Abstract
Neurons in the hypothalamic arcuate nucleus relay and translate important cues from the periphery into the central nervous system. However, the gene regulatory program directing their development remains poorly understood. Here, we report that the LIM-homeodomain transcription factor Isl1 is expressed in several subpopulations of developing arcuate neurons and plays crucial roles in their fate specification. Mice with conditional deletion of the Isl1 gene in developing hypothalamus display severe deficits in both feeding and linear growth. Consistent with these results, their arcuate nucleus fails to express key fate markers of Isl1-expressing neurons that regulate feeding and growth. These include the orexigenic neuropeptides AgRP and NPY for specifying AgRP-neurons, the anorexigenic neuropeptide αMSH for POMC-neurons, and two growth-stimulatory peptides, growth hormone-releasing hormone (GHRH) for GHRH-neurons and somatostatin (Sst) for Sst-neurons. Finally, we show that Isl1 directly enhances the expression of AgRP by cooperating with the key orexigenic transcription factors glucocorticoid receptor and brain-specific homeobox factor. Our results identify Isl1 as a crucial transcription factor that plays essential roles in the gene regulatory program directing development of multiple arcuate neuronal subpopulations.
KEY WORDS: Arcuate neurons, Bsx, GR, Hypothalamus, Isl1
Summary: During hypothalamus development in mice, Isl1 is expressed in several subpopulations of developing arcuate neurons and plays crucial roles in specifying their cell fate.
INTRODUCTION
In the central nervous system (CNS), the hypothalamus plays crucial roles in centrally integrating and processing sensory inputs, and subsequently coordinating appropriate responses to regulate homeostatic processes that are essential to survival and species propagation (Hill et al., 2008). Among the multiple nuclei of the hypothalamus, the arcuate nucleus (ARC) in the mediobasal hypothalamus is particularly important in sensing and processing various peripheral cues. The ARC is a key regulator of these processes because its diverse array of neuronal subpopulations that control energy balance, growth and reproductive behaviors are located adjacent to the third ventricle and the median eminence, making them ideally positioned to interact with both the peripheral blood stream and numerous regions of the brain (Bluet-Pajot et al., 2001; Biebermann et al., 2012; Hrabovszky, 2014). Despite recent advances in our understanding of the physiological roles of distinct arcuate neuronal subpopulations, the gene regulatory program that orchestrates the development of the ARC remains poorly understood.
GHRH-neurons in the ARC release growth hormone-releasing hormone (GHRH), which is then carried by the hypothalamo-hypophyseal portal system to the anterior pituitary gland. In turn, GHRH triggers secretion of growth hormone (GH) by stimulating the GHRH receptor (Bluet-Pajot et al., 2001). Activation of GH signaling leads to the hepatic expression of insulin-like growth factor 1 (IGF1), which controls bone epiphyses and growth plate development as well as muscle and adipose tissue development (Cohen and Rosenfeld, 1994). The actions of GHRH are antagonized by the growth-hormone-inhibiting hormone somatostatin (Sst), which is released from the neurosecretory nerve terminals of periventricular nucleus (PeV) Sst-neurons (Bluet-Pajot et al., 2001). These PeV Sst-neurons are distinct from the centrally projecting arcuate neurons that also express Sst. The physiological roles of the arcuate Sst-neurons or their Sst peptides remain poorly understood. Paradoxically, however, it has been suggested that these neurons trigger activation of GHRH-neurons (Slama et al., 1993; Lanneau et al., 2000; Bluet-Pajot et al., 2001), probably through Sst receptors in GHRH-neurons (Tannenbaum et al., 1998).
In addition, the ARC contains two subpopulations of neurons that control feeding behavior: pro-opiomelanocortin (POMC)-neurons and agouti-related protein (AgRP)-neurons (Biebermann et al., 2012). POMC-neurons co-express two anorexigenic peptides, cocaine and amphetamine-related transcript (CART) and the melanocyte-stimulating hormone-α (αMSH), which is produced from the large precursor peptide POMC. By contrast, AgRP-neurons co-express the orexigenic peptides neuropeptide Y (NPY) and AgRP. Fasting upregulates the expression of NPY and AgRP (Lewis et al., 1993; Stephens et al., 1995; Mizuno and Mobbs, 1999) and decreases αMSH expression (Cowley et al., 2001). Additionally, fasting increases plasma levels of glucocorticoid, a well-known peripheral orexigenic signal, which in turn activates the expression of NPY and AgRP in AgRP-neurons (Fehm et al., 2004). Thus, POMC- and AgRP-neurons play essential roles in coordinating the CNS response to peripheral metabolic cues by informing the brain on the peripheral status of energy homeostasis.
We have previously shown that glucocorticoid receptor (GR; also known as Nr3c1), the glucocorticoid-activated orexigenic transcription factor, is expressed in AgRP-neurons and directly regulates the expression of AgRP through a novel glucocorticoid response element (GRE) located in the promoter region of the Agrp gene (Lee et al., 2013). We have further demonstrated that, in response to fasting-elevated glucocorticoid levels, GR synergizes with another orexigenic transcription factor expressed in AgRP-neurons, brain-specific homeobox factor (Bsx) (Sakkou et al., 2007; Nogueiras et al., 2008), to direct the activation of AgRP transcription (Lee et al., 2013).
In this study, we investigated the gene regulatory program that governs the development of the ARC during embryogenesis. Islet-1 (Isl1), a member of the LIM-homeodomain (LIM-HD) transcription factor family, has been shown to regulate cell fate specification in multiple tissues and species (Thaler et al., 2002; Lee and Pfaff, 2003; Peng et al., 2005; Mazzoni et al., 2013; Cho et al., 2014; Li et al., 2014; Perdigoto et al., 2014). Here, we report that Isl1 plays crucial roles in the development of multiple ARC neurons, which control feeding and linear growth. We found that Isl1 is expressed in several developing arcuate neurons during their fate specification, and plays crucial roles in inducing the expression of central fate markers of those neurons, including AgRP and NPY in AgRP-neurons, αMSH in POMC-neurons, GHRH in GHRH-neurons, and Sst in Sst-neurons. We further show that Isl1 directly controls the expression of AgRP probably by forming a complex with two partner orexigenic transcription factors expressed in AgRP-neurons, GR and Bsx. Overall, our results demonstrate that Isl1 is an important regulator of the gene program that directs development of feeding- and growth-controlling arcuate neurons.
RESULTS
Expression of Isl1 in developing arcuate nucleus neurons
Our search of the public databases www.brain-map.org and www.genepaint.org suggested that, among many LIM-HD factors, Isl1 is highly expressed in the mediobasal hypothalamus where the ARC is located. This finding prompted us to systematically monitor the expression pattern of Isl1 throughout the development of the ventral hypothalamus. Nkx2.1 (Nkx2-1) is expressed in progenitor cells of the ventral hypothalamus and is required for development of this region (Marín et al., 2002). At embryonic day (E) 10.5, Isl1 is upregulated in a subset of Nkx2.1+ progenitor cells in the lateral diencephalon (Fig. 1A), suggesting that Isl1 expression is induced as neurons emerge from Nkx2.1+ progenitors in the ventricular zone. By E13.5, Isl1 is expressed relatively broadly in the mantle zone, in which postmitotic neurons reside (Fig. 1A). Isl1 becomes highly enriched in the ARC over time and continues to be expressed in the ARC across all postnatal stages (e.g. http://developingmouse.brain-map.org/gene/show/16165).
Fig. 1.
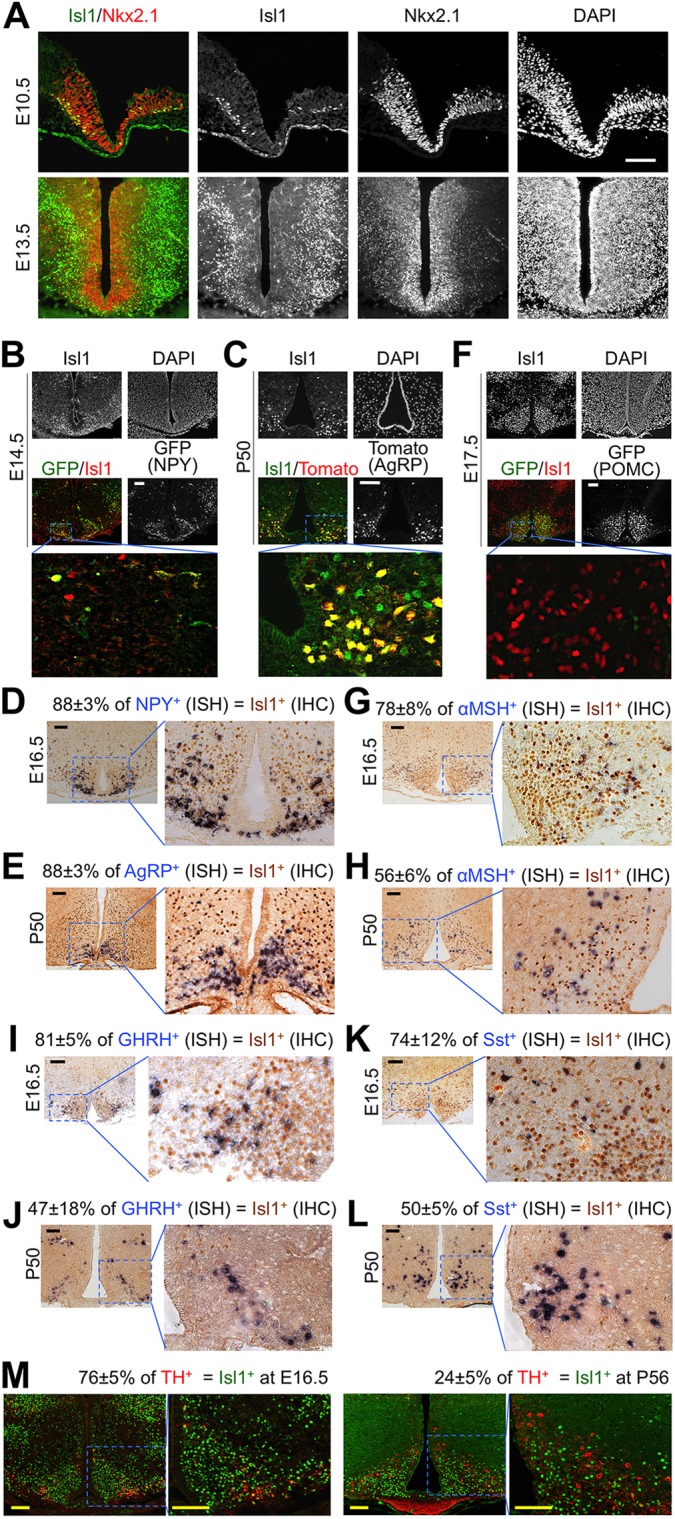
Expression of Isl1 in arcuate neurons. (A) Wild-type mouse embryonic hypothalamus was immunostained with anti-Isl1 and anti-Nkx2.1 antibodies. DAPI staining marks all nuclei in the section. (B,C,F) Representative images of the ARC from E14.5 Npy-hrGFP embryos (B), P50 Agrp-ires-Cre;Rosa26CAG-tdTomato mice (C) and E17.5 Pomc-eGFP embryos (F), which were immunostained with anti-Isl1 antibody. (D,E,G-L) ISH (blue) for the genes encoding NPY (D), AgRP (E), αMSH (G,H), GHRH (I,J) and Sst (K,L) was performed on either E16.5 or P50 wild-type mouse ARC, followed by immunostaining with anti-Isl1 antibody (brown). (M) Co-immunostaining of E16.5 and P56 wild-type mouse ARC with antibodies against TH and Isl1. Scale bars: 100 μm. (D,E,G-M) Values indicate the percentage of cells expressing the given marker that are also Isl1 positive (mean±s.d. of three hypothalami per panel).
To test whether Isl1 is expressed in specific classes of the ARC neurons, we performed immunohistochemistry and in situ hybridization (ISH) analyses. To label AgRP-neurons, we used two transgenic mouse models. The Npy-hrGFP transgenic mouse is an ideal model to label AgRP-neurons with GFP in developing embryos, because GFP expression is driven by the promoter of the Npy gene, which is turned on in embryonic AgRP-neurons prior to the expression of AgRP (van den Pol et al., 2009). To label AgRP-neurons postnatally, we generated Agrp-ires-Cre;Rosa26CAG-tdTomato mice, in which a red fluorescent protein (tdTomato) is specifically and postnatally expressed in AgRP-neurons by means of expression of Cre driven by the Agrp promoter and the resulting Cre-mediated recombination only in AgRP-neurons (Tong et al., 2008; Madisen et al., 2010). Isl1 was expressed in most GFP+ AgRP-neurons in the ARC of Npy-hrGFP mice at E14.5 (Fig. 1B). Likewise, Isl1 was expressed in most tdTomato+ AgRP-neurons of Agrp-ires-Cre;Rosa26CAG-tdTomato mice at postnatal day (P) 50 (Fig. 1C). Consistently, immunostaining with anti-Isl1 antibody, combined with ISH for either Npy or Agrp, revealed the co-expression of Isl1 with Npy at E16.5 and with Agrp at P50 (Fig. 1D,E). These results establish that Isl1 is expressed in the AgRP-neurons from embryonic to postnatal stages. In Pomc-eGFP mice (Cowley et al., 2001), ∼70-80% of GFP+ POMC-neurons co-expressed Isl1 in the ARC at E17.5 (Fig. 1F). Likewise, the combined analyses of ISH and immunostaining showed co-expression of αMSH and Isl1 at E16.5 and P50 (Fig. 1G,H). The combined ISH and immunohistochemical analyses also revealed that Isl1 is expressed in Ghrh+ and Sst+ neurons in the ARC at E16.5 and P50 (Fig. 1I-L). The ARC also contains tyrosine hydroxylase (TH)-positive neurons (Bluet-Pajot et al., 2001; Biebermann et al., 2012; Hrabovszky, 2014), and 15% of these cells are GHRH-neurons (Phelps et al., 2003). Interestingly, we also found that ∼76% of TH+ neurons in the ARC express Isl1 at E16.5 and ∼23% of TH+ neurons express Isl1 at P56 (Fig. 1M). Taken together, these results indicate that Isl1 is expressed in AgRP-, POMC-, GHRH-, Sst- and TH-neurons in the ARC during embryonic development and through subsequent postnatal stages.
Isl1cko mice show deficits in feeding and growth
To investigate whether Isl1 plays a role in the development of AgRP-, POMC-, GHRH- and Sst-neurons, we generated Isl1f/f;Nkx2.1-Cre mice (herein referred to as Isl1cko), in which the Isl1 gene is deleted in Nkx2.1+ neural progenitors, which give rise to Isl1+ neurons in the ARC (Fig. 1A). Confirming that Isl1+ cells do indeed derive from Nkx2.1+ progenitors in the developing ARC, Isl1 protein was no longer expressed in the ARC of Isl1cko mice (Fig. 2A).
Fig. 2.
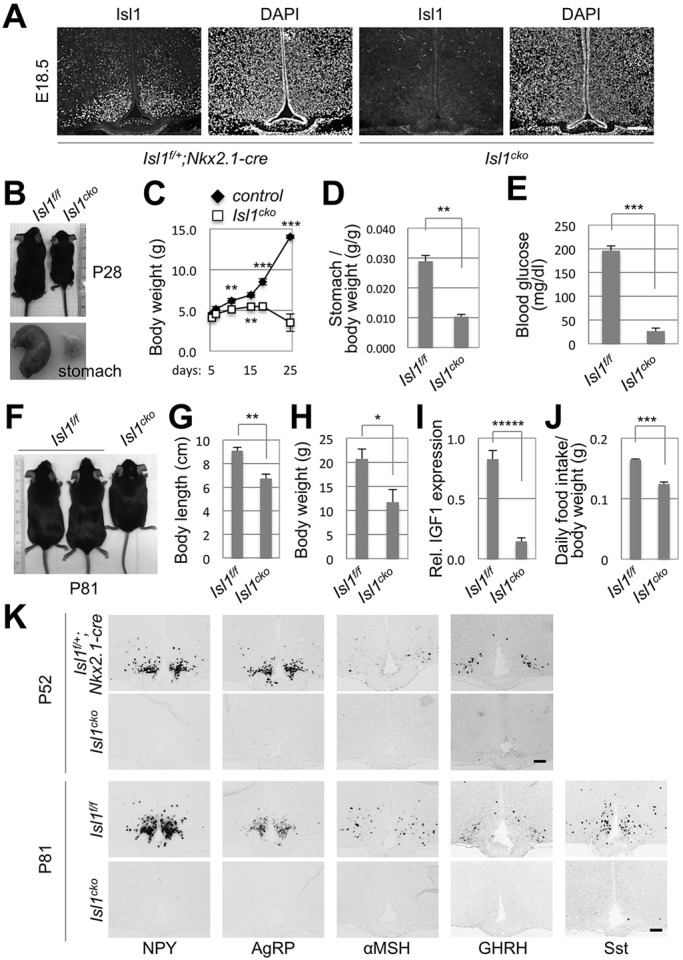
Feeding and growth deficits of Isl1cko mice. (A) The ARC from E18.5 control or Isl1cko embryos was immunostained with anti-Isl1 antibody. (B-J) Representative images of control and Isl1cko mice and their stomachs at P28 (B) and P81 (F). Their body weights were measured at P28 (C) and P81 (H). Their stomachs were dissected out to measure their contents (D) at P28. Blood glucose levels were measured from tail blood with a glucometer (E) and the body length was measured from the nose tip to the tail base (G). The expression level of Igf1 mRNA at P81 was determined by qRT-PCR (I). Daily food intake was measured for 4 weeks from P80 (J). All values are presented as mean±s.e.m. *P<0.05, **P<0.01, ***P<0.001, *****P<0.00001; Student's t-test. (K) ISH for the genes encoding NPY, AgRP, αMSH, GHRH and Sst was performed on the ARC of surviving escaper Isl1cko and their littermate control mice at P52 and P81. Three controls and three mutants were used for the data shown in C-E,G,H,J, and four controls and three mutants were used for the data shown in I. Scale bars: 100 μm.
The body weight of Isl1cko mice was similar to that of their littermate controls at birth. Interestingly, Isl1cko mice began to show retardation in both linear growth and body weight gain around P10, which became progressively worse (Fig. 2B,C). Isl1cko mice also showed a marked reduction in the size and the content weight of their stomachs (Fig. 2B,D) as well as in blood glucose levels (Fig. 2E). Approximately 64% of Isl1cko mice died between P21 and P30, probably owing to a decrease in feeding and the resulting low glucose levels. Notably, the remaining ∼36% of Isl1cko mice, which escaped the early death, continued to exhibit growth retardation at later stage of life, as shown by a significant decrease in the body length and weight (Fig. 2F-H). These Isl1cko escaper mice showed a drastic reduction in hepatic IGF1 levels, a well-defined surrogate marker for the growth hormone produced in the pituitary (Fig. 2I), suggesting defects in the GHRH-GH-IGF1 axis. In addition, the escaper mice also exhibited a substantial reduction in food intake (Fig. 2J), indicating that the feeding circuitry is disrupted in Isl1cko mice.
To investigate whether the hypothalamic dysregulation of growth and feeding contributes to the severe growth/feeding phenotypes of Isl1cko mice, we monitored the expression of key neuropeptides controlling growth and feeding in the ARC of adult Isl1cko escaper mice. Strikingly, the ISH analyses revealed that the expression of the genes encoding NPY, AgRP, αMSH, GHRH and Sst was eliminated in Isl1cko mice at P52 and P81 (Fig. 2K). These results suggest that Isl1 is required for the development and/or survival of AgRP-, POMC-, GHRH- and Sst-neurons in the ARC, which control feeding and growth.
Lost expression of POMC, AgRP, NPY, GHRH and Sst in the ARC of Isl1cko embryos
To test whether Isl1 is needed for development of arcuate neurons that control feeding and growth, we analyzed the expression of key neuropeptides that designate the identities of specific ARC subpopulations in Isl1cko embryos. The genes encoding NPY, αMSH, GHRH and Sst were not expressed in the ARC of E16.5 Isl1cko embryos, whereas their expression was readily detected in control littermate embryos (Fig. 3A), suggesting that Isl1 is required for the specification of neurons expressing these neuropeptides or their differentiation or survival during embryogenesis. To test whether development of the arcuate neuronal subpopulations that express these key neuropeptides was delayed in the absence of Isl1 but eventually recovered postnatally, we monitored the expression of the neuropeptides at P21, including AgRP, expression of which is known to increase postnatally in AgRP-neurons (Nilsson et al., 2005). Interestingly, the expression of NPY and AgRP in AgRP-neurons, αMSH in POMC-neurons, GHRH in GHRH-neurons, and Sst in Sst-neurons remained severely compromised in the ARC of P21 Isl1cko mice in comparison with their littermate controls (Fig. 3B). Of note, normal levels of NPY expression were detected in the amygdala region of P28 Isl1cko brains (Fig. 3D), proving that the ISH procedure was successfully carried out.
Fig. 3.
Impaired expression of αMSH, AgRP, NPY, GHRH and Sst in the ARC of Isl1cko mice. (A,B,D,E) ISH was performed for the genes encoding NPY/AgRP, αMSH, GHRH, Sst and Dlx1 (A,B) as well as NPY (D) and SF1, CRH and TRH (E) on the ARC and other nuclei of control and Isl1cko mice at E16.5 (A), P21 (B) and P28 (D,E). (C) Signal intensity for Dlx1 expression as shown in A and B was quantified using ImageJ. **P<0.01; ns, not significant; Student's t-test. Error bars indicate s.e.m. Three controls and three mutants were quantified at E16.5 and four controls and four mutants were analyzed at P21. (D,E) Two controls and two mutants were analyzed at P28. Scale bars: 100 μm.
To test the specificity of the loss of AgRP-, POMC-, GHRH- and Sst-neurons in Isl1cko mice, we examined the expression of several well-established markers of hypothalamic neurons in Isl1cko mice: SF1, a marker for a subregion of the ventromedial hypothalamus that expresses Nkx2.1 but not Isl1 (Dellovade et al., 2000; Davis et al., 2004); CRH, a marker for the PVN, which expresses neither Nkx2.1 nor Isl1 (Shimogori et al., 2010); and TRH, a marker for the dorsomedial hypothalamus, which expresses low levels of Isl1 but not Nkx2.1 (Davis et al., 2004). The expression of the genes encoding SF1, CRH and TRH (Fig. 3E), as determined by quantification of the ISH signals (data not shown), was not significantly altered in the hypothalamus of P28 Isl1cko mice, indicating that the SF1+, CRH+ and TRH+ neurons are formed in the normal manner in Isl1cko mice. Interestingly, although we observed that most TH+ cells express Isl1 at E16.5 (Fig. 1M), TH expression was comparable between control and mutant mice at this embryonic stage (data not shown).
Together, these results suggest that Isl1 plays important roles in specifically driving and maintaining the expression of the feeding- and growth-controlling neuropeptides NPY, AgRP, αMSH, GHRH and Sst in the ARC. Given that the loss of these key neuropeptides in the hypothalamus of Isl1cko mice would result in disruption of the central control of feeding and growth, the loss of NPY, AgRP, αMSH, GHRH and Sst is likely to be a major contributing factor to the severely compromised feeding and growth phenotypes of Isl1cko mice (Fig. 2).
Isl1 may direct the fate specification of the embryonic arcuate neuronal subpopulations
The neuropeptides NPY/AgRP, αMSH, GHRH and Sst represent key markers of the cell fates of AgRP-, POMC-, GHRH- and Sst-neurons, respectively, and also serve as central molecules that determine the functionality of these neurons. Thus, the loss of NPY, AgRP, αMSH, GHRH and Sst in the ARC of Isl1cko mice points to the possibility that Isl1 serves as a crucial factor that directs the fate determination of these arcuate neuronal subpopulations. Alternatively, Isl1 might be needed for the proliferation or neurogenesis of neural progenitors giving rise to these neurons or for their survival. Notably, although Isl1 is not expressed in the ARC neural progenitors (Fig. 1A), it is possible that defects in neuronal differentiation indirectly influence proliferation of neuronal progenitors. To monitor the proliferation status of neural progenitors, we performed bromodeoxyuridine (BrdU) labeling assays in E12.5 embryos, which were exposed to BrdU for 2 h in utero. The number of BrdU+ cells was comparable between Isl1cko and control embryos (Fig. 4A). In addition, our analysis of Ki67 (Mki67) expression at E14.5 did not find any significant difference between Isl1cko and littermate control mice either (data not shown). These results suggest that deletion of Isl1 does not affect the proliferation of neural progenitors in the developing ARC.
Fig. 4.

No apparent defects in proliferation and neurogenesis of the arcuate neurons in Isl1cko mice. (A,B) Immunohistochemistry with anti-BrdU antibody (A) and ISH for Mash1 (B) were performed on the ARC of control and Isl1cko mice at E12.5. BrdU was intraperitoneally injected to pregnant females 2 h before euthanasia. (C) ISH for Gad1, Gsx1 and Hmx2 on the ARC of control and Isl1cko mice at E14.5 or E18.5. Signal intensity for Gad1 was quantified using ImageJ. (D) ISH with Gad1 performed on the ARC of control and Isl1cko mice at P21. Dashed lines indicate the boundary of the ARC. Signal intensity for Gad1 was quantified using ImageJ. Double arrows indicate area expressing Gad1. (E) Immunohistochemistry with anti-Sox2 antibody performed on the ARC of control and Isl1cko mice at P28. (F) Immunohistochemistry with anti-NeuN antibody was performed on the ARC of control and Isl1cko mice at E16.5 and P15. The number of NeuN+ cells was quantified using ImageJ. Three controls and four mutants were used for all experiments. Scale bars: 100 μm. **P<0.01; Student's t-test. Error bars indicate s.e.m.
To investigate ARC neurogenesis, we examined the expression of a proneural basic helix-loop-helix factor, Mash1 (Ascl1), which is a well-known marker for neural progenitors that is also functionally important for neurogenesis (Lo et al., 1991; McNay et al., 2006). The number of Mash1+ cells was comparable between E12.5 Isl1cko and littermate control mice (Fig. 4B). Given the requirement of Ngn3 (Neurog3) in ARC neurogenesis (Pelling et al., 2011), we also evaluated the expression of Ngn3. We found that the number of Ngn3+ cells was comparable between E12.5 Isl1cko and littermate control mice (data not shown). To investigate further the role of Isl1 in neuronal differentiation in the ARC, we monitored the presence of GABAergic neurons using ISH with a probe detecting glutamate decarboxylase 1 (Gad1), which encodes an enzyme that converts glutamate to γ-aminobutyric acid (GABA) and CO2. Of note, most NPY+ AgRP-neurons are known to be GABAergic, whereas most POMC neurons are non-GABAergic (Horvath et al., 1997; Ovesjo et al., 2001; Hentges et al., 2004). There are also other types of GABAergic neurons in the ARC, including a subset of kisspeptin-neurons (Hrabovszky, 2014). Interestingly, more GABAergic neurons were observed in E14.5 Isl1cko mice in comparison with control mice (Fig. 4C). These results indicate that GABAergic neurons are still formed in Isl1cko mice, although they fail to upregulate NPY on time. Dlx1 is a homeodomain (HD) transcription factor expressed in a subset of dopaminergic neurons in the ARC (Yee et al., 2009). Interestingly, the number of Dlx1+ cells and the level of Dlx1 expression dramatically increased in E16.5 Isl1cko brains (Fig. 3A,C) but not in P21 Isl1cko brains (Fig. 3B,C). Because Dlx2, a functional homolog of Dlx1, has been shown to induce the expression of GAD1 in slice cultures of the mouse embryonic cerebral cortex (Stühmer et al., 2002), the increase in GAD1 expression probably reflects the increase in Dlx1 expression (Fig. 3A,C). To examine additional markers of differentiated arcuate neurons, we monitored the expression of the homeobox transcription factors Gsx1 and Hmx2/3, which have been shown to play essential roles in the expression of GHRH in the ARC (Wang et al., 2004; Li et al., 1996). The expression of Gsx1 and Hmx2 in the ARC was largely comparable between control and Isl1cko mice (slightly more Gsx1 and slightly less Hmx2 in Isl1cko) (Fig. 4C). Overall, these results suggest two prominent consequences of the lack of Isl1. First, AgRP-, POMC-, GHRH- and Sst-neurons appear not to be properly specified. Of note, presumptive GHRH-neurons may specifically lack the expression of only a limited number of factors, including their key fate marker GHRH (based on the expression of Gsx1 and Hmx2). Second, presumptive embryonic AgRP-, POMC-, GHRH- and Sst-neurons may show aberrant gene expression profiles such as upregulation of Dlx1, although it remains to be determined whether or not this reflects a switch in cell fate specification of presumptive embryonic ARC neurons.
To investigate further the effects of cell fate specification failure and aberrant gene expression in presumptive AgRP-, POMC-, GHRH- and Sst-neurons in the ARC, we examined P21-28 Isl1cko mice. Interestingly, the number of GABAergic neurons was significantly reduced in P21 Isl1cko (Fig. 4D). In addition, the ARC region becomes smaller in Isl1cko mice relative to control mice at P21 (Fig. 4D), whereas the size of the embryonic ARC structure was comparable between Isl1cko and control mice (e.g. DAPI in Fig. 2A, Dlx1 in Fig. 3A). The number of Sox2+ ependymal cells, which represent proliferating adult neural progenitor cells (Ming and Song, 2011), did not change in the ARC of Isl1cko mice at P28 (Fig. 4E), suggesting that the reduced ARC area in Isl1cko mice was not caused by a loss of neural progenitors. We also examined the raw number of neurons in the ARC using a neuronal marker, NeuN (Rbfox3). The number of NeuN+ cells was significantly decreased in the ARC of Isl1cko mice at P15, relative to their control littermates, whereas no significant changes were observed at E16.5 (Fig. 4F). These results suggest that the number of neurons decrease in the ARC of Isl1cko mice at postnatal stages, leading to a shrinkage in the overall size of the ARC structure.
To determine the underlying cause for the decrease in the ARC size, we first examined whether presumptive AgRP-, POMC-, GHRH- and Sst-neurons are converted to oligodendrocytes or astrocytes using immunostaining for the oligodendrocyte marker Olig2 and the astrocyte marker GFAP, respectively. Both immunostainings were comparable between Isl1cko and control mice (data not shown). To evaluate cell death, we performed immunostaining for activated caspase 3, an apoptotic cell marker, and found that very few cells die in either control or Isl1cko mice at E18.5 (Fig. 5A). The gross morphology of the ARC was again comparable between control and Isl1cko mice, as determined by the DAPI staining (Fig. 5A). Similar results were also obtained at P21, but DAPI staining again revealed that the ARC region becomes smaller in Isl1cko mice relative to control mice (Fig. 5B). Strikingly, we detected a significant increase in immunostaining for activated caspase 3 with Isl1cko mice relative to control mice at P15 (Fig. 5C, arrows). These results suggest that at least some of the miss-specified presumptive AgRP-, POMC-, GHRH- and Sst-neurons might be lost postnatally in Isl1cko mice, probably between P0 and P20.
Fig. 5.

Increased apoptosis in the arcuate neurons of Isl1cko mice. (A-C) The ARC of control and Isl1cko mice at E18.5 (A), P21 (B) and P15 (C) immunostained with anti-activated caspase 3 (Casp3) antibody. DAPI staining marks all nuclei in the section. Dashed lines indicate the boundary of the ARC (B). Double arrows in B indicate ARC region; arrows in C indicate Casp3+ cells. The number of Casp3+ cells was quantified using ImageJ (C). *P<0.05; Student's t-test. Error bars indicate s.e.m. Four controls and four mutants were used in all experiments. Scale bars: 100 μm.
Taken together, our data suggest that, during hypothalamic development, Isl1 plays an important role in the fate specification of AgRP-, POMC-, GHRH- and Sst-neurons and their postnatal survival rather than the proliferation of neural progenitors and their differentiation to neurons.
Isl1 directly regulates the expression of AgRP in AgRP-neurons
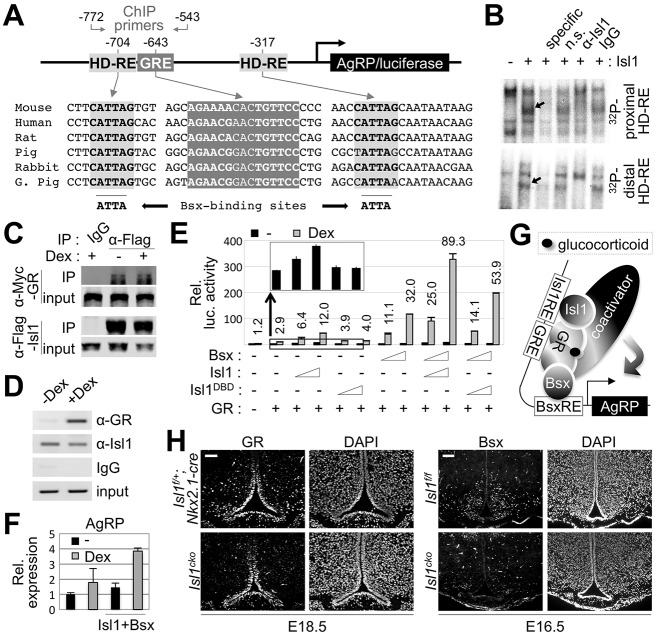
Our results raise the possibility that Isl1 is crucial to upregulate the expression of NPY, AgRP, POMC, GHRH and Sst during specification of arcuate neuronal subtypes in the developing hypothalamus. We have recently shown that Isl1 directly regulates cholinergic pathway genes, thereby determining the cholinergic neuronal identity of spinal motor neurons and forebrain cholinergic neurons (Cho et al., 2014). Likewise, it is possible that Isl1 directly controls the expression of key neuropeptides during ARC development. To test this possibility, we investigated the Agrp gene as a direct Isl1 target because we have previously identified critical gene regulatory regions in the Agrp gene, which drive gene expression specifically in AgRP-neurons (Lee et al., 2013). The Agrp promoter region was found to recruit two orexigenic transcription factors, the HD protein Bsx and the nuclear hormone receptor GR (Lee et al., 2013). Notably, the HD-binding motifs in the Agrp promoter, which have been shown to serve as Bsx-binding sites (Sakkou et al., 2007; Lee et al., 2013), are identical to the Isl1-binding consensus motif, which we defined by in vitro SELEX (systematic evolution of ligands by exponential enrichment) assays (i.e. CATTAG; Lee et al., 2008) (Fig. 6A). The CATTAG sequences in the distal HD response element (HD-RE) were completely conserved in multiple mammalian species (Fig. 6A). Similarly, the proximal HD-RE, located closer to the transcriptional initiation site, was also relatively well conserved (Fig. 6A). The evolutionarily conserved sequences of HD-REs in the Agrp promoter, along with the crucial role of Isl1 in inducing AgRP expression (Fig. 2K; Fig. 3B), led us to hypothesize that Isl1 directly upregulates the Agrp gene by binding to the HD-REs in the Agrp promoter. To test whether Isl1 directly binds to the HD-REs of Agrp, we performed electrophoresis mobility shift assays using 32P-labeled DNA probes and in vitro translated Isl1 protein (Fig. 6B). Incubation of Isl1 with DNA probes containing each HD-RE led to the formation of an Isl1–DNA complex (Fig. 6B, arrows) suggesting that Isl1 binds both HD-REs in the Agrp promoter. To test the specificity of the interaction between Isl1 and HD-RE sequences, we competed the association between Isl1 and 32P-labeled HD-RE probe with either unlabeled HD-RE oligonucleotides or unlabeled non-specific oligonucleotides. The formation of the Isl1–HD-RE–DNA complex was disrupted by HD-RE oligonucleotides, but less efficiently by non-specific oligonucleotides (Fig. 6B), supporting the idea that Isl1 specifically interacts with each HD-RE in the Agrp gene. To confirm the identity of the Isl1–HD-RE–DNA complex, we incubated the complex with either anti-Isl1 antibody or IgG and found that anti-Isl1 antibody, but not IgG, led to disappearance of the shifted bands. Thus, the electrophoresis mobility shift assays demonstrate that Isl1 directly binds to the Agrp promoter.
Fig. 6.
Direct regulation of AgRP expression by Isl1. (A) Schematic of the Agrp promoter as well as the evolutionary conservation of two HD-REs and GRE in mammals. (B) Electrophoretic mobility shift assays to test binding of Isl1 to both HD-REs. The specific bands (arrows) detected by the 32P-probe/Isl1–complex were extinguished by non-labeled specific competitor or anti-Is1 antibody but not by non-specific (n.s.) competitor or IgG. (C) Co-immunoprecipitation experiments with HEK293 cells transfected with the expression vectors for Myc-tagged GR and Flag-tagged Isl1 and treated with vehicle or 0.1 μM Dex. (D) ChIP experiments with P56 wild-type hypothalami using anti-GR and anti-Isl1 antibodies were performed following intraperitoneal injection of either vehicle or Dex (10 mg/kg). (E) Luciferase reporter assay with AgRP-1kb:Luc reporter was performed in HEK293 cells transfected with expression vectors for Bsx, Isl1, Isl1DBD (Isl1-N230S) and GR as indicated. Isl1DBD is a DNA-binding defective mutant form of Isl1. Inset shows the basal transactivation levels of the reporter (without Dex) in response to increasing amount of Isl1 or Isl1DBD (indicated by wedges below). The numbers above each column indicate activation folds by Dex relative to the reporter activity without Bsx, Isl1, Isl1DBD and GR. (F) In N42 embryonic mouse hypothalamus cells, Dex-induced expression of Agrp was measured by qRT-PCR upon ectopic expression of Isl1 and Bsx. (G) Model for a complex of Isl1–GR–Bsx to activate the transcription of Agrp by recruiting co-activators. (H) Immunohistochemistry with anti-GR and anti-Bsx was performed on E18.5 and E16.5 ARC, respectively. Three controls and three mutants were used in these experiments. Scale bars: 100 μm. Error bars indicate s.d.
Given our previous finding that GR directly binds to the GRE in close proximity to the distal HD-RE, and activates Agrp transcription in response to peripheral cue glucocorticoids (Fig. 6A; Lee et al., 2013), we considered the possibility that Isl1 forms a transcriptional activating complex with GR and cooperates with GR in inducing Agrp transcription in the hypothalamus. To test this possibility, we investigated the association between Isl1 and GR in HEK293 cells using co-immunoprecipitation assays. Isl1 interacted with GR in a GR ligand dexamethasone (Dex)-independent manner in cells (Fig. 6C), supporting a possible cooperative action between Isl1 and GR. Next, to determine whether Isl1 binds to the Agrp gene in the hypothalamus and whether this recruitment requires glucocorticoids, we intraperitoneally injected P56 mice with either Dex or vehicle, collected the hypothalami from the mice 2 h after injection, performed chromatin immunoprecipitation (ChIP) assays with anti-Isl1 or anti-GR antibody and P56 mouse hypothalamus lysates, and did PCR analyses with a primer set encompassing the distal HD-RE and the GRE (Fig. 6A). This analysis revealed that Isl1 constitutively binds the HD-RE in the Agrp gene irrespective of glucocorticoid signaling, whereas GR is recruited to the GRE of the Agrp gene only in Dex-treated hypothalamus (Fig. 6D).
The co-recruitment of Isl1 and GR to the Agrp promoter prompted us to investigate whether Isl1 is capable of transactivating the Agrp promoter in combination with GR in response to the orexigenic glucocorticoid signaling. Given the presence of the two well-conserved HD-REs in the Agrp promoter and the co-expression of Bsx1 with Isl1 and GR in the AgRP-neurons, we also asked whether Bsx participates in this transcriptional cooperation. To address these questions, we performed luciferase reporter assays with a reporter in which a 1 kb region of the Agrp promoter is linked to a luciferase gene (Lee et al., 2013; Fig. 6A). Isl1 alone, but not a DNA-binding defective mutant form of Isl1 (Isl1DBD), activates the reporter in a dose-dependent manner (Fig. 6E, inset, which is a magnified image of only the basal level of transcription in the absence of Dex), suggesting that Isl1 activates Agrp gene transcription by directly binding to the gene. Interestingly, Isl1 strongly synergized with GR in the presence of Dex, whereas DNA-binding defective Isl1 was not as effective as wild-type Isl1 in synergistically activating the reporter (Fig. 6E). Bsx further enhanced the transactivation of the Agrp promoter by Isl1 and GR. In addition, in an established embryonic mouse hypothalamus cell line, N42 (Cellutions Biosystems, Toronto, Canada), ectopic expression of Isl1 and Bsx increased the expression of AgRP in both the absence and presence of Dex (Fig. 6F). Taken together, these results suggest a model in which Isl1, Bsx and GR form a ternary complex on the Agrp promoter by binding to the HD-REs and GRE, and that this Isl1–Bsx–GR ternary complex then synergistically stimulates the glucocorticoid-directed transactivation of the Agrp promoter, probably by facilitating recruitment of transcriptional coactivators to the Agrp gene (Fig. 6G).
Isl1 is important for the expression of the orexigenic transcription factors GR and Bsx
Given the essential role of Isl1 in the fate specification of AgRP-neurons as well as in the transcription of the Agrp gene, we investigated whether Isl1 is important for establishing the expression of the two key orexigenic transcription factors GR and Bsx, which in turn cooperate with Isl1 to transactivate the Agrp gene during hypothalamus development. Interestingly, we found that the expression of both GR and Bsx was severely impaired in the ARC of Isl1cko mice, whereas GR expression in the ependymal layer remained intact (Fig. 6H). These results indicate that Isl1 plays a crucial role in the expression of GR and Bsx in developing AgRP-neurons. Although it remains to be determined whether the Gr and Bsx genes are direct or indirect targets of Isl1, our results demonstrate that Isl1 modulates the expression of multiple orexigenic genes (Gr, Bsx and Agrp) in AgRP-neurons. These results also establish Isl1 as a key transcription factor that orchestrates timely induction of the orexigenic neuropeptide AgRP in response to an increased level of the peripheral orexigenic cue glucocorticoid.
DISCUSSION
Our studies raise an interesting possibility that Isl1 may function as a central regulator of cell fate specification of arcuate neuronal cell types and their postnatal survival in the developing hypothalamus. Although our results uncover essential roles of Isl1 in the expression of key fate markers of several arcuate neurons, it remains to be determined whether this reflects requirement of Isl1 specifically for the expression of these fate markers or a broader role of Isl1 to regulate a wider spectrum of target genes in each arcuate neuronal type, and whether some of the presumptive arcuate neurons adopt a completely different cell fate. This is currently a difficult question to resolve, primarily because only a few markers are known for each arcuate neuronal type. At least for one key neuropeptide gene, Agrp, we show that Isl1 directly controls its expression by cooperating with two other orexigenic transcription factors (GR and Bsx), providing mechanistic insights into Isl1-directed gene networks in ARC development. In addition, our loss-of-function studies indicate that, in AgRP-neurons, Isl1 upregulates the expression of NPY, GR and Bsx. Given the cooperative actions of Isl1 with GR and Bsx, Isl1-dependent regulation of Gr and Bsx is expected to amplify the positive regulatory action of Isl1 in AgRP expression (Fig. 6G). Of note, the ability of a DNA-binding defective mutant form of Isl1 to still synergize with Bsx1 and GR in the activation of AgRP-luciferase reporter assays (Fig. 6E) indicates that Isl1 binding to the HD-RE is not absolutely required for AgRP expression, raising the possibility that protein-protein interactions among GR, Isl1 and/or Bsx may overcome the defective DNA binding of Isl1. Interestingly, while this manuscript was being prepared, Isl1 was reported to promote the expression of the anorexigenic neuropeptide POMC in the arcuate POMC-neurons by directly binding to two related Isl1RE-containing enhancer elements in the Pomc gene (Nasif et al., 2015). Combined with our Isl1cKO studies, this finding supports the idea that Isl1 may specify the identity of the arcuate POMC-neurons. In this regard, it is notable that both the Ghrh and Sst genes possess Isl1-bound ChIPseq peaks in a ChIPseq dataset from embryonic stem cells (Mazzoni et al., 2013), pointing to the possibility that Isl1 directly binds to Ghrh and Sst genes and induces their expression in developing arcuate GHRH- and Sst-neurons.
In this work, we demonstrated that Isl1 is required for the expression of key identity markers of several arcuate neuronal subpopulations: the orexigenic neuropeptides AgRP and NPY of AgRP-neurons, the anorexigenic neuropeptide αMSH of POMC-neurons, and the growth stimulatory peptides GHRH of GHRH-neurons and Sst of Sst-neurons. Our finding leads to the question of how a single transcription factor Isl1 might direct the specification of distinct neuronal types. Notably, Isl1 has been shown to drive fate specification of many cell types during multiple tissue development, often by forming cell type-specific complexes with various partner transcription factors. For instance, Isl1 partners with the LIM-HD factor Lhx3 in specifying motor neurons in the spinal cord (Thaler et al., 2002; Lee and Pfaff, 2003), the LIM-HD factor Lhx8 in directing the fate of forebrain cholinergic neurons in the ventral telencephalon (Cho et al., 2014), the HD factor Phox2a in cranial motor neurons (Mazzoni et al., 2013), the basic helix-loop-helix transcription factor Beta2 (Neurod1) in pancreas (Peng et al., 2005), the high-mobility group box factor Sox2 in epidermal Merkel cells (Perdigoto et al., 2014) and the POU-domain transcription factor Pou4f2 in retinal ganglion cells (Li et al., 2014). Given these results, it is probable that Isl1 forms distinct cell type-specific complexes in AgRP-, POMC-, GHRH- and Sst-neurons in the ARC, which enable Isl1 to control distinct sets of target genes in each neuronal type in the developing hypothalamus. Indeed, supporting this possibility, our study discovered a novel type of Isl1 complex consisting of Isl1, Bsx and GR in AgRP-neurons that directly binds and triggers the expression of the Agrp gene (Fig. 6G). In the future, it will be interesting to investigate whether Isl1 also forms one or more cell type-specific complexes with other partner transcription factors in regulating the expression of its target genes in AgRP-, POMC-, GHRH- and Sst-neurons. In light of this, it is noteworthy that the HD transcription factors Gsx1, Hmx2 and Hmx3 have been shown to be required for the expression of GHRH in the arcuate GHRH-neurons (Li et al., 1996; Wang et al., 2004). Gsx1 was also shown to bind directly to a Gsx-binding site in the Ghrh promoter region (Mutsuga et al., 2001). Thus, it will be interesting to examine whether Isl1 cooperates with Gsx1 and Hmx2/3 in inducing the expression of GHRH in GHRH-neurons and whether this involves formation of an Isl1-containing complex with Gsx1 and/or Hmx2/3. It would also be interesting to investigate the mechanistic basis of Isl1-directed ARC neuronal development by identifying a full range of direct target genes of Isl1 in distinct arcuate neurons using genome-wide approaches such as RNAseq and ChIPseq.
Deletion of Isl1 in part of the developing hypothalamus, including the ARC, led to marked defects in linear growth and feeding behavior in Isl1cko mice (Fig. 2). The loss of two growth-stimulating neuropeptides, GHRH of GHRH-neurons and Sst of Sst-neurons, probably contributes to the severe deficits in linear growth in our Isl1cko mice. Notably, GHRH-null mice showed stunted growth but survived to adulthood (Alba and Salvatori, 2004). Thus, although the stunted growth of the Isl1cko mice results from lack of both feeding and GHRH, their early death is likely to be caused by the defects in the hypothalamic feeding circuits and the resulting lack of feeding and starvation, rather than being an outcome of inadequate growth. The pathophysiological mechanism that underlies the strong feeding deficits of our Isl1cko mice remains to be determined. Neonatal inactivation of Npy and/or Agrp does not affect food intake in mice (Qian et al., 2002). Similarly, neonatal ablation of AgRP-neurons displays minimal effects on feeding, whereas their ablation in adults leads to rapid starvation and death (Bewick et al., 2005; Gropp et al., 2005; Luquet et al., 2005; Xu et al., 2005; Wu et al., 2009). These results led to a proposal that loss of AgRP expression or AgRP-neurons in neonatal stages triggers developmental compensation but such loss in adults is detrimental for survival. Of note, αMSH is a melanocortin-receptor agonist, whereas AgRP is an inverse agonist of melanocortin action (Biebermann et al., 2012; Myers and Olson, 2012). Between the two predominant CNS melanocortin receptors, MC3R and MC4R, the latter expressed in the PVN and amygdala is known to primarily control food intake (Myers and Olson, 2012). These results raise an interesting possibility that restoring AgRP-like activity, which antagonizes MC4R, might be a crucial developmental compensatory component in AgRP- and AgRP/NPY-null mice (Qian et al., 2002). Our Isl1cko mice may be defective in activating developmental compensation for loss of AgRP-like activity, thereby completely losing the antagonism function against MC4R. Alternatively, an additional MC4R-activating program might be inappropriately activated in our Isl1cko mice despite the loss of αMSH expression. These additional mechanisms may involve Isl1 expression in Nkx2.1+ neurons in either the ARC and/or other nuclei in hypothalamic feeding circuits. Finally, it should be noted that the growth and feeding deficits observed with our Isl1cko mice might also reflect function of Isl1 in Isl1, Nkx2.1 double-positive non-arcuate cell types. For instance, Nkx2.1 and Isl1 play roles in organogenesis of the thyroid and pituitary gland, both of which can affect linear growth (Ericson et al., 1998; Mullen et al., 2012; Pakarinen et al., 2002; Takuma et al., 1998; Westerlund et al., 2008).
In summary, our results identify Isl1 as a key transcription factor that functions in the development of multiple arcuate neuronal subpopulations in the developing hypothalamus. Further studies of Isl1 in developing hypothalamus should provide important insights into the gene regulatory program directing the development of Isl1-expressing arcuate neurons.
MATERIALS AND METHODS
Animals
All mice were maintained on a normal 12 h light, 12 h dark cycle with ad libitum access to normal chow and water, unless otherwise noted. The generation of Isl1f/f and Nkx2.1-Cre mice has been described previously (Mu et al., 2008; Xu et al., 2008). Isl1f/f mice were crossed with Isl1f/+;Nkx2.1Cre mice to generate Isl1f/f;Nkx2.1Cre mice (Isl1cko mice). Mice were intraperitoneally injected with Dex (10 mg/kg) and analyzed 2 h post-injection. For the BrdU assay, pregnant female mice were intraperitoneally injected with BrdU (150 mg/kg) and sacrificed 2 h later. All studies were approved by the Institutional Animal Care and Use Committee.
ChIP
Mouse hypothalami were dissected out and homogenized before being crosslinked with 1% formaldehyde for 10-15 min at room temperature, and then quenched by 125 mM glycine. The cells were washed in Buffer I (0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM HEPES, pH 6.5) and Buffer II (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM HEPES, pH 6.5) sequentially, lysed with lysis buffer (0.5% SDS, 5 mM EDTA, 50 mM Tris-HCl, pH 8.0, protease inhibitor cocktail), and then subjected to sonication. Cell lysates were diluted in ChIP buffer (0.5% Triton X-100, 2 mM EDTA, 100 mM NaCl, 50 mM Tris-HCl, pH 8.0, protease inhibitor cocktail) and incubated with IgG and protein A agarose beads for 1 h for immunoclearing. The supernatant was collected after quick spin-down, immunoprecipitated with anti-GR (SC-1004, Santa Cruz), our home-made anti-Isl1 antibody raised against the C-terminal 134-349 amino acid region of rat Isl1 and validated through immunoblotting (data not shown) and immunohistochemistry (Fig. 2A) or control IgG (Santa Cruz), and incubated with protein A agarose over night at 4°C. Next day, the beads were washed with TSE I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 150 mM NaCl), TSE II (same components as in TSE I except 500 mM NaCl) and Buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.0) sequentially for 10 min at each step. The beads were subsequently washed with TE buffer (10 mM Tris-HCl, 1 mM EDTA) three times. The protein/chromatin complexes were eluted in elution buffer (1% SDS, 1 mM EDTA, 0.1 M NaHCO3, 50 mM Tris-HCl, pH 8.0) and reverse cross-linked by incubating at 65°C overnight. The eluates were incubated at 50°C for more than 2 h with proteinase K. The DNA was purified with phenol/chloroform and ethanol precipitation and dissolved in water. The following primers were used for PCR: 5′-CAAGCTGATGAGGCCAGGCGTA-3′ and 5′-GCTCTCCCTCCTCTGTGCTTTC-3′.
Immunohistochemistry and ISH
Anesthetized mice were perfused transcardially with PBS first and then with 4% paraformaldehyde. Brains were removed and placed in 4% paraformaldehyde overnight, washed with PBS, and incubated with 30% sucrose. Embryos were fixed in 4% paraformaldehyde and incubated in 30% sucrose. Brain sections (12 μm thick) were prepared with a cryostat and incubated with rabbit anti-GR (SC-1004, Santa Cruz; 1:500), rabbit anti-Nkx2.1 (SC-13040, Santa Cruz; 1:500), rabbit anti-Isl1 (home-made; 1:3000), rabbit anti-Bsx (Lee et al., 2013; 1:2000), sheep anti-digoxigenin-AP (11093274910, Roche Diagnostics; 1:4000), rat anti-BrdU (ab6326, Abcam; 1:500), rabbit anti-NeuN (Ab177487, Abcam; 1:2000), rabbit anti-cleaved casp3 (#9661, Cell Signaling; 1:1000) or goat anti-Sox2 (SC-17320, Santa Cruz; 1:1000) antibodies at 4°C overnight and followed by 1-2 h incubation with fluorescence-conjugated secondary antibodies (1:500, Jackson ImmunoResearch; donkey Alexa488-conjugated anti-rabbit #711-545-152, anti-rat #712-545-153, anti-goat #705-545-147, Alexa594-conjugated anti-rabbit #711-585-152). For ISH, antisense RNA probes were labeled with digoxigenin-UTP (Roche Diagnostics) according to the manufacturer's protocol. Hybridization was performed at 68°C overnight. Hybridized sections were washed in 4× SSC and 0.2× SSC solution, and incubated with anti-digoxigenin-AP antibody (11093274910, Roche Diagnostics; 1:4000) overnight. The sections were subjected to color reaction with NBP/BCIP. The VECTASTAIN Elite ABC Kit (PK-6101, Vector Labs) was used according to the manufacturer's instruction for immunohistochemistry assay following ISH. cDNAs for mouse Agrp, somatostatin, Crh and Trh were cloned to pBluescript vector to generate digoxigenin-labeled riboprobes. Probes for Pomc, Ghrh, Mash1 and Dlx1 have been described previously (Nilaweera et al., 2002; McNay et al., 2006). The probes for Sf1 and Gad1 were kind gifts from Dr Holly Ingraham at the University of California, San Francisco (CA, USA) and Dr Jane E. Johnson at the University of Texas Southwestern (TX, USA), respectively. The Npy probe was obtained from the Microarray Core Facility (now the Genomic and RNA Profiling Core) at Baylor College of Medicine (TX, USA).
Co-immunoprecipitation
Co-immunoprecipitation was carried out as described previously (Cho et al., 2014).
Luciferase assay
HEK293 cells were maintained in DMEM supplemented with 10% fetal bovine serum and penicillin/streptomycin. For luciferase assays, cells were seeded into 48-well plates and transiently transfected with AgRP 1kb-luc reporter (Lee et al., 2013) and combinations of rat Isl1/pcs2, mouse Bsx/pcs2 and rat GR/pcs2 as described, using SuperFect (Qiagen) according to the manufacturer's instruction. The actin-β-galactosidase plasmid was co-transfected for normalization of the luciferase assay. Data are shown in relative luciferase units (mean±s.d.).
RNA extraction and quantitative RT-PCR analysis
Total RNAs were extracted from a piece of mouse liver using Trizol (Invitrogen) and reverse-transcribed using Thermo Scientific Maxima H Minus Reverse Transcriptase. N42 cells were maintained in DMEM supplemented with 10% fetal bovine serum and penicillin/streptomycin. For qRT-PCR assays, N42 cells were transiently transfected with the indicated vectors using Lipofectamine LTX (Invitrogen). Two days after transfection, cells were treated with either vehicle or Dex (100 nM) for 1 h. The following primers were used with SYBR Green kit (Thermo Scientific Luminaris Color Higreen, #K0371) for quantitative RT-PCR of Igf1: 5′-TCATGTCGTCTTCACACCTCT-3′ and 5′-TCCACAATGCCTGTCTGAGG-3′. The primers for Agrp were as previously described (Kim et al., 2013).
Electrophoretic mobility shift assay
Double-stranded oligonucleotides were end labeled with γ-32P-ATP using T4 polynucleotide kinase. Isl1 protein was synthesized using the Promega TNT Coupled Transcription-Translation kit. Binding mixtures for reaction included 10 mM HEPES (pH 7.7), 75 mM KCl, 2.5 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, 10% glycerol, 1 μg poly(dI-dC), labeled probe and in vitro translated protein. The reaction mixtures were incubated at room temperature for 30 min and analyzed with a non-denaturing 5% polyacrylamide gel. The following oligonucleotides were used: Isl1RE, 5′-AGCCATTAACACTAATGAAGCAGGC-3′; non-specific competitor, 5′-TCGAGACCCAGGGAACAGTTCGTTCTGTTTCCG-3′.
Statistical analysis
Statistical differences were determined by two-tailed Student's t-test. Statistical significance is displayed as *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 or *****P<0.00001.
Acknowledgements
We thank Drs Xiuqian Mu, Holly Ingraham and Jane Johnson for providing Isl1 floxed mice, SF1 probe and GAD1 probe, respectively. We also thank Dr Ashley Brown for critically reading this manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
B.L., S.L., S.-K.L. and J.W.L. designed experiments. B.L. and S.L. performed experiments. B.L., S.L., S.-K.L. and J.W.L. analyzed data. B.L., S.L. and J.W.L. wrote the manuscript.
Funding
This research was supported by the National Research Foundation of Korea Basic Science Research Program [NRF-2015R1A2A1A15055611] and Bio & Medical Technology Development Program [NRF-2012M3A9C6050508]; the Korean Ministry of Science, ICT and Future Planning [MSIP-2011-0030001 to S.L.]; the National Institute of Neurological Disorders and Stroke [R01 NS054941 to S.-K.L.]; and the National Institute of Diabetes and Digestive and Kidney Diseases [R01 DK064678 to J.W.L. and R01 DK103664 to J.W.L. and S.-K.L.]. Deposited in PMC for release after 12 months.
References
- Alba M. and Salvatori R. (2004). A mouse with targeted ablation of the GHRH gene: a new model of isolated GH deficiency. Endocrinology 145, 4134-4143. 10.1210/en.2004-0119 [DOI] [PubMed] [Google Scholar]
- Bewick G. A., Gardiner J. V., Dhillo W. S., Kent A. S., White N. E., Webster Z., Ghatei M. A. and Bloom S. R. (2005). Post-embryonic ablation of AgRP neurons in mice leads to a lean, hypophagic phenotype. FASEB J. 19, 1680-1682. 10.1096/fj.04-3434fje [DOI] [PubMed] [Google Scholar]
- Biebermann H., Kühnen P., Kleinau G. and Krude H. (2012). The neuroendocrine circuitry controlled by POMC, MSH, and AGRP. Handb. Exp. Pharmacol. 209, 47-75. 10.1007/978-3-642-24716-3_3 [DOI] [PubMed] [Google Scholar]
- Bluet-Pajot M.-T., Tolle V., Zizzari P., Robert C., Hammond C., Mitchell V., Beauvillain J.-C., Viollet C., Epelbaum J. and Kordon C. (2001). Growth hormone secretagogues and hypothalamic networks. Endocrine 14, 1-8. 10.1385/ENDO:14:1:001 [DOI] [PubMed] [Google Scholar]
- Cho H.-H., Cargnin F., Kim Y., Lee B., Kwon R.-J., Nam H., Shen R., Barnes A. P., Lee J. W., Lee S. et al. (2014). Isl1 directly controls a cholinergic neuronal identity in the developing forebrain and spinal cord by forming cell type-specific complexes. PLoS Genet. 10, e1004280 10.1371/journal.pgen.1004280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. and Rosenfeld R. G. (1994). Physiologic and clinical relevance of the insulin-like growth factor binding proteins. Curr. Opin. Pediatr. 6, 462-467. 10.1097/00008480-199408000-00019 [DOI] [PubMed] [Google Scholar]
- Cowley M. A., Smart J. L., Rubinstein M., Cerdan M. G., Diano S., Horvath T. L., Cone R. D. and Low M. J. (2001). Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature 411, 480-484. 10.1038/35078085 [DOI] [PubMed] [Google Scholar]
- Davis A. M., Seney M. L., Walker H. J. and Tobet S. A. (2004). Differential colocalization of Islet-1 and estrogen receptor alpha in the murine preoptic area and hypothalamus during development. Endocrinology 145, 360-366. 10.1210/en.2003-0996 [DOI] [PubMed] [Google Scholar]
- Dellovade T. L., Young M., Ross E. P., Henderson R., Caron K., Parker K. and Tobet S. A. (2000). Disruption of the gene encoding SF-1 alters the distribution of hypothalamic neuronal phenotypes. J. Comp. Neurol. 423, 579-589. [DOI] [PubMed] [Google Scholar]
- Ericson J., Norlin S., Jessell T. M. and Edlund T. (1998). Integrated FGF and BMP signaling controls the progression of progenitor cell differentiation and the emergence of pattern in the embryonic anterior pituitary. Development 125, 1005-1015. [DOI] [PubMed] [Google Scholar]
- Fehm H. L., Born J. and Peters A. (2004). Glucocorticoids and melanocortins in the regulation of body weight in humans. Horm. Metab. Res. 36, 360-364. 10.1055/s-2004-814568 [DOI] [PubMed] [Google Scholar]
- Gropp E., Shanabrough M., Borok E., Xu A. W., Janoschek R., Buch T., Plum L., Balthasar N., Hampel B., Waisman A. et al. (2005). Agouti-related peptide-expressing neurons are mandatory for feeding. Nat. Neurosci. 8, 1289-1291. 10.1038/nn1548 [DOI] [PubMed] [Google Scholar]
- Hentges S. T., Nishiyama M., Overstreet L. S., Stenzel-Poore M., Williams J. T. and Low M. J. (2004). GABA release from proopiomelanocortin neurons. J. Neurosci. 24, 1578-1583. 10.1523/JNEUROSCI.3952-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. W., Elmquist J. K. and Elias C. F. (2008). Hypothalamic pathways linking energy balance and reproduction. Am. J. Physiol. Endocrinol. Metab. 294, E827-E832. 10.1152/ajpendo.00670.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath T. L., Bechmann I., Naftolin F., Kalra S. P. and Leranth C. (1997). Heterogeneity in the neuropeptide Y-containing neurons of the rat arcuate nucleus: GABAergic and non-GABAergic subpopulations. Brain. Res. 756, 283-286. 10.1016/S0006-8993(97)00184-4 [DOI] [PubMed] [Google Scholar]
- Hrabovszky E. (2014). Neuroanatomy of the human hypothalamic kisspeptin system. Neuroendocrinology 99, 33-48. 10.1159/000356903 [DOI] [PubMed] [Google Scholar]
- Kim S.-G. Lee B., Kim D.-H., Kim J., Lee S., Lee S.-K. and Lee J. W. (2013). Control of energy balance by hypothalamic gene circuitry involving two nuclear receptors, NOR1 and GR. Mol. Cell. Biol. 33, 3826-3834. 10.1128/MCB.00385-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanneau C., Bluet-Pajot M. T., Zizzari P., Csaba Z., Dournaud P., Helboe L., Hoyer D., Pellegrini E., Tannenbaum G. S., Epelbaum J. et al. (2000). Involvement of the Sst1 somatostatin receptor subtype in the intrahypothalamic neuronal network regulating growth hormone secretion: an in vitro and in vivo antisense study. Endocrinology 141, 967-979. 10.1210/en.141.3.967 [DOI] [PubMed] [Google Scholar]
- Lee S.-K. and Pfaff S. L. (2003). Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron 38, 731-745. 10.1016/S0896-6273(03)00296-4 [DOI] [PubMed] [Google Scholar]
- Lee S., Lee B., Joshi K., Pfaff S. L., Lee J. W. and Lee S.-K. (2008). A regulatory network to segregate the identity of neuronal subtypes. Dev. Cell 14, 877-889. 10.1016/j.devcel.2008.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B., Kim S.-G., Kim J., Choi K. Y., Lee S., Lee S.-K. and Lee J. W. (2013). Brain-specific homeobox factor as a target selector for glucocorticoid receptor in energy balance. Mol. Cell. Biol. 33, 2650-2658. 10.1128/MCB.00094-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis D. E., Shellard L., Koeslag D. G., Boer D. E., McCarthy H. D., McKibbin P. E., Russell J. C. and Williams G. (1993). Intense exercise and food restriction cause similar hypothalamic neuropeptide Y increases in rats. Am. J. Physiol. 264, E279-E284. [DOI] [PubMed] [Google Scholar]
- Li H., Zeitler P. S., Valerius M. T., Small K. and Potter S. S. (1996). Gsh-1, an orphan Hox gene, is required for normal pituitary development. EMBO J. 15, 714-724. [PMC free article] [PubMed] [Google Scholar]
- Li R., Wu F., Ruonala R., Sapkota D., Hu Z. and Mu X. (2014). Isl1 and Pou4f2 form a complex to regulate target genes in developing retinal ganglion cells. PLoS ONE 9, e92105 10.1371/journal.pone.0092105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo L. C., Johnson J. E., Wuenschell C. W., Saito T. and Anderson D. J. (1991). Mammalian achaete-scute homolog 1 is transiently expressed by spatially restricted subsets of early neuroepithelial and neural crest cells. Genes Dev. 5, 1524-1537. 10.1101/gad.5.9.1524 [DOI] [PubMed] [Google Scholar]
- Luquet S., Perez F. A., Hnasko T. S. and Palmiter R. D. (2005). NPY/AgRP neurons are essential for feeding in adult mice but can be ablated in neonates. Science 310, 683-685. 10.1126/science.1115524 [DOI] [PubMed] [Google Scholar]
- Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., Ng L. L., Palmiter R. D., Hawrylycz M. J., Jones A. R. et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133-140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín O., Baker J., Puelles L. and Rubenstein J. L. (2002). Patterning of the basal telencephalon and hypothalamus is essential for guidance of cortical projections. Development 129, 761-773. [DOI] [PubMed] [Google Scholar]
- Mazzoni E. O., Mahony S., Closser M., Morrison C. A., Nedelec S., Williams D. J., An D., Gifford D. K. and Wichterle H. (2013). Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat. Neurosci. 16, 1219-1227. 10.1038/nn.3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNay D. E. G., Pelling M., Claxton S., Guillemot F. and Ang S.-L. (2006). Mash1 is required for generic and subtype differentiation of hypothalamic neuroendocrine cells. Mol. Endocrinol. 20, 1623-1632. 10.1210/me.2005-0518 [DOI] [PubMed] [Google Scholar]
- Ming G.-L. and Song H. (2011). Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron 70, 687-702. 10.1016/j.neuron.2011.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T. M. and Mobbs C. V. (1999). Hypothalamic agouti-related protein messenger ribonucleic acid is inhibited by leptin and stimulated by fasting. Endocrinology 140, 814-817. 10.1210/en.140.2.814 [DOI] [PubMed] [Google Scholar]
- Mu X., Fu X., Beremand P. D., Thomas T. L. and Klein W. H. (2008). Gene-regulation logic in retinal ganglion cell development: Isl1 defines a critical branch distinct from but overlapping with Pou4f2. Proc. Natl. Acad. Sci. USA 105, 6942-6947. 10.1073/pnas.0802627105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen R. D., Park S. and Rhodes S. J. (2012). A distal modular enhancer complex acts to control pituitary- and nervous system-specific expression of the LHX3 regulatory gene. Mol. Endocrinol. 26, 308-319. 10.1210/me.2011-1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutsuga N., Iwasaki Y., Morishita M., Nomura A., Yamamori E., Yoshida M., Asai M., Ozaki N., Kambe F., Seo H. et al. (2001). Homeobox protein Gsh-1-dependent regulation of the rat GHRH gene promoter. Mol. Endocrinol. 15, 2149-2156. 10.1210/mend.15.12.0747 [DOI] [PubMed] [Google Scholar]
- Myers M. G. Jr and Olson D. P. (2012). Central nervous system control of metabolism. Nature 491, 357-363. 10.1038/nature11705 [DOI] [PubMed] [Google Scholar]
- Nasif S., de Souza F. S. J., González L. E., Yamashita M., Orquera D. P., Low M. J. and Rubinstein M. (2015). Islet 1 specifies the identity of hypothalamic melanocortin neurons and is critical for normal food intake and adiposity in adulthood. Proc. Natl. Acad. Sci. USA 112, E1861-E1870. 10.1073/pnas.1500672112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilaweera K. N., Ellis C., Barrett P., Mercer J. G. and Morgan P. J. (2002). Hypothalamic bHLH transcription factors are novel candidates in the regulation of energy balance. Eur. J. Neurosci. 15, 644-650. 10.1046/j.1460-9568.2002.01894.x [DOI] [PubMed] [Google Scholar]
- Nilsson I., Johansen J. E., Schalling M., Hökfelt T. and Fetissov S. O. (2005). Maturation of the hypothalamic arcuate agouti-related protein system during postnatal development in the mouse. Brain Res. Dev. Brain Res. 155, 147-154. 10.1016/j.devbrainres.2005.01.009 [DOI] [PubMed] [Google Scholar]
- Nogueiras R., López M., Lage R., Perez-Tilve D., Pfluger P., Mendieta-Zerón H., Sakkou M., Wiedmer P., Benoit S. C., Datta R. et al. (2008). Bsx, a novel hypothalamic factor linking feeding with locomotor activity, is regulated by energy availability. Endocrinology 149, 3009-3015. 10.1210/en.2007-1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovesjo M.-L., Gamstedt M., Collin M. and Meister B. (2001). GABAergic nature of hypothalamic leptin target neurones in the ventromedial arcuate nucleus. J. Neuroendocrinol. 13, 505-516. 10.1046/j.1365-2826.2001.00662.x [DOI] [PubMed] [Google Scholar]
- Pakarinen P., Kimura S., El-Gehani F., Pelliniemi L. J. and Huhtaniemi I. (2002). Pituitary hormones are not required for sexual differentiation of male mice: phenotype of the T/ebp/Nkx2.1 null mutant mice. Endocrinology 143, 4477-4482. 10.1210/en.2002-220052 [DOI] [PubMed] [Google Scholar]
- Pelling M., Anthwal N., McNay D., Gradwohl G., Leiter A. B., Guillemot F. and Ang S.-L. (2011). Differential requirements for neurogenin 3 in the development of POMC and NPY neurons in the hypothalamus. Dev. Biol. 349, 406-416. 10.1016/j.ydbio.2010.11.007 [DOI] [PubMed] [Google Scholar]
- Peng S.-Y., Wang W.-P., Meng J., Li T., Zhang H., Li Y.-M., Chen P., Ma K.-T. and Zhou C.-Y. (2005). ISL1 physically interacts with BETA2 to promote insulin gene transcriptional synergy in non-beta cells. Biochim. Biophys. Acta. 1731, 154-159. 10.1016/j.bbaexp.2005.08.013 [DOI] [PubMed] [Google Scholar]
- Perdigoto C. N., Bardot E. S., Valdes V. J., Santoriello F. J. and Ezhkova E. (2014). Embryonic maturation of epidermal Merkel cells is controlled by a redundant transcription factor network. Development 141, 4690-4696. 10.1242/dev.112169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps C. J., Romero M. I. and Hurley D. L. (2003). Growth hormone-releasing hormone-producing and dopaminergic neurones in the mouse arcuate nucleus are independently regulated populations. J. Neuroendocrinol. 15, 280-288. 10.1046/j.1365-2826.2003.01009.x [DOI] [PubMed] [Google Scholar]
- Qian S., Chen H., Weingarth D., Trumbauer M. E., Novi D. E., Guan X., Yu H., Shen Z., Feng Y., Frazier E. et al. (2002). Neither agouti-related protein nor neuropeptide Y is critically required for the regulation of energy homeostasis in mice. Mol. Cell. Biol. 22, 5027-5035. 10.1128/MCB.22.14.5027-5035.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakkou M., Wiedmer P., Anlag K., Hamm A., Seuntjens E., Ettwiller L., Tschöp M. H. and Treier M. (2007). A role for brain-specific homeobox factor Bsx in the control of hyperphagia and locomotory behavior. Cell Metab. 5, 450-463. 10.1016/j.cmet.2007.05.007 [DOI] [PubMed] [Google Scholar]
- Shimogori T., Lee D. A., Miranda-Angulo A., Yang Y., Wang H., Jiang L., Yoshida A. C., Kataoka A., Mashiko H., Avetisyan M. et al. (2010). A genomic atlas of mouse hypothalamic development. Nat. Neurosci. 13, 767-775. 10.1038/nn.2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slama A., Bluet-Pajot M. T., Mounier F., Videau C., Kordon C. and Epelbaum J. (1993). 125l-Somatostatin-labeled cells in the anterior arcuate nucleus mediate somatostatin effects on growth hormone but not prolactin secretion. Neuroendocrinology 58, 178-184. 10.1159/000126530 [DOI] [PubMed] [Google Scholar]
- Stephens T. W., Basinski M., Bristow P. K., Bue-Valleskey J. M., Burgett S. G., Craft L., Hale J., Hoffmann J., Hsiung H. M., Kriauciunas A. et al. (1995). The role of neuropeptide Y in the antiobesity action of the obese gene product. Nature 377, 530-532. 10.1038/377530a0 [DOI] [PubMed] [Google Scholar]
- Stühmer T., Anderson S. A., Ekker M. and Rubenstein J. L. (2002). Ectopic expression of the Dlx genes induces glutamic acid decarboxylase and Dlx expression. Development 129, 245-252. [DOI] [PubMed] [Google Scholar]
- Takuma N., Sheng H. Z., Furuta Y., Ward J. M., Sharma K., Hogan B. L., Pfaff S. L., Westphal H., Kimura S. and Mahon K. A. (1998). Formation of Rathke's pouch requires dual induction from the diencephalon. Development 125, 4835-4840. [DOI] [PubMed] [Google Scholar]
- Tannenbaum G. S., Zhang W.-H., Lapointe M., Zeitler P. and Beaudet A. (1998). Growth hormone-releasing hormone neurons in the arcuate nucleus express both Sst1 and Sst2 somatostatin receptor genes. Endocrinology 139, 1450-1453. 10.1210/endo.139.3.5977 [DOI] [PubMed] [Google Scholar]
- Thaler J. P., Lee S.-K., Jurata L. W., Gill G. N. and Pfaff S. L. (2002). LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell 110, 237-249. 10.1016/S0092-8674(02)00823-1 [DOI] [PubMed] [Google Scholar]
- Tong Q., Ye C.-P., Jones J. E., Elmquist J. K. and Lowell B. B. (2008). Synaptic release of GABA by AgRP neurons is required for normal regulation of energy balance. Nat. Neurosci. 11, 998-1000. 10.1038/nn.2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Pol A. N., Yao Y., Fu L.-Y., Foo K., Huang H., Coppari R., Lowell B. B. and Broberger C. (2009). Neuromedin B and gastrin-releasing peptide excite arcuate nucleus neuropeptide Y neurons in a novel transgenic mouse expressing strong Renilla green fluorescent protein in NPY neurons. J. Neurosci. 29, 4622-4639. 10.1523/JNEUROSCI.3249-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Grimmer J. F., Van De Water T. R. and Lufkin T. (2004). Hmx2 and Hmx3 homeobox genes direct development of the murine inner ear and hypothalamus and can be functionally replaced by Drosophila Hmx. Dev. Cell 7, 439-453. 10.1016/j.devcel.2004.06.016 [DOI] [PubMed] [Google Scholar]
- Westerlund J., Andersson L., Carlsson T., Zoppoli P., Fagman H. and Nilsson M. (2008). Expression of Islet1 in thyroid development related to budding, migration, and fusion of primordia. Dev. Dyn. 237, 3820-3829. 10.1002/dvdy.21772 [DOI] [PubMed] [Google Scholar]
- Wu Q., Boyle M. P. and Palmiter R. D. (2009). Loss of GABAergic signaling by AgRP neurons to the parabrachial nucleus leads to starvation. Cell 137, 1225-1234. 10.1016/j.cell.2009.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu A. W., Kaelin C. B., Morton G. J., Ogimoto K., Stanhope K., Graham J., Baskin D. G., Havel P., Schwartz M. W. and Barsh G. S. (2005). Effects of hypothalamic neurodegeneration on energy balance. PLoS Biol. 3, e415 10.1371/journal.pbio.0030415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Tam M. and Anderson S. A. (2008). Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J. Comp. Neurol. 506, 16-29. 10.1002/cne.21529 [DOI] [PubMed] [Google Scholar]
- Yee C. L., Wang Y., Anderson S., Ekker M. and Rubenstein J. L. R. (2009). Arcuate nucleus expression of NKX2.1 and DLX and lineages expressing these transcription factors in neuropeptide Y(+), proopiomelanocortin(+), and tyrosine hydroxylase(+) neurons in neonatal and adult mice. J. Comp. Neurol. 517, 37-50. 10.1002/cne.22132 [DOI] [PMC free article] [PubMed] [Google Scholar]