Abstract
Purpose
We investigated whether mutations in the gene encoding the phosphatidylinositol 3-kinase (PI3K) catalytic subunit (PIK3CA) correlates with response to neoadjuvant human epidermal growth factor receptor 2 (HER2) –targeted therapies in patients with breast cancer.
Patients and Methods
Baseline tissue biopsies were available from patients with HER2-positive early breast cancer who were enrolled onto the Neoadjuvant Lapatinib and/or Trastuzumab Treatment Optimization trial (NeoALTTO). Activating mutations in PIK3CA were identified using mass spectrometry–based genotyping.
Results
PIK3CA mutations were identified in 23% of HER2-positive breast tumors, and these mutations were associated with poorer outcome in all of the treatment arms. Patients treated with a combination of trastuzumab and lapatinib who had wild-type PIK3CA obtained a total pathologic complete response (pCR) rate of 53.1%, which decreased to 28.6% in patients with tumors that carried PIK3CA activating mutations (P = .012).
Conclusion
Activating mutations in PIK3CA predicted poor pCR in patients with HER2-positive breast cancer treated with neoadjuvant therapies that target HER2. Consequently, the combination of anti-HER2 agents and PI3K inhibitors is being investigated.
INTRODUCTION
Human epidermal growth factor receptor 2 (HER2) –positive breast cancer is defined by overexpression and/or amplification of the HER2 gene (also known as ERBB2).1,2 HER2 is a receptor tyrosine kinase that does not bind ligand directly, but instead mediates signaling in concert with other epidermal growth factor receptor family members (heterodimers) or by forming homodimers. Blocking signaling through the receptor complex, either with monoclonal antibodies that bind the receptor at the cell surface3–5 or with small-molecule drugs that inhibit kinase activity directly,6,7 is associated with significant clinical benefit, and these agents have become standard of care for treating patients with HER2-positive breast cancer.
The phosphatidylinositol 3-kinase (PI3K) pathway sits at the nexus of cellular growth and metabolic signaling downstream for HER2 and is frequently aberrantly activated in cancer.8 The gene encoding the catalytic component p110α, PIK3CA, is one of the most frequently mutated genes in breast cancer,9 emphasizing the importance of this pathway for regulating growth in breast malignancies. Although PIK3CA activating mutations are present in all breast cancer subtypes, they are enriched in estrogen receptor (ER) –positive and HER2-positive disease and are less common in basal cancers. Although the role of this pathway in driving cellular transformation is well defined, knowledge regarding the influence of PI3K on the biology of disease and its role in governing therapy response in patients remains limited.
The PI3K pathway is a critical mediator of signaling downstream of receptor tyrosine kinases and has been implicated in mediating resistance to therapies that target HER2. Activating mutations in PIK3CA and loss of PTEN, which is a key negative regulator of PI3K signaling, drive resistance to both trastuzumab and lapatinib in breast cancer cell lines, and low levels of PTEN have been associated with worse outcome in patients.10–12
In this study, we assessed the correlation of PIK3CA mutation status with therapy response measured by pathologic complete response (pCR), event-free survival (EFS), and overall survival (OS) in 355 patients with HER2-positive breast cancer who had been recruited to the Neoadjuvant Lapatinib and/or Trastuzumab Treatment Optimization (NeoALTTO) trial (Breast International Group 1-06), a randomized, multicenter, open-label, neoadjuvant, phase III trial designed to assess the efficacy of dual inhibition of HER2.13
PATIENTS AND METHODS
Patient Population and Samples
NeoALTTO, a phase III parallel-group, open-label, randomized, neoadjuvant study of trastuzumab, lapatinib, or their combination, included patients with newly diagnosed HER2-positive invasive breast cancer amenable to surgery. Full eligibility criteria can be accessed elsewhere.13 Patients received anti-HER2 therapy for 6 weeks, and paclitaxel was then added to the regimen for a further 12-week period until definitive surgery, for a total period of 18 weeks of anti-HER2 therapy. The study primary end point was the rate of pCR in the breast. Secondary end points included rate of locoregional total pCR, EFS, and OS. Core biopsies were obtained before treatment (baseline), at 2 weeks on treatment, and at the time of surgery. Here, we report the results on baseline biopsies. Recently, EFS and OS were reported at a median clinical follow-up of 3.8 years.14
Laboratory Methods (sample processing and genotyping)
Baseline frozen biopsies were embedded in frozen tissue matrix (OCT; Sakura Finetek, Torrance, CA) and cut at the cryostat for tumor cellularity assessment by a pathologist. Genomic DNA was isolated from 20 × 10 μm sections using the DNeasy Kit (Qiagen, Hilden, Germany). The Sequenom (San Diego, CA) mass-spectrometry genotyping system was used to test for hotspot mutations in PIK3CA (E542A/K, E545A/K, and H1047R/L), AKT1 (E17K), KRAS (G12A/C/D/V and G13D), and BRAF (V600E). All polymerase chain reaction (PCR) primers were designed using the Sequenom online assay design system and are listed in Appendix Table A1 (online only). PCR reactions were prepared with the iPLEX-Gold PCR reaction mix (Sequenom) with 15 ng of input DNA. The PCR, shrimp alkaline phosphatase treatment, and extension reaction were performed according to the manufacturer's protocol (Sequenom). The results were analyzed using MassArrayTyper v 4.0.22.67 (Sequenom) using both automated analysis and visual inspection. Confirmation of kinase mutations was performed by next-generation sequencing on an Illumina Hiseq2000 (Illumina, San Diego, CA) with a paired-end 51 base protocol as described.15 DNA enrichment was performed with the human kinome DNA capture baits (Agilent, Santa Clara, CA).
Statistical Analysis
The differences in pCR rates by PIK3CA mutation status were calculated with a corresponding 95% CI and tested using a χ2 test of association. A logistic regression model was fitted to pCR and included ER status and treatment arm, as well as PIK3CA mutation status. Another model was fitted adding an interaction term for PIK3CA and treatment arm to assess whether the proportions of patients with pCR who have the PIK3CA mutation are statistically different between the treatment arms. As a hypothesis-generating, exploratory analysis, the treatment effect was further examined by performing χ2 tests of association on pCR and PIK3CA mutation status for each treatment arm separately.
The number and percentage of EFS and OS events by PIK3CA mutation status were calculated. Cox regression models were fitted to EFS and OS with treatment arm, hormone receptor status, and PIK3CA mutation status. Interaction tests were performed for treatment arm and PIK3CA mutation status.
RESULTS
Study Design
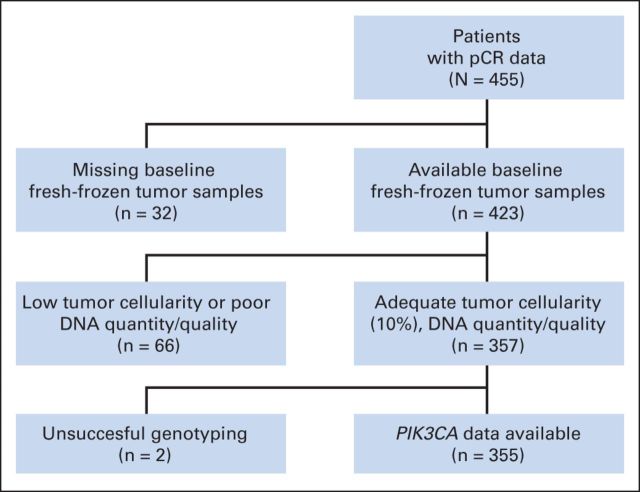
To study the effect of PIK3CA mutations on the response to HER2-targeted therapies in breast cancer, we used tumor biopsies from the NeoALTTO trial.13 pCR data were available for 455 patients with HER2-positive breast cancer enrolled onto the NeoALTTO trial, of whom 423 (93%) had baseline fresh-frozen tumor samples; a total of 355 samples (78%) underwent successful PIK3CA genotyping (Fig 1).
Fig 1.
Tumor sample disposition for analysis of PIK3CA mutation. pCR, pathologic complete response.
Somatic Mutation Profiling
For assessment of the impact of PIK3CA mutation on treatment outcome, we analyzed DNA from baseline samples. Samples with less than 10% tumor material or that produced low DNA yield were excluded from the analysis, which left a total of 357 samples (78.5%) for genotyping (Fig 1). Genotypes for hotspot mutations in PIK3CA, AKT1, KRAS, and BRAF were obtained for 355 samples (78%; Fig 1). As shown in Table 1, this subset of patients is representative of the whole NeoALTTO population.
Table 1.
Patient Clinical Characteristics
| Characteristic | NeoALTTO (N = 455) |
NeoALTTO PIK3CA Subset (n = 355) |
NeoALTTO Patients Not in PIK3CA Subset (n = 100) |
|||
|---|---|---|---|---|---|---|
| No. of Patients | % | No. of Patients | % | No. of Patients | % | |
| ER status | ||||||
| Negative | 239 | 52.5 | 186 | 52.4 | 53 | 53.0 |
| Positive | 216 | 47.6 | 169 | 47.6 | 47 | 47.0 |
| pCR | ||||||
| Yes | 138 | 31.4 | 108 | 31.6 | 30 | 30.6 |
| No | 302 | 68.6 | 234 | 68.4 | 68 | 69.4 |
| Missing | 15 | 13 | 2 | |||
| Treatment | ||||||
| Lapatinib | 154 | 33.8 | 124 | 34.9 | 30 | 30.0 |
| Trastuzumab | 149 | 32.7 | 112 | 31.6 | 37 | 37.0 |
| Lapatinib + trastuzumab | 152 | 33.4 | 119 | 33.5 | 33 | 33.0 |
| Grade | ||||||
| 1 | 12 | 2.6 | 8 | 2.2 | 4 | 4.0 |
| 2 | 172 | 37.8 | 127 | 35.8 | 45 | 45.0 |
| 3 | 205 | 45.1 | 170 | 47.9 | 35 | 35.0 |
| X | 65 | 14.3 | 49 | 13.8 | 16 | 16.0 |
| Unknown | 1 | 0.2 | 1 | 0.3 | 0 | 0.0 |
| Tumor size, cm | ||||||
| ≤ 5 | 274 | 60.2 | 212 | 59.7 | 62 | 62.0 |
| > 5 | 181 | 39.8 | 143 | 40.3 | 38 | 38.0 |
| Age, years | ||||||
| < 65 | 403 | 88.6 | 321 | 90.4 | 82 | 82.0 |
| ≥ 65 | 52 | 11.4 | 34 | 9.6 | 18 | 18.0 |
| N stage | ||||||
| N0 | 123 | 27.0 | 94 | 26.5 | 29 | 29.0 |
| N1 | 260 | 57.1 | 196 | 55.2 | 64 | 64.0 |
| N2a | 44 | 9.7 | 40 | 11.3 | 4 | 4.0 |
| N3a | 3 | 0.7 | 2 | 0.6 | 1 | 1.0 |
| N3b | 7 | 1.5 | 7 | 2.0 | 0 | 0.0 |
| N3c | 12 | 2.6 | 11 | 3.1 | 1 | 1.0 |
| Nx | 6 | 1.3 | 5 | 1.4 | 1 | 1.0 |
Abbreviations: ER, estrogen receptor; NeoALTTO, Neoadjuvant Lapatinib and/or Trastuzumab Treatment Optimization; pCR, pathologic complete response.
No activating mutations were detected in AKT1 or BRAF, and only a single activating mutation in KRAS (0.3%) was identified in the entire cohort. PIK3CA mutations were detected in 80 samples (23%), which included nine in codon 542 (2.5%), 13 in codon 545 (3.7%), and 58 in codon 1047 (16.3%). The overall proportion of helical and kinase domain mutations was consistent with those observed in breast cancers in the Catalogue of Somatic Mutations in Cancer (Appendix Table A2, online only). The PIK3CA mutation frequency was similar in each treatment arm (23% in lapatinib arm, n = 124; 19% in trastuzumab arm, n = 112; and 25% in combination arm, n = 119) and was not influenced by ER status (23% in ER-positive patients, n = 169; and 22% in ER-negative patients, n = 186; Appendix Table A3, online only).
To verify the Sequenom genotyping result, PIK3CA mutation status was assessed in a random selection of 96 samples using next-generation sequencing with capture of the relevant exons of the genes studied here. The presence of the PIK3CA mutation was confirmed in 56 (96.6%) of 58 samples. The signal intensity observed in the Sequenom assay was strongly correlated with the mutation frequency observed by sequencing (r = 0.944).
PIK3CA Mutation Status and pCR
We analyzed the correlation between tumor PIK3CA mutation status and locoregional total pCR, as defined by no invasive cancer in the breast and no pathologic involvement of axillary nodes. Patients with PIK3CA mutations were less likely to have a pCR. The rate of pCR decreased from 34.5% (92 of 267 patients) in wild-type patients to 21.3% (16 of 75 patients) in patients who carried PIK3CA mutations (Fig 2A). The difference was 13.1% (95% CI, 2.2% to 24.0%; P = .03 for the χ2 test of association). Likewise, there was also a lower breast pCR, defined as absence of invasive tumor cells in the breast, in tumors with PIK3CA mutations (Fig 2A). Patients with PIK3CA mutations had a lower pCR rate independent of ER status (Fig 2B, Appendix Table A4, online only).
Fig 2.
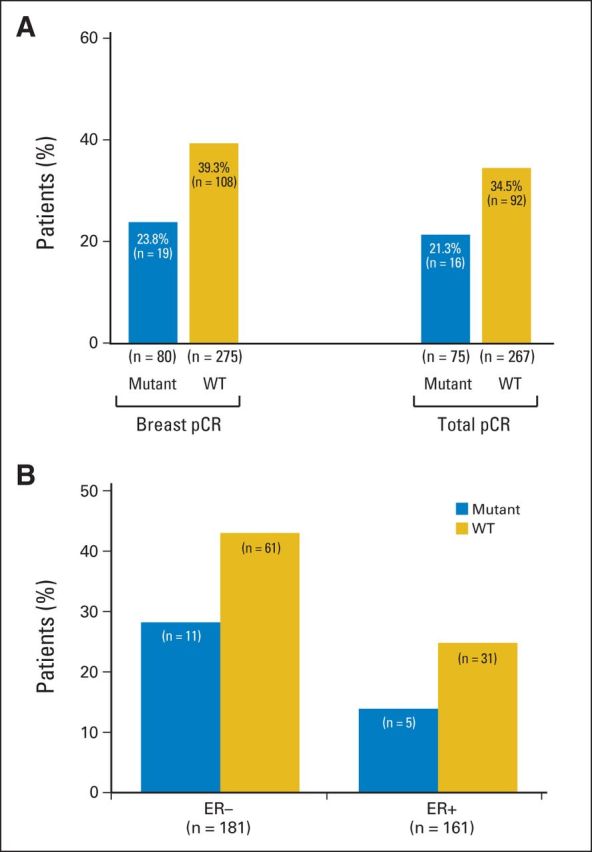
Influence of PIK3CA mutation and estrogen receptor (ER) status on pathologic complete response (pCR). (A) The proportion of patients who obtained breast and locoregional total pCR is shown for the entire patient cohort (gold, PIK3CA wild type [WT]; blue, PIK3CA mutant). (B) Total pCR rates for ER-negative and ER-positive tumors separately (gold, PIK3CA WT; blue, PIK3CA mutant). Patients were further subdivided based on ER status (negative or positive). In each of the bar graphs, the number of patients is being displayed. The total number of patients with available pCR data in each group is shown.
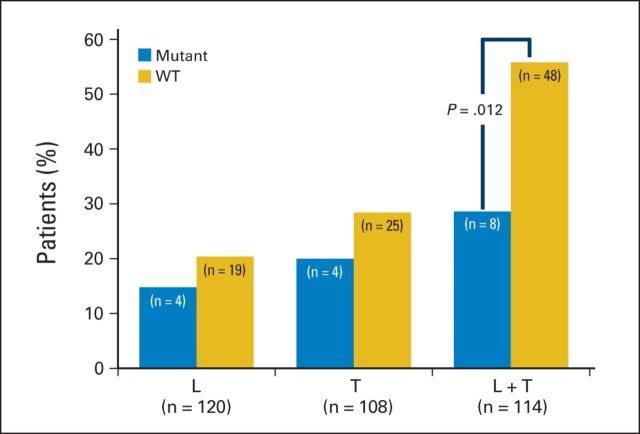
For each treatment arm, the pCR rate was lower for patients with PIK3CA mutations than for wild-type patients, although this difference was largest in the combination (lapatinib plus trastuzumab) treatment arm (Fig 3, Appendix Table A5, online only). The correlation between breast pCR and total pR is shown in Appendix Table A6 (online only). The logistic regression model is summarized in Table 2, showing that PIK3CA status, ER status, and treatment arm are all statistically significant. The interaction test for treatment arm and PIK3CA was not statistically significant (P = .5), but individual tests by treatment arm found significant differences in the combination arm (P = .012), but not for lapatinib only (P = .51) or trastuzumab only (P = .44). However, the test for interaction to demonstrate a significant value of the mutant to predict a better response to dual inhibition compared with the response to the single agents is underpowered.
Fig 3.
Influence of PIK3CA mutation on pathologic complete response (pCR) by treatment arm. The proportion of patients who obtained total pCR is shown for each treatment arm, separated based on the PIK3CA mutation status. The total number of patients in each group is shown (gold, PIK3CA wild type [WT]; blue, PIK3CA mutant). In each of the bar graphs the number of patients is being displayed. The total number of patients with available pCR data in each group is shown. L, lapatinib; T, trastuzumab.
Table 2.
Logistic Regression Model Results for pCR
| Variable | Odds Ratio | 95% CI | P |
|---|---|---|---|
| PIK3CA (mutant v WT) | 0.45 | 0.23 to 0.85 | .015 |
| ER status (negative v positive) | 2.44 | 1.45 to 4.03 | < .001 |
| Treatment arm | < .001 | ||
| L versus T | 0.67 | 0.36 to 1.27 | |
| L + T versus T | 2.97 | 1.66 to 5.33 |
Abbreviations: ER, estrogen receptor; L, lapatinib; pCR, pathologic complete response; T, trastuzumab; WT, wild type.
PIK3CA Mutation Status and EFS and OS
Using a simplified summary of EFS, we note that 15 patients (18.7%) with PIK3CA mutations and 64 patients (23.3%) with normal PIK3CA had EFS events. The Cox model of EFS, including treatment arm, ER status, and PIK3CA status, found that EFS does not seem to be affected by PIK3CA status (hazard ratio [HR], 0.78; 95% CI, 0.44 to 1.36; P = .38). A model testing the interaction term was nonsignificant (P = .27).
The analysis of OS events found that five patients (6.2%) with PIK3CA mutations had died compared with 35 patients (12.7%) with normal PIK3CA. As with EFS, the Cox model for OS found that OS does not seem to be affected by PIK3CA status (HR, 0.50; 95% CI, 0.19 to 1.23; P = .14). A model fitted to test a possible interaction between treatment arm and PIK3CA status was nonsignificant (P = .97).
DISCUSSION
In HER2-positive breast cancer, a recent meta-analysis of published neoadjuvant studies has shown a relationship between pCR and long-term outcome regardless of hormone receptor status (EFS: HR, 0.39; OS: HR, 0.34), with a stronger association in the hormone receptor–negative subgroup (EFS: HR, 0.25; OS: HR, 0.19).16 Therefore, neoadjuvant studies have been proposed to be suited to test biologic hypotheses in early HER-positive breast cancer.
In this study, we find that mutations in PIK3CA downstream of HER2 correlate with a lower response to HER2-targeted neoadjuvant therapy in breast cancer as measured by pCR. There is no significant difference for PIK3CA mutation status in survival follow-up (EFS and OS).
The finding that activating mutations in components of a signaling pathway downstream of a receptor tyrosine kinase are associated with reduced response to inhibitors of the receptor is not without precedent. Indeed, in colon cancer, the effectiveness of anti–epidermal growth factor receptor antibody drugs is limited by activating mutations in receptor downstream RAS genes.17,18 In the case of the closely related HER2 receptor, in vitro studies with HER2-targeted drugs in HER2-positive breast cancer have already provided preclinical evidence for a role of PI3K mutations in mediating resistance to HER2 therapy.10,12,19 Our present data may provide clinical support for these preclinical findings by suggesting that in HER2-positive tumors, the presence of PIK3CA mutations results in lower clinical benefit from trastuzumab, lapatinib, and especially dual HER2 blockade, as measured by pCR. Our findings are supported by similar reports from clinical trials in the advanced disease setting. In the phase III CLEOPATRA trial, the effects of the anti-HER2 monoclonal antibody pertuzumab, which prevents the formation of HER2-HER3 heterodimers, were studied when added to trastuzumab as first-line treatment for patients with HER2-positive metastatic breast cancer. This study compared the efficacy of pertuzumab, trastuzumab, and docetaxel versus placebo, trastuzumab, and docetaxel. As in NeoALTTO, this study demonstrated the superiority of dual HER2 blockade. The longest progression-free survival (PFS) was observed in patients whose tumors expressed wild-type versus mutated PIK3CA in both the placebo, trastuzumab, and docetaxel group (13.8 v 8.6 months, respectively) and pertuzumab, trastuzumab, and docetaxel group (21.8 v 12.5 months, respectively).20 Overall, patients with tumors with wild-type PIK3CA had a better outcome to therapy than those with PIK3CA mutations (HR, 0.63; P < .001).20
In a recently reported analysis of the neoadjuvant GeparSixto study of chemotherapy and dual HER2 blockade with trastuzumab and lapatinib, patients with PIK3CA-mutant tumors also had a significantly lower pCR rate compared with the wild-type population (17.8% v 36.8%, respectively; P = .015).21 Similarly, in the EMILIA trial, a phase III study that compared lapatinib and capecitabine versus the antibody-drug conjugate T-DM1, patients with tumors harboring a PIK3CA mutation had a shorter PFS with lapatinib when compared with patients with wild-type tumors (4.3 v 6.4 months, respectively). Interestingly, for patients treated with T-DM1, an antibody-drug conjugate that contains trastuzumab attached to a potent chemotherapeutic agent, PFS was unaffected by PIK3CA mutation status.22 The only published report that failed to find a correlation between PIK3CA status and response to therapy with trastuzumab was the adjuvant FinHER study, in which HER2-positive patients were randomly assigned to receive 9 weeks of trastuzumab or no trastuzumab.23 However, the study was small, with only a total of 157 patients with HER2-positive disease with available tumor samples and 34 patients (19.3%) with PIK3CA mutations. Furthermore they reported on distant disease-free survival despite only 9 weeks therapy with trastuzumab.
Although the number of events was too small to reach statistical significance, we did not observe worse EFS and OS in PIK3CA-mutant tumors. In this regard, it may be complex to dissect out the potential effect of HER2 therapy in PIK3CA-mutant tumors from the prognostic implications of PI3KCA mutations in breast cancer in the absence of a control arm without HER2 therapy. Indeed, PIK3CA mutations have been associated with improved outcome, including OS and breast cancer–specific survival.24,25 Although there are limited data, PIK3CA mutations may also confer an improved prognosis in HER2-positive tumors. For example, out of the 760 breast cancer samples from the Cancer Genome Atlas, 104 tumors (14%) had genomic amplification of HER2. In this subset of 104 HER2-positive tumors, 26 tumors (25%) had a known activating mutation in PIK3CA. The OS of the PIK3CA-mutant tumors was better than that of the PIK3CA wild-type tumors, although the data were not statistically significant (log-rank P = .1561).26
In HER2-amplified and PIK3CA-mutant breast cancer models, the observed resistance against HER2 therapies is reverted in full by the addition of PI3K inhibitors.10,12 Building from these observations, a number of clinical trials combining anti-HER2 agents with pan-PI3K inhibitors are under way, and clinical activity has already been reported in patients with trastuzumab-resistant disease.27,28 More recently, a new class of PI3K inhibitors that have a selective effect on the p110α isoform of PI3K, the product of PIK3CA, has entered the clinic. These PI3Kα-specific inhibitors are more potent and have been shown to be clinically active in PIK3CA-mutant breast tumors.29,30 Given the strong scientific rationale and the emerging clinical observations that PIK3CA-mutant tumors respond less to HER2 therapies, a strong case can be made to study these combinations in the clinic.
Acknowledgment
The Neoadjuvant Lapatinib and/or Trastuzumab Treatment Optimization trial was sponsored by GlaxoSmithKline and conducted in collaboration with the Breast International Group and by the Spanish Breast Cancer Cooperative Group SOLTI. The BrEAST Data Center was responsible for data management, and Frontier Science was responsible for statistical analysis.
Appendix
Table A1.
Sequenom Genotyping Primer Sequences
| Oligo Sequence (5′ to 3′) | Oligo Name | Identifier | Reaction |
|---|---|---|---|
| ACGTTGGATGTCTTCATGAAGACCTCACAG | BRAF-V600E-F | rs113488022 | W-1 |
| ACGTTGGATGTTCAAACTGATGGGACCCAC | BRAF-V600E-R | W-1 | |
| CCCACTCCATCGAGATTTC | BRAF-V600E-D | ||
| ACGTTGGATGTAGAGTGTGCGTGGCTCTCA | AKT1-E17K-F | rs121434592 | W-1 |
| ACGTTGGATGTTCTTGAGGAGGAAGTAGCG | AKT1-E17K-R | W-1 | |
| CGCCAGGTCTTGATGTACT | AKT1-E17K-D | ||
| ACGTTGGATGTCCATTTTTGTTGTCCAGCC | PIK3CA-H1047R/L-F | rs121913279 | W-1 |
| ACGTTGGATGAACTGAGCAAGAGGCTTTGG | PIK3CA-H1047R/L-R | W-1 | |
| GAAACAAATGAATGATGCAC | PIK3CA-H1047R/L-D | ||
| ACGTTGGATGAGGCCTGCTGAAAATGACTG | KRAS-G13D-F | rs112445441 | W-1 |
| ACGTTGGATGTAGCTGTATCGTCAAGGCAC | KRAS-G13D-R | W-1 | |
| GTCAAGGCACTCTTGCCTACG | KRAS-G13D-D | ||
| ACGTTGGATGTACACGAGATCCTCTCTCTG | PIK3CA-E545A-F | rs121913274 | W-1 |
| ACGTTGGATGTAGCACTTACCTGTGACTCC | PIK3CA-E545A-R | W-1 | |
| TCCATAGAAAATCTTTCTCCTGC | PIK3CA-E545A-D | ||
| ACGTTGGATGAGGCCTGCTGAAAATGACTG | KRAS-G12C-F | rs121913530 | W-2 |
| ACGTTGGATGTAGCTGTATCGTCAAGGCAC | KRAS-G12C-R | W-2 | |
| ACTCTTGCCTACGCCAC | KRAS-G12C-D | ||
| ACGTTGGATGTAGCACTTACCTGTGACTCC | PIK3CA-E542A-F | PIK3CA E542A | W-2 |
| ACGTTGGATGGCAATTTCTACACGAGATCC | PIK3CA-E542A-R | W-2 | |
| ACGAGATCCTCTCTCTG | PIK3CA-E542A-D | ||
| ACGTTGGATGTAGCACTTACCTGTGACTCC | PIK3CA-E545K-F | rs104886003 | W-3 |
| ACGTTGGATGTACACGAGATCCTCTCTCTG | PIK3CA-E545K-R | W-3 | |
| CCTCTCTCTGAAATCACT | PIK3CA-E545K-D | ||
| ACGTTGGATGTAGCTGTATCGTCAAGGCAC | KRAS-G12*-F | rs121913529 | W-3 |
| ACGTTGGATGAGGCCTGCTGAAAATGACTG | KRAS-G12*-R | W-3 | |
| CTTGTGGTAGTTGGAGCTG | KRAS-G12*-D | ||
| ACGTTGGATGGCAATTTCTACACGAGATCC | PIK3CA-E542K-F | rs121913273 | W-4 |
| ACGTTGGATGTAGCACTTACCTGTGACTCC | PIK3CA-E542K-R | ||
| CTCCTGCTCAGTGATTTC | PIK3CA-E542K-D |
NOTE. Sequences are included for the forward, reverse, and detection primers. Genotyping was performed in four separate multiplexed reactions (W-1 to W-4).
Abbreviations: D, detection; F, forward; R, reverse.
Assay measures multiple amino acid changes at that position.
Table A2.
PIK3CA Genotyping Results Compared With COSMIC Data Set
| Site | NeoALTTO |
COSMIC |
||
|---|---|---|---|---|
| No. of Cancers | % | No. of Cancers | % | |
| PIK3CA E542 | 9 | 2.5 | 227 | 2.5 |
| PIK3CA E545 | 13 | 3.7 | 437 | 4.8 |
| PIK3CA H1047 | 58 | 16.3 | 1,423 | 16.3 |
Abbreviations: COSMIC, Catalogue of Somatic Mutations in Cancer; NeoALTTO, Neoadjuvant Lapatinib and/or Trastuzumab Treatment Optimization.
Table A3.
PIK3CA Genotyping Results by ER Status and Treatment Arm
| Patient Group | Total No. of Patients |
PIK3CA Mutant |
|
|---|---|---|---|
| No. of Patients | % | ||
| ER status | |||
| Negative | 186 | 41 | 22.0 |
| Positive | 169 | 39 | 23.1 |
| Treatment arm | |||
| Lapatinib | 124 | 29 | 23.4 |
| Trastuzumab | 112 | 21 | 18.8 |
| Lapatinib + trastuzumab | 119 | 30 | 25.2 |
Abbreviation: ER, estrogen receptor.
Table A4.
PIK3CA Mutation Status and pCR by Hormone Receptor Status
| PIK3CA Status | Total pCR |
|||
|---|---|---|---|---|
| No |
Yes |
|||
| No. of Patients | % | No. of Patients | % | |
| ER negative | ||||
| Mutant | 28 | 71.8 | 11 | 28.2 |
| WT | 81 | 57.0 | 61 | 43.0 |
| ER positive | ||||
| Mutant | 31 | 86.1 | 5 | 13.9 |
| WT | 94 | 75.2 | 31 | 24.8 |
Abbreviations: ER, estrogen receptor; pCR, pathologic complete response; WT, wild type.
Table A5.
PIK3CA Mutation Status and pCR by Treatment Group
| Treatment and PIK3CA Status | Total pCR |
|||
|---|---|---|---|---|
| No |
Yes |
|||
| No. of Patients | % | No. of Patients | % | |
| Lapatinib | ||||
| Mutant | 23 | 85.2 | 4 | 14.8 |
| WT | 74 | 79.6 | 19 | 20.4 |
| Trastuzumab | ||||
| Mutant | 16 | 80.0 | 4 | 20.0 |
| WT | 63 | 71.6 | 25 | 28.4 |
| Lapatinib + trastuzumab | ||||
| Mutant | 20 | 71.4 | 8 | 28.6 |
| WT | 38 | 44.2 | 48 | 55.8 |
Abbreviations: pCR, pathologic complete response; WT, wild type.
Table A6.
Breast pCR and Total pCR
| Total pCR | Breast pCR (No.) |
|
|---|---|---|
| pCR | No pCR | |
| pCR | 108 | 0 |
| No pCR | 13 | 221 |
| Missing | 6 | 7 |
Abbreviation: pCR, pathologic complete response.
Footnotes
Supported in part by the Center for Cancer Systems Biology, the Rational Therapy for Breast Cancer (RATHER) FP7 project, and GlaxoSmithKline.
Presented at the 2013 European Cancer Congress, September 27-October 1, 2013, Amsterdam, the Netherlands.
Clinical trial information: NCT00553358.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) and/or an author's immediate family member(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: Catherine Ellis, GlaxoSmithKline (C) Consultant or Advisory Role: Holger Eidtmann, Roche (C); Nadia Harbeck, GlaxoSmithKline (C), Roche (C); Martine Piccart-Gebhart, Pharmamar (C); José Baselga, Novartis (C) Stock Ownership: Catherine Ellis, GlaxoSmithKline Honoraria: Holger Eidtmann, Roche, GlaxoSmithKline; Evandro de Azambuja, Roche, GlaxoSmithKline; Nadia Harbeck, Roche; Martine Piccart-Gebhart, Amgen, Astellas Pharma, AstraZeneca, Bayer AG, Invivis Pharmaceuticals, Merck, Novartis, Pfizer, Roche, Genentech, sanofi-aventis, Symphogen, Verastem Research Funding: Evandro de Azambuja, GlaxoSmithKline; Martine Piccart-Gebhart, Amgen, Astellas Pharma, AstraZeneca, Bayer AG, Eli Lilly, Invivis Pharmaceuticals, Merck, Novartis, Pfizer, Roche-Genentech, sanofi-aventis, Symphogen, Synthon, Verastem Expert Testimony: None Patents, Royalties, and Licenses: None Other Remuneration: Evandro de Azambuja, GlaxoSmithKline, Roche
AUTHOR CONTRIBUTIONS
Conception and design: Holger Eidtmann, Sherene Loi, Catherine Ellis, Martine Piccart-Gebhart, René Bernards, José Baselga
Administrative support: Holger Eidtmann
Provision of study materials or patients: Holger Eidtmann, Jose Jimenez, Claudia Aura, Maria Carmen Díaz-Delgado, Martine Piccart-Gebhart
Collection and assembly of data: Ian J. Majewski, Lorenza Mittempergher, Astrid J. Bosma, Ludmila Prudkin, Sherene Loi
Data analysis and interpretation: Ian J. Majewski, Paolo Nuciforo, Lorenza Mittempergher, Astrid J. Bosma, Holger Eidtmann, Eileen Holmes, Christos Sotiriou, Debora Fumagalli, Jose Jimenez, Claudia Aura, Maria Carmen Díaz-Delgado, Lorena de la Peña, Sherene Loi, Nikolaus Schultz, Evandro de Azambuja, Nadia Harbeck, José Baselga
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Slamon DJ, Clark GM, Wong SG, et al. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 2.Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 3.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 4.Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 5.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353:1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 6.Geyer CE, Forster J, Lindquist D, et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med. 2006;355:2733–2743. doi: 10.1056/NEJMoa064320. [DOI] [PubMed] [Google Scholar]
- 7.Blackwell KL, Burstein HJ, Storniolo AM, et al. Randomized study of lapatinib alone or in combination with trastuzumab in women with ErbB2-positive, trastuzumab-refractory metastatic breast cancer. J Clin Oncol. 2010;28:1124–1130. doi: 10.1200/JCO.2008.21.4437. [DOI] [PubMed] [Google Scholar]
- 8.Vivanco I, Sawyers CL. The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berns K, Horlings HM, Hennessy BT, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 11.Dave B, Migliaccio I, Gutierrez MC, et al. Loss of phosphatase and tensin homolog or phosphoinositol-3 kinase activation and response to trastuzumab or lapatinib in human epidermal growth factor receptor 2-overexpressing locally advanced breast cancers. J Clin Oncol. 2011;29:166–173. doi: 10.1200/JCO.2009.27.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eichhorn PJ, Gili M, Scaltriti M, et al. Phosphatidylinositol 3-kinase hyperactivation results in lapatinib resistance that is reversed by the mTOR/phosphatidylinositol 3-kinase inhibitor NVP-BEZ235. Cancer Res. 2008;68:9221–9230. doi: 10.1158/0008-5472.CAN-08-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baselga J, Bradbury I, Eidtmann H, et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): A randomised, open-label, multicentre, phase 3 trial. Lancet. 2012;379:633–640. doi: 10.1016/S0140-6736(11)61847-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Azambuja E, Holmes AP, Piccart-Gebhart M, et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO), a randomised, open-label, multicentre, phase 3 trial: Survival outcomes and their association with pathological complete response. Lancet Oncol. 2014;15:1137–1146. doi: 10.1016/S1470-2045(14)70320-1. [DOI] [PubMed] [Google Scholar]
- 15.Majewski IJ, Mittempergher L, Davidson NM, et al. Identification of recurrent FGFR3 fusion genes in lung cancer through kinome-centered RNA sequencing. J Pathol. 2013;230:270–276. doi: 10.1002/path.4209. [DOI] [PubMed] [Google Scholar]
- 16.Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet. 2014;384:164–172. doi: 10.1016/S0140-6736(13)62422-8. [DOI] [PubMed] [Google Scholar]
- 17.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 18.Douillard JY, Oliner KS, Siena S, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 19.Nagata Y, Lan KH, Zhou X, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 20.Baselga J, Cortes J, Im SA, et al. Biomarker analyses in CLEOPATRA: A phase III, placebo-controlled study of pertuzumab in HER2-positive, first-line metastatic breast cancer (MBC) Cancer Res. 2012;72:S5–1. doi: 10.1200/JCO.2013.54.5384. (abstr) [DOI] [PubMed] [Google Scholar]
- 21.Loibl S, Denkert C, Schneeweis A, et al. PIK3CA mutation predicts resistance to anti-HER2/chemotherapy in primary HER2-positive/hormone-receptor-positive breast cancer: Prospective analysis of 737 participants of the GeparSixto and GeparQuinto studies. Cancer Res. 2013;73:S4–06. (abstr) [Google Scholar]
- 22.Baselga J, Verma S, Ro J, et al. Relationship between tumor biomarkers (BM) and efficacy in EMILIA, a phase III study of trastuzumab emtansine (T-DM1) in HER2-positive metastatic breast cancer (MBC) Cancer Res. 2013;73:LB-63. doi: 10.1158/1078-0432.CCR-15-2499. (abstr) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loi S, Michiels S, Lambrechts D, et al. Somatic mutation profiling and associations with prognosis and trastuzumab benefit in early breast cancer. J Natl Cancer Inst. 2013;105:960–967. doi: 10.1093/jnci/djt121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalinsky K, Jacks LM, Heguy A, et al. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009;15:5049–5059. doi: 10.1158/1078-0432.CCR-09-0632. [DOI] [PubMed] [Google Scholar]
- 25.Cizkova M, Susini A, Vacher S, et al. PIK3CA mutation impact on survival in breast cancer patients and in ERalpha, PR and ERBB2-based subgroups. Breast Cancer Res. 2012;14:R28. doi: 10.1186/bcr3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broad Institute. Broad GDAC. http://gdac.broadinstitute.org/
- 27.Krop IE, Saura C, Rodon Ahnert J, et al. A phase I/IB dose-escalation study of BEZ235 in combination with trastuzumab in patients with PI3-kinase or PTEN altered HER2+ metastatic breast cancer. J Clin Oncol. 2012;(suppl):30. abstr 508. [Google Scholar]
- 28.Saura C, Bendell J, Jerusalem G, et al. PD09-03: Phase I/II study of BKM120 in combination with trastuzumab in patients with HER2 overexpressing metastatic breast cancer resistant to trastuzumab-containing therapy. Cancer Res. 2011;71:PD09–03. (abstr) [Google Scholar]
- 29.Juric D, Krop I, Ramanathan RK, et al. GDC-0032, a beta isoform-sparing PI3K inhibitor: Results of a first-in-human phase Ia dose escalation study. Cancer Res. 2013;73:LB-64. (abstr) [Google Scholar]
- 30.Juric D, Rodon J, Gonzalez-Angulo AM, et al. BYL719, a next generation PI3K alpha specific inhibitor: Preliminary safety, PK, and efficacy results from the first-in-human study. Cancer Res. 2012;72:CT-01. (abstr) [Google Scholar]