Abstract
Infection with HIV persists despite suppressive antiretroviral therapy (ART) and treatment interruption results in rapid viral rebound. Antibody-mediated CD8+ lymphocyte depletion in simian immunodeficiency virus (SIV)-infected rhesus macaques (RMs) shows these cells contribute to virus control in untreated animals. However, the contribution of CD8+ lymphocytes to maintaining virus suppression under ART remains unknown. We showed that in SIV-infected RMs treated with short-term ART (i.e., 8-32 weeks), depletion of CD8+ lymphocytes resulted in increased plasma viremia in all animals, and that repopulation of CD8+ T cells was associated with prompt reestablishment of virus control. Although the number of SIV-DNA-positive cells remained unchanged after CD8 depletion and reconstitution, the frequency of SIV-infected CD4+ T cells pre-depletion positively correlated with both peak and area-under-the-curve of viremia post-depletion. These results suggest a role for CD8+ T cells in controlling virus production during ART, thus providing rationale to explore immunotherapeutic approaches in ART-treated HIV-infected individuals.
Introduction
CD8+ T cells are important for immune defense against viruses, intracellular pathogens and cancers. CD8+ T cell responses to lentiviruses such as HIV and SIV can be detected as early as day 7 post infection (Reimann et al.; Yasutomi et al.). Several lines of evidence indicate that CD8+ lymphocytes inhibit virus replication during HIV or SIV infection. First, there is a temporal association between the expansion of antigen-specific CD8+ lymphocytes and the post-peak decline of plasma viremia (Borrow et al.; Koup et al.). Second, there is a clear association between certain MHC class-I alleles (i.e., HLA-B*57 and Mamu-B*08 and −B*17) and disease progression during both HIV infection of humans and SIV infection of rhesus macaques (RMs) (Altfeld et al.; Costello et al.; Evans et al.; Evans et al.; Haynes et al.; Kaslow et al.; Klein et al.). Third, during both acute and chronic HIV or SIV infection, immunologic pressure mediated by virus-specific CD8+ T cells is manifested by the emergence of viral escape mutations (Borrow et al.; Chen et al.; McMichael and Phillips). Fourth, HIV-1-infected individuals with the “elite controller” phenotype exhibit CD8+ lymphocyte responses characterized by polyfunctionality, greater proliferative capacity, and stronger in vitro killing potential than those observed in normal progressors (Betts et al.; Migueles et al.; Peris-Pertusa et al.). However, this CD8+ T cell response is unable to clear or even control the infection in the overwhelming majority of HIV-infected individuals.
In the experimental animal model of SIV infection of RMs, the most direct evidence for the role of CD8+ lymphocytes in viral control came from studies in which these cells were transiently depleted in vivo using CD8-specific monoclonal antibodies. This procedure results in (i) abrogation of post-peak decline of viremia when performed during acute SIV infection (Matano et al.; Schmitz et al.) and (ii) increased virus replication when performed during chronic SIV infection (Chowdhury et al.; Jin et al.; Metzner et al., 2000). The fact that viral loads rapidly return to pre-depletion levels upon reconstitution of the CD8+ lymphocyte pool further confirms the antiviral role of these cells. However, the contribution of CD8+ lymphocytes in controlling virus replication and/or production during continuous, highly active ART is unknown. Assessing the potential antiviral role of CD8+ lymphocytes under ART is important as it would provide key rationale to explore immunotherapeutic approaches such as therapeutic vaccines and checkpoint blockade-inhibitors as clinical interventions to reduce the virus reservoir in HIV-infected individuals. In this study, we directly assessed the function of CD8+ lymphocytes in a cohort of thirteen SIV-infected, ART treated RMs and found that depletion of CD8+ lymphocytes resulted in increased virus levels in both plasma and lymphoid tissues in all treated animals.
Results
Administration of anti-CD8 antibody MT-807R1 to SIV-infected ART-treated RMs
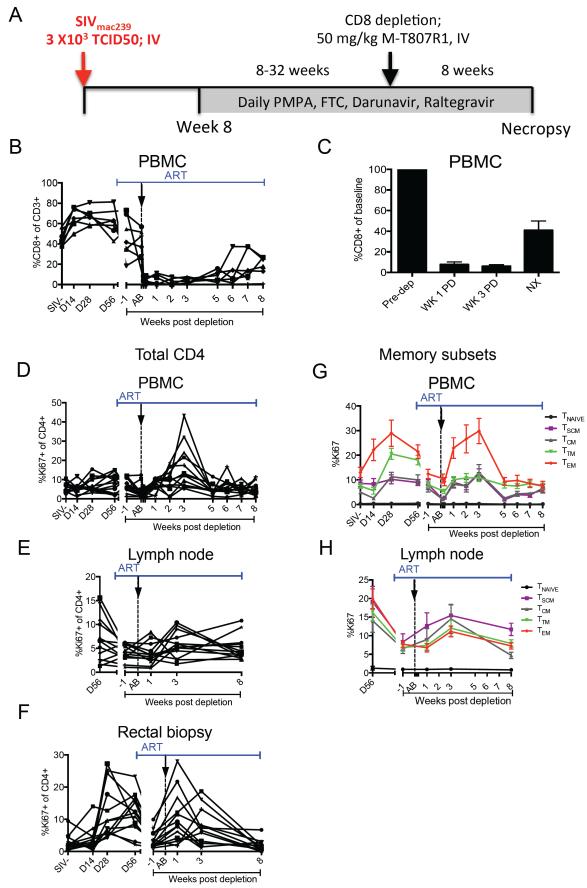
Sixteen Indian origin RMs infected intravenously (i.v.) with SIVmac239 started an ART regimen consisting of Tenofovir, Emtricitabine, Raltegravir, and Darunavir at week 8 post infection and were treated for 8 to 32 weeks prior to further intervention (Figure 1A, see also Table S1). Three RMs were euthanized prior to study completion due to rapid disease progression or side effects of ART, leaving 13 animals to complete the study. ART suppressed viremia to <60 copies/ml in 12/13 RMs that completed the protocol (see Experimental Procedures). Seven RMs showed at least four consecutive time-points with viremia below 60 copies/ml (mean= 6.9 weeks, range 4-10 weeks; i.e., “persistent suppressors”) while five showed a mix of undetectable and detectable levels (i.e., “intermittent suppressors”). The last animal never achieved undetectable viremia, even though ART decreased viremia by >5 logs. As expected, ART partially restored CD4+ T cells in PBMCs, lymph nodes (LN), and rectal biopsies, RB (data not shown). Despite variability in the duration of ART and kinetics of viremia suppression, at the time of CD8 depletion viral load had declined >99.97% as compared to pre-ART levels in all animals (Table S2). Once viral loads were consistently undetectable (persistent suppressors) or undetectable on at least 3 non-consecutive evaluations (intermittent suppressors), we administered one dose of the α-CD8 depleting antibody, MT-807R1, at 50 mg/kg iv. Animals were followed for 8 weeks after antibody administration and continued on ART (Figure 1A). Using this method, we were able to deplete >95% of CD8+ T cells in peripheral blood by day 1 post depletion (Figure 1B). This depletion was rapid and sustained (mean 92% SD=9 and mean 94% SD=5 at weeks 1 and 3, respectively) until approximately 5 weeks post depletion, at which time a variable degree of CD8+ lymphocyte reconstitution in the periphery was observed (Figure 1B, C). Since MT-807R1 is directed at the CD8α chain, CD3−CD8α+ NK cells were also depleted during this procedure (Figure S1A-C). Of note, CD8 depletion in LNs was less complete than in peripheral blood with an average of ~70% depletion (SD=26) at week 1 and ~85% depletion (SD=11) at week 3 post depletion (Figure S1D, E). CD8 depletion was least efficient in RB, with an average of ~62% depletion (SD=27) at week 1 post MT-807R1 administration and a more rapid reconstitution starting by week 3 (mean 56% depletion, SD=30) (Figure S1F, G). While MT-807R1 was less effective in depleting CD8+ lymphocytes in tissues as compared to blood, previous studies showed that CD8+ lymphocytes are functionally impaired following binding of the α-CD8 antibody even if they are not physically depleted (Schmitz et al.). CD8 depletion is followed by homeostatic proliferation of CD4+ T cells (Fukazawa et al.; Okoye et al.), and we observed increased CD4+ T cell proliferation starting at week 2 post depletion in PBMCs (Figure 1D), week 3 in LNs (Figure 1E), and week 1 in RBs (Figure 1F). This increased CD4+ T cell proliferation involved predominantly CD4+ effector memory (Tem) cells, with smaller and delayed increases observed in CD4+ stem-cell memory (Tscm) and central memory (Tcm) cells (Figure 1G, H). We also examined the frequency of CCR5+, HLA-DR+, and PD-1+ CD4+ T cells both prior to and during CD8 depletion. We find that CD8 depletion is associated with a moderate increase in the fraction of CD4+HLA-DR+ and CD4+CCR5+ T cells in LNs (Figure S2). Overall, these results indicated that administration of the MT-807R1 antibody was effective in depleting CD8+ T cells in blood and lymphoid tissues of ART-treated SIV-infected RMs.
Figure 1. Treatment of SIV-infected ART-treated RM with the depleting anti-CD8 antibody MT-807R1.
(A) Study design. (B) CD3+CD8+ T cells as percentage of CD3+ lymphocytes in PBMC. (C) CD8+ T cells as percentage of pre-depletion frequencies (“baseline”) in PBMC. Data shown is for 13 SIV-infected ART treated RM. Bars drawn at the mean and error bars represent standard error of mean. Proliferation (Ki67+) of total CD4+ T cells in (D) PBMC, (E) LN, and (F) RB. Proliferation (Ki67+) of CD4+ T cell subsets in (G) PBMC and (H) LN. Gap in x-axis represents 8-32 weeks of continuous ART. Black arrow and dotted vertical line indicate MT-807R1 administration. NX=necropsy.
CD8 depletion is followed by increase in plasma viral load and SIV RNA in LNs in 100% of ART-treated RMs
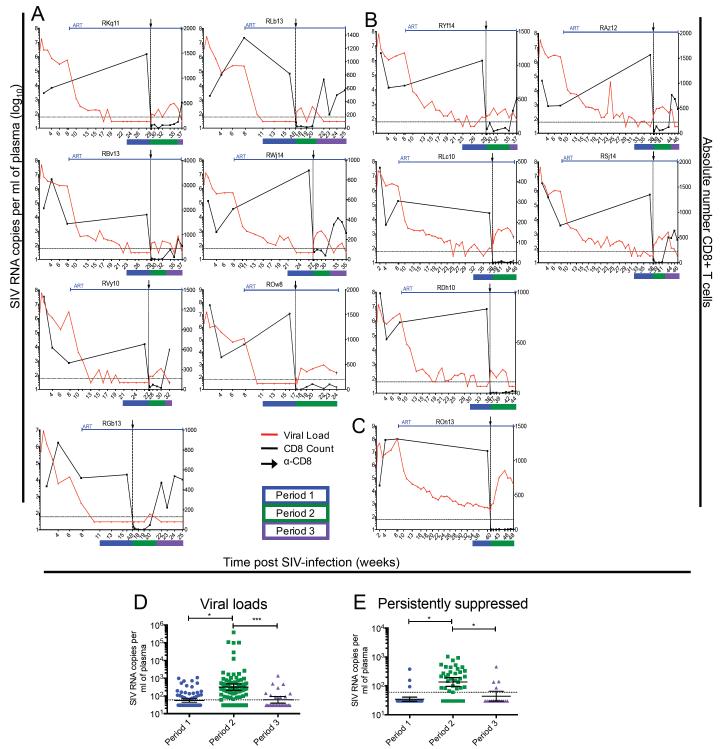
As shown in Figure 2, all 13 SIV-infected RMs showed a measureable increase in plasma viremia after CD8 depletion, with several animals rebounding as early as day 1 post-depletion, and only one animal remaining below 60 copies/ml until week 3 post-depletion, at which point virus was detectable (RGb13). The seven persistent suppressors all showed at least one time-point with detectable SIV-RNA post-depletion, with 6 out of 7 showing at least three time-points with viremia >60 copies/ml (Figure 2A). Similarly, the 5 intermittent suppressors had detectable viremia at all examined time-points post-depletion (Figure 2B). The one animal that never achieved undetectable viremia despite 32 weeks of ART and a decrease of viral load of >5 logs (i.e., >99.99% decline from baseline) showed a marked increase in viremia after CD8 depletion up to ~105 copies/ml (Figure 2C). Of note, CD8+ T cell reconstitution was consistently associated with a decrease in plasma viral loads to levels similar to those observed prior to CD8 depletion (Figure 2A-C). Interestingly, in 3 out of 4 cases where CD8+ T-cells did not reconstitute in peripheral blood during the 8 week follow up period post-depletion, plasma viral loads remained elevated above the limit of detection. To further analyze the impact of CD8 depletion and reconstitution on SIV viremia we next divided the experimental timeline into three periods defined as follows: period 1, the last 6 weeks of ART before CD8 depletion; period 2, the time-points after CD8 depletion in which the level of circulating CD8+ T cells was <10% of baseline; and period 3, the time-points during CD8+ lymphocyte reconstitution in which circulating CD8+ T cells were >20% of baseline (lines blue, green, and purple in Figure 2, respectively, also Table S3). Using a mixed linear effects model we compared viral loads in these three periods and found that the geometric mean of period 2 (275; 95% CI: 116-653) was significantly higher than either period 1 (53; 95% CI: 33-83, p<0.05) or period 3 (90; 95% CI: 45-179, p<0.001) (Fig. 2D). Due to the many non-detectable time points, we also conducted a binary analysis (detectable/non-detectable) which does not assume normality, and found that period 2 had significantly more detectable time points than either period 1 or 3 (data not shown). When we applied the mixed linear effects model to only the 7 ART-treated SIV-infected RMs with persistent suppression of viremia pre-depletion (Figure 2A), the geometric mean of period 2 (135; 95% CI: 96-193) was again significantly higher than either period 1 (34; 95% CI: 29-41, p<0.05) or period 3 (44; 95% CI: 29 −65, p<0.05) (Figure 2E).
Figure 2. CD8 depletion results in increased plasma viremia in ART-treated SIV-infected RMs.
Viral load (red) and CD8+ T cell counts (black) of SIV-infected RM that were (A) persistently suppressed, (B) intermittently suppressed, and (C) never fully suppressed on ART. Black arrow and dotted vertical line indicate anti-CD8 antibody MT-807R1 administration. (D) Geometric mean viremia for each statistical period for 13 RM. (Described fully in Table S3). Bars show the geometric mean with 95% CI, *p< 0.05, ***p<0.001. (E) Geometric mean viremia for each statistical period for 7 persistently suppressed RM. Bars show the geometric mean with 95% CI, *p <0.05. Statistical analysis was conducted using a linear, mixed effects model.
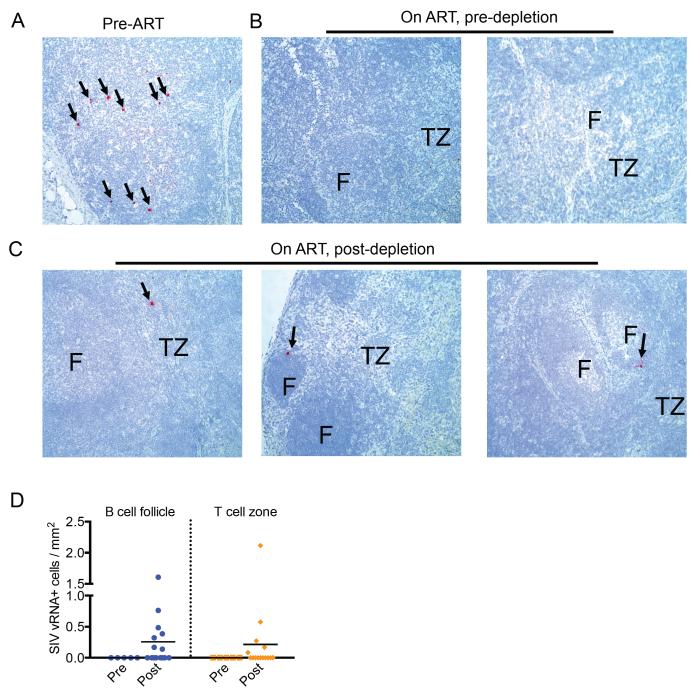
To determine whether the trends in viremia observed in our cohort of SIV-infected RMs were also present in lymphoid tissues, we next conducted a sequential analysis of the levels of SIV-RNA production in LNs using the RNAscope technology at 3 time-points, i.e., before initiation of ART, during ART but before CD8 depletion, and during ART after CD8 depletion. As shown in Figure 3, panels A-C, for representative animals, we observed a dramatic decrease in the number of LN SIV-RNA-positive cells after ART and a rebound of SIV-RNA production after CD8 depletion. A quantitative analysis of the number of SIV-RNA-positive cells per square millimeter in the B cell and T cell areas of LNs before and after CD8 depletion is shown in Figure 3D. Taken together, these data indicate that, in SIV-infected RMs, CD8+ lymphocytes contribute to maintenance of virus suppression during ART.
Figure 3. CD8 depletion results in increased SIV-RNA in LNs of ART-treated SIV-infected RMs.
Representative in situ hybridization of SIV-RNA in LNs of (A) chronically infected, (B) ART-treated, pre-CD8 depletion, and (C) ART-treated, post-CD8 depletion RMs. Data presented in (B) and (C) are from LNs of two and three different animals, respectively. Red indicates SIV-RNA positive, productively infected cell; F=follicle, TZ= T cell zone. Images are 20X magnification. (D) Number of SIV-RNA-positive cells per millimeter square in the B cell area (left) and T cell area (right) of the LN before and after CD8 depletion.
Ultrasensitive viral load assay reveals a 72 to 350 fold increase in virus production after CD8 depletion
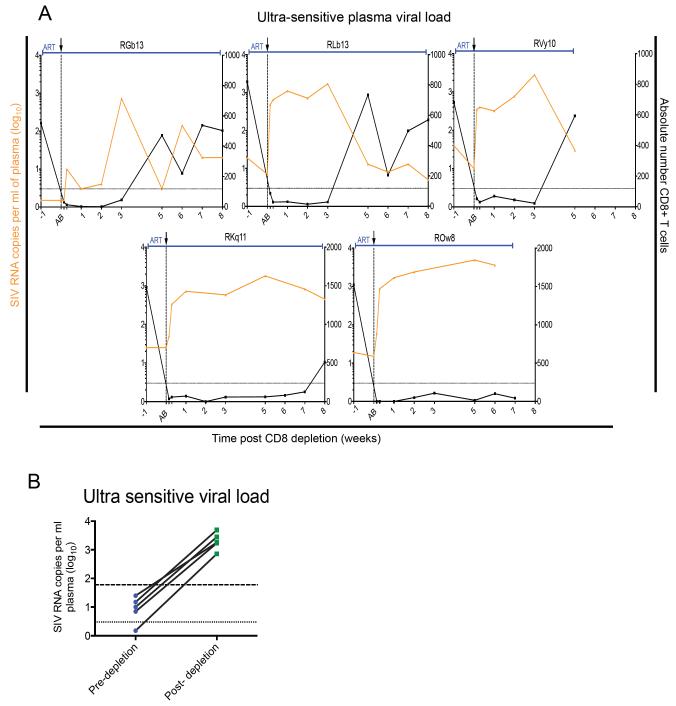
To better assess the level of residual viremia in the group of “persistently suppressed” SIV-infected ART-treated RMs at the time of CD8 depletion, we repeated the measurement of viral load using an ultrasensitive viral load assay with limit of detection of 3 copies/ml (Del Prete et al.). We found that viremia was <100 copies/ml in all these ART-treated RMs, with 5 out of the 7 animals showing levels of residual viremia ≤25 copies/ml (Table S2), thus in the range of levels of viremia measured in long-term ART-treated HIV-infected individuals,(Chun et al.; Maldarelli et al.). To more rigorously evaluate the impact of CD8 depletion on residual viremia during ART, we next used the same ultrasensitive assay to measure viral load at all post-CD8 depletion time points in the five RMs with the lowest residual viremia. As shown in Figure 4A, these data largely recapitulate the results shown in Figure 2, and indicate that the level of viremia increased from pre-depletion ranging from <3-25 copies/ml (geometric mean=8.3; 95% CI: 2-31) to post-depletion peak viremia ranging from 700-4,900 copies/ml (geometric mean=1977; 95% CI: 822-4754, p<0.05), for a 72 to 350-fold increase (Figure 4B). Of note, no correlation was found between pre- and post-CD8 depletion viral loads as measured using the ultrasensitive assay (data not shown). Overall, these data confirm that CD8+ lymphocytes contribute to the suppression of viremia observed in SIV-infected RMs treated with short-term (i.e., 8-32 weeks) ART.
Figure 4. Ultrasensitive viral load assay confirms the increase in viremia post-CD8 depletion.
(A) Viral load (orange) and CD8+ T cell counts (black) of SIV-infected RM that were persistently suppressed. Shaded area represents ART period. Black arrow and dotted vertical line indicate anti-CD8 antibody MT-807R1 administration. (B) Pre-depletion and peak post-depletion viremia (log10) for 5 RM. Dotted line is limit of detection of ultrasensitive viral load assay (3 copies/ml plasma). Dashed line is limit of detection of standard viral load assay (60 copies/ml plasma). Paired t-test, *p< 0.05.
Pre-depletion SIV-specific CD8+ T cells but not CD8+ NK cells positively correlate with the magnitude of on-ART plasma virus rebound post-depletion
The depleting MT-807R1 is directed at the α-chain of the CD8 molecule and depletes CD8α+ NK cells as well as CD8+ T cells in RMs. As expected, the frequency and absolute number of SIV-specific CD8+ T cells decreased substantially after initiation of ART (data not shown). However, we were able to detect low frequencies of Gag-CM9 specific CD8+ T cells (as measured by tetramer staining) in the PBMCs of Mamu-A*01 RMs at the time of CD8 depletion. First, we determined that the absolute number of pre-CD8 depletion circulating GAG-CM9-specific CD8+ T cells (but not their percentage of the total CD8+ T cells) directly correlated with viral load at both 1 and 3 weeks post-depletion (Figure S3A, B). However, we found no correlation between the numbers of circulating CD8+ NK cells pre-depletion and viral load post-depletion at either week 1 or 3 (Figure S3C, D). Second, we conducted the same analysis described in Figure 2D-E by dividing the study into three periods based on the kinetics of CD8+ NK cell numbers: period 1, the last 6 weeks of ART before CD8 depletion; period 2, the time-points after CD8 depletion in which the level of circulating CD8+ NK cells was <20% of baseline; and period 3, the time-points during CD8+ lymphocyte reconstitution in which circulating CD8+ NK cells were >20% of baseline (blue, green, and purple respectively in Figure S4). As shown in Figure S4A for one representative RM with persistent suppression of viremia, the CD8+ NK cells return to pre-depletion numbers around week 3 post-depletion while CD8+ T cells were still significantly depleted. Using the same mixed linear effects model of Figure 2, we found that while the geometric mean of the viral load in period 2 (199; 95% CI: 136-321) was significantly higher than period 1 (53; 95% CI: 44-70, p<0.05), the geometric mean of the viral load in period 3 (221; 95% CI: 122-382) was unchanged when compared to period 2 (Figure S4B). We found similar results when we analyzed only the persistently suppressed RMs (Figure 2A), with viral loads that remain elevated despite reconstitution of the CD8+ NK cell pool (Figure S4C). Finally, no correlation was observed between either the length of ART or the time to suppression with the change in viremia observed following CD8 depletion (data not shown). These data, together with those shown in Figure 2D-E, indicate that the depletion and repopulation of CD8+ T cells, but not the repopulation of CD8+ NK cells, is temporally associated with the control of viremia under ART.
Pre-depletion amounts of cell-associated SIV-DNA positively correlated with plasma SIV RNA post depletion
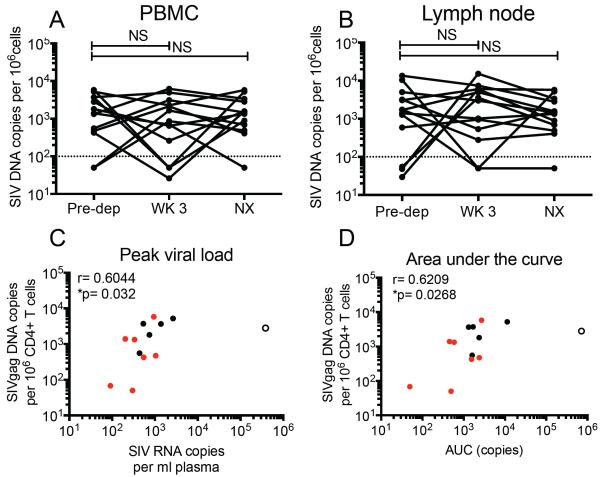
To determine whether CD8 depletion had an impact on the overall virus reservoir in ART-treated SIV-infected RMs, we sequentially measured the amount of total, cell-associated SIV-DNA by RT-PCR in bulk CD4+ T cells as well as in subsets of CD4+ Tscm , Tcm, transitional memory (Ttm), and effector memory (Tem) cells. As shown in Figure 5A and B, CD8 depletion did not induce any significant change in the number of SIV-DNA-positive CD4+ T cells in either blood or LNs. Similarly, we did not find any significant change in the amount of SIV-DNA in sorted CD4+ Tscm, Tcm, Ttm, and Tem in either blood or LNs following CD8 depletion (Figure S5A, B). While the analysis of SIV-DNA changes before and after CD8 depletion did not reveal a consistent pattern, we observed several RMs in which the cell-associated SIV DNA increased in either blood- or LN-derived CD4+ T cells, thus suggesting that CD8 depletion induced expansion of the virus reservoir under ART in a subset of animals. Interestingly, we observed a significant direct correlation between the amount of cell-associated SIV-DNA in CD4+ T cells before CD8 depletion and both the peak and the area-under-the-curve of plasma viremia post CD8 depletion (Figure 5C, D), suggesting that the size of the reservoir under ART is a key determinant of the resultant amount of virus production when CD8+ lymphocytes were removed from the system. Finally, we sought to determine whether the amount of virus production after CD8 depletion (i.e., peak and/or area-under-the curve) correlated with observed changes in the CD4+ T cell proliferation and activation, as measured by Ki67, CCR5, HLA-DR and PD-1 expression, in various cell subsets and tissues. As shown in Table S4, we found no significant correlations between the change in the fraction of proliferating (i.e., Ki-67+) cells in any of the examined CD4+ T cell subsets after CD8 depletion and the amount of viremia at the same time-point. We also found no correlation between the fraction of proliferating CD4+ T cells and the amounts of cell-associated SIV-DNA after CD8 depletion (data not shown). This lack of correlation suggests that the homeostatic proliferation of CD4+ T cells that is associated with CD8 depletion is unlikely to be a key factor responsible for the observed increase in SIV viremia. However, we found a direct correlation between changes in CD4+HLA-DR+ and CD4+PD-1+ T cells (but not CD4+Ki-67+ or CD4+CCR5+ T cells) and viremia that was significant only at the earliest time points post-depletion (i.e., days 1 and 2) (Table S5), suggesting that the initial activation of CD4+ T cells that occurs following CD8 depletion may contribute to the observed increase in SIV viremia.
Figure 5. Pre-depletion levels of SIV infection in CD4+ T cells predicts post CD8-depletion changes in viremia.
Fraction of SIV-infected sorted CD4+ T cells in (A) PBMC and (B) LN. WK 3= week 3 post-depletion, NX= necropsy. Kruskal-Wallis, NS=not significant. Correlation of cell-associated SIV-DNA in peripheral CD4+ T cells pre-depletion and the peak viral rebound (C) and area-under-the-curve (D) post depletion. Data is shown for all 13 animals. Spearman rank correlation. ( ) persistent suppressors, (◯) intermittent suppressors,(엯) never suppressed.
) persistent suppressors, (◯) intermittent suppressors,(엯) never suppressed.
Longitudinal virus sequencing by single genome analysis (SGA)
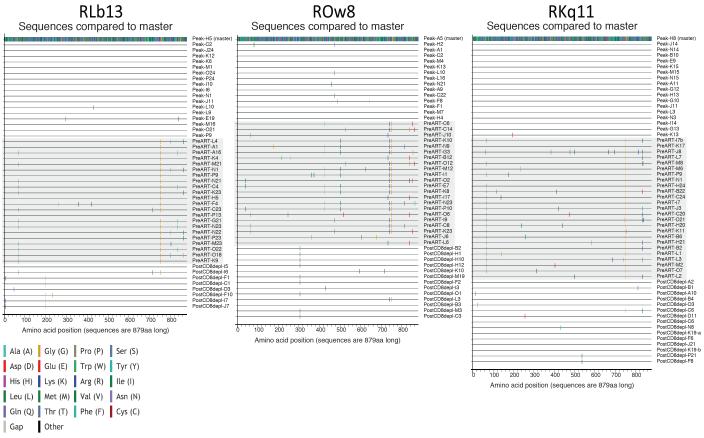
To gain insight into the dynamics of virus replication and/or reactivation after CD8 depletion, we performed a longitudinal study of plasma virus by single genome analysis (SGA) of the env gene of SIVmac239 at three time points: (i) day 10 post SIV-infection, i.e., peak viremia, (ii) day 56 post SIV-infection, i.e., immediately prior to ART initiation, and (iii) after CD8 depletion. We conducted this analysis on three representative RMs (RKq11, ROw8, and RLb13) with persistent SIV suppression under ART (Table S2). As shown in Figure 6, circulating viruses sequenced at day 10 post-infection were relatively homogenous within each RM (99.9-100% amino acid identity). In contrast, sequences isolated day 56 post-infection showed several fixed mutations that likely represent escape from either T cell-mediated or antibody responses. Interestingly, the viral sequences derived from plasma after CD8 depletion were more similar to those derived at peak viremia, compared to those derived immediately prior to ART (i.e., peak vs. pre ART amino acid identity = 99.5-99.7%, and peak vs. post CD8 depletion amino acid identity = 99.8-99.9). Importantly, the previously fixed escape mutations observed at the pre-ART time point represent only a small fraction of the circulating viruses after CD8 depletion. In addition, we conducted a MiSeq deep sequence analysis of the circulating cell-associated SIV-DNA immediately prior to ART initiation and immediately before CD8 depletion. As shown in Figure S6, this analysis revealed a consistent increase in inoculum-like sequences (i.e., similar to those derived at peak viremia) under ART. This latter observation is consistent with the finding that viruses similar to the initial infecting variants reemerge in plasma during the early stages of ART (data not shown), and suggests the presence of a pool of cells infected with inoculum-like virus that outlive cells infected with virus harboring escape mutations. Of note, when we conducted the same longitudinal sequence analysis of plasma virus by SGA in the SIV-infected RM that never achieved undetectable viremia (ROn13), we found a complex pattern of virus quasi-species diversity throughout the three examined time points that was consistent with continuous virus replication (data not shown). Overall, these data suggest that the plasma viruses that are present after CD8 depletion are produced from long-lived cells that were infected prior to ART initiation and before the generation of immune escape mutants.
Figure 6. Highlighter plots of Single Genome Amplification derived Env amino acid sequences.
Highlighter plots were generated to illustrate variation in amino acid composition within Env over the course of the experimental protocol in three animals: RLb13 (left), ROw8 (center), and RKq11 (right). Sequences were isolated at peak viral load, pre-ART initiation, and post-CD8 depletion (range week 1 to week 6). Colored ticks indicate deviation from master sequence.
Discussion
This study demonstrates that CD8+ lymphocytes can contribute to maintaining suppression of plasma viremia in SIV-infected RMs that are treated with short-term ART (i.e., 8-32 weeks). To date, the in vivo suppressive effect of CD8+ lymphocytes has primarily been characterized in studies involving untreated animals (Chowdhury et al.; Jin et al.; Matano et al.; Schmitz et al., 1999a) with only one report in three SIV-infected RMs treated with nonsuppressive tenofovir monotherapy (Van Rompay et al.). Indeed, in our knowledge, the current work represents the first experiment in which SIV-infected RMs underwent depletion of CD8+ lymphocyte in the setting of highly active ART (i.e., >99.97% suppression of viremia). While the majority of the RMs included in this study showed some residual virus production under ART, these levels of viremia were often within the range observed in long-term ART-treated HIV-infected individuals (Maldarelli et al.). Importantly, the magnitude of the viral load increase after CD8 depletion (i.e., 72 to 350 fold) and the fact that control of viremia is promptly reestablished upon CD8+ T cell reconstitution strongly support the hypothesis of an important (and yet previously unrecognized) role of CD8+ lymphocytes in cooperating with ART to maintain virus suppression. As such, the current set of data provides an evidence base to explore immune-based interventions, such as therapeutic vaccines and check-point blockade inhibitors, in ART-treated HIV-infected individuals. It should be noted, however, that interventions aimed at improving the antiviral CD8+ lymphocyte-mediated response under ART will be expected to be effective only against infected cells with sufficient expression of viral antigen, suggesting that combining viral induction and immune based approaches may enhance effectiveness of such interventions.
When using ultrasensitive viral load assays, residual viremia below clinical limit of detection is present in many ART-treated HIV-infected individuals (Chun et al., 2011; Dornadula et al.; Maldarelli et al.). Whether and to what extent this residual viremia is due to continuous, low-level de novo cycles of replication as opposed to either virus reactivation occurring in latently infected cells or the presence of persistently virus expressing cells remains an active area of research. Recent evidence suggests clonal expansion of virally infected cells during ART is another important contributor to HIV persistence (Maldarelli et al.; von Stockenstrom et al.; Wagner et al.). Similarly, at this time we do not know to what extent the increase in viremia observed following CD8 depletion in SIV-infected, ART-treated reflects de novo virus replication versus reactivation of latent virus versus increased virus production from persistently, nonlatently infected cells. Regardless of the mechanisms responsible for the very low viremia of ART-treated HIV-infected individuals and SIV-infected RMs, the current study suggests a direct in vivo role for CD8+ lymphocytes in maintaining very low to undetectable viral loads under ART. However, the current study does not define whether the antiviral role of CD8+ lymphocytes under ART involves classical cytotoxic T lymphocyte activity, non-cytolytic mechanisms (i.e., chemokine production and inhibition of virus transcription (Cocchi et al.; Walker et al.)), or both.
In this study, CD8 depletion was followed by various degrees of CD4+ T cell activation and proliferation, which could at least in part contribute to the observed increase in SIV viremia after CD8 depletion through reactivation of virus production from latently infected, previously resting CD4+ T cells. Interestingly, we observed a direct correlation between the changes in HLA-DR and PD-1 expression (but not Ki-67 or CCR5 expression) on CD4+ T cells and the level of viremia early after CD8 depletion. In particular, animal RGb13, who showed the greatest suppression of viremia with ART (<3 copies for three consecutive measurements), experienced a delayed rebound of viremia that was coincident with the peak of CD4+ T cell activation post CD8 depletion. In this context, however, we wish to emphasize that the relationship between CD8 depletion, increased SIV viremia, and increased CD4+ T cell activation is very complex, and that the exact contribution of this latter factor will need to be assessed in future in vivo interventional studies in which CD4+ T cell activation is selectively inhibited in SIV-infected ART-treated RMs at the time of CD8 depletion.
In this cohort of ART-treated SIV-infected RMs, the increased viremia that follows CD8 depletion was not consistently associated with specific changes (i.e., increase or decrease) in the levels of cell-associated SIV-DNA. However, a subset of CD8 depleted SIV-infected RMs showed an increase in SIV-DNA levels, thus consistent with de novo rounds of virus replication and/or homeostatic proliferation of SIV-infected cells. In this experimental setting, the change in the fraction of SIV-DNA-positive cells in blood and LNs after CD8 depletion is the result of factors such as (i) de novo infection of cells; (ii) lifespan of cells that have reactivated virus production, and (iii) proliferation levels and lifespan of cells that harbor integrated SIV-DNA but do not express the virus. An additional complication of these studies is that the level of cell-associated SIV-DNA is measured as number of copies per fixed amount of cells (typically one million) while the actual total number of SIV-DNA-positive cells in the body is virtually impossible to measure. Clarifying the relative contribution of these mechanisms to the changes in the size of virus reservoirs after CD8 depletion is a challenging scientific problem that is the object of intensive studies in our laboratories.
In an attempt to elucidate the impact of CD8 depletion on the viral dynamics in our cohort of ART-treated SIV-infected RMs we performed, in a subset of animals, a longitudinal sequence analysis of plasma SIV-RNA and cell-associated SIV-DNA by SGA and deep-sequencing, respectively. Our SGA analysis revealed a clear and consistent pattern of virus sequence dynamics in three of the SIV-infected RMs with the best virus suppression under ART. This analysis showed that (i) certain amino acid substitutions become “fixed” in the circulating population of viruses in the interval between peak viremia and ART initiation, and (ii) the virus that emerges after CD8 depletion shows sequences that largely overlap with the peak viremia quasi-species and not the pre-ART quasi-species. In addition, the SIV-DNA analysis revealed a concomitant increase in the fraction of “inoculum-like” sequences during ART treatment. The most parsimonious explanation for these findings is that the de novo virus production after CD8 depletion is mainly derived from a subset of long-lived cells that, at the time of ART initiation, were either latently infected or produced only a small minority of circulating virions. Potential mechanisms responsible for the increased virus production by these putatively long-lived SIV infected cells after CD8 depletion include absence of CD8+ lymphocyte-mediated cytolytic activity, lack of non-cytolytic virus suppression (Klatt et al., 2010; Wong et al., 2010), and increased CD4+ T cell activation. Of note, the striking similarity between the inoculum (i.e., peak viremia) sequences and the post-CD8 depletion sequences does not support the hypothesis that the observed increase of viremia after CD8 depletion is simply the consequence of an “amplification” of cellular and/or anatomic pockets of residual virus replication under ART.
While the effect of CD8 depletion on the levels of plasma viremia in ART-treated SIV-infected RMs is clear, there are important caveats to this study. The first is that the used antibody depletes both CD8+ T cells and CD8α-expressing NK cells. However, our analysis of the correlation between viremia after CD8 depletion and either the numbers of CD8+ T cells and CD8+ NK cells pre-depletion or their levels during depletion and repopulation strongly suggests that CD8+ T cells, rather than NK cells, are the main contributors of the control of viremia under ART. In addition, we observed reactivation of virus production in lymphoid tissues in which the levels of CD8+ NK cells were extremely low even before CD8 depletion. Further studies using a CD8-β-specific antibody that depletes only CD8αβ+ T cells will formally establish the specific contribution of CD8+ T cells in the control of viremia in ART-treated SIV-infected RMs. The second caveat is that the period of ART suppression was relatively short even in the SIV-infected RMs defined as persistent suppressors, and therefore the level of virus suppression may not be as complete as in long-term ART-treated HIV-infected humans. However, we wish to point out that the level of virus suppression was >99.97% in all treated RMs (average 99.99%), with five animals showing levels of viremia similar to those observed in long-term ART-treated HIV-infected humans (Maldarelli et al., 2007). Further studies in which SIV-infected RMs are treated for longer periods of time (i.e., >1 year) will determine whether and to what extent the virus suppression that occurs in the setting of long-term ART is affected by CD8 depletion. We wish to point out that even if future studies reveal that CD8+ cells are no longer needed to maintain suppression of viremia when ART is administered for years, the current results would still provide rationale for clinical interventions in which CD8 enhancing therapies (checkpoint blockade inhibitors, therapeutic vaccines, etc) are performed during the first six-to-eight months of ART. The third caveat of the current study is that the impact of CD8 depletion on the reservoir was measured using a relatively insensitive assay, i.e., total cell-associated SIV-DNA, which does not provide a functional analysis of the reservoir of replication competent virus. While interesting, analyses such as the quantitative viral outgrowth assay require a large number of cells and were not feasible in animals undergoing an already heavy schedule of blood collections. For these reasons, the current work can be considered as a pilot, hypotheses-generating study that should prompt additional in-depth investigation of this intriguing and previously unrecognized role of CD8 depletion in inducing increased virus production in ART-treated SIV-infected RMs.
Our current set of findings may seem to contrast with the observation that immunosuppressive treatment is not consistently associated with increased viremia in ART-treated HIV-infected individuals. However, these clinical instances are different from CD8 depletion as they may involve depletion of all T cells, as in the case of aggressive chemotherapy regimens, or markedly reduce CD4+ T cell activation, as in the case of several commonly used immunomodulators. For instance, cyclosporine and Tacrolimus (FK506) are calcineurin inhibitors that inhibit NFAT-mediated CD4+ T cell activation and have a well-characterized direct antiviral effect via inhibition of cyclophilin (Bartz et al., 1995; Braaten et al., 1996; Emmel et al., 1989; Franke and Luban, 1996; Franke et al., 1994; Schaller et al., 2011; Sokolskaja et al., 2010; Streblow et al., 1998). Similarly, mycophenolate induces apoptosis of activated CD4+ T lymphocytes and, in a time-dependent manner and inhibits HIV replication (Chapuis et al., 2000; Margolis et al., 2002) while rapamycin (sirolimus) decreases LTR-driven transcription of HIV (Roy et al., 2002) and reduces expression of the main HIV co-receptor CCR5 (Gilliam et al., 2007). These caveats notwithstanding, the current set of data has important implications for understanding how the host antiviral cellular immune response works in concert with antiretroviral drugs in suppressing SIV replication, and defining the rationale for therapeutic interventions aimed at boosting the virus-specific CD8+ lymphocyte response in ART-treated HIV-infected individuals.
Experimental Procedures
Animals
Sixteen Indian origin RMs were enrolled in this study. They were all infected i.v. with 3,000 TCID50 of SIVmac239. Three animals were euthanized due to severe weight loss prior to CD8 depletion and were thus excluded from analysis. The remaining 13 animals consisted of 6 females and 7 males and ranged in age from 4-13 years at time of infection. Nine were Mamu-A*01 positive, and all 16 were Mamu-B*08 and B*17 negative. All animals were housed at the Yerkes National Primate Research Center (YNPRC) of Emory University and maintained in accordance with US National Institutes of Health guidelines. Anesthesia was used for all blood and tissue collections. All studies were approved by the Emory University Institutional Animal Care and Usage Committee (IACUC).
Antiretroviral therapy
All RM were put on ART at 8 weeks post SIV infection. The drug regimen started as 20 mg/kg PMPA (Tenofovir) and 30 mg/kg FTC (Emtricitabine) administered once a day by subcutaneous injection along with 100 mg Raltegravir and 400 mg Darunavir orally twice daily. During the first 8 weeks, all RM received this treatment regimen. Three animals remained on this regimen for the duration of the study. In the remaining 10 animals, we increased the dose of Darunavir and Raltegravir as described in Supplemental Table 1, until suppression was achieved. Animals were given oral antiretroviral treatment via orogastric tube on days where anesthesia was also administered, due to fasting and nausea. Suppression of plasma viremia to below the limit of detection of our assay (60 copies/ml of plasma) was achieved in 12 out of 13 animals, with persistent suppression observed in 7 out of 13 animals. Of note, the original study design planned for CD8 depletion after two consecutive undetectable viremia measured 2 weeks apart. However, some animals who had not reached this definition show side effects of ART. As such, we decided to perform the depleting treatment in all enrolled animals to generate as much data as possible regarding CD8 depletion in ART-treated RMs.
Depletion of CD8+ lymphocytes
We administered one dose of MT-807R1 (from the NIH NHP Reagent Resource Program) i.v. at 50 mg/kg (<0.25 EU/ml of endotoxin by LAL gel-clot). The extent of CD8 depletion was determined (i) in peripheral blood by flow cytometric staining and complete blood counts (CBC) as both number of cells/cmm of blood and percentage of CD3+ T cells, and (ii) in tissues by flow cytometry and measuring the level of CD8+ T cells as fraction of their pre-depletion frequency.
Sample collection & tissue processing
Blood was collected in EDTA tubes. Plasma was obtained by centrifugation. Peripheral blood mononuclear cells (PBMC) were obtained by density-gradient centrifugation using 90% Lymphocyte Separation Media from Lonza. LN biopsies were taken at day 56 pre-ART initiation, 1 week pre-CD8 depletion, weeks 1 and 3 post-CD8 depletion, as well as at necropsy. LNs were cut in half and one half was put in 4% paraformaldehyde and then embedded in paraffin. LN-derived cells were obtained by grinding over a 70-μm cell-strainer. RBs were digested in 0.75 mg/ml Collagenase (Sigma-Aldrich) and 0.15 μL DNase in 10% fetal calf serum (FCS), 1% penicillin-streptomycin, and 1% L-glutamine at 37° C with gentle shaking. After 2 hours, tissues were mechanically separated using plastic cannula and run over a 70-μm filter.
Immunophenotype by flow cytometry
Multiparametric flow cytometry was performed on PBMC, LNMC, and cells isolated from RBs using fluorescently labeled monoclonal antibodies cross-reactive in RMs. The following antibodies were used: CD3-APC/Cy7 (SP34-2), CD4-PE-CF594 (L200) CD8-BV711 (RPA-T8), CCR7-FITC (150503), CD45RA-PE/Cy7 (L48), CD95-PE/Cy5 (DX2), Ki67-AlexaFluor700 (B56), CD14-BV650 (M5E2), CD56-BV605 (NCAM16.2), CD62L-PE (SK11), HLA-DR-PerCPCy5.5 (G46.6), CCR5-APC (3A9) from BD Biosciences; CD28-ECD (CD28-2) from Beckman Coulter; CD16-BV421 (3G8), CD4-BV650 (OK-T4), PD-1-BV421 (EH12.2H7), from Biolegend; CD8-APC (DK25) from DAKO. All specimens were acquired on an LSR II (BD Biosciences) and analysis of the acquired data was performed using FlowJo software (Tree Star).
Cell sorting
After isolation, cells were resuspended in PBS containing 2 mM EDTA and spun to remove contaminating platelets. Prior to sorting, CD4+ T cells were enriched using magnetic beads and column purification (Miltenyi Biotec). Enriched cells were then stained with previously determined volumes of CD3-APC/Cy7 (SP34-2), CD4-Brilliant Violet 650 (OKT-4) CD8-Brilliant Violet 421 (RPA-T8), Live/Dead-Aqua, CD45RA-APC (5H9), CCR7-PE/Cy7 (3D12), CD95-PE/Cy5 (DX2), CD28-ECD (CD28-2), CD62L-PE (SK11). Populations for sorting were defined as follows: Tscm (CD45RA+CCR7+CD95+CD62L+), Tcm (CD45RA−CD95+CD28+CCR7+CD62L+), Ttm (CD95+CCR7+CD62L−), and Tem (CD95+CCR7−CD62L−). Sorting was performed on a FACSAria LSR II (BD Biosciences) equipped with FACS Diva software.
Plasma viral load and cell-associated SIVgag DNA
Plasma viral quantification was performed as described previously (Taaffe et al.) Quantification of SIVmac gag DNA was performed as previously described (Chahroudi et al., 2014). Additional details can be found in the Supplemental Experimental Procedures.
In situ hybridization (ISH)
For ISH we used a next-generation, ultra-sensitive RNA in situ hybridization technology, RNAscope (Advanced Cell Diagnostics), as described previously (Smedley et al.).
Ultrasensitive viral load assay
Ultrasensitive plasma viral RNA measurements were performed essentially as described previously (Del Prete et al.).
Single genome analysis of circulating SIV
Single genome analysis of plasma-derived SIV was performed essentially as described previously (Burton et al., 2015; Smith et al., 2016). Additional details can be found in the Supplemental Experimental Procedures.
Deep sequencing of cell-associated SIV-DNA
Deep-sequencing of two amplicons covering nucleotides 6679-7103 and 8748-9196 of the wildtype SIVmac239 genome (NCBI accession#: M33262.1) and corresponding to Env amino acids 35-158 and 724-845 was performed essentially as described previously (Vanderford et al., 2011). Additional details can be found in the Supplemental Experimental Procedures.
Statistical analysis
The data presented in Figure 2D and E and Figure S4B, C were analyzed by fixing a mixed effects linear model that allows estimation of mean viral load by period. Comparisons between frequency of infection pre- and post-depletion were carried out using either Kruskal-Wallis (Figure 5A and B) or Wilcoxon matched-pairs signed rank test (Figure S5A and B). Correlations were determined using the non-Gaussian Spearman correlation. Significance was attributed at p<0.05. Analysis was done using GraphPad Prism 6.0, and R 3.1.3.
Supplementary Material
Highlights.
CD8+ lymphocyte depletion during ART increases SIV plasma viral load (72-350 fold)
Reconstitution of CD8+ T cells is associated with re-establishment of viral control.
Pre-depletion levels of SIV-DNA+ CD4+ T cells correlate with viremia post depletion.
Acknowledgements
We thank J. Levy, D.G. Carnathan, M. Mavigner, C.S. McGary and G. Mylvaganam for helpful discussions; B. Cervasi, K. P. Gill and J.P. Mackel for technical support; S. Ehnert for study organization and scheduling; YNPRC veterinary staff, especially S. Jean for caring for the animals and daily ART administration; Merck (I.MTA.14.232) Gilead Sciences (I.MTA.14.230), and Janssen R&D Ireland (I.MTA.14.231) for generously providing Raltegravir, PMPA and FTC, and Darunavir, respectively. We thank the Emory CFAR Virology core for viral loads and the Emory CFAR Immunology core for use of the MiSeq platform. This work was funded in part by R01-AI90797 (to G.S.), RR000165/OD011132 to the YNPRC, the Emory CFAR, NIH Grant # P30-AI-504, R01 AI-58706 and U19 AI-109633 (to C.D.) and supported in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E (J.L.).
Footnotes
Author Contributions
E.K.C. and G.S. designed and performed the experiments. L.S., S.A.S., S.P., and C.D. performed single genome sequencing of SIV-RNA. D.L., S.E.B., and T.H.V. performed deep sequencing of cell-associated SIV-DNA. R.F. and J.D.L performed ultrasensitive viral load analyses. B.O.L, M.N, and T.H.V. performed viral load and cell-associated DNA analyses. K.E performed statistical analyses. J.D.E. performed RNAscope analysis. J.E.S., M.P., A.C., C.D., and S.E.B. discussed the data. E.K.C., A.C., and G.S. wrote the manuscript.
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altfeld M, Addo MM, Rosenberg ES, Hecht FM, Lee PK, Vogel M, Yu XG, Draenert R, Johnston MN, Strick D, et al. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. Aids. 2003;17:2581–2591. doi: 10.1097/00002030-200312050-00005. [DOI] [PubMed] [Google Scholar]
- Bartz SR, Hohenwalter E, Hu MK, Rich DH, Malkovsky M. Inhibition of human immunodeficiency virus replication by nonimmunosuppressive analogs of cyclosporin A. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:5381–5385. doi: 10.1073/pnas.92.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. Journal of virology. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrow P, Lewicki H, Wei X, Horwitz MS, Peffer N, Meyers H, Nelson JA, Gairin JE, Hahn BH, Oldstone MB, Shaw GM. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nature medicine. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- Braaten D, Aberham C, Franke EK, Yin L, Phares W, Luban J. Cyclosporine A-resistant human immunodeficiency virus type 1 mutants demonstrate that Gag encodes the functional target of cyclophilin A. Journal of virology. 1996;70:5170–5176. doi: 10.1128/jvi.70.8.5170-5176.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton SL, Kilgore KM, Smith SA, Reddy S, Hunter E, Robinson HL, Silvestri G, Amara RR, Derdeyn CA. Breakthrough of SIV strain smE660 challenge in SIV strain mac239-vaccinated rhesus macaques despite potent autologous neutralizing antibody responses. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:10780–10785. doi: 10.1073/pnas.1509731112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahroudi A, Cartwright E, Lee ST, Mavigner M, Carnathan DG, Lawson B, Carnathan PM, Hashempoor T, Murphy MK, Meeker T, et al. Target cell availability, rather than breast milk factors, dictates mother-to-infant transmission of SIV in sooty mangabeys and rhesus macaques. PLoS pathogens. 2014;10:e1003958. doi: 10.1371/journal.ppat.1003958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuis AG, Paolo Rizzardi G, D'Agostino C, Attinger A, Knabenhans C, Fleury S, Acha-Orbea H, Pantaleo G. Effects of mycophenolic acid on human immunodeficiency virus infection in vitro and in vivo. Nature medicine. 2000;6:762–768. doi: 10.1038/77489. [DOI] [PubMed] [Google Scholar]
- Chen ZW, Craiu A, Shen L, Kuroda MJ, Iroku UC, Watkins DI, Voss G, Letvin NL. Simian immunodeficiency virus evades a dominant epitope-specific cytotoxic T lymphocyte response through a mutation resulting in the accelerated dissociation of viral peptide and MHC class I. Journal of immunology. 2000;164:6474–6479. doi: 10.4049/jimmunol.164.12.6474. [DOI] [PubMed] [Google Scholar]
- Chowdhury A, Hayes TL, Bosinger SE, Lawson BO, Vanderford T, Schmitz JE, Paiardini M, Betts M, Chahroudi A, Estes JD, Silvestri G. Differential impact of in vivo CD8+ T-lymphocyte depletion in controller versus progressor SIV-infected macaques. Journal of virology. 2015 doi: 10.1128/JVI.00869-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Murray D, Justement JS, Hallahan CW, Moir S, Kovacs C, Fauci AS. Relationship between residual plasma viremia and the size of HIV proviral DNA reservoirs in infected individuals receiving effective antiretroviral therapy. The Journal of infectious diseases. 2011;204:135–138. doi: 10.1093/infdis/jir208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- Costello C, Tang J, Rivers C, Karita E, Meizen-Derr J, Allen S, Kaslow RA. HLA-B*5703 independently associated with slower HIV-1 disease progression in Rwandan women. Aids. 1999;13:1990–1991. doi: 10.1097/00002030-199910010-00031. [DOI] [PubMed] [Google Scholar]
- Del Prete GQ, Shoemaker R, Oswald K, Lara A, Trubey CM, Fast R, Schneider DK, Kiser R, Coalter V, Wiles A, et al. Effect of suberoylanilide hydroxamic acid (SAHA) administration on the residual virus pool in a model of combination antiretroviral therapy-mediated suppression in SIVmac239-infected indian rhesus macaques. Antimicrobial agents and chemotherapy. 2014;58:6790–6806. doi: 10.1128/AAC.03746-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornadula G, Zhang H, Shetty S, Pomerantz RJ. HIV-1 virions produced from replicating peripheral blood lymphocytes are more infectious than those from nonproliferating macrophages due to higher levels of intravirion reverse transcripts: implications for pathogenesis and transmission. Virology. 1999;253:10–16. doi: 10.1006/viro.1998.9465. [DOI] [PubMed] [Google Scholar]
- Emmel EA, Verweij CL, Durand DB, Higgins KM, Lacy E, Crabtree GR. Cyclosporin A specifically inhibits function of nuclear proteins involved in T cell activation. Science. 1989;246:1617–1620. doi: 10.1126/science.2595372. [DOI] [PubMed] [Google Scholar]
- Evans DT, Jing P, Allen TM, O'Connor DH, Horton H, Venham JE, Piekarczyk M, Dzuris J, Dykhuzen M, Mitchen J, et al. Definition of five new simian immunodeficiency virus cytotoxic T-lymphocyte epitopes and their restricting major histocompatibility complex class I molecules: evidence for an influence on disease progression. Journal of virology. 2000;74:7400–7410. doi: 10.1128/jvi.74.16.7400-7410.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DT, Knapp LA, Jing P, Mitchen JL, Dykhuizen M, Montefiori DC, Pauza CD, Watkins DI. Rapid and slow progressors differ by a single MHC class I haplotype in a family of MHC-defined rhesus macaques infected with SIV. Immunology letters. 1999;66:53–59. doi: 10.1016/s0165-2478(98)00151-5. [DOI] [PubMed] [Google Scholar]
- Franke EK, Luban J. Inhibition of HIV-1 replication by cyclosporine A or related compounds correlates with the ability to disrupt the Gag-cyclophilin A interaction. Virology. 1996;222:279–282. doi: 10.1006/viro.1996.0421. [DOI] [PubMed] [Google Scholar]
- Franke EK, Yuan HE, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- Fukazawa Y, Lum R, Okoye AA, Park H, Matsuda K, Bae JY, Hagen SI, Shoemaker R, Deleage C, Lucero C, et al. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nature medicine. 2015;21:132–139. doi: 10.1038/nm.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliam BL, Heredia A, Devico A, Le N, Bamba D, Bryant JL, Pauza CD, Redfield RR. Rapamycin reduces CCR5 mRNA levels in macaques: potential applications in HIV-1 prevention and treatment. Aids. 2007;21:2108–2110. doi: 10.1097/QAD.0b013e3282f02a4f. [DOI] [PubMed] [Google Scholar]
- Haynes BF, Pantaleo G, Fauci AS. Toward an understanding of the correlates of protective immunity to HIV infection. Science. 1996;271:324–328. doi: 10.1126/science.271.5247.324. [DOI] [PubMed] [Google Scholar]
- Jin X, Bauer DE, Tuttleton SE, Lewin S, Gettie A, Blanchard J, Irwin CE, Safrit JT, Mittler J, Weinberger L, et al. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. The Journal of experimental medicine. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaslow RA, Carrington M, Apple R, Park L, Munoz A, Saah AJ, Goedert JJ, Winkler C, O'Brien SJ, Rinaldo C, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nature medicine. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- Klatt NR, Shudo E, Ortiz AM, Engram JC, Paiardini M, Lawson B, Miller MD, Else J, Pandrea I, Estes JD, et al. CD8+ lymphocytes control viral replication in SIVmac239-infected rhesus macaques without decreasing the lifespan of productively infected cells. PLoS pathogens. 2010;6:e1000747. doi: 10.1371/journal.ppat.1000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein MR, van der Burg SH, Hovenkamp E, Holwerda AM, Drijfhout JW, Melief CJ, Miedema F. Characterization of HLA-B57-restricted human immunodeficiency virus type 1 Gag- and RT-specific cytotoxic T lymphocyte responses. The Journal of general virology. 1998;79(Pt 9):2191–2201. doi: 10.1099/0022-1317-79-9-2191. [DOI] [PubMed] [Google Scholar]
- Koup RA, Safrit JT, Cao Y, Andrews CA, McLeod G, Borkowsky W, Farthing C, Ho DD. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. Journal of virology. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldarelli F, Palmer S, King MS, Wiegand A, Polis MA, Mican J, Kovacs JA, Davey RT, Rock-Kress D, Dewar R, et al. ART suppresses plasma HIV-1 RNA to a stable set point predicted by pretherapy viremia. PLoS pathogens. 2007;3:e46. doi: 10.1371/journal.ppat.0030046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis DM, Kewn S, Coull JJ, Ylisastigui L, Turner D, Wise H, Hossain MM, Lanier ER, Shaw LM, Back D. The addition of mycophenolate mofetil to antiretroviral therapy including abacavir is associated with depletion of intracellular deoxyguanosine triphosphate and a decrease in plasma HIV-1 RNA. Journal of acquired immune deficiency syndromes. 2002;31:45–49. doi: 10.1097/00126334-200209010-00006. [DOI] [PubMed] [Google Scholar]
- Matano T, Shibata R, Siemon C, Connors M, Lane HC, Martin MA. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. Journal of virology. 1998;72:164–169. doi: 10.1128/jvi.72.1.164-169.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael AJ, Phillips RE. Escape of human immunodeficiency virus from immune control. Annual review of immunology. 1997;15:271–296. doi: 10.1146/annurev.immunol.15.1.271. [DOI] [PubMed] [Google Scholar]
- Metzner KJ, Jin X, Lee FV, Gettie A, Bauer DE, Di Mascio M, Perelson AS, Marx PA, Ho DD, Kostrikis LG, Connor RI. Effects of in vivo CD8(+) T cell depletion on virus replication in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. The Journal of experimental medicine. 2000;191:1921–1931. doi: 10.1084/jem.191.11.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migueles SA, Osborne CM, Royce C, Compton AA, Joshi RP, Weeks KA, Rood JE, Berkley AM, Sacha JB, Cogliano-Shutta NA, et al. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity. 2008;29:1009–1021. doi: 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoye A, Park H, Rohankhedkar M, Coyne-Johnson L, Lum R, Walker JM, Planer SL, Legasse AW, Sylwester AW, Piatak M, Jr., et al. Profound CD4+/CCR5+ T cell expansion is induced by CD8+ lymphocyte depletion but does not account for accelerated SIV pathogenesis. The Journal of experimental medicine. 2009;206:1575–1588. doi: 10.1084/jem.20090356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris-Pertusa A, Lopez M, Rallon NI, Restrepo C, Soriano V, Benito JM. Evolution of the functional profile of HIV-specific CD8+ T cells in patients with different progression of HIV infection over 4 years. Journal of acquired immune deficiency syndromes. 2010;55:29–38. doi: 10.1097/QAI.0b013e3181e69609. [DOI] [PubMed] [Google Scholar]
- Reimann KA, Tenner-Racz K, Racz P, Montefiori DC, Yasutomi Y, Lin W, Ransil BJ, Letvin NL. Immunopathogenic events in acute infection of rhesus monkeys with simian immunodeficiency virus of macaques. Journal of virology. 1994;68:2362–2370. doi: 10.1128/jvi.68.4.2362-2370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy J, Paquette JS, Fortin JF, Tremblay MJ. The immunosuppressant rapamycin represses human immunodeficiency virus type 1 replication. Antimicrobial agents and chemotherapy. 2002;46:3447–3455. doi: 10.1128/AAC.46.11.3447-3455.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller T, Ocwieja KE, Rasaiyaah J, Price AJ, Brady TL, Roth SL, Hue S, Fletcher AJ, Lee K, KewalRamani VN, et al. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS pathogens. 2011;7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999a;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- Schmitz JE, Simon MA, Kuroda MJ, Lifton MA, Ollert MW, Vogel CW, Racz P, Tenner-Racz K, Scallon BJ, Dalesandro M, et al. A nonhuman primate model for the selective elimination of CD8+ lymphocytes using a mouse-human chimeric monoclonal antibody. The American journal of pathology. 1999b;154:1923–1932. doi: 10.1016/S0002-9440(10)65450-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley J, Turkbey B, Bernardo ML, Del Prete GQ, Estes JD, Griffiths GL, Kobayashi H, Choyke PL, Lifson JD, Keele BF. Tracking the luminal exposure and lymphatic drainage pathways of intravaginal and intrarectal inocula used in nonhuman primate models of HIV transmission. PloS one. 2014;9:e92830. doi: 10.1371/journal.pone.0092830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Kilgore KM, Kasturi SP, Pulendran B, Hunter E, Amara RR, Derdeyn CA. Signatures in Simian Immunodeficiency Virus SIVsmE660 Envelope gp120 Are Associated with Mucosal Transmission but Not Vaccination Breakthrough in Rhesus Macaques. Journal of virology. 2016;90:1880–1887. doi: 10.1128/JVI.02711-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolskaja E, Olivari S, Zufferey M, Strambio-De-Castillia C, Pizzato M, Luban J. Cyclosporine blocks incorporation of HIV-1 envelope glycoprotein into virions. Journal of virology. 2010;84:4851–4855. doi: 10.1128/JVI.01699-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streblow DN, Kitabwalla M, Malkovsky M, Pauza CD. Cyclophilin a modulates processing of human immunodeficiency virus type 1 p55Gag: mechanism for antiviral effects of cyclosporin A. Virology. 1998;245:197–202. doi: 10.1006/viro.1998.9155. [DOI] [PubMed] [Google Scholar]
- Taaffe J, Chahroudi A, Engram J, Sumpter B, Meeker T, Ratcliffe S, Paiardini M, Else J, Silvestri G. A five-year longitudinal analysis of sooty mangabeys naturally infected with simian immunodeficiency virus reveals a slow but progressive decline in CD4+ T-cell count whose magnitude is not predicted by viral load or immune activation. Journal of virology. 2010;84:5476–5484. doi: 10.1128/JVI.00039-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rompay KK, Singh RP, Pahar B, Sodora DL, Wingfield C, Lawson JR, Marthas ML, Bischofberger N. CD8+-cell-mediated suppression of virulent simian immunodeficiency virus during tenofovir treatment. Journal of virology. 2004;78:5324–5337. doi: 10.1128/JVI.78.10.5324-5337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderford TH, Bleckwehl C, Engram JC, Dunham RM, Klatt NR, Feinberg MB, Garber DA, Betts MR, Silvestri G. Viral CTL escape mutants are generated in lymph nodes and subsequently become fixed in plasma and rectal mucosa during acute SIV infection of macaques. PLoS pathogens. 2011;7:e1002048. doi: 10.1371/journal.ppat.1002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Stockenstrom S, Odevall L, Lee E, Sinclair E, Bacchetti P, Killian M, Epling L, Shao W, Hoh R, Ho T, et al. Longitudinal Genetic Characterization Reveals That Cell Proliferation Maintains a Persistent HIV Type 1 DNA Pool During Effective HIV Therapy. The Journal of infectious diseases. 2015;212:596–607. doi: 10.1093/infdis/jiv092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner TA, McLaughlin S, Garg K, Cheung CY, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345:570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CM, Moody DJ, Stites DP, Levy JA. CD8+ lymphocytes can control HIV infection in vitro by suppressing virus replication. Science. 1986;234:1563–1566. doi: 10.1126/science.2431484. [DOI] [PubMed] [Google Scholar]
- Wong JK, Strain MC, Porrata R, Reay E, Sankaran-Walters S, Ignacio CC, Russell T, Pillai SK, Looney DJ, Dandekar S. In vivo CD8+ T-cell suppression of siv viremia is not mediated by CTL clearance of productively infected cells. PLoS pathogens. 2010;6:e1000748. doi: 10.1371/journal.ppat.1000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasutomi Y, Reimann KA, Lord CI, Miller MD, Letvin NL. Simian immunodeficiency virus-specific CD8+ lymphocyte response in acutely infected rhesus monkeys. Journal of virology. 1993;67:1707–1711. doi: 10.1128/jvi.67.3.1707-1711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.