ABSTRACT
The rate and regulation of mRNA decay are major elements in the proper control of gene expression. Edc3 and Lsm4 are two decapping activator proteins that have previously been shown to function in the assembly of RNA granules termed P bodies. Here, we show that deletion of edc3, when combined with a removal of the glutamine/asparagine rich region of Lsm4 (edc3Δ lsm4ΔC) reduces mRNA stability and alters pathways of mRNA degradation. Multiple tested mRNAs exhibited reduced stability in the edc3Δ lsm4ΔC mutant. The destabilization was linked to an increased dependence on Ccr4-mediated deadenylation and mRNA decapping. Unlike characterized mutations in decapping factors that either are neutral or are able to stabilize mRNA, the combined edc3Δ lsm4ΔC mutant reduced mRNA stability. We characterized the growth and activity of the major mRNA decay systems and translation in double mutant and wild-type yeast. In the edc3Δ lsm4ΔC mutant, we observed alterations in the levels of specific mRNA decay factors as well as nuclear accumulation of the catalytic subunit of the decapping enzyme Dcp2. Hence, we suggest that the effects on mRNA stability in the edc3Δ lsm4ΔC mutant may originate from mRNA decay protein abundance or changes in mRNPs, or alternatively may imply a role for P bodies in mRNA stabilization.
KEY WORDS: P bodies, Deadenylation, Exosome, mRNA decapping, mRNA decay, mRNA stability
Summary: A strain mutated in two decapping activators, previously implicated in P body assembly, has reduced mRNA stability and increased dependence on decapping and Ccr4-dependent deadenylation for mRNA degradation.
INTRODUCTION
The degradation of mRNAs is a fundamental process in the control and modulation of gene expression (Pérez-Ortín et al., 2012). The mRNA degradation process commences with shortening of the poly(A) tail by the Ccr4/Not and Pan2/3 deadenylation complexes. In yeast and other eukaryotes, after mRNA is deadenylated, further decay can occur through one of two pathways: the 5′-to-3′ pathway, which is dependent on removal of the m7G cap from mRNA, and the 3′-to-5′ pathway, in which degradation occurs via the action of the exosome complex (Parker, 2012). Whereas one pathway can partially substitute for the absence of the other, in yeast most mRNAs are degraded by the decapping-dependent pathway (Beelman et al., 1996; Cao and Parker, 2001; Haimovich et al., 2013; Medina et al., 2014; van Dijk et al., 2011). This degradation pathway begins with the removal of the m7G cap from the mRNA by the decapping complex composed of Dcp1 and its catalytic subunit, Dcp2, which is followed by 5′-to-3′ exonucleolytic digestion via Xrn1; however, mRNA cap removal occurs at a slower rate in the absence of accessory proteins, termed mRNA decapping activators (Borja et al., 2011; Fromm et al., 2012; Nissan et al., 2010; Parker, 2012; Valkov et al., 2016). Most prominent among these factors are Dhh1, Pat1, Edc3 and the Lsm1-7 complex.
Edc3 is unusual among the decapping activators in that its absence has only a minor effect on general mRNA stability in yeast. The edc3 deletion does not affect mRNA abundance genome-wide, with the exception of the RPS28B and YRA1 mRNAs (Badis et al., 2004; Dong et al., 2007; Sun et al., 2013). In addition, in yeast, the absence of Edc3 does not affect the mRNA decapping and stability of individual mRNAs or the mRNA half-lives determined genome-wide (Decker et al., 2007; Kshirsagar and Parker, 2004; Parker, 2012; Sun et al., 2013). However, Edc3 has an additional structural role in the assembly of cytoplasmic foci of mRNA and proteins involved in mRNA degradation in other contexts (Decker et al., 2007; Teixeira and Parker, 2007). These structures, termed cytoplasmic processing bodies (P bodies), contain the Ccr4/Not and Pan2/3 deadenylase complexes in addition to decapping factors (Bett et al., 2013; Cougot et al., 2004; Teixeira and Parker, 2007). Cytoplasmic mRNA decay is thought to occur within P bodies since they contain decaying mRNA (Sheth and Parker, 2003).
The edc3Δ mutant's structural role results in the absence of microscopically observable P bodies only without aeration or in respiratory deficient yeast (Decker et al., 2007). To eliminate microscopically visible P bodies, the removal of the prion-like glutamine/asparagine rich domain of the Lsm4 protein is also required. Lsm4 is an essential protein that is a component of both the decapping activating Lsm1-7 complex and the Lsm2-8 complex, which is part of the U6 snRNP (Beggs, 2005; Tharun, 2009). Whereas depletion of the Lsm4 complex inhibits mRNA decapping (Tharun et al., 2000), the absence of the Q/N rich C-terminus of Lsm4 (lsm4ΔC) has been shown to cause a slight increase in mRNA half-lives (Decker et al., 2007; Reijns et al., 2008).
The edc3Δ and lsm4ΔC double mutant has been used by multiple groups to examine the role of P bodies in mRNA stability and additional cellular functions (Aronov et al., 2015; Decker et al., 2007; Lavut and Raveh, 2012; Simpson et al., 2014). In this study, we sought to characterize the effect of the edc3Δ lsm4ΔC mutant on mRNA metabolism by using yeast as a simple eukaryotic model that is tractable to multiple types of experimental manipulation. When the mutations were combined in the edc3Δ lsm4ΔC strain, we found that multiple mRNAs were reduced in stability, whereas the stability of other mRNAs was unaffected. To further examine the effect of the edc3Δ lsm4ΔC mutant on the mRNA stability, we used mutants in which the major pathways of mRNA degradation were compromised. Our results suggest that the edc3Δ lsm4ΔC mutant has a faster degradation rate, owing to greater reliance on deadenylation by the Ccr4/Not complex as well as mRNA decapping. We also provide evidence that links the edc3Δ lsm4ΔC mutant to altered mRNA translation and the subcellular localization of the mRNA decapping enzyme. Finally, we report that the edc3Δ lsm4ΔC mutation confers a survival advantage when yeast cells are exposed to long-term starvation.
RESULTS
mRNAs are destabilized by a synergistic effect of deletion of both EDC3 and the glutamine/asparagine rich domain of Lsm4
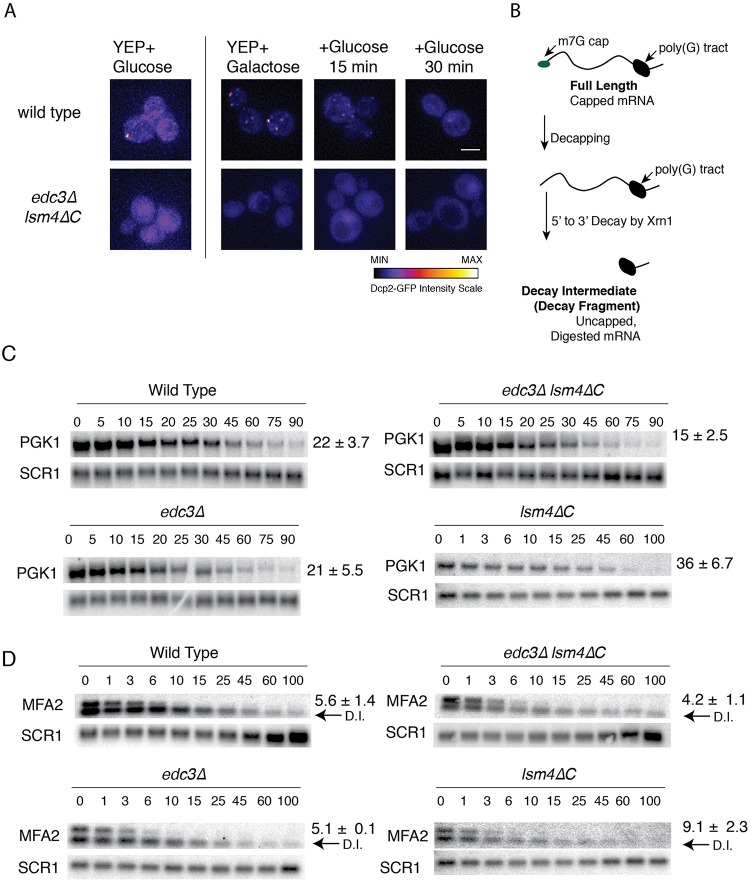
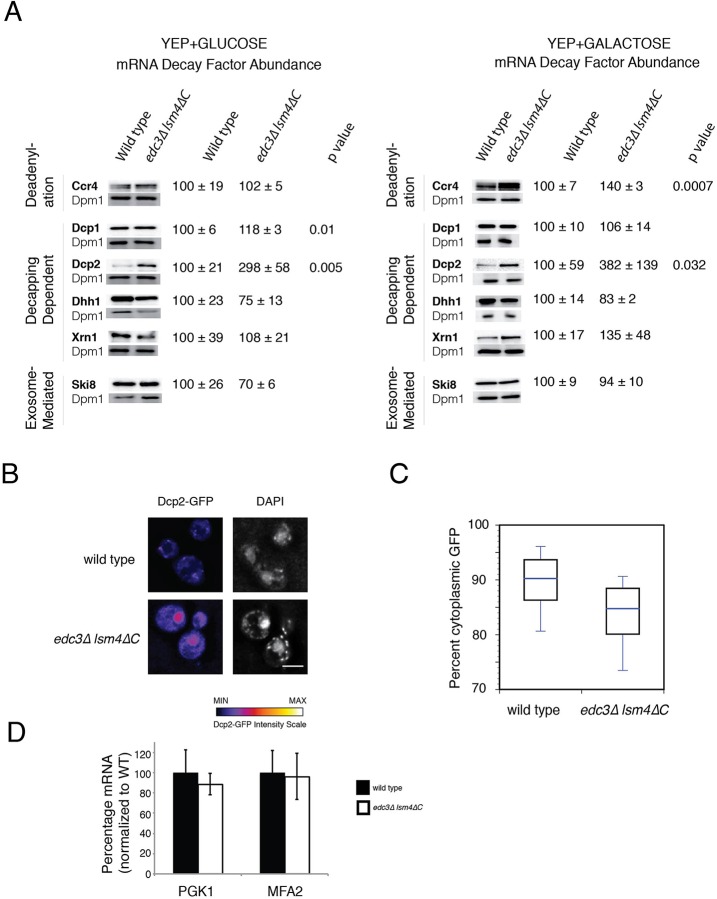
The edc3Δ lsm4ΔC mutant has been reported to be defective in the formation of P bodies (Decker et al., 2007). To confirm this result, we examined wild-type yeast and the edc3Δ lsm4ΔC mutant for the ability to form P bodies under unstressed growth. We defined P bodies as foci of the decapping enzyme, Dcp2-GFP. We observed P bodies in the wild-type strain when grown in glucose, but not in the edc3Δ lsm4ΔC mutant (Fig. 1A). The amount of P bodies was not consistently visible in all cells, as reported previously (Lui et al., 2014); however, the number of P bodies was consistently qualitatively greater in cells grown in galactose (Fig. 1A), whereas P bodies were absent from the edc3Δ lsm4ΔC mutant (Fig. 1A).
Fig. 1.
PGK1 and MFA2 mRNA stability in the edc3Δ lsm4ΔC mutant. (A) Wild type and edc3Δ lsm4ΔC mutant yeast cells expressing Dcp2-GFP from its endogenous locus grown in SC+2% glucose or 2% galactose as indicated. Later time points depict the cells grown in galactose after being washed, resuspended, and grown in medium containing glucose. (B) Depiction of the full length capped mRNA and the decay fragment generated by decapping and 5′ to 3′ degradation, which was ultimately blocked by a poly(G) tract inserted into the 3′ UTR. (C) Northern blots for the half-life determination of PGK1 mRNA in strains indicated. Time points (min) after transcriptional shut-off by glucose addition are shown. SCR1 is the loading control. Error=s.d., n=3-15 biological replicates. (D) As above for MFA2 mRNA, the half-life indicated to the right of the blots. Arrows labeled D.I. indicate the mRNA decay intermediate generated by decapping and 5′-to-3′ degradation blocked by the poly(G) tract in the 3′ UTR. Time points (min) are shown above the respective northern blots.
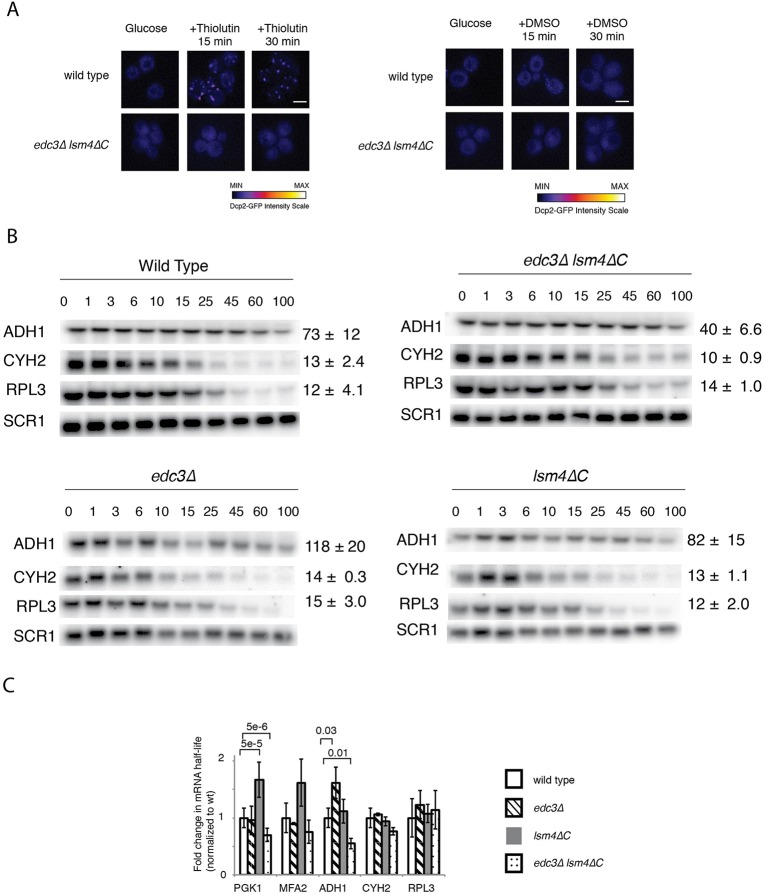
We next determined the stability of mRNAs in the wild-type edc3Δ lsm4ΔC strain and in the individual edc3Δ and lsm4ΔC mutants. We examined PGK1 and MFA2 mRNAs, which have well-characterized mechanisms of decay (Cao and Parker, 2001) and have been found to be localized to P bodies in multiple studies (Aronov et al., 2015; Brengues et al., 2005; Decker et al., 2007; Sheth and Parker, 2003; Simpson et al., 2014; Teixeira et al., 2005; Zid and O'Shea, 2014). PGK1 and MFA2 mRNA were engineered with poly(G) tracts in their 3′ UTRs, thus allowing for the generation of mRNA decay intermediates after the mRNA is decapped and degraded 5′-to-3′ by Xrn1 (Fig. 1B) (Decker and Parker, 1993). This decay intermediate is more stable than the full-length mRNA, because further degradation of this intermediate occurs primarily from the 3′ end by the exosome (Fig. 1C, the decay intermediate is indicated by the arrow labeled D.I.) (Anderson and Parker, 1998). The MFA2 and PGK1 mRNAs were placed under the control of the genomically integrated GAL1 promoter at the CUP1 locus (Hatfield et al., 1996). Transcription was induced by growth in galactose. Half-lives were determined after inhibition of transcription with the addition of glucose. PGK1 and MFA2 represent typical stable and unstable mRNAs. To provide another example of a longer-lived mRNA, we also examined ADH1 mRNA, which encodes an essential glycolytic enzyme. For shorter-lived mRNAs, we chose mRNAs encoding ribosomal proteins: CYH2/RPL28 and RPL3. We chose mRNAs encoding for ribosomal proteins additional mRNAs because several studies have indicated that they behave differently than other mRNAs (Arribere et al., 2011; Gasch et al., 2000). The ADH1, CYH2/RPL28 and RPL3 mRNA half-lives were determined after inhibition of transcription with the drug thiolutin. We observed that thiolutin, compared with a DMSO control, induced P body formation in the wild-type yeast, but not in the edc3Δ lsm4ΔC mutant (Fig. 2A).
Fig. 2.
Multiple mRNAs are destabilized in the edc3Δ lsm4ΔC mutant. (A) Left: wild type and edc3Δ lsm4ΔC mutant yeast cells expressing Dcp2-GFP from its endogenous locus grown in SC+2% glucose after transcription was inhibited by thiolutin. Right: cells treated with DMSO alone. The images were plotted to the same contrast range and the scale bars represent 3 µm. Intensity of the GFP signal is indicated below. (B) Northern blots for ADH1, CYH2/RPL28 and RPL3 mRNA performed after thiolutin transcriptional shut-off. Error=s.d., n=3 biological replicates. Quantification of mRNA half-lives for mRNA normalized to SCR1 as a loading control. Time points (min) are shown above the respective northern blots. (C) Quantification of mRNA half-lives for mRNA normalized to wild type half-life for each mRNA for the mRNA indicated. Statistically significant pairings according to a two-tailed t-test indicated with their P values.
The mRNA half-lives varied from 5 to 73 min in the wild-type strain (Figs 1C,D and 2B). We found that the PGK1 mRNA had a 46% shorter half-life in the edc3Δ lsm4ΔC mutant than in wild-type yeast (Fig. 1C), whereas MFA2's half-life, although shorter, was not significantly altered (Fig. 1D). We found similar trends after using thiolutin to inhibit transcription. The edc3Δ lsm4ΔC mutant had shorter mRNA half-lives by at least 1.4-fold, except for those mRNAs encoding the ribosomal protein genes CYH2/RPL28 and RPL3 (Fig. 2B), results which are consistent with this class of mRNAs behaving differently in decay from bulk mRNA (Arribere et al., 2011; Gasch et al., 2000). None of the individual mutants exhibited such an effect. The edc3Δ strain had half-lives similar to those expected, with the exception of the ADH1 mRNA (Decker et al., 2007; Kshirsagar and Parker, 2004). The ADH1 half-life was instead significantly longer, thus suggesting inhibition of decapping as observed for other mRNAs with the EDC3 decapping factor deletion (Dong et al., 2007; Sun et al., 2013). Half-lives in the lsm4ΔC background were similar to those of the wild-type or were modestly increased, as observed by other groups (Decker et al., 2007; Reijns et al., 2008), notably for the PGK1 mRNA. Together, these data suggest that mRNAs tend to be destabilized in the edc3Δ lsm4ΔC mutant. Finally, reduced mRNA stability in the edc3Δ lsm4ΔC mutant was not due to growth defects, because the mutant grew similarly to wild-type yeast under optimal growth conditions (Fig. S1), as observed previously (Aronov et al., 2015; Decker et al., 2007).
Reduction of mRNA stability in the edc3Δ lsm4ΔC mutant is consistent with increased deadenylation
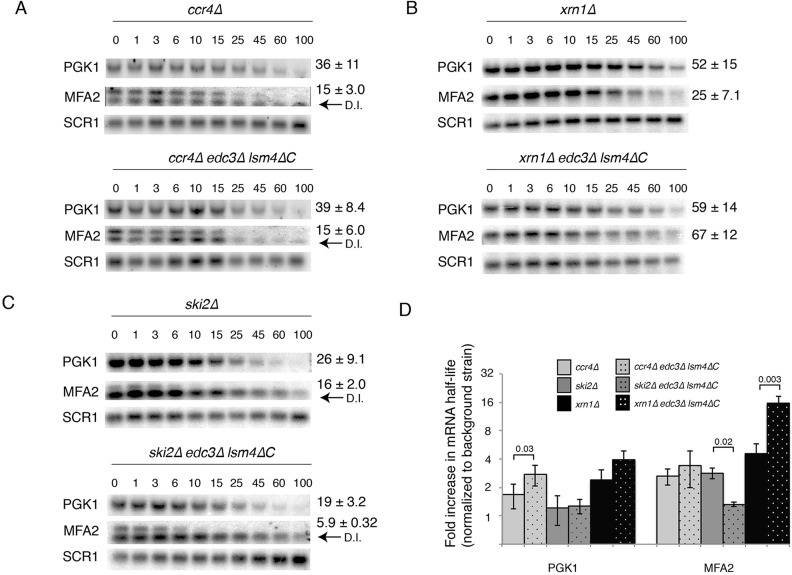
To investigate the mechanism regulating mRNA stability in the edc3Δ lsm4ΔC mutant, we examined strains in which genes encoding proteins required for the different pathways of mRNA decay had been deleted, and we compared the results with those for the wild type and edc3Δ lsm4ΔC mutant. We used mutants defective in deadenylation (ccr4Δ) as well as decapping-dependent (xrn1Δ) and exosome-mediated (ski2Δ) mRNA degradation (Anderson and Parker, 1998; Parker, 2012; Tucker et al., 2002).
Mutations affecting a decay pathway required for more rapid decay in the mutant should result in greater relative mRNA stability. In yeast, for example, decapping dependent 5′-to-3′ decay is the primary pathway of mRNA degradation. When the cytoplasmic 5′-to-3′ exonuclease XRN1 is deleted, mRNAs become more stable than when the cytoplasmic exosome (3′-to-5′ exonuclease) is inactivated, such as with a ski2 deletion (Cao and Parker, 2001; Parker, 2012).
We determined the relative fold-change of the deadenylation mutant compared with the corresponding background (i.e. half-life in the ccr4Δ edc3Δ lsm4ΔC strain compared with the edc3Δ lsm4ΔC mutant). The ccr4Δ mutant exhibited a 1.6- and 3-fold increase in the stability of the PGK1 and MFA2 mRNA, respectively (Fig. 3A,D). These results provide two insights into the mechanism of mRNA degradation: first, they suggest that the MFA2 mRNA deadenylates faster than the PGK1 mRNA, as reported previously (Tucker et al., 2001). Second, PGK1 was significantly threefold more stable in the ccr4Δ edc3Δ lsm4ΔC mutant than in the wild type, a result consistent with faster deadenylation in this strain (Fig. 3D); however, the fold stabilization for the MFA2 mRNA in the ccr4Δ mutant was comparable to that of the wild type. We attribute this result to the rapid deadenylation of the MFA2 mRNA (Decker and Parker, 1993), which may not be able to be significantly accelerated further in the edc3Δ lsm4ΔC mutant.
Fig. 3.
Destabilization of mRNA in the edc3Δ lsm4ΔC mutant is attributable to increased deadenylation and decapping-dependent degradation. (A) A CCR4 deletion mutant in the wild-type and edc3Δ lsm4ΔC mutant backgrounds expressing PGK1 and MFA2 mRNA under the control of the GAL promoter when grown in YEP+galactose. Time points (min) indicated are after transcriptional shut-off by addition of glucose. The loading control is SCR1. Error=s.d.; n=5-6 biological replicates. (B) As above, but with an xrn1Δ mutation, PGK1 n=2-3 biological replicates, MFA2 n=3 biological replicates. (C) As above, but with a ski2Δ mutation, PGK1 n=5 biological replicates, MFA2 n=2 biological replicates. (D) A bar graph of PGK1 and MFA2 mRNA half-lives in the strains indicated above in log2 scale. The PGK1 and MFA2 mRNA half-lives were normalized to the wild type or edc3Δ lsm4ΔC strain, respectively. Statistically significant pairings according to a two-tailed t-test indicated with their P values. Error bars indicate standard deviation.
The stabilization provided by the ski2Δ mutant was reduced in the edc3Δ lsm4ΔC mutant for the MFA2 mRNA. This result supports a model in which the degradation of full-length mRNA by the exosome is reduced in the edc3Δ lsm4ΔC mutant background. Concomitantly, there is an increased reliance on decapping-dependent degradation (Fig. 3C,D). Because the decay intermediate is primarily degraded by the cytoplasmic exosome, it is more stable and prominent in the ski2Δ mutant (indicated by an arrow labeled D.I. in Fig. 3C,D).
Similarly, in the xrn1 deletion mutant, which is deficient in decapping-dependent mRNA degradation (Muhlrad et al., 1994), the MFA2 and PGK1 mRNAs had increased stabilization in the xrn1Δ edc3Δ lsm4ΔC mutant background (Fig. 3B,D). Because of the deletion of XRN1, the lower band was absent in the MFA2 mRNA blot, thus indicating that the trapped mRNA decay intermediate was not formed. The results from the xrn1 deletion were consistent with increased deadenylation of mRNA. In yeast, deadenylated mRNA is preferentially directed to the decapping-dependent pathway, especially for MFA2 which specifically recruits decapping activators (Cao and Parker, 2003; Chowdhury et al., 2014; Sharif et al., 2013). The results for the xrn1Δ mutant therefore suggest that the relative contribution of mRNA decapping to decay may be increased. Together, these results suggest that the reduced mRNA stability in the edc3Δ lsm4ΔC mutant is attributable to increased deadenylation and increased targeting to the decapping-dependent decay pathway.
The mRNA decay systems are equally active in both the wild-type and edc3Δ lsm4ΔC mutant
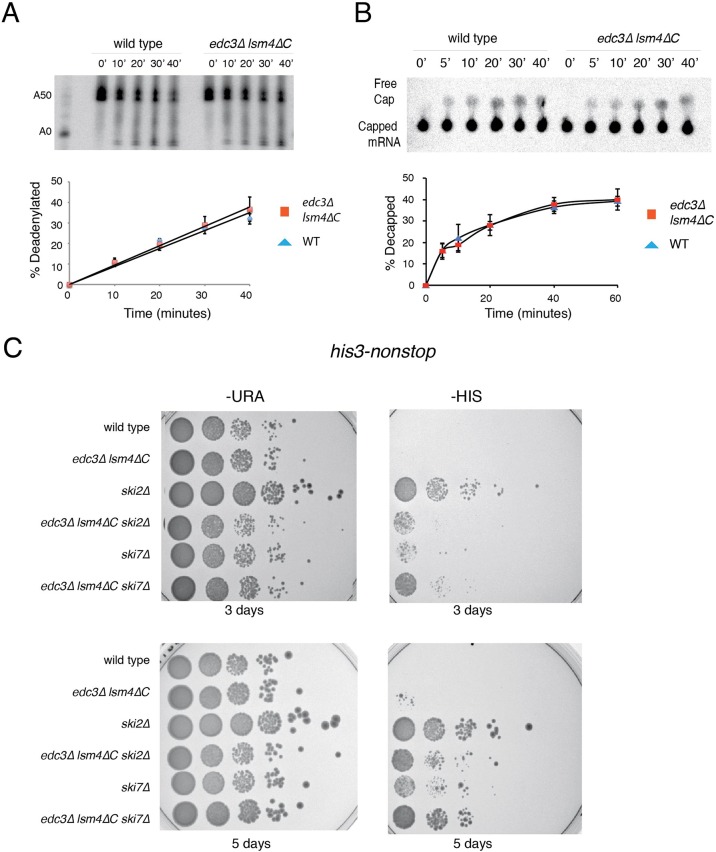
Our experimental evidence suggested that the more rapid decay observed in the edc3Δ lsm4ΔC mutant occurred via faster deadenylation by the Ccr4/Not deadenylation complex. One explanation for such an effect could be increased enzymatic activity. Using purified Ccr4 complexes from the wild-type and edc3Δ lsm4ΔC mutant strains, we observed no difference in the in vitro deadenylation rates (Fig. 4A). Because the dependence on the decapping pathway was significantly increased for MFA2 in vivo (Fig. 3D), we examined the in vitro mRNA decapping by using purified decapping complex (LaGrandeur and Parker, 1998). We observed similar decapping rates between the wild-type and edc3Δ lsm4ΔC mutants as assessed by the release of radiolabeled m7G cap from in vitro transcribed MFA2 mRNA (Fig. 4B). Finally, we considered whether the cytoplasmic exosome activity might be altered between the strains. We performed an in vivo assay to examine exosome activity, using an mRNA lacking a stop codon. Such mRNAs are recognized and destroyed by the cytoplasmic exosome (van Hoof et al., 2002). Active mRNA destruction by the exosome is assessed by growth on a medium lacking histidine using strains deleted for the HIS3 gene, with only a plasmid borne HIS3 lacking a stop codon (non-stop HIS3). Strains defective in exosome activity should not be able to grow on medium without added histidine, as observed for the ski2 and ski7 mutants (Fig. 4C) (Schaeffer et al., 2008). The inabilities of the wild-type and edc3Δ lsm4ΔC mutant strains to grow on medium lacking histidine indicate that the cytoplasmic exosome is active in both strains. Our evaluations of the decapping, deadenylation and the cytoplasmic exosome systems suggested that all three systems are equally active in both strains.
Fig. 4.
The major mRNA decay systems have similar activities in the wild-type and edc3Δ lsm4ΔC strains. (A) Deadenylation assay of transcribed MFA2 3′UTR with 50 3′ adenosines. Equal concentrations of purified TAP tagged Ccr4 from wild-type and edc3Δlsm4ΔC cells were used. Error=s.d.; n=3 biological replicates. (B) Decapping assay of m7G cap labeled mRNA. Equal concentrations of purified TAP tagged Dcp1 from wild-type and edc3Δlsm4ΔC cells were used. Error=s.d.; n=3 biological replicates. (C) Growth assay of yeast with a plasmid containing a non-stop variant of the HIS3 gene in strains deleted for the HIS3 gene as well as the gene indicated. Strains grown on synthetic complete plates lacking uracil or histidine respectively for both three and five days.
The edc3Δ lsm4ΔC mutant has alterations in mRNA decay protein levels, mRNA decapping enzyme localization and translation
We observed a differential effect of the deadenylase and decapping complex mutants on mRNA degradation in vivo between the wild-type and edc3Δ lsm4ΔC strains (Fig. 3D). Because the activities of the decapping enzyme and deadenylase complex in vitro and cytoplasmic exosome in vivo were similar (Fig. 4), we examined the levels of key mRNA decay proteins between these strains. We found that most mRNA decay proteins examined were present at similar levels when the yeast was grown in galactose or glucose normalized to the ER membrane protein Dpm1 (Fig. 5A). However, in the edc3Δ lsm4ΔC strain, Dcp2, the enzymatic subunit of the decapping enzyme, was significantly more abundant when cells were grown in either carbon source. The levels of the cytoplasmic deadenylase Ccr4 and the non-catalytic subunit of the decapping enzyme Dcp1 were elevated to a significant extent when cells were grown in galactose or glucose respectively (Fig. 5A).
Fig. 5.
Decay factor protein abundance, Dcp2 localization and mRNA levels in wild-type and edc3Δ lsm4ΔC mutant strains. (A) Quantification of C-terminal TAP tagged mRNA decay factors protein levels in wild-type and edc3Δ lsm4ΔC strains. Cells grown in either glucose or galactose containing media. Loading normalized to the Dpm1 protein abundance. (B) Wild-type and mutant strains depicting Dcp2-GFP intensity in unstressed live cells. To the right is the DAPI staining. (C) Percentage of Dcp2-GFP that is cytoplasmic as determined from sum-projected Z-stacks, masked and intensity within the cytoplasm, box showing quantification of cytoplasmic Dcp2-GFP (line, median; box 25th and 75th percentiles; whiskers, 10th and 90th percentiles, n=50 biological replicates). (D) The relative full-length mRNA was determined for MFA2 and PGK1 in the strains indicated as compared to the SCR1 loading control. The bars represent mean percentage of the indicated mRNA in the strain normalized to the WT background. Error bars indicate standard deviation, n=3 biological replicates. Significance determined in comparison to wild type by two-tailed t-test.
Whereas Dcp2 was elevated in the edc3Δ lsm4ΔC strain, we also observed that Dcp2 was concentrated in the nucleus in this mutant (Fig. 5C). Previous studies show a similar Dcp2 nuclear accumulation in the edc3Δ lsm4ΔC mutant, although this accumulation was not remarked upon (Decker et al., 2007; Grousl et al., 2009). We found that cytoplasmic Dcp2 was significantly reduced in the edc3Δ lsm4ΔC strain (P<1×10−9) by nuclear co-localization using DAPI (Fig. 5C). The greater nuclear accumulation may alter the effect of the elevated levels of Dcp2. Consistent with this model, we observed a similar level of MFA2 and PGK1 mRNA, normalized to SCR1 RNA, in the wild-type and edc3Δ lsm4ΔC mutants (Fig. 5D).
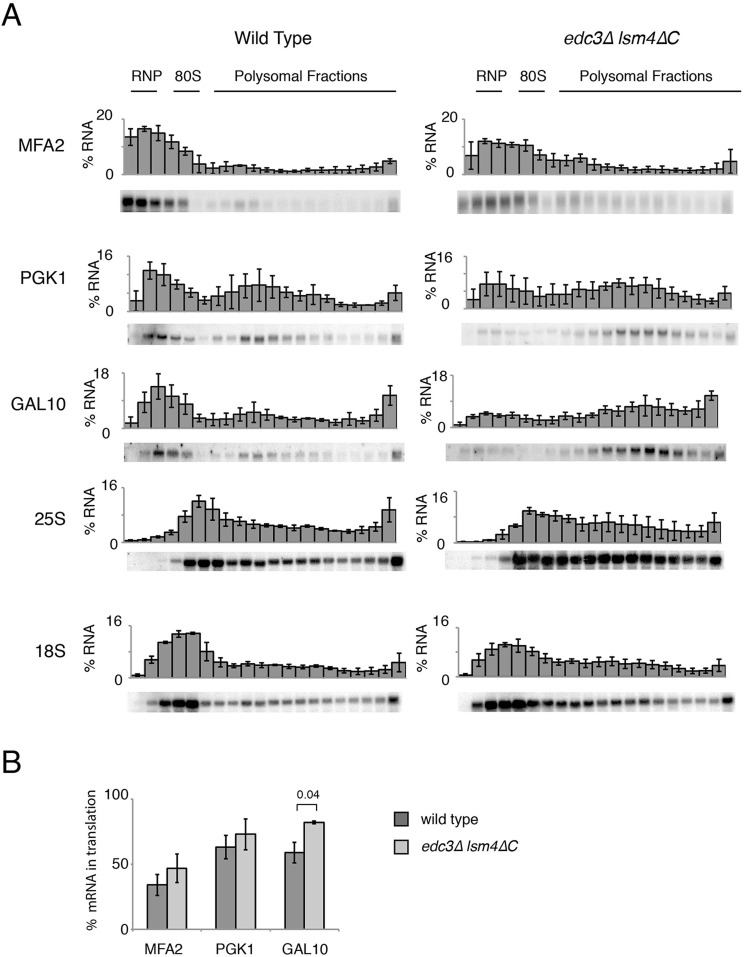
An alternative source of altered mRNA stability can come from changes in the translation of mRNAs (Drummond et al., 2011; Huch and Nissan, 2014; Jacobson and Peltz, 1996). We therefore examined the polysome profiles of extracts from yeast exponentially growing in glucose to gain insight into their translation. We found no differences between the edc3Δ lsm4ΔC strain, the wild type and the edc3Δ and lsm4ΔC mutants (data not shown), as reported previously (Decker et al., 2007). Because there were no gross alterations in translation as assessed by polysome analysis, we used northern blotting to examine the distribution of mRNAs separated on a sucrose gradient. We probed for the PGK1, MFA2 and GAL10 mRNAs after induction by growth in galactose for the wild-type and edc3Δ lsm4ΔC strains (Fig. 6A). In summary, we found a larger portion of these mRNAs in actively translating sucrose gradient fractions in the edc3Δlsm4ΔC mutant than in the wild type (Fig. 6B). These fractions are presumably associated with ribosomes and therefore are not localized in P bodies (Parker and Sheth, 2007).
Fig. 6.
Sucrose gradient distribution of mRNA and ribosomal subunits for wild-type and the edc3Δ lsm4ΔC strains. (A) Northern blot analysis of fractions from a sucrose gradient after ultracentrifugation for the wild-type and edc3Δ lsm4ΔC strains. Each panel indicates the mRNA probed, a representative northern and quantification of intensity in each fraction (error bars indicate standard deviation, n=3 biological replicates). Yeast grown in standard yeast extract/peptone medium (YEP) supplemented with 2% galactose. (B) Quantification of the percentage of the indicated mRNAs in the translating fractions (80S and polysomal). Error bars indicate standard deviation. Significant P value is indicated as determined by t-test.
Deadenylation and decapping dependent growth in the edc3Δ lsm4ΔC mutant
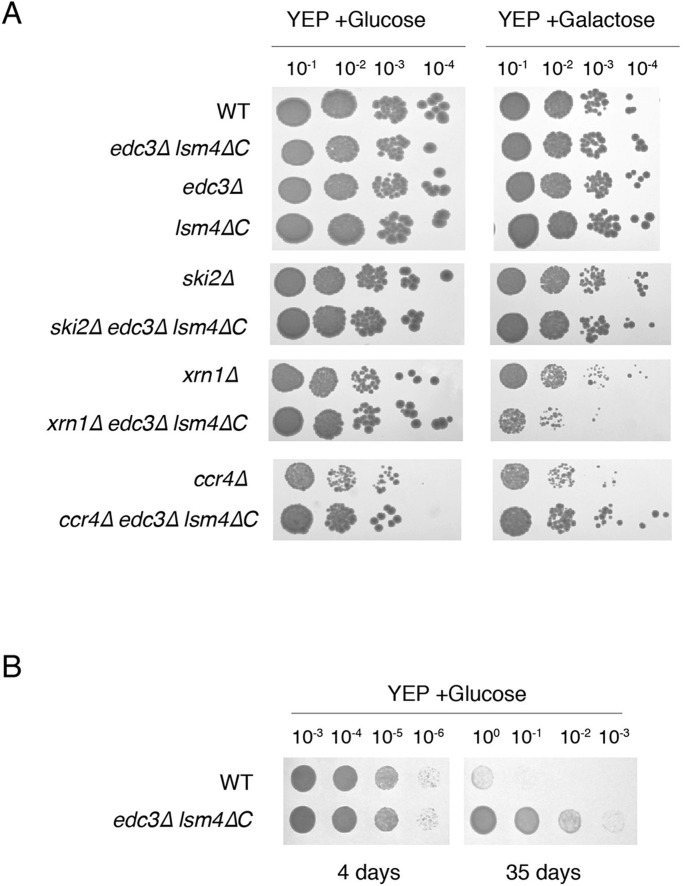
Because the mRNA degradation pathway and mRNA stability was altered for the edc3Δ lsm4ΔC mutant, we asked whether the growth might be affected for cells in which mRNA transcription was increased. We therefore examined whether P bodies would be important for adaptation to higher mRNA levels due to galactose induction (Lavut and Raveh, 2012). The growth rate was largely similar in the wild-type, edc3Δ lsm4ΔC mutant and single mutation strains with galactose as the carbon source, albeit with a faster doubling rate in the edc3Δ lsm4ΔC mutant (Fig. 7, Fig. S1).
Fig. 7.
The edc3Δ lsm4ΔC strain has altered growth in decay mutants and increased long-term survival viability. (A) Serial dilutions (1:10) of wild type (WT) and the edc3Δ lsm4ΔC mutant strain grown on YEP with glucose or galactose for carbon source as indicated (n=3 biological replicates). (B) Wild-type and edc3Δ lsm4ΔC mutant yeast grown in liquid YEP+2% glucose and plated on YEP+2% glucose plates after 4 or 35 days (n=3 biological replicates).
Despite the similar exponential growth in liquid culture, we postulated that the altered mechanism of mRNA decay might affect growth in the presence of elevated mRNA levels (Fig. 7, Fig. S1). We therefore examined decay pathway mutants expressing elevated mRNA levels in the wild-type and edc3Δ lsm4ΔC mutant strains and observed three patterns: first, the ski2Δ mutant did not show any difference in growth between the wild-type and edc3Δ lsm4ΔC mutant strain; therefore, there was no growth inhibition due to the overexpression of mRNA when the exosome was inactivated. Second, in the xrn1Δ mutant, the edc3Δ lsm4ΔC mutant was inhibited by the overexpression of mRNA and had tenfold less survival than that of the wild type. This finding suggests a greater dependence on the decapping-dependent pathway in the edc3Δ lsm4ΔC mutant, as indicated by our half-life data. Finally, in the ccr4Δ deadenylase mutant, the wild-type strain exhibited slower growth and a fivefold reduced survival rate.
If the mRNA destabilization in the edc3Δ lsm4ΔC mutant is related to P bodies, then these data suggest that P bodies stabilize mRNA. As such, they could also provide a competitive advantage to the cell. We therefore examined whether P bodies increased viability for yeast entering stationary phase. Surprisingly, we observed that yeast unable to form P bodies were more viable after extended incubation in stationary phase (Fig. 7B) suggesting that there may be a competitive cost to retaining the ability to form P bodies.
DISCUSSION
In this work, we examined the effects of the edc3Δ lsm4ΔC mutant in mRNA metabolism. Edc3 and Lsm4 have been well established as components of the decapping complex and the decapping activating Lsm1-7 complex, respectively. Mutations in decapping factors result in increased mRNA stability. In contrast, we found that the edc3Δ and lsm4ΔC mutations individually behave as expected by either increasing or not affecting mRNA stability (Fig. 2C). However, when the two mutations were combined in the edc3Δ lsm4ΔC mutant, we found that the mutant exhibits reduced mRNA stability for mRNAs (Fig. 2C). By combining the edc3Δ lsm4ΔC mutant with mutations in mRNA decay pathways, we obtained insights into the more rapid decay observed; specifically, the edc3Δ lsm4ΔC mutant has increased dependence on deadenylation by the Ccr4/Not complex and decapping with a concomitant reduction in 3′-to-5′ degradation mediated by the exosome (Fig. 3D). These results suggest that the importance of deadenylation and decapping is increased in the edc3Δ lsm4ΔC mutant. Furthermore, we found that the enzymatic decay systems had similar activity (Fig. 4). These results suggest an alternative source for the altered mRNA decay.
Whereas most of the mRNAs examined here had shorter half-lives, we asked why the shorter-lived mRNAs were not significantly destabilized. A similar effect has also been observed in previous examinations of the role of P bodies in mRNA degradation (Decker et al., 2007; Eulalio et al., 2007; Stoecklin et al., 2006). These studies have examined short-lived mRNAs on the basis of the presumption that the absence of P bodies would stabilize mRNA. Detection of faster decay in unstable mRNA molecules may be precluded by a limit on the rate of deadenylation as well as by the difficulty of assessing the difference between very short half-lives. In the edc3Δ lsm4ΔC mutant, the decrease in mRNA stability, whatever the source, might have had the same limitation as that in previous studies.
Whereas we found the individual mutations edc3Δ and lsm4ΔC significantly stabilized or had a neutral effect on mRNAs. The combination of the two mutations resulted in destabilization. How could mutations in two decapping factors result in reduced mRNA stability? We can envision three possibilities.
The first possibility is that the altered levels of mRNA decay proteins directly promote the faster degradation observed. Specifically, we found that the levels of the catalytic subunit of the decapping enzyme (Dcp2) were elevated in the edc3Δ lsm4ΔC mutant under growth in glucose and galactose (Fig. 5A). In addition, Dcp1 was significantly elevated in abundance in the edc3Δ lsm4ΔC mutant when grown in glucose. Similarly, when grown in galactose, Ccr4 had significantly increased levels in the edc3Δ lsm4ΔC strain.
These decay factor levels might not cause the increase in degradation observed in the edc3Δ lsm4ΔC mutant. For example, efficient Dcp2 activity depends on the presence of Dcp1 in yeast, which was not concomitantly increased when grown in galactose (Beelman et al., 1996; Floor et al., 2010; She et al., 2004; Tharun et al., 2000). Since we observe increased degradation in the edc3Δ lsm4ΔC mutant whether the cells are grown in galactose or glucose, the Ccr4 and Dcp1 levels, which are only elevated in specific carbon sources, also may not be the cause of faster degradation (Fig. 2C). However, we can not exclude that these elevated levels have some effect. For instance, overexpression of Ccr4 on a high copy plasmid has been reported to increase the decay rate of PGK1, but not MFA2 (Tucker et al., 2002), and elevated Dcp1 would allow the elevated Dcp2 to have greater activity. Finally, we found that much of the additional Dcp2 was located in the nucleus (Fig. 5B,C). At least three lines of evidence suggest that the excess nuclear Dcp2 protein might not be expected to decap mRNA. First the poly(A) tails of the nuclear-retained mRNA are longer than those of cytoplasmic mRNA, and this phenomenon has been reported to result in decapping inhibition (Decker and Parker, 1993; Hilleren and Parker, 2001; Jensen et al., 2001). Second, nuclear degradation of normal mRNA is slower than cytoplasmic decay and proportional to time spent in the nucleus, in which mRNAs are exported within seconds in yeast (Das et al., 2003; Kufel et al., 2004; Oeffinger and Zenklusen, 2012). Third, 5′-to-3′ nuclear degradation is carried out by Rat1 rather than Xrn1 (Bousquet-Antonelli et al., 2000; Das et al., 2003). We observed a substantially increased dependence on Xrn1 in the mutant defective in P body formation, a result consistent with cytoplasmic degradation (Fig. 3D). We cannot preclude the possibility that altered decay factor levels account for the reduced mRNA stability in edc3Δ lsm4ΔC mutant; however, these results, together with results from previous studies, suggest that the altered levels may not be the origin of the faster degradation observed in the edc3Δ lsm4ΔC mutant.
A second possibility for the reduced stability in the edc3Δ lsm4ΔC mutant could be a rearrangement of RNPs on mRNAs, perhaps through altered decay protein levels, as discussed above, or through the absence of both Edc3 and the glutamine/asparagine rich domain of Lsm4. Both Edc3 and Lsm4 have previously been shown to be involved in enhanced mRNA decay and decapping both in vivo and in vitro (Badis et al., 2004; Dong et al., 2007; Fromm et al., 2012; Harigaya et al., 2010; Kshirsagar and Parker, 2004; Nissan et al., 2010; Tharun et al., 2000). Although both individually promote mRNA decapping, it is possible that they could act in an antagonistic manner when both are absent, as suggested by the increased rate of decay observed only when the mutations were combined (Fig. 2C). This mechanism might be possible in the absence of some of the positive activators that bind to Dcp2 and normally promote both general and specific mRNA degradation. These findings suggest the possibility of a combinatorial code for mRNA decapping (He and Jacobson, 2015).
A third potential source for the reduced mRNA stability observed in the edc3Δ lsm4ΔC mutant could be linked to its inability to function in the assembly of P bodies (Decker et al., 2007). Whereas all cytoplasmic mRNAs are ultimately subject to destruction, in this hypothesis, mRNAs not bound in P bodies have a different mechanism of decay and decay more rapidly in their absence. P bodies have several roles in mRNA degradation by differentially acting on cytosolic, polysomal and P body bound mRNAs. Specifically, P bodies may sequester the mRNA decapping-dependent decay and deadenylation factors and alter the mechanism of mRNA degradation in the cell. In agreement with such a model, recent evidence supports the notion that mRNA degradation primarily occurs on translating mRNA (Hu et al., 2009; Pelechano et al., 2015). Non-P body bound mRNA may be degraded by a decay mechanism more similar to that for ribosome associated mRNA. P body associated mRNA may degrade differently.
A plausible model for the reduced mRNA stability in the edc3Δ lsm4ΔC mutant in the P body assembly can be constructed on the basis of multiple recent studies on the effect of mRNA poly(A) length on P bodies and translation. First, mRNAs with long poly(A) tails are engaged in translation and are not found in P bodies under non-stress conditions, as used in this study (Aizer et al., 2014; Brengues and Parker, 2007; Hoyle et al., 2007; Zheng et al., 2008). Second, deadenylated mRNA with short poly(A) tails persist until they are either decapped or degraded from their 3′ end (Hu et al., 2009; Muhlrad et al., 1995). Third, these deadenylated mRNAs have reduced translation and accumulate in P bodies (Brengues and Parker, 2007; Hoyle et al., 2007; Jacobson and Peltz, 1996; Zheng et al., 2008). Finally, deadenylated mRNAs have much longer half-lives than poly(A)+ mRNAs, thus suggesting that the longest portion of their lifespan is P body associated (Presnyak et al., 2015). Together, these results are consistent with both cytosolic- and ribosomal-associated mRNAs having similar access to decay factors as well as P bodies having a fundamentally different environment. That is, the mRNA decay factors involved in decapping and deadenylation accumulate in P bodies, while excluding the exosome (Jain and Parker, 2013). The accumulation of deadenylation factors within P bodies may limit the rate of deadenylation of ribosome-associated and non-P-body-associated mRNAs (Cougot et al., 2004; Teixeira and Parker, 2007; Zheng et al., 2008).
Finally, we found that Edc3 and the glutamine/asparagine rich region of Lsm4 are important when combined with other deletions in mRNA degradation factors. For example, different decay pathways are required for enhanced growth in in the edc3Δ lsm4ΔC mutant and wild-type yeast after induction of elevated transcription (Fig. 7A). Interestingly, we observed increased long-term survival in the edc3Δ lsm4ΔC mutant compared with wild-type (Fig. 7B). Another study found the opposite result for long-term survival by using a mutant defective in P body formation (pat1Δ) (Lavut and Raveh, 2012; Shah et al., 2013). However, this result is complicated because Pat1 strongly affects mRNA stability genome-wide in yeast (Sun et al., 2013). Although we found that long-term survival was compromised in the edc3Δ lsm4ΔC mutant, a previous study has demonstrated that Edc3 and glutamine/asparagine rich region of Lsm4 is important for mating in yeast (Aronov et al., 2015). These data suggest that Edc3 and the glutamine/asparagine rich region of Lsm4, and thus potentially P bodies, could be evolutionarily selected for their importance in mating, despite compromising the long-term survival ability of yeast.
MATERIALS AND METHODS
Yeast strains and growth conditions
The genotypes of all strains used in this study are listed in Table S1. Strains were grown on either yeast extract/peptone (YP) medium or synthetic complete (SC) medium lacking amino acids, as indicated. As the carbon source, the media contained 2% galactose for glucose transcriptional shut-off experiments, otherwise 2% glucose was used. Strains were grown at 30°C. Where indicated, transcription was halted by the addition of thiolutin (6 µg ml−1). The thiolutin concentration was titrated to be the minimum level to inhibit transcription (Pelechano and Pérez-Ortín, 2008). Yeast genomic knock-out strains were generated using homologous recombination with regions of homology approximately 50 nucleotides upstream of the ATG and 50 downstream of the stop codon, in which the gene of interest was replaced with the nourseothricin (natNT2) and hygromycin B (hphMX4) antibiotic resistance genes as previously described (Janke et al., 2004). The knockout strains were confirmed by PCR and sequencing The plasmids used in this study are indicated in Table S3.
Western blot analysis
TAP tags were detected by western blotting using rabbit Peroxidase-Anti-Peroxidase (Sigma) at 1:2500 (Lot #103M4822). Dpm1 was detected using mouse anti-Dpm1 (Life Technologies) at 1:2500 (Lot#1219807) and rabbit anti-mouse HRP (Agrisera, Vännäs, Sweden) at 1:2500 (Lot#1402).
Microscopy
Live yeast cells were resuspended in appropriate minimal medium and visualized on a DeltaVision Spectris microscope with an Olympus 60×1.4NA objective without binning. Microscopic images were deconvolved using the classical maximum likelihood estimation algorithm in Huygens Essential 4.4 (SVI, Hilversum, The Netherlands). Each of the resulting images was depicted by means of sum intensity projections of a Z series of 20 slices of 0.25 µm thickness displayed with Fiji (Schindelin et al., 2012). Images in each panel are within the same contrast range displayed using the Fire lookup table in Fiji. Quantification of nuclear and cytoplasmic fluorescence was accomplished with sum projected Z-stacks of GFP and DAPI channels of live cells. The DAPI channel was thresholded using the Huang method (Huang and Wang, 1995). Approximately 100 cells were counted for wild-type and the edc3Δ lsm4ΔC mutant after removal of outliers in the DAPI channel. The intensities of the fluorescence were quantified with reference to the non-thresholded sum projection.
Affinity protein purification
Two to six liters of yeast containing an integrated C terminal TAP tag attached to the protein of interest was grown to mid-log phase at 30°C in YPD medium. Cells were harvested and lysed using a pressure cell homogenizer (Stanstead Fluid Power). Cells were purified as previously described (Puig et al., 2001). Briefly, proteins were affinity-purified from cleared cell lysate using IgG resin (GE Healthcare), eluted with AcTEV (Invitrogen) and dialyzed into lysis buffer containing 50% glycerol for use in enzymatic assays.
Decapping assay
The decapping enzyme was purified from the wild-type and edc3∆ lsm4∆C mutant strains by protein A purification of the C-terminal tagged Dcp1 protein (yTN259 and yTN263). After purification as previously described (Puig et al., 2001), extracts were dialyzed overnight into lysis buffer with 50% glycerol. The protein concentrations used in the assay were normalized by western blot analysis. Capped mRNA was produced as previously described (LaGrandeur and Parker, 1998). After T7 transcription of the XbaI-linearized pTN226 plasmid, mRNA was capped with the Vaccinia Capping System (New England Biolabs) with radiolabeled GTP. Labeled mRNA was separated on a urea polyacrylamide gel and the full length mRNA was identified by phosphorimager; the band was cut out and eluted overnight. Decapping was performed as previously described (LaGrandeur and Parker, 1998). Briefly, the decapping reaction contained cap-radiolabeled mRNA, purified decapping enzyme from the wild-type and mutant strains, and 5 fmol m7G[32P]pppMFA2pG mRNA. The reaction was carried out in Mg buffer (50 mM Tris, pH 7.6, 5 mM MgCl2, 50 mM NH4Cl, 1 mM DTT). The released caps were monitored by PEI TLC plates developed in 0.75 M LiCl.
Deadenylation assay
The deadenylation complex was purified from the wild-type and P body mutant strains by protein A purification of the C-terminal tagged Ccr4 protein (yTN282 and yTN296). After purification as previously described (Puig et al., 2001), extracts were dialyzed overnight into lysis buffer with 50% glycerol. The protein concentrations used in the assay was normalized by western blot analysis. Deadenylation was performed on short mRNA containing the final 50 nucleotides of the MFA2pG 3′ UTR and 50 nucleotide poly(A) tail. Briefly, pTN229 was linearized by HindIII and mung bean nuclease, and then transcribed from the T7 promoter in the presence of radiolabeled nucleotides (Tucker et al., 2002). The deadenylation assay was performed similarly to previously published protocols (Tucker et al., 2001; Wilusz et al., 2001). The reaction was performed in TW buffer (20 mM HEPES, pH 7, 50 mM KCl, 1 mM MgOAc and 1 mM DTT) with approximately 10 µM RNA. The reaction was started by the addition of purified Ccr4 complex and incubated at 30°C. The reaction was quenched with a stop solution (20 mM EDTA and 300 mM NaOAc).
RNA analysis
RNA was purified by lysing yeast cells with glass beads, and this was followed by a phenol/chloroform/isoamyl alcohol extraction and ethanol precipitation. Northern blots were probed with the oligonucleotides listed in Table S2. The mRNA half-lives were determined using transcriptional shut-off with 6 µg ml−1 thiolutin or by use of an inducible GAL promoter integrated into the genome, which controls the MFA2pG and PGK1pG mRNAs. The optimal thiolutin concentration was examined by titration and half-life determination (Pelechano and Pérez-Ortín, 2008). The transcription from the GAL promoter was repressed by the addition of 4% dextrose to the cell medium after washing as indicated. Quantification of bands on northern blots was performed with Quantity One (Bio-Rad). The mRNA half-lives were determined by the best linear fits of mRNA band intensities normalized to the SCR1 loading control. The mRNA half-lives were subjected to two-tailed unpaired Student's t-tests. Statistical significance was determined with a P-value cut-off of 0.05.
Polysomal fractionation of cellular extracts
Sucrose gradient analysis was performed as previously described (Huch et al., 2016). Cells were grown in YEP+2% galactose to an OD ∼0.5. Harvesting was performed by centrifugation at room temperature for 1 min at 3000 g, and the cells were frozen in liquid nitrogen. Cells were lysed at 4°C using glass beads with incubation on ice for 5 min after 2 min pulses with a Disruptor Genie with 1× TN buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM DTT) supplemented with a 10 mM ribonucleoside-vanadyl complex, 0.5 mg ml−1 heparin and complete EDTA Free Protease Inhibitor (Roche). Cycloheximide was not used during cell harvesting due to its ability to eliminate P bodies (Coller and Parker, 2005; Sheth and Parker, 2003). Cellular debris was cleared by centrifugation at 1500 g for 2 min. The resulting supernatant was loaded onto a 15-50% sucrose gradient with an 80% sucrose cushion. After ultracentrifugation at 39,000 rpm for 90 min, the extract was aliquoted and frozen on dry ice for later RNA extraction.
Acknowledgements
We thank R. Parker (Univ. of Colorado, USA) for supplying strains and plasmids used in this study, S. Wilson for critical comments, and F. Salomons (Karolinska Inst., Sweden) and A. Köhler (Univ. of Vienna, Austria) for use of their microscopes.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: T.N., S.H., V.B. Investigation: S.H., T.N., M.Mü., M.Mu., J.G. Writing: T.N., S.H. Funding Acquisition: T.N. Supervision: T.N.
Funding
This work was supported by Vetenskapsrådet - The Swedish Research Council [621-2010-4602 to T.N.]; Umeå Universitet and the Carl Tryggers Stiftelse för Vetenskaplig Forskning [CTS 15:345 to T.N.]. The Carl Tryggers Stiftelse för Vetenskaplig Forskning provided funding for the open access charge.
Supplementary information
Supplementary information available online at http://bio.biologists.org/lookup/doi/10.1242/bio.020487.supplemental
References
- Aizer A., Kalo A., Kafri P., Shraga A., Ben-Yishay R., Jacob A., Kinor N. and Shav-Tal Y. (2014). Quantifying mRNA targeting to P-bodies in living human cells reveals their dual role in mRNA decay and storage. J. Cell Sci. 127, 4443-4456. 10.1242/jcs.152975 [DOI] [PubMed] [Google Scholar]
- Anderson J. S. J. and Parker R. (1998). The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17, 1497-1506. 10.1093/emboj/17.5.1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronov S., Dover-Biterman S., Suss-Toby E., Shmoish M., Duek L. and Choder M. (2015). Pheromone-encoding mRNA is transported to the yeast mating projection by specific RNP granules. J. Cell Biol. 209, 829-842. 10.1083/jcb.201408045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere J. A., Doudna J. A. and Gilbert W. V. (2011). Reconsidering movement of eukaryotic mRNAs between polysomes and P bodies. Mol. Cell 44, 745-758. 10.1016/j.molcel.2011.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G., Saveanu C., Fromont-Racine M. and Jacquier A. (2004). Targeted mRNA degradation by deadenylation-independent decapping. Mol. Cell 15, 5-15. 10.1016/j.molcel.2004.06.028 [DOI] [PubMed] [Google Scholar]
- Beelman C. A., Stevens A., Caponigro G., LaGrandeur T. E., Hatfield L., Fortner D. M. and Parker R. (1996). An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382, 642-646. 10.1038/382642a0 [DOI] [PubMed] [Google Scholar]
- Beggs J. D. (2005). Lsm proteins and RNA processing. Biochem. Soc. Trans. 33, 433-438. 10.1042/BST0330433 [DOI] [PubMed] [Google Scholar]
- Bett J. S., Ibrahim A. F. M., Garg A. K., Kelly V., Pedrioli P., Rocha S. and Hay R. T. (2013). The P-body component USP52/PAN2 is a novel regulator of HIF1A mRNA stability. Biochem. J. 451, 185-194. 10.1042/BJ20130026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borja M. S., Piotukh K., Freund C. and Gross J. D. (2011). Dcp1 links coactivators of mRNA decapping to Dcp2 by proline recognition. RNA 17, 278-290. 10.1261/rna.2382011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C., Presutti C. and Tollervey D. (2000). Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102, 765-775. 10.1016/S0092-8674(00)00065-9 [DOI] [PubMed] [Google Scholar]
- Brengues M. and Parker R. (2007). Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 2592-2602. 10.1091/mbc.E06-12-1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M., Teixeira D. and Parker R. (2005). Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310, 486-489. 10.1126/science.1115791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D. and Parker R. (2001). Computational modeling of eukaryotic mRNA turnover. RNA 7, 1192-1212. 10.1017/S1355838201010330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D. and Parker R. (2003). Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell 113, 533-545. 10.1016/S0092-8674(03)00353-2 [DOI] [PubMed] [Google Scholar]
- Chowdhury A., Kalurupalle S. and Tharun S. (2014). Pat1 contributes to the RNA binding activity of the Lsm1-7-Pat1 complex. RNA 20, 1465-1475. 10.1261/rna.045252.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J. and Parker R. (2005). General translational repression by activators of mRNA decapping. Cell 122, 875-886. 10.1016/j.cell.2005.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., Babajko S. and Séraphin B. (2004). Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165, 31-40. 10.1083/jcb.200309008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B., Butler J. S. and Sherman F. (2003). Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae. Mol. Cell. Biol. 23, 5502-5515. 10.1128/MCB.23.16.5502-5515.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C. J. and Parker R. (1993). A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 7, 1632-1643. 10.1101/gad.7.8.1632 [DOI] [PubMed] [Google Scholar]
- Decker C. J., Teixeira D. and Parker R. (2007). Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179, 437-449. 10.1083/jcb.200704147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S., Li C., Zenklusen D., Singer R. H., Jacobson A. and He F. (2007). YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol. Cell 25, 559-573. 10.1016/j.molcel.2007.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond S. P., Hildyard J., Firczuk H., Reamtong O., Li N., Kannambath S., Claydon A. J., Beynon R. J., Eyers C. E. and McCarthy J. E. (2011). Diauxic shift-dependent relocalization of decapping activators Dhh1 and Pat1 to polysomal complexes. Nucleic. Acids Res. 39, 7764-7774. 10.1093/nar/gkr474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Schweizer D. and Izaurralde E. (2007). P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 27, 3970-3981. 10.1128/MCB.00128-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floor S. N., Jones B. N., Hernandez G. A. and Gross J. D. (2010). A split active site couples cap recognition by Dcp2 to activation. Nat. Struct. Mol. Biol. 17, 1096-1101. 10.1038/nsmb.1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm S. A., Truffault V., Kamenz J., Braun J. E., Hoffmann N. A., Izaurralde E. and Sprangers R. (2012). The structural basis of Edc3- and Scd6-mediated activation of the Dcp1:Dcp2 mRNA decapping complex. EMBO J. 31, 279-290. 10.1038/emboj.2011.408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D. and Brown P. O. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241-4257. 10.1091/mbc.11.12.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grousl T., Ivanov P., Frydlova I., Vasicova P., Janda F., Vojtova J., Malínska K., Malcova I., Novákova L., Janoskova D. et al. (2009). Robust heat shock induces eIF2α-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J. Cell Sci. 122, 2078-2088. 10.1242/jcs.045104 [DOI] [PubMed] [Google Scholar]
- Haimovich G., Medina D. A., Causse S. Z., Garber M., Millán-Zambrano G., Barkai O., Chávez S., Pérez-Ortín J. E., Darzacq X. and Choder M. (2013). Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell 153, 1000-1011. 10.1016/j.cell.2013.05.012 [DOI] [PubMed] [Google Scholar]
- Harigaya Y., Jones B. N., Muhlrad D., Gross J. D. and Parker R. (2010). Identification and analysis of the interaction between Edc3 and Dcp2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 30, 1446-1456. 10.1128/MCB.01305-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield L., Beelman C. A., Stevens A. and Parker R. (1996). Mutations in transacting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 5830-5838. 10.1128/MCB.16.10.5830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F. and Jacobson A. (2015). Control of mRNA decapping by positive and negative regulatory elements in the Dcp2 C-terminal domain. RNA 21, 1633-1647. 10.1261/rna.052449.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P. and Parker R. (2001). Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3′-end formation of nascent transcripts. RNA 7, 753-764. 10.1017/S1355838201010147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle N. P., Castelli L. M., Campbell S. G., Holmes L. E. A. and Ashe M. P. (2007). Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 179, 65-74. 10.1083/jcb.200707010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Sweet T. J., Chamnongpol S., Baker K. E. and Coller J. (2009). Co-translational mRNA decay in Saccharomyces cerevisiae. Nature 461, 225-229. 10.1038/nature08265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L.-K. and Wang M.-J. J. (1995). Image thresholding by minimizing the measures of fuzziness. Pattern Recognition 28, 41-51. 10.1016/0031-3203(94)E0043-K [DOI] [Google Scholar]
- Huch S. and Nissan T. (2014). Interrelations between translation and general mRNA degradation in yeast. Wiley Interdiscip. Rev. RNA 5, 747-763. 10.1002/wrna.1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch S., Gommlich J., Muppavarapu M., Beckham C. and Nissan T. (2016). Membrane-association of mRNA decapping factors is independent of stress in budding yeast. Sci. Rep. 6, 25477 10.1038/srep25477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A. and Peltz S. W. (1996). Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem. 65, 693-739. 10.1146/annurev.bi.65.070196.003401 [DOI] [PubMed] [Google Scholar]
- Jain S. and Parker R. (2013). The discovery and analysis of P Bodies. Adv. Exp. Med. Biol. 768, 23-43. 10.1007/978-1-4614-5107-5_3 [DOI] [PubMed] [Google Scholar]
- Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E. et al. (2004). A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947-962. 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Jensen T. H., Patricio K., McCarthy T. and Rosbash M. (2001). A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell 7, 887-898. 10.1016/S1097-2765(01)00232-5 [DOI] [PubMed] [Google Scholar]
- Kshirsagar M. and Parker R. (2004). Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics 166, 729-739. 10.1534/genetics.166.2.729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel J., Bousquet-Antonelli C., Beggs J. D. and Tollervey D. (2004). Nuclear pre-mRNA decapping and 5′ degradation in yeast require the Lsm2-8p complex. Mol. Cell. Biol. 24, 9646-9657. 10.1128/MCB.24.21.9646-9657.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGrandeur T. E. and Parker R. (1998). Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 17, 1487-1496. 10.1093/emboj/17.5.1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavut A. and Raveh D. (2012). Sequestration of highly expressed mRNAs in cytoplasmic granules, P-bodies, and stress granules enhances cell viability. PLoS Genet. 8, e1002527 10.1371/journal.pgen.1002527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui J., Castelli L. M., Pizzinga M., Simpson C. E., Hoyle N. P., Bailey K. L., Campbell S. G. and Ashe M. P. (2014). Granules harboring translationally active mRNAs provide a platform for P-body formation following stress. Cell Rep. 9, 944-954. 10.1016/j.celrep.2014.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina D. A., Jordán-Pla A., Millán-Zambrano G., Chávez S., Choder M. and Pérez-Ortín J. E. (2014). Cytoplasmic 5′-3′ exonuclease Xrn1p is also a genome-wide transcription factor in yeast. Front. Genet. 5, 1 10.3389/fgene.2014.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Decker C. J. and Parker R. (1994). Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′-->3′ digestion of the transcript. Genes Dev. 8, 855-866. 10.1101/gad.8.7.855 [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker C. J. and Parker R. (1995). Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 15, 2145-2156. 10.1128/MCB.15.4.2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan T., Rajyaguru P., She M., Song H. and Parker R. (2010). Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol. Cell 39, 773-783. 10.1016/j.molcel.2010.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M. and Zenklusen D. (2012). To the pore and through the pore: a story of mRNA export kinetics. Biochim. Biophys. Acta. 1819, 494-506. 10.1016/j.bbagrm.2012.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R. (2012). RNA degradation in Saccharomyces cerevisae. Genetics 191, 671-702. 10.1534/genetics.111.137265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R. and Sheth U. (2007). P bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635-646. 10.1016/j.molcel.2007.02.011 [DOI] [PubMed] [Google Scholar]
- Pelechano V. and Pérez-Ortín J. E. (2008). The transcriptional inhibitor thiolutin blocks mRNA degradation in yeast. Yeast 25, 85-92. 10.1002/yea.1548 [DOI] [PubMed] [Google Scholar]
- Pelechano V., Wei W. and Steinmetz L. M. (2015). Widespread co-translational RNA decay reveals ribosome dynamics. Cell 161, 1400-1412. 10.1016/j.cell.2015.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Ortín J. E., de Miguel-Jiménez L. and Chávez S. (2012). Genome-wide studies of mRNA synthesis and degradation in eukaryotes. Biochim. Biophys. Acta. 1819, 604-615. 10.1016/j.bbagrm.2011.12.002 [DOI] [PubMed] [Google Scholar]
- Presnyak V., Alhusaini N., Chen Y.-H., Martin S., Morris N., Kline N., Olson S., Weinberg D., Baker K. E., Graveley B. R. et al. (2015). Codon optimality is a major determinant of mRNA stability. Cell 160, 1111-1124. 10.1016/j.cell.2015.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M. and Séraphin B. (2001). The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218-229. 10.1006/meth.2001.1183 [DOI] [PubMed] [Google Scholar]
- Reijns M. A. M., Alexander R. D., Spiller M. P. and Beggs J. D. (2008). A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 121, 2463-2472. 10.1242/jcs.024976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer D., Meaux S., Clark A. and van Hoof A. (2008). Determining in vivo activity of the yeast cytoplasmic exosome. Meth. Enzymol. 448, 227-239. 10.1016/S0076-6879(08)02612-8 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah K. H., Zhang B., Ramachandran V. and Herman P. K. (2013). Processing body and stress granule assembly occur by independent and differentially regulated pathways in Saccharomyces cerevisiae. Genetics 193, 109-123. 10.1534/genetics.112.146993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif H., Ozgur S., Sharma K., Basquin C., Urlaub H. and Conti E. (2013). Structural analysis of the yeast Dhh1-Pat1 complex reveals how Dhh1 engages Pat1, Edc3 and RNA in mutually exclusive interactions. Nucleic. Acids Res. 41, 8377-8390. 10.1093/nar/gkt600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She M., Decker C. J., Sundramurthy K., Liu Y., Chen N., Parker R. and Song H. (2004). Crystal structure of Dcp1p and its functional implications in mRNA decapping. Nat. Struct. Mol. Biol. 11, 249-256. 10.1038/nsmb730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U. and Parker R. (2003). Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300, 805-808. 10.1126/science.1082320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson C. E., Lui J., Kershaw C. J., Sims P. F. G. and Ashe M. P. (2014). mRNA localization to Pbodies in yeast is biphasic with many mRNAs captured in a late Bfr1pdependent wave. J. Cell Sci. 127, 1254-1262. 10.1242/jcs.139055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G., Mayo T. and Anderson P. (2006). ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 7, 72-77. 10.1038/sj.embor.7400572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Schwalb B., Pirkl N., Maier K. C., Schenk A., Failmezger H., Tresch A. and Cramer P. (2013). Global analysis of eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Mol. Cell 52, 52-62. 10.1016/j.molcel.2013.09.010 [DOI] [PubMed] [Google Scholar]
- Teixeira D. and Parker R. (2007). Analysis of P-body assembly in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 2274-2287. 10.1091/mbc.E07-03-0199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M. A., Brengues M. and Parker R. (2005). Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11, 371-382. 10.1261/rna.7258505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S. (2009). Lsm1-7-Pat1 complex: a link between 3′ and 5′ -ends in mRNA decay? RNA Biol. 6, 228-232. 10.4161/rna.6.3.8282 [DOI] [PubMed] [Google Scholar]
- Tharun S., He W., Mayes A. E., Lennertz P., Beggs J. D. and Parker R. (2000). Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404, 515-518. 10.1038/35006676 [DOI] [PubMed] [Google Scholar]
- Tucker M., Valencia-Sanchez M. A., Staples R. R., Chen J., Denis C. L. and Parker R. (2001). The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104, 377-386. 10.1016/S0092-8674(01)00225-2 [DOI] [PubMed] [Google Scholar]
- Tucker M., Staples R. R., Valencia-Sanchez M. A., Muhlrad D. and Parker R. (2002). Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 21, 1427-1436. 10.1093/emboj/21.6.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkov E., Muthukumar S., Chang C.-T., Jonas S., Weichenrieder O. and Izaurralde E. (2016). Structure of the Dcp2-Dcp1 mRNA-decapping complex in the activated conformation. Nat. Struct. Mol. Biol. 23, 574-579. 10.1038/nsmb.3232 [DOI] [PubMed] [Google Scholar]
- van Dijk E. L., Chen C. L., d'Aubenton-Carafa Y., Gourvennec S., Kwapisz M., Roche V., Bertrand C., Silvain M., Legoix-Né P., Loeillet S. et al. (2011). XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 475, 114-117. 10.1038/nature10118 [DOI] [PubMed] [Google Scholar]
- van Hoof A., Frischmeyer P. A., Dietz H. C. and Parker R. (2002). Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295, 2262-2264. 10.1126/science.1067272 [DOI] [PubMed] [Google Scholar]
- Wilusz C. J., Gao M., Jones C. L., Wilusz J. and Peltz S. W. (2001). Poly(A)-binding proteins regulate both mRNA deadenylation and decapping in yeast cytoplasmic extracts. RNA 7, 1416-1424. [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Ezzeddine N., Chen C.-Y. A., Zhu W., He X. and Shyu A.-B. (2008). Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J. Cell Biol. 182, 89-101. 10.1083/jcb.200801196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid B. M. and O, 'Shea E. K. (2014). Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature 514, 117-121. 10.1038/nature13578 [DOI] [PMC free article] [PubMed] [Google Scholar]