Abstract
In craniomaxillofacial (CMF) surgery, a reliable way of simulating the soft tissue deformation resulted from skeletal reconstruction is vitally important for preventing the risks of facial distortion postoperatively. However, it is difficult to simulate the soft tissue behaviors affected by different types of CMF surgery. This study presents an integrated bio-mechanical and statistical learning model to improve accuracy and reliability of predictions on soft facial tissue behavior. The Rubin-Bodner (RB) model is initially used to describe the biomechanical behavior of the soft facial tissue. Subsequently, a finite element model (FEM) computers the stress of each node in soft facial tissue mesh data resulted from bone displacement. Next, the Generalized Regression Neural Network (GRNN) method is implemented to obtain the relationship between the facial soft tissue deformation and the stress distribution corresponding to different CMF surgical types and to improve evaluation of elastic parameters included in the RB model. Therefore, the soft facial tissue deformation can be predicted by biomechanical properties and statistical model. Leave-one-out cross-validation is used on eleven patients. As a result, the average prediction error of our model (0.7035mm) is lower than those resulting from other approaches. It also demonstrates that the more accurate bio-mechanical information the model has, the better prediction performance it could achieve.
Keywords: Craniomaxillofacial surgery, soft facial tissue, Rubin-Bodner model, stress distribution, generalized regression neural network
1. Introduction
The goal of craniomaxillofacial (CMF) surgery [1, 2] is to reconstruct a normal facial appearance and function of patients suffering from CMF deformities. A predicting system capable of accurate simulation of soft-tissue changes after skeletal reconstruction has profound utilitarian value in orthodontics, plastic/general surgery, growth/aging prediction, and forensic science [3–5]. Currently we are able to accurately plan the osteotomies based on bone surgeries, whereas incapable to provide precise prediction of facial soft tissue deformation following the virtual osteotomy despite of multiple attempts for years. It is an urgent demand, from both doctors and patients, to develop a planning system that can accurately simulate soft-tissue changes after skeletal reconstruction.
Up to this point, various approaches of soft tissue deformation prediction have been developed. There are three most popular approaches, namely, mass spring model (MSM) [6], finite element model (FEM) [7], and mass tensor model (MTM) [8–10]. MSM [6] with easy architecture and low memory usage makes it very attractive for fast simulations. However, MSM lacks bio-mechanical relevance and clinical accuracy. FEM [7] divides the whole soft-tissue volume into massive geometrically discrete volumes and assigns material properties to them. One of its most obvious demerits is computation cost. MTM [8, 9] can be considered a hybrid of MSM and linear-FEM (LFEM) using homogenous tissue property. It was reported fast computation time and acceptable accuracy [10].
The major disadvantage of these methods is that they are individual-based, without considering population-based statistical information. Furthermore, many factors during CMF surgery contribute to the complicated response of living tissue, because the material behavior of living tissue is nonlinear, time dependent and anisotropic. In order to describe material behavior of soft facial tissue, Rubin and Bodner [11] developed nonlinear three dimensional constitutive equation for facial tissues valid for arbitrary deformations and achieved reasonable agreement with the experimental data of Har-Shai et al [12]. In the Rubin-Bodner model (RB model), the tissue is modeled as a composite material with a fully elastic component and a dissipative component containing both elastic and viscous elements. However, the elastic component also affected by specific CMF surgery was not considered in the original RB model. We modified the function of RB model to model the reduction in elastic stiffness caused by distortional deformations of the elastic component. Due to this modification, the elastic component of the present model is referred to as a “reductively elastic” component rather than a “fully elastic” component, which reflects soft tissue behaviors after specific CMF surgery. Therefore, it is necessary to improve the RB model to achieve better accuracy in simulations related to CMF surgery.
The object of our work is to develop a statistical model describing the relationship between the biomechanical features and the soft tissue deformations with specific CMF surgical types. In this study, we firstly classify patients into two groups: single surgery on mandible and mixed surgery on both mandible and maxilla, using an anatomic detailed template to map the detailed anatomic structures of soft tissue to each patient on both pre-operative and post-operative data. Secondly, finite element model (FEM) with RB model is applied to calculate the stress of each node in soft facial tissue mesh data resulted from bone displacement. Thirdly, a statistical model using Generalized Regression Neural Network (GRNN) [13] is implemented to obtain the relationship between the facial soft tissue deformation and the stress feature based on pre and post-operative data respectively. Subsequently, the elastic relevant parameters of the RB model are adaptively optimized by minimizing the difference between predictions and post-operative data with specific CMF surgery. Finally, cross-validation for the prediction of facial soft tissue deformation model is conducted by comparing the performance of different methods.
This paper completes our conference paper [14] by including more details of our methods, additional experiments and analysis.
2. Methods
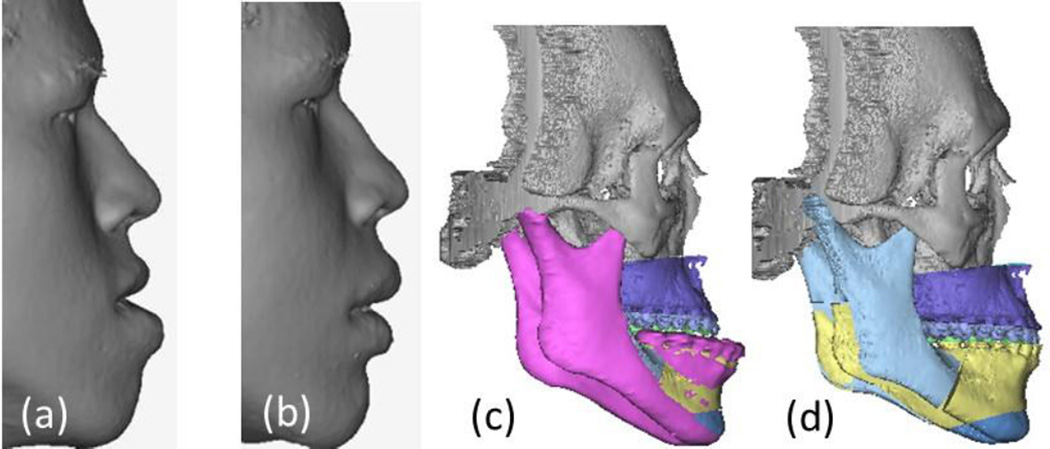
This retrospective study was approved by the Institutional Review Board of Wake Forest Baptist Medical Center (IRB00028345). It is based on existing image data for CMF surgery. Following data collection, patient identifying information were destroyed, consistent with data validation and study design, producing an anonymous analytical data set. We have eleven sets of patient’s pre-operative and post-operative CT data and 3D facial surface scans from a 3D surface camera. By using facial surface scans, we can prevent any unintended soft tissue strain during the CT scanning. The 3D camera was operated by a doctor while ensuring the patient’s facial expression was neutral. Both the preoperative and postoperative surface scans were rigidly registered to the preoperative CT images with the Mimics software (Materialise, Belgium). The facial surface in CT images was replaced with the surface scanned by 3D camera which provided a more faithful facial surface. The mandible which would open in CT scanning was moved to be closed since the mouth was close in 3D surface scanning. Muscles which connect the bones would be also moved accordingly. The bones of preoperative and postoperative CT images were also segmented in Mimics which would be further used to determine surgical plan. Fig. 1(a)–(b) shows the preoperative and postoperative surface scans of a patient. Fig. 1(c)–(d) illustrates the skeletal reconstruction in CMF surgery. To reduce the prominence of his chin and improve the overall appearance, the patient underwent a surgery to setback the mandible (bilateral sagittal split osteotomies) and advance the maxilla (Le Fort I osteotomy). The postoperative surface scan was acquired six months thereafter to calculate the facial soft tissue deformation.
Fig. 1.
(a) The preoperative surface scan. (b) The postoperative surface scan. (c) Skull structure before surgery. (d) Skull structure after surgery. Note that for this patient, all the bone parts were repositioned after CMF surgery except the gray one.
2. 1. Anatomic Detailed Template Generation
We consider the following muscles that contributed in facial soft tissue deformation: Buccinator, Depressor anguli oris, Depressor labii, Levator anguli oris, Levator labii, Levator labii alaeque nasi, Mentalis, Orbicularis oris, Zygomaticus major, Zygomaticus minor and Masseter [15]. Since it is difficult to segment these muscles from patients’ CT data, and manual segmentation of each patient is extremely labor intensive, we propose a method to locate the muscles for each patient by an anatomic detailed template.
The National Library of Medicine (NLM) Visible Female Data was used to generate an anatomic detailed template. The reasons for using NLM’s Visible Female data are that Female data has much higher resolution than Male data (2048×1216 pixels, 0.33mm interval and 24-bit color) [16], and there is no difference in facial anatomy between male and female [17–19]. However, if the model is anatomically over detailed, the combination of material models for each tissue and the sheer number of anatomical structures can be overwhelming for a detailed parameter study. Therefore, in this aim, we will further detail the facial tissues into 2 categories: muscle and the remaining soft tissues. The latter category is composed of soft tissue between skin and mucosa, grouped as homogenous material. This bifurcated soft tissue model is the best possible balance between maintaining the anatomical details related to facial tissue changes, and conserving computational cost [20].
Fig. 2 demonstrates different angles of the anatomic detailed template. In the next stage, we use this template to map the detailed anatomic structures of soft tissue to each individual patient.
Fig. 2.
Anatomic template from the Visible Human Female Dataset.
2. 2. Mapping by Anatomic Detailed Template
Once the template was generated, the detailed anatomic structures were then mapped from this model to each individual patient. In Visible Female Dataset, we find the location of each muscle in the mesh and identify the elements containing these muscles. Assuming the muscles are contained in the same elements for each patient, we map the muscles from Visible Human to the patients. It is noteworthy that the shapes of muscles vary for each patient when fitting the template to the facial geometries with defined anatomic cephalometric landmarks [21]. Landmarks are manually defined on the 3D surface to capture the facial shape mostly located at lips and nose. The 3D surface scan of the patient is imported into TrueGrid (XYZ Scientific Applications, Inc., Livermore, CA) as facial geometries. Then a surface projection technique of TrueGrid is applied on projecting the anatomic soft tissue structures to the 3D surface by matching the landmarks. It keeps both preoperative and postoperative data the same in number of finite elements with the mesh nodes. A natural correspondence between different mesh nodes was established for statistical analysis on soft tissue deformation prediction. It includes 48396 hexahedral elements (each element has eight mesh nodes) in Fig. 3. The right-most subfigure shows the shape of one element in the mesh object. To limit the number of elements and reduce the computational complexity, we restrict the calculation area to the zone below the nose (cut the nose tip) assuming the area above nose unaltered after surgery.
Fig. 3.
Hexahedral mesh elements of template.
2. 3. Determination of Displacement Boundary Condition
The mesh nodes can be classified into two categories: the boundary nodes and the free nodes. Boundary nodes located in skull parts would be repositioned during surgery. Remaining parts are free nodes subject to the displacement of boundary nodes.
The displacement boundary condition, consisting of the displacements of all the boundary nodes, can be determined from the paired preoperative and postoperative skulls by Iterative Closest Point (ICP) algorithm [22, 23]. The post-operative skull was preliminarily manually registered to the pre-operative one based on an unaltered part (usually the part above nose) in Mimics. Afterwards, the pre-operative skull was cut into parts according to the post-operative one. Then, the skull parts were separately matched by manual alignment to the post-operative counterparts. The displacements of skull parts were exported as STL (STereoLithography) files which were subsequently imported into Matlab (The MathWorks, Inc., Natick, Massachusetts). In order to obtain the displacement boundary condition, it is necessary to compute all boundary nodes in aforesaid STL files. The ICP algorithm was conducted to calculate rotation transformation R and translation transformation t between the pre-operative and post-operative skull parts. Assuming the coordinate of a boundary node as b, and its displacement as u, the displacement of this node was calculated as follows: u = (R·bpre + t) − bpost. Fig. 1 (c)–(d) shows the skeletal reconstruction in osteotomies.
Determination of displacement boundary conditions entails some manual operations to match different regions of the model. The same occurs for the matching of surface scans with visual template. The postoperative model was registered to the preoperative one at cranium, a surgically unaltered area. Afterwards, the preoperative model was virtually osteomized and bony segments were individually moved and matched to the postoperative counterparts. The errors entailed by the above operations were calculated as mean ± standard deviation (SD), which is 0.05 ±0.03 (mm).
2. 4. Improved Rubin-Bodner Model Corresponding to Different CMF Surgical Types
To establish a statistical model describing the relationship between the bio-mechanical features and the soft tissue deformations, an improved RB Model is developed to depict the biomechanical behavior of the soft facial tissue in the specific CMF surgery. Under the RB model, the tissue is simulated as a composite material with a fully elastic component and a dissipative component containing both elastic and viscous elements. Consequently it has accurate soft tissue bio-mechanical properties when processing viscoelastic materials such as facial soft tissue. Because the postoperative CTs will be taken six months after surgery to avoid surgical swelling, the short term soft tissue deformation is irrelevant. We only make use of the elastic terms in this work, ignoring the time dependent factors associated with the parts of the RB model which describe the transient and dissipative material behaviors.
RB Model [11] is characterized by a specific (per unit mass) strain energy function ρ. It is specified in the form
| (1) |
where κ0 is the mass density in the reference configuration, and ε0 and p represent material parameters. The function t characterizes the response of the elastic dilatation and distortion. The corresponding Cauchy stress tensor δ in the spatial description is given by
| (2) |
with E as the second order identity tensor, γ̅′ as the deviatoric component of the modified left Cauchy–Green deformation tensor and ε as shear modulus. S is a measure of dilatation (namely determination) of the matrix of deformation gradient of boundary nodes. The material parameters (κ0, p, ε0, η1, η2) are determined from literature [11, 24] authored by Rubin and Bodner. κ0 is the value of the mass density. p is an elastic hardening coefficient. ε0 is overall homogeneous scaling of the energy. η1 controls the isotropic bulk modulus, and η2 controls the isotropic shear modulus. Rubin and Bodner developed a nonlinear three-dimensional constitutive equation (δ (κ0, p, ε0, η1, η2)) to characterize elastic behavior of the biological tissues with these 5 parameters, where δ is the stress of the tissue given a particular dilatation. These parameters are obtained by minimizing the difference between the real stress measured by experimental data of Har-Shai et al[12] and the stress simulated by the δ(κ0, p, ε0, η1, η2) constitutive equation. ε = ε0ept. γ1 is a pure measure of the elastic distortion, given by
| (3) |
| (4) |
| (5) |
where H is the deformation gradient of boundary nodes, and γ̅ = S−2/3γ = S−2/3 HHT is the modified left Cauchy–Green deformation tensor. E is the second order identity tensor. γ̅′ is the deviatoric part of the modified left Cauchy–Green deformation tensor.
We use the parameter θ to modify the model by reducing the elastic stiffness caused by distortional deformations for two types of surgical plans. When θ=θ1, it refers to single surgery on mandible (MA, MB, MR, ML). When θ=θ2, it refers to mix surgery on both mandible and maxilla (MB+XB, MB+XA). Here, M indicates mandible, X indicates maxilla, A indicates skeleton shift forward, B indicates skeleton shift backward, R indicates skeleton shift right, L indicates skeleton shift left.
Therefore, we have 6 parameters in total to establish the improved RB model. Five parameters {κ0, p, ε0, η1, η2} are determined in RB model [11], and parameter θ is estimated from different surgical types with GRNN [13] respectively.
The GRNN based on kernel regression networks [25] is a variation of the radial basis neural networks. Without iterative training procedure as back propagation networks, GRNN approximates any arbitrary function between input and output vectors, drawing the function directly estimated from the training data. We use the GRNN to estimate the parameters θ for improving the RB model to describe the biomechanical behaviors of soft facial tissue with specific CMF surgery. We set cases 1,3,5,6,9,10 (in Table 1) as Group A corresponding θ=θ1, and cases 2,4,7,8,11 as Group B corresponding θ=θ2. The improved RB model algorithm is described below:
Initialize , where υ is the number of iteration, it always starts from 1 for each case.
Calculate , where is the mean of the post-operative soft tissue deformation di. Meanwhile RMSE is fed into a procedure which alters the parameter from time υ to (υ + 1).
Obtain , where, is prediction of soft tissue deformation, and is the post-operative soft tissue deformation, i = 1,2,3 …n. n indicates the number of cases included in Group A or Group B.
Get a template , n=1,2,…N, N denotes the numbers of mesh nodes from Visible Human Female Dataset. Use to do mapping process with Iterative Closest Point (ICP) algorithm to get skin-mesh-data of each case. For the ith case, pre-skin-point and post-skin-point are extracted from pre and post- operative CT data of each patient.
- An output estimate is calculated from GRNN. Build the GRNN by using all but one sample from the set. Designate βj(δ, η), calculated by the RB model, as the input vector of GRNN. The argument δ represents Cauchy stress tensor, while η is on behalf of the two material parameters η1 and η2 in Eq. (2). We designate di as training output vector of GRNN. Here i =1,2,…h−1, h stands for the case number in Group A or Group B. An output estimate yields to an input vector βj as below.
Where, . The optimal value of smoothing factor σ referring to the size of the neuron’s region is a vital parameter of GRNN. For optimization of the smoothing factor σ [26],the holdout method consists in removing one sample βj(δ, η) at a time and constructing the network base on all of the other samples . Then the network is used to estimate output for the removed sample. By repeating this process for each sample and storing each estimate, the root mean-squared error (RMSE) can be measured between the actual sample values di and the estimates . The optimal value of σ can be obtained by an approach hereinafter:(6) - Build the GRNN by using all but one sample from the training set of size n and assign an initial value for σ as 1.
- Apply the resulting GRNN to the holdout sample and then record the error. Repeat these procedure n times and compute the root mean squared error (RMSE). RMSE is a measure of the differences between values output by GRNN and the values actually observed, calculated as .
- Change the value of σ by a step length such as 0.01 and repeat the procedure. If the RMSE increases, change the value of σ in the other direction and repeat. If the RMSE decreases, increase the value of σ in the same direction. If σ changes only minutely like 0.005, select the current value.
Then repeat the procedure (1)–(5) q times to get the optimal until the RMSE reaches the minimum, such as RMSE ≤ 0.5.
By the same token, we get the optimal value of parameters θ2 for Group B.
Table 1.
Bmax (mm) is the maximal skull displacement during CMF surgery. ESDM, ELFEM, and ERBM mean the prediction difference of corresponding methods (Unit: mm). Surgical plans are described in the second column.
| Patient | Surgery | Bmax | ESDM | ELFEM | ERBM |
|---|---|---|---|---|---|
| 1 | MB | 11.6847 | 3.6352 | 1.3241 | 0.9484 |
| 2 | MB+XB | 6.1581 | 1.8381 | 0.8054 | 0.5632 |
| 3 | MB | 7.3624 | 5.0321 | 1.2456 | 0.7096 |
| 4 | MB+XA | 7.2343 | 2.9817 | 0.7389 | 0.6345 |
| 5 | MR | 8.3622 | 2.2543 | 0.8567 | 0.4521 |
| 6 | MA | 10.5866 | 3.3946 | 0.9391 | 0.7784 |
| 7 | MB+XB | 13.4578 | 3.8809 | 1.1490 | 0.7840 |
| 8 | MB+XA | 7.3898 | 4.5982 | 0.8845 | 0.4738 |
| 9 | ML | 10.5032 | 3.1190 | 0.9978 | 0.7914 |
| 10 | MR | 6.1608 | 2.1845 | 0.9531 | 0.8463 |
| 11 | MB+XA | 13.6617 | 3.3014 | 0.9821 | 0.7566 |
| Mean | 9.3238 | 3.2927 | 0.9888 | 0.7035 |
Currently we are able to precisely plan the osteotomies (bone surgeries) for new patients, the surgical plan could be determined from the paired preoperative skull and virtual postoperative skulls. According to the new patient’s virtual osteotomies, the elastic relevant parameters in the improved RB model can be chosen to provide the optimal results based on specific CMF surgical plan. Table 2 shows the material parameters assigned to the soft facial tissue in the improved RB model. Moreover, the improved RB Model is implemented into the commercial FEM software ABAQUS6.8 (SIMULIA, Providence, RI) to generate the stress as the bio-mechanical features to predict the soft facial tissue deformations.
Table 2.
Material Parameters of Improved Rubin-Bodner Model.
| κ0(g/cm3) | ε0(MPa) | p | η1 | η2 | θ1 | θ2 | |
|---|---|---|---|---|---|---|---|
| Muscles | 1 | 3.7 | 25 | 595 | 0.00216 | 0.95 | 0.9 |
| Remaining soft tissues |
1.1 | 1.7 | 36 | 1294 | 0.0008 | 0.95 | 0.9 |
3. Results
Leave-one-out cross-validation is implemented to assess the accuracy of our prediction. To be more specific, the validation opts out a single patient from the 11 patients as the test data, while regarding the remaining patients as training data to train the GRNN model. Then repeat aforesaid procedure until each patient has been treated as the test data for once.
We evaluate the performance based on the difference E between the prediction result and ground truth (post-operative data) of soft tissue deformation given by:
| (7) |
where Xk as the actual displacement of the kth node, as the predicted displacement of the kth node, and N denotes the numbers of mesh nodes for calculation. We selected 4530 nodes in total lying on the skin to compare actual displacements and finite element results.
Table 1 represents the prediction performance of different models namely, statistical deformation model (SDM) [27], linear FEM (LFEM) [10], and the improved Rubin-Bodner model (RBM) developed in this research.
4. Discussion
There is convincing evidence that RBM method provides more accurate prediction than any other methods in Table 1. Since the accuracy of SDM depends significantly on the quantity of sample data available, the insufficient sample size for training may result in low accuracy. By taking advantage of bio-mechanical information, the performance of LFEM has been substantially improved compared to SDM. However, both methods failed to consider the change of properties on facial soft tissues affected by specific CMF surgery. RBM approach significantly outperforms these algorithms. The reason is that RBM combines improved RB Model, statistical information, and individual bio-mechanical information. The results indicate that the more accurate bio-mechanical information the model has, the better prediction performance it could achieve.
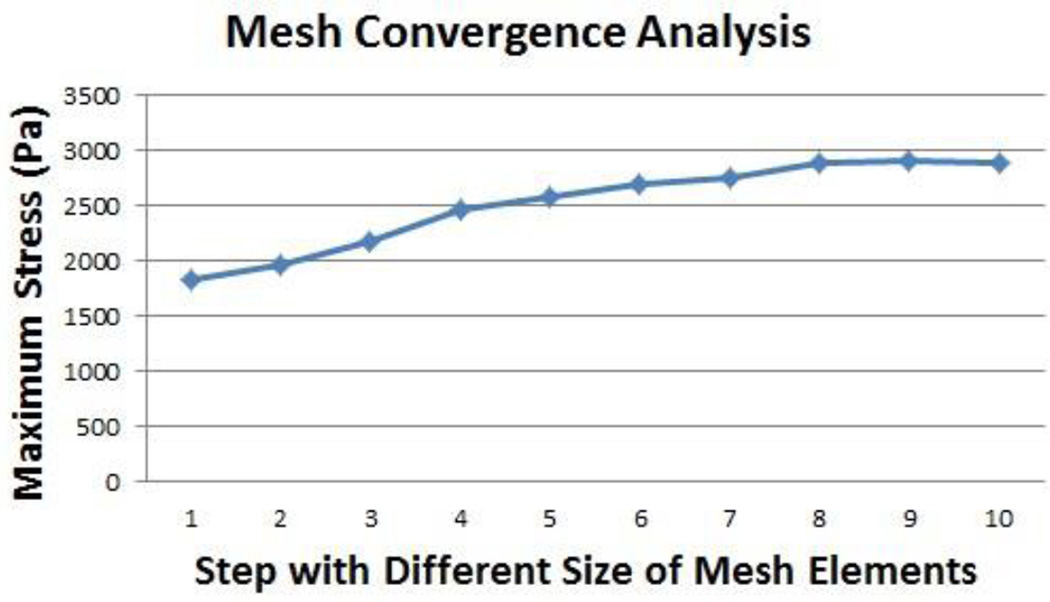
In finite element modeling, a finer mesh typically results in a more accurate solution for representing the 3D object. However, as a mesh is made finer, the computation time increases. Performing a convergence study [28] in maximum stress analysis refines the mesh and reduces the size of the elements, which will keep the fast computation times and acceptable accuracy. ABAQUS uses the maximum stress analysis to optimize the mesh quality by default. In this study, we conducted convergence analysis in ABAQUS to obtain mesh independent solutions. Mesh convergence was tested by increasing the size of the mesh elements in ten steps, from step 1 (3.4 mm element) to step 10 (1.6 mm element). The step size is 0.2 mm. We then selected the maximum stress of the whole model as the result for convergence test in the various mesh groups. The results of the convergence tests were shown in Fig. 4 with a mesh element of 2 mm (Step 8). Errors of all groups were within an accept range (<0.3%).
Fig. 4.
Mesh convergence analysis.
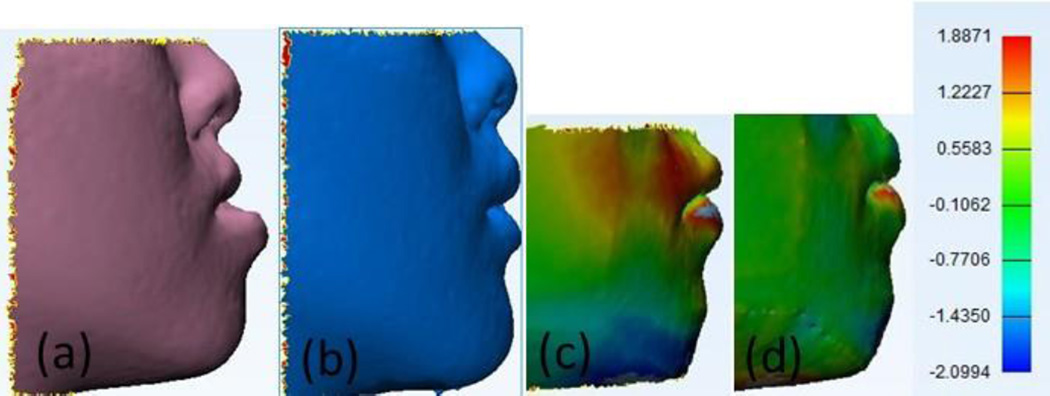
To make effective prediction of the surgery oriented in the appearance, the quality of prediction about facial soft tissue deformation is judged by the similarity to actual facial image after surgery. Because the 4th patient has acute profile and less fat, we take this paradigmatic case for visualization by using the Inverse Distance Weighted (IDW) Interpolation [29]. The actual preoperative and postoperative outlook of this patient, and the color maps of prediction errors by LFEM, RBM methods are illustrated in Fig. 5. In particular, RBM method in Fig. 5(d) is more faithful than LFEM method in Fig. 5(c) by comparing to the postoperative data. The red area stands for positive error and blue area means negative error. Finally the green area indicates negligible minor error. It is clearly demonstrated that RBM method is superior over LFEM with closer prediction to the postoperative image.
Fig. 5.
Prediction (No.4 patient in Table 2) using RBM method compare with LFEM method. (a) Pre-operative. (b) Post-operative. (c) LFEM. (d) RBM.
5. Conclusions
The challenging problem in craniomaxillofacial surgery is that we do not have a reliable way of simulating the soft tissue-change resulted from skeletal reconstruction. In current practice, doctors only plan osteotomies (improving hard tissue functions) and hope for the best for optimal facial soft tissue. In order to accurately simulate soft tissue changes following the virtual osteotomies, this study developed a reliable and novel systematic approach to predict soft facial tissue deformation; an integrated bio-mechanical and statistical learning model was proposed to meet the prediction accuracy prior to the CMF surgery. The key contribution of this approach includes (1) improving the Rubin-Bodner model as to better simulate soft tissue behavior by adaptively optimizing the elastic relevant parameters in its function for different surgical plans; (2) generating a new mesh data of each patient’s pre & postoperative faces by mapping from the anatomic detailed template; (3) constructing a 3D FEM of the soft facial tissue to extract biomechanical stress information based on bone displacement calculated from pre-operative and post-operative CT data; and (4) establishing the GRNN method to characterize the relationship between the biomechanical stress feature and the facial soft tissue deformation. Leave-one-out Cross-validation on all patients demonstrated the effectiveness and efficiency of our robust model. This model can achieve significantly high accuracy in soft facial tissue deformations prediction prior to clinical surgery to prevent the risks of facial distortion after CMF surgery.
Highlights.
RB model is improved to better simulate soft tissue behavior for CMF surgery.
Mesh data of each patient is generated by mapping an anatomic detailed template.
3D FE analysis extracts biomechanical information from bone displacement.
GRNN relates the biomechanical information and changes experienced by tissues.
Acknowledgments
This work is funded by The United States NIH/NIDCR grant R01DE021863 (Xia & Zhou).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
None declared.
Ethical Approval
This retrospective study was approved by the Institutional Review Board of Wake Forest Baptist Medical Center (IRB00028345).
References
- 1.Keeve E, Girod S, Kikinis R, Girod B. Deformable modeling of facial tissue for craniofacial surgery simulation. Comput Aided Surg. 1998;3:228–238. doi: 10.1002/(SICI)1097-0150(1998)3:5<228::AID-IGS2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 2.Kim H, Jurgens P, Weber S, Nolte LP, Reyes M. A new soft-tissue simulation strategy for cranio-maxillofacial surgery using facial muscle template model. Prog Biophys Mol Biol. 2010;103:284–291. doi: 10.1016/j.pbiomolbio.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Chatzis V, Pitas I. Interpolation of 3-D binary images based on morphological skeletonization. IEEE Trans Med Imaging. 2000;19:699–710. doi: 10.1109/42.875192. [DOI] [PubMed] [Google Scholar]
- 4.Stoyanov D, Yang GZ. Soft tissue deformation tracking for robotic assisted minimally invasive surgery. Conf Proc IEEE Eng Med Biol Soc. 2009;2009:254–257. doi: 10.1109/IEMBS.2009.5334010. [DOI] [PubMed] [Google Scholar]
- 5.Weichert F, Schroder A, Landes C, Shamaa A, Awad SK, Walczak L, et al. Computation of a finite element-conformal tetrahedral mesh approximation for simulated soft tissue deformation using a deformable surface model. Med Biol Eng Comput. 2010;48:597–610. doi: 10.1007/s11517-010-0607-0. [DOI] [PubMed] [Google Scholar]
- 6.Mollemans W, Schutyser F, Van Cleynenbreugel J, Suetens P. Tetrahedral mass spring model for fast soft tissue deformation. Surgery Simulation and Soft Tissue Modeling, Lect Notes Comput Sc. 2003;2673:145–154. [Google Scholar]
- 7.Joldes GR, Wittek A, Miller K. Suite of finite element algorithms for accurate computation of soft tissue deformation for surgical simulation. Med Image Anal. 2009;13:912–919. doi: 10.1016/j.media.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotin S, Delingette H, Ayache N. A hybrid elastic model for real-time cutting, deformations, and force feedback for surgery training and simulation. Visual Comput. 2000;16:437–452. [Google Scholar]
- 9.Schwartz JM, Denninger M, Rancourt D, Moisan C, Laurendeau D. Modelling liver tissue properties using a non-linear visco-elastic model for surgery simulation. Med Image Anal. 2005;9:103–112. doi: 10.1016/j.media.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Mollemans W, Schutyser F, Nadjmi N, Maes F, Suetens P. Predicting soft tissue deformations for a maxillofacial surgery planning system: From computational strategies to a complete clinical validation. Med Image Anal. 2007;11:282–301. doi: 10.1016/j.media.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Rubin MB, Bodner SR. A three-dimensional nonlinear model for dissipative response of soft tissue. Int J Solids Struct. 2002;39:5081–5099. [Google Scholar]
- 12.Har-Shai Y, Bodner SR, Egozy-Golan D, Lindenbaum ES, Ben-Izhak O, Mitz V, et al. Mechanical properties and microstructure of the superficial musculoaponeurotic system. Plast Reconstr Surg. 1996;98:59–70. doi: 10.1097/00006534-199607000-00009. discussion 1–3. [DOI] [PubMed] [Google Scholar]
- 13.Li CF, Bovik AC, Wu XJ. Blind Image Quality Assessment Using a General Regression Neural Network. IEEE Trans Neural Networks. 2011;22:793–799. doi: 10.1109/TNN.2011.2120620. [DOI] [PubMed] [Google Scholar]
- 14.Zhang G, Xia JJ, Zhang X, Zhou X. Prediction of facial soft tissue deformations with improved Rubin-Bodner model after craniomaxillofacial (CMF) surgery. Proceedings of the 2015 IEEE International Conference on Image Processing (ICIP) 2015;2015:2796–2800. [Google Scholar]
- 15.Schünke M, Schulte E, Schumacher U, Voll M, Wesker K. Prometheus Lernatlas der Anatomie. Georg Thieme Verlag; 2009. [Google Scholar]
- 16.Kim J, Kang DI, Soh KS, Kim S. Analysis on postmortem tissues at acupuncture points in the image datasets of Visible Human Project. J Altern Complement Med. 2012;18:120–129. doi: 10.1089/acm.2010.0850. [DOI] [PubMed] [Google Scholar]
- 17.Netter FH. Atlas of human anatomy. 5th. Philadelphia, PA: Elsevier; 2010. [Google Scholar]
- 18.Diogo R, Wood B. Soft-tissue anatomy of the primates: phylogenetic analyses based on the muscles of the head, neck, pectoral region and upper limb, with notes on the evolution of these muscles. J Anat. 2011;219:273–359. doi: 10.1111/j.1469-7580.2011.01403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kau CH, Richmond S. Three-dimensional imaging for orthodontics and maxillofacial surgery. Chichester, West Sussex, UK: Ames, Iowa: Wiley-Blackwell; 2010. [Google Scholar]
- 20.Barbarino GG, Jabareen M, Trzewik J, Nkengne A, Stamatas G, Mazza E. Development and validation of a three-dimensional finite element model of the face. J Biomech Eng. 2009;131:041006. doi: 10.1115/1.3049857. [DOI] [PubMed] [Google Scholar]
- 21.Proffit WR, Fields HW, Sarver DM. Contemporary Orthodontics. 4th. St. Louis, Mo: Mosby Elsevier; 2007. [Google Scholar]
- 22.De Groeve P, Schutyser F, Van Cleynenbreugel J, Suetens P. Registration of 3D photographs with spiral CT images for soft tissue simulation in maxillofacial surgery. Springer; 2001. pp. 991–996. [Google Scholar]
- 23.Almhdie A, Leger C, Deriche M, Ledee R. 3D registration using a new implementation of the ICP algorithm based on a comprehensive lookup matrix: Application to medical imaging. Pattern Recogn Lett. 2007;28:1523–1533. [Google Scholar]
- 24.Rubin MB, Bodner SR. Modeling nonlinear dissipative response of biological tissues. Int J Solids Struct. 2004;41:1739–1740. [Google Scholar]
- 25.Bishop CM. Pattern Recognition and Machine Learning. New York: Springer; 2006. [Google Scholar]
- 26.Specht DF. A General Regression Neural Network. IEEE Trans Neural Networks. 1991;2:568–576. doi: 10.1109/72.97934. [DOI] [PubMed] [Google Scholar]
- 27.Meller S, Nkenke E, Kalender WA. Statistical face models for the prediction of soft-tissue deformations after orthognathic osteotomies. Proceedings of Medical Image Computing and Computer-Assisted Intervention, Lect Notes Comput Sc. 2005;3750:443–450. doi: 10.1007/11566489_55. [DOI] [PubMed] [Google Scholar]
- 28.Zienkiewicz OC, Taylor RL, Zhu JZ. The finite element method : its basis and fundamentals. Seventh. Butterworth-Heinemann; 2005. [Google Scholar]
- 29.Watson DF, Philip GM. A refinement of inverse distance weighted interpolation. Geo-Processing. 1985;2:315–327. [Google Scholar]