Abstract
Monitoring the environment with serine/threonine protein kinases is critical for growth and survival of Mycobacterium tuberculosis, a devastating human pathogen. Protein kinase B (PknB) is a transmembrane serine/threonine protein kinase that acts as an essential regulator of mycobacterial growth and division. The PknB extracellular domain (ECD) consists of four repeats homologous to penicillin-binding protein and serine/threonine kinase associated (PASTA) domains, and binds fragments of peptidoglycan. These properties suggest that PknB activity is modulated by ECD binding to peptidoglycan substructures, however, the molecular mechanisms underpinning PknB regulation remain unclear. In this study, we report structural and genetic characterization of the PknB ECD. We determined the crystal structures of overlapping ECD fragments at near atomic resolution, built a model of the full ECD, and discovered a region on the C-terminal PASTA domain that has the properties of a ligand-binding site. Hydrophobic interaction between this surface and a bound molecule of citrate was observed in a crystal structure. Our genetic analyses in M. tuberculosis showed that nonfunctional alleles were produced either by deletion of any of single PASTA domain or by mutation of individual conserved residues lining the putative ligand-binding surface of the C-terminal PASTA repeat. These results define two distinct structural features necessary for PknB signal transduction, a fully extended ECD and a conserved, membrane-distal putative ligand-binding site.
Keywords: bacterial genetics, crystal structure, Mycobacterium tuberculosis, peptidoglycan, receptor protein serine/threonine kinase, PASTA domain
Introduction
In addition to the classical bacterial histidine kinases, Mycobacterium tuberculosis (Mtb) encodes 11 eukaryotic-like serine/threonine protein kinases (STPK)3 with functions in growth, virulence, and persistence (1). Although over 250 STPK phosphorylation targets have been identified, the environmental cues that regulate STPKs have remained largely unknown (2). Nonetheless, PknB has emerged as a strong candidate for coordination of cell wall peptidoglycan remodeling with multiple intracellular pathways. Following activation induced by formation of an asymmetric homodimer (3), PknB phosphorylates proteins involved in peptidoglycan synthesis, the tricarboxylic acid cycle, cell division, and the extracellular stress response (4, 5), consistent with its proposed role in cell-wall homeostasis.
PknB contains an intracellular N-terminal kinase domain followed by a 50-residue linker, a single transmembrane helix, and a peptidoglycan-interacting ECD that comprises four PASTA repeats, termed PASTA1–4. This architecture is found throughout Firmicutes and Actinobacteria with homologs reported in Streptococcus (6), Staphylococcus (7), Bacillus (8), and Corynebacterium (9), although the number of PASTA domains is variable and additional immunoglobulin-like domains are present in ECDs of some of the homologs (10). Phylogenetic analysis revealed position-dependent evolution indicative of specialization of individual PASTA domains and found that the PASTA domain composition of a particular protein is specific to each genus (11).
In Bacillus subtilis, protein kinase C, a PknB homolog, is dispensable for normal growth but is required for peptidoglycan fragment-induced spore reactivation, with the PASTA domains determining peptidoglycan species specificity (8). In mycobacteria, PknB deletion is lethal, knockdown results in elongated cells, and overexpression produces round bulbous cells (12). Direct interaction of muropeptides with PknB ECD is similarly dependent on fine chemical modifications and requires at least one sugar and three peptide residues of the peptidoglycan monomer (13). The ECD determines polar localization of PknB, a step that may be required for proper activation of kinase signaling.
Structure determination of PknB ECD using nuclear magnetic resonance, revealed an extended conformation of the PASTA domains in solution and suggested a ligand-dependent dimerization activation mechanism (14). Allelic substitution experiments with a C-terminal truncation series reported by Chawla et al. (15) indicated that all alleles lacking PASTA4 were nonfunctional. Conversely, overexpression of a C-terminal fragment of PknB that included its transmembrane domain and the ECD suppressed mycobacterial growth in a PASTA4-dependent manner (16). These genetic data argued for the presence of ligand-binding site in PASTA4. In this study we determined five x-ray crystal structures of the PknB ECD allowing us to build a complete atomic model from the two largest overlapping fragments. Binding of a citrate molecule to a highly conserved hydrophobic groove on PASTA4 in one of the structures identified a potential ligand-binding site. Allelic substitution experiments in M. tuberculosis confirmed the functional significance of this PASTA4 surface and revealed an unexpected requirement for each of the PASTA domains for proper PknB function.
Results
To determine the atomic structure of the PknB ECD and to create a platform for investigation of ligand binding, we expressed and purified multiple constructs corresponding to individual PASTA domains and domain combinations. Proteins representing PASTA1–2, PASTA2–4, and PASTA3–4 (domains are numbered from N to C terminus) as well as the full ECD were purified by glutathione affinity chromatography and gel filtration, and were tested for crystallization against a panel of sparse-matrix screens. Although diffraction quality crystals of the full ECD were never obtained, several shorter constructs yielded crystals that diffracted to ∼2-Å resolution.
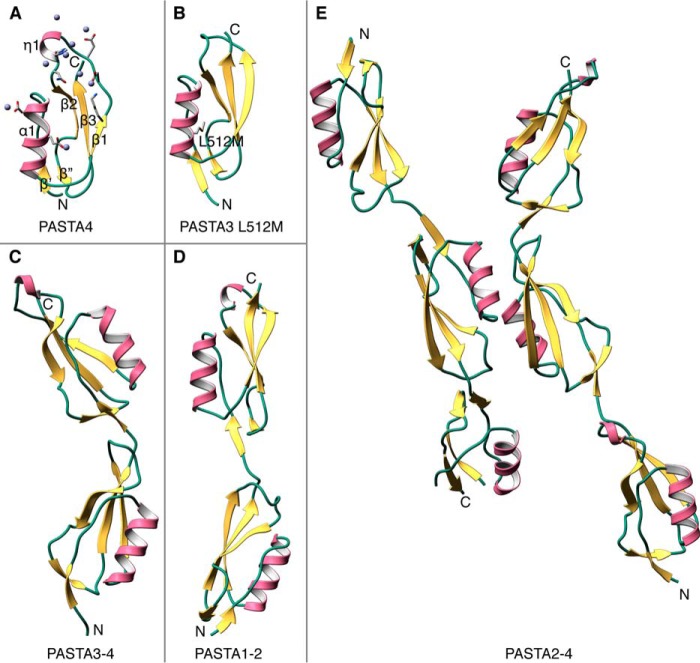
The PASTA4 structure was determined at 2.0-Å resolution using single-wavelength anomalous dispersion of Zn2+-bound protein. Zn2+ ions were present in the crystallization solution and 9 were found in the refined structure (Table 1, Fig. 1A). Of these, two were essential to forming crystal contacts. As reported previously, the defining structural features of a PASTA domain include a ψ-loop, which has one face exposed to the solvent, and an amphipathic α-helix that is nested on the concave side of this β-structure. In the case of PknB PASTA4, the β1-β2 loop is 13 residues long and includes a 310 helix (residues 597–599).
TABLE 1.
Data collection and refinement statistics
| PASTA4 | PASTA3 L512M | PASTA3–4 | PASTA1–2 | PASTA2–4 | |
|---|---|---|---|---|---|
| Data collection | |||||
| Wavelength (Å) | 1.282 | 1.116 | 1.116 | 1.116 | 1.116 |
| Temp (K) | 100 | 100 | 100 | 100 | 100 |
| Space group | P 65 2 2 | P 21 21 2 | P 21 | C 2 | P 21 |
| Unit cell parameters | |||||
| a/b/c (Å) | 41.7/41.7/122.5 | 24.4/73.4/32.6 | 40.5/24.5/64.9 | 80.7/43.1/43.7 | 101.3/24.5/73.3 |
| α/β/γ (o) | 90/90/120 | 90/90/90 | 90/99.17/90 | 90/106.3/90 | 90/89.35/90 |
| Resolution (Å)a | 34.61-2.00b (2.07-2.00) | 32.58-2.00 (2.08-2.00) | 39.95-2.00 (2.03-2.00) | 41.99-1.80 (1.87-1.80) | 41.90-2.21 (2.28-2.21) |
| Rsym (%) | 9.6 (30.8) | 12.9 (42.6) | 7.7 (47.9) | 9.0 (57.1) | 13.8 (65.5) |
| I/σI | 17.11 (3.87) | 8.0 (2.1) | 10.6 (1.9) | 14.8 (2.1) | 7.8 (1.9) |
| Completeness (%) | 92.48 (54.42) | 99.37 (95.11) | 97.5 (93.8) | 99.78 (98.00) | 99.43 (95.03) |
| Redundancy | 15.5 (3.9) | 3.6 (2.9) | 2.3 (2.1) | 3.9 (3.9) | 4.0 (4.0) |
| SAD/MR solution | |||||
| Protein per a.u. | 1 | 1 | 1 | 1 | 2 |
| Zn2+ sites per a.u | 9 | ||||
| Mean figure of merit | 0.450 | ||||
| Refinement | |||||
| Resolution (Å) | 34.61-2.00 | 32.58-2.00 | 39.95-2.00 | 41.99-1.80 | 41.90-2.21 |
| Number of reflections | 4363 | 4249 | 8586 | 13502 | 18769 |
| Rwork/Rfree (%) | 21.53/26.39 | 17.60/22.60 | 19.09/23.05 | 16.93/20.73 | 20.21/24.89 |
| Number of atoms | |||||
| Protein | 549 | 484 | 1025 | 1,028 | 2,898 |
| Ligands | 9 | 13 | |||
| Solvent | 50 | 51 | 100 | 135 | 141 |
| Average B-factors | |||||
| Protein (Å2) | 23.6 | 14 | 24.3 | 21.6 | 30.7 |
| Ligands (Å2) | 36.7 | 50.2 | |||
| Solvent (Å2) | 31.8 | 23.4 | 30.7 | 33.5 | 32.0 |
| Root mean square deviations | |||||
| Bond lengths (Å) | 0.005 | 0.007 | 0.003 | 0.008 | 0.003 |
| Bond angles (°) | 0.91 | 1.08 | 0.77 | 1.21 | 0.7 |
| Ramachandran plot | |||||
| Favored (%) | 97 | 100 | 100 | 99 | 98 |
| Outliers (%) | 0 | 0 | 0 | 0.72 | 0 |
| PDB ID | 5E0Y | 3OUV | 5E0Z | 5E10 | 5E12 |
a Values in parentheses are for the highest resolution shell.
b The effective resolution is lower due to low completeness.
FIGURE 1.
X-ray crystal structures PknB ECD. A, the carboxyl-terminal PASTA4 repeat in complex with Zn2+ (gray spheres). Zn2+-bound residues are highlighted. The key structural features include ψ-loop (β1β2β3) nestling the α1 helix and a β′/β″ brace close to the N terminus of the domain. B, PASTA3 domain of PknB. The selenomethionine derivative of the L512M mutant was solved by molecular replacement with the PASTA4 model. Selenomethionine 512 is represented as sticks. Unlike PASTA4, the shorter β1-β2 loop does not form a 310 helix. C, PASTA3–4 two-domain structure. D, PASTA1–2 two-domain structure. All four PASTA domains have similar structures with longer β1-β2 loops in PASTA2 and PASTA4. E, PASTA2–4 crystallized with 2 molecules in the asymmetric unit. The structure was determined by multiple rounds of molecular replacement. A loop in PASTA4 of the molecule shown on the left was disordered due to a close crystal contact.
An L512M mutant of PASTA3 labeled with selenomethionine (Fig. 1B) was purified and crystallized as an alternate approach for solving the phase problem. However, the phases were determined using molecular replacement with PASTA4 and the structure refined, also at 2.0-Å resolution, against data obtained at 1.116-Å wavelength, away from selenium edge. The overall-fold of PASTA3 domain is very similar to PASTA4 except that the loop between β-1 and β-2 is only 8 residues long and does not form a 310 helix.
We found that to achieve crystallization of multidomain constructs it was necessary to trim the unstructured N-terminal extensions. Our starting set of constructs contained N-terminal GHM linker residues that remained following tobacco etch virus (TEV) cleavage. When several of these failed to crystallize, we took advantage of the naturally occurring glycine residues of PASTA domains 2 (Gly-423) and 3 (Gly-491) to clone constructs that incorporated these residues as part of the TEV recognition site (ENLYFQ↓G). Although TEV cleavage efficiency was decreased, the resulting processed constructs contained no linker artifacts and crystallized readily. In the case of PASTA1–2, an analogous glycine (Gly-355) would have given a construct containing an unstructured N terminus. Instead, a single glycine linker residue was cloned upstream of amino acid Val-360 creating a construct that crystallized well.
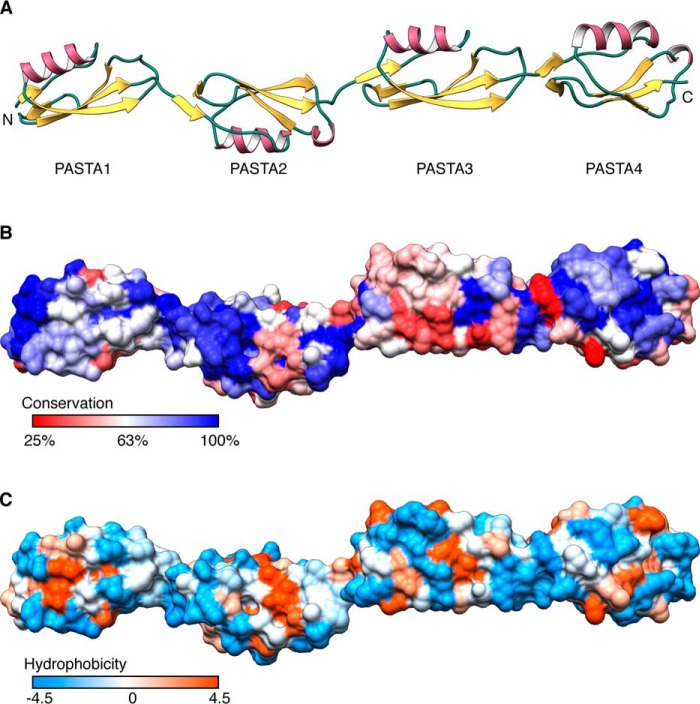
PASTA3–4 at 2.0 Å, PASTA2–4 at 2.2 Å, and last PASTA1–2 at 1.8-Å resolution (Fig. 1, C–E) were solved by molecular replacement with previously determined structures. Of these, PASTA2–4 proved the most challenging as search with the PASTA3–4 structure failed, likely due to a difference in inter-domain angles. Instead, sequential searches with the individual PASTA domains produced phases that resulted in an interpretable electron density map. The complete atomic model of the PknB ECD was then constructed based on this structure and the overlapping PASTA1–2 fragment (Fig. 2A).
FIGURE 2.
The PknB ECD comprises an elongated head to tail arrangement of four PASTA repeats with two highly conserved surfaces. A, ribbon diagram of a composite model created by superimposing PASTA2 of the PASTA1–2 and PASTA2–4 structures. B, surface residue conservation in mycobacterial PknB orthologs from least conserved (red) to most conserved (blue). C, the PknB ECD surface was colored according to Kyte-Doolittle hydrophobicity scale from most hydrophilic (blue) to most hydrophobic (orange).
The relationship between PASTA domains in the full ECD is that observed previously for this family of sensor kinases. The PASTA domains of PknB stack top to bottom rather than side to side resulting in an extended conformation. The linkers between PASTA domains are short and well ordered in every structure presented. To achieve bending angles much greater than those observed, the domains might have to undergo partial unfolding. Such conformations are thus likely to be transient.
To map the functionally significant surfaces in the PknB ECD, we undertook bioinformatic analysis. The pairwise identity of individual PASTA domains to each other is low, around 30%. In contrast, full-length alignment of PknB orthologs from the Mycobacterium family reveals a striking degree of surface conservation in the ECD that focuses on PASTA4 and the linker region between PASTA1 and PASTA2 (Fig. 2B). Of these two regions only the conserved patch in PASTA4 has partially hydrophobic character (Fig. 2C).
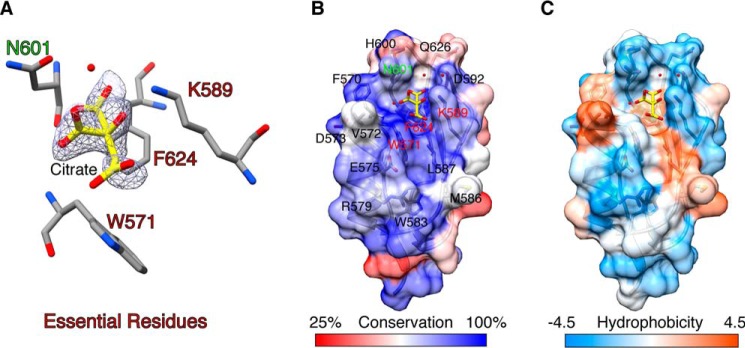
The conserved region in PASTA4 domain was bound to a molecule of citric acid in the PASTA2–4 structure (Fig. 3) suggesting it is a ligand-binding site. This citrate-binding site is formed between helix α1 and strand β1. Citrate contacts include side chain atoms of residues Phe-624, Asn-601, and Lys-589. The bottom of the groove in which the citrate molecule is bound is hydrophobic and in addition to Phe-624 contains partially exposed conserved tryptophan (Trp-571), a typical feature of protein-binding sites (Fig. 3). The citrate molecule is positioned to interact with this site through its hydrophobic face with all three charged carboxylic groups and the hydroxyl group pointing away from the protein surface.
FIGURE 3.
The PASTA4 domain of PknB ECD contains a highly conserved surface with features of a ligand-binding site. A, citrate molecule bound to PASTA4 domain of PASTA2–4 crystal structure showing a 2Fo − Fc omit map contoured at 1 I/σI. Residues essential for PknB function are labeled red, non-essential Asn-601 is labeled green. B and C, citrate binds in the center of a large conserved surface in PASTA4 domain that features a central hydrophobic groove surrounded by charged residues, a typical pattern for a ligand-binding site. Color scheme as described in the legend to Fig. 2, B and C.
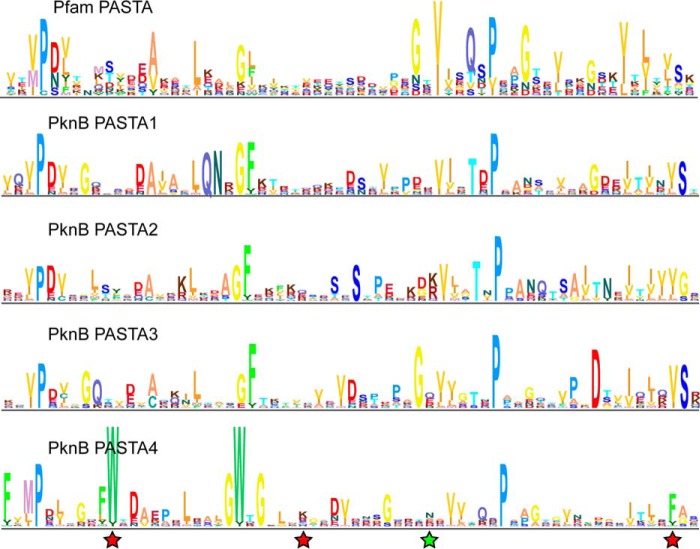
The residues lining the putative ligand-binding site in PASTA4 were specific to this domain. The differences in primary sequence between PASTA4 and PASTA1–3 domains are illustrated by their HMM logo representations (Fig. 4). Although PASTA1–3 produce logos similar to that of a typical PASTA domain (Pfam PASTA), PASTA4 contains an unusual pair of conserved tryptophans. The first of these (Trp-571) is found in a position that does not typically feature a bulky hydrophobic residue. It is therefore unlikely to be required for folding and might instead participate in ligand binding. Taken together, these analyses are consistent with a unique role for the PASTA4 domain of PknB.
FIGURE 4.
Mycobacterial PASTA4 orthologs contain unusual conserved tryptophans. Sequence logo representations of Hidden Markov Models of a typical PASTA domain (Pfam PF03793.15) and of the four Mycobacterial PASTA domains. The mycobacterial logos were derived from an alignment of 72 unique full-length sequences of PknB orthologs. Residues in PASTA4 that were tested by alanine substitution and found essential are marked with red stars. N601A substitution produced a functional allele and is marked with a green star.
To test the functional significance of the identified ligand-binding site on PASTA4 we determined if the native PknB of M. tuberculosis could be replaced with alleles encoding alanine in the place of four conserved residues (Fig. 4). These experiments were performed using a strain of M. tuberculosis in which the endogenous copy of pknB was deleted, and expression of this essential gene was supplied from a plasmid integrated at a distal chromosomal site. This integrating plasmid could be efficiently exchanged with a differentially marked plasmid containing a functional copy of pknB, but not with an empty vector, providing a simple assay to determine whether individual alleles of pknB were functional in vivo. Exchange was successful for a plasmid encoding an N601A mutation in PASTA4, indicating that this allele was functional and allowed bacterial growth. However, the N601A-expressing cells produced smaller colonies than bacteria containing a wild type allele, suggesting that this allele may be partially impaired. In contrast, plasmids expressing W571A, K589A, and F624A alleles of pknB behaved identically to an empty vector control, producing no viable bacterial colonies after the exchange (Table 2). Thus, conserved residues in the PASTA4 putative ligand-binding site were necessary for PknB function and M. tuberculosis viability.
TABLE 2.
Transformation efficiency of KanR pknB plasmids into the M. tuberculosis strain carrying ΔpknB::hyg+pknB_WT at attBL5-Zeo
| Plasmid | pknB variant | Transformation efficiency/μg DNA (± S.D.) n = 3 |
|---|---|---|
| pKP570 | MCK-pknB_WT | 5.03 ± 0.5 × 104 |
| pKP571 | MCK-pknB_ΔPASTA1 | 0 |
| pKP572 | MCK-pknB_ΔPASTA2 | 0 |
| pKP573 | MCK-pknB_ΔPASTA3 | 0 |
| pKP574 | MCK-pknB_ΔPASTA4 | 0 |
| pKP575 | MCK-pknB_W571A | 0 |
| pKP576 | MCK-pknB_K589A | 0 |
| pKP577 | MCK-pknB_N601A | 3.41 ± 0.3 × 104 |
| pKP578 | MCK-pknB_F624A | 0 |
| pMV306 | L5 integrating vector | 0 |
| pMV261 | Replicating plasmid | 1.87 ± 0.2 × 105 |
| pTTP1A | Tweety Integrating plasmid | 6.50 ± 0.5 × 104 |
To test whether PASTA4 is the only ECD subdomain required for function, we designed and tested four PASTA domain deletions in the allele swap experiment. Because the interdomain linkers make extensive contact with the β1-β2 loop of the preceding PASTA domain, we designed the internal PASTA deletion mutants with the aim of keeping these interactions intact. Thus, the ΔPASTA2 construct has PASTA1–2 linker followed by the first residue of β′ strand of PASTA3 and ΔPASTA3 has PASTA2–3 linker followed by PASTA4. Plasmid exchange studies with any of the four individual PASTA deletion alleles led to no viable colony formation in Mtb (Table 2), indicating that each of the four PASTA domains is independently essential for PknB function and M. tuberculosis growth.
To test expression and stability of PknB variants used in allele substitution experiments, the same integrating plasmid constructs (Table 2) were transformed into wild type M. smegmatis. Western blotting analysis revealed levels of expression even from the complementation-competent pknB constructs that were close to the limit of detection (data not shown). Instead, the same promoter-RBS-pknB variant combinations were cloned into the mycobacterial replicating plasmid expected to maintain ∼30-fold higher copy number (Table 3) and transformed into wild type M. smegmatis. Anti-myc Western blotting analysis of these strains confirmed that all the variant PknB proteins, including internal PASTA domain deletions, were stably expressed in mycobacteria (Fig. 5).
TABLE 3.
Plasmids used for Western blotting analysis
| Plasmid | pknB variant |
|---|---|
| pKP842 | MEK-pknB_WT |
| pKP843 | MEK-pknB_ΔPASTA1 |
| pKP844 | MEK-pknB_ΔPASTA2 |
| pKP845 | MEK-pknB_ΔPASTA3 |
| pKP846 | MEK-pknB_ΔPASTA4 |
| pKP847 | MEK-pknB_W571A |
| pKP848 | MEK-pknB_K589A |
| pKP849 | MEK-pknB_N601A |
| pKP850 | MEK-pknB_F624A |
| pDE43-MEK | Mycobacterial replicating vector control |
FIGURE 5.
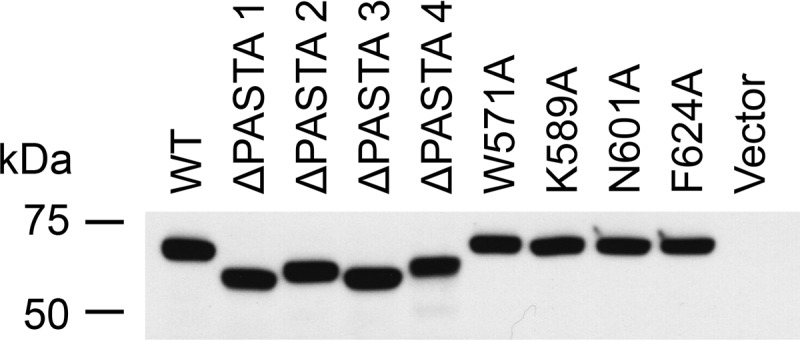
PASTA domain deletion constructs and point mutants are stably expressed in wild type M. smegmatis. Anti-myc Western blotting analysis of M. smegmatis lysates (40 μg of protein/lane) from cells expressing the indicated N-terminal myc-tagged PknB alleles shows comparable levels of expression of WT, four PASTA domain deletions, and PASTA4 point mutants.
Discussion
Serine/threonine protein kinases play central roles in shaping mycobacterial response to the environment and are critical for the survival and propagation of the bacterium within the host. In this work, we combined the x-ray crystallographic, bioinformatics, and mutational analysis to map the functional sites on the surface of the extracellular sensor domain of PknB, an essential mycobacterial kinase. We discovered a site in PASTA4 that was highly conserved among mycobacterial orthologs of PknB. Allele substitution experiments in Mtb demonstrated that residues lining the site are essential for PknB function in vivo. In addition, we found that each of the four PASTA domains is essential for the PknB function.
In our efforts to obtain an atomic model of PknB ECD we aimed to obtain multiple overlapping structures, an approach similar to the one used by Barthe et al. (14) to obtain the PknB ECD NMR structure. However, rather than avoiding overlap in chemical shifts, our motivation was obtaining diffraction quality crystals. Indeed, diffraction quality crystals of PASTA1–4 were never obtained, but we succeeded in crystallizing a number of shorter fragments. Unstructured N-terminal extensions in PASTA domain constructs are likely to inhibit crystallization of longer ECD fragments, because crystal packing of these narrow, extended structures necessitates crystal contacts near the two termini. Investigation of PASTA2–4, PASTA3–4, and PASTA1–2 models confirms this analysis. In each case the structured N-terminal residues lay within 5 Å of the symmetry-related molecule in the crystal contact.
The near atomic resolution model of PknB ECD presented here agrees well with the previous NMR structures of Mtb PknB (14) and x-ray crystal structures of the Staphylococcus aureus PrkC (10, 17). The constituent PASTA domains are stacked head to tail and are predicted to extend away from the cytoplasmic membrane into the periplasmic space. The linkers connecting the repeated units are short and well structured, suggesting that although the ECD can show some inter subunit flexibility it is likely to maintain this extended conformation. Residue Val-360 likely defines the beginning of the folded ECD, which is thus likely to be loosely tethered to the membrane by a six-residue peptide with amino acid sequence GGITRD. Similar arrangement of a well folded ECD tethered by a flexible linker were found during our prior investigations of PknD and PknH extracellular domains (18, 19), suggesting that ligand binding controls localization and oligomerization of the kinase domains in the membrane rather than directly signaling via conformational changes induced across the cytoplasmic membrane.
The PknB sensor domain comprises four PASTA repeats, the last of which, PASTA4, contains a hydrophobic groove with conserved partially exposed aromatic residues, a signature of a ligand-binding site. This site was bound to a molecule of citrate in PASTA2–4 structure (crystallized in presence of 50 mm citrate). Although citrate is not a physiological ligand, this observation strengthened our suspicion that the identified surface may contribute to ligand binding in vivo. Our alanine substitution experiments showed that three of the four residues lining this site are required for mycobacterial growth. Given that the PASTA domain composition of PknB homologs is genus specific (11), our findings can be extended to other mycobacteria with high confidence but cannot be immediately extended to all Firmicutes. Nonetheless, our approach of constructing genus-specific full-length alignments and testing highly conserved surface residues with alanine substitutions in vivo is broadly applicable.
Previous surface plasmon resonance data reported by Mir et al. (13) on the direct interaction of the ECD with muropeptides suggested a narrow peptidoglycan recognition specificity. Muramic acid, tripeptide l-Ala, iso-d-Glu, and meso-DAP, and amidation of carboxyl groups on iso-d-Glu and meso-DAP were necessary for optimal binding. To achieve such specificity, the peptidoglycan-binding site would need to be large (the optimal ligand and surface domain of PASTA areas are ∼1000 Å2 and 3000 Å2, respectively). The conserved putative ligand-binding site in PASTA4 is ∼530 Å2 and is consistent with this requirement. However, on the basis of the available evidence we cannot exclude that this site is used for interaction with a non-peptidoglycan ligand.
To further investigate the individual roles of the four PknB PASTA domains, we tested a series of internal PASTA domain deletions. Narrow peptidoglycan binding specificity demonstrated by Mir et al. (13) suggests that in Mtb the PknB PASTA domains have not evolved to recognize multiple peptidoglycan chemotypes. If the peptidoglycan-binding site was unique and limited to a single PASTA repeat, the role of other PASTA domains might have been redundant. Instead, we found that PASTA domains were each independently required for PknB function. Although we cannot exclude additional ligand binding functions in these domains, the lack of conserved binding sites on PASTA1–3 argue against this interpretation. Instead, these domains may be necessary to extend PASTA4 away from the membrane and/or promote the ligand-dependent dimerization that is necessary for regulation of the intracellular kinase domain. These structural insights should enable the in vivo functional assays and in vitro binding studies to definitively map the PknB peptidoglycan-binding site and understand the molecular basis of kinase regulation.
Experimental Procedures
Cloning, Expression, and Purification
PASTA domain constructs used for crystallization were cloned as glutathione S-transferase (GST)-tagged, TEV protease-cleavable fusions from the Mtb strain H37Rv genomic DNA into entry vector pDONR221 and then the expression vector pDEST15 via the Gateway system (Invitrogen) and expressed in Escherichia coli BL-21 Codon Plus cells by autoinduction (20). Cell pellets were resuspended in GSTA buffer (300 mm NaCl, 20 mm HEPES, pH 7.5, 0.5 mm tris(2-carboxyethyl)phosphine) and sonicated for 4 min (30 s on, 1 min off). Lysates were cleared by centrifugation at 15,000 rpm in Sorvall SS-34 rotor and filtered over 0.45-μm syringe filters. GST-tagged proteins were bound to glutathione affinity-Sepharose (GE Healthcare), washed, and eluted in GSTB (300 mm NaCl, 20 mm HEPES, pH 8, 0.5 mm tris(2-carboxyethyl)phosphine, 20 mm reduced glutathione). Following addition of TEV protease (1:50 ratio by protein weigh), samples were dialyzed against 2 liters of GSTA at room temperature overnight. Free GST and uncleaved fusion protein were rebound to glutathione-Sepharose and the flow-through fraction was concentrated and purified further by size exclusion chromatography over Superdex 75 resin (GE Healthcare).
Crystallization, Data Collection, and Analysis
PASTA4 domain constructs included a 3-amino acid N-terminal linker with sequence GHM and PknB residues 558–626. It was crystallized by vapor diffusion in a condition containing 0.2 m zinc acetate, 0.1 m imidazole, pH 8, and 20% PEG 3000. Crystals were cryoprotected with 20% xylitol in mother liquor, mounted, and frozen in liquid nitrogen. Data set at the zinc edge was collected at the peak wavelength 1.282 Å at Beamline 8.3.1 of the Advanced Light Source (21). Data were reduced using HKL2000 (22), and structure solution using single wavelength anomalous dispersion analysis, building, and refinement were carried out in Phenix (23) and Coot (24). Data collection and refinement statistics for this and the following structures are listed in Table 1.
PASTA3 L512M domain (N-terminal linker GHM and residues 491–558) was constructed by site-directed mutagenesis. Because the PASTA3 sequence lacks methionines, this mutation was used to introduce a methionine residue for phasing. Leucine 512 was chosen for mutation due to its similar size and its position within the hydrophobic core of the protein. Selenomethionine labeling was performed via the dedicated autoinduction protocol (20). The resulting protein crystallized from 0.2 m lithium sulfate, 0.1 m sodium acetate, pH 4.5, and 50% PEG 400. Data were collected at 1.116 Å and processed as described above. PASTA4 domain was used as a search model in molecular replacement.
PASTA3–4 (no linker, residues 491–626) crystallized from a condition containing 0.1 m sodium acetate, pH 4.5, 2 m ammonium sulfate. These crystals were cryoprotected in 3 m sodium malonate, pH 4.5. Data collection and processing were carried out as described above. PASTA3 and PASTA4 domains were used as search models in molecular replacement.
PASTA 1–2 construct (N-terminal linker G and residues 360–491) was crystallized from conditions containing 0.1 m CHES, pH 9.5, 30% isopropyl alcohol, 30% PEG 3350. This structure was solved by molecular replacement with PASTA2 from PASTA2–4 structure and PASTA4 as search models.
PASTA2–4 construct (no linker, residues 423–626) crystallized in 0.1 m citrate, pH 3.5, 25% PEG 3350. The structure could not be determined using molecular replacement with the PASTA3–4 search model likely due to the difference in inter-domain angles. Instead, multiple rounds of molecular replacement with individual PASTA domains as search models yielded phases that eventually produced an interpretable electron density map. Selection of partial solutions based on log likelihood gain scores allowed sequential identification of the six PASTA domains found in the asymmetric unit.
Analysis of Sequence and Surface Conservation
To accurately assess surface conservation patterns of PknB ECD, we gathered mycobacterial orthologs of PknB using a BLAST search (25) against the proteins from the genus Mycobacterium (Taxonomy ID 1763). The results were cut off at query coverage above 97%, resulting in a collection of 167 sequences ranging from 66 to 100% identity. The sequences were aligned using COBALT (26) and the alignment filtered to remove redundant sequences. The final alignment included 72 unique sequences of PknB orthologs and is included as supplemental File S1. Surface conservation was mapped onto the protein structures using Chimera (27) and sequence logos were generated using Skylign web server (28).
Design and Cloning of Point Mutants and Domain Deletions
To maximize the probability of proper folding of the PASTA internal deletion mutants, we performed structural alignment of PASTA domain junctions to select domain boundaries. ΔPASTA1 mutant was designed as a deletion of amino acids 359–425. Similarly, ΔPASTA2 was designed to exclude residues 426–493 and ΔPASTA3 residues 494–559. In each case the linker residues of the preceding domain were included to preserve their interaction with the preceding PASTA domain. These domain deletions were obtained in the Gateway donor vector pDO23A (Invitrogen) containing N terminally myc-tagged full-length PknB using a modification of site-directed mutagenesis protocol (Agilent Technologies). Deletion of PASTA4 was obtained in the same clone by introducing an early stop codon at amino acid 560 by site-directed mutagenesis.
Generation of an Mtb attBL5-pknB Strain and Vivo Allele Substitution
Because pknB is an essential gene in mycobacteria, a strain bearing a swappable allele at the chromosomal phage attachment site attBL5 was generated by transforming Mtb strain H37Rv with an attPL5 integrating StrR plasmid expressing a copy of the Mtb pknB gene. In this strain, the pknB gene at the attBL5 site was constitutively expressed from a weaker derivative of the Pmyc1tetO promoter (29). The native pknB locus (nucleotides 15,590–17,464 in the genome) was subsequently deleted from this merodiploid strain using phage Che9c-mediated recombineering by adopting the published strategies and protocols (30, 31). Deletion of the native pknB locus in the resultant pknB-attL5 strain was confirmed by amplifying and sequencing the 5′ and 3′ recombinant junctions in the chromosome and by performing the gene essentiality test by transforming with either a empty ZeoR attPL5 vector that resulted in no colonies, or with a ZeoR attPL5 plasmid expressing wild type pknB that resulted in viable colonies. These studies were performed essentially as described in Refs. 32 and 33. This functional test confirmed that the strain carried only the swappable allele of the WT pknB gene at the attBL5 site.
This ZeoR and KanS pknB strains (ΔpknB::hyg+pknB_attBL5-Zeo) were used as a parental strain in the in vivo allele substitution studies to assess the essentiality of the point mutations and truncations of PknB ECD expressed from a KanR attPL5 integrating plasmid (MCK). Gateway entry plasmids carrying the Pmyc1tetO promoter derivative, and the wild type, truncated, or mutant versions of pknB were cloned into the KanR attPL5 destination plasmid pDE43-MCK (34) using the LR recombinase reactions of the Gateway Cloning Technology (Invitrogen). The resultant KanR attPL5 plasmids expressing either the wild type pknB or the mutated alleles were electroporated into the competent cells of ZeoR pknB strain and plated on 7H10+Kan (25 μg/ml) to test in vivo essentiality of the point mutations and truncations of PknB ECD following complement switching at the attBL5 site. Expression of the mutant versions of PknB used in this study had no dominant negative effect when transformed into wild type M. tuberculosis that expresses pknB from its native chromosomal locus.
Overexpression and Western Blotting Analysis of PknB Alleles
For pknB overexpression, the N-terminal myc-tagged M. tuberculosis pknB wild type and the ECD deletion and point mutant alleles were cloned into the mycobacterial replicating expression plasmid pDE43-MEK and expressed from the same Pmyc1tetO promoter derivative used in the complement switching assay. Wild type Mycobacterium smegmatis harboring constructs were grown in 7H9 + kanamycin (25 μg/ml) to an A600 of 0.8 to 1.0. Bacteria were harvested by centrifugation, washed twice in 10% glycerol, and resuspended in PBS containing the cOmplete protease inhibitor (Roche Applied Science). Bacteria were lysed by bead beating in the presence of 0.1 mm silica beads in a FastPrep instrument (MPBio) at a speed setting of 6.5 for 4 × 30 s. The lysates were then incubated with 2.0% SDS solution for 30 min at 55 °C before centrifugation and collecting the cell lysates.
M. smegmatis lysates, each corresponding to 40 μg of protein quantified using the Pierce BCA protein kit (ThermoFisher Scientific) were subjected to electrophoresis through a 4–20% mini PROTEAN TGX gradient gel (Bio-Rad) under denaturing conditions and electroblotted onto a PVDF membrane. The blot was stained with Ponceau S solution (Boston Bioproducts) to confirm uniform loading and transfer of proteins across all lanes. The blot was blocked with 5% nonfat milk in TBST buffer (50 mm Tris-HCl, pH 7.4, 0.15 m NaCl, and 0.1% Tween 20), and probed first in 1:4000 dilution of the anti-myc monoclonal antibody clone 9E10 raised in mouse (Sigma M4439) and subsequently in 1:1000 dilution of the anti-mouse secondary antibodies conjugated to horseradish peroxidase (Abcam ab6728). Following TBST buffer washes, the myc-specific bands were detected using the Pierce ECL Plus chemiluminescent detection system (ThermoFisher Scientific).
Author Contributions
D. M. P., K. G. P., T. A., and C. M. S. conceived and coordinated the study and wrote the paper. D. M. P., C. E. B., A. M., and T. Y. C. designed, performed, and analyzed structure-determination experiments. K. G. P. and K. C. M. generated the attBL5-pknB Mtb strain and K. G. P. performed allelic substitution experiments. D. M. P., K. G. P., C. E. B., K. C. M., A. M., T. Y. C., and C. M. S. reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We are grateful to James Holton, George Meigs, and Jane Tanamachi for assistance with Beamline 8.3.1 at the Lawrence Berkeley National Laboratory Advanced Light Source. We thank Dirk Schnappinger for sharing the various mycobacterial promoter and expression plasmids used in this study. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the United States Department of Energy under Contract DE-AC02–05CH11231.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM70962 (to T. A.), P01 AI068135 (to T. A. and C. M. S.), and AI064282 (to C. M. S.), and the Bill and Melinda Gates Foundation. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental File S1.
- STPK
- serine/threonine protein kinase
- TEV
- tobacco etch virus
- PASTA domain
- penicillin-binding protein and serine/threonine kinase associated
- ECD
- extracellular domain
- CHES
- 2-(cyclohexylamino)ethanesulfonic acid.
References
- 1. Chao J., Wong D., Zheng X., Poirier V., Bach H., Hmama Z., and Av-Gay Y. (2010) Protein kinase and phosphatase signaling in Mycobacterium tuberculosis physiology and pathogenesis. Biochim. Biophys. Acta 1804, 620–627 [DOI] [PubMed] [Google Scholar]
- 2. Alber T. (2009) Signaling mechanisms of the Mycobacterium tuberculosis receptor Ser/Thr protein kinases. Curr. Opin. Struct. Biol. 19, 650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mieczkowski C., Iavarone A. T., and Alber T. (2008) Auto-activation mechanism of the Mycobacterium tuberculosis PknB receptor Ser/Thr kinase. EMBO J. 27, 3186–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Molle V., and Kremer L. (2010) Division and cell envelope regulation by Ser/Thr phosphorylation: Mycobacterium shows the way. Mol. Microbiol. 75, 1064–1077 [DOI] [PubMed] [Google Scholar]
- 5. Gee C. L., Papavinasasundaram K. G., Blair S. R., Baer C. E., Falick A. M., King D. S., Griffin J. E., Venghatakrishnan H., Zukauskas A., Wei J. R., Dhiman R. K., Crick D. C., Rubin E. J., Sassetti C. M., and Alber T. (2012) A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria. Sci. Signal. 5, ra7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Echenique J., Kadioglu A., Romao S., Andrew P. W., and Trombe M. C. (2004) Protein serine/threonine kinase StkP positively controls virulence and competence in Streptococcus pneumoniae. Infect. Immun. 72, 2434–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Débarbouillé M., Dramsi S., Dussurget O., Nahori M. A., Vaganay E., Jouvion G., Cozzone A., Msadek T., and Duclos B. (2009) Characterization of a serine/threonine kinase involved in virulence of Staphylococcus aureus. J. Bacteriol. 191, 4070–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shah I. M., Laaberki M. H., Popham D. L., and Dworkin J. (2008) A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135, 486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fiuza M., Canova M. J., Zanella-Cléon I., Becchi M., Cozzone A. J., Mateos L. M., Kremer L., Gil J. A., and Molle V. (2008) From the characterization of the four serine/threonine protein kinases (PknA/B/G/L) of Corynebacterium glutamicum toward the role of PknA and PknB in cell division. J. Biol. Chem. 283, 18099–18112 [DOI] [PubMed] [Google Scholar]
- 10. Ruggiero A., Squeglia F., Marasco D., Marchetti R., Molinaro A., and Berisio R. (2011) X-ray structural studies of the entire extracellular region of the serine/threonine kinase PrkC from Staphylococcus aureus. Biochem. J. 435, 33–41 [DOI] [PubMed] [Google Scholar]
- 11. Jones G., and Dyson P. (2006) Evolution of transmembrane protein kinases implicated in coordinating remodeling of Gram-positive peptidoglycan: inside versus outside. J. Bacteriol. 188, 7470–7476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kang C. M., Abbott D. W., Park S. T., Dascher C. C., Cantley L. C., and Husson R. N. (2005) The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 19, 1692–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mir M., Asong J., Li X., Cardot J., Boons G. J., and Husson R. N. (2011) The extracytoplasmic domain of the Mycobacterium tuberculosis Ser/Thr kinase PknB binds specific muropeptides and is required for PknB localization. PLoS Pathog. 7, e1002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barthe P., Mukamolova G. V., Roumestand C., and Cohen-Gonsaud M. (2010) The structure of PknB extracellular PASTA domain from Mycobacterium tuberculosis suggests a ligand-dependent kinase activation. Structure 18, 606–615 [DOI] [PubMed] [Google Scholar]
- 15. Chawla Y., Upadhyay S., Khan S., Nagarajan S. N., Forti F., and Nandicoori V. K. (2014) Protein kinase B (PknB) of Mycobacterium tuberculosis is essential for growth of the pathogen in vitro as well as for survival within the host. J. Biol. Chem. 289, 13858–13875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Turapov O., Loraine J., Jenkins C. H., Barthe P., McFeely D., Forti F., Ghisotti D., Hesek D., Lee M., Bottrill A. R., Vollmer W., Mobashery S., Cohen-Gonsaud M., and Mukamolova G. V. (2015) The external PASTA domain of the essential serine/threonine protein kinase PknB regulates mycobacterial growth. Open Biol 5, 150025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paracuellos P., Ballandras A., Robert X., Kahn R., Hervé M., Mengin-Lecreulx D., Cozzone A. J., Duclos B., and Gouet P. (2010) The extended conformation of the 2.9-Å crystal structure of the three-PASTA domain of a Ser/Thr kinase from the human pathogen Staphylococcus aureus. J. Mol. Biol. 404, 847–858 [DOI] [PubMed] [Google Scholar]
- 18. Good M. C., Greenstein A. E., Young T. A., Ng H. L., and Alber T. (2004) Sensor domain of the Mycobacterium tuberculosis receptor Ser/Thr protein kinase, PknD, forms a highly symmetric β propeller. J. Mol. Biol. 339, 459–469 [DOI] [PubMed] [Google Scholar]
- 19. Cavazos A., Prigozhin D. M., and Alber T. (2012) Structure of the sensor domain of Mycobacterium tuberculosis PknH receptor kinase reveals a conserved binding cleft. J. Mol. Biol. 422, 488–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Studier F. W. (2005) Protein production by autoinduction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 21. MacDowell A. A., Celestre R. S., Howells M., McKinney W., Krupnick J., Cambie D., Domning E. E., Duarte R. M., Kelez N., Plate D. W., Cork C. W., Earnest T. N., Dickert J., Meigs G., Ralston C., et al. (2004) Suite of three protein crystallography beamlines with single superconducting bend magnet as the source. J. Synchrotron Radiat. 11, 447–455 [DOI] [PubMed] [Google Scholar]
- 22. Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 23. Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., and Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Papadopoulos J. S., and Agarwala R. (2007) COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079 [DOI] [PubMed] [Google Scholar]
- 27. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 28. Wheeler T. J., Clements J., and Finn R. D. (2014) Skylign: a tool for creating informative, interactive logos representing sequence alignments and profile hidden Markov models. BMC Bioinformatics 15, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ehrt S., Guo X. V., Hickey C. M., Ryou M., Monteleone M., Riley L. W., and Schnappinger D. (2005) Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van Kessel J. C., and Hatfull G. F. (2007) Recombineering in Mycobacterium tuberculosis. Nat. Methods 4, 147–152 [DOI] [PubMed] [Google Scholar]
- 31. Murphy K. C., Papavinasasundaram K., and Sassetti C. M. (2015) Mycobacterial recombineering. Methods Mol. Biol. 1285, 177–199 [DOI] [PubMed] [Google Scholar]
- 32. Brown A. C. (2009) Gene switching and essentiality testing. Methods Mol. Biol. 465, 337–352 [DOI] [PubMed] [Google Scholar]
- 33. Pashley C. A., and Parish T. (2003) Efficient switching of mycobacteriophage L5-based integrating plasmids in Mycobacterium tuberculosis. FEMS Microbiol. Lett. 229, 211–215 [DOI] [PubMed] [Google Scholar]
- 34. Schnappinger D., O'Brien K. M., and Ehrt S. (2015) Construction of conditional knockdown mutants in mycobacteria. Methods Mol. Biol. 1285, 151–175 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.