Abstract
Testis morphogenesis is a highly orchestrated process involving lineage determination of male germ cells and somatic cell types. Although the origin and differentiation of germ cells are known, the developmental course specific for each somatic cell lineage has not been clearly defined. Here, we construct a comprehensive map of somatic cell lineage progression in the mouse testis. Both supporting and interstitial cell lineages arise from WT1+ somatic progenitor pools in the gonadal primordium. A subpopulation of WT1+ progenitor cells acquire SOX9 expression and become Sertoli cells that form testis cords, whereas the remaining WT1+ cells contribute to progenitor cells in the testis interstitium. Interstitial progenitor cells diversify through the acquisition of HES1, an indication of Notch activation, at the onset of sex determination. HES1+ interstitial progenitors, through the action of Sertoli cell-derived Hedgehog signals, become positive for GLI1. The GLI1+ interstitial cells eventually develop into two cell lineages: steroid-producing fetal Leydig cells and non-steroidogenic cells. The fetal Leydig cell population is restricted by Notch2 signaling from the neighboring somatic cells. The non-steroidogenic progenitor cells retain their undifferentiated state during fetal stage and become adult Leydig cells in post-pubertal testis. These results provide the first lineage progression map that illustrates the sequential establishment of somatic cell populations during testis morphogenesis.
KEY WORDS: Testis, Lineage specification, Sertoli cells, Leydig cells, Notch, Hedgehog
Highlighted Article: Somatic progenitor cell populations in the mouse testis are defined by progressive lineage-specific acquirement of WT1, HES1, SOX9 and GLI1 beginning at the time of sex determination.
INTRODUCTION
Morphogenetic transformation of gonadal primordium into a testis begins around embryonic day (E) 11.5 and 12.5 in mice (Brennan and Capel, 2004). Before this period, gonadal primordium is morphologically indistinguishable between XX and XY embryos (Brennan and Capel, 2004; DeFalco and Capel, 2009). Around E11.5, testis morphogenesis is initiated by the Y chromosome-linked Sry gene (Gubbay et al., 1990; Hawkins et al., 1992; Koopman et al., 1991; Lovell-Badge and Robertson, 1990), which is expressed in the supporting cell lineage Sertoli cells of the XY gonads (Albrecht and Eicher, 2001; Schmahl et al., 2000). SRY induces the differentiation of Sertoli cells through a positive-feedback loop between SOX9 and FGF9 (Chaboissier et al., 2004; Kim et al., 2006; Palmer and Burgoyne, 1991; Schmahl et al., 2004; Willerton et al., 2004). Sertoli cells then orchestrate formation of testis cords, a hallmark structure that separates Sertoli cells and germ cells from the interstitium (Brennan and Capel, 2004). The coelomic epithelium, which encloses the gonad and mesonephros, has been described as one source of Sertoli cells and interstitial cells (Brennan and Capel, 2004; Karl and Capel, 1998; Schmahl et al., 2000; Tanaka and Nishinakamura, 2014).
In contrast to Sertoli cells, which are a homogeneous population within testis cords, the cell types in the testis interstitium are diverse. The testis interstitium harbors the steroidogenic Leydig cells, peritubular myoid cells, macrophages, vasculature, and other uncharacterized cell types such as fibroblasts and vascular-associated cells (Brennan and Capel, 2004; DeFalco et al., 2014). In the mouse, steroidogenic Leydig cells consist of two populations based on the time of their appearance: fetal and adult Leydig cells (Benton et al., 1995; Huhtaniemi and Pelliniemi, 1992). Fetal Leydig cells serve as the primary source of androgens that virilize the embryos. The population of fetal Leydig cells declines after birth and is eventually replaced by the adult Leydig cells at puberty. Adult Leydig cells maintain androgen production throughout adulthood, functionally replacing fetal Leydig cells (Griswold and Behringer, 2009; Habert et al., 2001). Despite their similar functions in producing androgens, fetal and adult Leydig cells exhibit many differences in their transcriptomes (Dong et al., 2007; Shima et al., 2013), morphology (Haider, 2004) and regulation (Agelopoulou et al., 1984; Aubert et al., 1985; Baker and O'Shaughnessy, 2001; Dong et al., 2007; El-Gehani et al., 1998; Gangnerau and Picon, 1987; Ma et al., 2004; Majdic et al., 1998; O'Shaughnessy et al., 1998; Patsavoudi et al., 1985; Zhang et al., 2001). These differences between fetal and adult Leydig cells led to the hypothesis that the two Leydig cell populations are in fact distinct cell lineages arising from separate origins (Baker et al., 1999; Haider, 2004; Kerr and Knell, 1988; Lording and De Kretser, 1972; O'Shaughnessy et al., 2003; O'Shaughnessy and Fowler, 2011; Roosen-Runge and Anderson, 1959; Shima et al., 2013). In fact, multiple origins of fetal Leydig cells have been suggested, including Sf1+ non-steroidogenic interstitial cells originating from the gonadal primordium (Barsoum et al., 2013; Barsoum and Yao, 2010), mesonephros (Merchant-Larios and Moreno-Mendoza, 1998; Val et al., 2006), neural crest (Mayerhofer et al., 1996), coelomic epithelium (Karl and Capel, 1998), and cells residing in the border between gonad and mesonephros (DeFalco et al., 2011). By contrast, it has been suggested that adult Leydig cells stem from peritubular interstitial progenitor cells that arise in the adult testis (Davidoff et al., 2004; Ge et al., 2006; Li et al., 2016; Odeh et al., 2014; Stanley et al., 2012), or from COUP-TFII (Nr2f2)-positive interstitial cells in the fetal testis (Kilcoyne et al., 2014).
Although significant advancement has been made in identifying the molecular mechanisms underlying testis morphogenesis, how these molecular pathways integrate and the progenitor cells respond to them remain unclear. This deficiency in the field is largely attributed to a lack of clear definition of various cell types in the testis. In this study, we determined the lineage progression of somatic progenitor cells in the mouse testis by genetic lineage-tracing experiments in vivo. In addition, we explore the mechanisms of lineage assignment with a particular focus on the testis interstitium.
RESULTS
Wt1+ somatic cells are progenitors for both supporting and interstitial cells in the testis
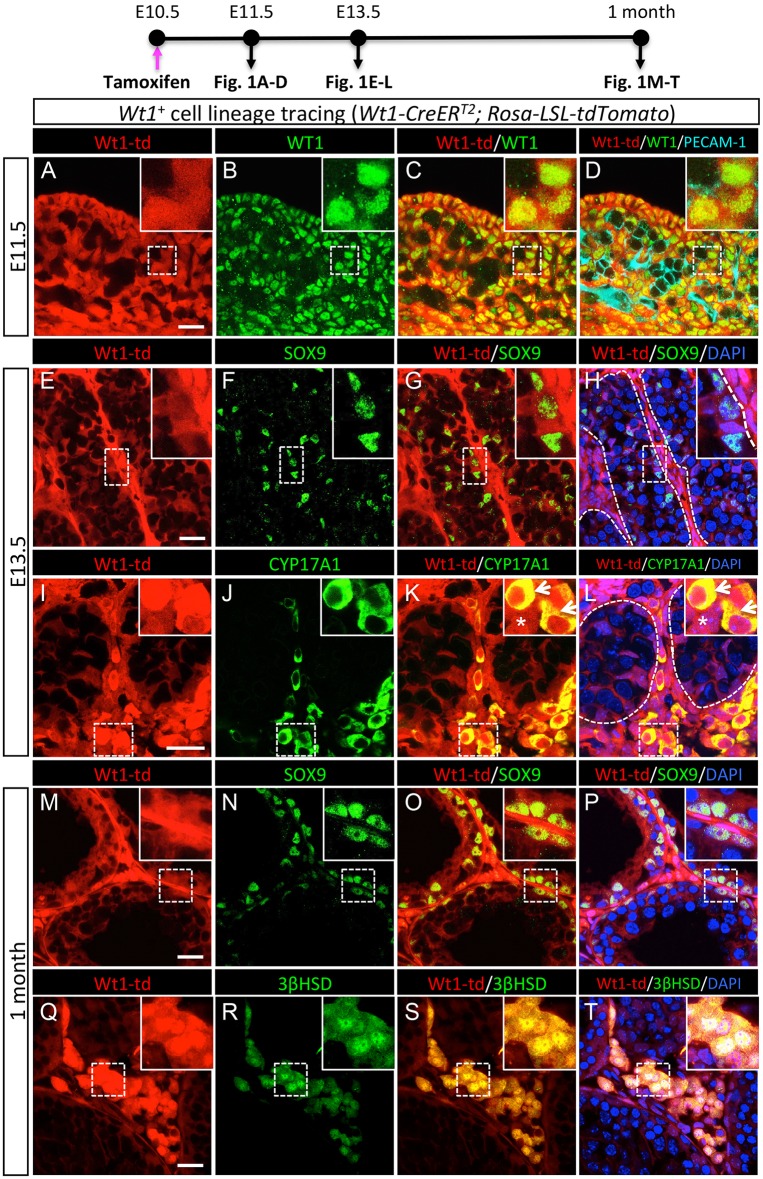
One of the earliest markers that define the somatic cell lineages in the gonads is Wilms' tumor 1 (WT1), a transcription factor essential for gonadogenesis (Armstrong et al., 1993; Kreidberg et al., 1993). We examined the expression of WT1 in the gonadal primordium by immunohistochemistry. At E11.5, when testis morphogenesis begins, endogenous WT1 protein was present in somatic cells but absent in the PECAM1-positive germ cells and endothelial cells of fetal testes (Fig. S1A-C). One day later, at E12.5, although its expression remained in coelomic epithelial cells, WT1 protein inside the testis became mostly restricted to AMH-positive Sertoli cells within the testis cords and its presence was significantly reduced in the interstitium surrounding testis cords (Fig. S1D-F). This pattern of WT1 expression, which is consistent with previous observations (Armstrong et al., 1993), implies that either WT1+ somatic progenitor cells in the gonadal primordium give rise only to Sertoli cells, or that WT1 expression is extinguished in the interstitial cell populations. To test these possibilities, we utilized a tamoxifen-induced Rosa-LSL-tdTomato lineage-tracing model, in which Wt1+ cells were labeled permanently with tdTomato fluorescent protein in the presence of tamoxifen (Liu et al., 2012). We administered a single tamoxifen injection to pregnant mice carrying Wt1-CreERT2; Rosa-LSL-tdTomato embryos at E10.5, before the onset of testis morphogenesis (Brennan and Capel, 2004; Eggers et al., 2014). The dose (1 mg/mouse) and frequency (one injection) of the tamoxifen treatment induced recombination for ∼24 h, so that all tdTomato-positive cells are derived specifically from the WT1+ cell population between E10.5 and E11.5 (Liu et al., 2015). At E11.5, or 24 h after tamoxifen treatment, the lineage-labeled Wt1+ cells were present in the somatic compartment of the fetal testis, colocalizing with the endogenous WT1 protein (Fig. 1A-D; Fig. S2A-D). At E13.5, the lineage-labeled Wt1+ cells became localized to not only the testis cords but also the interstitium (Fig. 1E,H,I). To identify what cell types the lineage-labeled Wt1+ cells become, we performed immunofluorescence for the Sertoli cell marker SOX9 and the Leydig cell marker CYP17A1. The lineage-labeled Wt1+cells within the testis cords were positive for SOX9, confirming their identity as Sertoli cells (Fig. 1E-H; Fig. S2E-H). By contrast, in the interstitium, the lineage-labeled Wt1+ cells gave rise to two subpopulations: CYP17A1+ steroidogenic fetal Leydig cells (Fig. 1K,L, arrows) and CYP17A1− non-steroidogenic interstitial cells (Fig. 1K,L, asterisks; Fig. S2I-L). This pattern of Wt1+ cell lineage contribution persisted in the postnatal testis: the linage-labeled Wt1+ cells derived from E10.5 fetal testis were located in both seminiferous tubules and the interstitial compartment delineated by laminin staining (Fig. S2M-T). In addition to becoming SOX9+ Sertoli cells within testis cords (Fig. 1M-P), lineage-labeled Wt1+ cells in the interstitium were also positive for 3βHSD, a marker for Leydig cells (Fig. 1Q-T). 3βHSD-positive adult Leydig cells all contained Wt1-tdTomato lineage labeling, strongly suggesting that E10.5 Wt1+ progenitor cells give rise to all steroidogenic cells, including adult Leydig cells. These results demonstrate that Wt1+ progenitor cells contribute to the supporting cell linage (Sertoli cells), the steroidogenic cell lineage (Leydig cells), and non-vasculature and non-steroidogenic interstitial cells in fetal and adult testes.
Fig. 1.
Lineage-tracing analysis of the Wt1+ cells in the fetal testis. (A-T) Lineage tracing of the fetal testis-derived Wt1+ cells in the Wt1-CreERT2; Rosa-LSL-tdTomato embryos was induced by tamoxifen administration at E10.5. The testes were analyzed at E11.5 (A-D), E13.5 (E-L) and 1 month of age (M-T) by fluorescence immunohistochemistry for Wt1-td (red; tdTomato-labeled fetal testis-derived Wt1+ cells), WT1 (green; endogenous protein), the germ cell marker PECAM1 (light blue), the Sertoli cell marker SOX9 (green), or the Leydig cell markers CYP17A1 or 3βHSD (green), and the nuclear counterstain DAPI (blue). The insets are higher magnifications of boxed areas in A-T. Arrows in the insets of K and L point to CYP17A1+ fetal Leydig cells. Asterisks in the insets of K and L indicate CYP17A1− non-steroidogenic cells. Dashed lines outline the testis cords. Scale bars: 25 μm.
Wt1+ progenitor cells give rise to Hes1+ cells that contribute to steroidogenic and non-steroidogenic interstitial cells in the testis
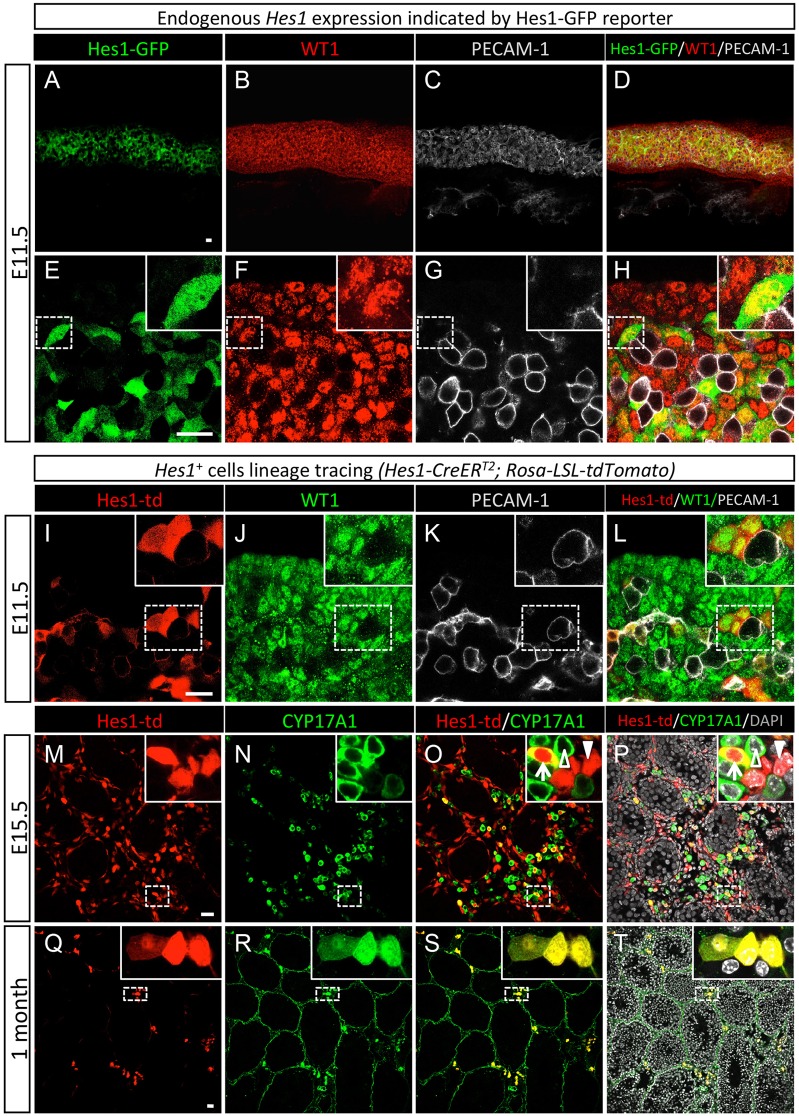
To define further the interstitial cell populations, we searched for factors that are involved in cell fate specification and maintenance. One such candidate is HES1, a downstream effector of Notch signaling (Kageyama et al., 2007) implicated in fetal Leydig cell differentiation (Tang et al., 2008). Hes1 mRNA expression is enriched in the interstitial cells in the differentiated fetal testis based on in situ hybridization (Tang et al., 2008) and sorted cell microarrays (Jameson et al., 2012). By analyzing fetal testes of Hes1-GFP reporter embryos, we uncovered that, as early as the onset of testis morphogenesis (E10.5-E11.5), Hes1-GFP expression (indicative of endogenous Hes1 expression) was already present in a subpopulation of Wt1+ somatic progenitor cells but absent in the PECAM1+ germ cell population (Fig. 2A-H; Fig. S3A-D). Co-immunostaining with the Sertoli cell marker SOX9 showed that Hes1-GFP was primarily located in the SOX9-negative somatic cells (Fig. S3E-H). In E12.5 and E16.5 testes, Hes1-GFP was predominantly expressed in the interstitium of the testis and located in a mainly CYP17A1− population (Fig. S3I-P). To investigate what Hes1+ cells at E10.5 become later in development, we used a lineage-labeling approach similar to that used in the Wt1+ model in Fig. 1. Instead of the Wt1+ cells, we lineage labeled the Hes1+ cells in Hes1-CreERT2; Rosa-LSL-tdTomato embryos at the onset of gonadal formation (E10.5) before the separation of testis cords and interstitium. One day after the lineage labeling at E11.5, we found that the lineage-marked Hes1+ cells represented a subpopulation of Wt1+ somatic progenitor cells, consistent with observations in the Hes1-GFP mouse model (Fig. 2I-L). At E15.5, we stained the lineage-labeled testes with the Leydig cell marker CYP17A1 and observed three cell populations in the interstitium (Fig. 2M-P; Fig. S4): (1) Hes1+/CYP17A1+ fetal Leydig cells (arrow in the inset); (2) Hes1−/CYP17A1+ fetal Leydig cells (empty arrowhead); and (3) Hes1+/CYP17A1− non-steroidogenic interstitial cells (filled arrowhead). Although some of the fetal Leydig cells originated from the Hes1+ progenitor cells (Hes1+/CYP17A1+), the majority of the fetal Leydig cells did not (Hes1−/CYP17A1+; Fig. 2M-P). This observation implies the presence of multiple sources for fetal Leydig cells. To rule out the possibility that the three cell populations in the interstitium are a consequence of insufficient lineage labeling, we enhanced the efficiency of lineage labeling of the Hes1+ cells by administering tamoxifen to embryos from E10.5 to E13.5 for four consecutive days (Fig. S5). Similar patterns of three interstitial cell populations were observed in the testis, confirming the presence of heterogeneous interstitial populations delineated by HES1 and CYP17A1. At one month of age, the Hes1+ cells derived from the E10.5 fetal testis were found in the interstitium and were positive for CYP17A1, indicating that these cells have become adult Leydig cells (Fig. 2Q-T).
Fig. 2.
Detection of Hes1 expression by the Hes1-GFP reporter and Hes1 lineage-tracing analysis. (A-H) Expression of Hes1-GFP in the E11.5 fetal testes was detected by fluorescence immunohistochemistry for GFP (Hes1+ cells), WT1 (red) and PECAM1 (gray), which marks germ cells and endothelial cells in low and high magnifications. (I-T) Lineage tracing of fetal testis-derived Hes1+ cells in the Hes1-CreERT2; Rosa-LSL-tdTomato embryos was induced by a single injection of tamoxifen at E10.5 (I-L), or at E10.5 and E11.5 for two consecutive days (M-T). The samples were examined at E11.5 (I-L), E15.5 (M-P) and 1 month of age (Q-T) for Hes1-td (red; tdTomato-labeled fetal testis-derived Hes1+ cells), WT1 (green), PECAM1 (gray), the Leydig cell marker CYP17A1 (green), and nuclear counterstain DAPI (gray). The insets are higher magnifications of the boxed areas. Scale bars: 25 μm. Arrows in insets of O and P point to Hes1+/CYP17A1+ fetal Leydig cells. Unfilled arrowheads in insets of O and P indicate Hes1−/CYP17A1+ fetal Leydig cells. Filled arrowheads in insets of O and P represent Hes1+/CYP17A1− non-steroidogenic interstitial cells.
Differentiation of steroidogenic cells in the interstitium is mediated by Notch2 signaling
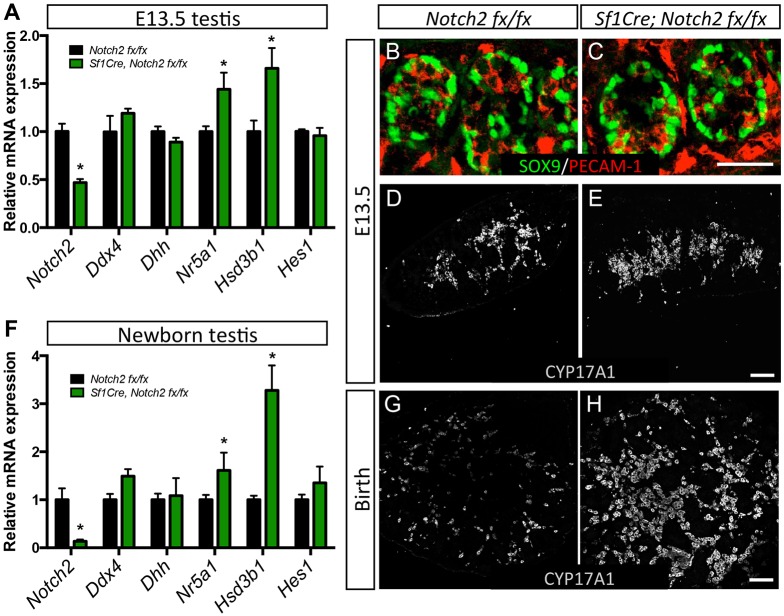
Steroidogenic fetal Leydig cell population is sensitive to the level of Notch activation: inhibition of general Notch signaling pathway with a γ-secretase inhibitor resulted in an increase in the number of fetal Leydig cells (Tang et al., 2008). Notch2, one of the Notch receptors, is a potential candidate for Notch activation based on its expression in the testis interstitium (Tang et al., 2008). We inactivated Notch2 in the testis using a gonadal somatic cell-specific Sf1-Cre (hereafter referred as Notch2 cKO). At E13.5, Sf1-Cre-mediated recombination decreased Notch2 expression to about 50% (Fig. 3A). The reduced Notch2 expression did not affect the development of Sertoli cells (Dhh expression in Fig. 3A and SOX9 immunolocalization in Fig. 3B,C) and germ cells (Ddx4 expression in Fig. 3A and PECAM1 immunolocalization in in Fig. 3B,C). However, the population of CYP17A1+ fetal Leydig cells was slightly expanded in the cKO testis compared with the controls (Fig. 3D,E; Fig. S6). Leydig cell-specific genes, such as Hsd3b1 and Nr5a1, were also significantly elevated (Fig. 3A). In the newborn testes, where expression of Notch2 was more effectively depleted (Fig. 3F), the number of CYP17A1+ fetal Leydig cells was greatly increased compared with that of control testes (Fig. 3G,H; Fig. S6), implicating Notch2 as the major Notch receptor involved in suppressing fetal Leydig cell differentiation.
Fig. 3.
Leydig cell phenotypes as a result of inactivation of Notch2 in Sf1-positive somatic cells. (A,F) Quantitative PCR analysis of gene expression for Notch2, Ddx4 (germ cell marker), Dhh (Sertoli cell marker), Nr5a1 and Hsd3b1 (Fetal Leydig cells) and Hes1 in control (n=5) and Notch2 cKO (n=3) E13.5 (A) and newborn (F) testes. *P<0.05. Student's t-test was used and values are presented as mean±s.e.m. (B-E,G,H) Immunofluorescence of SOX9 (green), PECAM1 (red) and CYP17A1 (gray) in control (Notch 2 fx/fx) and Sf1-Cre; Notch2 fx/fx or Notch2 cKO at E13.5 (B-E) and newborn (G,H) testes. Scale bars: 50 μm (B,C); 100 μm (D,E,G,H).
Hes1+ cells from the gonadal primordium become positive for Gli1 as a result of Hedgehog activation
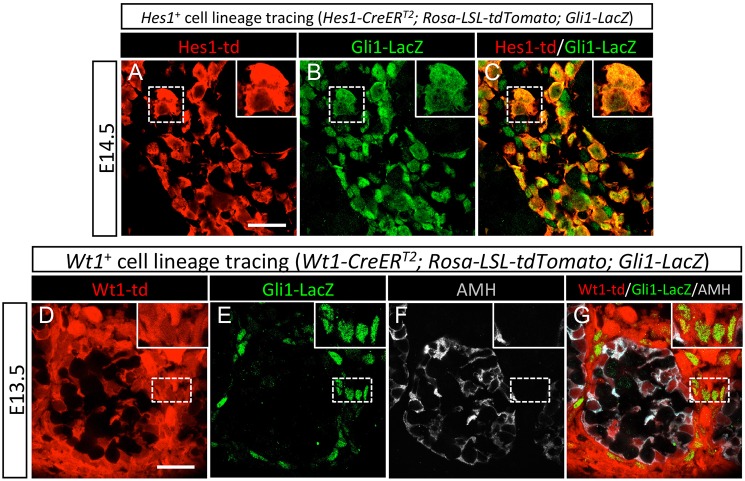
In addition to the Notch pathway, the Hedgehog (Hh) pathway is important for fetal Leydig cell differentiation (Yao et al., 2002). Unlike Notch signaling, which is activated soon after the gonadal formation around E10.5 (Fig. 2A-H; Fig. S3A-H) (Jameson et al., 2012; Tang et al., 2008), the Hh pathway does not become functional until after the onset of testis morphogenesis at E12.5 (Barsoum and Yao, 2011; Jameson et al., 2012; Yao et al., 2002). The Hh pathway, induced by the Hh ligand desert hedgehog (DHH) from Sertoli cells, was activated in the interstitium, which consequently expresses Hh-responsive gene Gli1 (Barsoum and Yao, 2011; Yao et al., 2002). Inactivation of Dhh, or inhibition of Gli1 along with another Hh downstream target Gli2, resulted in a decrease of fetal Leydig cells (Barsoum and Yao, 2011; Yao et al., 2002). The observations that both Hh and Notch signaling regulate fetal Leydig cell differentiation suggest that these signaling molecules may target the same type of cells in the testis. We developed the Hes1-CreERT2; Rosa-LSL-tdTomato lineage-tracing model, as described in Fig. 2, to label Notch-responsive cells at E10.5. These Hes1-CreERT2; Rosa-LSL-tdTomato mice were then bred to Gli1-lacZ mice, in which the Hh-responsive Gli1 expression is indicated by the presence of β-galactosidase. The resulting mice (Hes1-CreERT2; Rosa-LSL-tdTomato; Gli1-lacZ) allowed us to visualize Notch (tdTomato)- and Hh (lacZ)-responding cells at the same time. At E14.5, almost all lineage-labeled Hes1+ cells became positive for Gli1-lacZ expression (detected by β-galactosidase immunostaining; Fig. 4A-C; Fig. S7A-C), indicating that Hes1+ progenitor cells in the gonadal primordium give rise to Gli1+ cells in the fetal testis. Because Hes1+ cells in the gonadal primordium represent a subpopulation of Wt1+ progenitor cells (Fig. 2A-H), we anticipate that some Wt1+ cells would also give rise to Gli1+ cells. This idea was tested by the generation of Wt1-CreERT2; Rosa-LSL-tdTomato; Gli1-lacZ mice followed by lineage labeling of the Wt1+ cells. As expected, the fetal gonad-derived Wt1+ cells in the E14.5 testis interstitium were positive for Gli1-lacZ and negative for the Sertoli cell marker AMH, indicating that a subpopulation of Wt1+ cells become Gli1+ as they differentiate into the interstitial cell linage (Fig. 4D-G; Fig. S7D-F).
Fig. 4.
Contribution of Hes1+ and Wt1+ progenitor cells to the Gli1+ interstitial cell population in the fetal testis. (A-C) Lineage-tracing analysis of Hes1+ progenitor cells in the Hes1-CreERT2; Rosa-LSL-tdTomato; Gli1-lacZ embryos was induced by tamoxifen (TM) administration at E10.5 and E11.5. The samples were analyzed at E14.5 by fluorescence immunohistochemistry for Hes1-td (red; tdTomato-labeled fetal testis-derived Hes1+ cells), β-galactosidase (green for Gli1+ cells). (D-G) Lineage labeling of Wt1+ progenitor cells in the Wt1-CreERT2; Rosa-LSL-tdTomato; Gli1-lacZ embryos was induced by TM administration at E10.5. The samples were analyzed at E13.5 by fluorescence immunohistochemistry for Wt1-td (red; tdTomato-labeled fetal testis-derived Wt1+ cells), β-galactosidase (green for Gli1+ cells) and AMH (gray for Sertoli cells). The insets are higher magnification of the boxed areas. Scale bars: 25 μm.
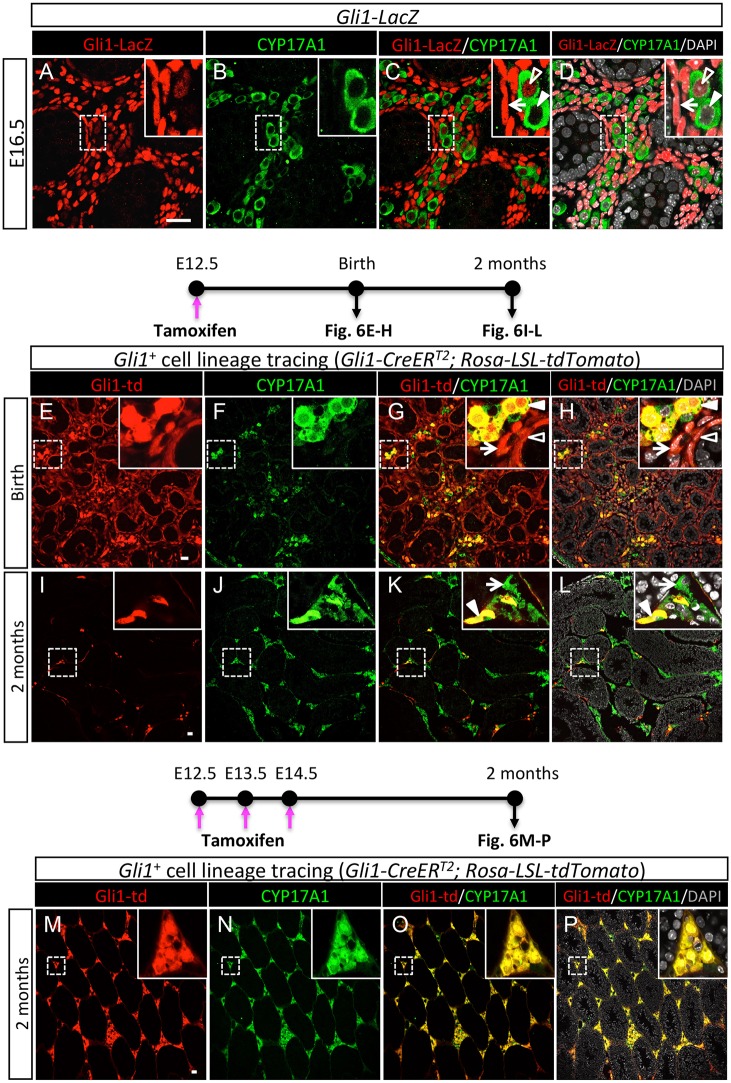
Next, we examined the composition of Gli1+ interstitial cell populations by characterizing endogenous Gli1 expression in detail by co-immunostaining of β-galactosidase (Gli1-lacZ expression) and various somatic cell markers (Fig. 5A-D; Fig. S8). Gli1-lacZ expression was present in a subpopulation of CYP17A1+ fetal Leydig cells (Fig. 5C,D insets, unfilled arrowheads) and CYP17A1− non-steroidogenic interstitial cells (Fig. 5C,D insets, arrows) in the fetal and newborn testes (Fig. 5A-D; Fig. S8E-H,M-P). In addition, Gli1-lacZ+ cells in proximity to testis cords were positive for the myoid peritubular cell marker αSMA, suggesting that these Gli1+ cells are myoid peritubular cells. This pattern of Gli1 expression resembles the lineage contributions of the Hes1+ progenitor cells (Fig. 2M-P; Fig. S7A-C), further supporting the notion that Hes1 and Gli1 mark the same interstitial cell populations.
Fig. 5.
Detection of Gli1 in the fetal testis by Gli1-lacZ reporter and genetic lineage-tracing analysis. (A-D) Expression of Gli1-lacZ in the E16.5 fetal testes was detected by fluorescence immunohistochemistry for β-galactosidase (red for Gli1+ cells), the Leydig cell marker CYP17A1 (green), and the nuclear counterstain DAPI (gray). In insets of C and D, arrows point to Gli1+/CYP17A1− non-steroidogenic interstitial cells, unfilled arrowheads indicate Gli1+/CYP17A1+ fetal Leydig cells, and filled arrowheads represent Gli1−/CYP17A1+ fetal Leydig cells. (E-L) Lineage-tracing analysis of the Gli1+ cells in the Gli1-CreERT2; Rosa-LSL-tdTomato embryos was induced by tamoxifen (TM) administration at E12.5. The testes were examined at different stages of development for Gli1-td (red; tdTomato labeled fetal testis-derived Gli1+ cells), the Leydig cell marker CYP17A1 (green), and DAPI (gray). Arrows in G,H insets indicate Gli1+/CYP17A1− interstitial cells, filled arrowheads in G,H insets indicate Gli1+/CYP17A1+ fetal Leydig cells and unfilled arrowheads in G,H insets indicate Gli1+/CYP17A1− peritubular myoid cells. Arrows in K,L insets indicate Gli1−/CYP17A1+ adult Leydig cells, filled arrowheads in K,L insets indicate Gli1+/CYP17A1+ adult Leydig cells. (M-P) Lineage-tracing analysis of the Gli1+ cells in the Gli1-CreERT2; Rosa-LSL-tdTomato embryos was induced by TM administration from E12.5 to E14.5 for three consecutive days. The testes were examined for Gli1-td (red; tdTomato labeled fetal testis-derived Gli1+ cells), the Leydig cell marker CYP17A1 (green), and DAPI (gray). The insets are higher magnifications of the boxed areas. Scale bars: 25 μm.
Fetal testis-derived Gli1+ cells give rise to adult Leydig cells in the testis
We and others have proposed that the non-steroidogenic interstitial cell populations in the fetal testis could be a source of steroidogenic cells in the adult testis (Barsoum and Yao, 2010; Kilcoyne et al., 2014). To test this hypothesis, we took advantage of Gli1 expression in the non-steroidogenic cell populations (Fig. S8) and developed a lineage-tracing model (Gli1-CreERT2; Rosa-LSL-tdTomato embryos), which allowed us to follow the fate of fetal-derived Gli1+ cells to adulthood. We labeled the Gli1+ cells by a single injection of tamoxifen at E12.5, when Gli1 expression just starts to appear in the testis interstitium (Barsoum and Yao, 2011). At the time of birth, or about 7 days after the lineage labeling by tamoxifen, fetal testis-derived Gli1+ cells contributed to CYP17A1− non-steroidogenic interstitial cells (Fig. 5E,G,H, arrows) and a subpopulation of CYP17A1+ fetal Leydig cells (Fig. 5G,H, solid arrowheads). The fetal testis-derived Gli1+ non-steroidogenic interstitial population also gave rise to cells that were tightly associated with testis cords (Fig. 5E,G,H, unfilled arrowheads), consistent with the endogenous Gli1-lacZ expression pattern in the fetal testis (Fig. 5A-D). At 2 months and 4 months of age when adult Leydig cells occupy the testis, the fetal testis-derived Gli1+ cells were positive for CYP17A1 in the testis interstitium, indicating that they have become adult Leydig cells in the adult testis (Fig. 5I-L). However, not all the adult Leydig cells were derived from the Gli1+ cells in this experimental model, as the presence of CYP17A1+/GLI1− adult Leydig cells was evident (Fig. 5I-L, arrows). We suspect that a single tamoxifen treatment was not sufficient to label all the Gli1+ cells. We therefore increased the efficiency of lineage labeling of the Gli1+ cells by administering tamoxifen to embryos from E12.5 to E14.5 for three consecutive days. Under this experimental scheme, all fetal testis-derived Gli1+ cells were positive for CYP17A1 expression at 2 months of age, (Fig. 5M-P), indicating that the fetal testis-derived Gli1+ progenitor pool is the sole source for the entire adult Leydig population in the testis.
DISCUSSION
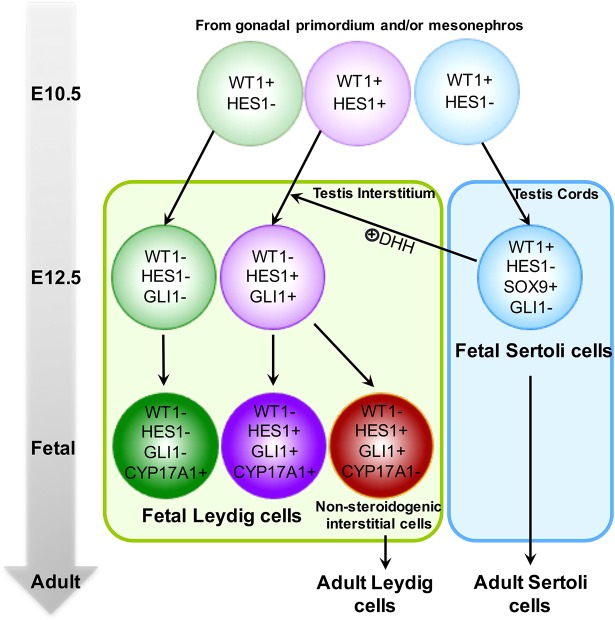
Using various lineage-tracing models, we constructed a lineage map (Fig. 6) that delineates the fate progression of heterogeneous somatic cell populations during testis morphogenesis in mice. At the onset of testis morphogenesis at E10.5, all progenitor cells in the gonadal primordium are positive for WT1 (Armstrong et al., 1993; Kreidberg et al., 1993; Wilhelm and Englert, 2002). At this stage, the WT1+ progenitor cell pool is already heterogeneous, consisting of at least three subpopulations: HES1− pre-Sertoli cells, HES1− interstitial progenitor cells and HES1+ interstitial progenitor cells (see model in Fig. 6). Between E10.5 and E12.5, WT1+ pre-Sertoli cells acquire SOX9 expression via the action of SRY, starting the organization of testis cords while maintaining WT1 expression. Meanwhile, HES1+ and HES1− interstitial progenitor cells lose WT1 expression and commit to interstitial cell lineages that are located outside testis cords. Although both restricted to the testis interstitium, HES1+ and HES1− interstitial progenitor cells exhibit distinct lineage contributions. HES1− interstitial progenitor cells give rise to a subpopulation of GLI1−/CYP17A1+ fetal Leydig cells, whereas HES1+ interstitial progenitor cells later acquire the expression of GLI1 via the action of DHH and contribute to two subsequent cell lineages: GLI1+/CYP17A1+ fetal Leydig cells and GLI1+/CYP17A1− non-steroidogenic cells. Thus, testis interstitium in the fetal testis comprises at least three distinct cell lineages: GLI1−/CYP17A1+ fetal Leydig cells, GLI1+/CYP17A1+ fetal Leydig cells and GLI1+/CYP17A1− non-steroidogenic cells. Because fetal Leydig cells (GLI1−/CYP17A1+ and GLI1+/CYP17A1+) do not give rise to adult Leydig cells (Shima et al., 2015), the GLI1+/CYP17A1− non-steroidogenic interstitial cells are likely to be the progenitor cells for the adult Leydig cell population (Fig. 6).
Fig. 6.
Model of the lineage progression of supporting cells and interstitial cells in the testis. At E10.5, the WT1+ progenitor cell pool consists of at least three subpopulations: HES1− pre-Sertoli cells, HES1− interstitial progenitor cells and HES1+ interstitial progenitor cells. Between E10.5 and E12.5, the three WT1+ subpopulations in the gonadal primordium give rise to SOX9+ Sertoli cells and SOX9− interstitial cells (HES1+ and HES1−) within and outside testis cords, respectively. Whereas HES1− interstitial cells give rise to a subpopulation of fetal Leydig cells (GLI1−/CYP17A1+), HES1+ interstitial cells further acquire expression of GLI1 at around E12.5 and differentiate into two distinct cell lineages: GLI1+ progenitor cells that contribute to a subpopulation of fetal Leydig cells (CYP17A1+), and GLI1+ non-steroidogenic interstitial cells that eventually give rise to adult Leydig cells. This model is based on the results of this study. Markers that were reported in other studies such as SF1, MAFB and ARX are not included for the sake of simplicity.
WT1+ populations in the gonadal primordium and/or mesonephros: progenitors of Sertoli and Leydig cells
At E10.5, WT1 already exhibits a broad spatiotemporal expression pattern as it expresses in the coelomic epithelium, the somatic compartment of the testis and the mesonephros (Armstrong et al., 1993; Kreidberg et al., 1993; Pelletier et al., 1991). When these WT1+ cells are lineage-marked, they give rise to not only Sertoli cells but also fetal and adult Leydig cells. It is unclear how a common WT1+ progenitor pool adapts two distinct cell fates (supporting cell lineage and interstitial cell lineage) at the bipotential stage (E10.5). The difference in the location of WT1+ positive cells (coelomic epithelium in the testis and mesonephros) is unlikely to be the cause because coelomic epithelial cells have been shown to give rise to both Sertoli cells and fetal Leydig cells (DeFalco et al., 2011; Karl and Capel, 1998), and the mesonephros only contributes to an almost exclusively endothelial cell population in the testis (Combes et al., 2009). Because endogenous WT1 expression in the somatic compartment of the testis becomes restricted to Sertoli cells within the testis cords after E12.5, WT1 could be involved in the maintenance of Sertoli cell identity. In line with this hypothesis, ablation of Wt1 gene in Sertoli cells results in the transformation of Sertoli cells into fetal Leydig-like cells, whereas overexpression of Wt1 in fetal Leydig cells promotes the expression of Sertoli cell-specific genes and suppresses steroidogenic genes (Wen et al., 2014; Zhang et al., 2015). WT1 appears to mediate the cell fate determination of the common progenitor cells to become either Sertoli cells or fetal Leydig cells. Continued presence of WT1 in the pre-Sertoli cells promotes the Sertoli cell fate, whereas diminishing Wt1 expression in the interstitial progenitor cells leads to the establishment of interstitial cell lineages. Although how WT1 delineates Sertoli cell fate is not clear, several binding partners of WT1 have been suggested to regulate its activities, such as the transcriptional co-suppressor BASP1 and the serine protease HtrA2 (Carpenter et al., 2004; Hartkamp et al., 2010). Interestingly, Basp1 and Htra2 are expressed at higher levels in pre-Sertoli cells than in interstitial cells at the onset of testis morphogenesis (Jameson et al., 2012). It remains to be determined how WT1 expression is extinguished in the Leydig cell lineage. Because the Sertoli cell lineage is established earlier than the Leydig cell lineage, factors suppressing WT1 expression in the fetal Leydig cells might come from Sertoli cells.
Testicular interstitium consists of HES1+ and HES1− cell populations that derive from the WT1+ somatic progenitors
Among the WT1+ progenitor cell pool in the gonadal primordium, a subpopulation of WT1+ cells acquires HES1 expression at the onset of sex determination. The HES1+ progenitor cells subsequently become positive for Leydig cell marker CYP17A1 in the fetal and post-pubertal testes. Hes1 apparently marks a progenitor population specific for not only fetal Leydig cells but also adult Leydig cells in the testis, suggesting that both adult Leydig cells and at least a subpopulation of fetal Leydig cells (GLI1+/CYP17A1+) could derive from a common progenitor population in the gonadal primordium.
It is unclear how the HES1+ and HES1− populations are established at early testis development. Because both populations are derived from WT1+ cells, the segregation of HES1+ and HES1− lineages might be the consequence of asymmetric division of the WT1+ progenitor cells (Lai, 2004; Rhyu et al., 1994). Two daughter cells of the WT1+ progenitor cells might inherit different signaling components that influence Notch signaling and therefore adopt different fates: HES1− cells in which Notch signaling is suppressed, and HES1+ cells in which the Notch pathway is activated (Lai, 2004; Rhyu et al., 1994).
Notably, both HES1+ and HES1− interstitial cells contribute to fetal Leydig cells, indicating that fetal Leydig cells are a heterogeneous population with progenitor cells pre-determined in nascent testes. It has been shown that fetal Leydig cells arise from two different origins: the coelomic epithelium and the gonadal-mesonephric border region (DeFalco et al., 2011). The precursor cells in these regions are associated with Mafb and Arx, putative markers for fetal Leydig cell progenitors (DeFalco et al., 2011; Miyabayashi et al., 2013). Interestingly, isolated Mafb-EGFP interstitial cells from fetal testes are highly enriched for not only Arx but also Hes1 expression (Jameson et al., 2012), implying that MAFB+ cells, ARX1+ cells and HES1+ cells probably represent a similar progenitor population in fetal testes. In addition to MAFB+/ARX1+/HES1+ cells as sources for fetal Leydig cells, another source of HES1− somatic progenitors also contribute to fetal Leydig cells. The mechanism that defines the HES1− progenitor cells remains to be determined.
Notch2 is the main Notch receptor responsible for the differentiation of fetal Leydig cells
Although the Notch pathway is known to control fetal Leydig cell population in the interstitium, the signaling components that facilitate its action are still unknown. We have provided genetic evidence that places Notch2 as the main Notch receptor. Among the four mammalian Notch receptors, involvement of Notch3 in testes determination is minimal because its expression is not detected until after E12.5 (Jameson et al., 2012; Tang et al., 2008), whereas strong expression of Hes1, the downstream target of Notch signaling, is detected with Hes1-GFP reporter at E11.5. Notch1 and Notch4 are specifically expressed in the endothelial cells in the gonads, whereas Notch2 is detected in both supporting and interstitial progenitor cells in the gonadal primordium (Brennan et al., 2002; Jameson et al., 2012). We inactivated Notch2 using the SF1-cre model, which targets all WT1+ somatic cell populations (SF1 and WT1 colocalize to gonadal somatic cells). This model did not induce deletion in endothelial cells that are enriched with Notch2 expression (Jameson et al., 2012). In the Notch2 conditional knockout newborn testes, the number of fetal Leydig cells is increased. Because Notch signaling is capable of restricting progenitor cells from taking on a specialized fate (Lai, 2004), the increased fetal Leydig cells are likely to be the consequence of compromised inhibitory Notch signaling. A previous study of the Hes1−/− testis revealed a loss of germ cells phenotype associated with disrupted Notch signaling (Tang et al., 2008). Although germ cells appeared to be affected in the absence of Hes1, inhibition of the entire Notch signaling in cultured testis did not affect either germ cells or Sertoli cells, suggesting that Notch signaling has a specific role on fetal Leydig cells (Tang et al., 2008). The difference in the germ cell phenotype between the Notch2 knockout and the Hes1 knockout could be due to the fact that the Notch2 conditional knockout only targets Sf1+ somatic cells, whereas the Hes1 conventional knockout not only targets the Sf1+ somatic cells in the gonadal primordium but also other cell types such as germ cells and endothelial cells. It is also possible that HES1 plays a Notch-independent role, as found in many other model systems (Ingram et al., 2008; Katoh and Katoh, 2007). Our results provide genetic evidence that implicates Notch2 as the major receptor of Notch signaling that restricts fetal Leydig cell population in the fetal testis.
GLI1+ non-steroidogenic interstitial cells in the fetal testis are the progenitor cells for adult Leydig cells
In our previous work that focused on fetal Leydig cells, we uncovered a unique cell population in the interstitium that are negative for any steroidogenic enzyme expression (Barsoum et al., 2013). We speculate that this non-steroidogenic interstitial population could be the progenitors for adult Leydig cells. Using the Gli1-lineage tracing model, we provide evidence that these non-steroidogenic cells are positive for GLI1+ and give rise to adult Leydig cells. The GLI1+ interstitial cells consist of two subpopulations: GLI1+/CYP17A1+ fetal Leydig cells and GLI1+/CYP17A1− non-steroidogenic interstitial cells. It has been clearly demonstrated that fetal Leydig cells do not contribute to adult Leydig cells in the postnatal testis (Shima et al., 2015). Instead the GLI1+/CYP17A1− non-steroidogenic interstitial cells are the primary progenitor source of the adult Leydig population. The existence of such non-steroidogenic interstitial progenitor cells was also observed in cell-sorting studies in which interstitial cells with high Sf1 expression represent fetal Leydig cells, whereas low Sf1-expressing interstitial cells are potential non-steroidogenic cell progenitors (Inoue et al., 2015; McClelland et al., 2015). These non-steroidogenic interstitial progenitor cells appear to be enriched for Ptch1, suggesting that these cells respond to Hedgehog signaling (Inoue et al., 2015; McClelland et al., 2015). Consistent with these observations, genetic manipulation of Hedgehog signaling in the fetal testis significantly influences the adult Leydig cell population (Barsoum et al., 2013), supporting the presence of a Hedgehog-responsive GLI1+ progenitor population for adult Leydig cells. This idea of fetal progenitors for adult Leydig cells was also noted in the studies that manipulations of COUP-TFII+ interstitial progenitor cells during the fetal stage result in changes in the number of adult Leydig cells in adult testis (Kilcoyne et al., 2014; Qin et al., 2008). It is not clear whether COUP-TFII and GLI1 mark a similar or distinct interstitial progenitor cell population in the fetal testis. Because COUP-TFII also appears to be a downstream target of Hedgehog signaling (Krishnan et al., 1997a,b; Lee et al., 2006; Takamoto et al., 2005), it is likely that COUP-TFII and GLI1 mark the same interstitial progenitor cell population in the fetal testis.
In conclusion, in contrast to the static snapshots of cell types at a certain developmental stage, our time course lineage-tracing analyses capture the cell fate-defining steps as somatic cells in the testis progress through the primordial stage to the differentiated stage in adulthood. In the testis primordium, where all somatic progenitor cells express WT1, the Sertoli cell lineage emerges as a result of SRY and SOX9 action. A distinct HES1+ cell population also appears at this primordial stage, and by downregulating WT1 they commit to the interstitial cell lineage. These HES1+ interstitial progenitors gain GLI1 expression in response to Sertoli cell-derived DHH signal, and are destined to become a subpopulation of fetal Leydig cells and future adult Leydig cells. Another subpopulation of fetal Leydig cells is also identified, as the descendants of the HES1−/GLI1− interstitial cell population. Although the processes that specify unique cell populations (i.e. non-steroidogenic interstitial population and HES1−/GLI1− fetal Leydig cells) remain to be determined, our findings provide an important foundation for the understanding of how various cell types gain their identities during testis organogenesis.
MATERIALS AND METHODS
Animals
Gli1-lacZ (Jax #008211), Gli1-CreERT2 (Jax #007913), Wt1-CreERT2 (Jax #010912), Rosa-LSL-tdTomato (Jax #007905) and Notch2 floxed mice (Jax #010525) were purchased from the Jackson Laboratory. Sf1-Cre mice (Bingham et al., 2006) were provided by the late Dr Keith Parker at UT Southwestern Medical Center. Hes1-CreERT2 mice (Kopinke et al., 2011) were provided by Dr Charles Murtaugh at the University of Utah. Hes1-GFP mice (Fre et al., 2011) and Hes1f/f mice (Revollo et al., 2013) were kindly provided by Dr Spyros Artavanis-Tsakonas at Harvard College and Dr John A. Cidlowski at National Institute of Environmental Health Sciences, respectively. Female mice were housed with male mice overnight and checked for the presence of a vaginal plug the next morning. The day when the vaginal plug was detected was considered embryonic day (E) 0.5. All animal procedures were approved by the National Institute of Environmental Health Sciences (NIEHS) Animal Care and Use Committee and are in compliance with a NIEHS-approved animal study proposal. All experiments were performed on at least three animals for each genotype.
Tamoxifen treatment
CreERT2 activity was induced by intraperitoneal injection of 1 mg tamoxifen (T-5648, Sigma-Aldrich) per mouse per day in corn oil. An equivalent volume of corn oil served as vehicle control (Liu et al., 2015).
Immunohistochemistry and histological analysis
Testes were collected and fixed in 4% paraformaldehyde overnight at 4°C. The samples were then dehydrated through a sucrose gradient, embedded in OCT compound and cryosectioned to 10 μm sections as described (Liu et al., 2012). The frozen sections were incubated with primary antibodies (Table S1) in PBS-Triton X-100 (0.1%) solution with 5% normal donkey serum at 4°C overnight. The sections were then washed and incubated in the appropriate secondary antibodies (1:500; Invitrogen) before mounting in Vector Mount with DAPI (Vector Laboratories). Slides were imaged under a Leica DMI4000 confocal microscope. For histological analysis, the samples were fixed in 4% paraformaldehyde in PBS at 4°C overnight, dehydrated through an ethanol gradient, and embedded in paraffin wax. Sections were stained with Hematoxylin & Eosin, and were scanned using Aperio ScanScope XT Scanner (Aperio Technologies) for digital image analysis.
Gene expression analysis
Total RNA was isolated from E13.5 and newborn testes using the PicoPure RNA Isolation Kit (Arcturus, Mountain View, CA, USA) according to the manufacturer's protocol. The cDNA preparation was synthesized using random hexamers and the Superscript II cDNA synthesis system (Invitrogen) following manufacturer's instruction. Gene expression was analyzed by real-time PCR using the Bio-Rad CFX96 Real-Time PCR Detection system (Liu et al., 2015). Taqman probes (ThermoFisher Scientific), were used to examine the fold changes of the transcripts. The following Taqman probes were used: Notch2 (Mm00803077), Ddx4 (Mm00802445_m1), Dhh (Mm01310203_m1), Ihh (Mm00439613_m1), Nr5a1 (Mm00446826_m1), Hsd3b1 (Mm01261921_mH), Hes1 (Mm01342805_m1) and 18S rRNA (Mm03928990_g1). Fold changes in gene expression were determined by quantification of cDNA from target (knockout) samples relative to a calibrator sample (control). All real-time PCR analyses were performed in duplicate, and the results were analyzed from a minimum of three biological replicates for each experiment. The relative fold change of transcript was normalized to 18S rRNA as an endogenous reference.
Statistical analysis
Data were analyzed using Prism (Version 6, GraphPad Software) by two-tailed Student's t-test. Values are expressed as mean±s.e.m. A minimum of three biological replicates was used for each experiment.
Acknowledgements
We are grateful to Dr Charles Murtaugh at the University of Utah for providing the Hes1-CreERT2 mice; Dr Spyros Artavanis-Tsakonas at Harvard College for the Hes1-GFP mice; and Dr Ken-Ichirou Morohashi at Kyushu University in Japan for the SOX9 antibody. We thank Dr Kathryn McClelland for her critical comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
C.L. and K.R. performed the experiments; C.L. and H.H.-C.Y. designed the study, analyzed data and wrote the paper.
Funding
This work was supported by the Intramural Research Program (ES102965) of the National Institutes of Health (NIH) National Institute of Environmental Health Sciences and NIH Graduate Partnerships Program. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.135756.supplemental
References
- Agelopoulou R., Magre S., Patsavoudi E. and Jost A. (1984). Initial phases of the rat testis differentiation in vitro. J. Embryol. Exp. Morphol. 83, 15-31. [PubMed] [Google Scholar]
- Albrecht K. H. and Eicher E. M. (2001). Evidence that Sry is expressed in pre-Sertoli cells and Sertoli and granulosa cells have a common precursor. Dev. Biol. 240, 92-107. 10.1006/dbio.2001.0438 [DOI] [PubMed] [Google Scholar]
- Armstrong J. F., Pritchard-Jones K., Bickmore W. A., Hastie N. D., Bard J. B. (1993). The expression of the Wilms’ tumour gene, WT1, in the developing mammalian embryo. Mech. Dev. 40, 85-97. 10.1016/0925-4773(93)90090-K [DOI] [PubMed] [Google Scholar]
- Aubert M. L., Begeot M., Winiger B. P., Morel G., Sizonenko P. C. and Dubois P. M. (1985). Ontogeny of hypothalamic luteinizing hormone-releasing hormone (GnRH) and pituitary GnRH receptors in fetal and neonatal rats. Endocrinology 116, 1565-1576. 10.1210/endo-116-4-1565 [DOI] [PubMed] [Google Scholar]
- Baker P. J. and O'Shaughnessy P. J. (2001). Role of gonadotrophins in regulating numbers of Leydig and Sertoli cells during fetal and postnatal development in mice. Reproduction 122, 227-234. 10.1530/rep.0.1220227 [DOI] [PubMed] [Google Scholar]
- Baker P. J., Sha J. A., McBride M. W., Peng L., Payne A. H. and O'Shaughnessy P. J. (1999). Expression of 3beta-hydroxysteroid dehydrogenase type I and type VI isoforms in the mouse testis during development. Eur. J. Biochem. 260, 911-917. 10.1046/j.1432-1327.1999.00245.x [DOI] [PubMed] [Google Scholar]
- Barsoum I. B. and Yao H. H.-C. (2010). Fetal Leydig cells: progenitor cell maintenance and differentiation. J. Androl. 31, 11-15. 10.2164/jandrol.109.008318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsoum I. and Yao H. H. C. (2011). Redundant and differential roles of transcription factors Gli1 and Gli2 in the development of mouse fetal Leydig cells. Biol. Reprod. 84, 894-899. 10.1095/biolreprod.110.088997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsoum I. B., Kaur J., Ge R. S., Cooke P. S. and Yao H. H.-C. (2013). Dynamic changes in fetal Leydig cell populations influence adult Leydig cell populations in mice. FASEB J. 27, 2657-2666. 10.1096/fj.12-225060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton L., Shan L.-X. and Hardy M. P. (1995). Differentiation of adult Leydig cells. J. Steroid Biochem. Mol. Biol. 53, 61-68. 10.1016/0960-0760(95)00022-R [DOI] [PubMed] [Google Scholar]
- Bingham N. C., Verma-Kurvari S., Parada L. F. and Parker K. L. (2006). Development of a steroidogenic factor 1/Cre transgenic mouse line. Genesis 44, 419-424. 10.1002/dvg.20231 [DOI] [PubMed] [Google Scholar]
- Brennan J. and Capel B. (2004). One tissue, two fates: molecular genetic events that underlie testis versus ovary development. Nat. Rev.Genet. 5, 509-521. 10.1038/nrg1381 [DOI] [PubMed] [Google Scholar]
- Brennan J., Karl J. and Capel B. (2002). Divergent vascular mechanisms downstream of Sry establish the arterial system in the XY gonad. Dev. Biol. 244, 418-428. 10.1006/dbio.2002.0578 [DOI] [PubMed] [Google Scholar]
- Carpenter B., Hill K. J., Charalambous M., Wagner K. J., Lahiri D., James D. I., Andersen J. S., Schumacher V., Royer-Pokora B., Mann M. et al. (2004). BASP1 is a transcriptional cosuppressor for the Wilms’ tumor suppressor protein WT1. Mol. Cell. Biol. 24, 537-549. 10.1128/MCB.24.2.537-549.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaboissier M.-C., Kobayashi A., Vidal V. I., Lutzkendorf S., van de Kant H. J., Wegner M., de Rooij D. G., Behringer R. R. and Schedl A. (2004). Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development 131, 1891-1901. 10.1242/dev.01087 [DOI] [PubMed] [Google Scholar]
- Combes A. N., Wilhelm D., Davidson T., Dejana E., Harley V., Sinclair A. and Koopman P. (2009). Endothelial cell migration directs testis cord formation. Dev. Biol. 326, 112-120. 10.1016/j.ydbio.2008.10.040 [DOI] [PubMed] [Google Scholar]
- Davidoff M. S., Middendorff R., Enikolopov G., Riethmacher D., Holstein A. F. and Müller D. (2004). Progenitor cells of the testosterone-producing Leydig cells revealed. J. Cell Biol. 167, 935-944. 10.1083/jcb.200409107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFalco T. and Capel B. (2009). Gonad morphogenesis in vertebrates: divergent means to a convergent end. Annu. Rev. Cell Dev. Biol. 25, 457-482. 10.1146/annurev.cellbio.042308.13350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFalco T., Takahashi S. and Capel B. (2011). Two distinct origins for Leydig cell progenitors in the fetal testis. Dev. Biol. 352, 14-26. 10.1016/j.ydbio.2011.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFalco T., Bhattacharya I., Williams A. V., Sams D. M. and Capel B. (2014). Yolk-sac-derived macrophages regulate fetal testis vascularization and morphogenesis. Proc. Natl. Acad. Sci. USA 111, E2384-E2393. 10.1073/pnas.1400057111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Jelinsky S. A., Finger J. N., Johnston D. S., Kopf G. S., Sottas C. M., Hardy M. P. and Ge R.-S. (2007). Gene expression during development of fetal and adult Leydig cells. Ann. N. Y. Acad. Sci. 1120, 16-35. 10.1196/annals.1411.016 [DOI] [PubMed] [Google Scholar]
- Eggers S., Ohnesorg T. and Sinclair A. (2014). Genetic regulation of mammalian gonad development. Nat. Rev. Endocrinol. 10, 673-683. 10.1038/nrendo.2014.163 [DOI] [PubMed] [Google Scholar]
- El-Gehani F., Zhang F. P., Pakarinen P., Rannikko A. and Huhtaniemi I. (1998). Gonadotropin-independent regulation of steroidogenesis in the fetal rat testis. Biol. Reprod. 58, 116-123. 10.1095/biolreprod58.1.116 [DOI] [PubMed] [Google Scholar]
- Fre S., Hannezo E., Sale S., Huyghe M., Lafkas D., Kissel H., Louvi A., Greve J., Louvard D. and Artavanis-Tsakonas S. (2011). Notch lineages and activity in intestinal stem cells determined by a new set of knock-in mice. PLoS ONE 6, e25785 10.1371/journal.pone.0025785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangnerau M.-N. and Picon R. (1987). Onset of steroidogenesis and differentiation of functional LH receptors in rat fetal testicular cultures. Arch. Androl. 18, 215-224. 10.3109/01485018708988486 [DOI] [PubMed] [Google Scholar]
- Ge R.-S., Dong Q., Sottas C. M., Papadopoulos V., Zirkin B. R. and Hardy M. P. (2006). In search of rat stem Leydig cells: identification, isolation, and lineage-specific development. Proc. Natl. Acad. Sci. USA 103, 2719-2724. 10.1073/pnas.0507692103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold S. L. and Behringer R. R. (2009). Fetal Leydig cell origin and development. Sex. Dev. 3, 1-15. 10.1159/000200077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbay J., Collignon J., Koopman P., Capel B., Economou A., Münsterberg A., Vivian N., Goodfellow P. and Lovell-Badge R. (1990). A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature 346, 245-250. 10.1038/346245a0 [DOI] [PubMed] [Google Scholar]
- Habert R., Lejeune H. and Saez J. M. (2001). Origin, differentiation and regulation of fetal and adult Leydig cells. Mol. Cell. Endocrinol. 179, 47-74. 10.1016/S0303-7207(01)00461-0 [DOI] [PubMed] [Google Scholar]
- Haider S. G. (2004). Cell biology of Leydig cells in the testis. Int. Rev. Cytol. 233, 181-241. 10.1016/S0074-7696(04)33005-6 [DOI] [PubMed] [Google Scholar]
- Hartkamp J., Carpenter B. and Roberts S. G. E. (2010). The Wilms’ tumor suppressor protein WT1 Is processed by the serine protease HtrA2/Omi. Mol. Cell 37, 159-171. 10.1016/j.molcel.2009.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins J. R., Taylor A., Berta P., Levilliers J., Van der Auwera B. and Goodfellow P. N. (1992). Mutational analysis of SRY: nonsense and missense mutations in XY sex reversal. Hum. Genet. 88, 471-474. 10.1007/BF00215684 [DOI] [PubMed] [Google Scholar]
- Huhtaniemi I. and Pelliniemi L. J. (1992). Fetal Leydig cells: cellular origin, morphology, life span, and special functional features. Exp. Biol. Med. 201, 125-140. 10.3181/00379727-201-43493 [DOI] [PubMed] [Google Scholar]
- Ingram W. J., Mccue K. I., Tran T. H., Hallahan A. R. and Wainwright B. J. (2008). Sonic Hedgehog regulates Hes1 through a novel mechanism that is independent of canonical Notch pathway signalling. Oncogene 27, 1489-1500. 10.1038/sj.onc.1210767 [DOI] [PubMed] [Google Scholar]
- Inoue M., Shima Y., Miyabayashi K., Tokunaga K., Sato T., Baba T., Ohkawa Y., Akiyama H., Suyama M. and Morohashi K. I. (2015). Isolation and characterization of fetal Leydig progenitor cells of male mice. Endocrinology, 157, 1222-1233. 10.1210/en.2015-1773 [DOI] [PubMed] [Google Scholar]
- Jameson S. A., Natarajan A., Cool J., DeFalco T., Maatouk D. M., Mork L., Munger S. C. and Capel B. (2012). Temporal transcriptional profiling of somatic and germ cells reveals biased lineage priming of sexual fate in the fetal mouse gonad. PLoS Genet. 8, e1002575 10.1371/journal.pgen.1002575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R., Ohtsuka T. and Kobayashi T. (2007). The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 134, 1243-1251. 10.1242/dev.000786 [DOI] [PubMed] [Google Scholar]
- Karl J. and Capel B. (1998). Sertoli cells of the mouse testis originate from the coelomic epithelium. Dev. Biol. 203, 323-333. 10.1006/dbio.1998.9068 [DOI] [PubMed] [Google Scholar]
- Katoh M. and Katoh M. (2007). Integrative genomic analyses on HES/HEY family: Notch-independent HES1, HES3 transcription in undifferentiated ES cells, and Notch-dependent HES1, HES5, HEY1, HEY2, HEYL transcription in fetal tissues, adult tissues, or cancer. Int. J. Oncol. 31, 461-466. 10.3892/ijo.31.2.461 [DOI] [PubMed] [Google Scholar]
- Kerr J. B. and Knell C. M. (1988). The Fate of Fetal Leydig-Cells during the Development of the Fetal and Postnatal Rat Testis. Development 103, 535-544. [DOI] [PubMed] [Google Scholar]
- Kilcoyne K. R., Smith L. B., Atanassova N., Macpherson S., McKinnell C., van den Driesche S., Jobling M. S., Chambers T. J. G., De Gendt K., Verhoeven G. et al. (2014). Fetal programming of adult Leydig cell function by androgenic effects on stem/progenitor cells. Proc. Natl. Acad. Sci. USA 111, E1924-E1932. 10.1073/pnas.1320735111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Kobayashi A., Sekido R., DiNapoli L., Brennan J., Chaboissier M.-C., Poulat F., Behringer R. R., Lovell-Badge R. and Capel B. (2006). Fgf9 and Wnt4 act as antagonistic signals to regulate mammalian sex determination. PLoS Biol. 4, 1000-1009. 10.1371/journal.pbio.0040187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman P., Gubbay J., Vivian N., Goodfellow P. and Lovell-Badge R. (1991). Male development of chromosomally female mice transgenic for Sry. Nature 351, 117-121. 10.1038/351117a0 [DOI] [PubMed] [Google Scholar]
- Kopinke D., Brailsford M., Shea J. E., Leavitt R., Scaife C. L. and Murtaugh L. C. (2011). Lineage tracing reveals the dynamic contribution of Hes1+ cells to the developing and adult pancreas. Development 138, 431-441. 10.1242/dev.053843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreidberg J. A., Sariola H., Loring J. M., Maeda M., Pelletier J., Housman D. and Jaenisch R. (1993). WT-1 is required for early kidney development. Cell 74, 679-691. 10.1016/0092-8674(93)90515-R [DOI] [PubMed] [Google Scholar]
- Krishnan V., Elberg G., Tsai M.-J. and Tsai S. Y. (1997a). Identification of a novel sonic hedgehog response element in the chicken ovalbumin upstream promoter-transcription factor II promoter. Mol. Endocrinol. 11, 1458-1466. 10.1210/mend.11.10.9992 [DOI] [PubMed] [Google Scholar]
- Krishnan V., Pereira F. A., Qiu Y., Chen C.-H., Beachy P. A., Tsai S. Y. and Tsai M.-J. (1997b). Mediation of Sonic hedgehog-induced expression of COUP-TFII by a protein phosphatase. Science 278, 1947-1950. 10.1126/science.278.5345.1947 [DOI] [PubMed] [Google Scholar]
- Lai E. C. (2004). Notch signaling: control of cell communication and cell fate. Development 131, 965-973. 10.1242/dev.01074 [DOI] [PubMed] [Google Scholar]
- Lee K., Jeong J., Kwak I., Yu C.-T., Lanske B., Soegiarto D. W., Toftgard R., Tsai M.-J., Tsai S., Lydon J. P. et al. (2006). Indian hedgehog is a major mediator of progesterone signaling in the mouse uterus. Nat. Genet. 38, 1204-1209. 10.1038/ng1874 [DOI] [PubMed] [Google Scholar]
- Li X., Wang Z., Jiang Z., Guo J., Zhang Y., Li C., Chung J., Folmer J., Liu J., Lian Q. et al. (2016). Regulation of seminiferous tubule-associated stem Leydig cells in adult rat testes. Proc. Natl. Acad. Sci. USA 113, 2666-2671. 10.1073/pnas.1519395113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Paczkowski M., Othman M. and Yao H. H.-C. (2012). Investigating the origins of somatic cell populations in the perinatal mouse ovaries using genetic lineage tracing and immunohistochemistry. Methods Mol. Biol. 825, 211-221. 10.1007/978-1-61779-436-0_16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Peng J., Matzuk M. M. and Yao H. H.-C. (2015). Lineage specification of ovarian theca cells requires multicellular interactions via oocyte and granulosa cells. Nat. Commun. 6, 6934 10.1038/ncomms7934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lording D. W. and De Kretser D. M. (1972). Comparative ultrastructural and histochemical studies of the interstitial cells of the rat testis during fetal and postnatal development. Reproduction 29, 261-269. 10.1530/jrf.0.0290261 [DOI] [PubMed] [Google Scholar]
- Lovell-Badge R. and Robertson E. (1990). XY female mice resulting from a heritable mutation in the primary testis-determining gene, Tdy. Development 109, 635-646. [DOI] [PubMed] [Google Scholar]
- Ma X., Dong Y., Matzuk M. M. and Kumar T. R. (2004). Targeted disruption of luteinizing hormone beta-subunit leads to hypogonadism, defects in gonadal steroidogenesis, and infertility. Proc. Natl. Acad. Sci. USA 101, 17294-17299. 10.1073/pnas.0404743101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdic G., Saunders P. T. and Teerds K. J. (1998). Immunoexpression of the steroidogenic enzymes 3-beta hydroxysteroid dehydrogenase and 17 alpha-hydroxylase, C17,20 lyase and the receptor for luteinizing hormone (LH) in the fetal rat testis suggests that the onset of Leydig cell steroid production is independent of LH action. Biol. Reprod. 58, 520-525. 10.1095/biolreprod58.2.520 [DOI] [PubMed] [Google Scholar]
- Mayerhofer A., Lahr G., Seidl K., Eusterschulte B., Christoph A. and Gratzl M. (1996). The neural cell adhesion molecule (NCAM) provides clues to the development of testicular Leydig cells. J. Androl. 17, 223-230. [PubMed] [Google Scholar]
- McClelland K. S., Bell K., Larney C., Harley V. R., Sinclair A. H., Oshlack A., Koopman P. and Bowles J. (2015). Purification and Transcriptomic Analysis of mouse fetal Leydig cells reveals candidate genes for specification of gonadal steroidogenic cells. Biol. Reprod. 92, 145 10.1095/biolreprod.115.128918 [DOI] [PubMed] [Google Scholar]
- Merchant-Larios H. and Moreno-Mendoza N. (1998). Mesonephric stromal cells differentiate into Leydig cells in the mouse fetal testis. Exp. Cell Res. 244, 230-238. 10.1006/excr.1998.4215 [DOI] [PubMed] [Google Scholar]
- Miyabayashi K., Katoh-Fukui Y., Ogawa H., Baba T., Shima Y., Sugiyama N., Kitamura K. and Morohashi K.-i. (2013). Aristaless related homeobox gene, Arx, is implicated in mouse fetal Leydig cell differentiation possibly through expressing in the progenitor cells. PLoS ONE 8, e68050 10.1371/journal.pone.0068050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odeh H. M., Kleinguetl C., Ge R., Zirkin B. R. and Chen H. (2014). Regulation of the proliferation and differentiation of Leydig stem cells in the adult testis. Biol. Reprod. 90, 123 10.1095/biolreprod.114.117473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shaughnessy P. J. and Fowler P. A. (2011). Endocrinology of the mammalian fetal testis. Reproduction 141, 37-46. 10.1530/REP-10-0365 [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy P. J., Baker P., Sohnius U., Haavisto A. M., Charlton H. M. and Huhtaniemi I. (1998). Fetal development of Leydig cell activity in the mouse is independent of pituitary gonadotroph function. Endocrinology 139, 1141-1146. 10.1210/en.139.3.1141 [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy P. J., Fleming L. M., Jackson G., Hochgeschwender U., Reed P. and Baker P. J. (2003). Adrenocorticotropic hormone directly stimulates testosterone production by the fetal and neonatal mouse testis. Endocrinology 144, 3279-3284. 10.1210/en.2003-0277 [DOI] [PubMed] [Google Scholar]
- Palmer S. J. and Burgoyne P. S. (1991). In situ analysis of fetal, prepuberal and adult XX↔XY chimaeric mouse testes: Sertoli cells are predominantly, but not exclusively, XY. Development 112, 265-268. [DOI] [PubMed] [Google Scholar]
- Patsavoudi E., Magre S., Castanier M., Scholler R. and Jost A. (1985). Dissociation between testicular morphogenesis and functional differentiation of Leydig cells. J. Endocrinol. 105, 235-238. 10.1677/joe.0.1050235 [DOI] [PubMed] [Google Scholar]
- Pelletier J., Schalling M., Buckler A. J., Rogers A., Haber D. A. and Housman D. (1991). Expression of the Wilms’ tumor gene WT1 in the murine urogenital system. Genes Dev. 5, 1345-1356. 10.1101/gad.5.8.1345 [DOI] [PubMed] [Google Scholar]
- Qin J., Tsai M.-J. and Tsai S. Y. (2008). Essential roles of COUP-TFII in Leydig cell differentiation and male fertility. PLoS ONE 3, e3285 10.1371/journal.pone.0003285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revollo J. R., Oakley R. H., Lu N. Z., Kadmiel M., Gandhavadi M. and Cidlowski J. A. (2013). HES1 is a master regulator of glucocorticoid receptor-dependent gene expression. Sci. Signal. 6, ra103 10.1126/scisignal.2004389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhyu M. S., Jan L. Y. and Jan Y. N. (1994). Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 76, 477-491. 10.1016/0092-8674(94)90112-0 [DOI] [PubMed] [Google Scholar]
- Roosen-Runge E. C. and Anderson D. (1959). The development of the interstitial cells in the testis of the albino rat. Cells Tissues Organs 37, 125-137. 10.1159/000141460 [DOI] [PubMed] [Google Scholar]
- Schmahl J., Eicher E. M., Washburn L. L. and Capel B. (2000). Sry induces cell proliferation in the mouse gonad. Development 127, 65-73. [DOI] [PubMed] [Google Scholar]
- Schmahl J., Kim Y., Colvin J. S., Ornitz D. M. and Capel B. (2004). Fgf9 induces proliferation and nuclear localization of FGFR2 in Sertoli precursors during male sex determination. Development 131, 3627-3636. 10.1242/dev.01239 [DOI] [PubMed] [Google Scholar]
- Shima Y., Miyabayashi K., Haraguchi S., Arakawa T., Otake H., Baba T., Matsuzaki S., Shishido Y., Akiyama H., Tachibana T. et al. (2013). Contribution of Leydig and Sertoli cells to testosterone production in mouse fetal testes. Mol. Endocrinol. 27, 63-73. 10.1210/me.2012-1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima Y., Matsuzaki S., Miyabayashi K., Otake H., Baba T., Kato S., Huhtaniemi I. and Morohashi K.-I. (2015). Fetal Leydig cells persist as an androgen-independent subpopulation in the postnatal testis. Mol. Endocrinol. 29, 1581-1593. 10.1210/me.2015-1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley E., Lin C.-Y., Jin S., Liu J., Sottas C. M., Ge R., Zirkin B. R. and Chen H. (2012). Identification, proliferation, and differentiation of adult Leydig stem cells. Endocrinology 153, 5002-5010. 10.1210/en.2012-1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamoto N., You L.-R., Moses K., Chiang C., Zimmer W. E., Schwartz R. J., DeMayo F. J., Tsai M.-J. and Tsai S. Y. (2005). COUP-TFII is essential for radial and anteroposterior patterning of the stomach. Development 132, 2179-2189. 10.1242/dev.01808 [DOI] [PubMed] [Google Scholar]
- Tanaka S. S. and Nishinakamura R. (2014). Regulation of male sex determination: genital ridge formation and Sry activation in mice. Cell. Mol. Life Sci. 71, 4781-4802. 10.1007/s00018-014-1703-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Brennan J., Karl J., Hamada Y., Raetzman L. and Capel B. (2008). Notch signaling maintains Leydig progenitor cells in the mouse testis. Development 135, 3745-3753. 10.1242/dev.024786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Val P., Jeays-Ward K. and Swain A. (2006). Identification of a novel population of adrenal-like cells in the mammalian testis. Dev. Biol. 299, 250-256. 10.1016/j.ydbio.2006.07.030 [DOI] [PubMed] [Google Scholar]
- Wen Q., Zheng Q.-S., Li X.-X., Hu Z.-Y., Gao F., Cheng C. Y. and Liu Y.-X. (2014). Wt1 dictates the fate of fetal and adult Leydig cells during development in the mouse testis. Am. J. Physiol. Endocrinol. Metab. 307, E1131-E1143. 10.1152/ajpendo.00425.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm D. and Englert C. (2002). The Wilms tumor suppressor WT1 regulates early gonad development by activation of Sf1. Genes Dev. 16, 1839-1851. 10.1101/gad.220102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willerton L., Smith R. A., Russell D. and Mackay S. (2004). Effects of FGF9 on embryonic Sertoli cell proliferation and testicular cord formation in the mouse. Int. J. Dev. Biol. 48, 637-643. 10.1387/ijdb.031778lw [DOI] [PubMed] [Google Scholar]
- Yao H. H.-H., Whoriskey W. and Capel B. (2002). Desert Hedgehog/Patched 1 signaling specifies fetal Leydig cell fate in testis organogenesis. Genes Dev. 16, 1433-1440. 10.1101/gad.981202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F.-P., Poutanen M., Wilbertz J. and Huhtaniemi I. (2001). Normal prenatal but arrested postnatal sexual development of luteinizing hormone receptor knockout (LuRKO) mice. Mol. Endocrinol. 15, 172-183. 10.1210/mend.15.1.0582 [DOI] [PubMed] [Google Scholar]
- Zhang L., Chen M., Wen Q., Li Y., Wang Y., Wang Y., Qin Y., Cui X., Yang L., Huff V. et al. (2015). Reprogramming of Sertoli cells to fetal-like Leydig cells by Wt1 ablation. Proc. Natl. Acad. Sci. USA 112, 4003-4008. 10.1073/pnas.1422371112 [DOI] [PMC free article] [PubMed] [Google Scholar]