Abstract
In the chloroplast of the green alga Chlamydomonas reinhardtii, two discontinuous group II introns, psaA-i1 and psaA-i2, splice in trans, and thus their excision process resembles the nuclear spliceosomal splicing pathway. Here, we address the question whether fragmentation of trans-acting RNAs is accompanied by the formation of a chloroplast spliceosome-like machinery. Using a combination of liquid chromatography-mass spectrometry (LC-MS), size exclusion chromatography, and quantitative RT-PCR, we provide the first characterization of a high molecular weight ribonucleoprotein apparatus participating in psaA mRNA splicing. This supercomplex contains two subcomplexes (I and II) that are responsible for trans-splicing of either psaA-i1 or psaA-i2. We further demonstrate that both subcomplexes are associated with intron RNA, which is a prerequisite for the correct assembly of subcomplex I. This study contributes further to our view of how the eukaryotic nuclear spliceosome evolved after bacterial endosymbiosis through fragmentation of self-splicing group II introns into a dynamic, protein-rich RNP machinery.
Keywords: chloroplast, evolution, gene regulation, RNA processing, RNA splicing, spliceosome, Chlamydomonas reinhardtii, group II intron, ribonucleoprotein complex, trans-splicing
Introduction
The spliceosome is a dynamic RNP machinery that participates in the excision of mRNA introns in eukaryotes. This machinery consists of five small nuclear RNAs (snRNAs; U1, U2, U4, U5, and U6) and a large number of spliceosomal proteins (1, 2). It is generally accepted that spliceosomal snRNAs were derived from ancient group II introns, which were introduced into the eukaryotic cell after endosymbiosis (3, 4). Group II introns occur frequently in bacteria and organelles of fungi, algae, and higher plants but are rare in archaea and absent from nuclear genomes. Comparable with spliceosomal introns, the splicing reaction includes two transesterification reactions, yielding spliced exons and an excised intron lariat RNA. Despite the lack of significant sequence similarities, group II introns share a common secondary structure with six helical domains (D1–D6) surrounding a central wheel. During intron excision, these domains take over specific functions, and remarkably, they show plenty of functional and structural similarities to spliceosomal snRNAs (5–7). These similarities led to the assumption that during evolution of the nuclear splicing apparatus, group II introns fragmented and degenerated to spliceosomal snRNAs (3, 8, 9).
In contrast to bacterial group II introns, most organelle group II introns display variant forms of degeneration and fragmentation (9). For example, many of the plant group II introns have mispaired domain structures (10, 11). Moreover, group II introns are able to split into autonomous fragments due to rearrangements of organelle genomes (12, 13). Consequently, they are transcribed independently, and association of precursor RNAs by base pairing generates a catalytically active group II intron structure that is finally processed by trans-splicing. Such degenerated and fragmented group II introns along with the interplay of discrete RNAs during the splicing reaction serve to demonstrate how snRNAs may have evolved into being part of the nuclear spliceosome.
In the green alga Chlamydomonas reinhardtii, exceptional examples of split group II introns were described as part of the chloroplast psaA gene that encodes an apoprotein of photosystem I. The psaA gene is split into three dispersed exons, which are flanked by truncated group II intron sequences (14). Whereas the second intron (psaA-i2) is bipartite, the first intron (psaA-i1) is tripartite, and the missing group II intron secondary structure is delivered in trans by the chloroplast-encoded tscA RNA (15). After separate transcription, two group II introns are built up by base pairing, followed by two trans-splicing reactions and, ultimately, formation of the mature psaA mRNA. Splicing of such variant group II introns relies on nucleus-encoded splicing factors to compensate for lack of functional motifs and to retain splicing activity (11, 16). However, it is still unknown and under scientific debate whether these splicing factors function in a spliceosomal-like yet intron-specific manner.
For C. reinhardtii, seven splicing factors, specific for group II introns, have been described at the molecular level (17–23). Whereas splicing factor Raa1 (RNA maturation of psaA RNA) is involved in splicing of both reactions, Raa3, Raa4, Raa8, and Rat2 (RNA maturation of psaA tscA RNA) are psaA-i1-specific. Raa2 and Raa7 act specifically on the splicing of psaA-i2. Recent results revealed that splicing of psaA-i1 and -i2 is assisted by at least two high molecular weight protein complexes (subcomplexes I and II) harboring the general splicing factor Raa1 together with intron-specific subunits (17, 24). Here, we provide the first evidence for a high molecular weight apparatus involved in trans-splicing of the psaA mRNA, which is the first report of a high molecular weight splicing apparatus from a eukaryotic organelle. The plastid splicing apparatus described here shows large similarities to the nuclear spliceosome, including formation of RNPs with in trans-acting RNAs, recruitment of related RNA-binding proteins, and dynamic association. Although both splicing machineries have evolved independently, our results imply that various evolutionary routes exist to convert a self-splicing ribozyme into a proteinogenic, spliceosome-like apparatus. Our data contribute to the current understanding about the evolutionary relationship between group II and spliceosomal intron splicing.
Results
Core Components of trans-Splicing Subcomplexes I and II
Previous studies have led to the identification of two trans-splicing subcomplexes (I and II) comprising described factors and several unknown proteins that are involved in the splicing of both chloroplast group II introns (17, 24). We aimed to elucidate core components of both subcomplexes. For this purpose, we performed tandem affinity chromatography and LC-MS analyses using two alternative bait proteins, Rat2 and Raa2, for each subcomplex, respectively.
The RAT2 gene was previously identified in the C. reinhardtii class C trans-splicing mutant TR72:75, which shows a defect in trans-splicing of the chloroplast psaA mRNA and is crucial for 3′-end processing of the chloroplast tscA RNA (22). By using Raa4 as a bait, Rat2 was found among subcomplex I proteins (24). RT-PCR analysis, data from LC-MS analyses (24), and a new gene model prediction of the recent C. reinhardtii database version 5.5 led to an extension of the previously predicted size of the proposed gene product to about 144 kDa. A reevaluation of the gene annotation was conducted with mutant L135F, which is allelic to the previously described mutant TR72:75 (22) (supplemental Fig. S1). Functional complementation analysis with a fusion of RAT2 and the tandem affinity purification (TAP)4 sequence (24), resulting in strain RT2T, finally verified that the full-length copy of RAT2 is sufficient to restore photosynthesis in mutant L135F (supplemental Fig. S2).
Three biological replicates of RT2T were applied to TAP and purified proteins were identified using multidimensional protein identification technology (MudPIT) (24). As a control to identify false positive proteins, we used strain RST-1, which carries a fusion of the RBCS1 gene with the TAP tag (24). In total, 20 proteins were identified in all three RT2T replicates (P1–P3, supplemental Table S1). A list of the LC-MS data of each TAP experiment can be found in supplemental Table S6.
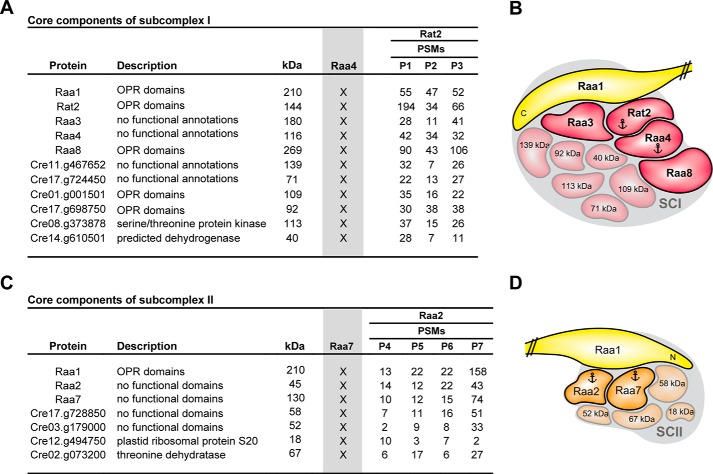
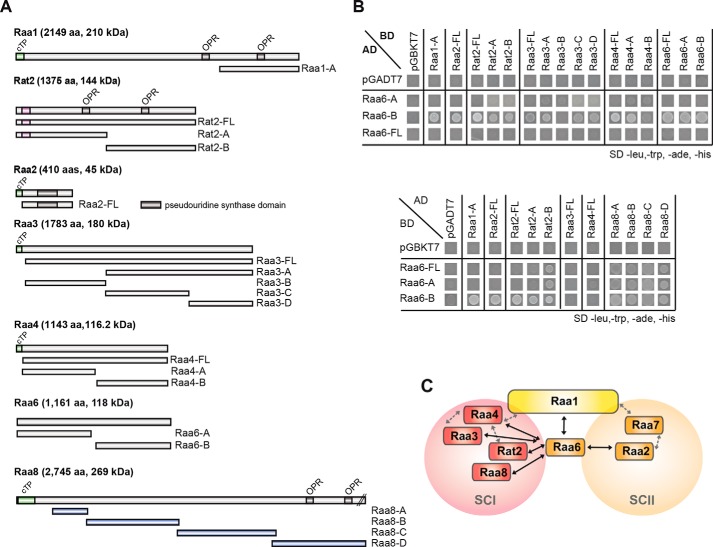
To identify core components of subcomplex I, we compared our TAP results of RT2T with the previously performed TAPs with bait Raa4. Using Raa4::TAP as a bait protein under different environmental conditions, a total of 32 proteins were identified. We uncovered a total of 11 proteins that appeared in data sets of Rat2 and Raa4 (Fig. 1). This analysis revealed the identification of five already known trans-splicing factors, Raa1, Rat2, Raa3, Raa4, and Raa8, with a high number of identified peptides (Fig. 1, column PSMs). In addition to the five trans-splicing factors, four uncharacterized proteins were detected with Rat2 and Raa4 bait proteins, including the previously identified octatricopeptide repeat (OPR) proteins Cre17.g698750 and Cre01.g001501. We further discovered Cre08.g373878, a protein with homologies to a serine/threonine protein kinase, and Cre14.g610500, a putative short chain dehydrogenase/reductase.
FIGURE 1.
Mass spectrometry data to identify core components of subcomplex I and II. A, core components of subcomplex I (SCI) were identified by comparison of TAP-MS results using Rat2 as bait with previous results with Raa4 as bait (gray column Raa4; X indicates the presence of the core component in the Raa4-TAP eluates) (24). Peptide spectral matches (PSMs) of identified proteins are listed. kDa, predicted molecular mass. B, schematic summary of results from A. C, core components of subcomplex II were identified by comparison of TAP-MS results using Raa2 as bait with previous results with Raa7 as bait (gray column Raa7; X indicates the presence of the core component in the Raa7-TAP eluates) (17). Peptide spectral matches of identified proteins are listed. D, schematic summary of results from C. Anchors indicate bait proteins used for TAP-MS.
To identify core components of subcomplex II, we used Raa2 (20) as a second bait protein. Raa2 was already found in TAP tag experiments with trans-splicing factor Raa7 as bait (17). Using mutant A18, which lacks a functional Raa2 protein, as recipient, we have constructed strain R2T, which generates a Raa2::TAP fusion. The Raa2::TAP protein and PsaA protein were detected in crude extracts of R2T, whereas A18 showed no signals (supplemental Fig. S3C). These analyses confirmed the restoration of photosynthetic deficiency in R2T and the functionality of Raa2::TAP.
Strain R2T was used in TAP experiments to identify subcomplex II components (P4–P7). Previous experiments have indicated that Raa2 is associated with thylakoid membranes (25). Consequently, detergent n-dodecyl β-d-maltoside was added to the lysis buffer. A total of four independent TAP experiments were performed. Proteins were considered as specific if they were found in four separate biological replicates and not in control experiments (C1–C4) (17). These analyses revealed 19 putative Raa2-interacting proteins (supplemental Table S2). Comparison of identified proteins in Raa2:: TAP and Raa7 ::TAP experiments revealed seven putative core components of subcomplex II, which were present in all replicates (Fig. 1C). Of note is that the characterized splicing factors Raa1, Raa2, and Raa7 are among these seven proteins. Furthermore, two uncharacterized proteins without functional annotations were identified in all independent TAP experiments. We found that the plastid ribosomal protein S20 and a protein annotated as threonine dehydratase were both among the seven putative core components of subcomplex II.
The results presented here define two core-splicing complexes, subcomplexes I and II. We further demonstrate that there is an interaction network of at least 11 and 7 interaction partners in subcomplex I and II, respectively. Several of the uncharacterized proteins are most probably further not yet identified trans-splicing factors. Finally, using two bait proteins, we show that tandem affinity purification is a powerful tool to define a core protein complex and to distinguish between highly specific and transient interaction partners.
Subcomplexes I and II Are Associated with Group II Intron RNA
Among the proteins purified in TAP experiments are various trans-splicing factors involved in maturation of the psaA mRNA, thus implying an association with RNA. Previously, in a purified Raa4::TAP protein complex, enrichment of tscA RNA was shown (24).
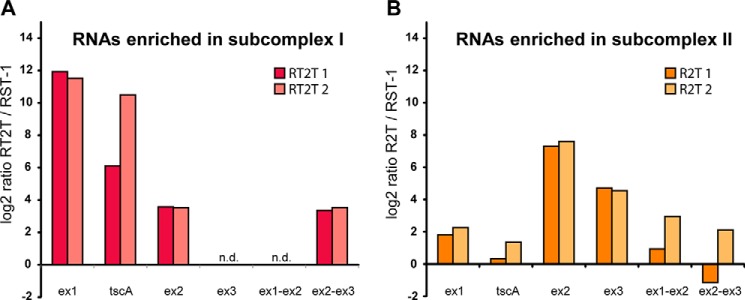
To prove association with intron RNA and formation of (RNP) complexes, we analyzed RNA precipitated from TAP eluates of Rat2 and Raa2 in a one-step qRT-PCR. As a reference gene for normalization, the plastid rrnL RNA was used, and two independent biological replicates of each strain were analyzed. The RST-1 strain expressing an RBCS1::TAP construct was applied for control experiments. This analysis confirmed the enrichment of intron RNA in RT2T TAP eluates, representing subcomplex I, relative to RST-1 TAP RNA (Fig. 2A). This enrichment includes the tscA RNA and exon 1 precursor RNA at about 6–10- and 12-fold the log2 ratio, respectively. Both RNAs contribute RNA structures to the first psaA group II intron, psaA-i1. We further show a slight enrichment of the exon 2 precursor RNA and exon 1-exon 2 splicing intermediate (about 3.5-fold the log2 ratio). The exon 3 precursor, which is part of psaA-i2, and the splicing intermediate of exon 1-exon 2 were not detectable in TAP eluates.
FIGURE 2.
RNAs associated with subcomplex I (A) and II (B). After TAP purification, RNA was precipitated, and RNA levels were analyzed with qRT-PCRs. Enrichment of RNAs was determined by calculating ratios of RT2T or R2T with results obtained with the control strain RST-1. The rrnL gene was used for normalization. In these qRT-PCRs, we used primer pairs specific for exon 1 precursor (ex1), exon 2 precursor (ex2), exon 3 precursor (ex3), tscA RNA, and partially spliced psaA exons (Ex1-Ex2 and Ex2-Ex3). n.d., not detected.
Enrichment of intron RNA was also shown for Raa2 TAP eluates, representing subcomplex II (Fig. 2B). As expected and in contrast to subcomplex I, a strong enrichment was only shown for psaA-i2-specific RNAs, namely the exon 2 and exon 3 precursor RNAs, which displayed enrichment in TAP eluates of 7- and 4-fold the log2 ratio, respectively. These RNAs are sufficient and required to form the whole psaA-i2 structure. However, psaA-i1-specific RNA and spliced variants (exon 1-exon 2 and exon 2-exon 3) were not significantly enriched in TAP eluates of Raa2.
Collectively, our findings demonstrate that subcomplexes I and II purified with Rat2:: TAP and Raa2 ::TAP are associated with specific intron RNA, namely psaA-i1 and psaA-i2 RNA, respectively, and thus form RNP complexes during psaA mRNA trans-splicing.
tscA RNA Is Essential for Core Formation of trans-Splicing Subcomplex I
The tscA RNA contains domains D2 and D3 as well as parts of D1 and D4 of the tripartite psaA-i1 group II intron sequence. Raa4 was previously shown to bind domains D2 and D3 of tscA RNA in vitro (19), supporting the notion that tscA is crucial for subcomplex I formation. To test this hypothesis, a deletion of the tscA locus in strains Raa4:: TAP (R4T) and Rat2 ::TAP (RT2T) was generated. The aadA gene conferring spectinomycin resistance was used to replace the main part of the tscA locus by homologous recombination (supplemental Fig. S4), and the resulting transformants, R4TΔtscA and RT2TΔtscA, were selected under low light conditions on Tris acetate-phosphate medium containing spectinomycin.
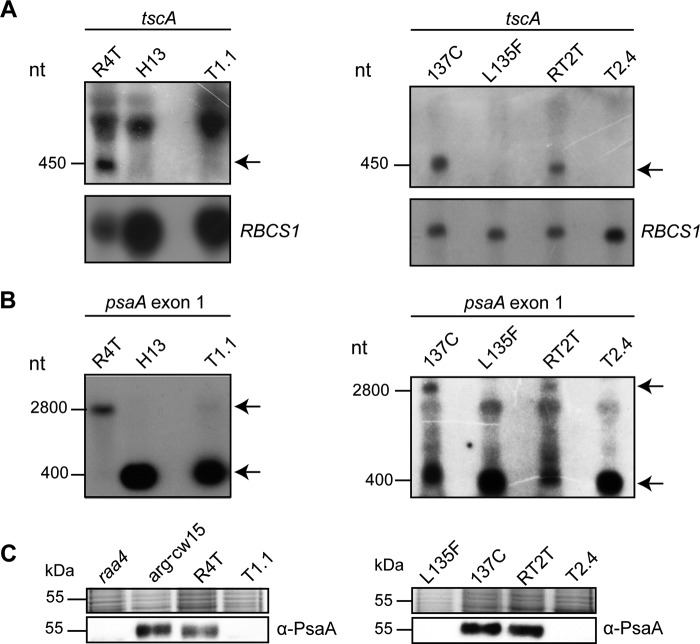
Transformants were characterized by Southern hybridization of BglII and NsiI restricted DNA with a radioactively labeled probe, carrying the aadA gene as well as parts of the tscA gene (supplemental Fig. S5). This analysis confirmed the deletion of tscA in selected transformants. Further proof for the absence of the tscA RNA was shown by Northern hybridizations (Fig. 3A). A tscA-specific probe detected the processed tscA RNA only in R4T and RT2T and not in transformants. The psaA exon 1-specific probe hybridized to the mature psaA mRNA in the host R4T as well as to the 400-nt psaA exon 1 transcript in transformants (Fig. 3B).
FIGURE 3.
Characterization of tscA deletion strains by Northern hybridization and immunodetection. Results for RT2T, carrying Rat2::TAP (left lane), and R4T, carrying Raa4::Tap (right lane) are shown. A, Northern hybridization with a labeled tscA probe. The tscA transcript corresponds to 450 nt, as indicated by an arrow. Rehybridization with an RBCS1 probe served as a loading control. B, RNA blot hybridization with a psaA exon 1 probe. The 2,800 nt signal detected with the psaA exon 1 probe represents the mature psaA transcript, whereas 400 nt corresponds to the primary psaA exon 1 transcript. C, immunodetection of the PsaA protein. RNA or crude cell extract of mutant strains H13, raa4, and L125F served as a negative control, whereas arg−cw15 and 137c as well as recipient strains R4T and RT2T represent positive controls. The chloroplast transformants T1.1 and T2.4 showed no PsaA protein. A Coomassie-stained gel served as a loading control.
A failure in psaA mRNA processing resulted in a lack of the PsaA protein and in photosynthesis deficiency. Thus, using crude cell extracts of R4TΔtscA and RT2TΔtscA for Western blot analysis, the absence of the PsaA protein in tscA deletion strains was confirmed (Fig. 3C). Furthermore, tscA deletion strains were not able to grow on medium without acetate and under high light conditions due to the defect in their photosynthetic apparatus (supplemental Fig. S6). Both R4TΔtscA and RT2TΔtscA were able to grow only under low light conditions.
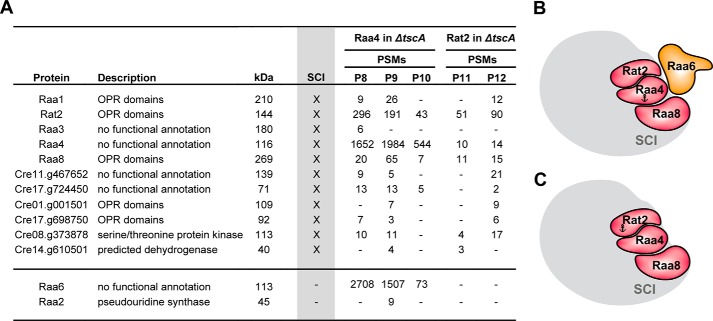
Three biological replicates of R4TΔtscA were used for TAP followed by LC-MS analysis. After exclusion of nonspecific proteins identified with control purifications (24), specific interaction partners were compared with the core components of subcomplex I (Fig. 4A). Surprisingly, only 3 of the 11 core proteins were found in the absence of the tscA RNA, namely Raa4, Rat2, and Raa8. In contrast, the trans-splicing factor Raa3, Raa1, and all further uncharacterized proteins were identified only with a low number of peptides and not in all three replicates of R4TΔtscA. Interestingly, in one replicate (P9), Raa1 was found together with Raa2. This observation suggests a transient association of both proteins with the remaining complex. The trans-splicing factor Raa2 is known to be essential for the second splicing reaction (20). Besides some core proteins of subcomplex I, we found trans-splicing factor Raa6 in all replicates of R4TΔtscA. We have experimental evidence that this protein is lacking in photosynthesis mutant Δraa6, which shows a defect in trans-splicing of psaA-i2. Raa6 was absent in all purifications with baits Raa4 and Rat2 with a functional tscA RNA. However, we obtained a large number of identified peptides of Raa6 with a tscA deletion background.
FIGURE 4.
Mass spectrometry data from TAP purifications using Raa4 or Rat2 as baits. A, both purifications were done with a recipient strain lacking the chloroplast tscA gene (ΔtscA). X, core components of subcomplex I (SCI), which were identified as described above. Peptide spectral matches (PSMs) of identified proteins are listed. Column kDa shows the predicted molecular mass in kDa. B and C, summary of MS data, when Raa4 or Rat2 was used as bait. Anchors indicate bait proteins used for TAP-MS.
To confirm the altered complex formation, two further tandem affinity purifications with strain RT2TΔtscA were conducted, and co-purified proteins were identified by LC-MS analysis (Fig. 4A, column Rat2 in ΔtscA). Again, nonspecific proteins were excluded, and this analysis revealed that besides the heterotrimer of Raa4, Rat2, and Raa8, further Cre08.g373878 was consistently co-purified with Rat2 with a deletion of the tscA RNA. The trans-splicing factor Raa6 was absent in TAP elutions of RT2TΔtscA, indicating a direct or more distinct association of Raa6 with Raa4.
We performed qRT-PCR analyses with both ΔtscA strains to elucidate whether the altered complex is still able to bind remaining intron RNAs. Strain RST-1 served as control strain, and expression of rrnL was analyzed as a reference for normalization. We showed that enrichment of psaA exon 1 primary transcript in TAP RNA of both tscA deletions was decreased compared with R4T and RT2T TAP RNA but was still enriched (7.0–7.7-fold the log2 ratio; supplemental Fig. S7). Moreover, the primary transcript of exon 2 still accumulates in TAP eluates but with no significant difference compared with RT2T and R4T TAP RNA. In addition, TAP RNAs of both tscA deletion strains were tested for the exon 1-exon 2 splicing intermediate as well as for tscA RNA, and, as expected, no RNA was measureable.
These TAP results indicate that the tscA RNA is crucial to form subcomplex I, and a deletion of this partial group II intron RNA results in a modified complex, consisting of at least Raa4, Rat2, and Raa8.
Interaction Network of the Plastid Splicing Factors Provides Evidence for the Formation of a Splicing Supercomplex
TAP analyses with bait Raa4::TAP in a tscA deletion strain revealed the association of Raa6 with the altered subcomplex I. Thus, these results imply a putative connection between both subcomplexes. To verify this, further TAP experiments with bait Raa6::TAP were conducted.
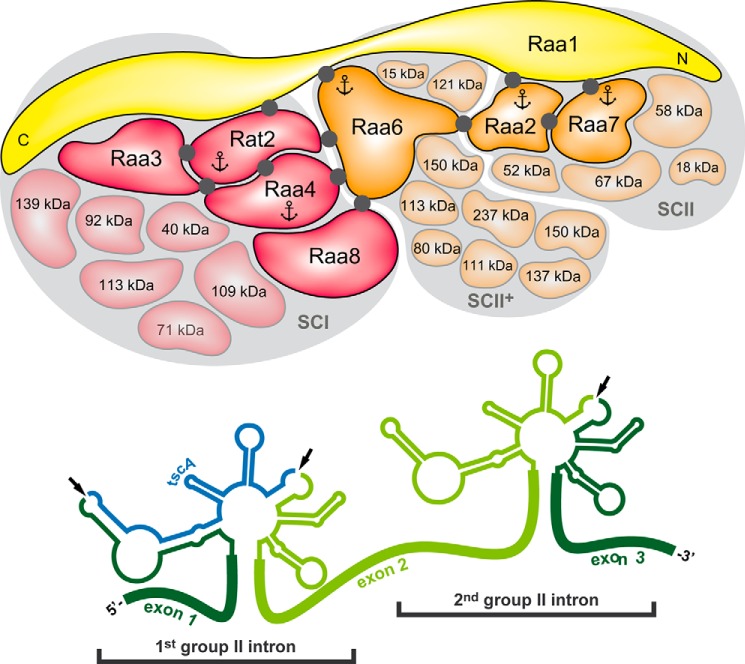
For TAP analyses, mutant T2-13, lacking a functional Raa6 protein, was transformed with a RAA6::TAP fusion. Transformants were selected under high light conditions, and genomic integration was verified via PCR in the positive transformant R6T (supplemental Fig. S8A). Furthermore, expression of the fusion protein Raa6::TAP and restoration of photosynthetic activity in strain R6T was shown by immunodetection, whereas mutant T2-13 showed no signals (supplemental Fig. S8B). Four biological replicates of R6T were used for three independent TAP experiments (P13–P15) and one single affinity chromatography purification (P16). Eluates were precipitated and analyzed with LC-MS analysis. TAP experiments with Raa6::TAP as bait exposed overlaps with the Raa2::TAP data set (Table 1). Proteins, found with bait Raa2 but not with the second subcomplex II bait Raa7, accumulated in Raa6::TAP eluates and included a protein with an RNA recognition motif (Cre03.g158950), two DEAD/DEAH box helicases (Cre01.g022350 and Cre07.g349300), and a tRNA-processing protein (Cre10.g428678). Moreover, core components of subcomplex I and II were purified with Raa6, but not in all replicates and with a low abundance (Table 1). This eluate contained Raa8 and Cre08.g373878, as well as the plastid ribosomal protein S20 (Cre12.g494750) and the threonine dehydratase (Cre02.g073200) of subcomplex II. Remarkably, the one-step purification (P16; Table 1) shows analogous results and reveals a co-purification of bait Raa6::TAP with Raa2, Rat2, and Raa4. Purification of subcomplex I and II components lacks consistency, and proteins were identified with a low number of peptides, thereby indicating a transient association of Raa6 with both subcomplexes during psaA mRNA trans-splicing. However, TAP results with Raa6 as bait indicate a connection of both subcomplexes and thus formation of a splicing supercomplex.
TABLE 1.
Raa6 co-purifies with Raa2::TAP-specific proteins and subunits of subcomplex I and II
Colors indicate subcomplex I (red), II (orange), and II+ (beige). Yellow marks Raa1, which is part of all subcomplexes.
To provide further evidence for such a supercomplex, we used SEC to determine the complex formation of each bait protein. Crude protein extracts of R4T, RT2T, R2T, and R6T were separated on a Superose 6 column. In total, 18 fractions corresponding to 20–2,500 kDa were collected; proteins were then separated via SDS-PAGE and finally analyzed using immunodetection with either α-RbcL as a control or α-calmodulin antibody for detection of the TAP fusion proteins. In C. reinhardtii, the holoenzyme of Rubisco has an estimated size of 560 kDa (26). Signals were observed in fractions corresponding to ∼600–200 kDa.
As shown in Fig. 5, Raa4:: TAP and Rat2 ::TAP fusion proteins were detected in fractions corresponding to a molecular mass of 1,200–1,800 kDa. Based on the predicted kDa values for the 11 proteins identified as core components in TAP experiments, the calculated complex size of 1,483 kDa is within the range obtained from the size exclusion chromatography. Raa2::TAP showed two distinct signal patterns. First, Raa2::TAP was detected in fractions corresponding to 200–1,000 kDa with a peak signal at about 500 kDa. Of note is that the calculated molecular mass of subcomplex II of about 580 kDa is consistent with this result. The signal pattern of this complex occurs in the same fractions as the signal distribution of RbcL, and thus, both complexes should have approximately the same size of about 500 kDa. Second, Raa2::TAP signals also occurred in fractions with high molecular masses of about 1,500–2,000 kDa. This complex might derive from interaction with psaA precursor RNAs, which were shown to elute with Raa2 in TAP eluates as well as with other proteins. Surprisingly, Raa6::TAP co-localized with Raa2, Raa4, and Rat2 in the fraction corresponding to ∼2,000 kDa, indicating an association with both subcomplexes. An additional Raa6 band was detected at a low molecular mass of 25 kDa that probably represents protein degradation products. Taken together, these findings provide further evidence for the formation of a splicing supercomplex connecting both subcomplexes and, thus, both psaA mRNA splicing reactions.
FIGURE 5.
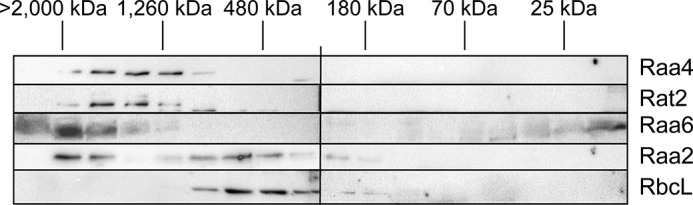
Splicing factors are part of high molecular weight complexes. Crude cell extracts of R4T, RT2T, R6T, and R2T were subjected to size exclusion chromatography, and 18 fractions were analyzed by immunoblotting with antiserum against the TAP sequence. As a control, the large subunit of Rubisco, which has an estimated size of 560 kDa, was detected using an α-rbcL antibody. Gel filtration molecular weight markers (Sigma) were used for determination of the molecular weight range of the fractions.
Yeast two-hybrid experiments were performed to ascertain whether Raa6 is also able to undergo direct protein-protein binding with splicing factors of both subcomplexes. In this analysis, we tested yeast strains carrying plasmid combinations encoding different trans-splicing factors either as bait or as prey proteins. Different fragments of Raa6 were used as bait and were shown to interact with subfragments of Raa1, Raa2, Rat2, Raa3, and Raa4 (Fig. 6). Interestingly (27, 28), Raa6-B interacted with several subfragments of the prey proteins, an observation that was described similarly by other investigators (27, 28). This further analysis confirms that Raa6 does indeed interact with subunits that belong to either subcomplex I or subcomplex II, thereby suggesting that Raa6 may be part of a splicing supercomplex.
FIGURE 6.
Direct protein-protein interactions between psaA-mRNA trans-splicing factors. A, full-length proteins and protein fragments of psaA mRNA splicing factors Raa1, Raa2, Rat2, Raa3, Raa4, Raa6, and Raa8 were tested for direct protein-protein interactions in yeast two-hybrid experiments. B, yeast strains carrying plasmids, comprising different protein fragments as shown in A fused to either the GAL4 activation (AD) or DNA binding domain (BD) were spotted onto SD medium lacking leucine, tryptophan, histidine, and adenine. Growth of yeast strains on SD medium lacking leucine, tryptophan, histidine, and adenine indicates interaction between proteins fused to the activation and DNA binding domains. To exclude the possibility of transactivation, all yeast strains were co-transformed with plasmids carrying either only the GAL4 DNA-binding domain (pGADT7) or the GAL4 activation domain (pGBKT7). Growth tests on SD medium lacking leucine and tryptophan to show plasmid uptake of both plasmids are shown in supplemental Fig. S9. C, scheme of direct protein-protein interactions identified in yeast two-hybrid experiments, indicated with black arrows. Gray, dashed arrows show direct protein-protein interactions identified previously (17, 24). cTP, chloroplast targeting peptide; OPR, octatricopeptide repeat.
Discussion
Group II introns are proposed to have an ancestral relationship to spliceosomal introns because splicing mechanisms and secondary intron structure share functional and structural similarities with nuclear pre-mRNA (3, 6, 7, 29). Decades ago, genetic investigations had already demonstrated the contribution of distinct proteins in organelle intron splicing (30). However, no RNA-protein complex comparable with the spliceosome has ever been described for any organelle.
Here we provide the first characterization of a high molecular RNP apparatus participating in psaA mRNA splicing. Using a total of five different bait proteins for TAP experiments, we were able to uncover core components of two trans-splicing subcomplexes. Our extended work finally discovered for the first time a supercomplex for group II intron splicing.
Subcomplex I is involved in splicing of psaA-i1 and is composed of Rat2, Raa4, Raa3, Raa8, and Raa1 as well as at least six further uncharacterized proteins. These proteins are putative trans-splicing factors and have highly specific interaction partners. Of particular importance are Cre01.g001501 and Cre17.g698750, two proteins with predicted OPR domains. In C. reinhardtii, 43 OPR proteins were predicted that participate in RNA metabolism-related processes (31). This includes the psaA mRNA splicing factors Raa1, Rat2, and Raa8 (18, 21, 31, 32). In most land plants, the genomes encode only a single OPR gene (33). However, interestingly, they exhibit tetratricopeptide repeat, pentatricopeptide repeat, and mTERF proteins, which also belong to the helical repeat superfamily and are involved in RNA processing (16, 34, 35), thereby demonstrating that despite the independent origin of group II introns in green algae and land plants, a co-evolution of helical repeat superfamily proteins as splicing factors has occurred.
Subcomplex II is involved in psaA-i2 splicing and consists of at least Raa1, Raa2, Raa7, and four further uncharacterized proteins. Interestingly, besides Raa1, no OPR proteins were found in subcomplex II. This finding can be explained with the different origin of the psaA group II introns. Both introns occur in the algal lineage of volvocales, and both are inserted at the same location of the psaA gene (36, 37). In the more distant green alga Scenedesmus obliquus, the psaA gene is solely disrupted by a single dipartite group II intron with an insertion site within psaA that is identical to the psaA-i2 of C. reinhardtii (38). This implies that psaA-i2 and its splicing machinery emerged first during evolution and were later accompanied by the splicing factors of psaA-i1.
Our study further shows that both subcomplexes are purified with target intron RNA, indicating the formation of RNP complexes during psaA mRNA splicing. Components of subcomplex I are co-purified with exon 1 precursor RNA and tscA RNA, both harboring subfragments of psaA-i1. The lack of tscA RNA prevents complex formation and leads to a modified complex with a heterotrimeric core composed of Raa4, Rat2, and Raa8. This finding is consistent with previous results showing that Raa3 together with psaA exon 1 transcript is found in a smaller 900-kDa complex in a tscA deletion strain (23). Together with our qRT-PCR data, we provide strong evidence that intron RNA is a prerequisite for the formation of subcomplex I. Although subcomplex II was purified with psaA-i2-specific intron RNA, an RNA-independent, proteinogenous variant was found to accumulate in the chloroplast. This finding is supported by SEC experiments, showing two different Raa2-associated complexes of 2,000 and 500 kDa. The 500-kDa complex was previously reported to be nonsensitive to RNase treatment, and its formation is completely abolished in mutants lacking functional Raa1 and Raa7 (25).
Although the catalytic activity of group II introns is provided by RNA, splicing is dependent on RNP formation. During splicing, the majority of bacterial group II introns form an RNP together with the intron-encoded maturase. Organellar group II introns usually are more degenerated then their bacterial ancestors (9). This might explain the complexity of organellar RNPs, which harbor more protein factors due to the lack of several functional RNA structures. Chloroplasts of land plants carry about 20 group II introns, and their excision is at least dependent on 16 nucleus-encoded factors (11, 39). Most of these factors were found in RNPs with a molecular size of 500–1,000 kDa (34, 40–42). The identification of psaA-mRNA splicing subcomplexes I and II as the most complex intermediate stage of group II intron fragmentation and protein-rich splicing RNPs provides further evidence for an evolutionary development from simple bacterial RNPs, consisting of a single RNA molecule and a maturase, to more complex RNPs that compensate fragmentation and degeneration of group II intron structures. This evolutionary development may have also occurred for the five protein-rich snRNPs, which are part of the nuclear spliceosome machinery (1). In particular, it has been hypothesized that the ancestral ribosomal group II intron fragmented into U2, U5, and U6 snRNA of the spliceosome (9). Furthermore Prp8, a nuclear key splicing factor, shows several similarities with group II intron maturases and, thus, may have evolved from an ancestral intron-encoded protein (43). It was also proposed that nuclear splicing factors and the U1 and U4 snRNAs, which lack similarities to group II introns and their maturases, were recruited or evolved de novo (9). Thus, the in trans-acting RNAs and the huge set of splicing factors, which we discovered in the splicing machinery of the psaA mRNA, furnish proof for such a progressive event during the evolution of the nuclear spliceosome.
In TAP experiments with Rat2:: TAP and Raa4 ::TAP, no trans-splicing factor was identified that acts solely on the second trans-splicing reaction. In contrast to this finding, the first trans-splicing reaction is prevented in tscA deletion strains, and an intermediate in the assembly pathway was purified, thereby presenting an altered complex that also contains factors Raa2 and Raa6. Both factors are essential for splicing of psaA-i2 (20). This finding suggests a connection between both splicing reactions (Fig. 7). Using Raa6 as bait in TAP experiments, we also showed that Raa6 was purified not only with subcomplex II-associated proteins but also with components of subcomplex I. Further, yeast two-hybrid experiments demonstrated a direct protein-protein interaction of Raa6 with Raa1, Rat2, Raa4, Raa3, and Raa2. Formation of a supercomplex during psaA mRNA splicing does not seem to be essential, because splicing of the first intron is not defective in class A mutants, and vice versa (17–19). Nonetheless, the association of both subcomplexes guarantees a coordination of both reactions and a more efficient splicing reaction. Furthermore, such a supercomplex might coordinate not only splicing but also processing of intron RNA, as was shown for nuclear supraspliceosomes. Recent analysis of the nuclear pre-mRNA processing machinery revealed that proteins involved in RNA 3′-end processing are integral components of supraspliceosomes (44). This is reminiscent of the function of psaA splicing factors Rat2 and Raa1, which are both involved in 3′-end processing of the tscA RNA (15, 22).
FIGURE 7.
trans-Splicing apparatus of the plastid psaA-mRNA. Depicted are both psaA-mRNA group II introns, psaA-i1 and psaA-i2, with a characteristic secondary RNA structure and identified splicing complexes. Comparative TAP-MS analyses with baits Raa4 and Rat2 led to the identification of subcomplex I (SCI), which is composed of at least 11 core proteins (red; see Fig. 1A). Further, using Raa7 and Raa2 as baits, subcomplex II (SCII) with at least seven core components (orange; see Fig. 1C) was identified. Both subcomplexes share the general splicing factor Raa1 (yellow). Complementation studies showed that the C terminus of Raa1 is involved in psaA-i1 splicing, whereas the central part of the Raa1 protein is essential for splicing of psaA-i2 (21). TAP-MS experiments with bait Raa6 revealed co-purification with Raa2-associated proteins (SCII+; see Table 1) and several core components of subcomplexes I and II. Black dots, direct protein-protein interactions, as detected in yeast two-hybrid analysis (this work; see Ref. 24). Anchors mark proteins that were used as baits in TAP-MS experiments. For clarification, intron RNAs and protein complexes are shown separately, and every splicing factor is present in 1:1 stoichiometry. Dark-colored proteins were previously identified as trans-splicing factors (17–20, 22, 23).
Mutant Δraa6 shows a defect in splicing of psaA-i2, and consequently, we predicted that Raa6 would form part of subcomplex II. However, Raa6 was not purified either with Raa2 or Raa7 as bait in TAP experiments. Thus, Raa6 is not a core component of subcomplex II. Instead, SEC experiments revealed that Raa6 is part of a complex with a size of about 2,000 kDa and co-localizes with Raa2 in protein fractions corresponding to this size. Further, TAP-MS experiments indicated an association of Raa6 with Raa2 and Raa2-interacting proteins. Collectively, these findings suggest a transient participation of Raa6 in splicing of the second psaA group II intron, which might occur in the late stage of splicing and after formation of an RNP. We thus propose that Raa6 and interacting proteins stabilize binding of subcomplex II with the target intron RNA to bring the dipartite intron into a catalytically active structure.
Our study advances our current understanding of group II intron splicing and the evolutionary relationship of group II introns with spliceosomal introns. In addition, this study also shows that two RNP subcomplexes are built up for each splicing step, which ultimately assemble into a supercomplex during psaA trans-splicing. Ongoing work will elucidate the role of the uncharacterized proteins that form part of the RNP complex as well as investigate the mode of time-dependent assembly of different complexes formed.
Experimental Procedures
Strains and Growth Conditions
C. reinhardtii strains and their growth conditions are listed in supplemental Table S3. When necessary, Tris acetate-phosphate medium or high salt minimal medium (45) was supplemented with spectinomycin (Sigma).
Transformation
Nuclear transformation was achieved by agitation with glass beads (46), and cells were treated with autolysin before transformation to remove cell walls.
Chloroplast transformation was performed with a homemade particle gun (47, 48). Before transformation, cells were plated on Tris acetate-phosphate medium and incubated for 16–24 h under high light conditions. After transformation, cells were grown for 16–24 h under low light conditions, transferred to Tris acetate-phosphate medium supplemented with spectinomycin (100 μg/ml), and placed under low light conditions for 3–4 weeks. To obtain homoplasmic lines, transformants were replated several times on Tris acetate-phosphate medium with spectinomycin.
Construction of Plasmids
Recombinant plasmids and oligonucleotides used for PCR experiments or generation of transgenic algal strains are listed in supplemental Table S4 and supplemental Table S5, respectively.
Generation of a RAT2::TAP construct was achieved by homologous recombination in yeast using linearized pRS426 (49). gDNA of RAT2 was amplified in four overlapping fragments (F1–F4). Fragments F1–F4 were amplified with oligonucleotides for_Rat2_IF and 815.3D, for_Rat2-F2 and rev_Rat2-F2, for_Rat2-F3 and rev_Rat2-F4, and for_Rat2-F4 and rev_Rat2-IF, respectively. For amplification of the artificial RBCS2/HSP70 tandem promoter (primers for_hsp70A_HR and rev_5′rbcS2_HR) and the TAP tag gene in fusion with the 3′-UTR of LHCB1 (primers for_TAP_3′lhcb_HR and rev_3′lhcb_HR), plasmid pCM10 was used as template. Fragments were amplified using a 25-bp overhang for recombination, resulting in plasmid pRat2::TAP.
For construction of pRAA2::TAP, gDNA of RAA2 was amplified using primers for_Raa2_TAP and rev_Raa2_TAP and cloned into pDrive, resulting in pCM6. After digestion of pCM6 and pCM10 with XbaI, the gDNA of RAA2 was cloned into pCM10, resulting in pRAA2::TAP.
For complementation analysis of mutant strain L135F, we constructed plasmid RAT1::3xHA. In the first step, the 3xHA antisense oligonucleotide was filled up by PCR using primer for_3xHA. After digestion with BglII, the fragment was introduced into BglII-linearized pCr1 (pIG3197-3). RAT1 was amplified using wild-type gDNA and primers for_Rat1-IF and rev_Rat1-IF. The PCR product was inserted in NheI-linearized pIG3197-3 using the In-Fusion® HD cloning kit (Clontech), which resulted in plasmid pCM51.
For generation of a spectinomycin-resistant cassette, the aadA gene encoding the enzyme aminoglycoside 3′-adenyltransferase (AAD) was amplified from pBatTL. To drive expression of the aadA gene, 5′- and 3′-UTRs of psbD and rbcL mRNAs were amplified from gDNA using primers with an overhang to linearized pRS426 (primers for_5′UTR_psbD and rev_psbD, for_aadA and rev_aadA, and for_rbcL and rev_3′UTR_rbcL). After recombination, plasmid pCM43 was digested with PacI and StuI to release the aadA resistance cassette. The tscA gene was amplified using oligonucleotides for_tscA and rev_tscA and cloned into pDrive, resulting in pCM45. After digestion with PacI and StuI, an internal 0.8-kb tscA fragment was replaced by the aadA resistance cassette (pCM49). The R12 fragment of pIG637.1 was digested with LguI and EheI and then integrated in pRS426 using homologous recombination in yeast. The resulting recombinant plasmid was assigned pCM57. After digestion of pCM49 with PacI, the tscA::aadA knock-out cassette was inserted into pCM57 by homologous recombination through flanking regions of tscA, resulting in pCM59.
For construction of pRaa6::TAP, the full-length gDNA of RAA6 was amplified with primers OOR_59 and OOR_60 and cloned into pDrive, resulting in pOR11. After digestion with NheI, the gDNA was further cloned into pCM10 upstream of the TAP tag sequence, resulting in pRaa6::TAP.
For the generation of yeast two-hybrid vectors carrying Raa6 subfragments, RAA6 fragments coding for amino acids 1–550 (pGADT7_Raa6-A) and 520–1118 (pGADT7_Raa6-B) were amplified from cDNA (primers pGAD_Raa6_Infu1, pGAD_Raa6_Infu2, pGAD_Raa6_Infu3, and pGAD_Raa6_Infu4) and inserted into EcoRI- and NdeI-restricted vector pGADT7 by in-fusion cloning (Clontech, Mountain View, CA). For construction of pGADT7_Raa6-FL, a fragment coding for amino acids 513–1118 was amplified from cDNA (primers for_29701_Fragment_2 and rev_pGAD_Raa6_Infu2) and inserted by in-fusion cloning (Clontech) in pGADT7_Raa6-A, which was linearized with EcoRI beforehand. The pGBKT7 vectors (pGBKT7_Raa6-A, pGBKT7_Raa6-B, and pGBKT7_Raa6-FL) were generated by excising the RAA6 fragments from the respective pGADT7 constructs with EcoRI and NdeI. The obtained fragments were then inserted into pGBKT7.
To construct Raa8 two-hybrid plasmids, cDNA fragments coding for amino acids 225–396 (pGADT7_Raa8-A), 389–844 (pGADT7_Raa8-B), 839–1323 (pGADT7_Raa8-C), and 1303–1615 (pGADT7_Raa8-D) were amplified (primers for-IF-Raa8-F1, rev-IF-Raa8-F1, for-IF-Raa8-F2, rev-IF-Raa8-F2, IF-42B-for, IF-42B-rev, IF-46A-for, and IF-46A-rev) and cloned into EcoRI- and NdeI-restricted vector pGADT7 by in-fusion cloning (Clontech).
Molecular Genetic Techniques
Standard molecular techniques were used as reported elsewhere (19, 24). C. reinhardtii total RNA isolation and RNA blot experiments were done as described previously (14, 19, 22). Transcripts were detected using radioactively labeled probes. For Southern analysis, total DNA isolation from C. reinhardtii was performed as described previously (50). For DNA blot experiments, 20 μg of DNA was digested and loaded on a 0.8% agarose gel. DNA samples were transferred to nylon membranes and hybridized with a radioactively labeled probe.
For PsaA immunoblot analysis, total protein extracts were loaded on 12% SDS-polyacrylamide gels with 6 m urea. After gel electrophoresis and blotting onto a PVDF membrane (Roche Applied Science), membrane was blocked with 5% nonfat dry milk in TBS (0.5 m Tris, 1.5 m NaCl). After incubation with rabbit polyclonal PsaA antibody (Agrisera) at 4 °C overnight, the membrane was decorated with peroxidase-linked anti-rabbit IgG for 1 h. Signals were detected using Western ECL substrate (Bio-Rad).
Tandem Affinity Purification
Tandem affinity purification was performed as described elsewhere (24). For purifications with Raa2, pelleted cells were resuspended in lysis buffer (100 mm Tris, 150 mm NaCl, pH 8.0) containing 0.5% n-dodecyl β-d-maltoside. Cells were lysed by sonication (four times for 60 s, 70–90% power) and incubated on ice for 20 min with agitation. After centrifugation (25,000 × g, 30 min, 4 °C), the supernatant was used for TAP purification.
LC-MS Analysis
For TAP experiments with strains Rat2::TAP and R4TΔtscA, MudPIT-MS/MS analyses were performed as described previously (24). TAP purifications with strain RT2TΔtscA were processed as described previously (17).
Data evaluation was carried out with Proteome Discoverer version 1.4 (Thermo Scientific), and the MS raw data were searched against the C. reinhardtii database 409_Creinhardtii_ed_Chlamydomonas reinhardtii_v5.5, which comprises 19,526 entries from the nuclear genome downloaded on September 18, 2014, and 69 entries from the C. reinhardtii chloroplast genome using two search algorithms Mascot version 2.4 (Matrix Science) and SEQUEST. Search settings were applied as described previously (17).
Quantitative RT-PCR
TAP eluates containing nucleic acid were purified as described elsewhere (24). For successful DNase I treatment, probes were cleaned with Amicon YM-30 columns (Millipore), followed by DNase I treatment for 30 min at 25 °C. For qRT-PCR, the One-Step qRT-PCR kit (KAPA Sybr Fast ABI Prism, Peqlab, Erlangen, Germany) was used. Of a total volume of 44 μl, 1 μl of each sample was subjected to DNase I treatment. The qRT-PCR and analysis of two independent replicates were performed as described previously (24). The following primer pairs were used for amplification of precursor RNAs and spliced variants: exon 1 precursor, OOR_113 and OOR_114; exon 2 precursor, OOR48 and rev_psaA_Ex2Int2; exon 3 precursor, OOR_99 and OOR_98; tscA RNA, tscA-RT_for and tscA-RT_rev; spliced psaA exons 1 and 2, OOR_113 and rev_psaA_Ex2Int2; spliced psaA exons 2 and 3, OOR_48 and OOR_99.
Sequence Analysis
Molecular masses of proteins in kDa were calculated by the program Clone Manager 9 Professional Edition (Scientific and Educational Software, Cary, NC). Sequences of all identified proteins and predicted protein domains were retrieved from the C. reinhardtii Joint Genome Institute database version 5.5 (51).
Size Exclusion Chromatography
A 1-liter culture at 2 × 106 cells/ml was used for size exclusion chromatography. After centrifugation, cells were resuspended in the same lysis buffer as for TAP experiments, followed by sonification and two successive centrifugation steps (15 min at 9,000 × g and 60 min at 35,000 × g). The supernatant was filtered with a 0.45-μm filter (Millipore), and then a 1-mg crude extract was loaded on a Superose 6 10/300 GL (GE Healthcare) using an ÄKTA purifier 100 (GE Healthcare). TAP lysis buffer was used for elution of 50 0.5-ml fractions with 0.4 ml/min. Fractions of R4T and RT2T were precipitated overnight at −20 °C using 6–8 volumes of 100% ethanol. After centrifugation, pellets were washed with 70% ethanol and resuspended in loading buffer. Fractions of R2T and R6T were precipitated by 10% TCA for 30 min on ice, pelleted by centrifugation, and washed twice with acetone. Protein extracts were loaded on an 8% (Rat2, Raa4, and Raa6) or 12% (Raa2 and RbcL) SDS-polyacrylamide gel. TAP-tagged proteins were detected by immunoblotting by incubation overnight with an α-calmodulin antibody (Millipore) and for 2 h with α-rabbit IgG HRP-linked antibody (Cell Signaling). Gel filtration molecular weight markers (Sigma) were used for determination of the molecular weight range of the fractions. To measure the void volume, dextran blue was used.
Yeast Two-hybrid Experiments
For yeast two-hybrid experiments, Saccharomyces cerevisiae strain PJ69-4A (52) was co-transformed with a bait and prey plasmid. Analyses and assays were performed as described previously (53).
Author Contributions
O. R., C. M., and J. J. performed the microbiological, biochemical, and genetic experiments. L. K. and D. W. performed the LC-MS. J. J., L. K., A. S., and D. W. made critical reading and editing of the manuscript. U. K., A. S., and D. W. supervised the research. O. R., C. M., and U. K. wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Katja Schmitt, Ivonne Schelberg, and Ingeborg Godehardt for excellent technical assistance.
This work was supported by Deutsche Forschungsgemeinschaft (Bonn Bad-Godesberg, Germany) Grants KU 517/13-1 (to U. K.) and JA 2296/1-1 (to J. J.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Tables S1–S6 and Figs. S1–S9.
- TAP
- tandem affinity purification
- MudPIT
- multidimensional protein identification technology
- OPR
- octatricopeptide repeat
- qRT-PCR
- quantitative RT-PCR
- nt
- nucleotide(s)
- gDNA
- genomic DNA
- AAD
- aminoglycoside 3′-adenyltransferase
- Rubisco
- ribulose-bisphosphate carboxylase/oxygenase.
References
- 1. Will C. L., and Lührmann R. (2011) Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3, a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen W., and Moore M. J. (2015) Spliceosomes. Curr. Biol. 25, R181–183 [DOI] [PubMed] [Google Scholar]
- 3. Cech T. R. (1986) The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell 44, 207–210 [DOI] [PubMed] [Google Scholar]
- 4. Cavalier-Smith T. (1991) Intron phylogeny: a new hypothesis. Trends Genet. 7, 145–148 [PubMed] [Google Scholar]
- 5. Robart A. R., Chan R. T., Peters J. K., Rajashankar K. R., and Toor N. (2014) Crystal structure of a eukaryotic group II intron lariat. Nature 514, 193–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Keating K. S., Toor N., Perlman P. S., and Pyle A. M. (2010) A structural analysis of the group II intron active site and implications for the spliceosome. RNA 16, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lambowitz A. M., and Zimmerly S. (2011) Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb. Perspect. Biol. 3, a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sharp P. A. (1991) Five easy pieces. Science 254, 663. [DOI] [PubMed] [Google Scholar]
- 9. Zimmerly S., and Semper C. (2015) Evolution of group II introns. Mob. DNA 6, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bonen L. (2008) Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion 8, 26–34 [DOI] [PubMed] [Google Scholar]
- 11. Brown G. G., Colas des Francs-Small C., and Ostersetzer-Biran O. (2014) Group II intron splicing factors in plant mitochondria. Front. Plant Sci. 5, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Glanz S., and Kück U. (2009) Trans-splicing of organelle introns: a detour to continuous RNAs. Bioessays 31, 921–934 [DOI] [PubMed] [Google Scholar]
- 13. Lemieux C., Otis C., and Turmel M. (2014) Six newly sequenced chloroplast genomes from prasinophyte green algae provide insights into the relationships among prasinophyte lineages and the diversity of streamlined genome architecture in picoplanktonic species. BMC Genomics 15, 857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kück U., Choquet Y., Schneider M., Dron M., and Bennoun P. (1987) Structural and transcription analysis of two homologous genes for the P700 chlorophyll a-apoproteins in Chlamydomonas reinhardtii: evidence for in vivo trans-splicing. EMBO J. 6, 2185–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goldschmidt-Clermont M., Choquet Y., Girard-Bascou J., Michel F., Schirmer-Rahire M., and Rochaix J. D. (1991) A small chloroplast RNA may be required for trans-splicing in Chlamydomonas reinhardtii. Cell 65, 135–143 [DOI] [PubMed] [Google Scholar]
- 16. de Longevialle A. F., Small I. D., and Lurin C. (2010) Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Mol. Plant 3, 691–705 [DOI] [PubMed] [Google Scholar]
- 17. Lefebvre-Legendre L., Reifschneider O., Kollipara L., Sickmann A., Wolters D., Kück U., and Goldschmidt-Clermont M. (2016) A pioneer protein is part of a large complex involved in trans-splicing of a group II intron in the chloroplast of Chlamydomonas reinhardtii. Plant J. 85, 57–69 [DOI] [PubMed] [Google Scholar]
- 18. Marx C., Wünsch C., and Kück U. (2015) The octatricopeptide repeat protein Raa8 is required for chloroplast trans splicing. Eukaryot. Cell 14, 998–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Glanz S., Jacobs J., Kock V., Mishra A., and Kück U. (2012) Raa4 is a trans-splicing factor that specifically binds chloroplast tscA intron RNA. Plant J. 69, 421–431 [DOI] [PubMed] [Google Scholar]
- 20. Perron K., Goldschmidt-Clermont M., and Rochaix J. D. (1999) A factor related to pseudouridine synthases is required for chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 18, 6481–6490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Merendino L., Perron K., Rahire M., Howald I., Rochaix J. D., and Goldschmidt-Clermont M. (2006) A novel multifunctional factor involved in trans-splicing of chloroplast introns in Chlamydomonas. Nucleic Acids Res. 34, 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Balczun C., Bunse A., Hahn D., Bennoun P., Nickelsen J., and Kück U. (2005) Two adjacent nuclear genes are required for functional complementation of a chloroplast trans-splicing mutant from Chlamydomonas reinhardtii. Plant J. 43, 636–648 [DOI] [PubMed] [Google Scholar]
- 23. Rivier C., Goldschmidt-Clermont M., and Rochaix J. D. (2001) Identification of an RNA-protein complex involved in chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 20, 1765–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jacobs J., Marx C., Kock V., Reifschneider O., Fränzel B., Krisp C., Wolters D., and Kück U. (2013) Identification of a chloroplast ribonucleoprotein complex containing trans-splicing factors, intron RNA, and novel components. Mol. Cell. Proteomics 12, 1912–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perron K., Goldschmidt-Clermont M., and Rochaix J. D. (2004) A multiprotein complex involved in chloroplast group II intron splicing. RNA 10, 704–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Spreitzer R. J., and Salvucci M. E. (2002) Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Annu. Rev. Plant Biol. 53, 449–475 [DOI] [PubMed] [Google Scholar]
- 27. Teichert I., Steffens E. K., Schnass N., Fränzel B., Krisp C., Wolters D. A., and Kück U. (2014) PRO40 is a scaffold protein of the cell wall integrity pathway, linking the MAP kinase module to the upstream activator protein kinase C. PLoS Genet. 10, e1004582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shahheydari H., Frost S., Smith B. J., Groblewski G. E., Chen Y., and Byrne J. A. (2014) Identification of PLP2 and RAB5C as novel TPD52 binding partners through yeast two-hybrid screening. Mol. Biol. Rep. 41, 4565–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Toor N., Keating K. S., Taylor S. D., and Pyle A. M. (2008) Crystal structure of a self-spliced group II intron. Science 320, 77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lazowska J., Jacq C., and Slonimski P. P. (1980) Sequence of introns and flanking exons in wild-type and box3 mutants of cytochrome b reveals an interlaced splicing protein coded by an intron. Cell 22, 333–348 [DOI] [PubMed] [Google Scholar]
- 31. Rahire M., Laroche F., Cerutti L., and Rochaix J. D. (2012) Identification of an OPR protein involved in the translation initiation of the PsaB subunit of photosystem I. Plant J. 72, 652–661 [DOI] [PubMed] [Google Scholar]
- 32. Eberhard S., Loiselay C., Drapier D., Bujaldon S., Girard-Bascou J., Kuras R., Choquet Y., and Wollman F. A. (2011) Dual functions of the nucleus-encoded factor TDA1 in trapping and translation activation of atpA transcripts in Chlamydomonas reinhardtii chloroplasts. Plant J. 67, 1055–1066 [DOI] [PubMed] [Google Scholar]
- 33. Kleinknecht L., Wang F., Stübe R., Philippar K., Nickelsen J., and Bohne A. V. (2014) RAP, the sole octotricopeptide repeat protein in Arabidopsis, is required for chloroplast 16S rRNA maturation. Plant Cell 26, 777–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hammani K., and Barkan A. (2014) An mTERF domain protein functions in group II intron splicing in maize chloroplasts. Nucleic Acids Res. 42, 5033–5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hsu Y. W., Wang H. J., Hsieh M. H., Hsieh H. L., and Jauh G. Y. (2014) Arabidopsis mTERF15 is required for mitochondrial nad2 intron 3 splicing and functional complex I activity. PLoS One 11, e112360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smith D. R., and Lee R. W. (2009) The mitochondrial and plastid genomes of Volvox carteri: bloated molecules rich in repetitive DNA. BMC Genomics 10, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Smith D. R., Lee R. W., Cushman J. C., Magnuson J. K., Tran D., and Polle J. E. W. (2010) The Dunaliella salina organelle genomes: large sequences, inflated with intronic and intergenic DNA. BMC Plant Biol. 10, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. de Cambiaire J. C., Otis C., Lemieux C., and Turmel M. (2006) The complete chloroplast genome sequence of the chlorophycean green alga Scenedesmus obliquus reveals a compact gene organization and a biased distribution of genes on the two DNA strands. BMC Evol. Biol. 6, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Germain A., Hotto A. M., Barkan A., and Stern D. B. (2013) RNA processing and decay in plastids. Wiley Interdiscip. Rev. RNA 4, 295–316 [DOI] [PubMed] [Google Scholar]
- 40. Watkins K. P., Kroeger T. S., Cooke A. M., Williams-Carrier R. E., Friso G., Belcher S. E., van Wijk K. J., and Barkan A. (2007) A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell 19, 2606–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kroeger T. S., Watkins K. P., Friso G., van Wijk K. J., and Barkan A. (2009) A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc. Natl. Acad. Sci. U.S.A. 106, 4537–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Khrouchtchova A., Monde R. A., and Barkan A. (2012) A short PPR protein required for the splicing of specific group II introns in angiosperm chloroplasts. RNA 18, 1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dlakić M., and Mushegian A. (2011) Prp8, the pivotal protein of the spliceosomal catalytic center, evolved from a retroelement-encoded reverse transcriptase. RNA 17, 799–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kotzer-Nevo H., de Lima Alves F., Rappsilber J., Sperling J., and Sperling R. (2014) Supraspliceosomes at defined functional states portray the pre-assembled nature of the pre-mRNA processing machine in the cell nucleus. Int. J. Mol. Sci. 15, 11637–11664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Harris E. H. (1989) The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use, pp. 25–64, Academic Press, Inc., San Diego, CA: [DOI] [PubMed] [Google Scholar]
- 46. Kindle K. L. (1990) High frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 87, 1228–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Boynton J. E., Gillham N. W., Harris E. H., Hosler J. P., Johnson A. M., Jones A. R., Randolph-Anderson B. L., Robertson D., Klein T. M., and Shark K. B. (1988) Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 240, 1534–1538 [DOI] [PubMed] [Google Scholar]
- 48. Klein R. M., Wolf E. D., Wu R., and Sanford J. C. (1992) High-velocity microprojectiles for delivering nucleic acids into living cells. Biotechnology 24, 384–386 [PubMed] [Google Scholar]
- 49. Christianson T. W., Sikorski R. S., Dante M., Shero J. H., and Hieter P. (1992) Multifunctional yeast high-copy-number shuttle vectors. Gene 110, 119–122 [DOI] [PubMed] [Google Scholar]
- 50. Newman S. M., Boynton J. E., Gillham N. W., Randolph-Anderson B. L., Johnson A. M., and Harris E. H. (1990) Transformation of chloroplast ribosomal RNA genes in Chlamydomonas: molecular and genetic characterization of integration events. Genetics 126, 875–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., Witman G. B., Terry A., Salamov A., Fritz-Laylin L. K., Maréchal-Drouard L., Marshall W. F., Qu L. H., Nelson D. R., Sanderfoot A. A., Spalding M. H., et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. James P., Halladay J., and Craig E. A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bloemendal S., Bernhards Y., Bartho K., Dettmann A., Voigt O., Teichert I., Seiler S., Wolters D. A., Pöggeler S., and Kück U. (2012) A homologue of the human STRIPAK complex controls sexual development in fungi. Mol. Microbiol. 84, 310–323 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.