ABSTRACT
CD24 expression has been implicated in the oncogenesis of multiple types of cancer and high tumor expression is considered a poor prognosis factor; however, the role of CD24 in oral cancer progression is unknown. Unlike other cancer types, we found that higher CD24 levels in human oral cancers are correlated to lower clinical stage and better overall survival. We then dissected the role of CD24 and mechanisms in oral cancer pathogenesis in mice using a genetic strategy and demonstrated that CD24 deficiency increased the oral cavity tumor burden in response to the carcinogen 4-nitroquioline 1-oxide (4-NQO). Immune profile analysis showed a significant expansion as well as increased suppressive function of myeloid-derived suppressor cells (MDSCs) in CD24−/− mice, but no apparent impairment in T cells, B cells, or dendritic cells. Further, studies with an orthotopically transplanted syngeneic squamous carcinoma model in the tongue of CD24−/− and CD24+/− mice confirmed the protective roles of CD24 against cancer. Moreover, the difference in tumor growth between CD24−/− and CD24+/− mice was blunted by immunodepletion of MDSCs. We conclude that CD24 expression impedes MDSC expansion and function, and thus slows oral cancer oncogenesis. This study is the first to examine the role of CD24 in a de novo oral cancer model, and it highlights the need to consider the immune regulatory roles of CD24 in the development of CD24-targeted therapy for cancer.
KEYWORDS: 4NQO, CD24, HNSCC, HSA, myeloid-derived suppressor cell (MDSC), oral cancer
Introduction
Cancer in the oral cavity (lips, cheeks, gums, tongue, palate, and floor of the mouth) is the sixth most common cancer worldwide, and the prognosis of cancer patients remains poor despite advances in therapy.1,2 In the United States in 2015, there were an estimated 45,780 new cases and over 8,000 deaths.3 Oral cancer carcinogenesis involves the accumulation of genetic alterations leading to the multistep malignant transformation of normal oral epithelial cells.4 Currently, the exact mechanism is unclear; thus, further understanding and identification of factors that initiate disease and modify disease outcomes, such as immune–cancer interactions and interplay between oral bacteria and host immune responses, will contribute to better prevention and patient treatment.
CD24 (also known as heat stable antigen) is a glycosyl-phosphatidyl-inositol (GPI) anchored sialoglycoprotein that is expressed by both haematopoietic and non-haematopoietic cells.5 By interacting with Siglec-G/10, an immunoreceptor tyrosine-based inhibition motif (ITIM)-containing negative regulator of inflammation, CD24 has been shown to serve as an essential brake for inflammation induced by damage-associated molecular patterns (DAMPs).6,7 Bacterial and viral-derived sialidases, which were previously considered virulence factors only in the context of pathogen growth, can disrupt the CD24-Siglec-G/10-mediated negative regulation mechanism by de-sialylation, thereby exacerbating bacteria-induced sepsis.6 CD24 plays important roles in immune regulation by providing co-stimulatory signals to T cells and B cells.5,8 For example, CD24 is critical for T cell homeostasis.9 Of further immunological relevance, dysregulation of CD24 has been implicated in the development of autoimmune diseases.10-12
Elevated CD24 expression has been observed on many types of cancer cells, including breast, ovary, bladder, prostate, and uterine cervical cancer, and is associated with tumor growth and progression.13-18
In the present study, we sought to gain more insight into the role of CD24 in oral cancer. In contrast to other cancers, we found that higher CD24 expression level in the tumor microenvironment confers better survival in oral cancer patients. Using both de novo 4-Nitroquinoline 1-oxide (4NQO) oral carcinogenesis model and an orthotopic transplantable tumor model, we found that CD24−/− mice were more prone to develop progressive oral cancer. Mechanistically, lack of surface CD24 led to the expansion of a highly immunosuppressive CD11b+Gr1+ myeloid cell population at baseline and during oncogenesis, which contributed to oral cancer progression. These findings highlight a new mechanism for immunomodulation of oncogenesis in the oral cavity.
Results
Low CD24 expression in oral cancer is associated with poor prognosis
Since elevated CD24 expression has been observed in many cancers, we wished to investigate the clinical association between CD24 expression and oral cancer progression. The role of CD24 expression in oral cancer has only been studied in the context of cancer stem cell marker and molecular signature.19,20 Oncomine® Power Tools21 was used to analyze the relationship between CD24 gene expression and oral cancer progression as well as patient outcome based on publically available datasets.22,23 We found that oral cavity squamous cell carcinoma cancer patients with regional lymph node metastasis showed lower expression of CD24 mRNA compared to patients without metastasis (Fig. 1A). Similarly, lower CD24 was also associated with a more advanced stage of nasopharyngeal carcinoma (Fig. 1B). Moreover, Kaplan–Meier analysis of 52 patients with oral cavity cancer (OCC) showed that higher expression of CD24 in the tumor microenvironment was associated with longer survival (Fig. 1C). Thus, unlike other cancer types, head and neck cancer patients carry a unique favorable prognosis with increased expression of CD24 in the cancer.
Figure 1.
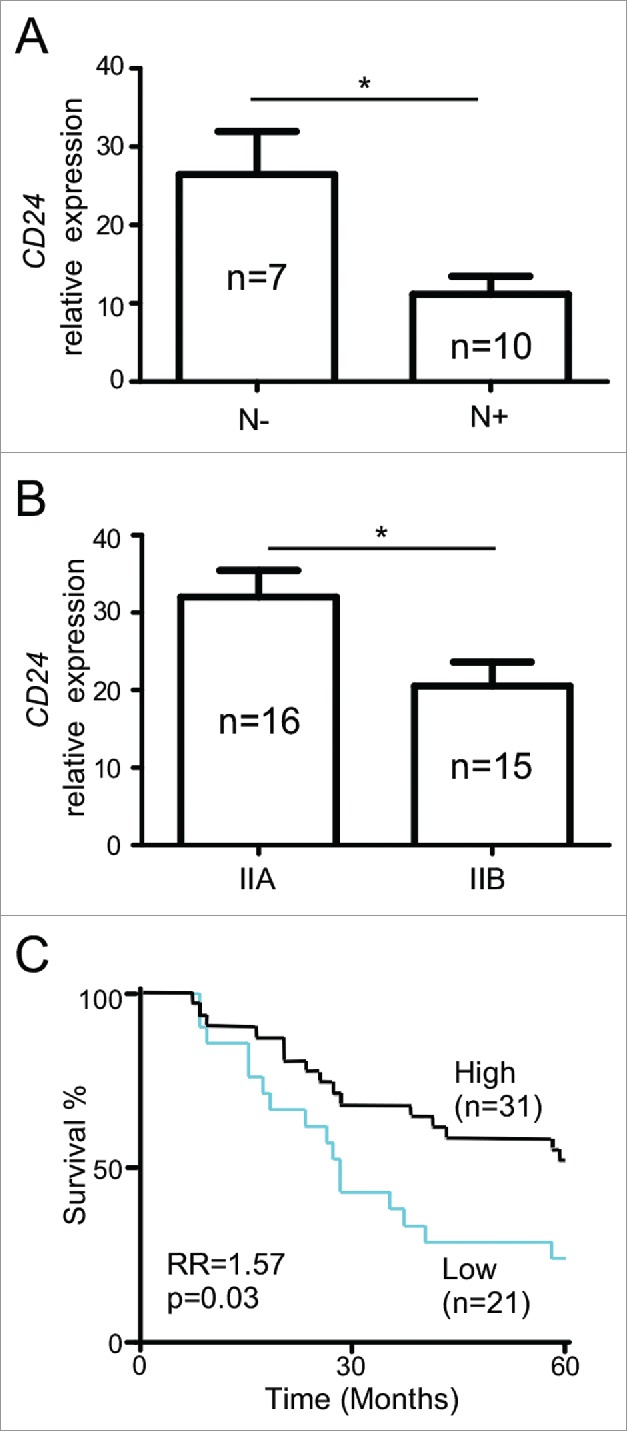
CD24 expression confers poor prognosis in oral cavity cancer. (A) Association of CD24 mRNA in squamous cell carcinoma of the oral cavity with regional lymph node metastasis. (N+, n = 10; N0, n = 7; p = 0.011). (B) Association of CD24 mRNA in nasopharyngeal carcinoma with the clinical stages (IIA, n = 16, IIB, n = 15; p = 0.019). (C) Impact of CD24 mRNA expression on overall survival of patients with oral cavity cancer patients (n = 52). Program X-tile was used to determine an optimal cut-off point for defining two groups of patients with different survival curves, based on relative CD24 mRNA expression. The vertical axis in panel A and B represents the normalized expression intensity of CD24 relative to the median intensity of all the probes in the microarray platform. Panels A and B were analyzed by two-tailed unpaired two-sample Student's t-test, error bars represent SEM. Panel C was analyzed by Kaplan-Meier analysis, and the log-rank test was used to determine whether the survival curves were significantly different; *p < 0.05.
CD24−/− mice are more susceptible to 4-NQO-induced oral carcinogenesis
Due to the apparent clinical connection between CD24 expression level in the tumor and oral cancer in patients, we wished to confirm the protective role of CD24 in oral cancer development using the carcinogenic 4-NQO oral cancer model.24,25 This model was selected because of the in situ development and similarities to carcinogenesis induction in humans. In this model, CD24+/− and CD24−/− mice were treated with 4-NQO (50 µg/mL) in the drinking water for 16 weeks and then changed to regular water for the remainder of the experiment. CD24−/− mice began to develop tongue lesions as early as 11 weeks in CD24 deficient mice; 100% of the mice developed lesions by 20 weeks (Fig. 2B). Mouse body weight change and tongue tumor lesions were examined weekly in a genotype-blinded manner after carcinogen treatment. CD24−/− and CD24+/− mice began to lose body weight 13 weeks after beginning 4-NQO administration; no significant difference was seen between the two groups (Fig. 2A). More strikingly, CD24−/− mice were more susceptible to cancer induction by the 4-NQO treatment as they develop tumor lesions earlier than CD24+/− mice (Fig. 2B). Histological analysis of the tongue lesions demonstrated that there were more high-grade dysplasia lesions in CD24−/− mice compared to CD24 heterozygous mice (Fig. 2C). To assess the total oral cavity tumor burden after 4-NQO treatment, the total number of tumors in the tongue and oral cavity, tumor size, and histological tumor grade were equally weighted. Using this five-category scoring system, the maximum score for any animal is five (see Methods section). Our data demonstrated that CD24−/− mice have a significantly higher tumor burden score than CD24+/− littermates (Fig. 2D).
Figure 2.
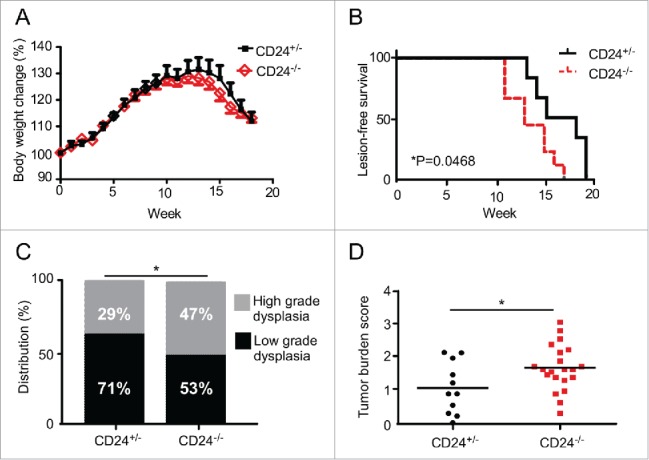
CD24 blunts 4-NQO-induced oral carcinogenesis. (A) CD24−/− and CD24+/− mice were subjected to 50 μg/mL 4-NQO in the drinking water for 16 weeks then switched to regular drinking water. (A) Mouse body weight and (B) tumor lesions of 4-NQO-treated mice (n = 14 CD24+/− and n = 17 CD24−/−) were monitored weekly. Data shown is combined data from two independent experiments with similar findings. (C) The pathology of tongue lesions from CD24−/− (n = 17) and CD24+/− mice (n = 14) was examined. Tumor lesions were classified as low-grade (no tumor or low-grade dysplasia) and high-grade (high-grade dysplasia or invasive cancer). (D) Tumor burden score in the mice was calculated by equally weighting the total number of tongue and oral cavity tumors, tumor size, and histological tumor grade as in Methods section. Using this five-category scoring system, the maximum score obtainable for any animal is five (p = 0.0297). Statistical significance was determined by Chi-square analysis, log-rank test, or two-tailed Student's t-test, where appropriate; *p < 0.05.
Oral microbiota do not influence 4-NQO-induced oral cancer progression in mice
We next sought to determine the mechanism behind the differences detected in the 4-NQO model of oral cancer. The oral cavity is unique in that it is continually exposed to external environment, pathogens, and dietary antigens and is home to a rich array of microflora comprising many different microbial species. The composition and quantity of this microflora varies from person to person and can change in response to tobacco use and alcohol consumption; the two risk factors that have known involvement in oral carcinogenesis.26,27,28 Although a significant number of oral cancer patients have altered bacterial flora, the link between intra-oral microflora alteration and oral carcinogenesis remains unclear.29-31 In the context of oral health, Ye et al.32 reported that CD24 is selectively and highly expressed in the gingival epithelial cells, where it maintains the expression of selected genes encoding tight junction components associated with a marginal barrier function of the oral epithelium. Knockdown of CD24 expression by RNA interference results in increased epithelial barrier disruption, which led to intercellular penetration of microbial products.32 Therefore, combining the knowledge that the presence of host CD24 modulates microbial activity in the oral cavity, and that patients with oral cancer have dysbiosis, it is possible that disruption of the CD24—oral microbiome axis alters cancer development.33,34
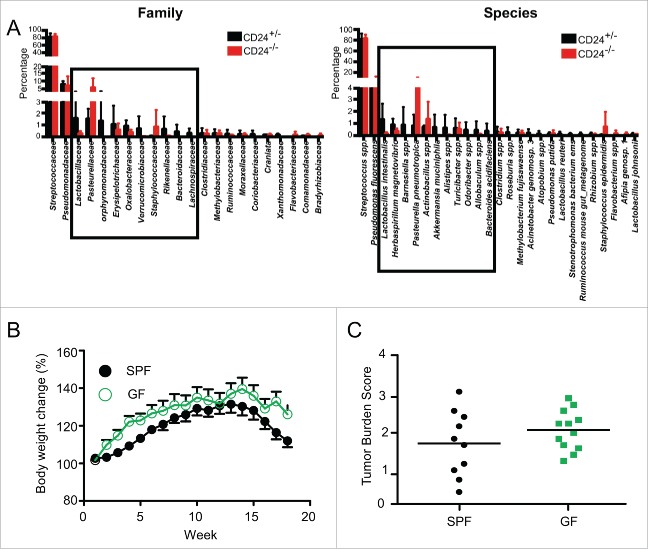
Zhang et al.35 demonstrated that probiotic L. salivarius REN treatment significantly damped 4-NQO-induced oral cancer development in rats; however, the significance of the oral microbiome in 4-NQO-induced oral carcinogenesis has never been addressed in mouse models. Further, due to the known role of bacteria-produced sialidase in disrupting the CD24-Siglec-G/10 connection, we investigated if CD24 deficiency alters intra-oral microflora in a manner that may contribute to oral carcinogenesis. To address these questions, we first analyzed the distribution of tongue microbiota in CD24+/− and CD24−/− mice by pyrosequencing. The data showed that the constitution of microbial flora between CD24+/− and CD24−/− mice in homeostatic conditions was similar. The predominant species of bacteria found on the tongues of both CD24−/− and CD24+/− mice were Streptococcus spp, and Pseudomonas fluorescens (Fig. 3A). To investigate definitively if there is any role of the oral microbiota in 4-NQO-induced oral carcinogenesis, we next compared 4-NQO-induced oral cancer development in both specific pathogen-free (SPF) and germ-free conditions. Upon 4-NQO treatment we found there were no significant differences in mouse body weight or oral tumor burden in germ-free conditions compared to SPF conditions (Figs. 3B and C). Our observations indicated that CD24 status does not greatly alter the commensal oral microbiota, and that oral cavity microbiota did not contribute significantly to the oral cancer progression in the 4-NQO mouse model.
Figure 3.
Oral microbiota do not influence 4-NQO induced oral cancer progression in mice. (A) Oral microbiota in mouse tongue from CD24+/− and CD24−/− mice were analyzed by pyrosequencing. Distribution of predominant bacterial family and species (>0.1%) were presented. Data shown is the Mean ± SD, n = 3 for each group. (B, C) Wild-type C57BL/6J mice were housed in GF (germ-free) or SPF (specific pathogen free) conditions. Both groups were subjected to 50 μg/mL 4-NQO for 16 weeks then switched to regular drinking water. (B) Mouse body weight change was monitored (SPF n = 14, GF, n = 18). (C) Total tumor burden of mice were quantified based on the combined factors including number of tongue lesions, primary lesion size, number of oral cavity lesions, primary oral cavity lesion size, and tumor diagnosis (p = 0.22 by two-tailed Student's t-test).
CD24−/− mice have an increase in myeloid-derived suppressor cells
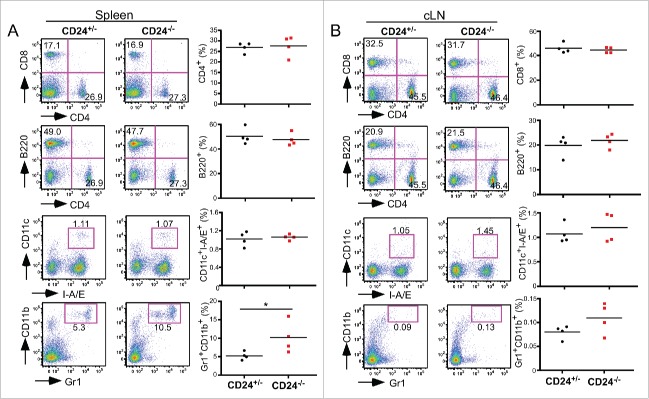
We next focused our attention on the possible immunological roles of CD24 in oral cancer development. Immune profiles of the spleen and draining cervical lymph nodes were analyzed in CD24+/− and CD24−/− mice, with and without 4-NQO treatment. In the steady state, no significant alterations in the T or B cell compartments were found in CD24+/− mice compared with CD24−/− mice. However, we observed a significant expansion of CD11b+Gr1+ myeloid cell population in the spleen of CD24−/− mice (Fig. 4A), but not in the cervical lymph node (Fig. 4B).
Figure 4.
Expansion of CD11b+Gr1+ myeloid cells but no apparent alteration of other immune compartments in the absence of CD24. CD4+ T cells, CD8+ T cells, B220+ B cells, CD11c+ I-A/E+ DC cells, CD4+CD25+FoxP3+ Treg, and CD11b+Gr1+ MDSC in CD24+/− and CD24−/− mice were analyzed by flow cytometry in (A) the spleen and (B) cervical lymph nodes of indicated mice. Multiple experiments were performed with similar findings. Shown is one experiment with four mice per group. Statistical analysis was determined by two-tailed Student's t-test, *p < 0.05.
After 4-NQO treatment, the percentages of CD4+ T cells, CD8+ T cells, B220+ B cells, and CD11c+ dendritic cells in the spleen did not change compared to untreated mice (data not shown). There remained a 2-fold expansion of the MDSC population in 4-NQO-treated CD24−/− mice compared to CD24+/− mice (Fig. 5A), indicating that MDSC expansion in the spleen of CD24−/− mice was not dependent on 4-NQO treatment. Interestingly, we observed a significant increase of FoxP3+CD25+ regulatory T cells (Tregs) in the spleen of mice after 4-NQO treatment, regardless of CD24 status (Fig. 5B). In the cervical lymph node, CD4+ T cells (Fig. 5C) dramatically decreased and B220+ B cells (Fig. 5D) increased in both CD24+/− and CD24−/− tumor-bearing mice. An increase in Tregs was also observed in the tumor-draining lymph nodes, analogous to the observation in the spleen (Fig. 5E). While there was no expansion of the MDSC population in the cervical lymph node of CD24−/− mice in homeostatic conditions, 4-NQO treatment induced an increase in MDSC in the draining lymph node (Fig. 5F).
Figure 5.
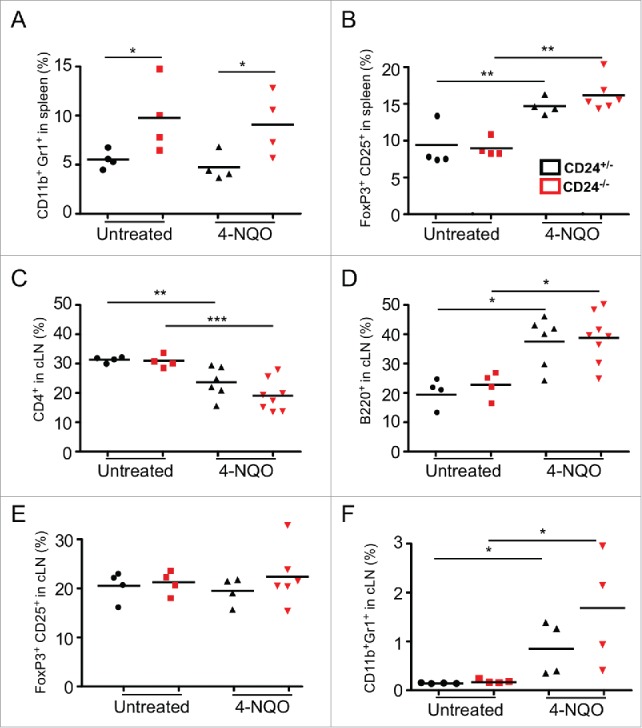
Immune profile of 4-NQO-treated CD24+/− and CD24−/− mice. CD24+/− and CD24−/− mice were treated with 4-NQO as described in the Methods. Immune profile alteration in 4-NQO treated mice was analyzed by flow cytometry at endpoint (20–30 weeks post initial 4NQO treatment) in the spleen ((A)and B) and tumor-draining cervical lymph node (C–F). n = 4–8 mice per group; statistical analysis was determined bytwo-tailed Student's t-test or Mann-Whitney test, *p < 0.05 and **p < 0.01.
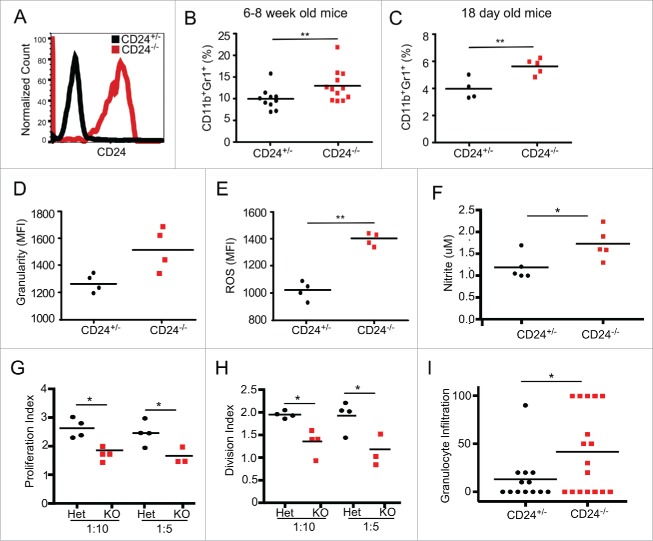
MDSCs expand and show more potent suppressive function in CD24−/− mice
Next, we further investigated the impact of CD24 on the expansion and suppressive function of MDSCs. MDSCs are a heterogeneous population of immature myeloid cells, and can be classified based on Gr1 and CD11b surface markers. There are two main sub-groups: monocytic MDSC (CD11b+Ly6G−Ly6Chigh) and granulocytic MDSC (CD11b+Ly6G+Ly6Clo).37,38 While the role of CD24 has been studied extensively in the lymphocytic lineage, the function of CD24 in MDSCs is less clear.9,11,39 Gr1+ CD11b+ MDSCs express CD24 on their cell surface in homeostatic conditions (Fig. 6A), thus the differences seen between CD24+/− and CD24−/− mice is likely MDSC cell intrinsic. Consistently, a higher percentage of bulk MDSCs (both monocytic and granulocytic MDSCs) was observed in the peripheral blood of CD24−/− mice than in CD24+/− mice (Fig. 6B). As early as 18 d of age, heightened MDSC expansion was already noticeable in CD24−/− mice (Fig. 6C), suggesting that MDSC expansion is intrinsically regulated by the presence of CD24. We assayed the potential suppressor function of MDSCs by quantifying their production of reactive oxygen species (ROS), which are the key suppressive mediators in MDSC function.40-42 We found that MDSCs isolated from CD24−/− mice exhibited higher granularity, richer ROS content, and more nitrite production, indicating increased suppressive capacity (Figs. 6D–F). Indeed, CD24−/− MDSCs was confirmed to have increased suppressive function against polyclonal CD8+ T cell proliferation comparing with WT MDSCs (Figs. 6G and H).
Figure 6.
CD24−/− MDSCs are increased in number and have greater functionality. (A) Cell surface expression of CD24 by peripheral blood CD11b+Gr1+ cells from CD24+/− and CD24−/− mice. (B) Frequency of CD11b+Gr1+ MDSC in the peripheral blood of 6–8 week old CD24−/− mice (n = 12) and CD24+/− mice (n = 10) were analyzed by flow cytometry; (C) Flow cytometric analysis of the frequency of CD11b+Gr1+ MDSC in the spleen of 18 d old CD24−/− mice (n = 5) and CD24+/− mice (n = 4) from the same litter. Flow cytometric analysis of granularity indicated by side scatter profile (SSC) (D), ROS production (E), and nitrite generation (F) by CD24−/− MDSCs. Data is representative of three independent experiments, n = 3–4 mice/experiment. (G-H) CD8+ and MDSCs were MACS isolated from CD24−/+ and CD24−/− mice. CD8+ cells were CFSE labeled and cultured in a 1:5 and 1:10 ratio (MDSC:CD8+) for 3 d. After analyzing CFSE dilution by flow cytometry, the proliferation index and division index were calculated using FlowJo software. (I) Granulocytic infiltration (numbers per 40× high power field) was enumerated in H&E stained tongue lesion sections histologically. Statistical analysis was determined by two-tailed Student's t-test, *p < 0.05 and **p < 0.01.
MDSC expansion and infiltration in tumor lesions is tightly associated with cancer progression.43,44 To determine whether the tumor-infiltrating MDSC population was increased in the CD24−/− 4-NQO-induced tumors compared with CD24+/− mice, we performed IHC on the tongue tumor sections. We found increased infiltration of MDSCs in tumor lesions from the CD24−/− mice compared to these from CD24+/− mice based on immune staining of CD11b, Gr1, and Arginase (Fig. 6I).42 These results suggest that CD24 is involved in oral cancer progression by controlling MDSC accumulation in tumor lesions.12
MDSCs contribute to increased growth of tongue squamous carcinoma in CD24−/− mice
To further investigate CD24-mediated MDSC expansion in oral cancer progression, an orthotopic cancer model was developed. An aggressive syngeneic squamous carcinoma cell (SCC) cell line, A223-LG, was injected to the tongue of mice. A223-LG was derived from a mouse model of SCC with oncogene Kras (G12D) activation and Smad4 deletion in the epithelial progenitors.45 We found that A223-LG-transplanted cancer exhibits significantly accelerated tumor growth in CD24−/− mice comparing with CD24+/− mice (Figs. 7A and B), which is consistent with the observation we made in the 4-NQO oral cancer model.
Figure 7.
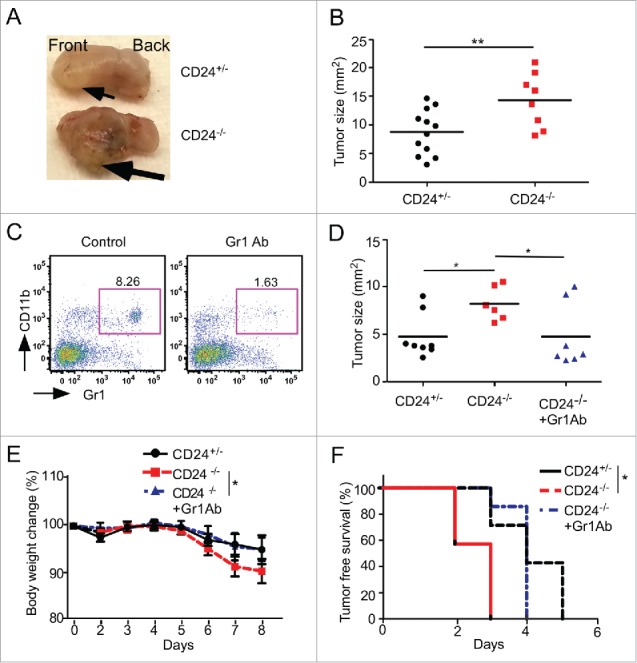
MDSC-dependent increase of oral squamous carcinoma in an orthotopic model of CD24 null mice. In an orthotopic oral cancer model, 0.15 × 105 A223-LG oral cancer cells were injected into the right lateral side of tongues of CD24+/− and CD24−/− mice. (A) A representative image of the tongues from 6–7 tumor-bearing mice; (B) Quantification of the tumor size (n = 8–12 mice/group, two independent experiments). (C) Confirmation of the depletion of the Gr1+CD11b+ cells in the peripheral blood by anti-Gr1 antibody by flow cytometry. (D–F) Impact of MDSC depletion on oral SCC tumor growth (D), body weight loss (E) and tumor-free survival (F) in CD24−/− tumor bearing mice. Statistical significance was determined by two-tailed Student's or log rank test, where appropriate, *p < 0.05 and **p < 0.01.
In order to validate the roles of MDSCs in tumor progression in this model, Gr1 neutralizing antibody (Clone RB6-8C5) was administered to deplete MDSCs (Fig. 7C). Clone RB6-8C5 was previously shown to deplete MDSCs in the peripheral blood and spleen, but not the liver.46 We found that anti-Gr1 antibody treatment significantly suppressed the A223-LG tumor growth in CD24−/− mice compared with CD24+/− mice, indicating that MDSCs contributed to the increased A223-LG tumor growth, as evidenced by tumor size, body weight change, and tumor-free survival (Figs. 7D–F). We conclude that CD24 protect against oral cancer in part via dampening MDSCs (Fig. 8).
Figure 8.
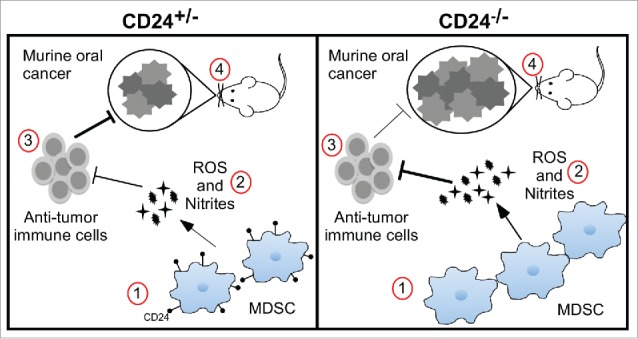
A proposed model of MDSC expansion and increased oral cancer in CD24−/− mice. For simplicity, only four factors are highlighted during oral cancer development (left and right panels): (1) MDSC homeostasis; (2) tolerogenic function of MDSC; (3) anti-tumor immunity; and (4) carcinogenesis. Comparing with the CD24 heterozygous mice (left panel), total loss of CD24 (right panel) leads to heightened carcinogenesis, as a result of diminished antitumor immunity secondary to increased tolerogenic function of MDSCs and enhanced MDSC expansion.
Discussion
Our attention was drawn to CD24 initially because that CD24 mRNA levels in the tumor were inversely correlated to the survival of patients with oral cancer. The roles of CD24 in oral cancer further intrigued us because it is a negative regulator of inflammation on the one hand, and its upregulation seemed to associate with cancer stemness on the other. By a genetic approach, we have addressed, for the first time, multiple factors in the pathogenesis of oral cancer in this study, including the roles of CD24, oral microbiota and the host immune system—particularly MDSCs. We discovered that (i) CD24 negatively regulates the cell number and functional hallmarks of MDSCs; (ii) CD24 protects mice from oral cancer progression that is tightly associated with constraining MDSCs in both the steady state and during oncogenesis; and (iii) contrary to the prediction, the presence of microbiota neither promotes nor inhibits oral cancer in 4-NQO treated wild-type mice. These observations have significantly enhanced our understanding of the complex pathogenesis of oral cancer.
In the oral cancer research field, several animal models for oral squamous cell carcinoma (OSCC) development have been established, including hamster, rat, and mouse models.47 Among them, the 4-NQO-induced oral cancer mouse model is one of the most widely used,24,25,47,48 as it recapitulates many features observed in human OSCC development including molecular expression of keratin, p16, and epidermal growth factor receptor (EGFR) which are associated with human oral carcinogenesis. Therefore, this model is suitable for the analysis of the OCC development in various mutant and transgenic strains.47 However, changes in the host immune system and oral microbiota induced by 4-NQO in mice have not been extensively characterized until now.
The role of CD24 in cancer cells is complex. For example, Saleem et al. found that lymph node metastatic OSCC express higher levels of CD44/CD24.49 It has been reported that downregulation of CD24 inhibits proliferation while inducing apoptosis in malignant cells.50 Further, CD24 may be a potential marker for cancer stem cells. In human nasopharyngeal carcinoma cell lines, cells with higher CD24+ expression have increased growth and tumor sphere formation in vitro.36 While previous studies look at the role of CD24 in the tumors, our investigation turned to analysis of CD24 in the immune system in regulation of tumor control.
In this study, we analyzed the immune cell profile in the spleen and the tumor-draining cervical lymph nodes in tumor-bearing mice. Similar to reports that Treg cells were increased in human OCC patients,51 and after 4-NQO treatment in rats,52 we also observed the expansion of Treg cells in both spleen and cervical lymph nodes in the 4NQO mouse model. The MDSC population is another immune suppressor cell type known to accumulate in the tumor and contribute to tumor progression. To date, there are no clinical reports of the MDSC upregulation in OCC patients specifically, although MDSCs have been shown to be increased in the peripheral blood of HNSCC patients.53 In the 4-NQO mouse model, MDSC accumulation in tumor lesions was observed. While we did not see a significant increase of CD11b+Gr1+ MDSCs in the spleen upon 4-NQO treatments in contrast to what was previously reported,54 we observed a significant increase of MDSCs in CD24−/− mice compared to CD24+/− mice in both 4-NQO treated and untreated conditions. The expansion of MDSC population in CD24−/− mice was observed as early as 18 d of age, suggesting that CD24 signaling intrinsically regulates the MDSC expansion. This may occur through CD24 binding to high mobility group box 1 (HMGB1). Loss of CD24 releases HMGB1, which is known to stimulate MDSC differentiation in circulation.12,55
In the 4-NQO model, CD24 was knocked out globally, including from the epithelial cells and the immune cells. We did not specifically address whether the increased oncogenesis was due to the lack of CD24 in the epithelium, the immune system, or both. However, this question was partially probed using the A223-LG syngeneic transplantable tumor model. We found that identical A223-LG cells also grew more progressively in CD24 KO mice comparing with CD24-sufficient mice. Moreover, by Gr1 antibody depletion of MDSCs during A223-LG orthotopic cancer progression, we demonstrated that the difference in tumor growth between CD24 sufficient and deficient mice is due to increased MDSCs in the latter. Taken together, we concluded that the expression of CD24 on the immune cells play more important protective roles against oral cancer via constraining MDSCs. It is possible that our results may also reflect cancer type or stage-specific roles of CD24, which warrants future studies.
MDSCs are able to regulate tumors through complex mechanisms. For example, in human ovarian carcinoma, MDSCs inhibited T cell activation in the tumor microenvironment and thus promoted cancer stem cell gene expression and metastasis. In this clinical study, a network involving MDSCs-miRNA101-CtBP2-cancer stem cell genes was identified.56 However, our results demonstrated that there is more likely a MDSC cell-intrinsic pathway that is altered by the loss of CD24. STAT signaling is essential for MDSC function: Stat3 is necessary for overall MDSC suppressive activity, Stat1 is necessary for iNOS and arginase production, and Stat5 is needed for MDSC survival.57 Deletion of CD24 removes the sterile inflammation brake and allows for Siglec-G/10 signaling. Further investigation is warranted to determine if there is a connection between Siglec-G/10 and the JAK/STAT pathways, and how CD24 regulated MDSC homeostasis and function.
In summary, using a genetic strategy, we examined a number of important factors in oral cancer, a poorly studied area, including the following: (1) oral microbiota; (2) CD24; and (3) MDSCs. We discovered for the first time, that CD24 constrains MDSCs and protect de novo oral carcinogenesis. We also found that, contrary to the prediction, oral microbiome neither promotes nor suppresses oral cancer. We have thus shed significant light on the roles of both the host immunity and microbiome in the pathogenesis of oral squamous carcinoma. Further studies are necessary to understand the underlying molecular mechanism of our findings and translate them into better management of OSCC. In particular, both cancer-intrinsic roles and the immune regulatory roles of CD24 must be considered in the development of CD24-targeted therapy for cancer.
Materials and methods
Animals
CD24−/− mice (a kind gift from Dr Yang Liu, Center for Cancer and Immunology Research, Children's National Medical Center, Washington, DC) were backcrossed to C57BL/6J mice for at least 10 generations. Mice were housed and handled in the Division of Laboratory Animal Resource facilities under the Medical University of South Carolina guidelines. Unless specified, mice were maintained under SPF conditions and allowed free access to food and water. GF mice were housed in the Medical University of South Carolina Gnotobiotic Animal Facility. All animal experiments were approved by the Institutional Animal Care and Use Committee at the Medical University of South Carolina.
4-NQO model
Male and female CD24−/−, CD24+/−, or wild-type C57BL/6 mice in SPF or GF conditions were treated with 4-NQO (CAS 56–57-5, Sigma-Aldrich) at 50 μg/mL in drinking water for a period of 16 weeks starting at 6–8 weeks of age. Fresh 4-NQO in the drinking water was prepared weekly during treatment. After carcinogen treatment, all animals were provided with regular drinking water; mouse body weight and presence of tongue lesions were monitored weekly. The experiment was terminated when mice developed advanced cancer with greater than 20% body weight loss. The development of oral lesions post 4-NQO treatment was assessed by a five-category scoring system, with five being the maximal score, including the number of the lesions (category score of 1, 0.75, 0.5, 0.25, and 0 for ≥5, 4–5, 2–3, 1–2, and 0 lesions, respectively) and the size of the lesions (category score of 1, 0.67, 0.33, and 0 with 15, 10–15, 5–10, 0–5 mm3, respectively) in the tongue and the oral cavity, and the pathological grade of the most advanced lesions (category score of 1, 0.75, 0.5, and 0 for invasive cancer, high-grade dysplasia, low-grade dysplasia, and normal, respectively).
A223-LG orthotopic oral cancer model
A223-LG squamous cancer cell line (syngeneic with C57BL/6J) was a kind gift from Dr Xiao-jing Wang's laboratory (University of Colorado, Denver). It was derived from transgenic mice that expressed a constitutively active KrasG12D mutant and simultaneous deletion of Smad4 in the epithelial progenitor cells.45 Cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. After two passages, 1.5 × 104 cells in 25 μL sterile 1X PBS were injected in the right lateral side of the tongue of CD24+/− and CD24−/− mice. All mice were anesthetized with ketamine before orthotopic injection. Mice were put on wet food to minimize pain associated with chewing; body weight and tumor development were monitored daily. In the MDSC depletion experiment, anti-Gr1 antibody (Clone RB6-8C5; Bio-X-Cell) was injected intraperitoneally at 150 μg/mouse 2 d before orthotopic tumor injection and continued every 2 d until the experiment endpoint.
Flow cytometry
Spleen and cervical lymph nodes from tumor-bearing and tumor-free mice were dissociated into a single-cell suspension and splenocytes were depleted of red blood cells with RBC lysis buffer (Sigma). After Fc-receptor blocking, cells were stained for surface markers in FACS buffer. Antibodies against CD4+ (RM4-5), FoxP3 (FJK-16), CD8a (53-6.7), B220 (RA3-6B2), Gr1 (RB6-8C5), CD11b (M1/70), CD11c (N418) were purchased from Affymetrix. FoxP3 intracellular staining was performed using commercial FoxP3/Transcription Factor Staining Buffer Set (Affymetrix). Samples were analyzed on a BD Verse cytometer and analyzed by FlowJo Software (Tree Star). Viability of the cells analyzed was ensured by gating on singlets and live cells, as indicated by the lack of 7-AAD or Viability Dye (Affymetrix).
Nitrite and ROS production analysis
Spleens from CD24+/− or CD24−/− mice were made into a single-cell suspension, RBC lysed, and MDSC were sorted using a MACS MDSC isolation kit (Miltenyi Biotec). Oxidation-sensitive dye DCFDA (Molecular Probes/Invitrogen) was used to measure ROS production by both PMN-MDSC and Mo-MDSC. Cells were incubated at 37°C in RPMI 1640 in the presence of 2.5 μM DCFDA for 30 min, followed by incubation with or without 30 ng/mL PMA (Sigma-Aldrich). After staining for the surface markers Gr1 and CD11b, the analysis was conducted by flow cytometry as described above on a BD Verse cytometer. To analyze nitrite production by bulk MDSC, the MACS sorted cells were plated at 1 × 105 cells per well in a 96-well plate with and without 1 μg/mL LPS (InvivoGen) for 24 h. Supernatants were harvested, and Greiss reaction to measure nitrite levels were performed per manufacturer's protocol (Life Technologies).
CD8+ suppression assay
MDSCs were isolated from spleens of CD24+/− or CD24−/− mice as described above. CD8+ cells were isolated from CD24+/− and labeled with 2.5 μM CFSE according to manufacturers protocol (Invitrogen). MDSC and CD8+ cells were plated at ratios of 1:1, 1:5, and 1:10 (MDSC:suppressor cell) at 1 × 105 in 96-well plate with soluble anti-CD3 (5 μg/mL). On day 3 of culture, cells were washed and CFSE dilution was read by flow cytometry. Division index was computed using Flowjo Software (Treestar).
Pathological examination and immunohistochemistry
Tongue sections were fixed in 4% paraformaldehyde and embedded in paraffin. After sectioning, the tongues were deparaffinized, rehydrated, and stained with H&E for histopathology. Gross lesions were identified and photographed. Histological determination of squamous neoplasia was performed by a clinical pathologist (S.S.) who was blinded to the genotype of the mice. The lesions observed were classified into four grades: non-cancerous, low-grade dysplasia, high-grade dysplasia, or invasive carcinoma. Dysplasia was defined as loss of polarity in the epithelial cells, nuclear polymorphism, hyperplasia, and increased or abnormal mitosis. Lesions with such changes involving the entire thickness of epithelium were considered as carcinoma in situ. Papilloma was defined as non-invasive exophytic growth of neoplastic cells, and invasive carcinoma was defined as a lesion with invasion into the sub-epithelial tissues. Immunohistochemistry was performed following standard procedures with anti-Gr1, anti-arginase (BD Biosciences), and anti-CD11b (Abcam).
Oral microbiota analysis
Amplicon pyrosequencing (bTEFAP) was performed as described.58 Samples were sequenced utilizing Roche 454 FLX titanium instruments and reagents, following manufacturer's guidelines. The Q25 sequence data derived from the sequencing process was processed using a proprietary analysis pipeline (www.mrdnalab.com, MR DNA, Shallowater, TX). Sequences were depleted of barcodes and primers. Sequences shorter than 200 bp, sequences with ambiguous base calls, and sequences with homopolymer runs exceeding 6 bp were removed. Sequences were then de-noised and chimeras were also discarded. Operational taxonomic units (OTUs) were defined after removal of singleton sequences, clustering at 3% divergence (97% similarity).58,64 OTUs were then taxonomically classified using BLASTn against a curated Green Genes database65 and compiled into each taxonomic level into both “counts” and “percentage” files. Counts files contain the actual number of sequences while the percent files contain the relative (proportion) percentage of sequences within each sample that map to the designated taxonomic classification.
Statistics
Data analysis was performed using GraphPad Prism software (GraphPad Software, Inc., California). In data analyzed by un-paired two-tailed Student's t-test or Mann-Whitney test for non-parametric data; where shown, error bars depict standard error of the mean (SEM). Kaplan–Meier survival curves were analyzed by Log-rank (Mantel-Cox test). Where stated, tumor curves were analyzed by two-way ANOVA. p-value less than 0.05 were considered significant (*p < 0.05; **p < 0.01, ***p < 0.001).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Dr Chenthamarakshan Vasu for advice on analysis of pyrosequencing data of 16s rRNAs from the oral microbiota of mice. We are grateful to all members of our laboratories and the L-COHR (Center for Oral Health Research Laboratory Core) for their assistance with this work.
Funding
This work was supported by MUSC COHR Pilot Grant 5P20RR017696 to B.L; Hollings Cancer Center's Cancer Center Support Grant P30 CA138313 at the Medical University of South Carolina; NIH extramural grants P01CA186866, R01CA188419, R01AI070603; R01AI077283 to Z.L, and T-COHR 5T32DE017551 to K.K and C.W.F.
References
- 1.Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol 2009; 45:309-16; PMID:18804401; http://dx.doi.org/ 10.1016/j.oraloncology.2008.06.002 [DOI] [PubMed] [Google Scholar]
- 2.Chaturvedi AK, Anderson WF, Lortet-Tieulent J, Curado MP, Ferlay J, Franceschi S, Rosenberg PS, Bray F, Gillison ML. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol 2013; 31:4550-9; PMID:24248688; http://dx.doi.org/ 10.1200/JCO.2013.50.3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Society AC. Cancer Facts & Figures 2015. Atlanta: American Cancer Society, 2015 [Google Scholar]
- 4.Dionne KR, Warnakulasuriya S, Zain RB, Cheong SC. Potentially malignant disorders of the oral cavity: current practice and future directions in the clinic and laboratory. Int J Cancer 2015; 136:503-15; PMID:24482244; http://dx.doi.org/ 10.1002/ijc.28754 [DOI] [PubMed] [Google Scholar]
- 5.Fang X, Zheng P, Tang J, Liu Y. CD24: from A to Z. Cell Mol Immunol 2010; 7:100-3; PMID:20154703; http://dx.doi.org/ 10.1038/cmi.2009.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen GY, Chen X, King S, Cavassani KA, Cheng J, Zheng X, Cao H, Yu H, Qu J, Fang D et al.. Amelioration of sepsis by inhibiting sialidase-mediated disruption of the CD24-SiglecG interaction. Nat Biotechnol 2011; 29(5):428-35; PMID:21478876; http://dx.doi.org/19264983 10.1038/nbt.1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen GY, Tang J, Zheng P, Liu Y. CD24 and Siglec-10 selectively repress tissue damage-induced immune responses. Science 2009; 323:1722-5; PMID:19264983; http://dx.doi.org/ 10.1126/science.1168988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Zheng P. CD24: a genetic checkpoint in T cell homeostasis and autoimmune diseases. Trends Immunol 2007; 28:315-20; PMID:17531534; http://dx.doi.org/ 10.1016/j.it.2007.05.001 [DOI] [PubMed] [Google Scholar]
- 9.Li O, Zheng P, Liu Y. CD24 expression on T cells is required for optimal T cell proliferation in lymphopenic host. J Exp Med 2004; 200:1083-9; PMID:15477346; http://dx.doi.org/ 10.1084/jem.20040779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bai XF, Li O, Zhou Q, Zhang H, Joshi PS, Zheng X, Liu Y, Wang Y, Zheng P, Liu Y. CD24 controls expansion and persistence of autoreactive T cells in the central nervous system during experimental autoimmune encephalomyelitis. J Exp Med 2004; 200:447-58; PMID:15314074; http://dx.doi.org/ 10.1084/jem.20040131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen CY, Kimura H, Landek-Salgado MA, Hagedorn J, Kimura M, Suzuki K, Westra W, Rose NR, Caturegli P. Regenerative potentials of the murine thyroid in experimental autoimmune thyroiditis: role of CD24. Endocrinology 2009; 150:492-9; PMID:18801910; http://dx.doi.org/ 10.1210/en.2008-0639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thaxton JE, Liu B, Zheng P, Liu Y, Li Z. Deletion of CD24 impairs development of heat shock protein gp96-driven autoimmune disease through expansion of myeloid-derived suppressor cells. J Immunol (Baltimore, Md : 1950) 2014; 192:5679-86; PMID:24808359; http://dx.doi.org/ 10.4049/jimmunol.1302755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JH, Kim SH, Lee ES, Kim YS. CD24 overexpression in cancer development and progression: a meta-analysis. Oncol Rep 2009; 22:1149-56; PMID:19787233; http://dx.doi.org/ 10.3892/or_00000548 [DOI] [PubMed] [Google Scholar]
- 14.Overdevest JB, Thomas S, Kristiansen G, Hansel DE, Smith SC, Theodorescu D. CD24 offers a therapeutic target for control of bladder cancer metastasis based on a requirement for lung colonization. Cancer Res 2011; 71(11):3802-11; PMID:21482678; http://dx.doi.org/18752058 10.1158/0008-5472.CAN-11-0519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagy B, Szendroi A, Romics I. Overexpression of CD24, c-myc and phospholipase 2A in prostate cancer tissue samples obtained by needle biopsy. Pathol Oncol Res 2009; 15:279-83; PMID:18752058; http://dx.doi.org/ 10.1007/s12253-008-9077-1 [DOI] [PubMed] [Google Scholar]
- 16.Tanaka T, Terai Y, Kogata Y, Ashihara K, Maeda K, Fujiwara S, Yoo S, Tanaka Y, Tsunetoh S, Sasaki H et al.. CD24 expression as a marker for predicting clinical outcome and invasive activity in uterine cervical cancer. Oncol Rep 2015; 34(5):2282-8; PMID:26351781; http://dx.doi.org/26444008 10.3892/or.2015.4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon MJ, Han J, Seo JH, Song K, Jeong HM, Choi JS, Kim YJ, Lee SH, Choi YL, Shin YK. CD24 overexpression is associated with poor prognosis in luminal A and triple-negative breast cancer. PloS One 2015; 10:e0139112; PMID:26444008; http://dx.doi.org/ 10.1371/journal.pone.0139112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu JX, Zhao YY, Wu X, An HX. Clinicopathological and prognostic significance of CD24 overexpression in patients with gastric cancer: a meta-analysis. PloS One 2014; 9:e114746; PMID:25503963; http://dx.doi.org/ 10.1371/journal.pone.0114746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biddle A, Gammon L, Liang X, Costea DE, Mackenzie IC. Phenotypic plasticity determines cancer stem cell therapeutic resistance in oral squamous cell carcinoma. EBio Med 2016; 4:138-45; PMID:26981578; http://dx.doi.org/26926234 10.1016/j.ebiom.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghuwalewala S, Ghatak D, Das P, Dey S, Sarkar S, Alam N, Panda CK, Roychoudhury S. CD44(high)CD24(low) molecular signature determines the cancer stem cell and EMT phenotype in oral squamous cell carcinoma. Stem Cell Res 2016; 16:405-17; PMID:26926234; http://dx.doi.org/ 10.1016/j.scr.2016.02.028 [DOI] [PubMed] [Google Scholar]
- 21.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004; 6:1-6; PMID:15068665; http://dx.doi.org/ 10.1016/S1476-5586(04)80047-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sengupta S, den Boon JA, Chen IH, Newton MA, Dahl DB, Chen M, Cheng YJ, Westra WH, Chen CJ, Hildesheim A et al.. Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma. Cancer Res 2006; 66:7999-8006; PMID:16912175; http://dx.doi.org/ 10.1158/0008-5472.CAN-05-4399 [DOI] [PubMed] [Google Scholar]
- 23.Rickman DS, Millon R, De Reynies A, Thomas E, Wasylyk C, Muller D, Abecassis J, Wasylyk B. Prediction of future metastasis and molecular characterization of head and neck squamous-cell carcinoma based on transcriptome and genome analysis by microarrays. Oncogene 2008; 27:6607-22; PMID:18679425; http://dx.doi.org/ 10.1038/onc.2008.251 [DOI] [PubMed] [Google Scholar]
- 24.Hawkins BL, Heniford BW, Ackermann DM, Leonberger M, Martinez SA, Hendler FJ. 4NQO carcinogenesis: a mouse model of oral cavity squamous cell carcinoma. Head Neck 1994; 16:424-32; PMID:7960739; http://dx.doi.org/ 10.1002/hed.2880160506 [DOI] [PubMed] [Google Scholar]
- 25.Young MR. Use of carcinogen-induced premalignant oral lesions in a dendritic cell-based vaccine to stimulate immune reactivity against both premalignant oral lesions and oral cancer. J Immunother (Hagerstown, Md : 1997) 2008; 31:148-56; PMID:18481384; http://dx.doi.org/ 10.1097/CJI.0b013e31815bdbf5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu J, Peters BA, Dominianni C, Zhang Y, Pei Z, Yang L, Ma Y, Purdue MP, Jacobs EJ, Gapstur SM et al.. Cigarette smoking and the oral microbiome in a large study of American adults. ISME J 2016; PMID:27015003; http://dx.doi.org/15014984 10.1038/ismej.2016.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujiki H, Takeuchi H, Nishitani N, Yamanaka H, Suzuki K, Kurusu M, Suganuma M. Carcinogenic potential of tobacco tar-resistant Staphylococcus aureus in buccal cavity. J Cancer Res Clin Oncol 2004; 130:301-5; PMID:15014984; http://dx.doi.org/ 10.1007/s00432-004-0554-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morita E, Narikiyo M, Yokoyama A, Yano A, Kamoi K, Yoshikawa E, Yamaguchi T, Igaki H, Tachimori Y, Kato H et al.. Predominant presence of Streptococcus anginosus in the saliva of alcoholics. Oral Microbiol Immunol 2005; 20:362-5; PMID:16238596; http://dx.doi.org/ 10.1111/j.1399-302X.2005.00242.x [DOI] [PubMed] [Google Scholar]
- 29.Gill G, Rice D. Radiation effects on microbiological flora of patients with head and neck cancer. Surg Forum 1975; 26:533-4; PMID:1216217 [PubMed] [Google Scholar]
- 30.Rice DH, Gill G. Abnormal microorganisms and cell-mediated immunity in patients with intraoral cancer. Arch Otolaryngol 1976; 102:99-100; PMID:1247427; http://dx.doi.org/ 10.1001/archotol.1976.00780070077010 [DOI] [PubMed] [Google Scholar]
- 31.Buckley DA, Murphy A, Dervan P, Hone R, O'Dwyer T, O'Loughlin S. Persistent infection of the chin with an unusual skin pathogen (Streptococcus milleri): a sign of intraoral carcinoma. Clin Exp Dermatol 1998; 23:35-7; PMID:9667108; http://dx.doi.org/ 10.1046/j.1365-2230.1998.00305.x [DOI] [PubMed] [Google Scholar]
- 32.Ye P, Nadkarni MA, Simonian M, Hunter N. CD24 regulated gene expression and distribution of tight junction proteins is associated with altered barrier function in oral epithelial monolayers. BMC Cell Biol 2009; 10:2; PMID:19138432; http://dx.doi.org/ 10.1186/1471-2121-10-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu X, Zhang Q, Hua H, Chen F. Changes in the salivary microbiota of oral leukoplakia and oral cancer. Oral Oncol 2016; 56:e6-8; PMID:27026576; http://dx.doi.org/ 10.1016/j.oraloncology.2016.03.007 [DOI] [PubMed] [Google Scholar]
- 34.Vasconcelos RM, Sanfilippo N, Paster BJ, Kerr AR, Li Y, Ramalho L, Queiroz EL, Smith B, Sonis ST, Corby PM. Host-Microbiome Cross-talk in Oral Mucositis. J Dental Res 2016; 95(7):725-33; PMID:27053118; http://dx.doi.org/ 10.1177/0022034516641890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang M, Wang F, Jiang L, Liu R, Zhang L, Lei X, Li J, Jiang J, Guo H, Fang B et al.. Lactobacillus salivarius REN inhibits rat oral cancer induced by 4-nitroquioline 1-oxide. Cancer Prevention Res (Philadelphia, Pa) 2013; 6:686-94; PMID:23658366; http://dx.doi.org/ 10.1158/1940-6207.CAPR-12-0427 [DOI] [PubMed] [Google Scholar]
- 36.Yang CH, Wang HL, Lin YS, Kumar KP, Lin HC, Chang CJ, Lu CC, Huang TT, Martel J, Ojcius DM et al.. Identification of CD24 as a cancer stem cell marker in human nasopharyngeal carcinoma. PloS One 2014; 9:e99412; PMID:24955581; http://dx.doi.org/ 10.1371/journal.pone.0099412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Motallebnezhad M, Jadidi-Niaragh F, Qamsari ES, Bagheri S, Gharibi T, Yousefi M. The immunobiology of myeloid-derived suppressor cells in cancer. Tumour Biol 2015; 37(2):1387–406; PMID:26611648; http://dx.doi.org/ 10.1007/s13277-015-4477-9 [DOI] [PubMed] [Google Scholar]
- 38.Qin H, Wei G, Gwak D, Dong Z, Xiong A, Kwak LW. Targeting tumor-associated myeloid cells for cancer immunotherapy. Oncoimmunology 2015; 4:e983961; PMID:25949898; http://dx.doi.org/ 10.4161/2162402X.2014.983761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wenger RH, Kopf M, Nitschke L, Lamers MC, Kohler G, Nielsen PJ. B-cell maturation in chimaeric mice deficient for the heat stable antigen (HSA/mouse CD24). Transgenic Res 1995; 4:173-83; PMID:7795661; http://dx.doi.org/ 10.1007/BF01968782 [DOI] [PubMed] [Google Scholar]
- 40.Kusmartsev S, Nefedova Y, Yoder D, Gabrilovich DI. Antigen-specific inhibition of CD8+ T cell response by immature myeloid cells in cancer is mediated by reactive oxygen species. J Immunol 2004; 172:989-99; PMID:14707072; http://dx.doi.org/19197294 10.4049/jimmunol.172.2.989 [DOI] [PubMed] [Google Scholar]
- 41.Sinha P, Clements VK, Ostrand-Rosenberg S. Reduction of myeloid-derived suppressor cells and induction of M1 macrophages facilitate the rejection of established metastatic disease. J Immunol 2005; 174:636-45; PMID:15634881; http://dx.doi.org/19197294 10.4049/jimmunol.174.2.636 [DOI] [PubMed] [Google Scholar]
- 42.Nagaraj D, Gabrilovich DI. Myeloid-derived-suppressor cells as regulators of the immune system. Nat Rev Immunol 2009; 9:162-74; PMID:19197294; http://dx.doi.org/ 10.1038/nri2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol 2012; 12:253-68; PMID:22437938; http://dx.doi.org/ 10.1038/nri3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646-74; PMID:21376230; http://dx.doi.org/ 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 45.White RA, Neiman JM, Reddi A, Han G, Birlea S, Mitra D, Dionne L, Fernandez P, Murao K, Bian L et al.. Epithelial stem cell mutations that promote squamous cell carcinoma metastasis. J Clin Invest 2013; 123:4390-404; PMID:23999427; http://dx.doi.org/ 10.1172/JCI65856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma C, Kapanadze T, Gamrekelashvili J, Manns MP, Korangy F, Greten TF. Anti-Gr-1 antibody depletion fails to eliminate hepatic myeloid-derived suppressor cells in tumor-bearing mice. J Leukocyte Biol 2012; 92:1199-206; PMID:23077247; http://dx.doi.org/ 10.1189/jlb.0212059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang XH, Knudsen B, Bemis D, Tickoo S, Gudas LJ. Oral cavity and esophageal carcinogenesis modeled in carcinogen-treated mice. Clin Cancer Res 2004; 10:301-13; PMID:14734483; http://dx.doi.org/ 10.1158/1078-0432.CCR-0999-3 [DOI] [PubMed] [Google Scholar]
- 48.Kanojia D, Vaidya MM. 4-nitroquinoline-1-oxide induced experimental oral carcinogenesis. Oral Oncology 2006; 42:655-67; PMID:16448841; http://dx.doi.org/ 10.1016/j.oraloncology.2005.10.013 [DOI] [PubMed] [Google Scholar]
- 49.Saleem S, Jamshed A, Faisal S, Hussain R, Tahseen M, Loya A, Sutton C. Patterns of cancer cell sphere formation in primary cultures of human oral tongue squamous cell carcinoma and neck nodes. Cancer Cell Int 2014; 14:542; PMID:25685059; http://dx.doi.org/ 10.1186/s12935-014-0143-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sagiv E, Starr A, Rozovski U, Khosravi R, Altevogt P, Wang T, Arber N. Targeting CD24 for treatment of colorectal and pancreatic cancer by monoclonal antibodies or small interfering RNA. Cancer Res 2008; 68:2803-12; PMID:18413748; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-6463 [DOI] [PubMed] [Google Scholar]
- 51.Lim KP, Chun NA, Ismail SM, Abraham MT, Yusoff MN, Zain RB, Ngeow WC, Ponniah S, Cheong SC. CD4+CD25hiCD127low Regulatory T cells are increased in oral squamous cell carcinoma patients. PloS One 2014; 9:e103975; PMID:25153698; http://dx.doi.org/ 10.1371/journal.pone.0103975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao J, Wang Z, Han J, Qiu X, Pan J, Chen J. Increased frequency of CD4+ CD25+ FOXP3+ cells correlates with the progression of 4-nitroquinoline1-oxide-induced rat tongue carcinogenesis. Clin Oral Invest 2014; 18:1725-30; PMID:24264641; http://dx.doi.org/ 10.1007/s00784-013-1146-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weed DT, Vella JL, Reis IM, De la Fuente AC, Gomez C, Sargi Z, Nazarian R, Califano J, Borrello I, Serafini P. Tadalafil reduces myeloid-derived suppressor cells and regulatory T cells and promotes tumor immunity in patients with head and neck squamous cell carcinoma. Clin Cancer Res 2015; 21:39-48; PMID:25320361; http://dx.doi.org/ 10.1158/1078-0432.CCR-14-1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chu M, Su YX, Wang L, Zhang TH, Liang YJ, Liang LZ, Liao GQ. Myeloid-derived suppressor cells contribute to oral cancer progression in 4NQO-treated mice. Oral Dis 2012; 18:67-73; PMID:21883708; http://dx.doi.org/ 10.1111/j.1601-0825.2011.01846.x [DOI] [PubMed] [Google Scholar]
- 55.Parker KH, Sinha P, Horn LA, Clements VK, Yang H, Li J, Tracey KJ, Ostrand-Rosenberg S. HMGB1 enhances immune suppression by facilitating the differentiation and suppressive activity of myeloid-derived suppressor cells. Cancer Res 2014; 74:5723-33; PMID:25164013; http://dx.doi.org/ 10.1158/0008-5472.CAN-13-2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cui TX, Kryczek I, Zhao L, Zhao E, Kuick R, Roh MH, Vatan L, Szeliga W, Mao Y, Thomas DG et al.. Myeloid-derived suppressor cells enhance stemness of cancer cells by inducing microRNA101 and suppressing the corepressor CtBP2. Immunity 2013; 39:611-21; PMID:24012420; http://dx.doi.org/ 10.1016/j.immuni.2013.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Condamine T, Gabrilovich DI. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol 2011; 32:19-25; PMID:21067974; http://dx.doi.org/ 10.1016/j.it.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, Edrington TS. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol 2008; 8:125; PMID:18652685; http://dx.doi.org/ 10.1186/1471-2180-8-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010; 26:2460-1; PMID:20709691; http://dx.doi.org/ 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 60.Dowd SE, Delton Hanson J, Rees E, Wolcott RD, Zischau AM, Sun Y, White J, Smith DM, Kennedy J, Jones CE. Survey of fungi and yeast in polymicrobial infections in chronic wounds. J Wound Care 2011; 20:40-7; PMID:21278640; http://dx.doi.org/ 10.12968/jowc.2011.20.1.40 [DOI] [PubMed] [Google Scholar]
- 61.Dowd SE, Sun Y, Wolcott RD, Domingo A, Carroll JA. Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned Salmonella-infected pigs. Foodborne Pathogens Dis 2008; 5:459-72; PMID:18713063; http://dx.doi.org/ 10.1089/fpd.2008.0107 [DOI] [PubMed] [Google Scholar]
- 62.Eren AM, Zozaya M, Taylor CM, Dowd SE, Martin DH, Ferris MJ. Exploring the diversity of Gardnerella vaginalis in the genitourinary tract microbiota of monogamous couples through subtle nucleotide variation. PloS One 2011; 6:e26732; PMID:22046340; http://dx.doi.org/ 10.1371/journal.pone.0026732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Capone KA, Dowd SE, Stamatas GN, Nikolovski J. Diversity of the human skin microbiome early in life. J Invest Dermatol 2011; 131:2026-32; PMID:21697884; http://dx.doi.org/ 10.1038/jid.2011.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Swanson KS, Dowd SE, Suchodolski JS, Middelbos IS, Vester BM, Barry KA, Nelson KE, Torralba M, Henrissat B, Coutinho PM et al.. Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME J 2011; 5:639-49; PMID:20962874; http://dx.doi.org/ 10.1038/ismej.2010.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied Environmental Microbiol 2006; 72:5069-72; PMID:16820507; http://dx.doi.org/ 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]