Abstract
During late gestation, villi extend into the intestinal lumen to dramatically increase the surface area of the intestinal epithelium, preparing the gut for the neonatal diet. Incomplete development of the intestine is the most common gastrointestinal complication in neonates, but the causes are unclear. We provide evidence in mice that Yin Yang 1 (Yy1) is crucial for intestinal villus development. YY1 loss in the developing endoderm had no apparent consequences until late gestation, after which the intestine differentiated poorly and exhibited severely stunted villi. Transcriptome analysis revealed that YY1 is required for mitochondrial gene expression, and ultrastructural analysis confirmed compromised mitochondrial integrity in the mutant intestine. We found increased oxidative phosphorylation gene expression at the onset of villus elongation, suggesting that aerobic respiration might function as a regulator of villus growth. Mitochondrial inhibitors blocked villus growth in a fashion similar to Yy1 loss, thus further linking oxidative phosphorylation with late-gestation intestinal development. Interestingly, we find that necrotizing enterocolitis patients also exhibit decreased expression of oxidative phosphorylation genes. Our study highlights the still unappreciated role of metabolic regulation during organogenesis, and suggests that it might contribute to neonatal gastrointestinal disorders.
KEY WORDS: Intestinal organogenesis, YY1, Villus, Oxidative phosphorylation, Necrotizing enterocolitis, Intestinal maturation, Mouse
Highlighted Article: The transcription factor YY1 plays a key role during intestinal morphogenesis in mice, affecting villi morphogenesis and the expression of mitochondrial genes.
INTRODUCTION
Necrotizing enterocolitis (NEC) is the most common gastrointestinal condition among newborns, affecting ∼7% of pre-term infants with a mortality rate of 20-30% (Neu and Walker, 2011; Zani and Pierro, 2015). The etiologies of NEC remain unclear, but they are expected to be multi-factorial and correspond closely with an underdeveloped intestine that is predisposed to deficiencies in digestion, barrier function and immune defenses. The immature intestine is susceptible to an excessive inflammatory response upon exposure to diet and colonization of microbiota, and ultimately manifests in necrosis of the tissue (Neu, 2014). Why a subset of pre-term babies may present with an immature intestine is unclear, necessitating a better understanding of intestinal development and maturation.
The intestine arises from a specified field of endodermal tissue that is shaped into a tube lined with a rapidly proliferating, pseudostratified epithelium by embryonic day (E) 9.5 in mouse (Grosse et al., 2011; Spence et al., 2011; Wells and Melton, 1999). Over the next 6 days of development, proliferation in the epithelium accompanies a tremendous expansion in the length and circumference of the intestine. By E14.5-E16.5, condensations of mesenchymal cells arise and serve as signaling centers that trigger the overlying epithelium to withdraw from the cell cycle, undergo a transition to a simple columnar morphology, and extend into luminal projections called villi (Burns et al., 2004; Kaestner et al., 1997; Madison et al., 2009; Mathan et al., 1976; Noah et al., 2011; Pabst et al., 1997, 1999; Shyer et al., 2015; Walton et al., 2012, 2016). Cells between villi remain proliferative and will ultimately undergo a morphological restructuring to form crypts of Lieberkhün. Meanwhile, villus cells are postmitotic and express genes appropriate to their differentiated function in secretion or absorption; epithelial cells transit towards the villus tips, where they ultimately delaminate into the lumen. A number of developmental signaling molecules and transcriptional regulators have been implicated in villus development (Choi et al., 2006; Kaestner et al., 1997; Karlsson et al., 2000; Kim et al., 2007; Lepourcelet et al., 2005; Madison et al., 2005; McLin et al., 2009; Ormestad et al., 2006; Shyer et al., 2013; Spence et al., 2011; van den Brink, 2007; Walker et al., 2014; Walton et al., 2012, 2016); however, the role of metabolic regulators in villus development has yet to be investigated.
Otto Warburg first recognized that cancer cells ferment much of their glucose supply into lactate regardless of the presence of oxygen, a phenomenon termed the Warburg effect or aerobic glycolysis (Warburg et al., 1924). Recently it has become better appreciated that metabolic shifts are also crucial in developmental processes. At the earliest stages of life glycolysis is preferred, as seen in the ESC-to-EpiSC transition (Zhou et al., 2012) and in the switch from oxidative phosphorylation to glycolysis that accompanies the conversion of differentiated cells into iPSCs (Folmes et al., 2011; Shyh-Chang and Daley, 2013). Metabolic processes have also recently been coupled to other developmental transitions, including macrophage activation (Tannahill et al., 2013), differentiation of mesenchymal and hematopoietic stem cells, and in the transition of satellite stem cells in muscle (Bracha et al., 2010; Chen et al., 2008; Qian et al., 2016; Takubo et al., 2013; Tormos et al., 2011; reviewed by Shyh-Chang et al., 2013). These studies highlight that metabolic processes are potentially driving regulators of development. In the adult intestine, a shift from glycolysis to oxidative phosphorylation correlates with the cell position along the crypt-to-villus axis (Stringari et al., 2012), and the suckling-weaning transition coincides with metabolic changes that are probably related to dietary changes (Mould et al., 2015). However, a role for metabolism has not been explored in intestinal organogenesis.
The transcription factor YY1 has been shown to play a significant role during cell differentiation, proliferation, and other major biological processes such as V(D)J rearrangement (Atchison, 2014), viral gene repression (Shi et al., 1991) and X inactivation (Jeon and Lee, 2011). YY1 also regulates mitochondrial genes, as it has been implicated in the maintenance of mitochondrial structure and function as well as in regulating oxidative phosphorylation and other ATP-yielding processes (Blattler et al., 2012; Perekatt et al., 2014). The molecular pathway through which YY1 acts to regulate these functions is unclear. Some studies have shown that YY1 functions downstream of the mTOR complex and PPARGC1α (also known as PGC1α) to control the transcription of these mitochondrial genes (Cunningham et al., 2007). mTOR plays a crucial role in intestinal regeneration after injury in adult mice, whereas mice with genetic ablation of mTOR components during mid-gestation have minimal deficiencies in normal adult homeostasis, particularly in the proximal gut, where villi are most pronounced (Faller et al., 2015; Sampson et al., 2016). However, the consequence of mTOR or YY1 disruption in early intestinal development has not been closely examined. Elucidating YY1 function in intestinal development therefore presents an opportunity to understand how the loss of a metabolic regulator might impact organogenesis in this tissue.
In the current study, we observe a regulatory shift in expression of oxidative phosphorylation genes that coincides with the process of villus formation. Genetic ablation of Yy1 in the developing intestinal epithelium results in a failure to elevate oxidative phosphorylation gene expression and in compromised villus elongation. To substantiate the link between aerobic metabolism and intestinal organogenesis, we show that pharmacological inhibition of the electron transport chain similarly compromises villus development. Finally, we demonstrate that expression of oxidative phosphorylation genes is decreased in NEC patient intestines, and propose that a deficient metabolic transition could underlie this prevalent neonatal condition.
RESULTS
YY1 expression in the developing endoderm is required for villus development
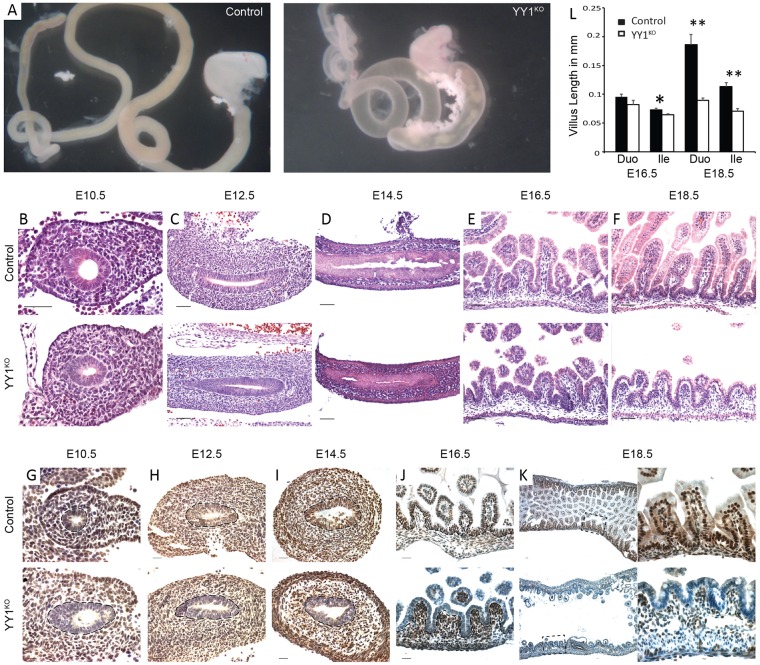
To characterize the role of YY1 during intestinal development, we employed the Shh-cre driver, which is expressed throughout the intestinal epithelium starting at ∼E9.5 (Harris-Johnson et al., 2009). At E18.5, Yy1f/f; Shh-cre pups (hereafter referred to as YY1KO) exhibited gross morphological defects with a translucent and distended intestine, indicating an underdeveloped intestine compared with littermate controls (Fig. 1A). Uniform, epithelial-specific YY1 loss was confirmed by immunostaining (Fig. 1G-K). Despite YY1 loss early in intestinal development, no morphological phenotype was observed until late gestation. We first observed reduced villus growth at E16.5 in Yy1 mutant embryos, although villus height at this stage was not significantly shorter (Fig. 1B-E). However, by E18.5, a dramatic failure of villus elongation was evident (Fig. 1F) and villus height in the mutants was significantly reduced compared with littermate controls (Fig. 1L), a pattern consistent from duodenum to ileum (Fig. S1).
Fig. 1.
YY1 expression in the developing endoderm is required for villus development. (A) YY1 loss leads to an underdeveloped intestine at E18.5, with mutant intestine being distended, translucent and devoid of villi. Yy1 mutants have similar morphology to control littermates at E10.5 (B), E12.5 (C) and E14.5 (D), subtle changes at E16.5 (E) and strikingly stunted villi at E18.5 (F) in the duodenum. YY1 immunostaining of duodenal sections shows specific and complete loss of YY1 immunoreactivity in the intestinal epithelium of Cre+ animals at E10.5 (G), E12.5 (H), E14.5 (I), E16.5 (J) and E18.5 (K). Dashed lines highlight the border between the epithelium and mesenchyme. (L) Villus height measurements are not dramatically altered in E16.5 duodenum (P=0.125, n=4) or ileum (P=0.026, n=4). However, Yy1 mutants have drastically reduced villus height at E18.5, a pattern observed from duodenum (**P=3×10−6, n=4) to ileum (**P=1×10−4, n=2). Error bars show s.e. Two-tailed t-tests were performed based on the results of an F-test for sample variances. Scale bars: 50 µm.
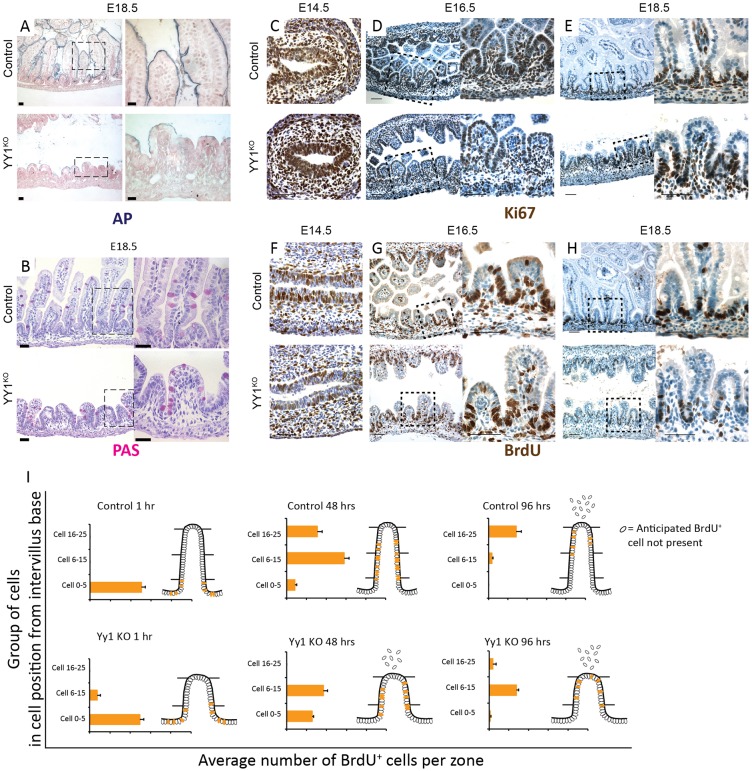
To further explore intestinal development upon YY1 loss, we examined proliferation and differentiation markers. Yy1 mutants exhibited an enterocyte differentiation defect as seen by reduced alkaline phosphatase staining (Fig. 2A, Fig. S2A), whereas periodic acid-Schiff staining suggested similar goblet cell development in mutants and controls (Fig. 2B, Fig. S2B). To evaluate whether the villus growth defect could be attributed to reduced proliferation or increased cell death, we assayed proliferation using Ki67 (Fig. 2C-E, Fig. S2C) and BrdU (Fig. 2F-H), and found proliferation to be unaffected upon YY1 loss (Fig. S2G). Also, we found no significant apoptosis upon YY1 loss, as measured by cleaved caspase 3 staining (Fig. S2D). The epithelium also had fewer cells (Fig. S2F), suggesting that the villus elongation deficit was not attributable to pressure accumulating in the lumen. To better understand why the Yy1 mutant epithelium had fewer cells, we traced epithelial cells over time using BrdU pulse-chase labeling at 1, 48 and 96 h of chase time, concluding at E18.5. In both Yy1 mutants and control littermates, BrdU cells migrated towards villi tips. However, BrdU-labeled cells were eliminated from the mutant epithelium more rapidly, possibly owing to the shorter villus length, which would shorten their transit path to the villus tip (Fig. 2I, Fig. S2E).
Fig. 2.
YY1 loss compromises enterocyte differentiation but not cell proliferation. (A) Yy1 mutant enterocytes exhibit diminished alkaline phosphatase (AP) staining at E18.5, whereas goblet cell differentiation appears normal, as seen by periodic acid-Schiff (PAS) staining (B). Yy1 mutants have similar numbers of proliferating cells, as seen by Ki67 (C-E) and BrdU (F-H) staining. (I) BrdU pulse-chase labeling suggests a more rapid elimination of mutant cells from the epithelium (1 h control, n=2; 1 h mutant, n=2; 48 h control, n=4; 48 h mutant, n=3; 96 h control, n=4; 96 h mutant, n=3). Error bars represent s.e. All histological staining and quantification were performed on duodenal regions. Scale bars: 50 µm.
Taken together, our results indicate that YY1 is dispensable for proliferation in the developing intestinal epithelium, but is clearly required for proper differentiation of enterocytes and elaboration of full villi.
Villogenesis initiates despite loss of YY1
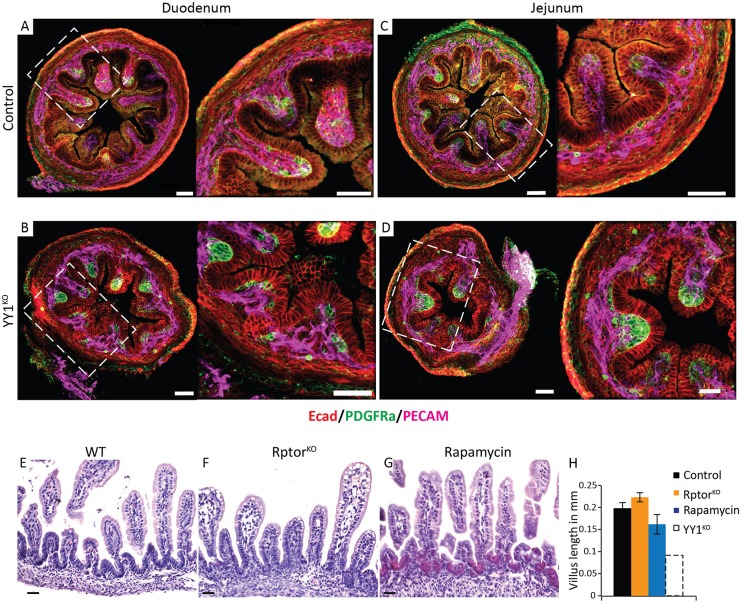
To better understand why villi are stunted in YY1KO animals, we evaluated whether initiation of villogenesis is disrupted. We stained for the presence of PDGFRα-expressing mesenchymal clusters, which form the signaling foci from which villus development proceeds (Walton et al., 2012). PDGFRα clusters were clearly present in the Yy1 mutants at E16.5 (Fig. 3A-D), suggesting that villogenesis initiates normally. These results thereby highlight that villogenesis initiates properly in the absence of YY1, yet extension of the nascent villi into the lumen is subsequently compromised.
Fig. 3.
YY1 is dispensable for the onset of villogenesis; YY1 acts independently of mTOR signaling in regulating villogenesis. (A-D) Immunofluorescent staining of E16.5 YY1KO intestine shows PDGFRα expression in mesenchymal clusters associated with villogenesis, similar to littermate controls. PECAM1 staining shows that Yy1 mutants have similar vascularization as the control. Villogenesis occurs normally upon inhibition of the mTOR pathway by genetic ablation of Rptor (F) or pharmacological inhibition using rapamycin (G), as compared with controls (E) at E18.5. mTOR pathway inhibition by RptorKO and rapamycin treatment was confirmed by western blot using pS6 antibody (Fig. S3A,B). E18.5 duodenal villi heights in mTOR-inhibited embryos were unchanged compared with controls (P=0.207, P=0.218). Control or mTOR-inhibited intestines were drastically longer than YY1KO villi heights (as seen in Fig. 1L). Error bars represent s.e. Two-tailed t-tests were performed based on the results of an F-test for sample variances. Scale bars: 50 µm.
YY1 acts independently of mTOR signaling in villus development
A recent study inactivating mTOR signaling during intestinal development did not compromise duodenal villus length in the adult (Sampson et al., 2016); however, villus formation was not investigated during early intestinal development. Since YY1 has been demonstrated to function downstream of mTOR (Cunningham et al., 2007), we queried whether disruption of mTOR signaling in the developing intestine would yield a similar villus elongation defect as observed upon YY1 loss. We inhibited mTOR signaling in the developing intestine using two approaches: pharmacologically, with the mTOR complex 1 (mTORC1) inhibitor rapamycin; and genetically, using conditional ablation of Rptor (RptorKO). Rptor is an essential component of mTORC1 that is required for mTOR-mediated nutrient sensing, and rapamycin destabilizes the mTOR-Rptor complex (Kim et al., 2002). We confirmed mTOR signaling inhibition via both approaches at E18.5 using western blot staining of phosphorylated S6 kinase (pS6), a canonical mTOR target, in whole gut tissues (Fig. S3A,B). However, villus length was unaffected in RptorKO and rapamycin-treated embryos compared with controls (Fig. 3E-H) and significantly longer than in YY1KO (Fig. 1L, Fig. 3H). Thus, although mTOR may be involved in regulating other intestinal processes (Faller et al., 2015; Sampson et al., 2016), our findings suggest a dispensable role of mTOR in villus development. This also indicates that, unlike YY1 function downstream of mTOR in other tissues (Blattler et al., 2012), YY1 acts independently of mTOR in regulating villus growth.
YY1 loss leads to mitochondrial dysfunction
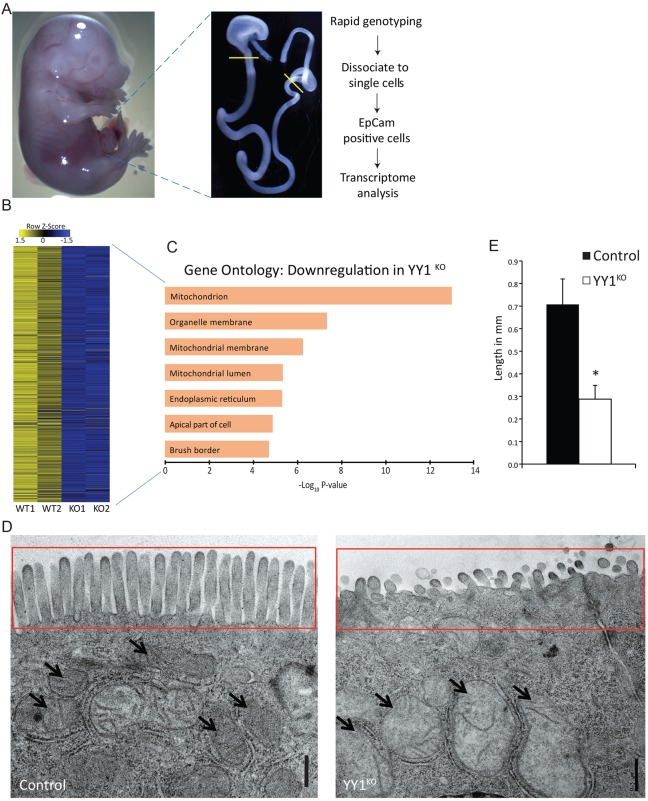
To elucidate the underlying cause of deficient villus elongation in the absence of Yy1, we performed transcriptome analysis at E15.5, early in villus formation, immediately prior to when morphological phenotypes were discernable in the mutant. Small intestinal epithelial cells from E15.5 embryos were enriched by dissection and purified using antibodies to the epithelial cell-specific marker Epcam (Fig. 4A). Transcriptome analysis identified 800 unique genes that were downregulated upon YY1 loss [log2 fold change (FC)<−1, Benjamini-Hochberg adjusted P<0.05; Fig. 4B], as confirmed by qRT-PCR for selected genes (Fig. S3C). Products of these genes were enriched for localization to the brush border, consistent with the deficient alkaline phosphatase activity ascribed to this structure (Fig. 2A, Fig. S2A), and supporting the idea that YY1 promotes maturation of the developing intestinal epithelium.
Fig. 4.
YY1 loss compromises mitochondrial ultrastructure and gene expression. (A) Experimental workflow to capture the E15.5 epithelial transcriptomes from control and YY1KO embryos. (B) Eight hundred genes were identified as downregulated (log2 FC>1 and Benjamini-Hochberg adjusted P<0.05) upon YY1 loss, and (C) their enriched ontologies, demonstrating reduced expression of nuclear mitochondrial genes upon YY1 loss. (D,E) Ultrastructural analysis of enterocytes shows deformed mitochondria (arrows) and significantly stunted microvilli (D, red box; E, *P=0.0467). Error bars represent s.e. Two-tailed t-tests were performed based on the results of an F-test for sample variances. Scale bars: 0.5 µm.
Mitochondrial function was the most strongly enriched attribute of genes downregulated in YY1KO epithelia (Fig. 4C), and many of these genes have been shown to bind YY1 via ChIP-seq (Table S1). We therefore examined mitochondrial ultrastructure using transmission electron microscopy (TEM), and revealed that Yy1 loss leads to defective mitochondrial morphology, with distended, electron-poor inner membrane space, and disrupted cristae (Fig. 4D). TEM also revealed that microvillus length was significantly reduced (Fig. 4E,D, red boxes), consistent with downregulation of genes associated with the brush border (Fig. 4C).
Together, transcriptomic analysis of E15.5 epithelium points to a role for YY1 regulation of mitochondrial function, and suggests that mitochondrial dysfunction disrupts proper villus growth and maturation in the developing intestine.
A metabolic shift to increased oxidative phosphorylation is required for villus development
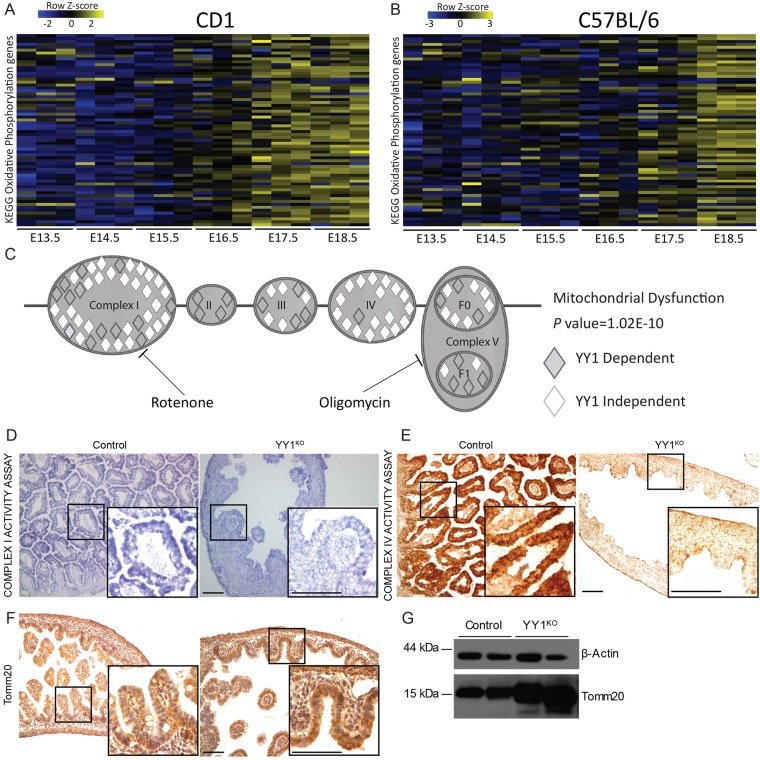
Although mitochondrial structure was clearly dependent upon YY1 in the developing gut (Fig. 4D), it was surprising to find that a phenotype only manifested itself ∼7 days after Shh-cre-mediated deletion of Yy1 at E9.5, especially considering that Yy1 transcripts (Fig. S4A) and protein levels (Fig. 1G-K) are fairly stable across developmental time. Consistent with mitochondrial localization, genes downregulated upon YY1 loss are also enriched for function in oxidative phosphorylation (OxPhos, ranked second after mitochondrial dysfunction in Ingenuity pathway analysis – canonical pathways; P=1.02×10−10) and involved in all complexes of the electron transport chain (ETC) (Fig. 5C). To understand why reduction of OxPhos genes does not cause a YY1KO phenotype prior to E16.5, we analyzed their expression during stages of intestinal development (Kaiser et al., 2007). Interestingly, OxPhos genes show a marked increase in expression at ∼E16.5 in mouse intestine (Fig. 5A,B), coincident with the time of the villus elongation phenotype in Yy1 mutants. Our analysis thus reveals a shift in metabolic gene expression to OxPhos that coincides with villus elongation. We further corroborated our findings by showing that genes downregulated upon YY1 loss are destined to increase at ∼E16.5 in the developing intestine (Fig. S4B,C) and are enriched for mitochondrial function (Fig. 4C).
Fig. 5.
Failure of oxidative phosphorylation gene expression at the onset of villus elongation in the absence of YY1. (A,B) Developmental timecourse showing expression of oxidative phosphorylation genes in mouse intestine in CD1 (A) and C57BL/6 (B) genetic backgrounds. (C) Genes downregulated upon YY1 loss (log2 FC>0.27 and Benjamini-Hochberg adjusted P<0.05, 2475 genes) were analyzed using IPA; the mitochondrial dysfunction canonical pathway (P=1.02×10−10) is enriched, highlighting YY1-dependent genes involved in all complexes (I-V) of the electron transport chain. Inhibitory targets of mitochondrial toxins are shown, with rotenone inhibiting complex I and oligomycin inhibiting complex V. (D,E) Mitochondrial activity assays for respiratory chain complex I (D, NADH-coenzyme Q reductase, blue) and complex IV (E, cytochrome C oxidase, brown) show dramatically reduced activity in Yy1 mutant epithelial cells. Non-epithelial cells exhibited similar staining in Yy1 mutant and controls; also see Fig. S4. (F,G) Tomm20 immunostaining (F) reveals increased but more diffuse immunoreactivity in the mutants. Elevated Tomm20 protein levels were also observed by immunoblot (G). β-actin is a loading control. Scale bars: 50 µm.
To determine whether mitochondrial function is compromised in the Yy1 mutant epithelium, we performed assays of mitochondrial activity, in situ, to separately query the activities of mitochondrial complexes I and IV. In both cases, epithelial cells in the Yy1 mutant were severely deficient in these activities compared with control epithelial cells, whereas cells in the developing muscle and stroma showed similar levels of mitochondrial complex activities (Fig. 5D,E, Fig. S4D,E). As an indicator of mitochondrial mass, we performed immunostaining and immunoblots for Tomm20. Consistent with the distended mitochondria observed by TEM (Fig. 4D), Tomm20 immunoreactivity was dispersed in Yy1 mutant epithelial cells (Fig. 5F). Additionally, western blot revealed that Tomm20 protein levels are elevated in the mutant (Fig. 5G). Increased Tomm20 levels might be a response to the energy demand sensed by the cells as a result of compromised respiratory transport chain function. Together, these assays are consistent with the gene expression analysis, and indicate that OxPhos is compromised in the Yy1 mutant epithelium.
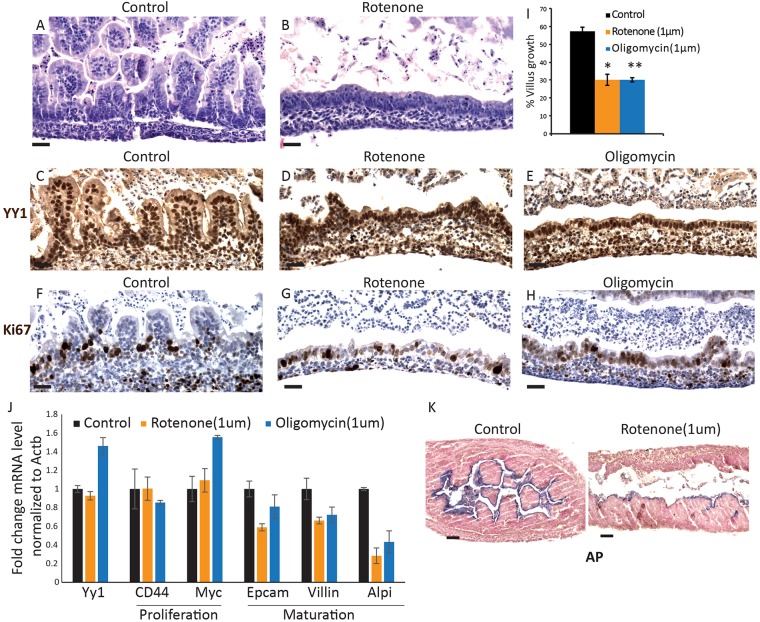
To test our hypothesis that OxPhos is indispensable to support the metabolic requirements of a budding villus structure, we utilized an explant culture system in which villus development can be observed ex vivo (Walton et al., 2012). Intestine segments of developing duodena were cultured in the presence or absence of the mitochondrial inhibitors rotenone (inhibits ETC complex 1) or oligomycin (inhibits complex 5) (Fig. 5C). Targeting these complexes reduced the formation of villus structures and revealed that ETC function is required for villus elongation (Fig. 6A-I), akin to YY1 activity in vivo. These results suggest that increased aerobic respiration is crucial to intestinal epithelial development, specifically during late gestation and villus development. Notably, the concentrations of ETC inhibitors used in this experiment did not compromise cell proliferation (Fig. 6F-H) or cause significant apoptosis (Fig. S5A-C), suggesting that the effects of the rotenone and oligomycin treatments were not generally toxic but were more specifically disruptive in the manifestation of mature villi (Fig. S5D). Indeed, RT-qPCR on explant tissues reveals that inhibition of the ETC reduces expression of villus maturation genes but does not affect proliferating cell markers (Fig. 6J). This is corroborated by reduced alkaline phosphatase staining upon ETC inhibition (Fig. 6K). Together, these results indicate that increased aerobic respiration is indispensable for villus elongation and maturation of intestinal cells.
Fig. 6.
Mitochondrial activity is required for villus development. (A,B) E14.5 explants cultured for 96 h in control and rotenone-supplemented media exhibit a failure to elaborate villi. Mitochondrial inhibitors did not affect YY1 levels (C-E) or proliferation (F-H), similar to the YY1KO phenotype (Figs 1, 2). (I) Quantification of villus formation in the explants. *P=0.00096, **P=0.001705, versus control. (J) qRT-PCR shows reduced levels of enterocyte marker transcripts upon treatment of explants with mitochondrial inhibitors. For statistical analysis, inhibitor treatments were grouped (n=4) and tested against the control (n=3). Epcam, P=0.0564; villin, P=0.0866; Alpi (intestinal alkaline phosphatase), P=0.00329. (K) Explants treated with rotenone also exhibited decreased alkaline phosphatase activity compared with controls. Error bars represent s.e. Two-tailed t-tests were performed based on the results of an F-test for sample variances. Scale bars: 50 µm.
NEC patients exhibit a transcriptome deficient in YY1-dependent genes and diminished in OxPhos gene expression
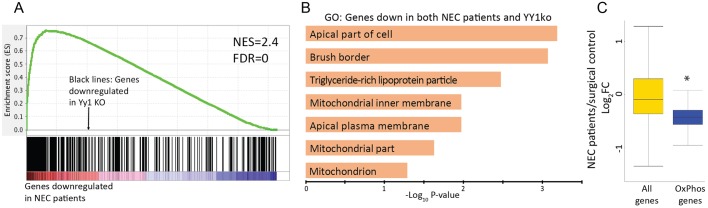
Underdeveloped intestines are suspected to be a primary cause of NEC (Neu and Walker, 2011; Zani and Pierro, 2015), but the molecular etiologies of NEC remain unclear. Given that loss of YY1 in the developing gut leads to a phenotype reminiscent of that of NEC patient bowels, we explored the possibility that NEC patients exhibit deficiencies in the YY1-dependent transcriptome. We analyzed expression data from NEC patients compared with surgical control patients (Chan et al., 2014) using Gene Set Enrichment Analysis (GSEA). A strong correlation was observed between genes downregulated in NEC patients and genes downregulated upon YY1 loss (Fig. 7A). Genes enriched at the leading edge of this correlation between NEC patients and YY1 loss were enriched in mitochondrial and brush border gene ontologies (Fig. 7B), consistent with altered metabolism and poorly matured intestine in both systems.
Fig. 7.
Necrotizing enterocolitis patients exhibit transcriptomes similar to those of Yy1 mutants. (A) GSEA analysis reveals that genes downregulated upon YY1 loss strongly correlate with genes of reduced expression in NEC patients. (B) Genes at the leading edge of the GSEA, representing those downregulated in both NEC patients and YY1KO transcriptomes, are enriched in the indicated gene ontologies. (C) Box plot showing that NEC patients exhibit an overall reduction in oxidative phosphorylation gene expression (Wilcoxon test, *P=2.20×10−10), suggesting a potential contribution to this disease etiology. Data in C are derived from four NEC patients and four surgical controls (GSE46619).
To corroborate our findings, we analyzed whether OxPhos genes were underexpressed in NEC patient samples. We indeed observed that OxPhos genes are significantly reduced in NEC patients (Fig. 7C, P=2.20×10−10, Wilcoxon test). Although YY1 target genes were underexpressed in the NEC patient group, YY1 itself was unaffected (average expression value of 10.41±0.04 in controls versus 10.40±0.05 in patients, ±s.d.), suggesting that multiple pathways are responsible for the diminished OxPhos gene expression in these patients. Our study thus highlights deficiencies in aerobic metabolism as a possible underlying etiology of NEC.
DISCUSSION
There has been a renewed interest in cellular metabolism in recent years, particularly in the cancer field, where the observation of Warburg metabolism has undergone a renaissance. Warburg observed that cancer cells ferment sugar (an inefficient source of 2 ATPs per molecule of glucose), even in the presence of oxygen (conditions in which differentiated cells can produce 36 molecules of ATP by metabolizing sugar to CO2 through oxidative phosphorylation) (Warburg et al., 1924). Recent hypotheses to explain the Warburg effect have centered upon the fact that biomass is a key requirement for a growing tissue, and possibly more important than ATP. Glycolysis provides glycolytic intermediates, which are the building blocks for amino acids and nucleotides, and acetyl-CoA for fatty acids (Vander Heiden et al., 2009). Thus, a growing tissue, be it in a tumor or in a developing embryo, benefits from the fermentation of sugar to acquire biomass. Our study identifies an increase in OxPhos genes as the intestine takes on a more differentiated role. We propose that glycolysis is the preferred metabolism from E9.5 through E16.5 in order to increase biomass as the gut is rapidly expanding and as nearly all cells are proliferative. Then, as cells begin to differentiate on elongated villi at E17.5, we believe that a shift to oxidative metabolism is favored, increasing ATP availability for transporters and enzymes associated with digestion. Interestingly, we find that enterocyte differentiation appears more affected than goblet cell differentiation; this finding might suggest different metabolic demands for these epithelial cell lineages.
Several recent studies have observed an association between metabolic state and developmental processes, including in hematopoietic, mesenchymal and neural lineages (Shyh-Chang et al., 2013; Ufer and Wang, 2011). In the mature intestine, where rapid growth in intestinal crypts feeds differentiated cells onto the villi, it was recently observed that the proliferating progenitor cells exhibit a more glycolytic metabolism than their differentiated counterparts (Stringari et al., 2012; Xiong et al., 2015). A key role of future studies will be to determine whether metabolic states are dependent on developmental signaling processes, whether developmental signaling processes depend upon metabolic states, or most likely an interdependence of the two processes. Along these lines, interesting recent analyses have placed metabolic control under the important intestinal WNT signaling pathway (Pate et al., 2014). We have previously shown that the WNT-dependent adult intestinal stem cells rely upon YY1 for renewal; whether there is a relationship between WNT and YY1 in mitochondrial regulation has not been explored, and YY1-dependent control of stem cell markers in the developing gut is less clear (Fig. S5E). Another metabolic regulator, PPARGC1α, has been shown to work as a co-factor with YY1 in other contexts (Cunningham et al., 2007). PPARGC1α and its family members are downregulated in the Yy1 embryonic intestinal mutant (Fig. S5F) and could play a role in mitochondria-dependent villus growth as well.
NEC is a major cause of neonatal mortality but the etiologies behind NEC are unknown (Neu and Walker, 2011; Zani and Pierro, 2015). Our study found a strong correlation between the transcriptomes of Yy1 mutants and NEC patients, raising the possibility that metabolic dysfunction could contribute to NEC. We also observed that OxPhos genes are downregulated in NEC patients, which could reflect the often-observed microvascular defects in NEC intestines, the underdeveloped state of NEC guts, or could be a primary deficit leading to the NEC phenotype. Our work, showing that intestinal maturation is compromised by disrupting a regulator of OxPhos genes or by directly inhibiting the ETC, demonstrates that a primary disruption of a metabolic shift can lead to an NEC-like phenotype. Additionally, our study highlights the potential impact of environmental toxins on NEC, as we demonstrate that mitochondrial toxins that are commonly used as pesticides can elicit an NEC phenotype. Future studies could explore whether there is a link between the environmental use of these toxins and NEC incidence.
MATERIALS AND METHODS
Mice
Yy1f/f mice (Affar et al., 2006) and Shh-cre mice (Harfe et al., 2004; Harris-Johnson et al., 2009) were purchased from Jackson Labs and male Shh-cre; Yy1f/+ mice were bred to Yy1f/+ and Yy1f/f mice for experimental litters. All mouse protocols and experiments were approved by Rutgers Institutional Animal Care and Use Committee.
Tissue preparation
For paraffin processing, duodenum, ileum or full small intestine was fixed overnight in 4% paraformaldehyde at 4°C, washed with PBS, and passed through increasing concentrations of an ethanol series and paraffin before embedding. For explant cultures, Histogel (ThermoFisher Scientific, HG-4000-012) was used during processing and embedding. In the case of tissue prepared for BrdU immunohistochemistry, the mice were injected with 1 mg BrdU 1 h, 48 h or 96 h before euthanasia. For immunofluorescent stains, E16.5 small intestines were fixed overnight in 4% paraformaldehyde at 4°C, washed with PBS, then embedded in 7% agarose and Vibratome sectioned at 100 µm.
Rapamycin (LC laboratories, R-5000) treatment was for 5 days beginning at E13.5 at 5 mg/kg. Rapamycin was dissolved in 100% ethanol, diluted in 5% Tween 80 and 5% PEG 400 (final concentrations). To induce Rptor knockout, pregnant dams were injected with tamoxifen (1 mg) for 4 days at beginning E13.5 and harvested at E18.5.
Rapid genotyping
Pregnant dams were sacrificed and dissected embryos were kept in ice-cold PBS. Embryo tail tissue was used for genotyping using KAPA Mouse Genotyping Kits (Kapa Biosystems, KK7352).
Immunohistochemistry
Intestinal sections (5 µm cut from paraffin blocks) were processed for immunostaining with the indicated primary antibodies, developed using the Vectastain ABC Kit (Vector Laboratories, PK6101) and counterstained with Hematoxylin. A 1 h antigen-retrieval step in 10 mM sodium citrate solution under 15 psi pressure was used for all stains. Slides were incubated in primary antibody overnight at 4°C. Primary antibodies and dilutions used for staining were: YY1 (Santa Cruz, E0511; 1:500), cleaved caspase 3 (CC3; Cell Signaling, 9661S; 1:200), BrdU (AbD Sterotec, MCA2060GA; 1:500), Tomm20 (Santa Cruz, sc-11415; 1:50) and Ki67 (Abcam, ab15580; 1:300). For cleaved caspase 3, the primary antibody was diluted in the Signal Stain Ab Diluent (Cell Signaling, 8112S). For periodic acid-Schiff staining, slides were incubated in 0.5% periodic acid and stained with Schiff's reagent (J612171, Alfa Aesar). For alkaline phosphatase staining, 1-Step NBT/BCIP (Thermo Scientific, 34042) was used along with Neutral Red counterstain. Before developing the alkaline phosphatase, the slides were incubated in 0.1 M Tris-HCl buffer pH 8.
Images were taken using a Retiga 1300 CCD (Q-Imaging) camera and a Nikon Eclipse E800 microscope with the QC-Capture imaging software. Oil was used for 60×/1.4 N.A. magnification and air for 10×/0.45 N.A., 20×/0.75 N.A. and 40×/0.75 N.A. Adjustments in contrast and brightness, when made, were applied uniformly to complete figure panels in Adobe Photoshop.
Vibratome sections were permeabilized in 0.1% Triton X-100 for 25 min, washed three times for 5 min each with PBS, and blocked with 5% normal goat serum before primary antibody was added overnight at 4°C. Primary antibodies were E-cadherin (BD Transduction Labs, 610181; 1:1000), PECAM1 (CD31; BD Transduction Labs, 557355; 1:1000) and PDGFRα (Santa Cruz, sc338; 1:200). Sections were then washed three times for 15 min each in blocking solution before DAPI and donkey anti-mouse Alexa 555, goat anti-rabbit Alexa 488 or goat anti-rat Alexa 647 (all Invitrogen; 1:1000) secondary antibodies were added for 1 h at room temperature. Finally, sections were washed three times for 15 min each in PBS and mounted on slides with ProLong Gold (ThermoFisher Scientific) for confocal imaging on a Nikon A1 confocal microscope with Nikon Elements software. Three-dimensional images were reconstructed using Imaris 8.0.2 software (Bitplane).
Quantification
ImageJ software was used to calculate villi and microvilli length. Only villi that were complete and protruding from the basement membrane were considered. Only microvilli with complete longitudinal sections were measured. For epithelial cell counts (Fig. S2F), 0.2 mm duodenal regions (E14.5) and 0.663 mm duodenal regions (E18.5) were quantified along one side of the intestine and then normalized to 1 mm. For explant experiments, only sections of tissue containing intact epithelium, mesenchyme and the basement membrane were measured. Regions of growth in explant assays were characterized as areas with villi protruding above the flat epithelial layer. Explant experiments were performed five times and multiple counts were averaged for each biological replicate to measure the region of villus growth (control n=3, rotenone n=6, oligomycin n=2) and biological replicates were analyzed for significance using a two-tailed t-test based on the results of an F-test for sample variances between biological replicates.
For the BrdU pulse-chase experiment (Fig. 2I), the average number of BrdU-positive cells per villus was determined using immunohistochemistry and grouped according to BrdU-positive cell position relative to the intervillus base. (group1, cells up to position 5; group 2, cells in positions 6-15; group 3, cells in positions 16-25). Averages were taken from up to four biological replicates of each condition.
Western blot
Whole cell lysate (50 μg) from whole small intestine tissue of control, rapamycin-treated or RptorKO mice was heat denatured in 5×SDS sample buffer, separated by 4-12% gradient SDS-PAGE, and transferred onto a PVDF membrane (Millipore, ISEQ00010). The membrane was blocked with 5% skimmed milk in PBS containing 0.1% Tween 20 for 1 h, and then pS6 (Cell Signaling; #4858; 1:1000), Tomm20 (as above; 1:1000) and β-actin (Abcam; ab8227; 1:5000) antibodies were used to detect proteins.
Microarray
To measure transcriptome differences between E15.5 epithelial cells of Shh-cre; Yy1f/f and Cre-negative Yy1f/f embryos, two biological replicates were used for each genotype. Each biological replicate included either cells isolated from one fetal intestine, or cells pooled from two intestines of the same genotype. Replicates all came from the same litter. Small intestines (proximal stomach to distal caecum) were dissected and cut into small pieces. Tissue pieces were then trypsinized with trypsin-EDTA 0.05% (ThermoFisher Scientific, 25300-054) for 5 min and then neutralized with 10% FBS to isolate single cells. Single cells were incubated with PE-conjugated anti-CD326 (Epcam clone G8.8, eBiosciences, 12-5791-81) for 15 min on ice. PE-stained cells were then incubated for 15 min with magnetic bead-conjugated anti-PE antibody (Miltenyi Biotec, Anti-PE MicroBeads, 130-048-801). Enriched epithelium cells were obtained by passing cells through a column (Miltenyi Biotec, MS Columns, 130-042-201) in a magnetic field to obtain Epcam-positive, magnetic antibody-conjugated cells. Cells were then dissolved in Trizol and RNA prepared by ethanol precipitation using 10 μg linear polyacrylamide (LPA; Sigma-Aldrich). RNA was then processed for microarray on Affymetrix Mouse 430_2 arrays according to manufacturer's instructions. Data were analyzed using R Bioconductor (Gentleman et al., 2004) with gcrma normalization (Wu et al., 2004) and the limma package (Smyth, 2004). 1495 unique genes were differentially expressed with log2 FC >1 and Benjamini-Hochberg adjusted P<0.05 (Table S1); 800 genes were identified as downregulated (log2 FC<−1 and Benjamini-Hochberg adjusted P<0.05) and were analyzed for gene ontology using DAVID (Dennis et al., 2003).
TEM
Intestinal tissues were dissected, flushed in cold PBS, minced into 1-3 mm fragments and fixed overnight at 4°C in 0.1 M sodium cacodylate buffer (pH 7.4) containing 2.5% glutaraldehyde and 2.0% (vol/vol) paraformaldehyde. Fixed tissues were washed briefly with 0.1 M sodium cacodylate buffer and postfixed with 1% osmium tetroxide in 0.1 M sodium cacodylate buffer for 1 h at 4°C. Tissues were washed with excess distilled water and en bloc-stained with 1% aqueous uranyl acetate for 30 min in the dark. Tissues were then washed with distilled water and dehydrated through a series of ethanol and propylene oxide. Tissues were then transferred into EMBed 812 (Electron Microscopy Sciences, 14120) and propylene oxide (mixed at 1:1) overnight at room temperature in tightly capped vials on a shaker. Tissues were then transferred into 100% EMBed 812 (Electron Microscopy Sciences) overnight at room temperature. Tissues were embedded in embedding molds at 65°C for two nights. Ultrathin sections (70 nm) were cut; grids were stained with uranyl acetate and lead citrate and observed under an FEI Tecnai 12 transmission electron microscope with a Gatan cooled CCD camera. Multiple grids were analyzed for each intestinal sample. The image was adjusted in Adobe Photoshop using the hue/saturation adjustment tool to +7, the brightness and contrast adjustment tool to +28 and +7, respectively, and finally selecting ‘increase contrast 1' using the levels adjustment tool.
Informatics analysis
Genes involved in oxidative phosphorylation (135 genes) were obtained from the GSEA-KEGG oxidative phosphorylation list (Liberzon et al., 2011; Kanehisa et al., 2016; Kanehisa and Goto, 2000). Those involved in lysosomal ontologies (23 genes), not directly implicated in oxidative phosphorylation, were omitted to obtain a final oxidative phosphorylation gene list (112 genes) (Table S1). A custom R script was used to prepare heatmaps of oxidative phosphorylation gene expression in mouse colon of CD1 and C57BL/6 genetic backgrounds (GSE5204) (Kaiser et al., 2007).
NEC patient microarray CEL files (GSE46619) were obtained from a previous study in which whole-genome microarray expression profiling was performed on bowel tissue from neonates diagnosed with NEC and surgical controls (Chan et al., 2014). We processed the CEL files using R Bioconductor (Gentleman et al., 2004) with gcrma normalization (Wu et al., 2004) and the limma package (Smyth, 2004). Principal component analysis and hierarchical clustering were performed as quality control, and one replicate found to be an outlier was omitted from further analysis. Our study analyzed four biological NEC replicates and four surgical controls. Genes downregulated in NEC patients were analyzed using DAVID (Dennis et al., 2003). Correlation study of NEC patients and YY1 downregulated genes was performed using GSEA (scoring scheme, weighted; permutations, 10,000) (Subramanian et al., 2005). NEC patient log2 FC of oxidative phosphorylation genes and all genes was calculated and Wilcoxon test used to evaluate statistical significance using a custom R script. A schematic of oxidative phosphorylation genes dependent on YY1 was prepared by modifying the pathway workflow obtained from IPA (www.qiagen.com/ingenuity) using genes downregulated upon YY1 loss (log2 FC<−0.27 and Benjamini-Hochberg adjusted P<0.05, 2475 genes).
YY1 ChIP-seq peaks were obtained from a previous study that identified YY1 binding sites in adult intestinal epithelium (GSM1295000) (Perekatt et al., 2014). Genes within 30 kb regions of YY1 binding sites were overlapped with genes downregulated in intestinal epithelium upon YY1 loss (log2 FC<−0.27 and Benjamini-Hochberg adjusted P<0.05, Table S1).
Mitochondrial activity assays
Complex I (NADH dehydrogenase-NADH coenzyme Q reductase complex) activity was assessed for mouse cryosections using previously described methods (Christensen and Diemer, 2003; Hench et al., 2011). In brief, flash-frozen tissues were prepared for cryosection (4 μm). Sections were covered with reaction medium [0.2 M Tris buffer pH 7.4 containing 1 mg/ml tetranitroblue tetrazolium (NBT), 5 mM MgCl2, 25 mM CoCl2 and 2 mg/ml NADH] at room temperature. After development of the blue color (8-10 min) the reaction was stopped using 4% formalin (10-15 min), then washed twice with distilled water and mounted in water-based mounting medium. Complex IV (cytochrome C oxidase) activity was assessed as previously described (Seligman et al., 1968). Briefly, cryosections (4 μm) were incubated in reaction buffer [2.5 ml 0.2 M phosphate buffer pH 7.6, 20 mg catalase, 10 mg cytochrome c, 5 mg 3,3′-diaminobenzidine tetrahydrochloride hydrate (DAB) and 750 mg sucrose in 7.5 ml deionized water] for 1 h, rinsed three times with water, dehydrated and mounted in xylene-based mounting medium.
Explants
Proximal duodena were dissected from E14.5 and E15.5 embryos, connective tissue was separated, and intestines were placed on Transwell membrane (Costar, 3428) in BGJb medium (Invitrogen, 12591-038) supplemented with 1% pen/strep (vol/vol) and 0.1 mg/ml ascorbic acid (Sigma, A4403). For mitochondrial inhibition, base medium was supplemented with 1 µM rotenone (Sigma, 45656) or 1 µM oligomycin (Sigma, O4876). Intestines were cultured for 72 h (starting at E15.5) or 96 h (starting at E14.5) at 37°C with 5% CO2 with medium changes every 24 h. For qPCR analysis, intestinal explants were cultured for 48 h (starting at E15.5). Tissue was dissolved in Trizol and RNA prepared using the RNeasy Micro Kit (Qiagen, 74004). The RNA was reverse transcribed using the SuperScript III First-Strand Synthesis System (Invitrogen, 18080051) to prepare cDNA for qPCR analysis using gene-specific primers and SYBR Green PCR Master Mix (Applied Biosystems, 4309155).
Acknowledgements
We thank Dr B. J. Aronow, Cincinnati Children's Hospital Medical Center, for insights on the mouse intestinal timecourse expression study and Dr Karen Schindler, Rutgers University, for providing access to the dissecting microscope with camera mount. Vibratome confocal imaging and three-dimensional reconstructions were performed in the Microscopy and Image Analysis Laboratory at the University of Michigan Medical School.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
N.K., M.S., S.J., K.D.W., A.Z., W.J.F., A.O.P., E.M.B. performed the research; N.K., M.S., S.J., K.D.W., O.J.S., D.L.G., J.X., E.M.B., N.G., E.W., M.P.V. interpreted data; N.K., M.S., M.P.V. drafted the manuscript; N.K. and M.P.V. conceived the study.
Funding
Work in the M.P.V. lab was supported by a grant from the National Institutes of Health (NIH) [R03DK099251] and the Human Genetics Institute of New Jersey. N.G. and M.P.V. were supported by the Rutgers Faculty Research Program. Work at the University of Michigan was funded by an NIH grant [R01 DK089933] and the University of Michigan Gastrointestinal Peptide Research Center. The E.W. lab was supported through NIH grants [R01 CA130893, R01 CA188096, R01 CA193970 and R01 CA163591]. This research was also supported by the Flow Cytometry Shared Resource of the Rutgers Cancer Institute of New Jersey [P30CA072720]. Deposited in PMC for release after 12 months.
Data availability
Microarray datasets for Yy1f/f and Yy1f/f; Shh-cre small intestine are available at Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE85602) under accession number GSE85602.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.137992.supplemental
References
- Affar E. B., Gay F., Shi Y., Liu H., Huarte M., Wu S., Collins T., Li E. and Shi Y. (2006). Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol. Cell. Biol. 26, 3565-3581. 10.1128/MCB.26.9.3565-3581.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchison M. L. (2014). Function of YY1 in long-distance DNA interactions. Front. Immunol. 5, 45 10.3389/fimmu.2014.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattler S. M., Verdeguer F., Liesa M., Cunningham J. T., Vogel R. O., Chim H., Liu H., Romanino K., Shirihai O. S., Vazquez F. et al. (2012). Defective mitochondrial morphology and bioenergetic function in mice lacking the transcription factor Yin Yang 1 in skeletal muscle. Mol. Cell. Biol. 32, 3333-3346. 10.1128/MCB.00337-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha A. L., Ramanathan A., Huang S., Ingber D. E. and Schreiber S. L. (2010). Carbon metabolism-mediated myogenic differentiation. Nat. Chem. Biol. 6, 202-204. 10.1038/nchembio.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns R. C., Fairbanks T. J., Sala F., De Langhe S., Mailleux A., Thiery J. P., Dickson C., Itoh N., Warburton D., Anderson K. D. et al. (2004). Requirement for fibroblast growth factor 10 or fibroblast growth factor receptor 2-IIIb signaling for cecal development in mouse. Dev. Biol. 265, 61-74. 10.1016/j.ydbio.2003.09.021 [DOI] [PubMed] [Google Scholar]
- Chan K. Y. Y., Leung K. T., Tam Y. H., Lam H. S., Cheung H. M., Ma T. P. Y., Lee K. H., To K. F., Li K. and Ng P. C. (2014). Genome-wide expression profiles of necrotizing enterocolitis versus spontaneous intestinal perforation in human intestinal tissues: dysregulation of functional pathways. Ann. Surg. 260, 1128-1137. 10.1097/SLA.0000000000000374 [DOI] [PubMed] [Google Scholar]
- Chen C.-T., Shih Y.-R. V., Kuo T. K., Lee O. K. and Wei Y.-H. (2008). Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells 26, 960-968. 10.1634/stemcells.2007-0509 [DOI] [PubMed] [Google Scholar]
- Choi M. Y., Romer A. I., Hu M., Lepourcelet M., Mechoor A., Yesilaltay A., Krieger M., Gray P. A. and Shivdasani R. A. (2006). A dynamic expression survey identifies transcription factors relevant in mouse digestive tract development. Development 133, 4119-4129. 10.1242/dev.02537 [DOI] [PubMed] [Google Scholar]
- Christensen T. and Diemer N. H. (2003). Reduction of mitochondrial electron transport complex activity is restricted to the ischemic focus after transient focal cerebral ischemia in rats: a histochemical volumetric analysis. Neurochem. Res. 28, 1805-1812. 10.1023/A:1026111506307 [DOI] [PubMed] [Google Scholar]
- Cunningham J. T., Rodgers J. T., Arlow D. H., Vazquez F., Mootha V. K. and Puigserver P. (2007). mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 450, 736-740. 10.1038/nature06322 [DOI] [PubMed] [Google Scholar]
- Dennis G. Jr, Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C. and Lempicki R. A. (2003). DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
- Faller W. J., Jackson T. J., Knight J. R. P., Ridgway R. A., Jamieson T., Karim S. A., Jones C., Radulescu S., Huels D. J., Myant K. B. et al. (2015). mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature 517, 497-500. 10.1038/nature13896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes C. D. L., Nelson T. J., Martinez-Fernandez A., Arrell D. K., Lindor J. Z., Dzeja P. P., Ikeda Y., Perez-Terzic C. and Terzic A. (2011). Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 14, 264-271. 10.1016/j.cmet.2011.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J. et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse A. S., Pressprich M. F., Curley L. B., Hamilton K. L., Margolis B., Hildebrand J. D. and Gumucio D. L. (2011). Cell dynamics in fetal intestinal epithelium: implications for intestinal growth and morphogenesis. Development 138, 4423-4432. 10.1242/dev.065789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe B. D., Scherz P. J., Nissim S., Tian H., McMahon A. P. and Tabin C. J. (2004). Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517-528. 10.1016/j.cell.2004.07.024 [DOI] [PubMed] [Google Scholar]
- Harris-Johnson K. S., Domyan E. T., Vezina C. M. and Sun X. (2009). beta-Catenin promotes respiratory progenitor identity in mouse foregut. Proc. Natl. Acad. Sci. USA 106, 16287-16292. 10.1073/pnas.0902274106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hench J., Bratic Hench I., Pujol C., Ipsen S., Brodesser S., Mourier A., Tolnay M., Frank S. and Trifunovic A. (2011). A tissue-specific approach to the analysis of metabolic changes in Caenorhabditis elegans. PLoS ONE 6, e28417 10.1371/journal.pone.0028417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y. and Lee J. T. (2011). YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146, 119-133. 10.1016/j.cell.2011.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner K. H., Silberg D. G., Traber P. G. and Schutz G. (1997). The mesenchymal winged helix transcription factor Fkh6 is required for the control of gastrointestinal proliferation and differentiation. Genes Dev. 11, 1583-1595. 10.1101/gad.11.12.1583 [DOI] [PubMed] [Google Scholar]
- Kaiser S., Park Y.-K., Franklin J. L., Halberg R. B., Yu M., Jessen W. J., Freudenberg J., Chen X., Haigis K., Jegga A. G. et al. (2007). Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 8, R131 10.1186/gb-2007-8-7-r131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. and Goto S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27-30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Kawashima M., Furumichi M. and Tanabe M. (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457-D462. 10.1093/nar/gkv1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson L., Lindahl P., Heath J. K. and Betsholtz C. (2000). Abnormal gastrointestinal development in PDGF-A and PDGFR-(alpha) deficient mice implicates a novel mesenchymal structure with putative instructive properties in villus morphogenesis. Development 127, 3457-3466. [DOI] [PubMed] [Google Scholar]
- Kim D.-H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P. and Sabatini D. M. (2002). mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163-175. 10.1016/S0092-8674(02)00808-5 [DOI] [PubMed] [Google Scholar]
- Kim B.-M., Mao J., Taketo M. M. and Shivdasani R. A. (2007). Phases of canonical Wnt signaling during the development of mouse intestinal epithelium. Gastroenterology 133, 529-538. 10.1053/j.gastro.2007.04.072 [DOI] [PubMed] [Google Scholar]
- Lepourcelet M., Tou L., Cai L., Sawada J.-I., Lazar A. J. F., Glickman J. N., Williamson J. A., Everett A. D., Redston M., Fox E. A. et al. (2005). Insights into developmental mechanisms and cancers in the mammalian intestine derived from serial analysis of gene expression and study of the hepatoma-derived growth factor (HDGF). Development 132, 415-427. 10.1242/dev.01579 [DOI] [PubMed] [Google Scholar]
- Liberzon A., Subramanian A., Pinchback R., Thorvaldsdottir H., Tamayo P. and Mesirov J. P. (2011). Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739-1740. 10.1093/bioinformatics/btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison B. B., Braunstein K., Kuizon E., Portman K., Qiao X. T. and Gumucio D. L. (2005). Epithelial hedgehog signals pattern the intestinal crypt-villus axis. Development 132, 279-289. 10.1242/dev.01576 [DOI] [PubMed] [Google Scholar]
- Madison B. B., McKenna L. B., Dolson D., Epstein D. J. and Kaestner K. H. (2009). FoxF1 and FoxL1 link hedgehog signaling and the control of epithelial proliferation in the developing stomach and intestine. J. Biol. Chem. 284, 5936-5944. 10.1074/jbc.M808103200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathan M., Moxey P. C. and Trier J. S. (1976). Morphogenesis of fetal rat duodenal villi. Am. J. Anat. 146, 73-92. 10.1002/aja.1001460104 [DOI] [PubMed] [Google Scholar]
- McLin V. A., Henning S. J. and Jamrich M. (2009). The role of the visceral mesoderm in the development of the gastrointestinal tract. Gastroenterology 136, 2074-2091. 10.1053/j.gastro.2009.03.001 [DOI] [PubMed] [Google Scholar]
- Mould A. W., Morgan M. A. J., Nelson A. C., Bikoff E. K. and Robertson E. J. (2015). Blimp1/Prdm1 functions in opposition to Irf1 to maintain neonatal tolerance during postnatal intestinal maturation. PLoS Genet. 11, e1005375 10.1371/journal.pgen.1005375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu J. (2014). Necrotizing enterocolitis: the mystery goes on. Neonatology 106, 289-295. 10.1159/000365130 [DOI] [PubMed] [Google Scholar]
- Neu J. and Walker W. A. (2011). Necrotizing enterocolitis. N. Engl. J. Med. 364, 255-264. 10.1056/NEJMra1005408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noah T. K., Donahue B. and Shroyer N. F. (2011). Intestinal development and differentiation. Exp. Cell Res. 317, 2702-2710. 10.1016/j.yexcr.2011.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormestad M., Astorga J., Landgren H., Wang T., Johansson B. R., Miura N. and Carlsson P. (2006). Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development 133, 833-843. 10.1242/dev.02252 [DOI] [PubMed] [Google Scholar]
- Pabst O., Schneider A., Brand T. and Arnold H.-H. (1997). The mouse Nkx2-3 homeodomain gene is expressed in gut mesenchyme during pre- and postnatal mouse development. Dev. Dyn. 209, 29-35. [DOI] [PubMed] [Google Scholar]
- Pabst O., Zweigerdt R. and Arnold H. H. (1999). Targeted disruption of the homeobox transcription factor Nkx2-3 in mice results in postnatal lethality and abnormal development of small intestine and spleen. Development 126, 2215-2225. [DOI] [PubMed] [Google Scholar]
- Pate K. T., Stringari C., Sprowl-Tanio S., Wang K., TeSlaa T., Hoverter N. P., McQuade M. M., Garner C., Digman M. A., Teitell M. A. et al. (2014). Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer. EMBO J. 33, 1454-1473. 10.15252/embj.201488598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perekatt A. O., Valdez M. J., Davila M., Hoffman A., Bonder E. M., Gao N. and Verzi M. P. (2014). YY1 is indispensable for Lgr5+ intestinal stem cell renewal. Proc. Natl. Acad. Sci. USA 111, 7695-7700. 10.1073/pnas.1400128111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian P., He X. C., Paulson A., Li Z., Tao F., Perry J. M., Guo F., Zhao M., Zhi L., Venkatraman A. et al. (2016). The Dlk1-Gtl2 locus preserves LT-HSC function by inhibiting the PI3K-mTOR pathway to restrict mitochondrial metabolism. Cell Stem Cell 18, 214-228. 10.1016/j.stem.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson L. L., Davis A. K., Grogg M. W. and Zheng Y. (2016). mTOR disruption causes intestinal epithelial cell defects and intestinal atrophy postinjury in mice. FASEB J. 30, 1263-1275. 10.1096/fj.15-278606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligman A. M., Karnovsky M. J., Wasserkrug H. L. and Hanker J. S. (1968). Nondroplet ultrastructural demonstration of cytochrome oxidase activity with a polymerizing osmiophilic reagent, diaminobenzidine (DAB). J. Cell Biol. 38, 1-14. 10.1083/jcb.38.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Seto E., Chang L.-S. and Shenk T. (1991). Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell 67, 377-388. 10.1016/0092-8674(91)90189-6 [DOI] [PubMed] [Google Scholar]
- Shyer A. E., Tallinen T., Nerurkar N. L., Wei Z., Gil E. S., Kaplan D. L., Tabin C. J. and Mahadevan L. (2013). Villification: how the gut gets its villi. Science 342, 212-218. 10.1126/science.1238842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyer A. E., Huycke T. R., Lee C., Mahadevan L. and Tabin C. J. (2015). Bending gradients: how the intestinal stem cell gets its home. Cell 161, 569-580. 10.1016/j.cell.2015.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N. and Daley G. Q. (2013). Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell 12, 395-406. 10.1016/j.stem.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N., Daley G. Q. and Cantley L. C. (2013). Stem cell metabolism in tissue development and aging. Development 140, 2535-2547. 10.1242/dev.091777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- Spence J. R., Lauf R. and Shroyer N. F. (2011). Vertebrate intestinal endoderm development. Dev. Dyn. 240, 501-520. 10.1002/dvdy.22540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringari C., Edwards R. A., Pate K. T., Waterman M. L., Donovan P. J. and Gratton E. (2012). Metabolic trajectory of cellular differentiation in small intestine by Phasor Fluorescence Lifetime Microscopy of NADH. Sci. Rep. 2, 568 10.1038/srep00568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S. et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545-15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takubo K., Nagamatsu G., Kobayashi C. I., Nakamura-Ishizu A., Kobayashi H., Ikeda E., Goda N., Rahimi Y., Johnson R. S., Soga T. et al. (2013). Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 12, 49-61. 10.1016/j.stem.2012.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannahill G. M., Curtis A. M., Adamik J., Palsson-McDermott E. M., McGettrick A. F., Goel G., Frezza C., Bernard N. J., Kelly B., Foley N. H. et al. (2013). Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature 496, 238-242. 10.1038/nature11986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormos K. V., Anso E., Hamanaka R. B., Eisenbart J., Joseph J., Kalyanaraman B. and Chandel N. S. (2011). Mitochondrial complex III ROS regulate adipocyte differentiation. Cell Metab. 14, 537-544. 10.1016/j.cmet.2011.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ufer C. and Wang C. C. (2011). The roles of glutathione peroxidases during embryo development. Front. Mol. Neurosci. 4, 12 10.3389/fnmol.2011.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink G. R. (2007). Hedgehog signaling in development and homeostasis of the gastrointestinal tract. Physiol. Rev. 87, 1343-1375. 10.1152/physrev.00054.2006 [DOI] [PubMed] [Google Scholar]
- Vander Heiden M. G., Cantley L. C. and Thompson C. B. (2009). Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029-1033. 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker E. M., Thompson C. A., Kohlnhofer B. M., Faber M. L. and Battle M. A. (2014). Characterization of the developing small intestine in the absence of either GATA4 or GATA6. BMC Res. Notes 7, 902 10.1186/1756-0500-7-902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton K. D., Kolterud A., Czerwinski M. J., Bell M. J., Prakash A., Kushwaha J., Grosse A. S., Schnell S. and Gumucio D. L. (2012). Hedgehog-responsive mesenchymal clusters direct patterning and emergence of intestinal villi. Proc. Natl. Acad. Sci. USA 109, 15817-15822. 10.1073/pnas.1205669109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton K. D., Whidden M., Kolterud A., Shoffner S. K., Czerwinski M. J., Kushwaha J., Parmar N., Chandhrasekhar D., Freddo A. M., Schnell S. et al. (2016). Villification in the mouse: Bmp signals control intestinal villus patterning. Development 143, 427-436. 10.1242/dev.130112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O., Posener K. and Negelein E. (1924). Über den stoffwechsel der carcinomzelle [The metabolism of tumors]. Biochem. Z. 152, 319-344. [Google Scholar]
- Wells J. M. and Melton D. A. (1999). Vertebrate endoderm development. Annu. Rev. Cell Dev. Biol. 15, 393-410. 10.1146/annurev.cellbio.15.1.393 [DOI] [PubMed] [Google Scholar]
- Wu Z., Irizarry R. A., Gentleman R., Martinez-Murillo F. and Spencer F. (2004). A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99, 909-917. 10.1198/016214504000000683 [DOI] [Google Scholar]
- Xiong X., Yang H., Tan B., Yang C., Wu M., Liu G., Kim S. W., Li T., Li L., Wang J. et al. (2015). Differential expression of proteins involved in energy production along the crypt-villus axis in early-weaning pig small intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 309, G229-G237. 10.1152/ajpgi.00095.2015 [DOI] [PubMed] [Google Scholar]
- Zani A. and Pierro A. (2015). Necrotizing enterocolitis: controversies and challenges. F1000Res 4, 1373 10.12688/f1000research.6888.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Choi M., Margineantu D., Margaretha L., Hesson J., Cavanaugh C., Blau C. A., Horwitz M. S., Hockenbery D., Ware C. et al. (2012). HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 31, 2103-2116. 10.1038/emboj.2012.71 [DOI] [PMC free article] [PubMed] [Google Scholar]