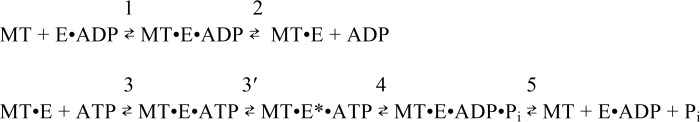Abstract
Mammalian KIF3AB is an N-terminal processive kinesin-2 that is best known for its roles in intracellular transport. There has been significant interest in KIF3AB to define the key principles that underlie its processivity but also to define the mechanistic basis of its sensitivity to force. In this study, the kinetics for entry into the processive run were quantified. The results show for KIF3AB that the kinetics of microtubule association at 7 μm−1 s−1 is less than the rates observed for KIF3AA at 13 μm−1 s−1 or KIF3BB at 11.9 μm−1 s−1. ADP release after microtubule association for KIF3AB is 33 s−1 and is significantly slower than ADP release from homodimeric KIF3AA and KIF3BB, which reach 80–90 s−1. To explore the interhead communication implied by the rate differences at these first steps, we compared the kinetics of KIF3AB microtubule association followed by ADP release with the kinetics for mixtures of KIF3AA plus KIF3BB. Surprisingly, the kinetics of KIF3AB are not equivalent to any of the mixtures of KIF3AA + KIF3BB. In fact, the transients for each of the mixtures overlay the transients for KIF3AA and KIF3BB. These results reveal that intermolecular communication within the KIF3AB heterodimer modulates entry into the processive run, and the results suggest that it is the high rate of microtubule association that drives rebinding to the microtubule after force-dependent motor detachment.
Keywords: ATPase, cooperativity, cytoskeleton, kinesin, microtubule, neuron, processivity, cargo transport, intraflagellar transport, presteady-state kinetics
Introduction
The kinesin-2 subfamily members are ubiquitously expressed and act as major transporters of intracellular cargoes (for reviews, see Refs. 1–5). In mammals, KIF3A, KIF3B, and KAP4 expression yields a heterotrimeric complex (6–9). The motor polypeptides KIF3A and KIF3B form a heterodimeric motor, KIF3AB, and the nonmotor polypeptide KAP associates at the C terminus of KIF3AB. KAP is a distinctive adaptor protein in that it is largely composed of armadillo repeats (10, 11), and it is these motifs that provide the specificity of interaction between KIF3AB and KAP and between KIF3AB-KAP and its specific cargo. A heterotrimeric kinesin-2 complex with KAP has been described in many species since its discovery in sea urchin eggs including Mus musculus, Chlamydomonas, Caenorhabditis elegans, Drosophila, Xenopus, and Tetrahymena (8–10, 12–19).
KIF3A, KIF3B, and KAP are essential genes (20–25). Knock-out mice for KIF3A or KIF3B have revealed the absence of cilia and a randomized left-right body axis (20–22). These nodal cilia are crucial for the proper mesodermal patterning during embryogenesis to establish the left-right body asymmetry. Other studies have linked KIF3AB-KAP to cilia-dependent signal transduction cascades including the Hedgehog signaling pathway (26, 27). The role of intraflagellar transport in ciliogenesis is considered the basis for KIF3AB-KAP to be an essential protein for development. KIF3AB-KAP is also implicated in a variety of cytoplasmic transport events including organelles, melanosomes, mRNA granules, and membrane-bound vesicles (2, 4, 25, 28–30). KIF3AB-KAP is also essential for axon elongation, and its transport of fodrin-associated vesicles may provide membrane components to the tips of neurites (28).
Kinesin-2 KIF3AB is clearly implicated in diverse but specific transport events as is conventional kinesin-1, suggesting that the mechanochemical properties of these two kinesins are in some way tuned differently and specifically for their cellular roles because one is not redundant for the other. One distinctive characteristic of KIF3AB and its orthologs is their response to hindering loads (18, 31–34). Conventional kinesin-1 continues to step processively until approaching its stall force at 6 pN (35–39), but Xenopus Xklp3a/Xklp3b (32) and C. elegans KLP11/KLP20 (18) tended to detach at hindering loads of ∼4 pN, shortening their run length capability dramatically. More recently, Andreasson et al. (34) reported that the processivity of murine KIF3AB is strongly force-dependent and dropped precipitously when hindering force was applied such that runs consisted of less than 25 steps at 4 pN. However, the other property observed for each of these kinesin-2 motors regardless of species was that upon detachment they were able to rebind the microtubule quickly and initiate another processive run. These observations have resulted in the hypothesis that kinesin-2 motors are tuned specifically to navigate microtubule (MT) roadblocks such as microtubule-associated proteins like tau in neurons (40) and cross-links along the transition zone when loading and transporting intraflagellar particles into cilia (41).
A presteady-state ATPase kinetics analysis of the steps for entry into the processive run was pursued to ask whether there are intrinsic characteristics within KIF3AB that account for its rapid rebinding to the microtubule after detachment. For these studies, we used motors that were co-expressed from mouse KIF3A and KIF3B constructs that included the native sequence for the motor domain and native helix α7 to initiate coiled coil formation followed by a synthetic heterodimerization domain (SHD) as an extension of the native helix to stabilize the native dimer (42, 43).
Our recent single molecule studies (43) revealed that in the absence of load KIF3AB with its native neck linker and helix α7 is highly processive with run lengths of 1.62 μm, which exceeded those of kinesin-1 K560 at 1.26 μm. The run lengths of KIF3AA and KIF3BB showed that both were highly processive, although the run length of KIF3BB at 1.51 μm is greater than KIF3AA at 1 μm and more similar to the run length of KIF3AB (Table 1). Homodimers of KIF3AA and KIF3BB do not appear to occur in vivo, but they provide a tool to assess the motor properties of KIF3A and KIF3B separately. Therefore, we engineered similarly designed constructs for their expression (43). Note that expression of KIF3AA and KIF3BB required a different dimerization motif. The dimerization motif used is a portion of the homodimeric coiled coil and four-helix bundle motif of EB1 (44, 45) and was reported previously not to interact with MTs (43, 46, 47). Control experiments with KIF3AC also confirmed that the dimerization motif (SHD or EB1) used to stabilize the helix α7 coiled coil did not alter the processivity (43). The experiments presented here include an analysis of KIF3AB, KIF3AA, and KIF3BB to ask specifically whether heterodimerization alters the intrinsic properties of KIF3A and KIF3B and to define the characteristics of KIF3AB that enable this kinesin to rebind the microtubule so quickly after detachment.
TABLE 1.
KIF3AB, KIF3AA, and KIF3BB experimentally determined constants
ND, not determined.
| KIF3AB | KIF3AA | KIF3BB | |
|---|---|---|---|
| Microtubule association | k+1 = 7.0 ± 0.4 μm−1 s−1 | k+1 = 13.0 ± 0.5 μm−1 s−1 | k+1 = 11.9 ± 0.1 μm−1 s−1 |
| k−1 = 0.8 ± 0.4 s−1 | k−1 = not observed | k−1 = not observed | |
| ADP release | k+2 = 33.5 ± 0.6 s−1 | k+2 = 89.9 ± 3.4 s−1 | k+2 = 80.2 ± 2.5 s−1 |
| K1/2,MT = 3.1 ± 0.2 μm | K1/2,MT = 5.3 ± 0.5 μm | K1/2,MT = 4.0 ± 0.4 μm | |
| MantATP bindinga | k+3 = 7.5 ± 0.5 μm−1 s−1 | ND | ND |
| k−3 = 46.1 ± 5.5 s−1 | |||
| Steady-state parametersa | kcat = 14.8 ± 0.8 s−1 | kcat = 34.7 ± 0.5 s−1 | kcat = 32.1 ± 0.4 s−1 |
| Km,ATP = 118 ± 7.7 μm | Km,ATP = 47.7 ± 0.1 μm | Km,ATP = 71.4 ± 4.0 μm | |
| K1/2,MT = 0.12 ± 0.1 μm | K1/2,MT = 0.19 ± 0.002 μm | K1/2,MT = 0.14 ± 0.005 μm | |
| Velocityb | 246.2 ± 11.1 nm/s | 239.2 ± 4.2 nm/s | 327.6 ± 7.2 nm/s |
| Run lengthb | 1.62 ± 0.11 μm | 0.98 ± 0.05 μm | 1.51 ± 0.16 μm |
Results
Scheme 1 represents the KIF3AB ATPase cycle, and Fig. 1 presents the proposed steps for entry into the processive run. These were used to design the presteady-state experiments presented. In Table 1, the experimentally determined kinetic and equilibrium constants are reported for heterodimeric KIF3AB and homodimeric KIF3AA and KIF3BB.
SCHEME 1.
FIGURE 1.
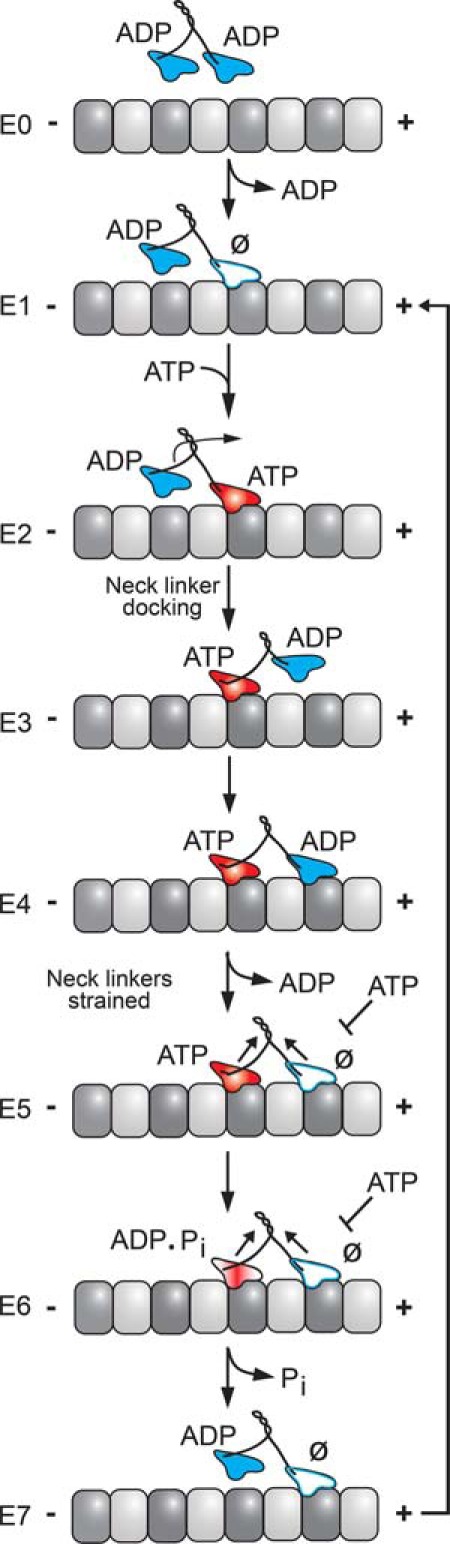
KIF3AB stepping model. KIF3AB in solution and detached from the microtubule holds ADP tightly bound at each head (E0). The processive run begins upon MT collision followed by ADP release (E0 and E1), and it is assumed that there is an equal probability of either KIF3A or KIF3B initiating the processive run. After MT collision, the leading head is nucleotide-free and the trailing head is detached from the MT with ADP tightly bound. ATP binding at the leading head triggers a series of structural transitions including neck linker docking that promotes the trailing head to move forward to its next MT binding site (E2–E4). ADP released upon MT collision of the second head results in the E5 two-head-bound state. Strain develops between the two heads and inhibits ATP binding to the nucleotide-free leading head (E5). ATP hydrolysis on the trailing head followed by phosphate (Pi) release results in the weak affinity of the trailing head to the MT and its subsequent detachment (E7). This series of events relieves the interhead tension (E4–E7). The leading head can now bind ATP, which initiates the next step coupled to ATP turnover without detachment from the MT. In this model, no intrinsic bias is assumed for KIF3A versus KIF3B to start the processive run. Ø, no nucleotide.
Microtubule Association Followed by MantADP Release
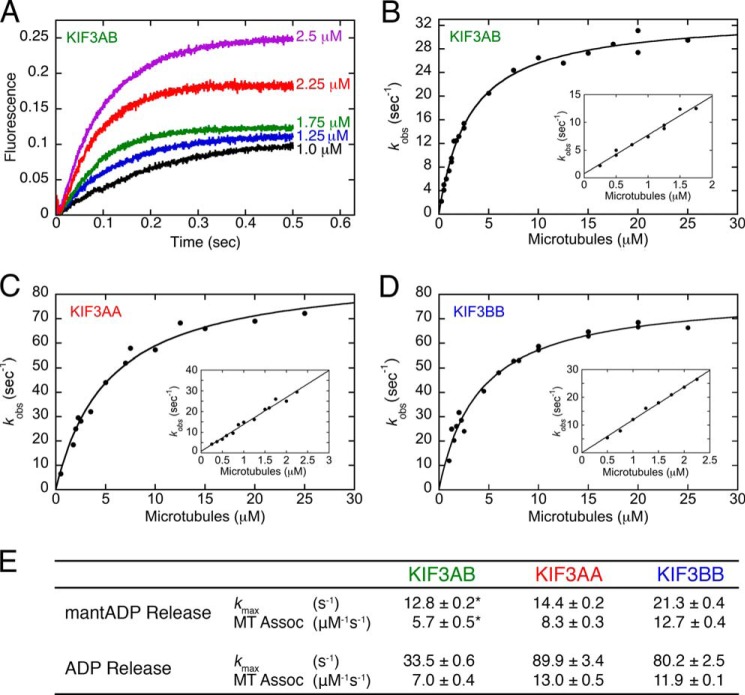
Albracht et al. (42) published a presteady-state kinetic analysis of KIF3AB, but at the time it was not known that KIF3AB was as processive as we discovered soon thereafter (43). Therefore, we wanted to take a fresh look at the ATPase cycle and design different experiments to explore entry into the processive run. The first experiments tested the hypothesis of whether there is an intrinsic bias resulting from heterodimerization of KIF3A and KIF3B that will lead to one head preferentially starting the processive run. We began by measuring the presteady-state kinetics of MT collision followed by mantADP release for homodimeric KIF3AA and KIF3BB in comparison with results for KIF3AB (42). For these experiments ADP at the active site was exchanged with mantADP (1:6 ratio of KIF3 nucleotide sites to mantADP). The KIF3-mantADP complex was rapidly mixed in the stopped-flow instrument with varying concentrations of MTs plus 2 mm MgATP. The transients in Fig. 2A show a biphasic decrease in fluorescence as a function of time due to fluorescence quenching when mantADP is released from the active site to the aqueous buffer. The observed rates of the initial fast exponential phase were plotted as a function of MT concentration, and the results in Fig. 2B reveal that the maximum rate constant for mantADP from KIF3BB at 21.3 s−1 is faster than this rate constant for KIF3AA at 14.4 s−1 with mantADP release for KIF3AB reported at 12.8 s−1 (42). At low MT concentrations, mantADP release is limited by MT collision; therefore, the observed rates plotted as a function of increasing MT concentrations can provide the second-order rate constant for MT association. Fig. 2C shows that the MT association constant for KIF3BB at 12.7 μm−1 s−1 is faster than this constant observed for KIF3AA at 8.3 μm−1 s−1 in comparison with 5.7 μm−1 s−1 observed for KIF3AB using the same experimental approach (42).
FIGURE 2.
Presteady-state kinetics of MT association and MantADP release upon MT collision. The KIF3AA·mantADP or KIF3BB·mantADP complex was preformed and rapidly mixed in the stopped-flow instrument with varying concentrations of MTs plus MgATP. A, representative transients indicate a biphasic decrease in mantADP fluorescence as a function of time. Final concentrations were 0.5 μm KIF3AA nucleotide sites/3.75 μm mantADP for 0.9–2.5 μm MTs. B, observed rates of the initial exponential fluorescence decay were plotted as a function of MT concentration. Final concentrations were 0.5 μm KIF3AA/BB nucleotide sites/3.75 μm mantADP for 0.5–1.5 μm MTs, 2.0 μm KIF3AA/BB nucleotide sites/7.5 μm mantADP for 1.5–4 μm MTs, and 2.5 μm KIF3AB nucleotide sites/15 μm mantADP for 4–30 μm MTs plus 1 mm ATP. Hyperbolic fits provide the maximum rate constant for mantADP release; k+2 = 14.4 ± 0.2 s−1 and K1/2,MT = 0.8 ± 0.05 μm for KIF3AA and k+2 = 21.3 ± 0.4 s−1 with K1/2,MT = 1.1 ± 0.1 μm for KIF3BB. C, at MT-limiting conditions, the linear fit provides kon,MT = 8.3 ± 0.3 μm−1 s−1 and koff = 2.3 ± 0.2 s−1 for KIF3AA, and for KIF3BB kon,MT = 12.7 ± 0.4 μm−1 s−1 and koff = 0.3 ± 0.3 s−1.
Subsequently, using equilibrium approaches Chen et al. (48) reported that mantADP binds more tightly than ADP to their homodimeric KIF3AA motor that was fused to the Drosophila kinesin-1 stalk (KIF3AA-KHC). Therefore, we designed a new experiment to test the hypothesis that mantADP may not capture the kinetics of native ADP release from KIFAB, KIF3AA, and KIF3BB. A similar experimental design was reported by Zhang et al. (49) for KIF3AC.
Microtubule Association and Release of Native ADP Are Significantly Faster than Determined by MantADP
To measure the kinetics of MT collision and ADP release (Fig. 1, E0 and E1), we took advantage of the fluorescence signal of mantATP binding as a readout of MT collision followed by native ADP release. For this experiment, the KIF3 motors with ADP tightly bound were rapidly mixed in the stopped-flow instrument with varying MT concentrations plus 50 μm mantATP for a final concentration of 25 μm mantATP. The second-order rate constant for KIF3AB mantATP binding is 7.5 μm−1 s−1 (42); therefore, mantATP binding at these conditions is >150 s−1 and significantly faster than ADP release at the conditions of the assay.
Fig. 3A includes representative transients for KIF3AB at varying MT concentrations, which reveal an exponential rise in fluorescence as mantATP binds the hydrophobic environment of the active site. Fig. 3B shows the observed rates of the initial exponential phase plotted as a function of MT concentration, and the hyperbolic fit to the data provides the maximum rate constant of ADP release k+2 at 33.5 s−1, which was significantly faster than reported previously for mantADP release (Fig. 3E and Table 1). Note too that at low MT concentrations (Fig. 3B, inset) the second-order rate constant k+1 for MT association at 7 μm−1 s−1 was somewhat faster than the constant determined previously by mantADP at 5.7 μm−1 s−1 (42). This experiment was repeated for KIF3AA (Fig. 3C) and KIF3BB (Fig. 3D). The maximum rate constant for ADP release for KIF3AA was somewhat faster at 89.9 s−1 than k+2 for KIF3BB at 80.2 s−1, but both constants were significantly faster than k+2 determined for mantADP release. The insets for Fig. 3, C and D, show that MT association was significantly faster for KIF3AA at 13 μm−1 s−1 and KIF3BB at 11.9 μm−1 s−1 than determined for KIF3AB at 7.0 μm−1 s−1 (Fig. 3E and Table 1). Therefore, these results support the interpretation of the mantADP equilibrium binding results reported by Chen et al. (48) for Drosophila KIF3AA-KHC and show conclusively that mantADP release did not capture the kinetics of MT association and ADP release from KIF3AB, KIF3AA, and KIF3BB. Moreover, the MT association data for KIF3AA and KIF3BB do not reveal a significant difference. Therefore, we conclude that there is an equal probability such that either KIF3A or KIF3B can initiate the processive run. Note too that the KIF3AB rate constant for ADP release at 33.5 s−1 is consistent with the Vmax for KIF3AB at 31 s−1 based on the velocity of single molecule stepping at 246 nm/s with one ATP turnover per 8-nm step (Table 1 and Ref. 43).
FIGURE 3.
Presteady-state kinetics of MT association followed by ADP release. KIF3 was rapidly mixed in the stopped-flow instrument with varying concentrations of MTs plus mantATP. Final concentrations were 0.25 or 0.5 μm KIF3 sites/25 μm mantATP for 0.25–2.5 μm MTs or 2.5 μm KIF3 sites and 50 μm mantATP for 2.5–25 μm MTs. A, representative KIF3AB transients ranging from 1.0 to 2.5 μm MTs show a biphasic increase in mantATP fluorescence as a function of time. B, the observed rates of each initial fast phase were plotted as a function of MT concentration. The hyperbolic fit provided the maximum rate constant of ADP release at k+2 = 33.5 ± 0.6 s−1 with a K1/2,MTs = 3.1 ± 0.2 μm for KIF3AB. Inset, the initial linear part at low MT concentrations provides the rate constant for MT association: k+1 = 7.0 ± 0.4 μm−1 s−1 and k−1 = 0.8 ± 0.4 s−1. C, KIF3AA maximum rate constant of ADP release at k+2 = 89.9 ± 3.4 s−1 with a K1/2,MTs = 5.3 ± 0.5 μm. Inset, KIF3AA k+1 = 13.0 ± 0.5 μm−1 s−1. D, KIF3BB maximum rate constant of ADP release k+2 = 80.2 ± 2.5 s−1 and K1/2,MTs = 4.0 ± 0.4 μm. Inset, KIF3BB k+1 = 11.9 ± 0.1 μm−1 s−1. E, table summarizing the results. *, the mantADP release kinetics for KIF3AB were reported previously (42) and are from Fig. 2 for KIF3AA and KIF3BB. Assoc, association.
Heterodimerization of KIF3AB Controls Entry into the Processive Run
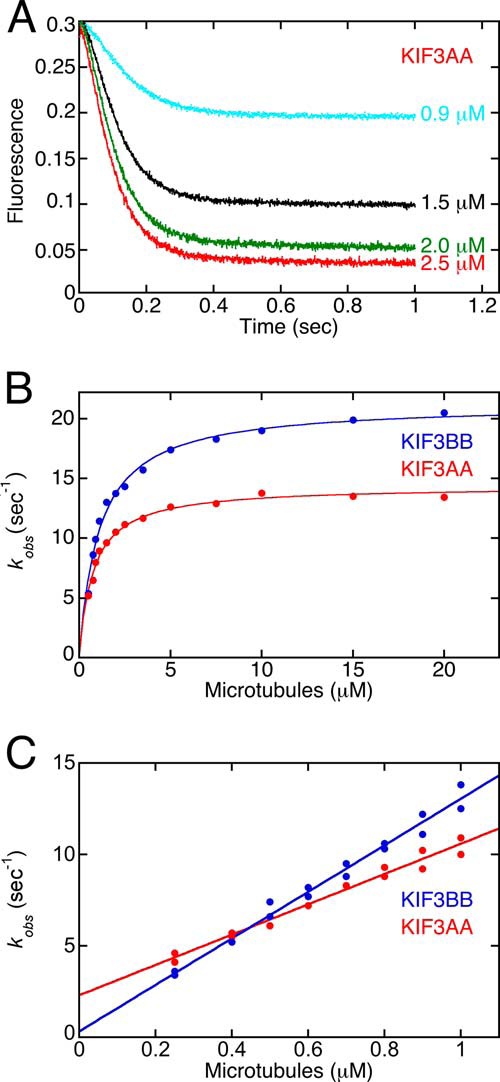
To test the hypothesis that heterodimerization of KIF3AB alters the intrinsic kinetics of KIF3A and KIF3B as revealed by their homodimers, we pursued additional experiments measuring MT association followed by native ADP release (Fig. 4). KIF3AB, KIF3AA, KIF3BB, and mixtures of KIF3AA + KIF3BB at 5 μm KIF3 site concentration were rapidly mixed in the stopped-flow instrument with 40 μm MTs plus 50 μm mantATP (syringe concentrations). The final concentrations were 2.5 μm KIF3 sites, 20 μm MTs, and 25 μm mantATP. Fig. 4A shows the comparison of the transients for 100% KIF3AA, 100% KIF3BB, and 100% KIF3AB. The transients of KIF3AA and KIF3BB overlay each other as one would expect because their kinetics are so similar as documented in Fig. 3. In contrast, however, the transient for KIF3AB is clearly displaced and reveals a slower observed rate of ADP release. The kinetics of ADP release were also measured with mixtures of KIF3AA plus KIF3BB at varying ratios (Fig. 4B). Note that the transients for each of the mixtures, regardless of composition, were equivalent to the transients of 100% KIF3AA or 100% KIF3BB, which we attribute to the kinetics of KIF3AA and KIF3BB being so similar (Figs. 3 and 4). The fits for each transient revealed an observed ADP release rate at 20 μm MTs that was very similar for each mixture at 61–65 s−1 (Fig. 4C). In contrast, the kinetics were significantly slower for ADP release from KIF3AB at 31 s−1. This analysis supports the conclusion that MT collision by KIF3AA and KIF3BB in the mixture experiments represents independent events, and it is the heterodimerization of KIF3A and KIF3B that alters the intrinsic kinetics of each head as observed for entry into the processive run.
FIGURE 4.
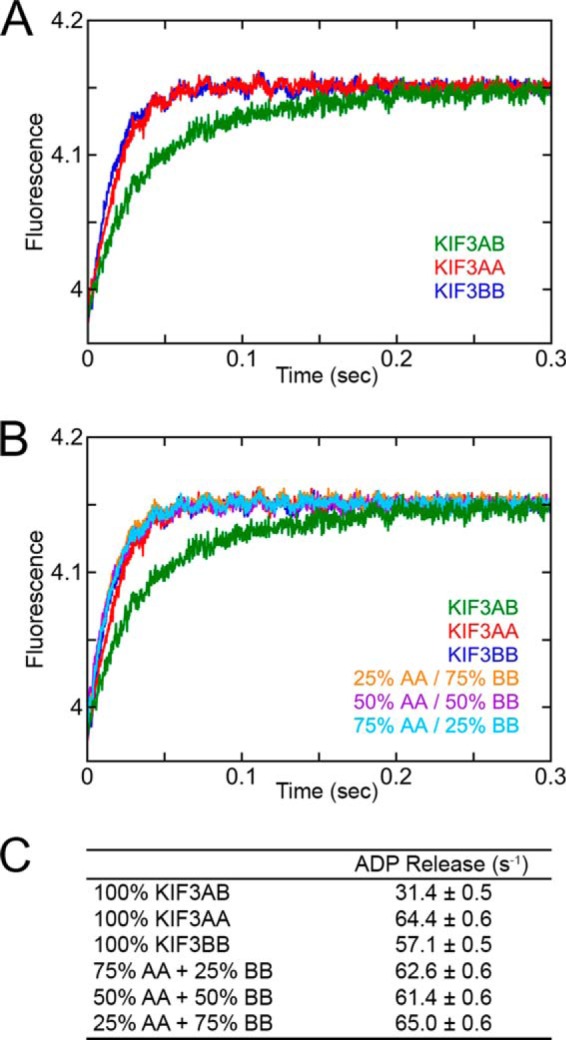
ADP release upon MT collision for KIF3AB, KIF3AA, KIF3BB, and KIF3AA + KIF3BB. KIF3AB, KIF3AA, KIF3BB, or mixtures of KIF3AA plus KIF3BB at total 5 μm site concentration were rapidly mixed in the stopped-flow instrument with 40 μm MTs plus 50 μm mantATP (syringe concentrations). The mixtures contain either 25% KIF3AA + 75% KIF3BB (1.25 μm KIF3AA and 3.75 μm KIF3BB), 50% KIF3AA + 50% KIF3BB (2.5 μm KIF3AA and 2.5 μm KIF3BB), or 75% KIF3AA + 25% KIF3BB (3.75 μm KIF3AA and 1.25 μm KIF3BB). Final concentrations were 2.5 μm KIF3 site concentration, 20 μm MTs, and 25 μm mantATP. A, individual transients show a biphasic increase in mantATP fluorescence as a function of time. B, the experimental transients for the mixtures of KIF3AA + KIF3BB in comparison with the transients of KIF3AB, KIF3AA (AA), and KIF3BB (BB). C, table of the observed rates of ADP release.
Discussion
The investigation of microtubule association followed by ADP release has revealed important insights. First, the fluorescent analog mantADP, which has been used successfully for many kinesins (50–70), did not accurately capture the kinetics of MT association and ADP release for KIF3AB, KIF3AA, and KIF3BB (Figs. 2 and 3). The data in Fig. 3 clearly show that MT association followed by ADP release is significantly faster when quantifying native ADP release. If one viewed the mantADP release kinetics in isolation, one may have assumed that it was KIF3B that initiated the processive run because both MT association and mantADP release were faster than the rates observed for KIF3AA. In contrast, the kinetics of KIF3AA and KIF3BB are similar and support the hypothesis that either KIF3A or KIF3B can begin the processive run through MT collision.
The second important insight was the impact of KIF3A on KIF3B and of KIF3B on KIF3A that occurs because of heterodimerization. The rate constants of MT association for homodimeric KIF3AA (13 μm−1 s−1) and KIF3BB (11.9 μm−1 s−1) were similar, but these were significantly faster than MT association for KIF3AB at 7 μm−1 s−1, indicating that the intrinsic kinetics of KIF3A and KIF3B as captured by the homodimers are altered due to intermolecular communication that results upon heterodimerization. The mixture experiments of KIF3AA + KIF3BB in Fig. 4 reinforce this conclusion because no combination of KIF3AA + KIF3BB homodimers was able to recapitulate the ADP release kinetics of KIF3AB.
There was another intriguing observation when the kinetics of MT association for KIF3AB were compared with the MT association kinetics for kinesin-2 KIF3AC published previously (49). MT association for KIF3AC was reported at 6.6 μm−1 s−1, and this constant for KIF3AC is comparable with the second-order rate constant for KIF3AB at 7 μm−1 s−1 reported here. Although Zhang et al. (49) reported a similar KIF3AA MT association rate constant at 11.4 μm−1 s−1, MT association by KIF3CC was exceedingly slow at 2.1 μm−1 s−1 followed by ADP release at 7.6 s−1 and very different from MT association and ADP release for KIF3BB, supporting the argument that the motor heads of KIF3B and KIF3C are very different catalytically.
These results indicate that it is the heterodimerization of KIF3AB and KIF3AC that modulates the rate of MT association for entry into a processive run rather than the intrinsic catalytic capability of KIF3A, KIF3B, or KIF3C. Additional evidence in support of this hypothesis is that the sequence of the neck linker followed by helix α7 for KIF3B and KIF3C are identical with the exception of a single residue. Furthermore, the structures of the neck linkers and α7 for KIF3A and KIF3C, and hence KIF3B, are very similar (71). Thus, it is likely that the modulation arises from interactions between the motor domains themselves that cannot be achieved by the homodimers despite how catalytically similar KIF3AA and KIF3BB are. Note too that even though the kinetics of KIF3A and KIF3B are very similar there is only 69% sequence identity between these proteins, suggesting that KIF3AB has evolved its particular set of kinetic properties. Therefore, we conclude that heterodimerization of KIF3A with KIF3B as well as KIF3A and KIF3C results in intermolecular communication outside of the neck linker that controls the rate constant of MT association regardless of which head initiates the processive run.
Although these results inform us of entry into the processive run and the regulation by heterodimerization, they cannot predict the impact of heterodimerization on other steps in the ATPase cycle as outlined in the stepping model in Fig. 1. For example, the single molecule velocity for KIF3AB is 246.2 nm/s or 30.8 s−1 based on one ATP turnover per 8-nm step; however, for KIF3AC it is 186.5 nm/s or 23.3 s−1 (Ref. 43 and Table 1). The difference in the stepping velocity at saturating ATP implies that later steps in the ATPase cycle are different for KIF3AB and KIF3AC and controlled by the catalytic capability of the motor domains as concluded by Andreasson et al. (34) for KIF3AB under load.
One aim of this study was to define the mechanistic basis of the ability of KIF3AB to rebind to the MT after detachment. This question is compelling for KIF3AB in particular because of its sensitivity to force, resulting in motor detachment rather than stalling as observed for kinesin-1. Our interpretation of the KIF3AB rapid reassociation is based on the high second-order rate constant for MT association at 7 μm−1 s−1. The rate of rebinding by KIF3·ADP is determined by the local MT concentration, which was estimated previously at 1 mm near the MT lattice (72). At this concentration, KIF3AB could rebind to the MT at 7,000 s−1, resulting in a very short detached state of ∼143 μs. Therefore, the catalytic capability of KIF3AB is optimized for reassociation with the MT to initiate another processive run. Based upon the comparisons of KIF3AB with KIF3AA and KIF3BB whose second-order rate constants for MT association are even higher than KIF3AB, one may ask why heterodimerization? Note, however, that homodimeric KIF3AA and KIF3BB are not native motors. Therefore, what is significant in these results is the very fast MT association rate constant for the physiologically relevant KIF3AB that results in rapid rebinding to the MT for intracellular transport.
Experimental Procedures
Constructs of Homodimeric and Heterodimeric KIF3
The M. musculus KIF3A and KIF3B plasmids for expression of KIF3AB heterodimer (42) and the KIF3AA and KIF3BB homodimers (43) were described previously in detail as well as their expression and purification. Briefly, to generate a stable heterodimer of KIF3AB, an SHD motif containing either an acidic (AHD) or basic fusion helix (BHD) was used (73, 74). The KIF3A-AHD polypeptide consisted of the KIF3A motor domain, neck linker, and three heptads of native helix sequence followed by the AHD (bold font), a TEV protease site (italic font), and StrepII tag (underlined font). Linker residues are in plain font. The expressed sequence included KIF3A(Met1–Glu376)-LEKEIAALEKEIAALEKTTSENLYFQGASNWSHPQFEK (predicted molecular weight, 46,341). The KIF3B-BHD polypeptide contained the KIF3B motor domain, neck linker, and three heptads of native helix sequence followed by the BHD (bold font), a TEV protease site (italic font) with linker residues (plain font), and a His8 tag (underlined font): KIF3B(Met1–Lys371)-LKEKIAALKEKIAALKETTSENLYFQGASHHHHHHHH (predicted molecular weight, 45,790).
For the KIF3AA and KIF3BB homodimers, each native N-terminal motor domain sequence, neck linker, and α7 helix were C-terminally fused to an in-register segment of the dimerization motif from EB1 (bold font) followed by the TEV protease site (italic font) with linker residues (plain font) and a His8 tag (underlined font): for KIF3A, KIF3A(Met1–Leu374)-DFYFGKLRNIELICQENEGENDPVLQRIVDILYATDETTSENLYFQGASHHHHHHHH (predicted molecular weight, 48,502), and for KIF3B, KIF3B(Met1–Leu369)-DFYFGKLRNIELICQENEGENDPVLQRIVDILYATDETTSENLYFQGASHHHHHHHH (predicted molecular weight, 48,011). Note that the EB1 motif is a dimerization domain only and not an MT binding domain as shown previously (43, 46, 47).
KIF3 Protein Expression and Purification
KIF3 motors were expressed in Escherichia coli BL21-CodonPlus (DE3)-RIL cell line (Stratagene, La Jolla, CA) for purification as described previously (42, 43). The predicted molecular weights based on amino acid sequence of KIF3AB, KIF3AA, and KIF3BB are 97,118, 97,004, and 96,022, respectively. Experiments were designed by using the KIF3 dimer concentration but converted to the ATP site concentration (two sites per dimer) to report here.
Experimental Conditions
The experiments were performed at 25 °C in ATPase buffer: 20 mm Hepes, pH 7.2 with KOH, 5 mm magnesium acetate, 0.1 mm EDTA, 0.1 mm EGTA, 50 mm potassium acetate, 1 mm dithiothreitol, 5% sucrose. The morning of each experiment bovine brain tubulin was cold depolymerized, clarified, and polymerized with 1 mm MgGTP at 37 °C. The MTs were stabilized with 40 μm paclitaxel, and the MT concentrations reported represent those of the paclitaxel-stabilized tubulin polymer. The reported concentrations of ATP, GTP, and the nucleotide analogs mantATP and mantADP include an equivalent concentration of magnesium acetate. Note that the fluorescent analogs mantATP and mantADP were purchased from Invitrogen as the isomeric mixture.
Experimental Approach for Data Analysis
The presteady-state kinetics experiments were repeated 8–12 times at varying motor and microtubule concentrations as well as time domains for data collection to acquire experimental results that merge to common data sets. For Figs. 2A and 3A, each representative transient is an average of 8–10 transients and is from a single experimental day. The plots of observed rates as a function of MT concentration (Figs. 2, B and C, and 3, B–D) include data from multiple experiments on different days to cover the entire MT concentration range. The mean of three to five data sets is reported with errors as S.E. For Fig. 4, the experiments were repeated four to six times on different days, and each transient represents the average of 8–10 individual transients. The transients in Fig. 4 are from a single day of experiments with the rates of ADP release reported in Fig. 4C as the mean of four data sets ±S.E.
MantADP Release upon MT Collision
To measure the kinetics of microtubule association followed by mantADP release (Fig. 1, E0 and E1), ADP tightly bound at the active sites of KIF3 was exchanged with the fluorescent analog mantADP at a concentration of 5 μm KIF3 nucleotide sites and 30 μm mantADP (1:6 ratio). The KIF3·mantADP complex was subsequently mixed in the stopped-flow instrument (KinTek SF2003, KinTek Corp., Austin, TX) with varying MT concentrations plus 2 mm MgATP (syringe concentration). The change in fluorescence was monitored as a function of time (λex = 360 nm, λem = 450 nm, detected using a 409-nm long pass filter). A double exponential function was fit to the biphasic transients, and the observed rates of the initial fast exponential phase were plotted as a function of increasing MT concentrations.
At low MT concentrations (0.25–2.25 μm tubulin polymer), the rate of mantADP release is limited by MT collision. Therefore, in this MT concentration range, the linear fit to the data (Equation 1) provides the second-order rate constant for MT association k+1 with the KIF3 off-rate, k−1, as defined by the y-intercept.
At higher concentrations of MTs, mantADP release after MT collision becomes rate-limiting, and the hyperbolic fit to the data provides the maximum rate constant of mantADP release, k+2 (see Fig. 2 and Scheme 1).
Release of ADP upon MT Collision
To measure the kinetics of native ADP release upon MT collision, the experimental design mimicked the reaction condition at the beginning of the cycle (Fig. 1, E0 and E1). KIF3 motors were rapidly mixed in the stopped-flow instrument with MTs plus mantATP. After MT collision, under these assay conditions, mantATP binding was significantly faster than ADP release from the KIF3 active site. Therefore, the enhanced fluorescence upon mantATP binding was used as a readout of ADP release. KIF3 at varying site concentrations (0.5, 1, or 5 μm) was rapidly mixed with 0.5–50 μm MTs plus 50 or 100 μm mantATP in the stopped-flow instrument. Final concentrations were 0.25 or 0.5 μm KIF3 sites/25 μm mantATP for 0.25–2.25 μm MTs or 2.5 μm KIF3 sites/50 μm mantATP for 2.5–25 μm MTs. A double exponential function was fit to each transient. The observed rates of the initial exponential phase were plotted as a function of MT concentration, and the hyperbolic fit to the data provided the maximum rate constant of ADP release. At low concentrations of MTs, the observed rate of ADP release is limited by MT collision; therefore, the linear fit (Equation 1) to these data provided the second-order rate constant of MT association (see Fig. 3, Scheme 1, and Table 1).
Does Heterodimerization Modulate the Kinetics for KIF3AB Entry into the Processive Run?
To test this hypothesis, additional ADP release experiments were performed as described in the section above. KIF3AB, KIF3AA, KIF3BB, and mixtures of KIF3AA + KIF3BB at 5 μm KIF3 site concentration were rapidly mixed with 40 μm MTs plus 50 μm mantATP (syringe concentrations). Final concentrations were 2.5 μm KIF3 site concentration, 20 μm MTs, and 25 μm mantATP. The mixtures corresponded to 25% KIF3AA + 75% KIF3BB, 50% KIF3AA + 50% KIF3BB, and 75% KIF3AA + 25% KIF3BB. A double exponential function fit to each transient provided the rate constants of each reaction. See Fig. 4.
Author Contributions
C. D. A., S. G.-L., I. R., and S. P. G. designed research. C. D. A. and S. G.-L. performed research. C. D. A., S. G.-L., I. R., and S. P. G. analyzed the data and wrote the manuscript.
Acknowledgments
We thank Brandon Bensel, Sean Quinn, and Rebecca Phillips for thoughtful discussions and input on these experiments and analyses.
This work was supported by National Institutes of Health Grant R37-GM054141 (to S. P. G). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- KAP
- kinesin-associated protein
- KIF3AB
- mouse kinesin-2 KIF3AB heterodimer
- KIF3AC
- mouse kinesin-2 KIF3AC heterodimer
- MT
- microtubule
- mantATP/mantADP
- 2′-(or 3′)-O-(N-methylanthraniloyl)ATP/ADP
- pN
- piconewtons
- SHD
- synthetic heterodimerization domain
- AHD
- SHD motif containing an acidic fusion helix
- BHD
- SHD motif containing a basic fusion helix
- TEV
- tobacco etch virus.
References
- 1. Hirokawa N., Noda Y., Tanaka Y., and Niwa S. (2009) Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10, 682–696 [DOI] [PubMed] [Google Scholar]
- 2. Hirokawa N., Niwa S., and Tanaka Y. (2010) Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68, 610–638 [DOI] [PubMed] [Google Scholar]
- 3. Verhey K. J., Kaul N., and Soppina V. (2011) Kinesin assembly and movement in cells. Annu. Rev. Biophys. 40, 267–288 [DOI] [PubMed] [Google Scholar]
- 4. Scholey J. M. (2013) Kinesin-2: a family of heterotrimeric and homodimeric motors with diverse intracellular transport functions. Annu. Rev. Cell Dev. Biol. 29, 443–469 [DOI] [PubMed] [Google Scholar]
- 5. Maday S., Twelvetrees A. E., Moughamian A. J., and Holzbaur E. L. (2014) Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron 84, 292–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aizawa H., Sekine Y., Takemura R., Zhang Z., Nangaku M., and Hirokawa N. (1992) Kinesin family in murine central nervous system. J. Cell Biol. 119, 1287–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kondo S., Sato-Yoshitake R., Noda Y., Aizawa H., Nakata T., Matsuura Y., and Hirokawa N. (1994) KIF3A is a new microtubule-based anterograde motor in the nerve axon. J. Cell Biol. 125, 1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yamazaki H., Nakata T., Okada Y., and Hirokawa N. (1995) KIF3A/B: a heterodimeric kinesin superfamily protein that works as a microtubule plus end-directed motors for membrane organelle transport. J. Cell Biol. 130, 1387–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamazaki H., Nakata T., Okada Y., and Hirokawa N. (1996) Cloning and characterization of KAP3: a novel kinesin superfamily-associated protein of KIF3A/3B. Proc. Natl. Acad. Sci. U.S.A. 93, 8443–8448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Doodhi H., Ghosal D., Krishnamurthy M., Jana S. C., Shamala D., Bhaduri A., Sowdhamini R., and Ray K. (2009) KAP, the accessory subunit of kinesin-2, binds the predicted coiled-coil stalk of the motor subunits. Biochemistry 48, 2248–2260 [DOI] [PubMed] [Google Scholar]
- 11. Gindhart J. G. Jr., and Goldstein L. S. (1996) Armadillo repeats in the SpKAP115 subunit of kinesin-II. Trends Cell Biol. 6, 415–416 [DOI] [PubMed] [Google Scholar]
- 12. Cole D. G., Chinn S. W., Wedaman K. P., Hall K., Vuong T., and Scholey J. M. (1993) Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature 366, 268–270 [DOI] [PubMed] [Google Scholar]
- 13. Wedaman K. P., Meyer D. W., Rashid D. J., Cole D. G., and Scholey J. M. (1996) Sequence and submolecular localization of the 115-kD accessory subunit of the heterotrimeric kinesin-II (KRP85/95) complex. J. Cell Biol. 132, 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tuma M. C., Zill A., Le Bot N., Vernos I., and Gelfand V. (1998) Heterotrimeric kinesin II is the microtubule motor protein responsible for pigment dispersion in Xenopus melanophores. J. Cell Biol. 143, 1547–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Le Bot N., Antony C., White J., Karsenti E., and Vernos I. (1998) Role of xklp3, a subunit of the Xenopus kinesin II heterotrimeric complex, in membrane transport between the endoplasmic reticulum and the Golgi apparatus. J. Cell Biol. 143, 1559–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brown J. M., Marsala C., Kosoy R., and Gaertig J. (1999) Kinesin-II is preferentially targeted to assembling cilia and is required for ciliogenesis and normal cytokinesis in Tetrahymena. Mol. Biol. Cell 10, 3081–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mueller J., Perrone C. A., Bower R., Cole D. G., and Porter M. E. (2005) The FLA3 KAP subunit is required for localization of kinesin-2 to the site of flagellar assembly and processive anterograde intraflagellar transport. Mol. Biol. Cell 16, 1341–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brunnbauer M., Mueller-Planitz F., Kösem S., Ho T. H., Dombi R., Gebhardt J. C., Rief M., and Okten Z. (2010) Regulation of a heterodimeric kinesin-2 through an unprocessive motor domain that is turned processive by its partner. Proc. Natl. Acad. Sci. U.S.A. 107, 10460–10465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pesavento P. A., Stewart R. J., and Goldstein L. S. (1994) Characterization of the KLP68D kinesin-like protein in Drosophila: possible roles in axonal transport. J. Cell Biol. 127, 1041–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nonaka S., Tanaka Y., Okada Y., Takeda S., Harada A., Kanai Y., Kido M., and Hirokawa N. (1998) Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95, 829–837 [DOI] [PubMed] [Google Scholar]
- 21. Takeda S., Yonekawa Y., Tanaka Y., Okada Y., Nonaka S., and Hirokawa N. (1999) Left-right asymmetry and kinesin superfamily protein KIF3A: new insights in determination of laterality and mesoderm induction by kif3A−/− mice analysis. J. Cell Biol. 145, 825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marszalek J. R., Ruiz-Lozano P., Roberts E., Chien K. R., and Goldstein L. S. (1999) Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc. Natl. Acad. Sci. U.S.A. 96, 5043–5048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marszalek J. R., Liu X., Roberts E. A., Chui D., Marth J. D., Williams D. S., and Goldstein L. S. (2000) Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors. Cell 102, 175–187 [DOI] [PubMed] [Google Scholar]
- 24. Lin F., Hiesberger T., Cordes K., Sinclair A. M., Goldstein L. S., Somlo S., and Igarashi P. (2003) Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proc. Natl. Acad. Sci. U.S.A. 100, 5286–5291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Teng J., Rai T., Tanaka Y., Takei Y., Nakata T., Hirasawa M., Kulkarni A. B., and Hirokawa N. (2005) The KIF3 motor transports N-cadherin and organizes the developing neuroepithelium. Nat. Cell Biol. 7, 474–482 [DOI] [PubMed] [Google Scholar]
- 26. Huangfu D., Liu A., Rakeman A. S., Murcia N. S., Niswander L., and Anderson K. V. (2003) Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426, 83–87 [DOI] [PubMed] [Google Scholar]
- 27. Drummond I. A. (2012) Cilia functions in development. Curr. Opin. Cell Biol. 24, 24–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Takeda S., Yamazaki H., Seog D. H., Kanai Y., Terada S., and Hirokawa N. (2000) Kinesin superfamily protein 3 (KIF3) motor transports fodrin-associating vesicles important for neurite building. J. Cell Biol. 148, 1255–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hendricks A. G., Perlson E., Ross J. L., Schroeder H. W. 3rd, Tokito M., and Holzbaur E. L. (2010) Motor coordination via a tug-of-war mechanism drives bidirectional vesicle transport. Curr. Biol. 20, 697–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carpenter B. S., Barry R. L., Verhey K. J., and Allen B. L. (2015) The heterotrimeric kinesin-2 complex interacts with and regulates GLI protein function. J. Cell Sci. 128, 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shastry S., and Hancock W. O. (2011) Interhead tension determines processivity across diverse N-terminal kinesins. Proc. Natl. Acad. Sci. U.S.A. 108, 16253–16258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schroeder H. W. 3rd, Hendricks A. G., Ikeda K., Shuman H., Rodionov V., Ikebe M., Goldman Y. E., and Holzbaur E. L. (2012) Force-dependent detachment of kinesin-2 biases track switching at cytoskeletal filament intersections. Biophys. J. 103, 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arpağ G., Shastry S., Hancock W. O., and Tüzel E. (2014) Transport by populations of fast and slow kinesins uncovers novel family-dependent motor characteristics important for in vivo function. Biophys. J. 107, 1896–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Andreasson J. O., Shastry S., Hancock W. O., and Block S. M. (2015) The mechanochemical cycle of mammalian kinesin-2 KIF3A/B under load. Curr. Biol. 25, 1166–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Svoboda K., and Block S. M. (1994) Force and velocity measured for single kinesin molecules. Cell 77, 773–784 [DOI] [PubMed] [Google Scholar]
- 36. Meyhöfer E., and Howard J. (1995) The force generated by a single kinesin molecule against an elastic load. Proc. Natl. Acad. Sci. U.S.A. 92, 574–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hyeon C., and Onuchic J. N. (2007) Internal strain regulates the nucleotide binding site of the kinesin leading head. Proc. Natl. Acad. Sci. U.S.A. 104, 2175–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yildiz A., Tomishige M., Gennerich A., and Vale R. D. (2008) Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell 134, 1030–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dogan M. Y., Can S., Cleary F. B., Purde V., and Yildiz A. (2015) Kinesin's front head is gated by the backward orientation of its neck linker. Cell Rep. 10, 1967–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hoeprich G. J., Thompson A. R., McVicker D. P., Hancock W. O., and Berger C. L. (2014) Kinesin's neck-linker determines its ability to navigate obstacles on the microtubule surface. Biophys. J. 106, 1691–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Prevo B., Mangeol P., Oswald F., Scholey J. M., and Peterman E. J. (2015) Functional differentiation of cooperating kinesin-2 motors orchestrates cargo import and transport in C. elegans cilia. Nat. Cell Biol. 17, 1536–1545 [DOI] [PubMed] [Google Scholar]
- 42. Albracht C. D., Rank K. C., Obrzut S., Rayment I., and Gilbert S. P. (2014) Kinesin-2 KIF3AB exhibits novel ATPase characteristics. J. Biol. Chem. 289, 27836–27848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guzik-Lendrum S., Rank K. C., Bensel B. M., Taylor K. C., Rayment I., and Gilbert S. P. (2015) Kinesin-2 KIF3AC and KIF3AB can drive long-range transport along microtubules. Biophys. J. 109, 1472–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Frye J., Klenchin V. A., and Rayment I. (2010) Structure of the tropomyosin overlap complex from chicken smooth muscle: insight into the diversity of N-terminal recognition. Biochemistry 49, 4908–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sen I., Veprintsev D., Akhmanova A., and Steinmetz M. O. (2013) End binding proteins are obligatory dimers. PLoS One 8, e74448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bu W., and Su L. K. (2003) Characterization of functional domains of human EB1 family proteins. J. Biol. Chem. 278, 49721–49731 [DOI] [PubMed] [Google Scholar]
- 47. Komaki S., Abe T., Coutuer S., Inzé D., Russinova E., and Hashimoto T. (2010) Nuclear-localized subtype of end-binding 1 protein regulates spindle organization in Arabidopsis. J. Cell Sci. 123, 451–459 [DOI] [PubMed] [Google Scholar]
- 48. Chen G. Y., Arginteanu D. F., and Hancock W. O. (2015) Processivity of the kinesin-2 KIF3A results from rear head gating and not front head gating. J. Biol. Chem. 290, 10274–10294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang P., Rayment I., and Gilbert S. P. (2016) Fast or slow, either head can start the processive run of kinesin-2 KIF3AC. J. Biol. Chem. 291, 4407–4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sadhu A., and Taylor E. W. (1992) A kinetic study of the kinesin ATPase. J. Biol. Chem. 267, 11352–11359 [PubMed] [Google Scholar]
- 51. Gilbert S. P., Moyer M. L., and Johnson K. A. (1998) Alternating site mechanism of the kinesin ATPase. Biochemistry 37, 792–799 [DOI] [PubMed] [Google Scholar]
- 52. Rice S., Lin A. W., Safer D., Hart C. L., Naber N., Carragher B. O., Cain S. M., Pechatnikova E., Wilson-Kubalek E. M., Whittaker M., Pate E., Cooke R., Taylor E. W., Milligan R. A., and Vale R. D. (1999) A structural change in the kinesin motor protein that drives motility. Nature 402, 778–784 [DOI] [PubMed] [Google Scholar]
- 53. Kallipolitou A., Deluca D., Majdic U., Lakämper S., Cross R., Meyhöfer E., Moroder L., Schliwa M., and Woehlke G. (2001) Unusual properties of the fungal conventional kinesin neck domain from Neurospora crassa. EMBO J. 20, 6226–6235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rosenfeld S. S., Xing J., Jefferson G. M., Cheung H. C., and King P. H. (2002) Measuring kinesin's first step. J. Biol. Chem. 277, 36731–36739 [DOI] [PubMed] [Google Scholar]
- 55. Hackney D. D. (2002) Pathway of ADP-stimulated ADP release and dissociation of tethered kinesin from microtubules. Implications for the extent of processivity. Biochemistry 41, 4437–4446 [DOI] [PubMed] [Google Scholar]
- 56. Auerbach S. D., and Johnson K. A. (2005) Alternating site ATPase pathway of rat conventional kinesin. J. Biol. Chem. 280, 37048–37060 [DOI] [PubMed] [Google Scholar]
- 57. Clancy B. E., Behnke-Parks W. M., Andreasson J. O., Rosenfeld S. S., and Block S. M. (2011) A universal pathway for kinesin stepping. Nat. Struct. Mol. Biol. 18, 1020–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lockhart A., Cross R. A., and McKillop D. F. (1995) ADP release is the rate-limiting step of the MT activated ATPase of non-claret disjunctional and kinesin. FEBS Lett. 368, 531–535 [DOI] [PubMed] [Google Scholar]
- 59. Pechatnikova E., and Taylor E. W. (1999) Kinetics processivity and the direction of motion of Ncd. Biophys. J. 77, 1003–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Foster K. A., Mackey A. T., and Gilbert S. P. (2001) A mechanistic model for Ncd directionality. J. Biol. Chem. 276, 19259–19266 [DOI] [PubMed] [Google Scholar]
- 61. Chen C. J., Rayment I., and Gilbert S. P. (2011) Kinesin Kar3Cik1 ATPase pathway for microtubule cross-linking. J. Biol. Chem. 286, 29261–29272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen C. J., Porche K., Rayment I., and Gilbert S. P. (2012) The ATPase pathway that drives the kinesin-14 Kar3Vik1 powerstroke. J. Biol. Chem. 287, 36673–36682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang P., Dai W., Hahn J., and Gilbert S. P. (2015) Drosophila Ncd reveals an evolutionarily conserved powerstroke mechanism for homodimeric and heterodimeric kinesin-14s. Proc. Natl. Acad. Sci. U.S.A. 112, 6359–6364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lockhart A., and Cross R. A. (1996) Kinetics and motility of the Eg5 microtubule motor. Biochemistry 35, 2365–2373 [DOI] [PubMed] [Google Scholar]
- 65. Cochran J. C., Gatial J. E. 3rd, Kapoor T. M., and Gilbert S. P. (2005) Monastrol inhibition of mitotic kinesin Eg5. J. Biol. Chem. 280, 12658–12667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Krzysiak T. C., Grabe M., and Gilbert S. P. (2008) Getting in sync with dimeric Eg5. Initiation and regulation of the processive run. J. Biol. Chem. 283, 2078–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Behnke-Parks W. M., Vendome J., Honig B., Maliga Z., Moores C., and Rosenfeld S. S. (2011) Loop L5 acts as a conformational latch in the mitotic kinesin Eg5. J. Biol. Chem. 286, 5242–5253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rosenfeld S. S., van Duffelen M., Behnke-Parks W. M., Beadle C., Corrreia J., and Xing J. (2009) The ATPase cycle of the mitotic motor CENP-E. J. Biol. Chem. 284, 32858–32868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sardar H. S., and Gilbert S. P. (2012) Microtubule capture by mitotic kinesin centromere protein E (CENP-E). J. Biol. Chem. 287, 24894–24904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cochran J. C., Sindelar C. V., Mulko N. K., Collins K. A., Kong S. E., Hawley R. S., and Kull F. J. (2009) ATPase cycle of the nonmotile kinesin NOD allows microtubule end tracking and drives chromosome movement. Cell 136, 110–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Phillips R. K., Peter L. G., Gilbert S. P., and Rayment I. (July 26, 2016) Family-specific kinesin structures reveal neck-linker length based on initiation of the coiled-coil. J. Biol. Chem. 10.1074/jbc.M116.737577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gilbert S. P., Webb M. R., Brune M., and Johnson K. A. (1995) Pathway of processive ATP hydrolysis by kinesin. Nature 373, 671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lindhout D. A., Litowski J. R., Mercier P., Hodges R. S., and Sykes B. D. (2004) NMR solution structure of a highly stable de novo heterodimeric coiled-coil. Biopolymers 75, 367–375 [DOI] [PubMed] [Google Scholar]
- 74. Rank K. C., Chen C. J., Cope J., Porche K., Hoenger A., Gilbert S. P., and Rayment I. (2012) Kar3Vik1, a member of the kinesin-14 superfamily, shows a novel kinesin microtubule binding pattern. J. Cell Biol. 197, 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]