Abstract
Bacterial endotoxin can induce inflammatory and metabolic changes in the host. In this study, we revealed a molecular mechanism by which a stress-inducible, liver-enriched transcription factor, cAMP-responsive element-binding protein hepatic-specific (CREBH), modulates lipid profiles to protect the liver from injuries upon the bacterial endotoxin lipopolysaccharide (LPS). LPS challenge can activate CREBH in mouse liver tissues in a toll-like receptor (TLR)/MyD88-dependent manner. Upon LPS challenge, CREBH interacts with TNF receptor-associated factor 6 (TRAF6), an E3 ubiquitin ligase that functions as a key mediator of TLR signaling, and this interaction relies on MyD88. Further analysis demonstrated that TRAF6 mediates K63-linked ubiquitination of CREBH to facilitate CREBH cleavage and activation. CREBH directly activates expression of the gene encoding Apolipoprotein A4 (ApoA4) under LPS challenge, leading to modulation of high-density lipoprotein (HDL) in animals. CREBH deficiency led to reduced production of circulating HDL and increased liver damage upon high-dose LPS challenge. Therefore, TLR/MyD88-dependent, TRAF6-facilitated CREBH activation represents a mammalian hepatic defense response to bacterial endotoxin by modulating HDL.
Keywords: cAMP response element-binding protein (CREB), endotoxin, gene transcription, high-density lipoprotein (HDL), toll-like receptor (TLR), ubiquitylation (ubiquitination)
Introduction
Bacterial infection can induce a variety of physiological changes in the host. The response of the host to bacterial infection includes not only inflammatory changes but also comprises metabolic changes in the host. Lipopolysaccharide (LPS),2 one of the key component of bacterial cell wall produced by exogenous bacterial infection or physiologically from small intestines, is capable of triggering host metabolic alterations (1, 2). Bacterial infections and inflammation can lead to hyperlipidemia (2). Bacterial infection increases triglyceride (TG) levels due to the increase in lipoprotein production as well as defective clearance of lipoproteins from circulation (3). The metabolic disorder is one of the hallmarks of a patient with sepsis. Plasma lipoproteins, especially HDL, are markedly reduced during sepsis, and a number of clinical studies have pointed out that low plasma HDL is a prognostic factor in severe sepsis (4, 5). It has been established that HDL plays a protective role in bacterial infection by scavenging circulatory levels of LPS (6, 7). Hence, enhancing HDL levels in serum can be an attractive strategy to counter the bacterial infection in patients.
Previously we have identified an endoplasmic reticulum (ER) anchored liver-specific transcription factor, named CREBH (cAMP-responsive element-binding protein, hepatic specific), which can be activated by ER stress, inflammatory stimuli, or metabolic signals (8, 9). Under inflammatory challenge, metabolic stress or circadian regulation (9–11), CREBH is released from ER membrane by Regulated Intramembrane Proteolysis (RIP) and translocates to Golgi where it undergoes proteolytic cleavage by S1P and S2P proteases to release its activated form. Activated CREBH functions as a potent transcription factor that induces expression of the genes involved in hepatic acute-phase response, lipogenesis, fatty acid (FA) oxidation, lipolysis, and gluconeogenesis (8–10, 12, 13). Therefore, CREBH is critical to the maintenance of energy homeostasis under stress conditions. However, whether CREBH plays a role in protecting the liver from injuries associated with acute inflammation or endotoxins remained to be elucidated.
In this study, we demonstrate the role and molecular mechanism through which CREBH acts as a key player in mounting an acute-phase response by modulating the apolipoprotein ApoA4, the key component of HDL cholesterol particles, in response to the bacterial endotoxin LPS. We found that the E3 ligase TRAF6 interacts with CREBH, under the regulation of TLR signaling, to facilitate CREBH cleavage and activation through an ubiquitination modification process. Functionally, activated CREBH, under LPS stimulation, regulates expression of the ApoA4 gene and therefore promotes production of HDL as a part of the host response to the challenge of bacterial infection. Our study revealed a novel mammalian hepatic defense response to bacterial endotoxin by modulating plasma lipid profiles.
Results
LPS Induces CREBH Cleavage in the Liver in a TLR/MyD88-dependent Manner
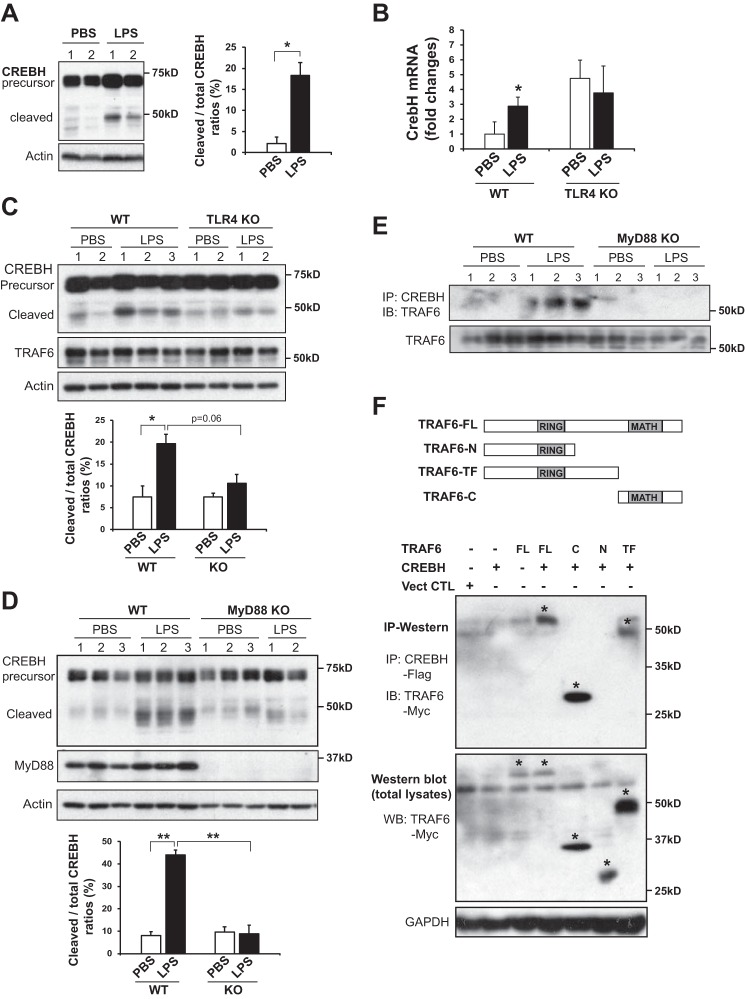
To validate the effect of LPS challenge in CREBH activation, wild-type C57BL/6J mice were injected intraperitoneally with LPS (2 μg/gm of body weight) for 18 h to induce acute endotoxemia. After LPS injection, levels of CREBH precursor and the cleaved/activated form of CREBH were increased in the livers of mice after LPS challenge (Fig. 1A). To understand LPS-induced activation of CREBH in the liver, we evaluated the presence of TLR4, the receptor for LPS, in mouse liver primary hepatocytes since the presence of TLR receptors on hepatocytes is a subject of debate (14). Semi-quantitative PCR analysis indicated that mouse primary hepatocytes express TLR2 and TLR3 as well as low levels of TLR4 (supplemental Fig. S1A). Further, primary hepatocytes produced higher levels of both CREBH precursor and activated CREBH upon LPS challenge, compared with primary hepatocyte challenged with PBS (supplemental Fig. S1B), thus confirming the response of primary hepatocytes to LPS challenge in CREBH induction. To elucidate the regulatory mechanism for CREBH activity in the liver in response to LPS, the TLR4 ligand, we first examined induction of the CrebH mRNA in the livers of WT and TLR4 knock-out (TLR4−/−) mice. Consistent with the elevation of total CREBH protein in the liver or primary hepatocytes under LPS treatment, expression levels of the CrebH mRNA were increased in the WT liver under LPS challenge (Fig. 1B). While the basal levels of the CrebH mRNA in the TLR4−/− liver were elevated, LPS treatment failed to further increase the CrebH mRNA transcripts in the absence of TLR4. These results suggest that TLR4 unlikely plays a dominant role in regulating CrebH gene transcription, although TLR4 was required to augment CrebH mRNA expression upon LPS challenge (Fig. 1B). The elevation in basal levels of CrebH mRNA in TLR4−/− mice may be due to alterative pathways triggered by TLR4 defect as an adaption or feedback regulation of CrebH transcription in TLR4−/− mice.
FIGURE 1.
LPS activates CREBH in the liver through the TLR4-MyD88-TRAF6 regulatory axis. A, Western blotting analysis of CREBH cleavage in the liver tissues from wild-type mice injected with either PBS (vehicle control) or LPS (2 μg/gm body weight) for 18 h. Western blotting analysis was performed to detect CREBH cleavage by using the anti-CREBH polyclonal antibody. Levels of β-actin were determined as loading controls. The graph on the right side shows the ratios (%) of the activated form versus total CREBH protein signals in the livers of mice challenged by LPS or PBS vehicle. The cleaved and total CREBH protein signals, determined by Western blot densitometry, were normalized to that of β-actin. Each bar denotes the mean ± S.E. *, p < 0.05. B, qRT-PCR analysis of CrebH mRNA in the livers of WT or TLR4−/− mice injected with LPS (2 μg/gm body weight) or PBS for 18 h. Expression values were normalized to β-actin mRNA levels. Fold changes of mRNA levels are shown by comparing to one of the control mice treated with PBS. Each bar denotes the mean ± S.E. (n = 3 mice per group). *, p < 0.05. C, Western blotting analysis of CREBH, TRAF6, and β-actin in the livers of WT or TLR4−/− mice injected with LPS (2 μg/gm body weight) or PBS for 18 h. The graph below the image shows the ratios (%) of the activated form versus total CREBH protein signals, determined by Western blot densitometry, in the livers of WT and TLR4−/− mice under PBS or LPS challenge. D, Western blotting analysis of CREBH, MyD88, and β-actin in the livers of WT or MyD88−/− mice injected with LPS (2 μg/gm body weight) or PBS for 18 h. The graph below the image shows the ratios (%) of the activated form versus total CREBH protein signals, determined by Western blot densitometry, in the livers of WT and MyD88−/− mice under PBS or LPS challenge. Each bar denotes the mean ± S.E. (n = 3 mice per group). *, p < 0.05; **, p < 0.01. E, IP-Western blotting analysis of CREBH and TRAF6 interaction in liver tissues from WT and MyD88−/− mice challenged with either PBS or LPS (2 μg/gm body weight) for 18 h. Mouse liver protein lysates was immunoprecipitated with the polyclonal anti-CREBH antibody, followed by immunoblotting with the anti-TRAF6 antibody. As a control, Western blotting analysis was performed to determine the levels of TRAF6 in liver tissues of WT and MyD88−/− mice. F, schematic representation of TRAF6 and its truncated mutants. TRAF6 carries an N-terminal RING finger domain and a C-terminal MATH domain (top panel). FL: full-length structure, N: N-terminal RING finger domain, TF: trans-membrane domain, C: C-terminal MATH domain. IP-Western blotting analysis of CREBH interactions with different TRAF6 variants. Flag-tagged CREBH and Myc-tagged TRAF6 plasmids were co-transfected into HEK293T cells. CREBH protein in the lysates of transfected cells was immunoprecipitated with an anti-Flag antibody, followed by Western blotting using an anti-Myc antibody, The expression of the full-length and truncated TRAF6 proteins were indicated by the symbol *. Levels of TRAF6 and GAPDH in total cellular protein lysates were determined by Western blotting analysis.
Next, we examined activation of CREBH protein in the liver of TLR4−/− mice in response to LPS. The levels of the cleaved/activated form of CREBH were reduced in the TLR4−/− liver, compared with that in WT control mice, after 18-hour LPS challenge (Fig. 1C). Myd88 is an essential signal transduction adaptor protein used by almost all TLRs to mediate the downstream TLR signaling pathways. To further elucidate the involvement of TLR signaling pathway in CREBH activation, we examined CREBH expression and cleavage in the liver of MyD88-null (MyD88−/−) mice. Upon LPS challenge, levels of the cleaved/activated CREBH protein in the liver of MyD88−/− mice were significantly reduced, compared with those in the WT control mice (Fig. 1D), suggesting that LPS-induced CREBH activation relies on TLR signaling through MyD88.
LPS Stimulates CREBH-TRAF6 Interaction and CREBH Cleavage in a TLR-dependent Manner
To address the mechanism underlying TLR-associated CREBH activation under LPS challenge, we examined the potential interaction between CREBH and TLR signaling components. Previously, we demonstrated that TRAF6, an E3 ubiquitin-protein ligase that mediate signal transduction from TLRs and TNF receptors (15), can interact with the ER-resident stress sensor IRE1α to facilitate pro-inflammatory response (16). We tested the potential interaction between CREBH and TRAF6 in mouse liver tissues upon LPS challenge. In response to LPS, endogenous CREBH interacts with TRAF6 in the livers of WT mice, but not MyD88−/− mice (Fig. 1E), indicating that LPS induces CREBH-TRAF6 interaction in the liver in a MyD88-dependent manner.
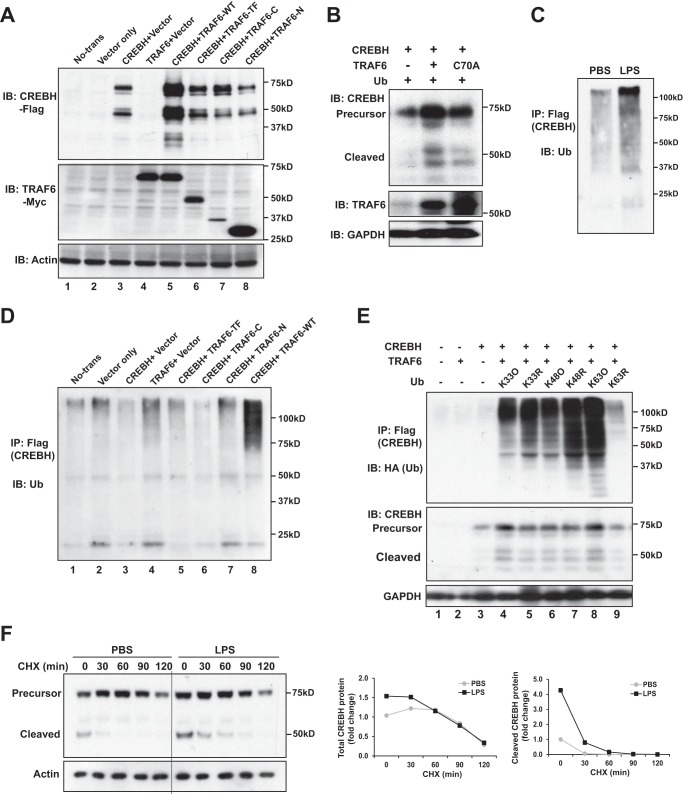
To delineate the regulatory mechanism by which TRAF6 interacts with CREBH, we investigated CREBH-TRAF6 interaction and CREBH cleavage in HEK293T cells transiently expressing CREBH and TRAF6 variants. To map the interaction domains, we generated truncated mutations for TRAF6 protein, including TRAF6 trans-membrane domain (TF), N-terminal RING finger domain (N), and C-terminal meprin-associated TRAF homology (MATH) domain (C) (Fig. 1F). IP-Western blotting analysis with the cells co-expressing CREBH and TRAF6 variants revealed that the C-terminal MATH and its proximal linker domains of TRAF6 are required for the interaction between CREBH and TRAF6 (Fig. 1F). Importantly, immunoblotting analysis indicated that expression of wild-type TRAF6 increased the levels of both total and cleaved CREBH proteins (Fig. 2A, lane 5; supplemental Fig. 1C). However, deletion of the TRAF6-MATH domain, which abolished the interaction between TRAF6 and CREBH, led to a significant reduction in CREBH cleavage (Fig. 2A, lanes 6, 8). Moreover, deletion of the N-terminal RING finger domain of TRAF6 significantly decreased CREBH cleavage (Fig. 2A, lane 7), although this deletion did not abolish the interaction between CREBH and TRAF6 (Fig. 1F). These observations implicated a critical role of the RING finger domain, where the TRAF6 E3 ligase activity resides, in CREBH cleavage. Supporting this notion, expression of TRAF6 with an E3 ligase catalytic-inactive mutation (C70A) failed to promote CREBH cleavage (Fig. 2B). Together, these results suggested that TRAF6 plays an important role in LPS-induced CREBH activation process possibly through TRAF6-CREBH interaction and the E3 ligase activity of TRAF6.
FIGURE 2.
TRAF6-mediates K63-linked ubiquitination of CREBH and promotes CREBH activation upon LPS treatment. A, HEK293T cells were co-transfected with the plasmid vectors expressing Flag-tagged full-length CREBH and different TRAF6 variants, including the full-length TRAF6 (TRAF6-WT), the truncated TRAF6 containing the trans-membrane domain (TRAF6-TF), the truncated TRAF6 containing the C-terminal MATH domain (TRAF6-C), and the truncated TRAF6 containing the N-terminal RING finger domain (TRAF6-N). Western blotting analyses were performed to determine the levels of CREBH, TARF6, and β-actin. B, HEK293T cells were co-transfected with the plasmid vectors expressing CREBH, Ub, and TRAF6 or its mutant C70A. Levels of CREBH, TRAF6, and GAPDH were analyzed by Western blotting analysis. C, IP-Western blotting analysis of CREBH ubiquitination in Huh7 cells infected with adenovirus expressing Flag-tagged human full-length CREBH. The Huh7 cells expressing full-length CREBH were treated with LPS (1 μg/ml) for 4 h. The Huh7 cellular lysates were immunoprecipitated with the anti-Flag antibody, followed by Western blotting using the anti-Ub antibody. D, IP-Western blotting analysis of CREBH ubiquitination in HEK293T cells co-expressing Flag-tagged full-length CREBH and different TRAF6 variants as described in panel A. The cellular lysates were immunoprecipitated with the anti-Flag antibody, followed by Western blotting analysis using the anti-Ub antibody. The different TRAF6 truncated mutants were described in Fig. 3A. E, HEK 293 T cells were co-transfected with Flag-tagged human full-length CREBH, TRAF6, and HA-tagged specific ubiquitin expression plasmids, including K33 only (K33O), K63 only (K63O), K48 only (K48O), K33 mutant (K33R), K48 mutant (K48R), and K63 mutant (K63R). Ubiquitination was determined by immunoprecipitation using the anti-Flag antibody and Western blotting analysis using the anti-HA antibody. Levels of CREBH and GAPDH were determined by Western blotting analysis. F, protein stability of CREBH was determined by the protein half-life examination after cycloheximide (CHX) (100 μm) was added. Huh7 cells infected with adenovirus expressing Flag-tagged human CREBH. At 48 h postinfection, the cells were treatment with LPS (1 μg/ml) for 4 h followed by adding CHX into the culture medium for the indicated time periods. CREBH protein levels were assessed by Western blotting analysis. The graphs on the right shows the quantification of total CREBH (precursor plus cleaved form) and cleaved CREBH protein signals in the PBS- or LPS-treated cells under the CHX treatment. The CREBH protein signals, determined by Western blot densitometry, were normalized to that of β-actin. Fold changes of CREBH protein levels are determined by comparing to that of PBS-treated cells under 0 min of CHX treatment (defined as 1).
TRAF6 Mediates the Ubiquitination of CREBH and Facilitates CREBH Cleavage upon LPS Challenge
Next, we determined the molecular mechanism by which TRAF6 regulates CREBH cleavage and activation under LPS challenge. Our recent study showed that TRAF6 can interact with the ER stress sensor IRE1α, leading to IRE1α ubiquitination and activation in macrophages (16). As shown in Fig. 2, A and B, deletion or mutation of the E3 ubiquitin protein ligase activity of TRAF6 significantly decreased CREBH cleavage. Therefore, we hypothesized that TRAF6 may mediate CREBH ubiquitination. Supporting this hypothesis, LPS challenge increased ubiquitination of CREBH in Huh7 cells expressing full-length CREBH (Fig. 2C). Furthermore, overexpression of TRAF6 induced a dramatic increase in CREBH ubiquitination in HEK293T cells transiently expressing Flag-tagged, full-length CREBH (Fig. 2D, lane 8). However, when the TRAF6 truncated mutants, including TRAF6 trans-membrane domain (TF), N-terminal RING finger domain (N), and C-terminal MATH domain (C), were co-expressed, levels of CREBH ubiquitination were significantly reduced, compared with that under wild-type TRAF6 expression (Fig. 2D, lanes 5–7), thus confirming the role of TRAF6 in mediating CREBH ubiquitination.
TRAF6 Mediates K63-linked Ubiquitination of CREBH and Stabilizes CREBH Proteins at the Early Time Points of LPS Stimulation
TRAF6 is known to catalyze K63-linked polyubiquitin conjugation onto its substrates (17, 18). To characterize TRAF6-mediated ubiquitination of CREBH, we co-expressed CREBH with a mutant ubiquitin isoform that carries a single lysine residue at the position 63 (K63O), 48 (K48O), or 33 (K33O) in HEK293T cells. The K63O, K48O, or K33O ubiquitin mutant carries a single lysine residue, which allow us to determine the topology of polyubiquitin chains. Expression of K63O led to the highest levels of CREBH ubiquitination, compared with expression of the other Ub variants (Fig. 2E, lane 8). In contrast, when K48O or K33O was expressed, lower levels of CREBH ubiquitination were detected, compared with that in the cells expressing K63O (Fig. 2E). To further delineate the type of polyubiquitination occurring on CREBH, we co-expressed CREBH and one of three different Ub mutants, including Ub in which lysine 48 was mutated to arginine (R or Arg) but all other lysine residues were intact (K48R), lysine 63 was mutated to Arg but all other lysine residues were intact (K63R), and lysine 33 was mutated to Arg but all other lysine residues were intact (K33R), in HEK293T cells. IP-Western blotting analysis indicated that expression of K63R diminished CREBH ubiquitination while expression of K48R or K33R did not affect CREBH ubiquitination (Fig. 2E, lanes 5, 7, 9). In fact, expression of K48R mutant increased levels of CREBH ubiquitination, possibly due to the stimulation of other forms of ubiquitination by the defect of K48-linked Ub. Together, these data suggested that TRAF6 mediates K63-linked, but not K48- or K33-linked, ubiquitination of CREBH.
Distinct from K48-linked ubiquitination, K63-linked ubiquitination stabilizes proteins and regulates the activities of target proteins by functioning as a prerequisite signal for subsequent modification or serving as a docking site for recruiting other proteins (18–20). As LPS can induce TRAF6-medaited, K63-linked CREBH ubiquitination, we tested whether LPS treatment can modulate CREBH protein stability. For this purpose, we determined CREBH half-life in hepatocytes under LPS or PBS treatment through cycloheximide (CHX) chase assay. The stability of CREBH protein under the LPS treatment, reflected by the levels of the cleaved/activated CREBH or total CREBH proteins, was enhanced at the early time point, within 60 min after the CHX treatment, compared with that under PBS treatment (Fig. 2F). In particular, at the early time window post the CHX treatment, the levels of cleaved/activated CREBH protein in the LPS-treated cells were significantly higher than those in PBS-treated cells (Fig. 2F). These data suggested that LPS treatment may stabilize CREBH protein, possibly through K63-linked ubiquitination, and lead to increased levels of activated CREBH upon acute LPS challenge.
CREBH Regulates HDL Levels by Activating Expression of the ApoA4 Gene upon LPS Challenge
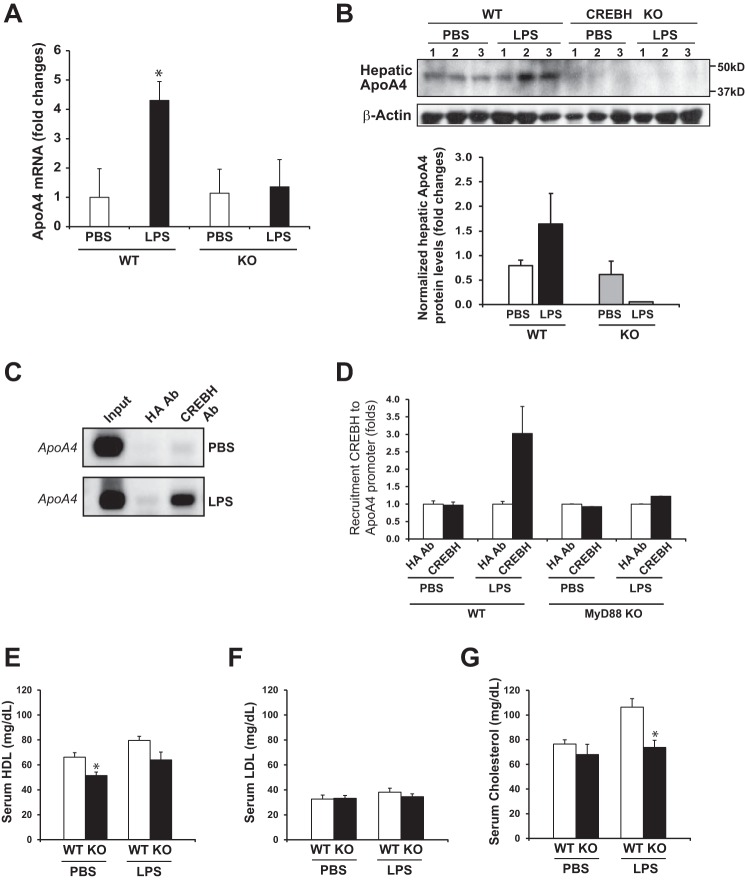
Previously, CREBH was shown to be associated with activating the genes involved in acute phase response (10) and lipid metabolism (8, 9). To define the CREBH target genes in the liver under LPS treatment, we performed quantitative real-time PCR (qPCR) analysis of genes involved in inflammation and lipid metabolism in the livers of mice challenged with LPS. Expression of the gene encoding serum amyloid protein (SAP) was decreased in CREBH−/− livers, compared with that in WT livers, upon LPS challenge (supplemental Fig. S2A), consistent with our previous study with CREBH-knockdown animals (10). Expression of the genes encoding the pro-inflammatory cytokine IL6 and the chemokines CCL2 and RANTES were also decreased in CREBH−/− livers under the LPS treatment (supplemental Fig. S2B). Furthermore, expression of the metabolic genes that were previously identified to be regulated by CREBH (9), such as ApoA4, ApoA5, ApoC2, and ApoB, was altered in response to LPS (supplemental Fig. S2B). However, expression of these genes, except ApoA4, was only marginally affected by CREBH deletion under LPS challenge. As for ApoA4, the mRNA levels in CREBH−/− liver were lower than that in WT liver. Consistently, expression levels of the ApoA4 mRNA in primary hepatocytes isolated from CREBH−/− mice were significantly reduced, compared with those in WT mouse primary hepatocytes (Fig. 3A). Further, at the protein level, while hepatic ApoA4 levels were elevated in WT mice treated with LPS, expression of hepatic ApoA4 in CREBH−/− mice was diminished (Fig. 3B).
FIGURE 3.
CREBH regulates expression of ApoA4 upon LPS treatment. A, qPCR analysis of expression levels of ApoA4 mRNA in primary hepatocytes isolated from WT and CREBH−/− mice. The primary hepatocytes were treated with LPS (100 ng//ml) or PBS for 4 h before total RNA was prepared from the cells. Each bar denotes the mean ± S.E. (n = 3). *, p < 0.05. B, Western blotting analysis of ApoA4 protein levels in the livers of WT and CREBH−/− mice injected with LPS (2 μg/gm body weight) or PBS for 18 h. Levels of β-actin were determined as the loading controls. The graph below the image shows the quantification of ApoA4 protein signals in the livers of WT or CREBH−/− mice injected with LPS or PBS. The ApoA4 protein signals, determined by Western blot densitometry, were normalized to that of β-actin. Fold changes of ApoA4 protein levels are determined by comparing to that in the PBS-treated WT mouse liver. Each bar denotes the mean ± S.E. (n = 3 mice per group). C, ChIP analysis of CREBH recruitment to the ApoA4 gene promoter. The WT mice were injected with LPS (2 μg/gm body weight) or PBS for 18 h. Chromatin isolated from the LPS- or PBS-challenged WT mouse liver tissues were subjected to immuoprecipitation with the anti-CREBH or anti-HA antibody. Semi-quantitative PCR analysis was performed to identify potential CREBH-binding regions in the ApoA4 promoter. ChIP with the anti-HA antibody was included as a control (HA Ab). The PCR reactions with the genomic DNA isolated from sonicated cell lysates were included as positive controls. D, ChIP analysis of CREBH recruitment to the ApoA4 gene promoter in WT and MyD88−/− mice after LPS or PBS challenge. Quantification of CREBH enrichment (fold changes) in the ApoA4 gene promoter was determined by comparing ChIP-qPCR signals from the samples pulled down by the anti-CREBH antibody to that pulled down by a rabbit anti-HA antibody. Each bar donates mean ± S.E. (n = 3 mice per group). E–G, serum levels of HDL, LDL, and total cholesterols in CREBH−/− and WT control mice after LPS (2 μg/gm body weight) or PBS challenge for 18 h. Each bar denotes the mean ± S.E. (n = 5). *, p < 0.05.
To determine whether CREBH, as a transcription factor, can directly target on the ApoA4 gene, we performed chromatin immunoprecipitation (ChIP) analysis to test the potential of CREBH binding to the promoter region of the ApoA4 gene. ChIP analysis indicated that endogenous CREBH strongly bound to the ApoA4 gene promoter in the liver tissues of mice challenged with LPS (Fig. 3C). Importantly, in the livers of MyD88−/− mice, the binding activity of CREBH to the ApoA4 gene promoter was diminished under the LPS treatment (Fig. 3D), suggesting that TLR/MyD88-mediated CREBH activation is necessary for ApoA4 gene transcriptional activation under LPS challenge. Additionally, we confirmed the role of CREBH in transcriptional activation of the ApoA4 gene by reporter assays. CREBH can activate transcription of the ApoA4 gene in the Huh7 cells co-expressing full-length CREBH and luciferase reporter driven by the ApoA4 gene promoter (supplemental Fig. S3A). When TRAF6 was co-expressed with the full-length CREBH, an increase in the ApoA4 reporter activity was observed, compared with expression of CREBH alone (supplemental Fig. S3B), thus confirming the role of TLR signaling through TRAF6 in facilitating CREBH transcriptional activity.
ApoA4 is the key component of HDL cholesterols (21). Since CREBH was shown to regulate ApoA4 expression, we determined the levels of serum cholesterols, including HDL and LDL, in CREBH−/− mice. Upon LPS challenge, levels of serum HDL and total cholesterols in the WT control mice were increased (Fig. 3, E–G). In contrast, the levels of serum HDL and total cholesterol in CREBH−/− mice were lower than those of WT mice, which is consistent with the role of CREBH in regulating ApoA4 expression. ApoA4 has been ascribed an acute-phase responsive protein under endotoxin injection (22). The decreased levels of HDL in CREBH−/− mice confirmed the role of CREBH in scavenging circulating endotoxins by regulating HDL production.
CREBH Plays an Important Role in Protecting the Liver from Endotoxin-induced Damage
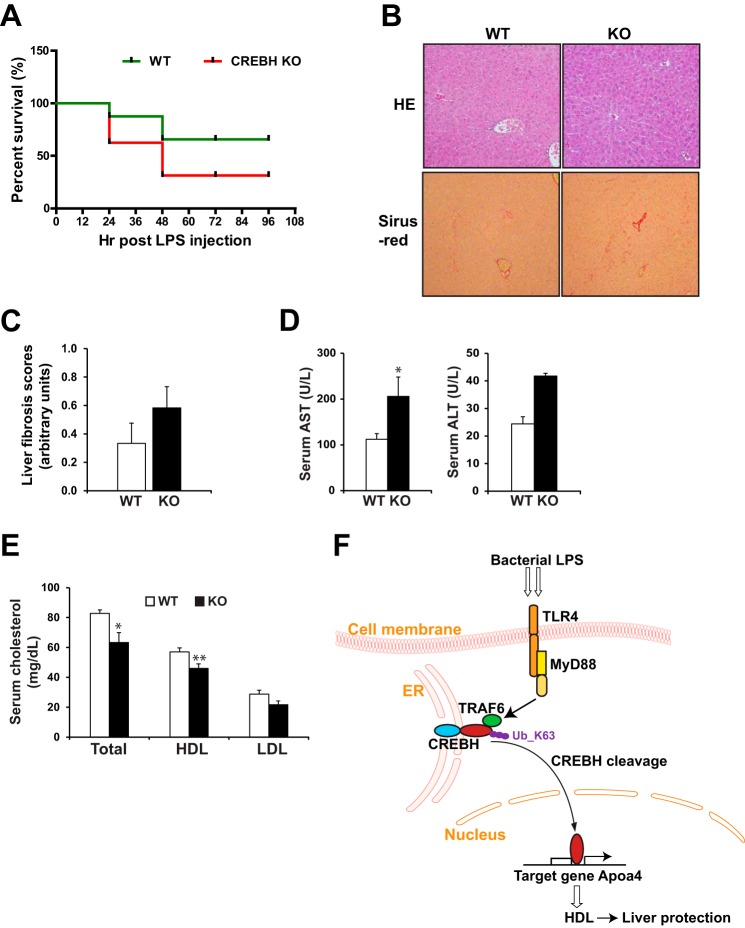
High-dose LPS challenge to the animals can cause endotoxemia or septic shock, a condition characterized by pronounced immune response, tissue or organ damage, and possibly death (23, 24). To determine the pathophysiological significance of CREBH-mediated regulation of HDL upon endotoxin challenges, we challenged CREBH−/− and WT control mice with high-dose LPS. CREBH−/− and WT control mice were administrated with intraperitoneal injection of LPS at 20 μg/gram body weight for up to 96 h. Under the LPS-induced endotoxemia, CREBH−/− and WT animals were euthanized at the end point of moribund state that reflects a point at which the animals will not recover. The criteria for end point determination was based on body posture, body hygiene, skin dryness, eyes and head position, cage movement, food intake, body weight, and body temperature as previously established (23, 24). In response to high-dose LPS challenge, 9 of 12 WT mice survived from the endotoxin shock at 96 h after the LPS injection (Fig. 4A). In comparison, 6 of 12 CREBH−/− mice died within 48 h after the LPS injection. Additionally, we evaluated body temperature and body weight reduction of CREBH−/− and WT control mice upon the LPS challenge. CREBH−/− mice displayed lower body temperature from 18 to 96 h after the challenge, and tended to lose more body weight, compared with the control mice (supplemental Fig. S4, A and B).
FIGURE 4.
CREBH regulates production of HDL and prevents liver from injuries upon acute endotoxemia. A, survival rates of CREBH−/− and WT control mice under the different time points after the LPS injection. CREBH−/− and WT mice were IP injected with LPS of 20 μg/gram body weight. Health assessment based on body posture, body hygiene, skin dryness, eyes and head position, cage movement, food intake, body weight, and body temperature were conducted for 96 h post injection. The animals at the end point of moribund state that reflects a point which the animals will not recover were euthanized according to the established guidelines shock research (23, 24). The graph shows the percentage of survived animals during the time course post the treatment. 12 mice per group (n = 12) were culled for the survival experiment. B, HE and Sirius-red staining of the liver tissue sections of the CREBH−/− and WT control mice at 96 h after the LPS injection. Magnification: 200×. C, quantification of hepatic fibrosis scores of the CREBH−/− and WT control mice at 96 h after the LPS injection, based on Sirius-red staining of collagen. D and E, serum levels of AST, ALT, total cholesterol, HDL, and LDL in the CREBH−/− and WT control mice at 96 h after the LPS injection. Each bar denotes the mean ± S.E. (n = 9 WT or 4 KO). *, p < 0.05; **, p < 0.01. F, illustration for the role and mechanism by which TLR signaling regulates CREBH activity to protect the liver from injuries caused by acute endotoxima. Ub_K63, K63-linked ubiquitin.
We examined tissue damage in CREBH−/− and WT control mice after 96-hour LPS challenge. While the liver of CREBH−/− mice did not exhibit significant morphological change, compared with the control mice, collagen staining indicated increased hepatic fibrosis in CREBH−/− mice after the LPS treatment (Fig. 4, B and C). The modest increase of hepatic fibrosis in the absence of CREBH may reflect more severe liver damage in CREBH−/− mice upon endotoxemia. This was confirmed by significantly elevated serum levels of aspartate transaminase (AST) and alanine transaminase (ALT), the indicators of liver damage or hepatotoxicity, in the CREBH−/− mice after LPS challenge (Fig. 4D). These results indicated the importance of CREBH in counteracting hepatotoxicity caused by endotoxemia. Further, we determined the levels of circulating cholesterols in CREBH−/− mice after the high-dose LPS challenge. Consistent with the role of CREBH in promoting the host response to endotoxin by regulating secreted cholesterols, serum levels of HDL and total cholesterols in CREBH−/− mice were significantly lower than those in the control mice after the LPS treatment (Fig. 4E). Additionally, we examined the potential injuries to the kidney of CREBH−/− and WT mice after the LPS treatment. Histological staining analysis did not identified significant difference in morphological change or fibrosis between CREBH−/− and WT mice (supplemental Fig. S4C).
Discussion
In this study, we concluded that the liver metabolic regulator CREBH plays a critical role in mounting a host defense response against endotoxins based on the following findings (Fig. 4F): 1) The bacterial endotoxin LPS are capable of inducing expression and cleavage of CREBH; 2) The TLR4-MyD88-TRAF6 signaling pathway is required for CREBH cleavage and activation in hepatocytes under LPS treatment; 3) TRAF6 interacts with CREBH to mediate CREBH ubiquitination upon LPS challenge; 4) TRAF6-mediated, K63-linked ubiquitination of CREBH stabilizes CREBH protien and promotes its activation; 5) CREBH regulates expression of the ApoA4 gene, but not the classical UPR or metabolic genes, under LPS challenge; 6) CREBH regulates HDL levels by activating the ApoA4 gene upon LPS challenge; and 7) CREBH plays an important role in endotoxin-triggered cholesterol modification and protects the liver from endotoxin-induced liver injuries. Our study revealed the molecular mechanism underlying the link between innate immunity and hepatic metabolism upon acute endotoxemia. We demonstrate that host response against bacterial infection involves a highly sophisticated cross communication between immune signaling and hepatic metabolism mediated through the stress-inducible transcription factor CREBH. The increases in both CREBH expression and activation upon the endotoxin LPS challenge confirmed that CREBH is a potent inflammatory-responsive transcriptional activator in hepatic acute-phase responses. The results also suggested that the inflammatory response has pronounced impact not only in innate immunity but also in modulation of lipid profiles.
Abnormality in lipid metabolism is a critical issue in sepsis patients. Plasma proteins like apolipoproteins are markedly modulated during the sepsis. HDL is thought to play a protective role in sepsis and endotoxemia (3). Elevation in HDL levels is an important response mounted by the body to counter the sepsis or endotoxin levels. Our study revealed one of the mechanisms underlying elevation in HDL cholesterol post endotoxin treatment, which is mediated through CREBH in a TLR/MyD88-dependent manner. TLR-mediated activation of CREBH increases transcription of the gene encoding ApoA4, a component of HDL cholesterol that plays an important role in body protection against sepsis. We demonstrated that the loss of CREBH represses the capability of the animal model to mount an HDL protective response against bacterial endotoxin LPS and thus led to increased liver injuries under endotoxemia.
Our studies provided novel insights into the role of the post-translational modification, namely ubiquitination, in CREBH cleavage and activation mechanism. As a stress-responsive transcription factor, CREBH has been connected to a variety of pathophysiological processes (9, 10). The ability of the same protein to function differentially in response to the wide types of stress cues may be attributed to post-translational modifications. TRAF6, an E3 ligase under the TLR signaling pathway, is involved in activation or function of downstream target molecules through ubiquitination (18). Our study expanded and confirmed the roles of TRAF6 from being an inflammatory regulator to an essential player of metabolic signaling. The role of bacterial infection in metabolic disorders, such as atherosclerosis, is well studied. Inflammation is known to contribute significantly to the atherosclerotic process and is associated with proatherogenic changes in lipoprotein metabolism that is characterized by increased VLDL and reduced HDL levels (22). Considering the protective role of HDL, it has been proposed that administration of recombinant HDL may prevent bacterial infection-associated pathogenesis inflammatory effects (25). Given the critical role of CREBH in HDL production, boosting CREBH activity may be beneficial to combating bacterial infection and preventing atherosclerosis.
The CREBH−/− mouse model provided a valuable tool to validate the functional impact of LPS-induced, TLR-mediated CREBH activation in the liver. CREBH−/− mice had abrogated levels of serum HDL upon relatively low-dose of LPS challenge. Our studies using the CREBH−/− mouse model implicated that TLR4-MyD88-TRAF6-CREBH regulatory axis represent an important pathway leading to hepatic acute-phase response upon the challenge of bacterial endotoxin. We previously showed that CREBH−/− mice displayed hepatic steatosis and hyperlipidemia under the AHF diet (9). Interestingly, CREBH−/− mice under the AHF diet produced low levels of serum HDL (data not shown). It is possible that the metabolic diet may also activate the same pathway mediated through TLR, an intriguing question to be elucidated in the future.
Experimental Procedures
Animal Experiments
CREBH-null mice with exons 4–7 of the CrebH gene deleted were previously described (26). The TLR4-null and Myd88-null mice were from the Jackson Laboratory (Bar Harbor, ME). CREBH-null, TLR4-null, Myd88-null, and wild-type control mice on a C57Bl/6J background of 3-month old were used for the experiments. All the animal experiments were approved by the Wayne State University IACUC committee and carried out under the institutional guidelines for ethical animal use.
Determination of Mouse Survival Rates under LPS Endotoxic Shock
CREBH−/− and wild-type (WT) control mice were intraperitoneally (IP) injected with LPS of 20 μg/gram body weight. Health assessment based on body posture, body hygiene, skin dryness, eyes and head position, cage movement, food intake, body weight, and body temperature were conducted for 96 h post injection by following the established rodent morbidity and moribund score and end point guidelines (23, 24). The animals at the end point of moribund state that reflects a point which the animals will not recover were euthanized and counted as endotoxic shock-associated death. The end point sign for euthanasia and endotoxemia-caused death count is that the animals displayed signs of the moribund state, including impaired mobility (unable to reach food and water), inability to maintain upright position, prolonged decreased food and water intake, labored breathing and cyanosis, extreme or prolonged weight loss/emaciation, prolonged diarrhea or constipation, bleeding from an orifice, repeated or severe self-mutilation, and unconsciousness (23). At the end point of the experiments, the mice were euthanized and tissue/blood samples were collected.
Co-immunoprecipitation and Western Blotting Analysis
Liver tissue or cell lysates in Nonidet P-40 lysis buffer were incubated with an antibody (1 μg) for overnight at 4 °C, followed by the addition of 30 μl recombinant protein G-Sepharose beads (Invitrogen) for 4 h at room temperature. Immunoprecipitates were washed four times with Nonidet P-40 lysis buffer and boiled in Laemmle buffer. Samples were subjected to 10% or 12% SDS-PAGE analysis and electrotransferred onto PVDF membrane (0.45 μm). Membranes were probed with primary antibodies, followed by incubation with horseradish peroxidase-conjugated secondary antibodies. Membranes were washed and visualized with a chemiluminescence detection system.
Luciferase Reporter Analysis
For luciferase reporter analysis, we used Dual-Luciferase Reporter Assay System (Promega, Madison, WI). Huh7 cells were co-transfected with the luciferase reporter vector, TK-Renilla luciferase vector, and the vector expressing full-length CREBH or vector control. Luciferase assay was performed at 24, 36, and 48 h. Data graphs were presented after normalization of Firefly, luciferase reporter activities to the control Renilla luciferase activities. For a full description of materials and methods used in this work, see supplemental information.
Author Contributions
Study concept and design: K. Z., A. D., F. S.; acquisition of data, analysis and interpretation of data: A. D., K. Z., Y. Q., H. K., J. W., X. H., X. Z., Z. Z., R. M., F. Y., A. K., D. F., F. S.; drafting of the manuscript: K. Z., A. D.; critical revision of the manuscript for important intellectual content: K. Z., A. D.; statistical analysis: A. D., J. W., X. Z., Z. Z., K. Z.; obtained funding: K. Z.; administrative, technical, or material support: A. K., D. F., F. S., K. Z.; study supervision: K. Z.
Supplementary Material
Acknowledgment
We thank Dr. Kenji Fukudome for critical comments on this work.
This work was supported by National Institutes of Health (NIH) Grants DK090313 and ES017829 (to K. Z.), AR066634 (to K. Z. and D. F.), and American Heart Association Grant 09GRNT2280479 (to K. Z.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Figs. S1–S4 and methods..
- LPS
- lipopolysaccharide
- CREBH
- cAMP-responsive element-binding protein, hepatic specific
- ER
- endoplasmic reticulum
- TLR
- Toll-like receptor
- TRAF6
- TNF receptor-associated factor 6
- MyD88
- Myeloid Differentiation Primary Response 88
- HDL
- high-density lipoprotein
- ApoA4
- apolipoprotein A4
- AHF
- atherogenic high-fat
- IP
- immunoprecipitation
- ChIP
- chromatin immunoprecipitation
- Ub
- ubiquitin
- KO
- knockout
- AST
- aspartate transaminase
- ALT
- alanine transaminase.
References
- 1. Harris K., Kassis A., Major G., and Chou C. J. (2012) Is the gut microbiota a new factor contributing to obesity and its metabolic disorders? J. Obes. 2012, 879151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Feingold K. R., Wang Y., Moser A., Shigenaga J. K., and Grunfeld C. (2008) LPS decreases fatty acid oxidation and nuclear hormone receptors in the kidney. J. Lipid Res. 49, 2179–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Feingold K. R., and Grunfeld C. (2011) The role of HDL in innate immunity. J. Lipid Res. 52, 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tsai M. H., Peng Y. S., Chen Y. C., Lien J. M., Tian Y. C., Fang J. T., Weng H. H., Chen P. C., Yang C. W., and Wu C. S. (2009) Low serum concentration of apolipoprotein A-I is an indicator of poor prognosis in cirrhotic patients with severe sepsis. J. Hepatol. 50, 906–915 [DOI] [PubMed] [Google Scholar]
- 5. Chien J. Y., Jerng J. S., Yu C. J., and Yang P. C. (2005) Low serum level of high-density lipoprotein cholesterol is a poor prognostic factor for severe sepsis. Crit. Care Med. 33, 1688–1693 [DOI] [PubMed] [Google Scholar]
- 6. van Leeuwen H. J., van Beek A. P., Dallinga-Thie G. M., van Strijp J. A., Verhoef J., and van Kessel K. P. (2001) The role of high density lipoprotein in sepsis. Netherlands J. Med. 59, 102–110 [DOI] [PubMed] [Google Scholar]
- 7. Säemann M. D., Poglitsch M., Kopecky C., Haidinger M., Horl W. H., and Weichhart T. (2010) The versatility of HDL: a crucial anti-inflammatory regulator. Eur. J. Clin. Investig. 40, 1131–1143 [DOI] [PubMed] [Google Scholar]
- 8. Kim H., Mendez R., Zheng Z., Chang L., Cai J., Zhang R., and Zhang K. (2014) Liver-enriched transcription factor CREBH interacts with peroxisome proliferator-activated receptor α to regulate metabolic hormone FGF21. Endocrinology 155, 769–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang C., Wang G., Zheng Z., Maddipati K. R., Zhang X., Dyson G., Williams P., Duncan S. A., Kaufman R. J., and Zhang K. (2012) Endoplasmic reticulum-tethered transcription factor cAMP responsive element-binding protein, hepatocyte specific, regulates hepatic lipogenesis, fatty acid oxidation, and lipolysis upon metabolic stress in mice. Hepatology 55, 1070–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang K., Shen X., Wu J., Sakaki K., Saunders T., Rutkowski D., Back S., and Kaufman R. (2006) Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 124, 587–599 [DOI] [PubMed] [Google Scholar]
- 11. Zheng Z., Kim H., Qiu Y., Chen X., Mendez R., Dandekar A., Zhang X., Zhang C., Liu A. C., Yin L., Lin J. D., Walker P. D., Kapatos G., and Zhang K. (2016) CREBH couples circadian clock with hepatic lipid metabolism. Diabetes doi: 10.2337/db16-0298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee M. W., Chanda D., Yang J., Oh H., Kim S. S., Yoon Y. S., Hong S., Park K. G., Lee I. K., Choi C. S., Hanson R. W., Choi H. S., and Koo S. H. (2010) Regulation of hepatic gluconeogenesis by an ER-bound transcription factor, CREBH. Cell Metab. 11, 331–339 [DOI] [PubMed] [Google Scholar]
- 13. Kim H., Mendez R., Chen X., Fang D., and Zhang K. (2015) Lysine acetylation of CREBH regulates fasting-induced hepatic lipid metabolism. Mol. Cell. Biol. 35, 4121–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Machida K., Tsukamoto H., Mkrtchyan H., Duan L., Dynnyk A., Liu H. M., Asahina K., Govindarajan S., Ray R., Ou J. H., Seki E., Deshaies R., Miyake K., and Lai M. M. (2009) Toll-like receptor 4 mediates synergism between alcohol and HCV in hepatic oncogenesis involving stem cell marker Nanog. Proc. Natl. Acad. Sci. U.S.A. 106, 1548–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yin Q., Lin S. C., Lamothe B., Lu M., Lo Y. C., Hura G., Zheng L., Rich R. L., Campos A. D., Myszka D. G., Lenardo M. J., Darnay B. G., and Wu H. (2009) E2 interaction and dimerization in the crystal structure of TRAF6. Nat. Struct. Mol. Biol. 16, 658–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qiu Q., Zheng Z., Chang L., Zhao Y. S., Tan C., Dandekar A., Zhang Z., Lin Z., Gui M., Li X., Zhang T., Kong Q., Li H., Chen S., Chen A., Kaufman R. J., Yang W. L., Lin H. K., Zhang D., Perlman H., Thorp E., Zhang K., and Fang D. (2013) Toll-like receptor-mediated IRE1α activation as a therapeutic target for inflammatory arthritis. EMBO J. 32, 2477–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lamothe B., Campos A. D., Webster W. K., Gopinathan A., Hur L., and Darnay B. G. (2008) The RING domain and first zinc finger of TRAF6 coordinate signaling by interleukin-1, lipopolysaccharide, and RANKL. J. Biol. Chem. 283, 24871–24880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., and Chen Z. J. (2000) Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103, 351–361 [DOI] [PubMed] [Google Scholar]
- 19. Nathan J. A., Kim H. T., Ting L., Gygi S. P., and Goldberg A. L. (2013) Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? EMBO J. 32, 552–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kanayama A., Seth R. B., Sun L., Ea C. K., Hong M., Shaito A., Chiu Y. H., Deng L., and Chen Z. J. (2004) TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol. Cell 15, 535–548 [DOI] [PubMed] [Google Scholar]
- 21. Ghiselli G., Crump W. L. 3rd, Musanti R., Sherrill B. C., and Gotto A. M. Jr. (1989) Metabolism of apolipoprotein A-IV in rat. Biochim. Biophys. Acta 1006, 26–34 [DOI] [PubMed] [Google Scholar]
- 22. Khovidhunkit W., Duchateau P. N., Medzihradszky K. F., Moser A. H., Naya-Vigne J., Shigenaga J. K., Kane J. P., Grunfeld C., and Feingold K. R. (2004) Apolipoproteins A-IV and A-V are acute-phase proteins in mouse HDL. Atherosclerosis 176, 37–44 [DOI] [PubMed] [Google Scholar]
- 23. Nemzek J. A., Xiao H. Y., Minard A. E., Bolgos G. L., and Remick D. G. (2004) Humane endpoints in shock research. Shock 21, 17–25 [DOI] [PubMed] [Google Scholar]
- 24. Nemzek J. A., Hugunin K. M., and Opp M. R. (2008) Modeling sepsis in the laboratory: merging sound science with animal well-being. Comp. Med. 58, 120–128 [PMC free article] [PubMed] [Google Scholar]
- 25. Gordon S. M., Hofmann S., Askew D. S., and Davidson W. S. (2011) High density lipoprotein: it's not just about lipid transport anymore. Trends Endocrinol. Metab. 22, 9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luebke-Wheeler J., Zhang K., Battle M., Si-Tayeb K., Garrison W., Chhinder S., Li J., Kaufman R. J., and Duncan S. A. (2008) Hepatocyte nuclear factor 4α is implicated in endoplasmic reticulum stress-induced acute phase response by regulating expression of cyclic adenosine monophosphate responsive element-binding protein H. Hepatology 48, 1242–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.