Abstract
Acinetobacter baumannii is a Gram-negative coccobacillus found primarily in hospital settings that has recently emerged as a source of hospital-acquired infections. A. baumannii expresses a variety of virulence factors, including type IV pili, bacterial extracellular appendages often essential for attachment to host cells. Here, we report the high resolution structures of the major pilin subunit, PilA, from three Acinetobacter strains, demonstrating that A. baumannii subsets produce morphologically distinct type IV pilin glycoproteins. We examine the consequences of this heterogeneity for protein folding and assembly as well as host-cell adhesion by Acinetobacter. Comparisons of genomic and structural data with pilin proteins from other species of soil gammaproteobacteria suggest that these structural differences stem from evolutionary pressure that has resulted in three distinct classes of type IVa pilins, each found in multiple species.
Keywords: cell adhesion, glycosylation, microbial pathogenesis, protein folding, type IV pili, x-ray crystallography
Introduction
Type IV pili are extracellular adhesive appendages primarily comprising a single protein subunit, called the major pilin, which is assembled into a narrow (∼6–9-nm) helical fiber of variable length (up to 2.5 μm) (1). One or more other proteins, called minor pilins, are also incorporated into the fiber at low levels. All pilins contain an N-terminal signal sequence followed by an ∼30-amino acid hydrophobic α-helix resembling a transmembrane domain (the α1-N domain). This is, in turn, followed by a soluble ∼15-kDa globular domain referred to as the pilin headgroup; the hydrophobic helical regions are buried together in the center of the fiber, whereas portions of the C-terminal headgroup are exposed (2, 3).
Type IV pili are found in both Gram-negative (4, 5) and Gram-positive (6–8) bacteria as well as Archea (9). They are involved in a wide range of processes, including twitching motility (10), horizontal gene transfer (11), host-cell adhesion (12), and microcolony/biofilm formation (13). This functional diversity is reflected in the sequence of the pilin proteins that typically have little or no sequence identity beyond the hydrophobic portion of the N-terminal α-helix. This lack of sequence identity is apparent even in cases where there is high structural similarity.
In contrast, within a given species, the minor pilins are typically well conserved. Only the major pilin is highly variable (14–16) and then only in those regions left exposed in the assembled pilus (17). This sequence diversity may result from diversifying selection as a mechanism by which to avoid detection by the host immune system (18). However, such diversity can also be found in species whose life cycle is primarily environmental (19). Glycosylation is an additional source of variability in some type IV pilins; O-linked glycosylation has been observed in multiple strains of both Pseudomonas aeruginosa (20–22) and Neisseria (23, 24). Additional glycosylation sites have been found in class II strains of Neisseria meningitidis where they have been hypothesized to play a role in immune evasion (25).
Among the many genera of Gram-negative bacteria that express type IV pili is Acinetobacter, a coccobacillus that is widely distributed in nature and can be isolated from the environment in the soil and in water as well as from a variety of mammalian hosts (26, 27). Several species of Acinetobacter, chiefly Acinetobacter baumannii, Acinetobacter calcoaceticus, Acinetobacter nosocomialis, and Acinetobacter pittii, are collectively referred to as the A. calcoaceticus-baumannii (Acb)4 complex and constitute an increasingly common source of nosocomial infections (28–30). Although reports of infections by A. baumannii predominate in the literature, phenotypic similarity makes it difficult to differentiate between related Acinetobacter species (31), and in model organisms, all four species have been found to be infectious (32).
Like other Acinetobacter species, A. baumannii lacks flagella but exhibits twitching motility, which is dependent on type IV pili (33). Type IV pili are also required for its natural transformation (33, 34), but their role in other biological processes is unclear. Virstatin, a known inhibitor of type IV pilus formation in Vibrio cholerae, was shown to both reduce type IV pilus expression and inhibit biofilm formation in A. baumannii (35). In another study, no correlation could be demonstrated between antigenic variation in the A. baumannii major pilin, pilA, and biofilm formation in vitro (36). More recently, Oh and Choi (37) reported that deletion of a LuxR-type regulator, AnoR, reduces both biofilm formation and surface motility in A. nosocomialis ATCC 17903. In addition to variation in the sequence of pilA, some Acinetobacter strains utilize an O-oligosaccharyltransferase to specifically glycosylate the major pilin at a C-terminal serine with the major capsule polysaccharide repeat unit (38, 39). These post-translational modifications are independent of the more general protein O-glycosylation system common to many gammaproteobacteria (including P. aeruginosa and Dichelobacter nodosus) (40–43). However, Harding et al. (38) reported that pilin C-terminal glycosylation is not required for either competence or twitching motility.
To understand the basis for the variability in sequence and glycosylation of Acinetobacter PilA, we have resolved the x-ray crystal structures of the major type IV pilin from three members of the Acb complex, strains ACICU and BIDMC 57 of A. baumannii and strain M2 of A. nosocomialis. In these three structures, we observe structural divergence independent of species within Acinetobacter. We demonstrate that Acinetobacter type IV pili promote host-cell adhesion in a manner independent of C-terminal glycosylation. We also provide evidence that the structural variation of Acinetobacter pilins is underpinned by functional differentiation.
Experimental Procedures
Protein Expression and Purification
Codon-optimized sequences of PilA from A. baumannii ACICU and A. nosocomialis M2, starting with alanine 23, were cloned into a pETM44 vector with an N-terminal His6 tag. These clones were transformed into BL21 (DE3) pLysS cells and grown to saturation overnight with shaking at 37 °C in LB medium with 50 μg/ml ampicillin. These saturation cultures were then diluted into fresh LB-ampicillin and grown to an optical density (OD) of 0.4–0.6 at 37 °C. These flasks were transferred to a refrigerated orbital shaker and cooled to 18 °C before induction with 30 mm isopropyl β-d-1-thiogalactopyranoside. These flasks were allowed to grow overnight before being harvested by centrifugation at 7500 × g for 10 min. The cells were then lysed using lysozyme (0.25 mg/ml final concentration) for 10 min, and the resulting lysate was centrifuged again, this time at 20,000 × g for 30 min. The supernatant was purified using a nickel-nitrilotriacetic acid column, and the elution was further purified by size exclusion chromatography over a GE Healthcare S200 Superdex column using an ÅKTA Purifier FPLC.
For crystallization, MBP-PilAACICU, MBP-PilABIDMC57, and MBP-PilAM2 were cloned and expressed as described previously (7). Briefly, the sequences of PilA from A. baumannii ACICU and A. nosocomialis M2, starting with alanine 23, were cloned into a maltose-binding protein fusion vector, making use of surface entropy reduction mutations (pMal E) described previously (44). A C-terminal His6 tag was included for ease of purification. These clones were transformed, expressed, and purified as described above.
Structure Determination and Refinement
All MBP fusion proteins were initially screened by sitting drop vapor diffusion at a concentration of 20 mg/ml in 20 mm Bis-Tris, pH 6.0, 50 mm maltose.
MBP-PilAACICU
MBP-PilAACICU was initially crystallized in the Hampton Research Index screen, condition D6: 0.1 m Bis-Tris, pH 5.5, 25% (w/v) polyethylene glycol 3350. These conditions were optimized to 0.1 m Bis-Tris, pH 5.5, 22.5% (w/v) polyethylene glycol 3000, 0.3 m 1,6-hexanediol, 5 mm maltotriose in place of maltose. Crystals were grown in sitting drops at room temperature and took ∼48 h to grow at a protein concentration of 5 mg/ml. They were then harvested and flash cooled in the mother liquor supplemented with 20% glycerol. Data were collected at the Stanford Radiation Source, the National Light Source beam line X25 in Brookhaven, NY, and eventually the data set used to resolve the structure was collected at the Advanced Photon Source, General Medical Sciences and Cancer Institutes Structural Biology Facility, beam line 23-ID-D.
MBP-PilABIDMC57
MBP-PilABIDMC57 was initially crystallized in the Molecular Dimensions Morpheus screen, condition A12: 0.1 m Bicine/Trizma (Tris base), pH 8.5, 12.5% (w/v) PEG 1000, 12.5% (w/v) PEG 3350, 12.5% (v/v) MPD, 0.03 m CaCl2, 0.03 m MgCl2. The optimal conditions were 0.1 m Bicine/Trizma, pH 8.0, 12.5% (w/v) PEG 1000, 12.5% (w/v) PEG 3350, 12.5% (v/v) MPD, 0.06 m CaCl2, 0.06 m MgCl2, 50 mm NaCl, 3% EtOH. Crystals were grown in hanging drops at room temperature and took ∼72 h to grow at a protein concentration of 10 mg/ml. They were then harvested and flash cooled in the mother liquor supplemented with 20% glycerol. Data were collected at the Stanford Synchrotron Radiation Lightsource, beam line 12-2.
MBP-PilAM2
MBP-PilAM2 was initially crystallized in the Molecular Dimensions Morpheus screen, condition A12: 0.1 m Bicine/Trizma base, pH 8.5, 12.5% (w/v) PEG 1000, 12.5% (w/v) PEG 3350, 12.5% (v/v) MPD, 0.03 m CaCl2, 0.03 m MgCl2. The optimal conditions were 0.1 m Bicine/Trizma, pH 8.0, 12.5% (w/v) PEG 1000, 12.5% (w/v) PEG 3350, 12.5% (v/v) MPD, 0.06 m CaCl2, 0.06 m MgCl2. Crystals were grown in sitting drops at room temperature and took ∼72 h to grow at a protein concentration of 10 mg/ml. They were then harvested and flash cooled in the mother liquor supplemented with 20% glycerol. Data were collected at the Advanced Photon Source, GM/CA, beam line 23-ID-B.
The ACICU and M2 data sets were processed with XDS (45); the BIDMC 57 data set was processed with HKL2000 (46). Molecular replacement was carried out by Phaser (47) using a sequential search of 1) maltose-binding protein and 2) PilA from P. aeruginosa strain K (PAK). Phenix and Coot were used for phasing, building, and refinement (48–51). The crystallographic parameters of the refined data are summarized in Table 1.
TABLE 1.
Crystallographic parameters
Values in parentheses are for the highest resolution shell. r.m.s., root mean square. CC, correlation coefficient; CC1/2, Pearson correlation coefficient between half-datasets.
| MBP- PilAACICU | MBP-PilABIDMC57 | MBP-PilAM2 | |
|---|---|---|---|
| Resolution range (Å) | 41.02–1.975 (2.046–1.975) | 43.93–2.2 (2.279–2.2) | 29.44–1.801 (1.865–1.801) |
| Space group | P 1 21 1 | C 1 2 1 | C 1 2 1 |
| Unit cell (Å) | 41.018, 128.3, 92.505, 90, 90, 90 | 175.07, 56.636, 49.997, 90, 91.6, 90 | 173.883, 55.334, 49.67, 90, 91.52, 90 |
| Total reflections | 105,740 (2,931) | 125,933 (5,583) | 150,644 (8,909) |
| Unique reflections | 53,305 (1,577) | 23,793 (2,036) | 42,472 (3,492) |
| Multiplicity | 2.0 (1.9) | 5.3 (2.7) | 3.5 (2.6) |
| Completeness (%) | 79.68 (23.63) | 94.85 (82.26) | 96.82 (79.43) |
| Mean I/σ(I) | 12.43 (2.50) | 11.1 (1.70) | 14.90 (1.77) |
| Wilson B-factor | 19.31 | 35.78 | 27.29 |
| Rmerge | 0.04968 (0.6135) | 0.341 (1.10) | 0.05086 (0.5152) |
| Rmeas | 0.07026 | 0.375 | 0.06005 |
| CC1/2 | 0.992 (0.184) | 0.966 (0.314) | 0.998 (0.791) |
| CC* | 0.998 (0.558) | 0.991 (0.691) | 1 (0.94) |
| Rwork | 0.1807 (0.2635) | 0.2109 (0.2950) | 0.1889 (0.3638) |
| Rfree | 0.2408 (0.3457) | 0.2440 (0.3336) | 0.2280 (0.3943) |
| r.m.s. (bonds) | 0.008 | 0.013 | 0.008 |
| r.m.s. (angles) | 1.09 | 1.33 | 1.18 |
| Ramachandran favored (%) | 98 | 97 | 96 |
| Ramachandran allowed (%) | 2 | 2 | 1 |
| Ramachandran outliers (%) | 0 | 0.62 | 0.83 |
| Clashscore | 5.03 | 2.43 | 9.08 |
| Average B-factor | 26.3 | 28.10 | 37.1 |
| Macromolecules | 25.9 | 27.30 | 37 |
| Ligands | 19.7 | 53.10 | 34.1 |
| Solvent | 30.6 | 41.90 | 40.20 |
Differential Scanning Fluorometry
Melting curves for pilin headgroups were obtained by the addition of SYPRO Orange protein stain (Sigma-Aldrich) and thermal denaturation in an iQ5 RT-PCR cycler. The buffer conditions were 10 mm NaPO4, 150 mm NaCl, 25 mm DTT. All measurements were made in triplicate. The resulting curves were normalized and fit using the Boltzmann equation to determine the melting temperature for each protein. Similar Tm values were obtained by circular dichroism (supplemental Fig. 6) (6, 52).
Pilus Modeling
Full-length PilAACICU and PilAM2 were modeled based on the structure of the full-length PAK pilin (53). The initial models of the pili were created by superimposition onto a model of the Neisseria gonorrhoeae type IV pilus filament (Protein Data Bank code 2HIL) (54), and the N-terminal helix position was adjusted to eliminate clashes between subunits. The resulting models were then minimized as rigid bodies using UCSF Chimera (55).
Glycan Modeling
The glycans attached to PilAACICU and PilAM2 (supplemental Fig. 2) were modeled using the PyRosetta protein structural modeling suite (56) with extensions for non-protein polymer units for the glycans (57). The initial models were generated in the Discovery Studio Visualizer (BIOVIA, Discovery Studio Modeling Environment, Release 4.5, 2015, Dassault Systèmes, San Diego, CA). Then the glycans were modeled using the FloppyTail algorithm (58) altered for use in glycans. The protocol consists of two parts. In the low resolution part, a random perturbation of the torsion angles was applied. The high resolution parts of the structures were refined by applying a smaller perturbation of the torsion angles, side-chain packing, and minimization. With this protocol, 6000 structures of PilAACICU and 6000 structures of PilAM2 were generated.
Surface Area Calculations
The 10 best scoring models were used for surface area calculations for each pilus. Exposed surface areas for pilins with and without glycans were calculated using the pilus and glycan models described above and the AreaIMol module of CCP4 (59) using a 1-nm spherical probe. C-terminal glycosylation reduces the exposed surface area from 3078 to 1801 Å (PilAACICU) and from 2938 to 2218 Å (PilAM2).
Cell Adhesion
A549 human airway adenocarcinoma cells (52) (CCL 185, American Type Culture Collection (ATCC), Manassas, VA) or Detroit 562 pharyngeal carcinoma cells (53) (ATCC CCL 138) were seeded in 24-well culture plates and cultured at 37 °C in 5% CO2 to 2.0 × 105 cells/well in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 2.0 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. The cells were washed twice with PBS, pH 7.2; fixed for 10 min at room temperature with 2.5% (v/v) glutaraldehyde in PBS, pH 7.2; and washed three times with PBS, pH 7.2 as described (60, 61). A. nosocomialis M2 was cultured overnight in Luria-Bertani broth; washed twice with PBS, pH 7.2; resuspended in PBS, pH 7.2 containing 2.0 mg/ml glucose; and quantified spectrophotometrically at A600. Fixed A549 or Detroit 562 cells (2.0 × 105/well) were incubated with 2.0 × 107 colony-forming units (cfu)/well A. nosocomialis M2 in 0.5 ml for 40 min at 37 °C and washed three times with PBS, pH 7.2. Bound bacteria were released with 0.05% trypsin, EDTA, and bound cfu were quantified on Luria-Bertani agar plates as described (60, 61). Significance was determined by Student's t test.
Figures
All depictions of protein structures were created using PyMOL (Schrödinger LLC) or UCSF Chimera (55). Sequence logos were created using WebLogo 3.4.
Results
Acinetobacter Species Produce Type IV Pilins with Diverse C Termini
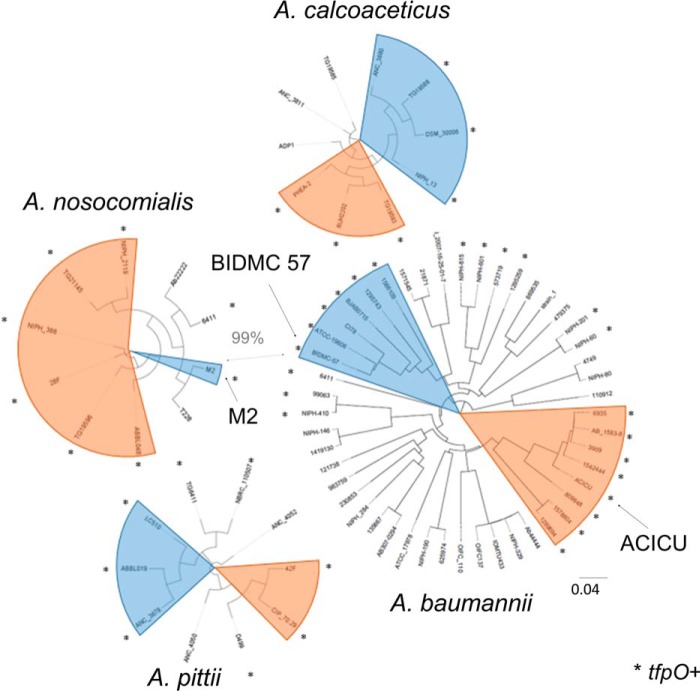
Although all strains of Acinetobacter carry the genes necessary to produce type IV pili, and the majority of those genes are highly conserved, the protein sequence of the major pilin, PilA, exhibits considerable variation in the soluble domain. In addition to this variation in amino acid sequence, Harding et al. (38) noted that strains from multiple Acinetobacter species with a serine residue at the C terminus of the major pilin also expressed an O-OTase similar to TfpO/PilO O-OTase from P. aeruginosa in addition to the PglL-like O-OTase found in all Acinetobacter strains. Analysis of pilA in 49 publically available A. baumannii genomes, 25 with a tfpO-like O-oligosaccharyltransferase gene immediately after pilA and 24 without, shows considerable variation in both groups. All 25 tfpO+ strains also have pilA genes with C-terminal serine residues.
Fig. 1 shows dendrograms of PilA sequences for each of the four Acinetobacter species in the Acb complex. Those strains that contain a tfpO gene and end their PilA sequence with a serine residue are marked with an asterisk to indicate putative glycosylation at the C terminus. In A. baumannii, the majority of these strains can be divided into two large clusters. The first (highlighted in orange) corresponds to the international clone II group, which is responsible for 50% of hospital infections worldwide (62). The second cluster (highlighted in blue) contains the type strain ATCC 19606. The largest cluster of tfpO− strains includes the international clone I group. Importantly, representatives of these two clusters can be found in each of the other members of the Acb complex. In particular, PilA proteins from A. baumannii PilABIDMC57 and A. nosocomialis PilAM2 are highlighted because their nucleotide sequences are over 99% identical.
FIGURE 1.
Dendrogram of Acinetobacter PilA. Dendrograms of PilA from A. baumannii, A. nosocomialis, A. calcoaceticus, and A. pittii are shown. Highlighted in orange are strains similar to PilAACICU, and highlighted in blue are those similar to PilABIDMC57. Strains with a gene homologous to tfpO following their pilA gene and a C-terminal serine in PilA are marked with an asterisk.
To better understand why such diversity exists in the type IV pili of A. baumannii, we resolved high resolution structures of PilA proteins representative of these two major clusters. From the largest cluster, A. baumannii ACICU (also known as H34) is an epidemic, multidrug-resistant strain belonging to the European clone II group that was isolated in an outbreak in Rome in 2005 (63). From the other large cluster, A. baumannii BIDMC 57 was isolated in 2013 at Beth Israel Deaconess Medical Center (Boston, MA) and sequenced at the Broad Institute (Cambridge, MA), and A. nosocomialis M2 (referred to as A. baumannii M2 in some earlier publications) (33, 64, 65) was isolated in 1996 from a hip infection of a patient at Cleveland MetroHealth Systems (Cleveland, OH).
High Resolution Structure of PilAACICU
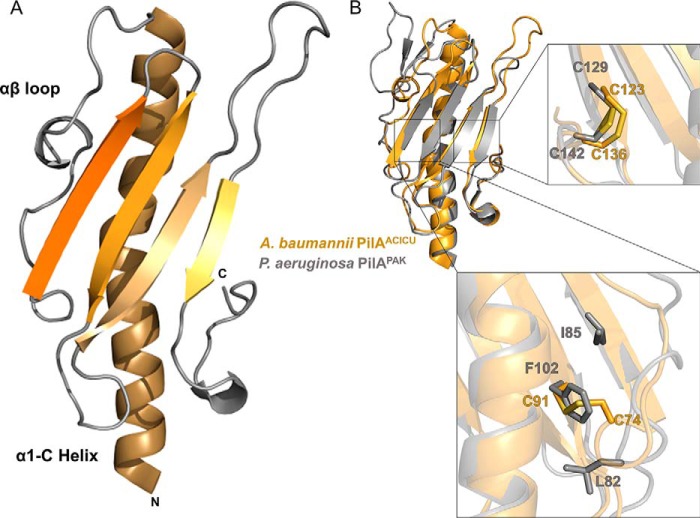
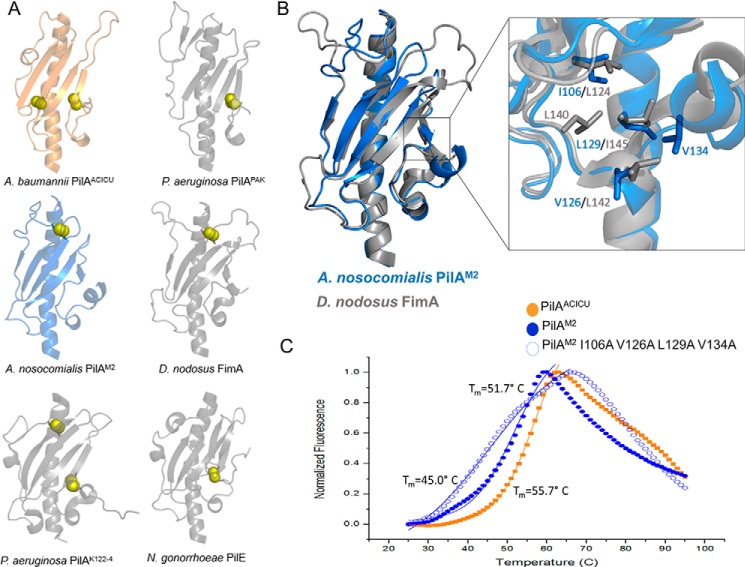
We determined the x-ray crystal structure of PilA from A. baumannii ACICU as a C-terminal fusion to maltose-binding protein to a resolution of 2.0 Å (Table 1). As depicted in Fig. 2, the overall fold of PilAACICUis very similar (r.m.s.d. = 2.19 Å) to PilA from PAK, differing primarily in the αβ-loop (the loop beginning with the end of the initial α-helix and ending with the beginning of the first β-strand) (53). Among the differences between PilA from PAK and from ACICU are the α-helical character of the ACICU αβ-loop and the longer length of the loop between the third and fourth strands of the central β-sheet. Combined, these features give PilAACICU an axis of pseudosymmetry running diagonally across the molecule from the helix in the αβ-loop to the small helical region in the C terminus, a feature also found in the structure of PilA from P. aeruginosa K122-4 (66). Notably, contacts between pilin headgroups in a pilus typically occur primarily between these same regions of the protein (2).
FIGURE 2.
Structure of PilAACICU. A, schematic representation of PilAACICU beginning with alanine 23. B, superimposition of PilAACICU (gold) with PilAPAK (gray); the inset panels show the two regions containing disulfide bonds in PilAACICU.
PilAACICU contains two disulfide bonds. One, between residues 123 and 136, is analogous to the C-terminal disulfide bond also found in PilAPAK, which is nearly universal in type IV pili from Gram-negative bacteria. The other, between residues 74 and 91, spans the first two strands of the central β-sheet (Fig. 2B). However, we note that this additional disulfide bond in PilAACICU (relative to PilAPAK) does not result in any substantial rearrangement of the protein backbone.
High Resolution Structures of PilABIDMC57 and PilAM2
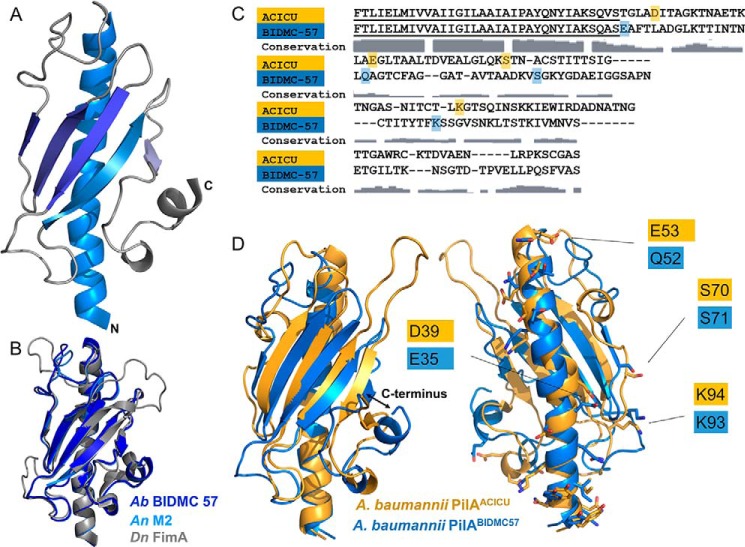
Although all Acinetobacter PilA sequences are nearly identical in the N-terminal hydrophobic α-helix, they diverge substantially beyond that point (e.g. PilAACICU and PilABIDMC57 are 35% identical from alanine 23 onward). However, the sequence variability of type IV pilins is such that homologs sharing only 30–40% sequence identity commonly have strikingly similar folds (the headgroups of PilAPAK and PilAACICU are 30% identical in sequence). To determine whether those pilins from the type strain cluster represented a distinct fold from those from the predominant international clone II cluster, we determined the x-ray crystal structures of PilA from A. baumannii BIDMC 57 and A. nosocomialis M2 as C-terminal fusions to MBP to resolutions of 2.2 and 1.8 Å, respectively (Table 1).
Although retaining the typical type IV pilin fold, the structure of PilABIDMC57 differs notably from that of PilAACICU and PilAPAK with its fold showing somewhat of an inversion along the previously described axis of pseudosymmetry (Fig. 3A), possessing an α-helix at the C terminus rather than the N terminus. The PilABIDMC57 αβ-loop contains 1) a short β-strand rather than an α-helix at its C terminus and 2) a seven-residue α-helix not found in PilAACICU. This rearrangement results in the C terminus of PilABIDMC57 being shifted 13 Å relative to PilAACICUwhen the two pilin headgroups are superimposed (Fig. 3D). Although there are few prior examples of multiple high resolution structures being solved from a single species, this degree of structural variation (r.m.s.d. = 4.17 Å) is greater than is expected within a given species. All three known structures of Clostridium difficile PilA1 are within 1-Å r.m.s.d. of each other (7), and PilAPAK and PilAK122-4 from P. aeruginosa are within 2 Å r.m.s.d. of each other and have superimposable C termini (53, 66). As expected from their sequence identity of over 99%, A. baumannii PilABIDMC57 and A. nosocomialis PilAM2 have nearly identical structures as well, differing only in conformations of the αβ-loop and the loop between the fourth and fifth β-strands (Fig. 3B).
FIGURE 3.
Structure of PilABIDMC57. A, schematic representation of PilABIDMC57 beginning with alanine 23. B, superimposition of PilABIDMC57 (dark blue) with PilAM2 (light blue) and FimA (gray). C, sequence alignment of PilAACICU and PilABIDMC57; conservation of amino acid sequence in PilA across A. baumannii (Ab) from a global alignment is indicated below. D, superimposition of PilAACICU (gold) and PilABIDMC57 (blue).
Structural Implications for PilABIDMC57 and PilAACICU Assembly
Of all the component proteins of a type IV pilus system, including major and minor pilins, extension and retraction ATPases, and other pilus biogenesis machinery, the major pilin protein is always the least conserved within a given species. However, the major pilin must retain the ability to assemble into a pilus, slowing the rate of variation in regions involved in intersubunit interactions. Although in some species the sequence variability of the major pilin is confined to hypervariable regions that are solvent-exposed in the assembled pilus (14), only the hydrophobic α1-N helix is well conserved in A. baumannii PilA (Fig. 3C). To understand how surface polymorphisms in PilA might impact pilin polymerization, we created models of A. baumannii pilus fibers from PilAACICU and PilABIDMC57 based on the N. gonorrhoeae pilus (54) and compared the interactions between the pilin headgroups. Despite a sequence identify of only 35% between the PilAACICU and PilABIDMC57 proteins (residues 23–136), many chemical moieties can be found in similar positions (Fig. 3D). These relationships are not obvious from a sequence alignment of the two protein sequences (Fig. 3C) but become clear upon superimposition of the two structures. One implication of these data is that despite their divergence in sequence the major pilins of the various Acinetobacter strains may be assembled through similar networks of non-covalent interactions.
Type IV Pili Promote Adhesion to A549 Cells
The resemblance of A. baumannii type IV pili to other type IVa pilus systems (particularly P. aeruginosa and D. nodosus) led us to hypothesize that they may have overlapping functions. Previously, Acinetobacter type IV pili have been shown to be essential for natural transformation and twitching motility, but few data are available about their roles in infection-associated processes such as adherence to host cells or biofilm formation. To determine whether type IV pili play a role in bacterial host-cell adhesion, we measured the ability of wild type A. nosocomialis M2 and mutants with altered type IV pili biogenesis phenotypes to bind to immortalized lung (A549) and nasopharyngeal (Detroit 562) epithelial cells in vitro. We found that the ΔpilA strain, which produces no type IV pili, exhibited reduced adhesion to A549 cells in vitro and that wild type adhesion was restored after complementation with the wild type pilA gene (Fig. 4). To probe the importance of C-terminal glycosylation in this process, we also tested a ΔpilA strain complemented with pilA point mutant S136A, which cannot be glycosylated. Wild type PilA and PilA(S136A) are equally capable of complementing the adhesion defect of the ΔpilA mutant strain, indicating that pilin glycosylation plays no significant role in host cell adhesion. Increased binding by the ΔpilT mutant of M2, which is known to be hyperpiliated (33, 65), shows both that increased piliation increases adhesion and that the ability to retract type IV pili is not a component of type IV pilus-mediated bacterial host-cell adhesion. We found that universally M2 binds Detroit 562 cells much more weakly than A549 cells with only the ΔpilT strain showing any adhesion above background; this finding is similar to several strains tested by Eijkelkamp et al. (36). We also tested the ability of A. nosocomialis M2 to form a biofilm in vitro and found no significant difference among the wild type strain, the ΔpilA mutant, and the complemented mutant (supplemental Fig. 1).
FIGURE 4.
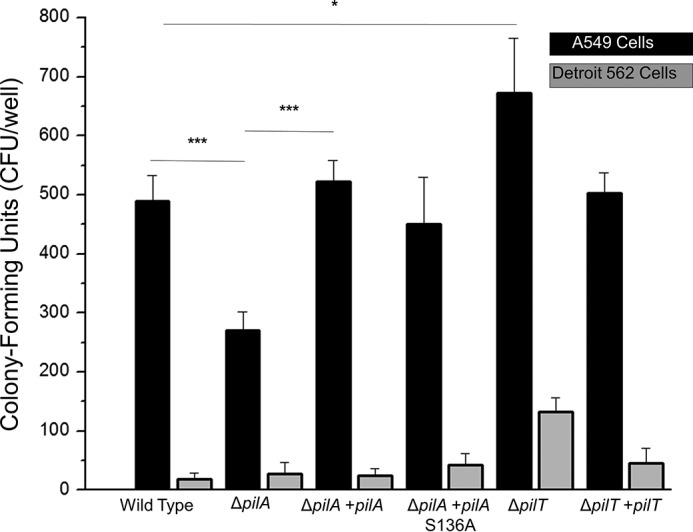
Acinetobacter nosocomialis M2 adherence to host cells. The average number of colony-forming units of A. nosocomialis recovered from a binding experiment with either A549 cells (black) or Detroit 562 cells (gray) is shown. Significance is marked as follows: *, p < 0.05; ***, p < 0.001. Error bars represent S.D.
C-terminal Glycans Mask the PilA Protein in Models of Pilus Assembly
There are four canonical functions for type IV pili: (i) twitching motility, (ii) horizontal gene transfer, (iii) host cell adhesion, and (iv) bacterial self-association (biofilm formation). As described above, none of these functions are dependent on C-terminal glycosylation of the major pilin in Acinetobacter in vitro. However, previous studies of C-terminal glycosylation in the major pilin of P. aeruginosa 1244 demonstrated a significant phenotype in vivo. Smedley et al. (21) demonstrated that deletion of the O-oligosaccharyltransferase tfpO (referred to by the authors as pilO) reduced survival of P. aeruginosa 1244 in a mouse model of lung infection. More recently, the P. aeruginosa 1244 ΔtfpO mutant was found to be more vulnerable to phagocytosis mediated by opsonization (67, 68).
Because opsonization is mediated by the binding of host immune proteins (commonly antibodies), the latter studies support the notion that the purpose of C-terminal glycosylation could be to interfere with immune recognition in vivo. Similarly, Gault et al. (25) recently showed that hypervirulent N. meningitidis strains with class II pilins are more heavily glycosylated than their class I counterparts.
To measure the extent to which C-terminal glycosylation of PilAACICU and PilAM2 would mask the pilin protein from binding, we modeled the full-length pilins and pilus fibers and measured the effect of glycosylation on the accessible surface area of each protein in its native context. We modeled an ensemble of each glycan based on the repeating unit of the major polysaccharide glycan and minimized each structure using Rosetta (69). The 10 best scoring glycan conformations were then combined to approximate the native conformational ensemble.
We then measured the differences in accessible surface area using a 10-Å particle probe to approximate the surface area needed for protein binding. The resulting models are shown in Fig. 5, and the change in accessible surface area for each protein is displayed in the inset panel. In both cases, C-terminal glycosylation reduces the exposed surface area (by 42% for PilAACICU and by 25% for PilAM2). The greater coverage of the PilAACICU protein stems from the greater flexibility of the linear PilAACICU glycan (in contrast to the branched PilAM2 glycan) (supplemental Fig. 2), which results in a more diverse conformational ensemble.
FIGURE 5.
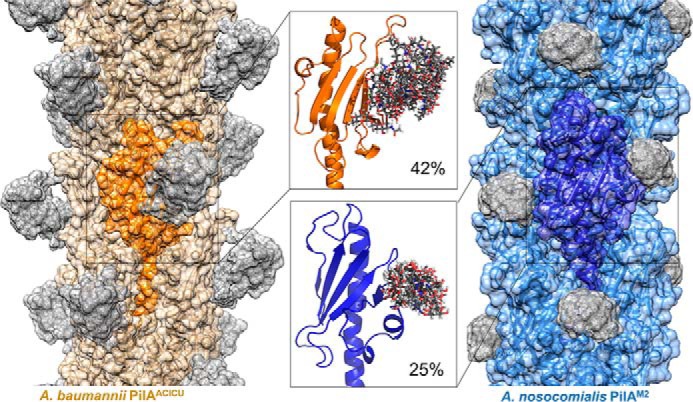
Models of glycosylated Acinetobacter type IV pili. Models of assembled type IV pili from A. baumannii ACICU (orange) and A. nosocomialis M2 (blue) are depicted with semitransparent surfaces; glycan residues are shown in gray. Inset panels show detail of the top 10 computed glycan conformations. The percentages note the change in surface area exposed to a 10-Å probe for each pilin monomer upon addition of the C-terminal glycan cloud.
If C-terminal glycosylation in Acinetobacter type IV pili reduces recognition by host immune proteins, one might expect that the pilins from strains lacking a C-terminal glycan would face greater pressure to diversify as has been shown in N. meningitidis (25). Using our alignment of 49 PilA sequences, we separated protein sequences into tfpO− and tfpO+ groups and compared the variability of their surface-exposed residues. For the tfpO− group, we modeled the structure of PilAAYE (an international clone I strain) based on the structure of PilAACICU. Supplemental Fig. 3A shows that, although overall the surface residue variability follows a similar pattern in the two groups, there is a region near the C terminus that is more conserved in the tfpO+ strains. Supplemental Fig. 3B shows sequence logos of this region for both groups. These data support a model in which C-terminal glycosylation in type IV pili exists, at least in part, as a countermeasure to the host humoral immune response.
Variation in Acinetobacter pilA Is Driven by Evolutionary Pressure
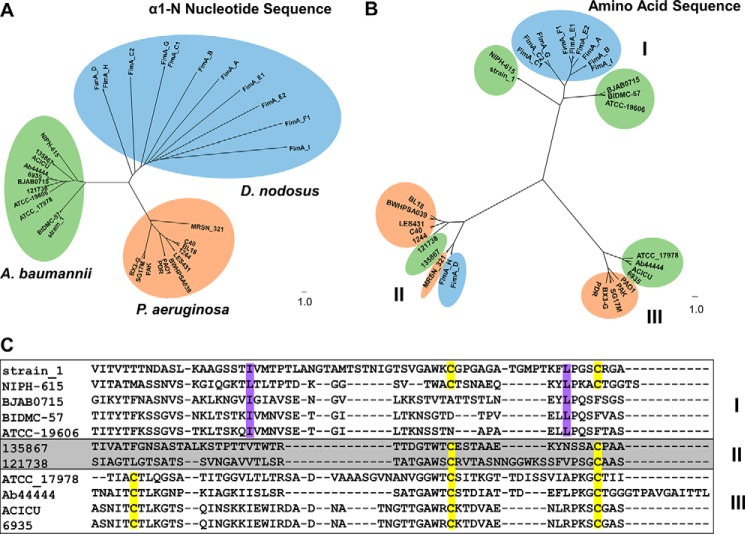
To understand the basis for the structural resemblance of PilAACICU and PilABIDMC57 of A. baumannii to P. aeruginosa PilAPAK and D. nodosus FimA, respectively, we compared nucleotide and amino acid sequences for the major pilin from 11 representative genomes from each of the three species (Fig. 6). To evaluate the possibility that the cross-species similarities in pilA arose from horizontal gene transfer, we aligned the sequences of 54 nucleotides encoding the first 18 residues of the mature protein product, which is identical (FTLIELMIVVAIIGILAA) in all 33 amino acid sequences.
FIGURE 6.
Divergent evolution of Acinetobacter PilA. A, dendrogram of the nucleotide sequence of the a1-N helix. Sequences from A. baumannii are highlighted in green, D. nodosus is in blue, and P. aeruginosa is in orange. B, dendrogram of the pilin amino acid sequence; sequences are colored identically to A. C, comparison of the three branches of A. baumannii PilA. Cysteine residues are highlighted in yellow, and the conserved C-terminal hydrophobic residues in cluster I are highlighted in violet.
Fig. 6A shows that, based on silent variations in these nucleotide sequences, the pilin genes can be separated neatly into three clusters based on species. That is, despite their dissimilarity in amino acid sequence, the nucleotide sequences for the α1-N domain of pilAACICU and pilABIDMC57 more closely resemble each other than their equivalents from P. aeruginosa and D. nodosus. However, when mature PilA amino acid sequences are aligned, all three of the resulting clusters contain representatives from multiple sequences. As they are labeled in Fig. 6B, cluster I contains the majority of the D. nodosus FimA serotypes (including serotype A, the sequence of Protein Data Bank code 3SOK) as well as A. baumannii PilABIDMC57 and PilAATCC 19606. Cluster II contains PilAACICU as well as P. aeruginosa PilAPAK and PilAPAO1. Cluster III consists of two FimA sequences that contain C-terminal disulfide bonds, A. baumannii PilA135867 and PilA121738, and the P. aeruginosa PilA sequences with C-terminal serine residues.
The A. baumannii sequences from each of the three clusters are shown in Fig. 6C. Cluster II contains species from the international clone I group, whereas cluster III contains species from the international clone II group. Cysteine residues are highlighted in yellow and indicate differential disulfide bonding patterns between the three clusters; cluster III PilA sequences contain two disulfide bonds, and the majority of cluster I sequences do not contain a disulfide bond at the C terminus. However, all sequences in cluster I, from both Acinetobacter and Dichelobacter, contain hydrophobic residues aligned to isoleucine 106 and leucine 129 (highlighted in violet). These data support the hypothesis that variation in Acinetobacter pilA is the result of common evolutionary pressures that are common to A. baumannii BIDMC 57 and D. nodosus (serotype A) and conversely A. baumannii ACICU and P. aeruginosa PAK but not to A. baumannii as a whole, resulting in a structural divergence of A. baumannii PilA.
Hydrophobic Interactions Stabilize the PilAM2 C Terminus
One notable aspect of the PilABIDMC57 and PilAM2 structures is that, unlike PilAACICU, they contain only a single disulfide bond between residues 56 and 86 in the αβ-loop and the first strand of the β-sheet, respectively, rather than a disulfide bond at the C terminus of the pilin headgroup (Fig. 7A). The addition of covalent disulfide bonds is typically understood to be a mechanism of stabilization in polypeptides, and hence the C-terminal disulfide bond, which is nearly ubiquitous in type IV pilins, is thought to be conserved to stabilize the pilin fold (3).
FIGURE 7.
Hydrophobic interactions in the PilAM2 C terminus. A, schematic representations of various major pilins; disulfide bonds are marked with yellow spheres. B, superimposition of PilAM2 (blue) and FimA (gray); an inset panel shows the C-terminal region where a disulfide bond is typically found in type IV pilins. C, differential scanning fluorometry curves showing stability measurements for PilAACICU, PilAM2, and PilAM2(I106A,V126A,L129A,V134A).
However, the lack of a C-terminal disulfide bond is not unique to Acinetobacter and, in fact, was observed previously in the structure of FimA from D. nodosus (serotype A) that PilABIDMC57 and PilAM2 closely resemble (Fig. 7B). The principal difference between the two folds is that the shorter loops of Acinetobacter PilABIDMC57 and PilAM2 result in a more compact structure. The lone disulfide bond in PilABIDMC57 and PilAM2 is found in a position identical to the disulfide bond in FimA and the N-terminal disulfide bond in PilAK122-4 (Fig. 7A). As PilABIDMC57, PilAM2, and FimA have such similar folds and both lack a disulfide bond at the C terminus, we compared the structures of their C termini for common structural features that might explain the absence of a C-terminal disulfide bond. Because D. nodosus is an obligate anaerobe and cysteine residues are more likely to be reduced in anaerobic environments, one explanation for the structural convergence of PilAM2 and FimA (serotype A) is that they use pilin folds, which are less reliant on a C-terminal disulfide bond for stability.
Hartung et al. (70) noted three non-covalent interactions in FimA that are in a similar position to the disulfide bond found in other pilins from Gram-negative bacteria: a backbone hydrogen bond between tyrosine 133 and valine 149, a hydrogen bond between the lysine 132 side chain and the backbone oxygen of lysine 150, and a van der Waals interaction between the tyrosine 133 phenyl ring and the aliphatic portion of the lysine 150 side chain. As no equivalents to these interactions can be found in the PilABIDMC57 and PilAM2 structures, we turned our attention to potentially stabilizing interactions between pairs of aliphatic side chains. In FimA, leucine 124, leucine 140, leucine 142, and isoleucine 145 can potentially form several such pairs, and several would be superimposable with those formed by isoleucine 106, valine 126, leucine 129, and valine 134 in PilAM2 and PilABIDMC57 (Fig. 7B).
As noted above, this pattern is conserved in all 14 sequences in cluster I with two of these positions, Ile-106 and Leu-129, being universally isoleucine, leucine, or valine. Conversely, although Ile-106 is conserved in PilAACICU, the other three positions are occupied by glutamate, arginine, and glycine.
To test our hypothesis that solvent exclusion from these aliphatic contacts stabilized PilAM2 in place of the canonical C-terminal disulfide bond, we measured the thermal stability of the PilAACICU and PilAM2 headgroups as well as a PilAM2 mutant with these four hydrophobic side chains truncated (I106A,V126A,L129A,V134A) using differential scanning fluorometry (Fig. 7C) (71). We found that although the wild type PilAM2 (Tm = 51.7 ± 0.2 °C) was less thermostable than PilAACICU (Tm = 55.7 °C ± 0.1), the hydrophobic C terminus of PilAM2 did contribute to the stability of the fold as evidenced by the lower melting temperature of the alanine mutant (Tm = 45.0 ± 0.5 °C).
Discussion
From an evolutionary standpoint, the x-ray crystal structures reported here pose three questions for us. Why have the major pilins of A. baumannii diverged? Why are some, but not all, PilA proteins C-terminally glycosylated? And why do the major pilins from A. baumannii ACICU and BIDMC 57 resemble their counterparts from other bacterial species (P. aeruginosa and D. nodosus, respectively) more closely than they do each other?
The presence of close homologs to both PilAACICU and PilABIDMC57 in all four species that make up the Acb complex strongly implies that the divergence in pilA predates the divergence of A. baumannii and A. nosocomialis. This, combined with the similarities between PilAACICU and PilAPAK and between PilABIDMC57 and FimA (serotype A), suggest that the divergence in Acinetobacter pilA is not due to functionally neutral diversifying selection, as is thought to be the case in Neisseria pilE, but instead due to functionally divergent evolution.
Determining which selective pressures favor a PilAACICU/PilAPAK-like structure over that of PilABIDMC57 and FimA (or vice versa) is more difficult, but possibilities include altered binding specificity and stability under different environmental conditions. We note that both Acinetobacter and Pseudomonas inhabit a wide range of environments and that both genera as well as Dichelobacter can be isolated from soil. Differing types of soil or solid surfaces may favor one structure over another.
Another possibility is that some Acinetobacter type IV pilus systems are optimized for one function (horizontal gene transfer, twitching motility, or adherence) over another. Direct comparisons of twitching motility between A. nosocomialis M2 (cluster I) and A. baumannii ATCC 17978 (cluster III) do show somewhat greater motility for M2 (supplemental Fig. 4), but this complex process is impacted by many factors in addition to the sequence, structure, and function of PilA.
The related question of why Acinetobacter pilA genes have diverged into glycosylated and non-glycosylated forms is complicated by the fact that no functional gain or defect has been attributed to the C-terminal glycan in Acinetobacter, and both tfpO− and tfpO+ strains have been shown to be infectious (72, 73). Similar results were obtained for the ΔtfpO mutant of P. aeruginosa 1244, which was also found to be equally susceptible to phage attachment (21). Also arguing against a functional role for C-terminal glycans is the lack of correlation between polysaccharide and polypeptide composition; for example, PilA proteins from A. baumannii ATCC 19606 and A. nosocomialis M2 are 93% identical, but the major polysaccharide glycans from these strains are completely unrelated (supplemental Fig. 5) (38, 74). Taken together, these findings imply that even gross alterations to the exposed surface of the major pilin have little functional impact and suggest that some or all of these binding events may occur not through the major pilin but rather through the minor pilins.
It was this lack of observable phenotype that led us to search for alternative explanations for the prevalence of tfpO-mediated glycosylation in Acinetobacter. Previous work in P. aeruginosa 1244, demonstrating that a ΔtfpO mutant was more vulnerable to phagocytosis mediated by opsonization (67, 68), implied that C-terminal glycosylation formed an obstacle to binding by host immune proteins. Our quantification of the ability of Acinetobacter C-terminal glycans to mask their conjugate polypeptides shows that over 25% of the PilA surface area available for binding is occluded. C-terminal glycosylation should, therefore, offer an advantage provided that the glycan surface is less vulnerable to binding by antibodies or other opsonins.
The evolutionary distance between A. baumannii BIDMC 57 (and A. nosocomialis M2) and D. nodosus suggests that the close resemblance between their respective pilin proteins is the result of convergent evolution. Although the functional benefit of this fold to the soil gammaproteobacteria found in class I of the alignment in Fig. 6 remains to be determined, it seems unlikely that the absence of a C-terminal disulfide bond in 12 of the 14 cluster I sequences is due to chance. We speculate that the cluster I fold may be advantageous in an anaerobic environment.
A further implication of the diversity in Acinetobacter type IV pilins is the challenge it poses for vaccine development. Because of their abundance in the extracellular space, type IV pili are obvious candidates for subunit vaccines and have been successfully used as such for other bacteria, including D. nodosus (75, 76). However, in Acinetobacter, the combination of variability in the PilA polypeptide with variation in polysaccharide structure in many strains may present a significant barrier to inducing a robust and durable immune response.
In conclusion, the results presented here reveal that type IV pili in Acinetobacter have diverged in a manner unrelated to the genetic divergence of species within the Acb complex and that similarities in type IV pili cross species, genus, and family lines. These data reinforce the principle that functional requirements determine protein structure while allowing considerable variation in sequence. These data also imply that three distinct functional classes of type IV pili exist in Acinetobacter and other soil gammaproteobacteria.
Author Contributions
K. H. P. conducted most of the experiments, analyzed the results, and wrote the paper. E. L. conducted the cell binding measurements. C. M. H. created the mutant Acinetobacter strains used in this study (with the aid of R. S. M.) and provided valuable experimental input and commentary during writing. J. W. L. and X. Z. performed the glycan conformational simulations. C. A. R. conducted the in vitro biofilm formation experiments. S. E. G., M. F. F., J. J. G., and E. J. S. helped to coordinate the study and write the paper.
Supplementary Material
Acknowledgments
We thank the staff at Argonne National Laboratory Advanced Photon Source, General Medical Sciences and Cancer Institutes Structural Biology Facility, beam lines 23ID-D and 23ID-B, and the staff at Stanford Synchrotron Radiation Lightsource, beam line 12-2, for technical assistance with x-ray data collection. We also thank Dr. Angela Wilks for the use of the circular dichroism spectrophotometer.
This work was supported in part by National Institutes of Health Grant R01 AI114902 (to E. J. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Figs. 1–6.
The atomic coordinates and structure factors (codes 4XA2, 5IHJ, and 5CFV) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Acb
- A. calcoaceticus-baumannii
- MBP
- maltose-binding protein
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- MPD
- 2-methyl-2,4-pentanediol
- PAK
- P. aeruginosa strain K
- OTase
- oligosaccharyltransferase
- r.m.s.d.
- root mean square deviation.
References
- 1. Strom M. S., and Lory S. (1993) Structure-function and biogenesis of the type IV pili. Annu. Re. Microbiol. 47, 565–596 [DOI] [PubMed] [Google Scholar]
- 2. Giltner C. L., Nguyen Y., and Burrows L. L. (2012) Type IV pilin proteins: versatile molecular modules. Microbiol. Mol. Biol. Rev. 76, 740–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Craig L., Pique M. E., and Tainer J. A. (2004) Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2, 363–378 [DOI] [PubMed] [Google Scholar]
- 4. Stone B. J., and Abu Kwaik Y. (1998) Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect. Immun. 66, 1768–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taniguchi T., Fujino Y., Yamamoto K., Miwatani T., and Honda T. (1995) Sequencing of the gene encoding the major pilin of pilus colonization factor antigen III (CFA/III) of human enterotoxigenic Escherichia coli and evidence that CFA/III is related to type IV pili. Infect. Immun. 63, 724–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Piepenbrink K. H., Maldarelli G. A., de la Peña C. F., Mulvey G. L., Snyder G. A., De Masi L., von Rosenvinge E. C., Günther S., Armstrong G. D., Donnenberg M. S., and Sundberg E. J. (2014) Structure of Clostridium difficile PilJ exhibits unprecedented divergence from known type IV pilins. J. Biol. Chem. 289, 4334–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Piepenbrink K. H., Maldarelli G. A., Martinez de la Peña C. F., Dingle T. C., Mulvey G. L., Lee A., von Rosenvinge E., Armstrong G. D., Donnenberg M. S., and Sundberg E. J. (2015) Structural and evolutionary analyses show unique stabilization strategies in the type IV pili of Clostridium difficile. Structure 23, 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Melville S., and Craig L. (2013) Type IV pili in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 77, 323–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lassak K., Ghosh A., and Albers S. V. (2012) Diversity, assembly and regulation of archaeal type IV pili-like and non-type-IV pili-like surface structures. Res. Microbiol. 163, 630–644 [DOI] [PubMed] [Google Scholar]
- 10. Wall D., and Kaiser D. (1999) Type IV pili and cell motility. Mol. Microbiol. 32, 1–10 [DOI] [PubMed] [Google Scholar]
- 11. Seifert H. S., Ajioka R. S., Marchal C., Sparling P. F., and So M. (1988) DNA transformation leads to pilin antigenic variation in Neisseria gonorrhoeae. Nature 336, 392–395 [DOI] [PubMed] [Google Scholar]
- 12. Takahashi H., Yanagisawa T., Kim K. S., Yokoyama S., and Ohnishi M. (2012) Meningococcal PilV potentiates Neisseria meningitidis type IV pilus-mediated internalization into human endothelial and epithelial cells. Infect. Immun. 80, 4154–4166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bieber D., Ramer S. W., Wu C. Y., Murray W. J., Tobe T., Fernandez R., and Schoolnik G. K. (1998) Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280, 2114–2118 [DOI] [PubMed] [Google Scholar]
- 14. Cehovin A., Winterbotham M., Lucidarme J., Borrow R., Tang C. M., Exley R. M., and Pelicic V. (2010) Sequence conservation of pilus subunits in Neisseria meningitidis. Vaccine 28, 4817–4826 [DOI] [PubMed] [Google Scholar]
- 15. Criss A. K., Kline K. A., and Seifert H. S. (2005) The frequency and rate of pilin antigenic variation in Neisseria gonorrhoeae. Mol. Microbiol. 58, 510–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Toma C., Kuroki H., Nakasone N., Ehara M., and Iwanaga M. (2002) Minor pilin subunits are conserved in Vibrio cholerae type IV pili. FEMS Immunol. Med. Microbiol. 33, 35–40 [DOI] [PubMed] [Google Scholar]
- 17. Blank T. E., Zhong H., Bell A. L., Whittam T. S., and Donnenberg M. S. (2000) Molecular variation among type IV pilin (bfpA) genes from diverse enteropathogenic Escherichia coli strains. Infect. Immun. 68, 7028–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maldarelli G. A., De Masi L., von Rosenvinge E. C., Carter M., and Donnenberg M. S. (2014) Identification, immunogenicity, and cross-reactivity of type IV pilin and pilin-like proteins from Clostridium difficile. Pathog. Dis. 71, 302–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aagesen A. M., and Häse C. C. (2012) Sequence analyses of type IV pili from Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus. Microb. Ecol. 64, 509–524 [DOI] [PubMed] [Google Scholar]
- 20. Allison T. M., Conrad S., and Castric P. (2015) The group I pilin glycan affects type IVa pilus hydrophobicity and twitching motility in Pseudomonas aeruginosa 1244. Microbiology 161, 1780–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smedley J. G. 3rd, Jewell E., Roguskie J., Horzempa J., Syboldt A., Stolz D. B., and Castric P. (2005) Influence of pilin glycosylation on Pseudomonas aeruginosa 1244 pilus function. Infect. Immun. 73, 7922–7931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Voisin S., Kus J. V., Houliston S., St-Michael F., Watson D., Cvitkovitch D. G., Kelly J., Brisson J. R., and Burrows L. L. (2007) Glycosylation of Pseudomonas aeruginosa strain Pa5196 type IV pilins with Mycobacterium-like α-1,5-linked d-Araf oligosaccharides. J. Bacteriol. 189, 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aas F. E., Vik A., Vedde J., Koomey M., and Egge-Jacobsen W. (2007) Neisseria gonorrhoeae O-linked pilin glycosylation: functional analyses define both the biosynthetic pathway and glycan structure. Mol. Microbiol. 65, 607–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Power P. M., Seib K. L., and Jennings M. P. (2006) Pilin glycosylation in Neisseria meningitidis occurs by a similar pathway to wzy-dependent O-antigen biosynthesis in Escherichia coli. Biochem. Biophys. Res. Commun. 347, 904–908 [DOI] [PubMed] [Google Scholar]
- 25. Gault J., Ferber M., Machata S., Imhaus A. F., Malosse C., Charles-Orszag A., Millien C., Bouvier G., Bardiaux B., Péhau-Arnaudet G., Klinge K., Podglajen I., Ploy M. C., Seifert H. S., Nilges M., et al. (2015) Neisseria meningitidis type IV pili composed of sequence invariable pilins are masked by multisite glycosylation. PLoS Pathog. 11, e1005162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Falagas M. E., and Kopterides P. (2006) Risk factors for the isolation of multi-drug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa: a systematic review of the literature. J. Hosp. Infect. 64, 7–15 [DOI] [PubMed] [Google Scholar]
- 27. Peleg A. Y., Seifert H., and Paterson D. L. (2008) Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dijkshoorn L., Nemec A., and Seifert H. (2007) An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5, 939–951 [DOI] [PubMed] [Google Scholar]
- 29. Jones A., Morgan D., Walsh A., Turton J., Livermore D., Pitt T., Green A., Gill M., and Mortiboy D. (2006) Importation of multidrug-resistant Acinetobacter spp infections with casualties from Iraq. Lancet Infect. Dis. 6, 317–318 [DOI] [PubMed] [Google Scholar]
- 30. Harding C. M., Kinsella R. L., Palmer L. D., Skaar E. P., and Feldman M. F. (2016) Medically relevant Acinetobacter species require a type II secretion system and specific membrane-associated chaperones for the export of multiple substrates and full virulence. PLoS Pathog. 12, e1005391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chang H. C., Wei Y. F., Dijkshoorn L., Vaneechoutte M., Tang C. T., and Chang T. C. (2005) Species-level identification of isolates of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex by sequence analysis of the 16S-23S rRNA gene spacer region. J. Clin. Microbiol. 43, 1632–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Antunes L. C., Visca P., and Towner K. J. (2014) Acinetobacter baumannii: evolution of a global pathogen. Pathog. Dis. 71, 292–301 [DOI] [PubMed] [Google Scholar]
- 33. Harding C. M., Tracy E. N., Carruthers M. D., Rather P. N., Actis L. A., and Munson R. S. Jr. (2013) Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. MBio 4, e00360–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wilharm G., Piesker J., Laue M., and Skiebe E. (2013) DNA uptake by the nosocomial pathogen Acinetobacter baumannii occurs during movement along wet surfaces. J. Bacteriol. 195, 4146–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nait Chabane Y., Mlouka M. B., Alexandre S., Nicol M., Marti S., Pestel-Caron M., Vila J., Jouenne T., and Dé E. (2014) Virstatin inhibits biofilm formation and motility of Acinetobacter baumannii. BMC Microbiol. 14, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Eijkelkamp B. A., Stroeher U. H., Hassan K. A., Papadimitrious M. S., Paulsen I. T., and Brown M. H. (2011) Adherence and motility characteristics of clinical Acinetobacter baumannii isolates. FEMS Microbiol. Lett. 323, 44–51 [DOI] [PubMed] [Google Scholar]
- 37. Oh M. H., and Choi C. H. (2015) Role of LuxIR homologue AnoIR in Acinetobacter nosocomialis and the effect of virstatin on the expression of anoR gene. J. Microbiol. Biotechnol. 25, 1390–1400 [DOI] [PubMed] [Google Scholar]
- 38. Harding C. M., Nasr M. A., Kinsella R. L., Scott N. E., Foster L. J., Weber B. S., Fiester S. E., Actis L. A., Tracy E. N., Munson R. S. Jr, and Feldman M. F. (2015) Acinetobacter strains carry two functional oligosaccharyltransferases, one devoted exclusively to type IV pilin, and the other one dedicated to O-glycosylation of multiple proteins. Mol. Microbiol. 96, 1023–1041 [DOI] [PubMed] [Google Scholar]
- 39. Hu D., Liu B., Dijkshoorn L., Wang L., and Reeves P. R. (2013) Diversity in the major polysaccharide antigen of Acinetobacter baumannii assessed by DNA sequencing, and development of a molecular serotyping scheme. PLoS One 8, e70329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Iwashkiw J. A., Seper A., Weber B. S., Scott N. E., Vinogradov E., Stratilo C., Reiz B., Cordwell S. J., Whittal R., Schild S., and Feldman M. F. (2012) Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathog. 8, e1002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lees-Miller R. G., Iwashkiw J. A., Scott N. E., Seper A., Vinogradov E., Schild S., and Feldman M. F. (2013) A common pathway for O-linked protein-glycosylation and synthesis of capsule in Acinetobacter baumannii. Mol. Microbiol. 89, 816–830 [DOI] [PubMed] [Google Scholar]
- 42. Cagatay T. I., and Hickford J. G. (2008) Glycosylation of type-IV fimbriae of Dichelobacter nodosus. Vet. Microbiol. 126, 160–167 [DOI] [PubMed] [Google Scholar]
- 43. DiGiandomenico A., Matewish M. J., Bisaillon A., Stehle J. R., Lam J. S., and Castric P. (2002) Glycosylation of Pseudomonas aeruginosa 1244 pilin: glycan substrate specificity. Mol. Microbiol. 46, 519–530 [DOI] [PubMed] [Google Scholar]
- 44. Moon A. F., Mueller G. A., Zhong X., and Pedersen L. C. (2010) A synergistic approach to protein crystallization: combination of a fixed-arm carrier with surface entropy reduction. Protein Sci. 19, 901–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 47. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Echols N., Headd J. J., Hung L. W., Jain S., Kapral G. J., Grosse Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R. D., Read R. J., Richardson D. C., et al. (2011) The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., and Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 51. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 52. Borbulevych O. Y., Piepenbrink K. H., and Baker B. M. (2011) Conformational melding permits a conserved binding geometry in TCR recognition of foreign and self molecular mimics. J. Immunol. 186, 2950–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Craig L., Taylor R. K., Pique M. E., Adair B. D., Arvai A. S., Singh M., Lloyd S. J., Shin D. S., Getzoff E. D., Yeager M., Forest K. T., and Tainer J. A. (2003) Type IV pilin structure and assembly: x-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol. Cell 11, 1139–1150 [DOI] [PubMed] [Google Scholar]
- 54. Craig L., Volkmann N., Arvai A. S., Pique M. E., Yeager M., Egelman E. H., and Tainer J. A. (2006) Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell 23, 651–662 [DOI] [PubMed] [Google Scholar]
- 55. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 56. Chaudhury S., Lyskov S., and Gray J. J. (2010) PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 26, 689–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Drew K., Renfrew P. D., Craven T. W., Butterfoss G. L., Chou F. C., Lyskov S., Bullock B. N., Watkins A., Labonte J. W., Pacella M., Kilambi K. P., Leaver-Fay A., Kuhlman B., Gray J. J., Bradley P., et al. (2013) Adding diverse noncanonical backbones to Rosetta: enabling peptidomimetic design. PLoS One 8, e67051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kleiger G., Saha A., Lewis S., Kuhlman B., and Deshaies R. J. (2009) Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell 139, 957–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 60. Lillehoj E. P., Hyun S. W., Feng C., Zhang L., Liu A., Guang W., Nguyen C., Luzina I. G., Atamas S. P., Passaniti A., Twaddell W. S., Puché A. C., Wang L. X., Cross A. S., and Goldblum S. E. (2012) NEU1 sialidase expressed in human airway epithelia regulates epidermal growth factor receptor (EGFR) and MUC1 protein signaling. J. Biol. Chem. 287, 8214–8231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lillehoj E. P., Hyun S. W., Liu A., Guang W., Verceles A. C., Luzina I. G., Atamas S. P., Kim K. C., and Goldblum S. E. (2015) NEU1 sialidase regulates membrane-tethered mucin (MUC1) ectodomain adhesiveness for Pseudomonas aeruginosa and decoy receptor release. J. Biol. Chem. 290, 18316–18331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Doi Y., Murray G. L., and Peleg A. Y. (2015) Acinetobacter baumannii: evolution of antimicrobial resistance-treatment options. Semin. Respir. Crit. Care Med. 36, 85–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Iacono M., Villa L., Fortini D., Bordoni R., Imperi F., Bonnal R. J., Sicheritz-Ponten T., De Bellis G., Visca P., Cassone A., and Carattoli A. (2008) Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob. Agents Chemother. 52, 2616–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Carruthers M. D., Harding C. M., Baker B. D., Bonomo R. A., Hujer K. M., Rather P. N., and Munson R. S. Jr. (2013) Draft genome sequence of the clinical isolate Acinetobacter nosocomialis strain M2. Genome Announc. 1, e00906–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Clemmer K. M., Bonomo R. A., and Rather P. N. (2011) Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology 157, 2534–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Audette G. F., Irvin R. T., and Hazes B. (2004) Crystallographic analysis of the Pseudomonas aeruginosa strain K122-4 monomeric pilin reveals a conserved receptor-binding architecture. Biochemistry 43, 11427–11435 [DOI] [PubMed] [Google Scholar]
- 67. Tan R. M., Kuang Z., Hao Y., and Lau G. W. (2014) Type IV pilus of Pseudomonas aeruginosa confers resistance to antimicrobial activities of the pulmonary surfactant protein-A. J. Innate Immun. 6, 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tan R. M., Kuang Z., Hao Y., Lee F., Lee T., Lee R. J., and Lau G. W. (2015) Type IV pilus glycosylation mediates resistance of Pseudomonas aeruginosa to opsonic activities of the pulmonary surfactant protein A. Infect. Immun. 83, 1339–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Simons K. T., Kooperberg C., Huang E., and Baker D. (1997) Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J. Mol. Biol. 268, 209–225 [DOI] [PubMed] [Google Scholar]
- 70. Hartung S., Arvai A. S., Wood T., Kolappan S., Shin D. S., Craig L., and Tainer J. A. (2011) Ultrahigh resolution and full-length pilin structures with insights for filament assembly, pathogenic functions, and vaccine potential. J. Biol. Chem. 286, 44254–44265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Huynh K., and Partch C. L. (2015) Analysis of protein stability and ligand interactions by thermal shift assay. Curr. Protoc. Protein Sci. 79, 28.9.1–28.9.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jacobs A. C., Thompson M. G., Black C. C., Kessler J. L., Clark L. P., McQueary C. N., Gancz H. Y., Corey B. W., Moon J. K., Si Y., Owen M. T., Hallock J. D., Kwak Y. I., Summers A., Li C. Z., et al. (2014) AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. MBio 5, e01076–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jones C. L., Clancy M., Honnold C., Singh S., Snesrud E., Onmus-Leone F., McGann P., Ong A. C., Kwak Y., Waterman P., Zurawski D. V., Clifford R. J., and Lesho E. (2015) Fatal outbreak of an emerging clone of extensively drug-resistant Acinetobacter baumannii with enhanced virulence. Clin. Infect. Dis. 61, 145–154 [DOI] [PubMed] [Google Scholar]
- 74. Scott N. E., Kinsella R. L., Edwards A. V., Larsen M. R., Dutta S., Saba J., Foster L. J., and Feldman M. F. (2014) Diversity within the O-linked protein glycosylation systems of Acinetobacter species. Mol. Cell. Proteomics 13, 2354–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bhardwaj V., Dhungyel O., de Silva K., and Whittington R. J. (2014) Investigation of immunity in sheep following footrot infection and vaccination. Vaccine 32, 6979–6985 [DOI] [PubMed] [Google Scholar]
- 76. Korpi F., Irajian G., Mahadavi M., Motamedifar M., Mousavi M., Laghaei P., Raei N., and Behrouz B. (2015) Active immunization with recombinant PilA protein protects against Pseudomonas aeruginosa infection in a mouse burn wound model. J. Microbiol. Biotechnol. 10.4014/jmb.1507.07044 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.