHR−; Sigma-Aldrich) were resuspended in dimethyl sulfoxide (DMSO; Sigma-Aldrich) and administered intraperitoneally. Ficz was injected 1 d after DSS administration (1 μg/mouse). For the AHR− treatment, WT→GF and Card9−/−→GF mice (4- to 5-week-old females) were treated (100 μg/mouse) three times per week until euthanization (Fig. 5c). Controls consisted of mice injected with DMSO vehicle alone for the Ficz and AHR− treatment groups. Three bacteria with strong AHR activity and that were isolated in feces of WT mice were identified by sequencing the 16S rDNA gene as previously described36. The resulting sequences were aligned, inspected by eye, and compared with the online tool BLAST. Strains were identified based on the highest hit scores. These strains were deposited in the Collection Nationale de Cultures de Microorganismes (CNCM) of the Institut Pasteur and named L. murinus CNCM I-5020, L. reuteri CNCM I-5022, and L. taiwanensis CNCM I-5019. Bacterial suspensions containing these three strains (109 colony-forming units (c.f.u.) of each strain in 500 μl of PBS) were administered three times per week for a period of 3 weeks to WT→GF and Card9−/−→GF mice (4- to 5-week-old females) by intragastric gavage (Fig. 5c). Oral gavage with PBS was performed in control mice. For the antifungal treatment, mice were fed 0.5 mg/ml fluconazole in drinking water (Sigma-Aldrich) 1 week before DSS administration and every day thereafter, as previously described19 (Supplementary Fig. 6c). For the IL-22 treatment, WT and Card9−/− mice were injected intraperitoneally three times per week with mouse IL-22–Fc (50 μg/mouse) (Genentech, South San Francisco, CA, USA) (WT IL-22 and Card9−/− IL-22) or an equivalent amount of isotype control (IgG2a) (Genentech) (WT isotype and Card9−/− isotype) for a period of 3 weeks. 3 d after the last injections, colitis was induced by DSS treatment (Supplementary Fig. 13c). In all treatments, body weight, blood in stool, and stool consistency were analyzed daily. The severity of colitis was assessed using the disease activity index (DAI) as previously described8.
Cytokine quantification
MLNs and spleens were sieved through a 70-μm cell strainer (BD Difco) in complete RPMI 1640 medium (10% heat-inactivated FCS, 2 mM L-glutamine, 50 IU/ml penicillin, and 50 μg/ml streptomycin; Sigma-Aldrich), and 1 × 106 cells per well were cultured (37 °C, 10% CO2) for 48 h with stimulation by phorbol 12-myristate 13-acetate (PMA, 50 ng/ml; Sigma-Aldrich) and ionomycin (1 μM; Sigma-Aldrich). The culture supernatant was frozen at –80 °C until processing. To measure cytokine levels in the colonic explants, tissues from the medial colon were isolated and rinsed in phosphate-buffered saline (PBS; Gibco, Paisley, UK). The colonic explants were cultured (37 °C, 10% CO2) overnight in 24-well tissue culture plates (Costar, Corning, Amsterdam, the Netherlands) in l ml of complete RPMI 1640 medium. The culture supernatants were collected and stored at –80 °C until processing. ELISAs were performed on the supernatants to quantify the following mouse cytokines according to the manufacturer’s instructions: IL-10, IL-17A, and IFN-γ (Mabtech, Nacka Strand, Sweden); IL-22 (eBioscience, San Diego, CA, USA); and IL-6 (R&D Systems, Minneapolis, MN, USA). For the colonic explants, cytokine concentrations were normalized according to the dry weight of each colonic explant.
Lamina propria cell isolation and flow cytometry
Cells from the colon and small intestine lamina propria were isolated as previously described8. The cells were stimulated and stained as previously described8. The following antibodies were used for surface staining of: CD3 (145-2C11, eBioscience); CD4 (L3T4, BD Difco); CD11b (M1/70, eBioscience); CD11c (N418, eBioscience); F4/80 (BM8, eBioscience); CD103 (M290, BD Difco); major histocompatibility complex (MHC) II (M5/114.15.2, BD Dico); TCR-γδ (eBioGL3, eBioscience); and NKp46 (29A1.4, eBioscience). Intracellular cytokine staining was performed using IL-17A (TC11-18H10, BD Difco) and IL-22 (IL-22JOP, eBioscience) antibodies. All antibodies were used at final concentration of 1 μg/ml. The cells were analyzed using a Gallios flow cytometer (Beckman Coulter, Brea, CA, USA). Leukocytes were gated using forward scatter (FSC) and side scatter (SSC), and within the leukocyte gates, the innate immune cells were identified as macrophages (MHCII+F4/80+CD103−CD11b+CD11c−) or dendritic cells (MHCII+F4/80− CD103+/−CD11b−CD11c+). For the lymphoid compartment, the leukocytes were gated using FSC and SSC. Within the lymphocyte gate, the populations were identified as TH17 cells (CD3+CD4+IL-17+IL-22+), TH22 cells (CD3+CD4+ IL-17−IL-22+), NKp46+ ILCs (including ILC3 and NK cells; CD3−CD4−NKp46+), LTi cells (CD3−CD4+NKp46−), γδ T cells (CD3+CD4−TCRγδ+) or CD3−CD4−NKp46− cells.
Histology
Colon samples for histological studies were maintained at 4 °C in 4% paraformaldehyde and then embedded in paraffin. 4-μm sections (three sections per sample) were stained with hematoxylin and eosin (H&E) and then examined in a blinded manner using a BX43 Olympus microscope, to determine the histological score according to previously described methods8 (Supplementary Table 2). The samples were also processed using a Starr Trek kit (Biocare Medical, Concord, CA, USA) or a Novolink Polymer Detection System (Leica Biosystems, Heidelberg, Germany) to stain two mouse cell markers via immunohistochemistry, according to the manufacturer’s instructions, using mouse monoclonal anti-Ki67 antibody (MM1, Leica Biosystems; 1:50) for cell proliferation and rabbit polyclonal anti–caspase-3 (cleaved at Asp175) antibody (Abcam, Cambridge, United Kingdom; cat.# ab52293; 1:150) for apoptosis. The number of cleaved caspase-3+ cells in 100 μm of analyzed colon was counted. Ki67 was quantified as a percentage of the total height of each crypt. For each sample, ten areas or crypts were analyzed.
Gene expression analysis using quantitative reverse-transcription PCR (qRT–PCR)
Total RNA was isolated from colon samples using an RNeasy Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. Quantitative RT–PCR was performed using SuperScript II Reverse Transcriptase (Life Technologies, Saint Aubin, France) and then a Takyon SYBR Green PCR kit (Eurogentec, Liège, Belgium) in a StepOnePlus apparatus (Applied Biosystems, Foster City, CA, USA) with specific mouse oligonucleotides. The oligonucleotides used were as follows: Gapdh (sense) 5′-AACTTTGGCATTGTGGAAGG-3′ and (antisense) 5′-ACACATTGGGGGTAGGAACA-3′; Il17a (sense) 5′-TTTAACTCCCTTGGCGCAAAA-3′ and (antisense) 5′-CTTTCCCTCCGCATTGACAC-3′; Il22 (sense) 5′-CATGCAGGAGGTGGTACCTT-3′ and (antisense) 5′-CAGACGCAAGCATTTCTCAG-3′; Reg3g (sense) 5′-TTCCTGTCCTCCATGATCAAAA-3′ and (antisense) 5′-CATCCACCTCTGTTGGGTTCA-3′; and Reg3b (sense) 5′-ATGCTGCTCTCCTGCCTGATG-3′ and (antisense) 5′-CTAATGCGTGCGGAGGGTATATTC-3′. We used the 2−ΔΔCt quantification method with mouse Gapdh as an endogenous control and the WT or WT→GF group as a calibrator.
Fecal DNA extraction and fungal quantification via quantitative PCR (qPCR)
Fecal DNA was extracted from the weighted stool samples as previously described37. More precisely, the feces samples were weighed and then resuspended for 10 min at room temperature in 250 μl of 4 M guanidine thiocyanate in 0.1 M Tris (pH 7.5) (Sigma) and 40 μl of 10% N-lauroyl sarcosine (Sigma). After the addition of 500 μl of 5% N-lauroyl sarcosine in 0.1 M phosphate buffer (pH 8.0), the 2-ml tubes were incubated at 70 °C for 1 h. One volume (750 ml) of a mixture of 0.1- and 0.6-mm-diameter silica beads (Sigma) (previously sterilized by autoclaving) was added, and the tube was shaken at 6.5 Meter/second three times for 30 s each in a FastPrep (MP Biomedicals) apparatus. Polyvinylpolypyrrolidone (15 mg) was added to the tube, which was then vortexed and centrifuged for 5 min at 20,000g. After recovery of the supernatant, the pellets were washed with 500 μl of TENP (50 mM Tris (pH 8), 20 mM EDTA (pH 8), 100 mM NaCl, 1% polyvinylpolypyrrolidone) and centrifuged for 5 min at 20,000g, and the new supernatant was added to the first supernatant. The washing step was repeated two times. The pooled supernatant (about 2 ml) was briefly centrifuged to remove particles and then split into two 2-ml tubes. Nucleic acids were precipitated by the addition of 1 volume of isopropanol for 10 min at room temperature and centrifugation for 10 min at 20,000g. Pellets were resuspended and pooled in 450 μl of 100 mM phosphate buffer, pH 8, and 50 ml of 5 M potassium acetate. The tube was placed on ice overnight and centrifuged at 20,000g for 30 min. The supernatant was then transferred to a new tube containing 20 μl of RNase (1 mg/ml) and incubated at 37 °C for 30 min. Nucleic acids were precipitated by the addition of 50 μl of 3 M sodium acetate and 1 ml of absolute ethanol. The tube was incubated for 10 min at room temperature, and the nucleic acids were recovered by centrifugation at 20,000g for 15 min. The DNA pellet was finally washed with 70% ethanol, dried, and resuspended in 100 μl of Tris–EDTA (TE) buffer. DNA suspensions were stored at –20 °C for real-time qPCR analysis of the 16S rDNA or ITS2 sequences. DNA was then subjected to qPCR by using a Takyon SYBR Green PCR kit (Eurogentec) for quantification of all fungal sequences or by using TaqMan Gene Expression Assays (Life Technologies) for quantification of all bacterial sequences. The probes and primers for the bacterial 16S rDNA genes and primers for the fungal 18S rDNA genes were used as described previously19,37. The threshold cycle for each sample was determined for each gene normalized to the CT value of the all-bacteria 16S ribosomal RNA gene. Data were calculated using the 2−ΔΔCt method.
16S rDNA gene sequencing
DNA was isolated from the feces of mice before and after DSS treatment using the protocol described above. Microbial diversity was determined for each sample by targeting a portion of the ribosomal genes. A 16S rDNA gene fragment comprising the V3 and V4 hypervariable regions (16S (sense) 5′-TACGGRAGGCAGCAG-3′ and (antisense) 5′-CTACCNGGGTATCTAAT-3′) was amplified using an optimized and standardized 16S-amplicon-library preparation protocol (Metabiote, GenoScreen, Lille, France). Briefly, 16S rDNA gene PCR was performed using 5 ng genomic DNA according to the manufacturer’s protocol (Metabiote) using 192 bar-coded primers (Metabiote MiSeq Primers) at final concentrations of 0.2 μM and an annealing temperature of 50 °C for 30 cycles. The PCR products were purified using an Agencourt AMPure XP-PCR Purification system (Beckman Coulter, Brea, CA, USA), quantified according to the manufacturer’s protocol, and multiplexed at equal concentrations. Sequencing was performed using a 300-bp paired-end sequencing protocol on an Illumina MiSeq platform (Illumina, San Diego, CA, USA) at GenoScreen, Lille, France. Raw paired-end reads were subjected to the following process: (1) quality-filtering using the PRINSEQ-lite PERL script38 by truncating the bases from the 3′ end that did not exhibit a quality <30 based on the Phred algorithm; (2) paired-end read assembly using FLASH39 (fast length adjustment of short reads to improve genome assemblies) with a minimum overlap of 30 bases and a 97% overlap identity; and (3) searching and removing both forward and reverse primer sequences using CutAdapt, with no mismatches allowed in the primers sequences. Assembled sequences for which perfect forward and reverse primers were not found were eliminated.
16S rDNA gene sequence analysis
The sequences were demultiplexed, quality-filtered using the ‘quantitative insights into microbial ecology’ (QIIME, version 1.8.0) software package40, and the forward and reverse Illumina reads were joined using the fastq-join method (http://code.google.com/p/ea-utils). The sequences were assigned to OTUs using the UCLUST algorithm41 with a 97% threshold of pairwise identity and classified taxonomically using the Greengenes reference database42. Rarefaction was performed (39,048–84,722 sequences per sample) and used to compare the abundances of OTUs across samples.
ITS2 rDNA gene sequencing
DNA was isolated from the feces of mice before and after DSS treatment using the protocol described above. Microbial diversity was determined for each sample by 454 pyrosequencing of the ribosomal genes. An ITS2 rDNA gene fragment of approximately 350 bases was amplified using the primers ITS2 (sense) 5′-GTGARTCATCGAATCTTT-3′ and (antisense) 5′-GATATGCTTAAGTTCAGCGGGT-3′ and the optimized and standardized ITS2-amplicon-library preparation protocol (Metabiote, GenoScreen). Briefly, for each sample, diluted genomic DNA was used for a 25-μl PCR reaction conducted under the following conditions: 94 °C for 2 min; 35 cycles of 15 s at 94 °C, 52 °C for 30 s, and 72 °C for 45 s; followed by 7 min at 72 °C. The PCR products were purified using AmpureXP beads (Beckman Coulter) and quantified using a PicoGreen staining kit (Molecular Probes, Paris, France). A second PCR of nine cycles was then conducted under similar conditions with the purified PCR products and 10-bp multiplex identifiers (SIM identifiers) added to the primers at the 5′ position to specifically identify each sample and avoid PCR biases. Finally, the PCR products were purified and quantified as described above. Sequencing was then performed using a Gs-FLX Titanium Sequencing System (Roche Life Science, Mannheim, Germany).
ITS2 sequence analysis
The sequences were demultiplexed and quality-filtered using the QIIME version 1.8.0 software package40. The sequences were trimmed for bar codes and PCR primers, and were binned for a minimal sequence length of 150 bp, a minimal base quality threshold of 25, and a maximum homopolymer length of 7. The sequences were then assigned to OTUs using the UCLUST algorithm41 with a 97% threshold of pairwise identity and classified taxonomically using the UNITE ITS database (alpha version 12_11)43. Rarefaction was performed (2,696–9,757 sequences per sample) and used to compare the abundances of OTUs across samples. For both 16S and ITS2 sequences, principal component analyses (PCA) based on genus composition were performed using the R package Ade4 (ref. 44) and used to assess the variations among experimental groups. The number of observed species and the Shannon diversity index were calculated using rarefied data (depth = 2,675 sequences/sample for ITS2 and depth = 39,931 sequences/sample for 16S) and were used to characterize species diversity in a community. The sequencing data were deposited in the European Nucleotide Archive under accession number PRJEB9079.
Gene expression by microarray analyses
Total RNA was isolated using the protocol described above. RNA integrity was verified using a Bioanalyser 2100 with RNA 6000 Nano chips (Agilent Technologies, Palo Alto, CA, USA). Transcriptional profiling was performed on mouse colon samples using the SurePrint G3 Mouse GE 8 × 60 K Microarray kit (design ID: 028005, Agilent Technologies). Cyanine-3 (Cy3)-labeled cRNAs were prepared with 100 ng of total RNA using a One-Color Low Input Quick Amp Labeling kit (Agilent Technologies), following the recommended protocol. The specific activities and cRNA yields were determined by using a NanoDrop ND-1000 (Thermo Fisher Scientific, Waltham, MA, USA). For each sample, 600 ng of Cy3-labeled cRNA (specific activity > 11.0 pmol Cy3/μg of cRNA) were fragmented at 60 °C for 30 min and hybridized to the microarrays for 17 h at 65 °C in a rotating hybridization oven (Agilent Technologies). After hybridization, the microarrays were washed and then immediately dried. After washing, the slides were scanned using a G2565CA Scanner System (Agilent Technologies) at a resolution of 3 μm and a dynamic range of 20 bits. The resulting TIFF images were analyzed using the Feature Extraction Software v10.7.3.1 (Agilent Technologies) according to the GE1_107_Sep09 protocol. The microarray data were submitted to GEO under accession number GSE67577.
Microarray analysis
Agilent Feature Extraction software was used to convert scanned signals into tab-delimited text that could be analyzed using third-party software. The R package ‘agilp’ was used to pre-process the raw data. Box plots and PCAs were used to obtain a general overview of the data in terms of the within-array distributions of signals and the between-sample variability. Agilent Feature Extraction software computed a P value for each probe in each array to test whether the scanned signals were significantly higher than the background signal. The null hypothesis was ‘the measured signal is equal to background signal’. Probes were considered to be detected if the P value was <0.05. The probes must have been present in at least 60% of samples per group and under at least one condition to be considered for analysis. To compare data from multiple arrays, the data were normalized to minimize the effect of nonbiological differences. Quantile normalization45 is a method that can quickly normalize within a set of samples without using a reference base. After normalization, spike-in, positive and negative control probes were removed from the normalized data. For differential expression analysis, we used the limma eBayes test46, which finds a compromise between the variance estimate for the gene under consideration and the average variance of all of the genes. The Benjamini–Hochberg correction method was used to control the false-discovery rate (FDR). All significant gene lists were annotated for enriched biological functions and pathways using the DAVID platform47,48 for gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) terms. Significant canonical pathways had adjusted P values, according to Benjamini’s method, below 0.05. We used Venn diagrams to globally visualize the overlap between all significant genes in the WT and Card9−/− comparisons. Thus, DAVID was performed to test for the biological pathway enrichment of Venn’s elements.
Luciferase assay
The H1L1.1c2 cell line containing a stably integrated dioxin response elements (DRE)-driven firefly luciferase reporter plasmid pGudLuc1.1 has been described previously49,50. The cells were seeded in 96-well plates at 7.5 × 104 cells/well in 100 μl of complete Dulbecco’s modified Eagle’s medium (DMEM) (with 10% heat-inactivated FCS, 50 IU/ml penicillin, and 50 μg/ml streptomycin; Sigma-Aldrich) and cultured (37 °C, 10% CO2) for 24 h before treatment. This cell line tested negative for mycoplasma contamination and was used in this study to determine AHR activity of bacterial and stool samples. Fresh stool samples from healthy subjects and patients with IBD in remission, and from mice, were collected, weighed, and stored at –80 °C until processing. Stool samples were suspended, diluted to 100 mg/ml in PBS, centrifuged (5,000 g, 15 min, 4 °C), and filtered (0.2 μm; VWR, Fontenay-sous-Bois, France). Lactobacillus and Bifidobacterium spp. were grown in MRS medium (BD Difco, Le Pont De Claix, France) supplemented with 10% cysteine (Sigma-Aldrich) at 37 °C under aerobic and anaerobic conditions, respectively. Allobaculum stercoricanis (Deutsche Sammlung von Mikroorganismen und Zellkulturen, DSMZ, 13633) was cultivated under the recommended culture conditions listed in the DSMZ. Bacterial strains isolated from feces of WT and Card9−/− mice were cultivated under the culture conditions described above. Culture supernatants of these bacteria were stored at –80 °C until processing. To assess agonistic activity, the cells were treated with stool suspensions diluted at 1:10 in complete DMEM with 0.1% DMSO or with culture supernatants diluted to 2%, 10%, or 20% in complete DMEM. Controls consisted of cells treated with DMEM with 0.1% DMSO or bacterial culture medium as the negative control, or 10 nM of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD; Sigma-Aldrich) diluted in DMEM with 0.1% DMSO as the positive control. After 24 h of incubation, wells were washed with 100 μl PBS, and 50 μl Promega lysis buffer was added to each well. The plates were shaken for 30 min to lyse the cells. After adding 100 μl of luciferase reagent (Promega), luciferase activity was measured using a luminometer. The results were normalized based on the negative luciferase activity of the control.
HPLC–HRMS analysis
Thawed stools from healthy subjects and patients with IBD in remission, or from mice, were extracted as previously described51. L-tryptophan (L-Trp) and L-kynurenine (L-Kyn) were measured via HPLC using a coulometric electrode array (ESA Coultronics, ESA Laboratories, Chelsford, MA, USA)52. Quantifications were performed by referencing calibration curves obtained with internal standards. Other compounds (tryptamine and IAA) were quantified via liquid chromatography coupled to mass spectrometry (LC-MS) by using a Waters ACQUITY ultraperformance liquid chromatography (UPLC) system equipped with a binary solvent delivery manager and sample manager (Waters Corporation, Milford, MA, USA) and that was coupled to a tandem quadrupole-time-of-flight (Q-TOF) mass spectrometer equipped with an electrospray interface (Waters Corporation). Compounds were identified by comparing with the accurate mass and the retention time of reference standards in our in-house library, and the accurate masses of the compounds were obtained from web-based resources, such as the Human Metabolome Database (http://www.hmdb.ca) and the METLIN database (http://metlin.scripps.edu).
IBD cohort
All individuals with IBD were recruited in the Gastroenterology Department of the Saint Antoine Hospital (Paris, France) and provided informed consent. Approval for human studies was obtained from the local ethics committee (Comité de Protection des Personnes Ile-de-France IV, IRB 00003835 Suivitheque study; registration number 2012/05NICB). Remission was defined by global physician assessment and normal serum C-reactive protein (CRP) [levels. Among the 112 patients with IBD included, 101 were genotyped for the rs10781499, rs2066844, rs2066845, rs2066847, rs12994997, and rs11564258 SNPs using Fluidigm (UMR CNRS 8199, Lille, France) (Supplementary Table 1).
NanoString
NanoString analysis was performed and analyzed according to the manufacturer’s recommendations.
Statistical analyses
GraphPad Prism version 6.0 (San Diego, CA, USA) was used for all analyses and preparation of graphs. For all data displayed in graphs, results are expressed as the mean ± s.e.m. For comparisons between two groups, two-tailed Student’s t-test for unpaired data or nonparametric Mann–Whitney test was used. For comparisons between more than two groups, one-way analysis of variance (ANOVA) and post hoc Tukey test or nonparametric Kruskal–Wallis test followed by a post hoc Dunn’s test was used. The Kolmogorov–Smirnov test of normality was applied to all data sets, and in cases where the data did not demonstrate a normal distribution, nonparametric tests were used to analyze statistical differences. An F or Bartlett’s test was performed to determine differences in variances between groups for t-tests and ANOVAs, respectively. An unpaired Student’s t-test with Welch’s correction was applied when variances were not equal.
No samples, mice, or data points were excluded from the reported analysis. Animals were randomly assigned to experimental groups. All analyses were performed unblended, except for the histological analyses, and were adjusted for multiple comparisons. Sample size was estimated according previous experience using the models described. No statistical method was used to predetermine sample size. For the human data (Fig. 6a), given the sample size of the two groups compared (n = 37 for healthy subjects and n = 102 for individuals with IBD) the magnitude of the effect that can be detected is 0.54 (with power = 0.8, alpha type I error = 0.05, and two-sided test), which is below the difference we observed (0.71). Moreover, based on the characteristics of the two groups compared (n value, mean, and s.d.), the power of the comparison was calculated to be 95.1% with an alpha type I error set at 0.05. Differences corresponding to P < 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank the members of the ANAXEM germ-free platform, the members of the animal facilities of INRA, and T. Ledent of the animal facilities of Saint-Antoine Hospital for their assistance in mouse care; M. Moroldo and J. Lecardonnel from the CRB GADIE core facility for technical assistance in performing the microarray analyses; S. Dumont for technical help in histology and immunochemistry; and C. Aubry, N. M. Breyner, F. Chain, S. Le Guin, C. Cherbuy, N. Lapaque, and D. Skurnik for fruitful discussions and technical help. We also thank E. Drouet and the Clinical Research Assistant team of Unité de Recherche Clinique de l’Est Parisien for their help in obtaining samples from patients with IBD. Ido1−/− and Il22−/− mice were provided by Soraya Taleb (Inserm Unit 970, Paris, France) and Bernhard Ryffel (CNRS, UMR7355, Orléans, France), respectively. The H1L1.1c2 cell line was provided by Michael S. Denison (University of California, Davis, CA, USA). Funding was provided by Equipe ATIP–Avenir 2012 (H.S.), INSERM–ITMO SP 2013 (H.S.) and ECCO grant 2012 (H.S.).
Footnotes
AUTHOR CONTRIBUTIONS
B.L., M.L.R., and H.S. conceived and designed the study, performed data analysis, and wrote the manuscript; B.L. designed and conducted all experiments, unless otherwise indicated; V.L. designed and performed the AHR activity experiments; G.D, C.B., S.J., T.W.H., J.M.N., L. Brot, F.M., and M.-L.M. provided technical help for the in vitro and in vivo experiments; H.-P.P. conducted the bioinformatics studies and analyzed the microarray experiments; J.-M.L. performed and analyzed HPLC experiments; S.T. provided material from the Ido1−/− mice and discussed the results; A.C.-M. and B.R. provided material from the Il22−/− mice and discussed the results; H.S., J.C., I.N.-L., A.B., L. Beaugerie, and P.S. provided data and samples for the patients with IBD; B.L., M.L.R., R.J.X., P.L., and H.S. discussed the experiments and results.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Silva MJ, et al. The multifaceted role of commensal microbiota in homeostasis and gastrointestinal diseases. J Immunol Res. 2015;2015:321241. doi: 10.1155/2015/321241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molodecky NA, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54.e42. doi: 10.1053/j.gastro.2011.10.001. quiz e30. [DOI] [PubMed] [Google Scholar]
- 3.Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. 2015;12:205–217. doi: 10.1038/nrgastro.2015.34. [DOI] [PubMed] [Google Scholar]
- 4.Lanternier F, et al. Inherited CARD9 deficiency in otherwise healthy children and adults with Candida species–induced meningoencephalitis, colitis, or both. J Allergy Clin Immunol. 2015;135:1558–68.e2. doi: 10.1016/j.jaci.2014.12.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsu YMSM, et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat Immunol. 2007;8:198–205. doi: 10.1038/ni1426. [DOI] [PubMed] [Google Scholar]
- 6.Goodridge HS, et al. Differential use of CARD9 by dectin-1 in macrophages and dendritic cells. J Immunol. 2009;182:1146–1154. doi: 10.4049/jimmunol.182.2.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hara H, et al. Cell-type-specific regulation of ITAM-mediated NF-κB activation by the adaptors CARMA1 and CARD9. J Immunol. 2008;181:918–930. doi: 10.4049/jimmunol.181.2.918. [DOI] [PubMed] [Google Scholar]
- 8.Sokol H, et al. CARD9 mediates intestinal epithelial cell restitution, T helper 17 responses, and control of bacterial infection in mice. Gastroenterology. 2013;145:591–601. doi: 10.1053/j.gastro.2013.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darfeuille-Michaud A, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 10.Sokol H, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn’s disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atarashi K, et al. Treg induction by a rationally selected mixture of clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 12.Zelante T, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Rutz S, Eidenschenk C, Ouyang W. IL-22, not simply a TH17 cytokine. Immunol Rev. 2013;252:116–132. doi: 10.1111/imr.12027. [DOI] [PubMed] [Google Scholar]
- 14.Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation, and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 15.Stelter C, et al. Salmonella-induced mucosal lectin RegIII-β kills competing gut microbiota. PLoS One. 2011;6:e20749. doi: 10.1371/journal.pone.0020749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Luca A, et al. IL-22 defines a novel immune pathway of antifungal resistance. Mucosal Immunol. 2010;3:361–373. doi: 10.1038/mi.2010.22. [DOI] [PubMed] [Google Scholar]
- 17.Ishigame H, et al. Differential roles of interleukin (IL)-17A and IL-17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity. 2009;30:108–119. doi: 10.1016/j.immuni.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 18.Wu W, Hsu YMSM, Bi L, Songyang Z, Lin X. CARD9 facilitates microbe-elicited production of reactive oxygen species by regulating the LyGDI–Rac1 complex. Nat Immunol. 2009;10:1208–1214. doi: 10.1038/ni.1788. [DOI] [PubMed] [Google Scholar]
- 19.Iliev ID, et al. Interactions between commensal fungi and the C-type lectin receptor dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richard ML, Lamas B, Liguori G, Hoffmann TW, Sokol H. Gut fungal microbiota: the yin and yang of inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:656–665. doi: 10.1097/MIB.0000000000000261. [DOI] [PubMed] [Google Scholar]
- 21.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim K, et al. Interleukin-22 promotes epithelial cell transformation and breast tumorigenesis via MAP3K8 activation. Carcinogenesis. 2014;35:1352–1361. doi: 10.1093/carcin/bgu044. [DOI] [PubMed] [Google Scholar]
- 23.Andoh A, et al. Interleukin-22, a member of the IL-10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterology. 2005;129:969–984. doi: 10.1053/j.gastro.2005.06.071. [DOI] [PubMed] [Google Scholar]
- 24.Sabat R, Ouyang W, Wolk K. Therapeutic opportunities of the IL-22–IL-22R1 system. Nat Rev Drug Discov. 2014;13:21–38. doi: 10.1038/nrd4176. [DOI] [PubMed] [Google Scholar]
- 25.Pickert G, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206:1465–1472. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spits H, et al. Innate lymphoid cells—a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 27.Chung KTT, Gadupudi GS. Possible roles of excess tryptophan metabolites in cancer. Environ Mol Mutagen. 2011;52:81–104. doi: 10.1002/em.20588. [DOI] [PubMed] [Google Scholar]
- 28.Jin UHH, et al. Microbiome-derived tryptophan metabolites and their aryl-hydrocarbon-receptor-dependent agonist and antagonist activities. Mol Pharmacol. 2014;85:777–788. doi: 10.1124/mol.113.091165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JS, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on, and independent of, Notch. Nat Immunol. 2012;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zenewicz LA, et al. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J Immunol. 2013;190:5306–5312. doi: 10.4049/jimmunol.1300016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jostins L, et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wikoff WR, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Behnsen J, et al. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity. 2014;40:262–273. doi: 10.1016/j.immuni.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin L, Xu X. Indole-3-acetic acid production by endophytic Streptomyces sp En-1 isolated from medicinal plants. Curr Microbiol. 2013;67:209–217. doi: 10.1007/s00284-013-0348-z. [DOI] [PubMed] [Google Scholar]
- 35.Hara H, et al. The adaptor protein CARD9 is essential for the activation of myeloid cells through ITAM-associated and Toll-like receptors. Nat Immunol. 2007;8:619–629. doi: 10.1038/ni1466. [DOI] [PubMed] [Google Scholar]
- 36.Suau A, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;65:4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomas J, et al. Primocolonization is associated with colonic epithelial maturation during conventionalization. FASEB J. 2013;27:645–655. doi: 10.1096/fj.12-216861. [DOI] [PubMed] [Google Scholar]
- 38.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 42.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kõljalg U, et al. Toward a unified paradigm for sequence-based identification of fungi. Mol Ecol. 2013;22:5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 44.Thioulouse J, Chessel D, Dolédec S, Olivier J. ADE-4: a multivariate analysis and graphical display software. Stat Comput. 1997;7:75–83. [Google Scholar]
- 45.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high-density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 46.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:e3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 47.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 48.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao B, et al. Common commercial and consumer products contain activators of the aryl hydrocarbon (dioxin) receptor. PLoS One. 2013;8:e56860. doi: 10.1371/journal.pone.0056860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He G, Zhao B, Denison MS. Identification of benzothiazole derivatives and polycyclic aromatic hydrocarbons as aryl hydrocarbon receptor agonists present in tire extracts. Environ Toxicol Chem. 2011;30:1915–1925. doi: 10.1002/etc.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao X, et al. Metabolite analysis of human fecal water by gas chromatography–mass spectrometry with ethyl chloroformate derivatization. Anal Biochem. 2009;393:163–175. doi: 10.1016/j.ab.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 52.Maneglier B, et al. Simultaneous measurement of kynurenine and tryptophan in human plasma and supernatants of cultured human cells by HPLC with coulometric detection. Clin Chem. 2004;50:2166–2168. doi: 10.1373/clinchem.2004.037465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.