ABSTRACT
Biomarkers that facilitate the prediction of disease recurrence in prostate cancer (PCa) may enable physicians to personalize treatment for individual patients. In the current study, PD-1 (PDCD1) promoter methylation was assessed in a cohort of 498 PCa patients included in The Cancer Genome Atlas (TCGA) and a second cohort of 300 PCa cases treated at the University Hospital of Bonn.
In the TCGA cohort, the PD-1 promoter was significantly hypermethylated in carcinomas versus normal prostatic epithelium (55.5% vs. 38.2%, p < 0.001) and PD-1 methylation (mPD-1) inversely correlated with PD-1 mRNA expression in PCa (Spearman's ρ = −0.415, p < 0.001). In both cohorts, mPD-1 significantly correlated with preoperative prostate specific antigen (PSA). In univariate Cox Proportional Hazard analysis, mPD-1 served as a significant prognostic factor for biochemical recurrence (BCR)-free survival (Hazard ratio: HR = 2.35 [1.35–4.10], p = 0.003, n = 410) in the TCGA cohort. In multivariate analysis, mPD-1 was shown to add significant independent prognostic information adjunct to pathologic tumor category (pT) and Gleason grading group (HR = 2.08 [1.16–3.74], p = 0.014, n = 350).
PD-1 promoter methylation analyses could thus potentially aid the identification of patients which might benefit from adjuvant treatment after radical prostatectomy. Moreover, our data suggest an intrinsic role of PD-1 in PCa carcinogenesis and disease progression, which needs to be addressed in future studies.
KEYWORDS: DNA methylation, PD-1, PDCD1, prognostic biomarker, prostate cancer
Abbreviations
- AR
androgen receptor
- BCR
biochemical recurrence
- 95%CI
95% confidence interval
- HR
Hazard ratio
- ISUP
International Society of Urological Pathology
- mPD-1
PD-1 (PDCD1) promoter methylation
- pN
pathologic nodal stage
- preopPSA
preoperative serum PSA
- PSA
prostata specific antigene
- pT
pathologic tumor stage
- R
resection state
- TCGA
The Cancer Genome Atlas
Introduction
Oncologic treatment is on the verge of an important breakthrough with emerging cancer immunotherapies. Within the last couple of years, new insight on the interaction between tumor and host immune response has particularly been focusing on programmed death-1 receptor (PD-1)/programmed death-1 ligand (PD-L1) as major immune modulators in various tumor entities. Meanwhile, PD-1 checkpoint inhibitors have gained regulatory approval for the treatment of metastasized malignant melanoma, Hodgkin lymphoma, non-small cell lung carcinoma, and bladder cancer.1,2 However, data on the efficacy of PD-1 checkpoint inhibition in prostate cancer (PCa) are sparse.3 At present, there are two ongoing clinical phase II trials evaluating the sensitivity of solid tumors including PCa to PD-1 immune checkpoint inhibition with results still pending (ClinicalTrials.gov Identifier: NCT02312557 and NCT02458638).
Recently, Gevensleben et al. have shown that the PD-1 receptor ligand PD-L1 is differentially expressed among primary PCa patients.4 In two large independent cohorts, high PD-L1 expression was significantly associated with adverse outcome.4 While PD-L1 expression has been recognized in various types of cancers,5 PD-1 receptor, also known as CD279 and PDCD1, is an immune inhibitory receptor belonging to the extended CD28/CTLA-4 family that is stably expressed on T cells which are exposed to a chronic antigen.6,7 PD-1 expression has further been shown to be regulated by promoter methylation.8 Very recently, however, Kleffel et al. demonstrated that PD-1 expression is not restricted to lymphoid cells but can also be found on malignant melanoma cells. Intrinsic PD-1 expression has been linked to tumor immune evasion and failure to anti-PD-1 treatment.9 Encouraged by Kleffel et al., we aimed to further complete the insight of the PD-1/PD-L1 pathway's activity in PCa.
Results
PD-1 expression in prostate samples (TCGA cohort)
The results shown for the TCGA cohort are entirely based upon data generated by The Cancer Genome Atlas (TCGA) Research Network: http://cancergenome.nih.gov/. Normalized PD-1 mRNA expression was highly variable in both normal and PCa samples and revealed no significant difference (16.4 [95%CI: 12.3–20.5] for native; 23.3 [95%CI: 20.5–26.1] for PCa; p = 0.19 (Mann–Whitney U test), n = 50/497). Since PD-1 expression has so far mainly been observed in tumor infiltrating lymphocytes (TIL), the association with immune cell invasion was analyzed in detail. PD-1 mRNA content neither correlated with TILs (ρ = −0.075; p = 0.21), nor with the stromal content of tumor samples (ρ = −0.047; p = 0.40). The majority of tumors evaluated in TCGA further revealed only sparse lymphoid infiltration (3.8% [95%CI: 2.7–5.0%]). However, low numbers of infiltrating T cells expressing high amounts of PD-1 may potentially distort PD-1 mRNA levels in the tumor probes which prompted us to analyze PD-1 promoter methylation.
PD-1 promoter methylation in prostate cancer (TCGA cohort)
Five beads of the Illumina Infinium HumanMethylation450 BeadChip that are located in the putative promoter region of PD-1 were included in the analysis (Fig. 1). The methylation level (mPD-1) obtained after mean averaging the methylation levels of each of the five single beads was used as a robust measure. As expected, mPD-1 significantly and inversely correlated with PD-1 mRNA expression (ρ = −0.415; p < 0.001). Additionally, mean mPD-1 significantly differed between normal and carcinomatous epithelium (38.2% [95%CI: 35.1–41.4%] for normal; 55.5% [95%CI: 54.4–56.6%] for PCa; p < 0.001 (Mann–Whitney U test); n = 50/498). For detailed clinicopathologic correlation and survival analyses, mPD-1 values were calculated as continuous as well as dichotomized variables to obtain qualitative results. In order to avoid overfitting, in a first step, mPD-1 was evaluated dichotomized choosing median methylation as cut-off. In a second step, an optimal mPD-1 cut-off was elaborated by an iterative approach (57.50%, Fig. 2) and accordingly stratified patients into PD-1 hypo- (mPD-1low) and hypermethylated (mPD-1high) cases.
Figure 1.

Genomic organization of the PD-1 (PDCD1) gene and location of the analyzed Infinium HumanMethylation450 BeadChip probes. Gene organization is based on the Genome Reference Consortium Human Build 38 (GRCh38) which is illustrated using the Ensemble genome browser (http://www.ensembl.org). Shown are three transcript variants (PDCD1-001, PDCD1-002, PDCD1-003). The PDCD1-002 transcript is assumed to undergo non-sense mediated decay, a process which prevents the expression of erroneous or truncated proteins. The methylation specific beads analyzed in the TCGA cohort are located in the annotated promoter region upstream of a CTCF binding site. The assay performed in the University Hospital Bonn cohort is also depicted. The gene contains an enhancer element and a region of open chromatin. Percentage of CG content is depicted indicating a high CpG content throughout the whole gene (promoter and gene body).
Figure 2.
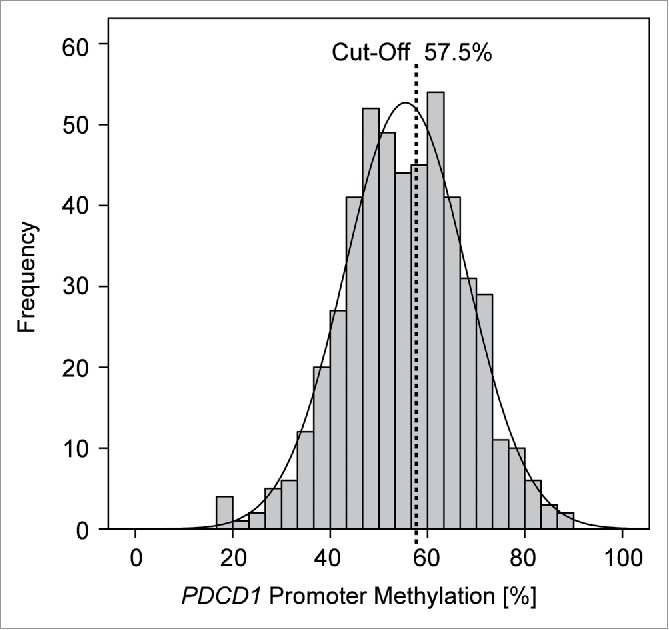
Frequency of PD-1 methylation (mPD-1) in 498 prostate carcinoma samples. PD-1 promoter methylation analysis revealed a symmetric, almost Gaussian distribution (black line) covering a broad spectrum of values (18–88%) with a bifurcation at 55%. An optimal mPD-1 cut-off was elaborated by an iterative approach (57.50%) stratifying patients into PD-1 hyper- (mPD-1high) and hypomethylated (mPD-1low) cases.
Clinicopathologic correlation (TCGA cohort)
PD-1 mRNA negatively correlated with androgen receptor (AR) activity scores when applied as a continuous variable (ρ = −0.28; p < 0.001), while mPD-1 revealed a significant positive correlation with AR activity scores (ρ = 0.20; p < 0.001). Analogously, preoperative prostate-specific antigen (preopPSA) and PD-1 mRNA had a negative correlation (ρ = −0.11; p = 0.015), while preopPSA and mPD-1 were positively correlated (ρ = 0.11; p = 0.018). Advanced tumor (pT) categories were by trend associated with higher mPD-1 (Table 1, p = 0.10).
Table 1.
Patients' baseline characteristics and association/correlation of PD-1 DNA methylation with clinicopathological parameters—TCGA cohort.
| Variable | All patients | [%] | Mean PD-1 methylation | p-value |
|---|---|---|---|---|
| Patient number | 498 | 100 | 55.5 [54.4–56.6] | |
| Patient with follow-up | 430 | 86.3 | ||
| Mean follow-up [Mo] | 22 [20–24] | |||
| Median follow-up [Mo] | 16 | |||
| Range follow-up [Mo] | 1–115 | |||
| Age [y] | ||||
| Mean | 61.0 [60.4–61.6] | |||
| Median | 61.0 | |||
| n ≤ Median | 251 | 50.4 | 55.2 [53.7–56.8] | |
| n > Median | 247 | 49.6 | 55.8 [54.2–57.3] | 0.58* |
| Pathological tumor category | ||||
| pT1/2 | 188 | 37.8 | 54.3 [52.5–56.1] | |
| pT3/4 | 303 | 60.8 | 56.3 [54.9–57.7] | 0.10* |
| Unknown | 7 | 1.4 | ||
| Preoperative PSA [ng/mL] | ||||
| Range | 0.7–107.0 | |||
| Mean | 11.0 [9.9–12.1] | |||
| Median | 7.5 | |||
| ≤Median | 252 | 50.6 | 54.1 [52.5–55.6] | |
| >Median | 243 | 48.8 | 56.8 [55.3–58.4] | 0.025* |
| Unknown | 3 | 0.6 | ||
| ISUP Gleason grading group | ||||
| 1 (<7 ) | 45 | 9.0 | 53.9 [50.3–57.5] | |
| 2 (3+4) | 147 | 29.5 | 54.0 [52.0–55.9] | |
| 3 (4+3) | 101 | 20.3 | 57.1 [54.9–59.3] | |
| 4 (8) | 64 | 12.9 | 55.3 [51.5–59.1] | |
| 5 (9−10) | 141 | 28.3 | 56.5 [54.4–58.6] | 0.30ε |
| Nodal category | ||||
| pN0 | 346 | 69.5 | 55.5 [54.1–56.7] | 0.45* |
| pN1 | 79 | 15.9 | 56.2 [53.2–59.1] | |
| Unknown | 73 | 14.7 | ||
| Surgical margin | ||||
| R0 | 316 | 63.5 | 55.3 [53.9–56.7] | |
| R1 | 147 | 29.5 | 55.7 [53.8–57.7] | 0.81* |
| unknown | 35 | 7.0 | ||
| ERG-translocation1 | ||||
| ERG+ | 152 | 30.5 | 57.9 [56.0–59.7] | 0.006* |
| ERG− | 181 | 36.3 | 54.6 [52.9–56.4] | |
| Unknown | 165 | 33.1 | ||
| Androgen receptor (AR) score1 | ||||
| AR+ | 166 | 33.3 | 57.6 [55.9–59.3] | |
| AR- | 167 | 33.5 | 54.7 [52.8–56.5] | 0.021* |
| Unknown | 165 | 33.1 |
Mann–Whitney U test
Kruskal–Wallis test
adopted from the Cancer Genome Atlas Research Network (2015)10.
Significant features are printed in bold.
High AR activity scores were more frequently detected in ERG-translocation negative (ERG−) PCa (χ2 = 12.0; p < 0.001, n = 333) as adopted from TCGA Research Network (2015).10 In contrast, mPD-1 was significantly associated with the presence of the ERG-gene fusion (ERG-translocation positive, ERG+) in PCa.
PD-1 expression and mPD-1 and biochemical recurrence-free survival analyses (TCGA cohort)
Subsequently, we analyzed whether PD-1 mRNA expression and mPD-1 allowed for the stratification of patients at risk for biochemical recurrence (BCR) in the TCGA cohort. In the univariate Cox proportional hazards model, continuous PD-1 expression failed to classify as a prognostic factor (HR = 0.99 [95%CI: 0.99–1.00], p = 0.17), while continuous mPD-1 values served as a prognostic factor (HR = 1.03 [95%CI: 1.0–1.05], p = 0.021, Table 2). Accordingly, patients with mPD-1high tumors showed a significantly reduced BCR-free survival compared to patients with mPD-1low tumors (Table 2). The prognostic value of mPD-1low and mPD-1high was further confirmed by Kaplan–Meier analyses using an optimized cut-off for mPD-1 dichotomization (p = 0.002, n = 417; Fig. 3).
Table 2.
Univariate and multivariate Cox Proportional Hazard analyses on biochemical recurrence-free survival in a cohort of prostate cancer cases treated by radical prostatectomy—TCGA cohort.
| Univariate |
Multivariate (n = 350) |
||||
|---|---|---|---|---|---|
| Variable | n | p | Hazard Ratio [95% CI] | p | Hazard Ratio [95% CI] |
| pT3–4 vs. pT1–2 | 404 | < 0.001 | 5.37 [2.14–13.5] | 0.013 | 1.67 [1.12–2.51] |
| pN1 vs. pN0 | 356 | 0.049 | 1.84 [1.00–3.35] | 0.50 | 0.80 [0.41–1.54] |
| Gleason grade group (ISUP) | 410 | < 0.001 | 1.70 [1.34–2.13] | 0.024 | 1.39 [1.04–1.85] |
| Preoperative PSA | 408 | < 0.001 | 1.04 [1.02–1.05] | 0.16 | 1.02 [0.99–1.04] |
| R1 vs. R0 | 385 | 0.13 | 1.52 [0.89–2.61] | ||
| ERG-rearrangement | 272 | 0.51 | 0.80 [0.41–1.57] | ||
| PD-1 methylation (dichotomized, cut-off 57.50%) | 410 | 0.003 | 2.35 [1.35–4.10] | 0.014 | 2.08 [1.16–3.74] |
| PD-1 methylation (dichotomized, median cut-off) | 410 | 0.029 | 1.87 [1.07–3.30] | ||
| PD-1 methylation (continous variable) | 410 | 0.021 | 1.03 [1.00–1.05] | ||
Figure 3.
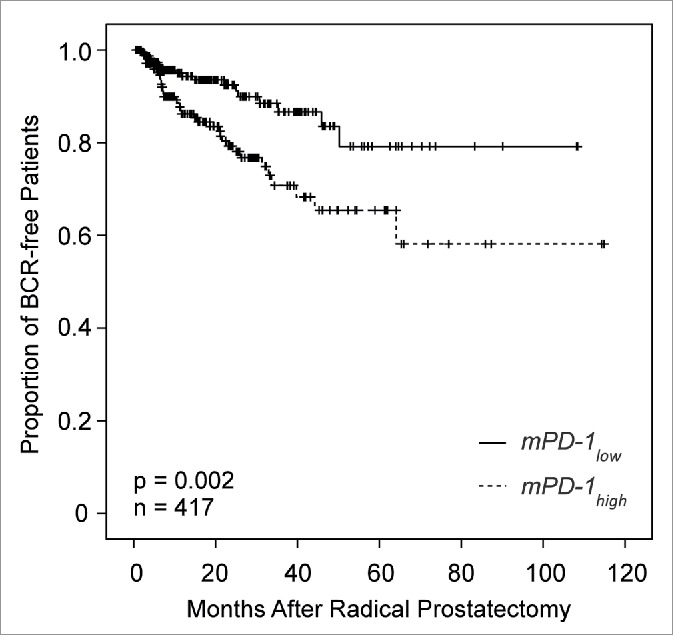
Kaplan–Meier analysis of biochemical recurrence (BRC)-free survival in 417 prostate cancer patients stratified by PD-1 DNA methylation status. Patient classification into mPD-1high and mPD-1low cases was based on an optimized cut-off (57.50% methylation) as shown in Fig. 2. Approximate mean BCR-free survival: 92 months (mPD-1low, 95%CI: 84–100 months, n = 239) and 79 months (mPD-1high, 95%CI: 67–90, n = 178; p = 0.002), respectively.
Since mPD-1 did not correlate with major clinicopathologic parameters, we hypothesized that it might serve as an independent prognostic factor in PCa. In multivariate Cox Proportional Hazards analysis, mPD-1 remained an independent significant prognostic factor when tested together with pT category, nodal status, Gleason grading group (grouped according to the latest International Society of Urological Pathology (ISUP) consensus),11 preopPSA, and surgical margins (Table 2).
PD-1 promoter methylation in prostate samples (University Hospital Bonn cohort)
For the confirmation of the mPD-1 biomarker performance, a quantitative methylation-specific real-time PCR (qMSP) was designed targeting a segment of the PD-1 promoter (GRCh38.p7, Chr2:241859868–241859951) as probed by the Infinium HumanMethylation450 BeadChip beads cg00795812, cg27051683, and cg03889044 (Fig. 1).
Clinicopathologic correlation (University Hospital Bonn cohort)
Due to the small size of the cohort available for the confirmation of our findings, an analysis of BCR-free survival intervals would have been underpowered and was therefore not performed. In the University Hospital Bonn cohort, average mPD-1 amounted to 91.3% [95%CI: 85.7–96.9%]. It significantly correlated with preoperative PSA values (r = 0.187; p = 0.001). Furthermore, advanced pT categories were associated with higher mPD-1 (mean mPD-1: 87.1% [95%CI: 80.4–93.8%] for pT1/2 (n = 205), 100.4% [95%CI: 90.5–110.3%] for pT3/4 (n = 95); p = 0.022). In addition, a strong trend toward higher mPD-1 in ERG+ PCa was observed (mean mPD-1: 95.0% [95%CI: 86.5–103.5%] for ERG− (n = 145), 103.7% [95%CI: 94.3–113.2%] for ERG+ (n = 65); p = 0.052).
Discussion
Epigenetic alterations are involved in the regulation of gene expression in key biological processes, i.e. development, differentiation, alternative splicing, and genetic imprinting (for review: 12,13,14). In addition, promoter hypermethylation also plays a major role in carcinogenesis altering the expression levels of e.g. critical growth regulators. Thereby, gene methylation has turned out to be a potential prognostic biomarker in different tumor entities (reviewed in: 15). Of note, PD-1 DNA methylation had a significant impact on the course of prostate adenocarcinoma in the current study. While PD-1 mRNA expression was highly variable in PCa and did not serve as a prognostic factor in the cohort under investigation, high PD-1 DNA methylation was linked to a BCR 7 months earlier compared to patients with low mPD-1 levels. In multivariate Cox proportional hazards analysis, PD-1 DNA methylation furthermore served as a significant prognostic factor in PCa.
In the TCGA cohort, PD-1 DNA methylation was found to be significantly higher in ERG-translocation positive PCa and in tumors with high AR activity scores. This may result from the fact that 5′-fusion partners of ERG, such as TMPRSS2, are highly androgen-responsive as shown by previous studies.16,17 AR has a crucial influence on PCa progression, especially in ERG fusion-positive PCa, since AR-activated TMPRSS2, a major fusion partner of ERG, directly regulates the expression of the oncogenic fusion product, thereby controlling cell viability, protein transcription, protein degradation, and cytoskeletal reorganization.18 In addition, PD-1 promoter methylation and consecutive downregulation of PD-1 protein expression may be influenced by ERG and its oncogenic fusion product in an AR dependent manner. This is in keeping with the observation that PD-L1, PD-1s major receptor ligand, is also significantly associated with AR expression in PCa.4 Analogously, in breast carcinoma, PD-L1 is more commonly expressed in cancers with lymphocytic infiltrates and AR expression.19 This implies that sex hormones may significantly influence the subcellular interaction between PD-1 and PD-L1, which may associate with the early observation that estrogen modulates autoimmune responses via the PD-1 receptor.20,21 In order to fully characterize the role of PD-1 expression in prostatic adenocarcinoma, however, further mechanistic studies are warranted.
In the present study, PD-1 mRNA content in PCa was independent from lymphocytic infiltration and from the stromal amount of the sample, which may potentially be due to a distinctive yet undefined role of PD-1 in PCa glands themselves. Tumor-intrinsic PD-1 expression, however, has only recently been characterized in malignant melanoma cells.9,22 This study shows that prostatic adenocarcinoma contains PD-1 mRNA and that PD-1 mRNA is subject to epigenetic PD-1 promoter control in the same way as it has been described for human T-lymphocytes, whose PD-1 expression underlies epigenetic control and is regulated by promoter methylation.7,8 This finding may be of significance for the future application of immunotherapies in PCa patients, since intrinsic PD-1 expression has been linked to tumor immune evasion and failure to anti-PD-1 treatment.9 The experience with PD-1 checkpoint inhibition in PCa is still limited,3 however, tumor intrinsic PD-1 DNA methylation interfering with tumor-intrinsic PD-1 functions may have a predictive value for future treatment with checkpoint inhibitors in PCa. The present study bears the strength to combine the analysis of PD-1 promoter methylation in two independent cohorts and by two different molecular assays, indicating that the PD-1 gene is differentially methylated in local and advanced PCa. The University Hospital Bonn cohort revealed a significant association between lower mPD-1 and organ confined PCa, which may be due to the altogether higher number of localized tumors compared to the TCGA cohort. In addition, mPD-1 correlated with preoperative serum PSA levels in both cohorts. Both parameters are well known to determine the outcome of PCa patients.23 The latter fact supports the notion that PD-1 promoter methylation qualified as a prognostic biomarker in PCa. However, the prognostic power of PD-1 still needs to be validated in an independent cohort with sufficient follow-up information. Furthermore, the prognostic value should be analyzed with regard to more clinically relevant endpoints, i.e. PCa -specific survival which was unfortunately not available for the analyzed cohorts. In addition, the retrospective nature of the present study further adds to its major limitations.
While methylation values obtained from the Illumina Infinium HumanMethylation450 BeadChip are generally uncalibrated, the use of our qMSP assay has the advantage that it achieves calibrated results. Our qMSP assay revealed that most patient samples presented with a virtually completely methylated PD-1 promoter. This is in concordance with the overall low expression of PD-1 in tumor cells compared to TILs.9
In the last decades, widespread PSA screening has increasingly led to the diagnosis of localized PCa which presents with highly variable outcomes.24 Some tumors slowly advance over decades with only little growth and are at risk of being over-treated by extensive surgery, whereas others progress rapidly leading to systemic disease after short periods of time.25 Of note, the prognostic power of PD-1 promoter methylation was found to be independent from that assigned to the Gleason grading group. Accordingly, PD-1 DNA methylation seems to be an eligible candidate biomarker guiding the decision-making process when determining whether a patient diagnosed as having PCa at intermediate risk might benefit from a radical prostatectomy or could instead be monitored by active surveillance. In qualification, it should however be noted that the clinical performance of the mPD-1 assay in a biopsy cohort of suited patients (i.e. an active surveillance cohort) needs to be evaluated in further prospective studies.
Conclusion
The present study reveals that PD-1 promoter methylation strongly modulates the expression of the immune checkpoint regulator PD-1 in PCa in that it inversely correlates with PD-1 mRNA expression. In addition, the significant positive correlation of mPD-1 and negative correlation of PD-1 mRNA with AR might indicate an intrinsic role of PD-1 in PCa carcinogenesis and progression. In this study, PD-1 promoter methylation serves as a significant predictor of BCR -free survival in PCa patients and adds significant independent prognostic information adjunct to pT-stage and Gleason grade. Hence, mPD-1 might help to personalize individual therapies. Furthermore, the presented data suggest a yet unknown role in PCa cells, which might be exploited to develop new therapy strategies for PCa in the future.
Methods
Subjects - TCGA cohort
The TCGA cohort comprises 498 patients with histologically confirmed adenocarcinoma of the prostate collected from centers involved in the TCGA project. In addition, transcription data were available from additional 50 specimens obtained from patients with simultaneous PCa. BCR-free survival was considered as the primary endpoint of the study. Informed consent has been obtained from all patients that were included in the cohort in accordance with the Helsinki Declaration of 1975.
Subjects—University Hospital Bonn cohort
The cohort from the University Hospital Bonn comprised of 300 patients with histologically confirmed PCa who underwent radical prostatectomy at the University Hospital Bonn between 1998 and 2008. The Institutional Review Board at the University Hospital of Bonn approved the study (board number 071/14) and waived the need for written informed consent.
PD-1 mRNA expression and choice of Illumina HumanMethylation450 beads (TCGA cohort)
PD-1 mRNA expression data were available from 50 normal and 497 carcinomatous prostate samples. For evaluation of PD-1 mRNA, we used the RNA Seq V2 (normalized counts) as provided by the TCGA Research Network. mPD-1 was available from 498 PCa specimens and 35 normal tissues. Clinical follow-up was assessable in 430 individuals (mean follow-up period 22 mo, range 1–115 mo). The TCGA Research Network has created methylation data by means of the Infinium HumanMethylation450 BeadChip (Illumina, Inc., San Diego, CA, USA). Methylation values for each probe pair comprised of a variant specific for the methylated and the unmethylated status, respectively, were calculated by the formula 100%*bead_M/(bead_M+bead_U). The five beads (cg20805133, cg00795812, cg27051683, cg17322655, cg03889044) located within the upstream CpG-island of the PD-1 promoter were selected and the mean methylation value was computed.
AR activity scores (TCGA cohort)
The AR activity scores were obtained from TCGA Research Network (2015),10 who assessed the AR output of tumors by calculating an AR activity score from the expression pattern of 20 genes that are experimentally validated AR transcriptional targets.26
Sample preparation (University Hospital Bonn cohort)
For the analysis of mPD-1, bisulfite converted DNA from patient samples was prepared using to the innuCONVERT Bisulfite All-In-One Kit (Analytik Jena, Germany).
PD-1 quantitative methylation real-time PCR (University Hospital Bonn cohort)
The DNA methylation of PD-1 was determined by means of a methylation-specific qPCR assay targeting the PD-1 promoter region. The methylation-specific qPCR assay was duplexed with a second assay which targets a CpG-free region within the ACTB gene locus and allows for the quantification of the total DNA irrespective of methylation. PCR conditions (buffers, temperature cycling program, real-time PCR instrument) were applied as previously described.27 The following primers and probes were used: PDCD1 forward primer, tcgaagcgaggttagaaatcgtt; PDCD1 reverse primer, ccttcaaaaccgaaccgaatat; PDCD1 probe, 6-FAM-ttggcgcggttgtttggtttcgaga-BHQ-1; ACTB forward primer, cccttaaaaattacaaaaaccacaa; ACTB reverse primer, ggaggaggtttagtaagttttttg; ACTB probe, Atto 647N-accaccacccaacacacaataacaaacaca-BHQ-2. Each sample was measured in triplicate with an input of 25 ng of bisulfite-converted formalin-fixed paraffin-embedded tissue (FFPET) DNA as quantified via UV. As a calibrator 3 ng of bisulfite-converted DNA from human sperm (NW Andrology & Cryobank Inc., Spokane, WA, USA) was used. Human sperm DNA showed 95% DNA methylation of the respective eight CpG dinucleotides within the analyzed region (GRCh38.p7, Chr2:241859868-241859951). Percent human sperm DNA methylation was obtained from Illumina HiSeq 2000 data provided by http://nihroadmap.nih.gov/epigenomics/ (data accessible at NCBI GEO database, accession GSM1127119). mPD-1 was calculated with the ΔΔCT method: , where and . Percentage methylation was calculated using the following formula: .
Immunohistochemical staining
ERG-immunohistochemical staining was applied as a surrogate marker for ERG-translocation as previously described.28
Statistical analyses
If not stated otherwise, PD-1 mRNA and mPD-1 values are given as means and 95% confidence intervals in square brackets. Comparisons were performed using the Wilcoxon–Mann–Whitney test (for two groups) and the Kruskal–Wallis test (for more than two groups), and the χ2-test. Correlations were calculated using the Spearman-rho (r) coefficient. BRC-free survival analyses were performed using the Kaplan–Meier method, and differences between the patient groups were testes by the log rank test (likelihood ratio assumed as Χ2). Hazard ratios (HR) were calculated using univariate and multivariate Cox proportional hazards models. For multivariate testing all parameters that were tested significant in univariate modeling, were entered the multivariate procedure. SPPS 22.0 (IBM Corp., NY, USA) were used for processing of data. p values less than 0.05 were considered as statistically significant.
Disclosure of potential conflicts of interest
Dimo Dietrich has been an employee and is a stockholder of Epigenomics AG, a company that aims to commercialize the DNA methylation biomarkers. Dimo Dietrich is co-inventor and owns patents on methylation biomarkers and related technologies. These patents are commercially exploited by Epigenomics AG. Dimo Dietrich receives inventor's compensation from Epigenomics AG. Dimo Dietrich is a consultant for AJ Innuscreen GmbH (Berlin, Germany), a 100% daughter company of Analytik Jena AG (Jena, Germany), and receives royalties from product sales.
References
- 1.Sundar R, Cho BC, Brahmer JR, Soo RA. Nivolumab in NSCLC: latest evidence and clinical potential. Ther Adv Med Oncol 2015; 7:85-96; PMID:25755681; http://dx.doi.org/ 10.1177/1758834014567470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov 2015; 14:561-84; PMID:26228759; http://dx.doi.org/ 10.1038/nrd4591 [DOI] [PubMed] [Google Scholar]
- 3.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB et al.. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012; 366:2443-54; PMID:22658127; http://dx.doi.org/ 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gevensleben H, Dietrich D, Golletz C, Steiner S, Jung M, Thiesler T, Majores M, Stein J, Uhl B, Müller S et al.. The immune checkpoint regulator PD-L1 is highly expressed in aggressive primary prostate cancer. Clin Cancer Res 2015; 22:1969-77; PMID:26573597; http://dx.doi.org/ 10.1158/1078-0432.CCR-15-2042 [DOI] [PubMed] [Google Scholar]
- 5.He J, Hu Y, Hu M, Li B. Development of PD-1/PD-L1 Pathway in Tumor Immune Microenvironment and Treatment for Non-Small Cell Lung Cancer. Sci Rep 2015; 5:13110; PMID:26279307; http://dx.doi.org/ 10.1038/srep13110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francisco LM, Sage PT, Sharpe AH. The PD-1 pathway in tolerance and autoimmunity. Immunol Rev 2010; 236:219-42; PMID:20636820; http://dx.doi.org/ 10.1111/j.1600-065X.2010.00923.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McPherson RC, Konkel JE, Prendergast CT, Thomson JP, Ottaviano R, Leech MD, Kay O, Zandee SE, Sweenie CH, Wraith DC et al.. Epigenetic modification of the PD-1 (Pdcd1) promoter in effector CD4(+) T cells tolerized by peptide immunotherapy. Elife 2014; 3; PMID:25546306; http://dx.doi.org/21943489 10.7554/eLife.03416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Youngblood B, Oestreich KJ, Ha SJ, Duraiswamy J, Akondy RS, West EE, Wei Z, Lu P, Austin JW, Riley JL et al.. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells. Immunity 2011; 35:400-12; PMID:21943489; http://dx.doi.org/ 10.1016/j.immuni.2011.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleffel S, Posch C, Barthel SR, Mueller H, Schlapbach C, Guenova E, Elco CP, Lee N, Juneja VR, Zhan Q et al.. Melanoma Cell-Intrinsic PD-1 Receptor Functions Promote Tumor Growth. Cell 2015; 162:1242-56; PMID:26359984; http://dx.doi.org/ 10.1016/j.cell.2015.08.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research Network . Electronic address scmo, cancer genome atlas research N. The molecular taxonomy of primary prostate cancer. Cell 2015; 163:1011-25; PMID:26544944; http://dx.doi.org/ 10.1016/j.cell.2015.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA, Grading Committee . The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am J Surg Pathol 2016; 40:244-52; PMID:26492179; http://dx.doi.org/ 10.1097/PAS.0000000000000530. [DOI] [PubMed] [Google Scholar]
- 12.Lev Maor G, Yearim A, Ast G. The alternative role of DNA methylation in splicing regulation. Trends Genet 2015; 31:274-80; PMID:25837375; http://dx.doi.org/ 10.1016/j.tig.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 13.Bestor TH, Edwards JR, Boulard M. Notes on the role of dynamic DNA methylation in mammalian development. Proc Natl Acad Sci U S A 2015; 112:6796-9; PMID:25368180; http://dx.doi.org/ 10.1073/pnas.1415301111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012; 13:484-92; PMID:22641018; http://dx.doi.org/ 10.1038/nrg3230 [DOI] [PubMed] [Google Scholar]
- 15.Dietrich D, Meller S, Uhl B, Ralla B, Stephan C, Jung K, Ellinger J, Kristiansen G. Nucleic acid-based tissue biomarkers of urologic malignancies. Crit Rev Clin Lab Sci 2014; 51:173-99; PMID:24878394; http://dx.doi.org/ 10.3109/10408363.2014.906130 [DOI] [PubMed] [Google Scholar]
- 16.Nelson PS, Clegg N, Arnold H, Ferguson C, Bonham M, White J, Hood L, Lin B. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc Natl Acad Sci U S A 2002; 99:11890-5; PMID:12185249; http://dx.doi.org/ 10.1073/pnas.182376299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin B, Ferguson C, White JT, Wang S, Vessella R, True LD, Hood L, Nelson PS. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res 1999; 59:4180-4; PMID:10485450 [PubMed] [Google Scholar]
- 18.Tan SH, Furusato B, Fang X, He F, Mohamed AA, Griner NB, Sood K, Saxena S, Katta S, Young D et al.. Evaluation of ERG responsive proteome in prostate cancer. Prostate 2014; 74:70-89; PMID:24115221; http://dx.doi.org/ 10.1002/pros.22731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tung N, Garber JE, Hacker MR, Torous V, Freeman GJ, Poles E, Rodig S, Alexander B, Lee L, Collins LC et al.. Prevalence and predictors of androgen receptor and programmed death-ligand 1 in BRCA1-associated and sporadic triple-negative breast cancer. Npj Breast Cancer 2016; 2:16002; http://dx.doi.org/ 10.1038/npjbcancer.2016.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bodhankar S, Vandenbark AA, Offner H. Oestrogen treatment of experimental autoimmune encephalomyelitis requires 17beta-oestradiol-receptor-positive B cells that up-regulate PD-1 on CD4+ Foxp3+ regulatory T cells. Immunology 2012; 137:282-93; PMID:23039230; http://dx.doi.org/ 10.1111/imm.12013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polanczyk MJ, Hopke C, Vandenbark AA, Offner H. Treg suppressive activity involves estrogen-dependent expression of programmed death-1 (PD-1). Int Immunol 2007; 19:337-43; PMID:17267414; http://dx.doi.org/ 10.1093/intimm/dxl151 [DOI] [PubMed] [Google Scholar]
- 22.Schatton T, Schutte U, Frank NY, Zhan Q, Hoerning A, Robles SC, Zhou J, Hodi FS, Spagnoli GC, Murphy GF et al.. Modulation of T-cell activation by malignant melanoma initiating cells. Cancer Res 2010; 70:697-708; PMID:20068175; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bostwick DG, Grignon DJ, Hammond ME, Amin MB, Cohen M, Crawford D, Gospadarowicz M, Kaplan RS, Miller DS, Montironi R et al.. Prognostic factors in prostate cancer. College of American Pathologists Consensus Statement 1999. Arch Pathol Lab Med 2000; 124:995-1000; PMID:10888774; http://dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 24.Sohn E. Screening: Diagnostic dilemma. Nature 2015; 528:S120-2; PMID:26672781; http://dx.doi.org/ 10.1038/528S120a [DOI] [PubMed] [Google Scholar]
- 25.Loeb S, Bjurlin MA, Nicholson J, Tammela TL, Penson DF, Carter HB, Carroll P, Etzioni R. Overdiagnosis and overtreatment of prostate cancer. Eur Urol 2014; 65:1046-55; PMID:24439788; http://dx.doi.org/ 10.1016/j.eururo.2013.12.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hieronymus H, Schultz N, Gopalan A, Carver BS, Chang MT, Xiao Y, Heguy A, Huberman K, Bernstein M, Assel M et al.. Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci U S A 2014; 111:11139-44; PMID:25024180; http://dx.doi.org/ 10.1073/pnas.1411446111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dietrich D, Hasinger O, Banez LL, Sun L, van Leenders GJ, Wheeler TM, Bangma CH, Wernert N, Perner S, Freedland SJ et al.. Development and clinical validation of a real-time PCR assay for PITX2 DNA methylation to predict prostate-specific antigen recurrence in prostate cancer patients following radical prostatectomy. J Mol Diagn 2013; 15:270-9; PMID:23266319; http://dx.doi.org/ 10.1016/j.jmoldx.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braun M, Goltz D, Shaikhibrahim Z, Vogel W, Bohm D, Scheble V, Sotlar K, Fend F, Tan SH, Dobi A et al.. ERG protein expression and genomic rearrangement status in primary and metastatic prostate cancer–a comparative study of two monoclonal antibodies. Prostate Cancer Prostatic Dis 2012; 15:165-9; PMID:22231490; http://dx.doi.org/ 10.1038/pcan.2011.67 [DOI] [PubMed] [Google Scholar]


