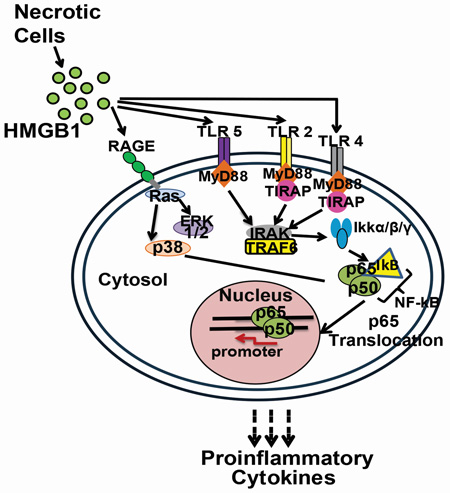
SUMMARY
Infectious and sterile inflammatory diseases are correlated with increased levels of high mobility group box-1 (HMGB1) in tissues and serum. Extracellular HMGB1 is known to activate toll-like receptors (TLRs) 2, 4 and RAGE (receptor for advanced glycation endproducts) in inflammatory conditions. Here we find that TLR5 is also an HMGB1 receptor that was previously overlooked due to lack of functional expression in the cell lines usually used for studying TLR signaling. HMGB1 binding to TLR5 initiates NF-κB signaling pathway activation in a MyD88-dependent manner, resulting in proinflammatory cytokine production and pain enhancement in vivo. Biophysical and in vitro results highlight an essential role for the C-terminal tail region of HMGB1 in facilitating interactions with TLR5. These results suggest that HMGB1-modulated TLR5 signaling is responsible for pain hypersensitivity.
Graphical abstract
INTRODUCTION
The hallmark of almost all pathogenic infections or tissue injuries involves a primary host inflammatory response mediated by receptors of the innate immune system (Mogensen, 2009). Increasing evidence suggests that the central nervous system (CNS) also mounts an organized innate immune response during systemic infection and neuronal injuries caused by various diseases (Aravalli et al., 2007, Muzio et al., 2007). When peripheral nerves are damaged, neurotransmitters, neuropeptides, and cellular debris initiate an innate immune response that drives peripheral and central sensitization, subsequently causing chronic pain (Grace et al., 2014, Ren and Dubner, 2010).
Toll-like receptors (TLRs) are important components of the innate immune system that regulate detection of pathogen associated molecular patterns (PAMPs) and damage associated molecular patterns (DAMPs; a.k.a “alarmins”) (Akira and Takeda, 2004). These receptors are evolutionarily conserved type I transmembrane proteins localized either at the plasma membrane (TLR 1, 2, 4, 5, 6, and 10) or within the endosomal compartment (TLR 3, 7, 8, 9), protecting the host against threats present either in the extracellular or intracellular environment (Akira and Takeda, 2004). In sterile brain injury (Liesz et al., 2011) and neuropathic pain (Tanga et al., 2005, Kim et al., 2007), TLR2- and TLR4-mediated proinflammatory signaling plays a pivotal role in orchestrating the response to neural traumas. In CNS damage, DAMPs, such as nucleic acids, high mobility group box 1 (HMGB1) and S100 proteins are passively released from necrotic brain cells. These DAMPs bind to cell surface TLRs of microglia, the first responder resident immune cell of the CNS (Takeuchi and Akira, 2010, Grace et al., 2014). Once activated, microglia transition to a state of reactive gliosis, release high levels of a wide array of proinflammatory mediators that can exacerbate tissue damage, and stimulate the recruitment of peripheral immune cells, such as neutrophils and monocytes/macrophages, to the site of injury (Ren and Dubner, 2010, Liesz et al., 2015). Similar proinflammatory cascades follow peripheral tissue and nerve injuries (Watkins et al., 2007). Together, peripheral and central immune responses induce neuroinflammatory cascades, which can exacerbate the initial injury as collateral damage (Watkins et al., 2007).
HMGB1 is a non-histone nuclear protein that is well known in binding DNA and in stabilizing DNA interactions with transcription factors to regulate gene transcription (Goodwin et al., 1973). HMGB1 has recently emerged as a DAMP that can activate innate and adaptive immunity and drive host inflammatory responses (Lotze and Tracey, 2005). Release of HMGB1 from necrotic cells into the extracellular milieu is a primary driver of alarmin-mediated inflammatory responses across many model systems (Kim et al., 2006, Park et al., 2006). In contrast to the retention of HMGB1 within the nucleus of healthy cells, HMGB1 is released into the extracellular space surrounding the necrotic and pyroptotic cells upon acetylation of its lysine residues that are present in the nuclear localization sequence (NLS) sites (Scaffidi et al., 2002). HMGB1 can then either act alone in initiating immune signaling or can bind to, and act in concert with, other inflammatory proteins (e.g., IL1β), thereby increasing their combined inflammatory properties (Muller et al., 2001).
Previous studies suggest that HMGB1 released from necrotic cells can signal through three cell surface receptors: receptor for advanced glycation endproducts (RAGE), TLR2 and TLR4 (Hori et al., 1995, Park et al., 2004). These receptors are ubiquitously expressed on cells of peripheral nerves and tissues and can coordinate diverse responses to insults (Farina et al., 2005, Mishra et al., 2006). While multiple prior studies have identified the role of TLR2 and TLR4 in pain amplification via actions at peripheral nerve terminals and/or the spinal cord (Qi et al., 2011), the work of Ji and colleagues raises the potential of TLR5 being involved in pain processing when they discovered that TLR5 is co-expressed with neurofilament-200, a marker of Aβ fiber neurons of the dorsal root ganglia (DRG) long associated with pain amplification under inflammatory or injury conditions (Xu et al., 2015).
The current study finds that an interaction between HMGB1 and TLR5 activates the proinflammatory signaling cascade leading to pain hypersensitivity. We took a tripartite approach to investigate this interaction both in vitro and in vivo. First, we demonstrated that the downstream signaling pathway of the HMGB1-TLR5 interaction is MyD88-dependent. Upon TLR5 activation by HMGB1 the NF-κB p65 subunit translocates from the cytosol to the nucleus resulting in the transcription and release of proinflammatory cytokines and nitric oxide (NO) in the extracellular milieu of engineered and primary peripheral blood mononuclear cells (PBMCs). In PBMCs, both HMGB1 and flagellin (the classical TLR5 agonist) induced NO production and the effects were significantly suppressed by the co-application of TH1020, a TLR5-specific inhibitor (Yan et al., 2016), thereby further supporting a role for HMGB1 as a TLR5 agonist. Second, we characterized this interaction biophysically using fluorescence polarization and solution NMR spectroscopy, which revealed that the C-terminal acidic tail region of HMGB1 is critical for binding to the extra cellular domain of TLR5 in vitro. Third, we demonstrated the functional role of this interaction in an animal model. Subcutaneous injection of HMGB1 induced allodynia (i.e. decreased pain threshold) in rats, which was attenuated by the application of the TLR5-specific inhibitor TH1020. Taken together, our study identifies HMGB1 as a TLR5 ligand that activates the downstream proinflammatory signaling cascade with the functional consequence of pain.
RESULTS
HMGB1 activates TLR5 signaling in vitro
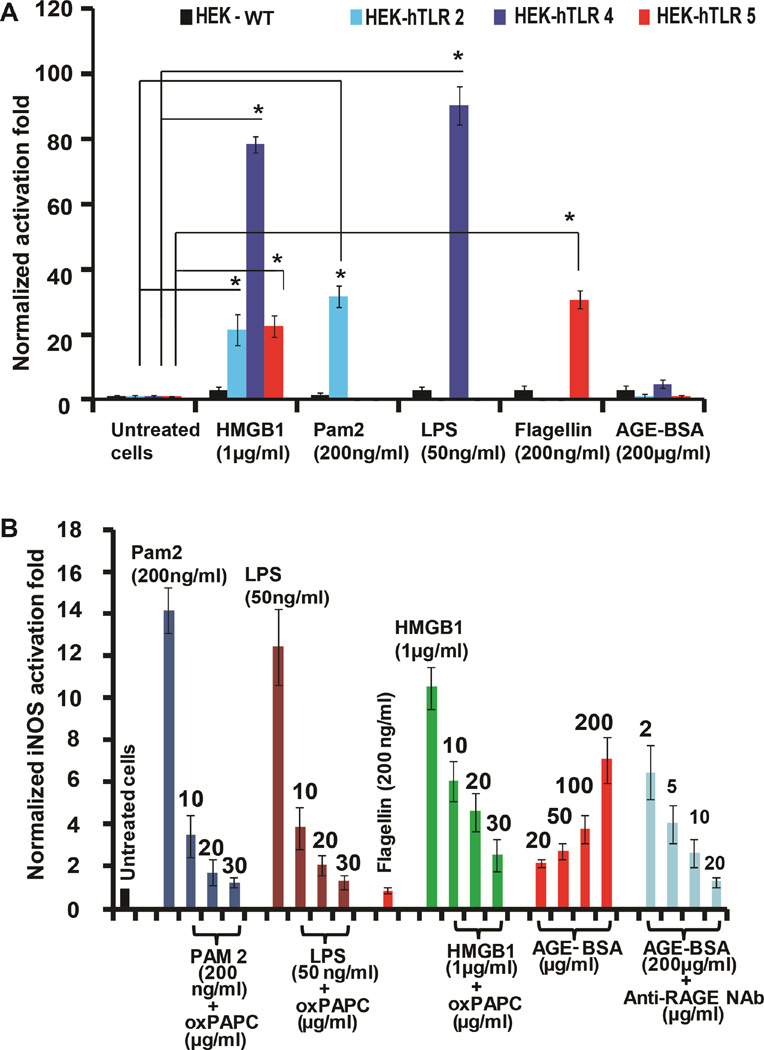
To investigate the cytokine-like effect of HMGB1 in TLR activation, we expressed and purified three different forms of HMGB1 from E. coli. The purified HMGB1 protein was applied to HEK-293 cells overexpressing various human (h) TLRs (HEK-hTLRs). TLR activation was determined by a previously established SEAP signaling assay (Cheng et al., 2015). Interestingly, we obtained significant stimulation of hTLR2, hTLR4 and hTLR5 by full-length rat HMGB1 (Figure 1A) in HEK cells. By contrast, no NF-κB-driven SEAP activation was observed upon HMGB1 addition to HEK-hTLR3, -hTLR7, -hTLR8 or -hTLR9 cells (Figure S1A). We also probed the involvement of the RAGE receptor in hTLR-overexpressing cells as HMGB1 is a known RAGE ligand. We did not observe any SEAP activation in the HEK-hTLR2, -hTLR4 or -hTLR5 cells by the RAGE ligand, AGE-BSA (Figure 1A). These results, while suggesting the absence/inactivity of the RAGE receptor in HEK-hTLR cells, encouraged us to perform several control experiments to probe HMGB1-mediated TLR5 activation. First, we confirmed that no bacterial lipopolysaccharide (LPS) contamination was present in the HMGB1 protein sample using the standard chromogenic limulus amebocyte lysate assay (van Zoelen et al., 2009). Second, we performed experiments in the presence of polymyxin B (PMB), a known inhibitor of an LPS-inflammatory response (Sakharwade et al., 2013). PMB effectively neutralized LPS-mediated activation of TLR4 in HEK-hTLR4 cells, but failed to inhibit HMGB1-mediated TLR4 and TLR5 activation in HEK-hTLR4 and -hTLR5 cells (Figure S1B and S1C), ruling out LPS contamination as the cause of the observed proinflammatory effects. Next, we utilized wild type (WT) HEK cells, which express only low endogenous levels of TLRs. As shown in Figure 1A, we did not observe any activation of the SEAP reporter in these cells upon stimulation by TLR2, TLR4, TLR5 and RAGE ligands. Additionally, HMGB1 did not exhibit any NF-κB activation in these cells, either. These results confirm the absence of RAGE in HEK cells and highlight the involvement of TLR2, TLR4 and TLR5 in HMGB1-mediated NF-κB activation.
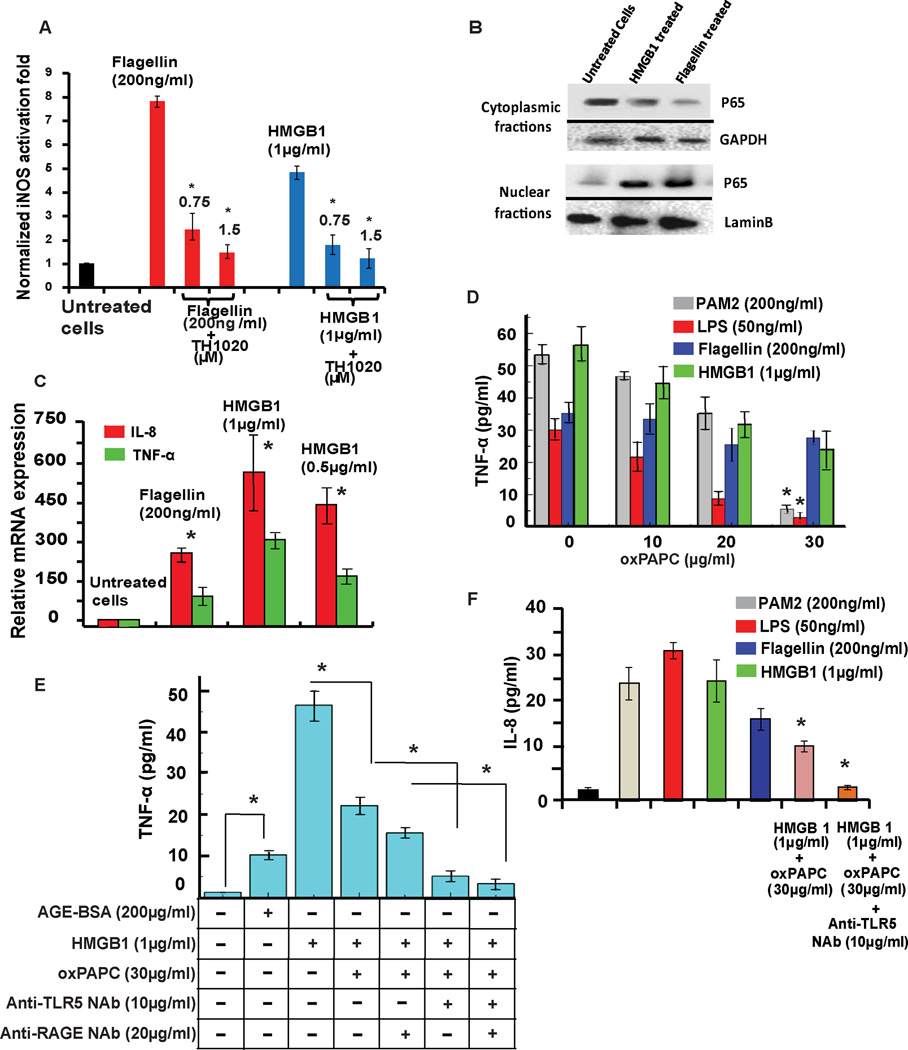
Figure 1. Activation of TLR5 by HMGB1.
(A) TLR ligands activate SEAP signaling in HEK cells. WT, TLR2-, TLR4-, and TLR5-overexpressing HEK cells were incubated with TLR ligands, HMGB1 or AGE-BSA for 24 h. NF-κB activation was evaluated by SEAP secretion in the culture supernatant by the QUANTI-Blue SEAP reporter assay. See also Figure S1A. (B) RAW 264.7 cells were incubated with TLR ligands with HMGB1, AGE-BSA and anti-RAGE NAb for 24 h. oxPAPC was added at varying doses. NF-κB-driven iNOS activation was evaluated by the NO produced. All data are represented as mean ± SD, n=3. *P< 0.05 (unpaired student’s t-test) relative to untreated cells.
Previous studies with RAW 264.7 cells suggested a role for HMGB1 in activating NF-κB signaling through TLR2 and TLR4 receptors with RAGE only playing a minor role (Park et al., 2004). Others have reported that RAW 264.7 cells lack a functional TLR5 (Means et al., 2003). We probed the activating effect of TLR ligands along with HMGB1 in RAW 264.7 cells by performing a NO assay as previously described (Csakai et al., 2014). HMGB1 significantly activated the NF-κB-driven inducible NO synthase (iNOS) gene in comparison to other TLR ligands such as PAM2 (TLR2), LPS (TLR4) and the RAGE ligand AGE-BSA (Figure 1B). To further probe the role of HMGB1 in activating RAGE, TLR2 and TLR4 in RAW 264.7 cells, we utilized oxidized 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine (oxPAPC), an inhibitor specific to both TLR2 and TLR4 (Erridge et al., 2008). oxPAPC efficiently inhibited both PAM2 and LPS-induced activation of TLR2 and TLR4 (Figure 1B) without significant cellular toxicity at 30 µg/ml (Figure S2A). Additionally, an anti-RAGE neutralizing antibody (NAb) significantly decreased the level of iNOS activity induced by AGE-BSA in RAW 264.7 cells. The inhibition of PAM2- and LPS-induced TLR2 and TLR4 signaling by oxPAPC was also demonstrated in HEK-hTLR2 and HEK-hTLR4 cells (Figure S2B–C), where a significant decrease in NF-κB-mediated SEAP activation was observed at a 30 µg/ml dosage. No such oxPAPC inhibitory effect was observed in HEK-hTLR5 cells treated with 200 µg/ml flagellin (Figure S2D). Together, these results indicated that HMGB1 activates SEAP signaling in hTLR5-overexpressing HEK cells but not in wild-type HEK cells. Furthermore, in comparison to previous in vitro results, RAW 264.7 cells that lack functional TLR5 still display HMGB1-mediated activation of TLR2 and TLR4, with RAGE playing a minor role.
HMGB1 activates TLR5 signaling via the MyD88 pathway
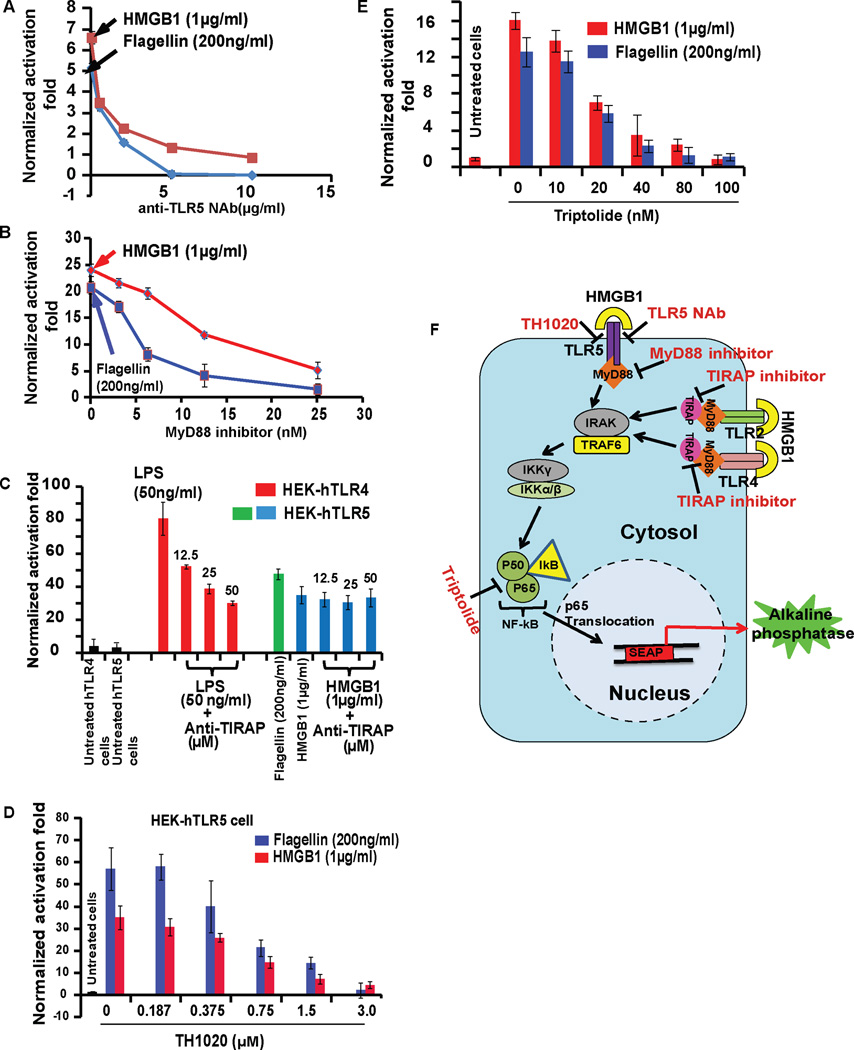
To dissect the mechanism of TLR5 activation in HEK-hTLR5 cells by full-length rat HMGB1, we utilized five independent methods to inhibit the signaling cascade at different stages of the downstream NF-κB signaling pathway. First, an anti-TLR5 NAb was used to block direct activation by flagellin or HMGB1, followed by a small molecule inhibitor of MyD88 (Davis et al., 2006) and a peptide inhibitor of the TIRAP pathway (Brown and McIntyre, 2011) to target the downstream adaptor proteins (Figure 2). Finally, two more inhibitors, triptolide (NF-κB inhibitor) (Qiu et al., 1999) and TH1020 (TLR5 inhibitor) (Yan et al., 2016) were applied to decipher how the HMGB1-TLR5 signaling activates the NF-κB-driven proinflammatory pathway.
Figure 2. HMGB1 activates TLR5 signaling through MyD88 pathway.
(A) HEK-hTLR5 cells were treated with HMGB1 or flagellin followed by addition of anti-TLR5 neutralizing Ab (anti-TLR5 NAb), (B) MyD88 inhibitor, (C) TIRAP inhibitor peptide, (D) TH1020, TLR5 inhibitor or (E) Triptolide, an NF-κB inhibitor for 24 h. (F) Schematic overview of the treatments of anti-TLR5 NAb, MyD88 inhibitor, TH1020 and Triptolide in the TLR5 signaling cascade. NF-κB activation was evaluated by SEAP secretion in the culture supernatant by the QUANTI-Blue SEAP reporter assay. Both HMGB1 (1 µg/ml) and flagellin (200 ng/ml) strongly activated NF-κB-mediated SEAP signaling, followed by dose-dependent inhibition by anti-TLR5 NAb, MyD88 inhibitor, TH1020 and Triptolide, but not the TIRAP inhibitor peptide. For the TIRAP inhibitor assay, we confirmed that the peptide decreased SEAP secretion from LPS-treated HEK-hTLR4 cells in a dose-dependent manner. All data are represented as mean ± SD, n=3.
Anti-TLR5 NAb caused almost complete inhibition of both flagellin- and full-length rat HMGB1-induced activation of TLR5 signaling in HEK-hTLR5 cells, supporting an interaction between HMGB1 and TLR5 (Figure 2A). Furthermore, the MyD88 peptide inhibitor also displayed significant inhibition of TLR5 activation by flagellin and HMGB1 at 25 µM (Figure 2B) while the TIRAP peptide inhibitor failed to prevent signaling (Figure 2C). Neither inhibitor showed any cellular toxicity (Figure S2E for MyD88 inhibitor, TIRAP inhibitor data not shown). Additional control experiments showed that anti-TLR4 NAb and TIRAP peptide inhibitor suppressed TLR4 activation by HMGB1 (Figure S2F) or LPS (Figure 2C) in HEK-hTLR4 cells. Thus, our results are consistent with previous reports of TLR4 activation by HMGB1 (Yang et al., 2010) or LPS (Chow et al., 1999).
Next, a dose-dependent inhibition of HMGB1-activated HEK-hTLR5 cells was observed with TH1020 (Figure 2D). Control experiments confirmed that TH1020 also inhibited flagellin-induced TLR5 activation in HEK-hTLR5 cells as reported previously (Yan et al., 2016) with no apparent cytotoxicity (Figure S2G). Furthermore, NF-κB activation was inhibited by treating HMGB1 stimulated HEK-hTLR5 cells with triptolide, a small molecule inhibitor of NF-κB (Qiu et al., 1999) (Figure 2E). As a positive control, the same assay was also performed on flagellin-treated HEK-hTLR5 cells (Figure 2E) showing a similar inhibitory effect. Summary of all these treatments targeted at downstream adaptor proteins are presented in Figure 2F.
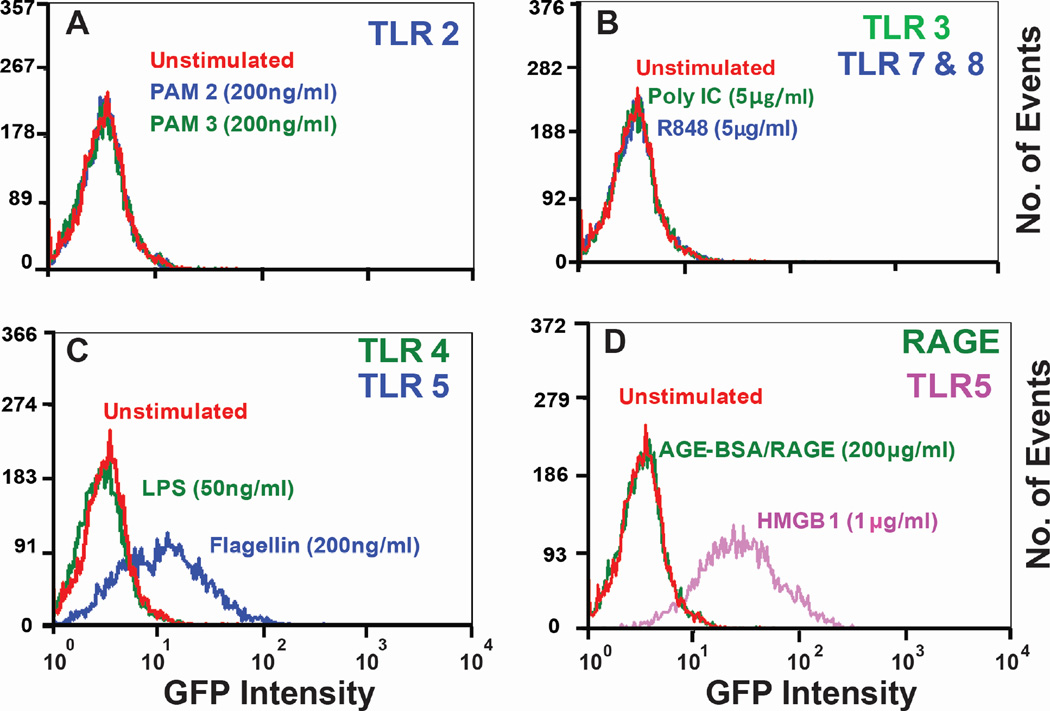
Finally, we tested the activation of the NF-κB pathway in Jurkat T cells stably transfected with a green fluorescent protein (GFP) NF-κB reporter gene (Figure 3). Because these cells do not respond to TLR2 and TLR4 agonists, but are by flagellin (TLR5 agonist) (Thibault et al., 2009), they were used as a valid platform to demonstrate the HMGB1-TLR5 interaction. Flow cytometric analysis showed a clear shift in the GFP signal of Jurkat cells treated with flagellin and HMGB1 compared to all other treatments with TLR/RAGE ligands (TLR2- PAM2/PAM3, TLR3- Poly I:C, TLR4- LPS, TLR7/8- R848 and RAGE- AGE BSA), which showed no change in GFP signal.
Figure 3. HMGB1 activates downstream NF-κB signaling through TLR5.
Flow cytometric analysis of NF-κB activation in human Jurkat TLR5 sensitive T cells with various TLR ligands: (A) PAM2 and PAM3 for TLR2, (B) Poly I:C for TLR3, R848 for TLR7/8, (C) LPS for TLR4, flagellin for TLR5 and (D) AGE-BSA for RAGE, HMGB1 for TLR5. HMGB1 and flagellin both triggered NF-κB activation upon binding to TLR5 while other ligands failed to activate NF-κB. Human Jurkat-T cell line was stably transfected with a GFP-labeled NF-κB reporter gene. Cells sensitive to TLR5 activation were sorted using a MoFlo cytomation fluorescence-activated cell sorter. 10% of activated cells were collected and used for this experiment.
HMGB1 enhances NO production in primary cells
We employed a previously reported nitric oxide (NO) assay (Csakai et al., 2014) on primary rat PBMCs. Treatment of the PBMCs with flagellin and HMGB1 resulted in significant production of NO that was decreased upon TH1020 treatment at 0.75 and 1.5 µM (Figure 4A).
Figure 4. TLR5-HMGB1 interaction produces NO in primary cells and releases proinflammatory cytokines in THP-1 cells.
(A) Normalized iNOS activation folds of the inhibitory effect of TH1020 on flagellin- and HMGB1-induced TLR5 activation in rat primary PBMCs. *P<0.05 for TH1020 relative to the positive controls. Values are the mean ±SD, n=2. (B) Western blot analysis of HEK-hTLR5 cells treated with flagellin (200ng/ml) or HMGB1 (1µg/ml) for 4 h at 37°C, 5% CO 2, shows significant translocation of p65 (NF-ΚB subunit) from the cytosolic to nuclear fraction. GAPDH and LaminB are shown as internal controls. (C) In HEK-hTLR5 cells, HMGB1 and flagellin induced IL-8 and TNF-α mRNA expression at 16 h post-treatment. Changes in gene expression levels are shown relative to untreated controls. IL-8 and TNF-α gene expression levels were determined using the expression ratio of the gene of interest to GAPDH. (D) ELISA assays conducted by inhibition of TLR2 and TLR4 in THP-1 cells by oxPAPC. oxPAPC displayed significant inhibition of TNF-α release in THP-1 cells upon activation by PAM2, LPS and HMGB1, whereas no inhibition was observed for flagellin-induced TNF-α release in comparison to uninhibited cells. (E) Treatment of THP-1 cells by AGE-BSA and HMGB1 induced significant TNF-α secretion. Inhibition of TLR2 and TLR4 by oxPAPC along with HMGB1 treatment displayed significant inhibition of TNF-α release. To elucidate the roles of TLR5 or RAGE in HMGB1-mediated NF-κB signaling, we combined anti-TLR5 NAb alone/or with anti-RAGE NAb with oxPAPC and HMGB1 samples. Significant inhibition of TNF-α release was observed with anti-TLR5 NAb, suggesting the role of TLR5 in mediating TNF-α release via HMGB1. See also Figure S2H. (F) oxPAPC (inhibitor of TLR2 and TLR4) and oxPAPC/TLR5-NAb displayed significant inhibition of IL-8 release in THP-1 cells upon activation by HMGB1. qPCR and ELISA data are represented as mean ± SD, n=3. *P<0.05-unpaired student’s t-test.
HMGB1 activates NF-κB translocation and proinflammatory cytokine production
Activation of the TLR5 receptor by flagellin from both Gram-positive and Gram-negative bacteria culminates in NF-κB translocation to the nucleus and production of proinflammatory cytokines, such as TNF-α and IL-8 (Hayashi et al., 2001). Western blot analysis of the nuclear translocation of the p65 subunit of NF-κB was performed in HMGB1 treated HEK-hTLR5 cells. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) and enzyme-linked immunosorbent assay (ELISA) assays were performed in a human monocytic leukemia cell line, THP-1, to monitor proinflammatory cytokine production.
Western blot results showed a significant increase of the p65 subunit in the nucleus upon exposure to flagellin and HMGB1 compared to the untreated cells (Figure 4B). This result supports the requirement of NF-κB translocation for downstream proinflammatory cytokine release.
To investigate the mRNA expression levels of downstream proinflammatory cytokines, qRT-PCR was performed on post-ligand stimulated HEK-hTLR5 cells (Gewirtz et al., 2004, Chamberlain et al., 2012). TLR5 activation by flagellin led to significant upregulation of the mRNA levels for both TNF-α and IL-8 in comparison to untreated cells (Figure 4C). Similarly, treating the cells with full-length HMGB1 at 0.5 or 1 µg/ml concentrations also resulted in a significant increase in the mRNA levels of both TNF-α and IL-8.
Furthermore, the production of TNF-a and IL-8 as proinflammatory cytokines upon TLR5 activation by HMGB1 was determined by two sets of ELISA experiments in the THP-1 cell line. In the first set (Figure 4D), TLR2, TLR4, and TLR5 were stimulated by their respective ligands, PAM2, LPS and flagellin or full-length rat HMGB1, respectively. We then coupled these TLR stimulations with varying concentrations of oxPAPC to measure TNF-α release upon TLR2 and TLR4 inhibition. With 30 ug/ml oxPAPC, we observed a significant inhibition of TNF-α secretion upon PAM2- and LPS-mediated TLR2 and TLR4 activation but no effect on flagellin-induced TLR5 stimulation, suggesting the specificity of oxPAPC to TLR2 and TLR4. Stimulation of cells with HMGB1 displayed a significant release of TNF-α, which was not completely blocked by 30 µg/ml of oxPAPC (Figure 4D). This result suggested that although HMGB1’s signaling through TLR2 and TLR4 was inhibited by 30 µg/ml oxPAPC, contributions from other putative receptors (i.e RAGE and TLR5) were still present.
In the second set, the role of TLR5 and RAGE in HMGB1-mediated cytokine release in THP-1 cells was determined. Treatment of THP-1 cells with AGE-BSA to activate RAGE, and HMGB1 to activate TLRs and RAGE caused significant TNF-α release in comparison to untreated cells (Figure 4E). Using 30 µg/ml oxPAPC, we observed ~ 50% inhibition of TNF-α release in HMGB1 stimulated samples due to the inhibition of TLR2 and TLR4. Furthermore, significant inhibition of TNF-α secretion was observed upon addition of an anti-TLR5 NAb, thereby confirming the role of HMGB1 in activating immune signaling through TLR5. Additionally, combining an anti-RAGE NAb with oxPAPC or anti-TLR5 NAb samples did not exhibit any significant inhibition in TNF-α level. Furthermore, no apparent cellular toxicity was observed for samples subjected to various treatment conditions (Figure S2H). Similar trends were observed for IL-8 secretion in THP-1 cells. When cells were treated with an anti-TLR5 NAb, significant inhibition of IL-8 production was observed in comparison to the HMGB1-activated and oxPAPC-inhibited cells (Figure 4F).
C-terminal tail of HMGB1 interacts with TLR5
To determine the binding affinity and structural basis of the HMGB1-TLR5 interaction, fluorescence anisotropy and NMR spectroscopy were performed on the complex of full-length HMGB1 and the recombinant zebrafish Danio rerio ortholog of TLR5 (drTLR5) extracellular domain (ECD). We utilized drTLR5 because the functionally active mammalian TLR5 cannot be purified in sufficient quantities. In the fluorescence anisotropy assay, we first validated binding between HMGB1 and the TLR4 accessory protein MD-2 (positive control) with an apparent Kd of 0.6 ± 0.2 µM (Figure S3A), which agrees with previously reported results (Yang et al., 2015). Next, unlabeled drTLR5 ECD was titrated into fluorescein-labeled HMGB1 (Figure S3B). Using a 1:1 protein-ligand model, a binding constant (Kd) of 2.9 ± 0.6 µM was obtained for HMGB1-drTLR5 interaction. The quality of the fit was demonstrated by an adjusted R2 value of 0.98. By contrast, no interaction was observed between HMGB1 and the control protein, bovine serum albumin.
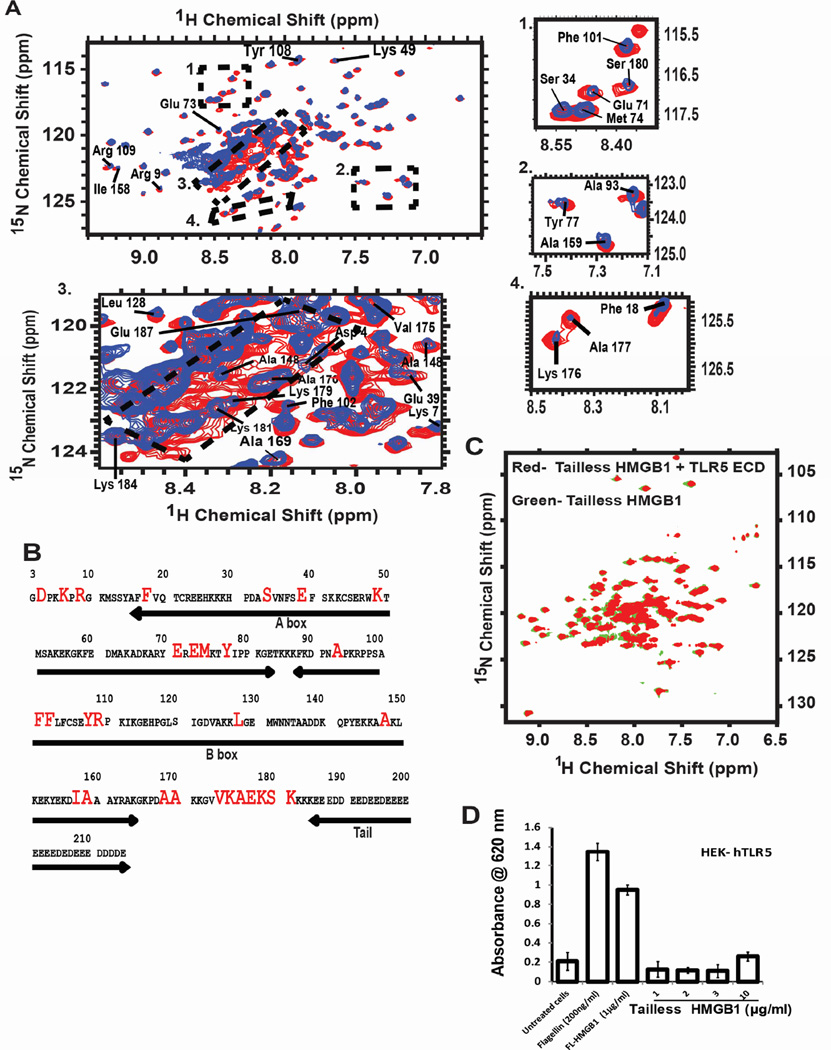
The binding mode of the drTLR5-HMGB1 interaction was further elucidated by solution NMR spectroscopy. 1H-15N heteronuclear single quantum correlation (HSQC) spectra were collected with the full-length 15N-uniform labeled HMGB1 (apo-HMGB1) and 15N-uniform labeled HMGB1 bound to drTLR5 ECD. Sequence-specific backbone residue assignments of the apo-HMGB1 were performed using previously published HMGB1 spectra (Watson et al., 2007) (Table S1, Figure S4A and S4B). We observed various chemical shift changes in the HMGB1-drTLR5 complex spectra (Figure 5A, Inset 1–4). The random coil region of the spectra (1H: 8.1–8.6 ppm, 15N: 120.5 ppm-123.5 ppm) corresponding to the “acidic tail” (residues 185 to 215) of HMGB1 showed maximum chemical shift broadening and peak shifts, suggesting that it was the primary binding site of HMGB1 and drTLR5 in solution (Figure 5A, Inset 3). The chemical shifts broadening pattern of this spectra also suggests that the tail region lacks a defined secondary structure in its ligand-bound state (Figure 5A, Inset 3). Further, residues from the linker connecting the B-box to the tail, such as Ala170, Val175, Ala177, Lys179, Lys181, and Lys184 to Glu187 (Figure 5A, Inset 3 and 4) of the tail region, were also broadened, sharpened and some shifted indicating potential interaction sites with drTLR5. Similarly, the chemical shifts of the Asp90-Pro91-Asn92-Ala93 residues in the spectra (Table S1) suggested a previously reported type-I β turn at the beginning of the B-box of HMGB1. Apart from the tail and basic linkers, some residues of the N-terminus, A-box and B-box were also affected by drTLR5 interaction (Figure 5A, Inset 1 and 2). All residues shifted upon drTLR5 addition are highlighted in the HMGB1 amino acid sequence shown in Figure 5B.
Figure 5. Characterization of binding sites of HMGB1 with TLR5.
(A) Two-dimensional 1H-15N HSQC spectra of 15N-uniformly labeled CBP-HMGB1 (blue) superimposed on 15N-uniformly labeled CBP-HMGB1 bound with unlabeled TLR5 ECD (red). Changes in chemical shifts, peak broadening and sharpening upon TLR5 addition are marked as dotted boxes, subsets 1–4. The subsets include HMGB1 N-terminal residues, the basic linker region (residue 170–187), and A-and B-box helices. The “random coil region” of the spectra (subset 3: 1H: 8.1–8.6 ppm, 15N: 120.5 ppm-123.5 ppm) has the largest chemical shift changes upon TLR5 ECD addition. All spectra were collected in 800 MHz proton frequency at 25°C, using 84 µM of each protein at pH 7.8. See al so Figure S4. (B) HMGB1 sequence highlighting chemical shift changes upon TLR5 addition. Different segments of the protein are shown by arrows. (C) HSQC spectra of the 1:1 complex of TLR5 ECD and 15N tailless HMGB1 (red) superimposed onto the 15N tailless HMGB1 spectra (green) confirms that the HMGB1 tail is the primary TLR5 ECD interaction site. See also Figure S4. (D) HEK-TLR5 cells treated with flagellin (200ng/ml), full length HMGB1 (1µg/ml) showed TLR5-induced SEAP activation while the tailless HMGB1 mutant (1–10 µg/ml) could not do so. Data are represented as mean ± SD, n=3. *P<0.05 (unpaired student’s t-test) relative to untreated cells.
The HSQC spectra of 1:1 and 1:2 (mol:mol) HMGB1-drTLR5 interactions were essentially the same as Figure 5A (red spectra) showing no new peak shift or new peak appearance in HMGB1 (data not shown). To confirm our conclusion that the acidic tail of HMGB1 plays a critical role in this interaction, we collected HSQC spectra of the 1:1 complex of drTLR5 and 15N-tailless HMGB1 (red spectra) and compared with the 15N-tailless HMGB1 spectrum (green spectra) (Figure 5C). Both spectra overlapped significantly on each other confirming that tailless HMGB1 does not interact with drTLR5. The importance of the C-terminal tail region of HMGB1 in binding and activating TLR5 signaling was also confirmed by in vitro SEAP assay (Figure 5D). We found that TLR5 is activated by full-length HMGB1 or flagellin (positive control), but not by the tailless mutant of HMGB1.
HMGB1-mediated TLR5 signaling causes allodynia in vivo
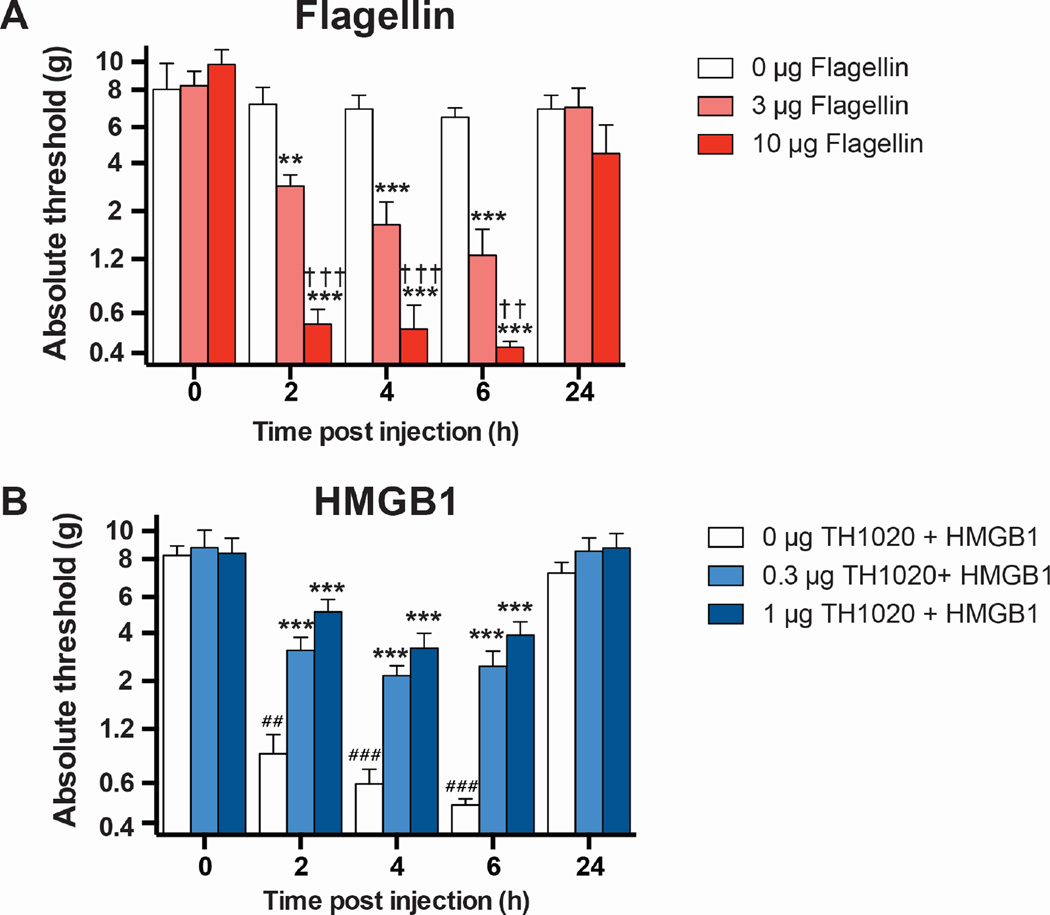
To test whether TLR5 was functionally expressed at peripheral nerve terminals and could transduce nociceptive signals, flagellin was subcutaneously administered into the hindpaw of rats. Flagellin induced dose-dependent allodynia within 2 h, which resolved within 24 h of administration (Figure 6A). This study demonstrated a functional role of TLR5 via this route of administration, providing a rationale to explore the activation/inhibition of TLR5 by HMGB1 and TH1020 in vivo. Next, functional activation of TLR5 by HMGB1 was assessed, where TH1020 (0 (vehicle control), 0.3, or 1 µg) was subcutaneously administered into the hindpaw of rats, followed by 10 µg of HMGB1, subcutaneously administered into the same site 30 min later. Compared to baseline, HMGB1 induced significant allodynia (Figure 6B), which was attenuated by TH1020. Post hoc tests did not reveal a significant difference between these two doses of TH1020.
Figure 6. TLR5-HMGB1 interaction causes allodynia in vivo.
(A) Dose-dependent effect of subcutaneous flagellin on hindpaw withdrawal thresholds to von Frey hairs (allodynia). N = 4/group, mean ± SEM. (B) Dose-dependent effect of subcutaneous TLR5 antagonist TH1020 on HMGB1-dependent allodynia (10 µg, subcutaneous). N = 6/group, mean ± SEM. **P < 0.01, ***P < 0.001, relative to 0 µg; ††P < 0.01, †††P < 0.001, relative to 3 µg; ##P < 0.01, ###P < 0.001, relative to baseline (BL).
DISCUSSION
The recognition of DAMPs and PAMPs by various TLRs is a critical determinant of its immunomodulatory properties in neuroinflammation. Given the fact that HMGB1 has been shown to activate multiple TLRs (Park et al., 2004), we were motivated to further examine what other receptors are involved in HMGB1-induced activation of inflammatory signaling apart from RAGE, TLR2 and TLR4. Here we successfully characterized HMGB1 as a TLR5 ligand (Figure 1 & 5) and demonstrated HMGB1-induced TLR5 activation is NF-κB-driven (Figure 2 & 3) and releases downstream proinflammatory cytokines (Figure 4) Notably, we also showed that activation of peripheral TLR5 by HMGB1 and flagellin induces pain following subcutaneous administration in rats (Figure 6A & B).
Our findings raised the question why TLR5 has been overlooked previously in the HMGB1-mediated TLR signaling. Previously, Means et al. showed that the functional expression of some TLRs including TLR5 was absent in the widely used platform cell lines such as RAW 264.7 murine macrophages, murine bone marrow-derived DCs, and resident/thioglycollate peritoneal macrophages (Means et al., 2003). Also it was reported that in endometrial epithelial cell lines and gastric epithelial cell lines (MKN45) the proinflammatory role of TLRs is predominantly via TLR5 rather than TLR2 or TLR4 (Young et al., 2004), despite the expression of TLR2 and TLR4 in these cells. As a result, the down-regulation or a lack of functionally expressed TLR2 and/or TLR4 can explain the ability of HMGB1 to interact with TLR5 to exert its proinflammatory effects. Similarly, Ji and coworkers showed expression of TLR1-TLR9 in mouse DRG neurons, but reported the unique distribution pattern of TLR5 among other TLRs in sensory neurons with Aβ-fibers (Xu et al., 2015).
To highlight the need of probing the functional expression of TLR5 receptor in different cell lines, we performed this study in a variety of cell lines, including human T-lymphocytes, human monocytes, engineered TLR HEK cells, and primary PBMCs (Figure 1–4). Altogether, these cell-based assays illustrated the ability of extracellular HMGB1 to amplify the proinflammatory signaling cascade through TLR5, which can be blocked partially by TH1020 inhibitor (Figure 4A). During the NF-κB-induced cytokine release, elevated levels of downstream effectors, such as TNF-α, IL-8 and iNOS (Figure 4), with negligible cytotoxicity (Figure S2) were observed confirming the immunostimulatory effects of HMGB1 when bound to TLR5.
Structurally, HMGB1 is composed of three domains: A-box, B-box and the 30-residue C-terminal acidic tail primarily composed of aspartic acid and glutamic acid residues (Watson et al., 2007). The A- and B-box domains of HMGB1 have been characterized and their role in DNA bending elucidated (Teo et al., 1995), but the structure and function of the acidic tail in modulating immune signaling by TLRs are poorly understood (Watson et al., 2007, Banerjee et al., 2010). Our NMR data suggest that the C-terminal acidic tail (residues 185-215), the basic linker-region (residues 170-184), few residues of the N-terminus and the A-(residues 14-83) and B- (residues 88-164) boxes of full-length HMGB1 are critical for binding and activation of TLR5 by HMGB1 (Figure 5). Although a few residues of the A- and B-boxes of the HMGB1 tailless mutant show significant chemical shift perturbation, the rest of the residues in these domains are almost structurally identical with the full-length protein (Figure S4A, green labels and Table S1) confirming that the tail region is primarily interacting with the drTLR5 ECD. Due to the inherent challenge to express and purify the highly acidic tail region of HMGB1, we do not have the evidence that supports if the C-terminal tail of HMGB1 alone is sufficient to activate the TLR5 signaling cascade. Nonetheless, our SEAP assay results using a tailless mutant of HMGB1 clearly demonstrate that the tailless mutant is indeed unable to trigger TLR5 activation (Figure 5D).
HMGB1 plays a role at the brain-immune interface, where binding of HMGB1 to the RAGE receptor leads to activation of NF-κB, ERK1/2, p38, and SAPK/JNK kinases during neurite outgrowth (Hori et al., 1995). Glial activation via TLR2 and TLR4 signaling results in production of NF-κB-driven proinflammatory cytokines/chemokines, contributing to the initiation and maintenance of pain hypersensitivity (Ren and Dubner, 2010, Grace et al., 2014). Adding to this list, this study shows that subcutaneous HMGB1 and flagellin can signal through TLR5 and create pain amplification in vivo (Figure 6A & B). These results can be explained by two possible mechanisms. As Aβ-fiber neurons express TLR5 (Xu et al., 2015), they may be directly engaged by HMGB1 or flagellin to mediate mechanical allodynia. A complementary mechanism may also involve activation of peripheral innate immune cells that express TLR5, leading to release of inflammatory mediators that sensitize peripheral terminals of sensory neurons (Takeuchi and Akira, 2010, Ren and Dubner, 2010). These data extend a prior report that allodynia induced by peripheral nerve injury is attenuated in Tlr5−/− mice (Stokes et al., 2013), by demonstrating a fundamental role for TLR5 in nociception, and identifying HMGB1 as an endogenous TLR5 ligand that may mediate nociceptive hypersensitivity (Grace et al., 2014).
In conclusion, our results support the ability of HMGB1 in activating TLR5-mediated proinflammation of which the C-terminal tail region of HMGB1 plays an important role in binding the TLR5 receptor. Additional evidence suggests a noceceptive role of TLR5 in the development of chronic pain, of which allodynia can be a symptom. Thus, the HMGB1-TLR5 interaction could provide a novel therapeutic target for the development of new pain management strategies (Grace et al., 2014, Ji et al., 2014).
EXPERIMENTAL PROCEDURES
QUANTI-Blue SEAP assay
HEK-293 cells stably transfected with human TLRs 2, 3, 4, 5, 7, 8 and 9 (HEK-hTLR) (InvivoGen, CA) with an inducible secreted embryonic alkaline phosphatase (SEAP) reporter gene were cultured in 200 µl of Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, and 1% L-glutamine (complete growth medium). Briefly, cells were seeded at 70,000 cells per well in 96-well plates (Thermo Scientific, MA) and incubated for 24 h at 37°C in complete growth medium. The media was removed from the 96-well plate and replaced with DMEM supplemented with 10% FBS (neither 1% penicillin/streptomycin nor 1% L-glutamine) and treated with the appropriate concentration of compounds. Then the cells were incubated overnight at 37°C for 18–24 h. To detect SEAP activity, 20 µl of media was removed from each well and transferred to a transparent 96-well plate containing 180 µl of QUANTI-Blue reagent (InvivoGen, CA) and incubated at 37°C (in the dark) for 30 min to 2 h ( See the supplemental experimental procedures for the detailed procedure).
NO activation assay with RAW 264.7 cells
RAW 264.7 cells were seeded at 70,000 cells/well in a 96-well plate with RPMI-1640 medium (10% fetal bovine serum, 1% L-gluamine, 1% penicillin/streptomycin) (complete growth media) and incubated at 37°C over night. Next day, the media was replaced with unsupplemented RPMI media followed by the treatment of appropriate concentration of compounds. On day 3, 90 µL media was transferred to a black 96-well plate and 10 µL of 0.05 mg/mL 2, 3-diaminonapthalene (Sigma Aldrich, MO) in 0.62 M HCl were added to the media. The plate was covered in aluminum foil and shaken at room temperature for 15 min and data were collected by a Beckman Coulter DTX 880 Multimode detector plate reader (See the supplemental experimental procedures for more detail).
Peptide Synthesis
The TIRAP inhibitor peptide was synthesized by a CEM Liberty microwave-assisted peptide synthesizer using standard solid phase Fmoc chemistry. The Fmoc-protected amino acids (0.5 mmol), HATU (0.5 mmol) and DIPEA (0.5 mmol) in 4 mL DMF were mixed with Rink Amide resin (0.1 mmol) with a substitution level of 0.45 mmol/g for the coupling step. Fmoc deprotection required 7 mL of 5% piperazine and 0.1 M HOBt in DMF. All coupling and deprotection processes were duplicated to ensure complete loading and deprotection. The peptide was cleaved from the resin using a water/trifluoroacetic acid (TFA)/triisopropylsilane (TIPS) cocktail of 2.5%/95%/2.5% for 1h. Chilled Et2O was used to precipitate the peptide. The peptide was purified by reverse phase high performance liquid chromatography (HPLC) using a semi-prep C18 column. Fractions were characterized by MALDI mass spectrometry and lyophilized. The purity of the peptide was analyzed by HPLC.
WST cell viability assay
The supernatant from compound-treated HEK-hTLR, RAW 264.7 or THP-1 cells described above in SEAP assay were removed. The adhered cells were treated with a 1:10 dilution of cell proliferation reagent WST-1 (Roche, IN). Cells were incubated at 37°C until a color change was observed (30 min-2h). Data were quantified on a Beckman-Coulter DTX 880 Multimode Detector using absorbance at 450 nm and normalized as (raw data-20% DMSO)/ (untreated cells-20% DMSO) such that untreated cells corresponded to 100% survival, and 20% DMSO to 0% survival.
Jurkat-T cell transfection and NF-κB–GFP reporter assay
Human Jurkat cells [American Type Culture Collection (ATCC) TIB-152] containing NF-κB–GFP reporter gene sensitized only for TLR5 ligand (flagellin) were grown and maintained in RPMI-1640 medium containing 10% FBS and 1% penicillin/streptomycin (See the supplemental experimental detail for the preparation of these cells). The TLR5 ligand sensitized cells were seeded in six-well plates at 1×106 cells per well with 3 ml of complete growth medium [RPMI 1640 medium, supplemented with 10% FBS, 1% penicillin/streptomycin] and TLR ligands (200 ng/ml of PAM2 and PAM3, 5µg/ml Poly I:C, 50 ng/ml LPS, 200 ng/ml flagellin, 5µg/ml R848, 1 µg/ml HMGB1 and 200 µg/ml AGE-BSA) for 24 h at 37°C in a 5% CO 2 humidified incubator. After 24 h, the cells in each well were mixed and 200 µl cells containing medium were stained by propidium iodide for 10 min before flow cytometry analysis performed by MoFlo Cytomation (Beckman Coulter, Brea).
Primary peripheral blood mononuclear cell (PBMC) isolation and NO activation assay
On day of treatment, rats were anesthetized with sodium pentobarbital and blood collected via cardiac puncture into EDTA-containing tubes. Approximately, 7ml blood was collected and 0.625ml Optiprep (Sigma Aldrich, MO) added with gentle mixing for every 5ml of blood collected. Careful layering of 1ml of PBS over the blood/Optiprep mixure was performed followed by harvesting the cells by centrifugation at 1300xg for 30 mins at 20°C. Th e cell pellet was washed two times by resuspending in 10ml cold RPMI 1640 media and centrifuging at 300xg for 10 min at 4°C. The isolated PBMCs were then dil uted to 1×106 cells/ml in enriched RPMI1640 (10% FBS and 1% penicillin) and plated into 96-well plates (100µl/well) for NO assay. Cells were treated with flagellin (200ng/ml) and HMGB1 (1µg/ml) for 1 h at 37°C, 5% CO2, followed by treatment of TH1020 at 0.75 µM or 1.5µM.
Western blot
HEK-hTLR5 cells at 80% confluency were treated with flagellin (100 ng/ml) and HMGB1 (1 µg/ml) in 6-well plate and incubated at 37°C, 5% CO 2 humidity. After 4 h, supernatant was aspirated, cells were washed 3 times with PBS, and total cell lysate was extracted using lysis buffer [25 mM Tris-Cl, pH 7.5, 250 mM NaCl, 5 mM EDTA, 1% Nonidet P-40, 0.1 mM sodium orthovanadate, 2 µg/ml leupeptin, 100 µg/ml phenylmethylsulfonyl flouride (PMSF), and protease inhibitor cocktail tablets] (Roche Diagnostics, IN). Cytosolic and nuclear fractions were extracted using P65 Translocation Assay Kit (Fivephoton Biochemicals, San Diego, CA, USA) according to the manufacturer’s instructions. Protein concentration was determined by a Bradford assay. Samples were heated to 75°C for 10 min and loaded into a 10% Tris-glycine gel (Invitrogen, CA). SDS-PAGE was performed in MOPS running buffer at 175V for 1.25 h at 4°C. Protein was transferred onto a nitro-cellulose membrane by electroblotting and probed with the primary p65 antibody (1:400) provided in the kit. Anti-rabbit IgG-HRP antibody (Cell Signaling Technology, MA) at 1:2000 dilution was used followed by visualization using enhanced chemiluminescence (ECL) method. GAPDH and Lamin B were used as internal controls for cytosolic and nuclear fractions, respectively.
Quantitative RT-PCR
HEK-hTLR5 cells were seeded in 6-well plates at 1×106 cells per well with DMEM complete growth medium and incubated at 37°C in a 5% CO2 humidity for 24 h. Next, the nonadherent cells and medium were aspirated and replaced with fresh unsupplemented medium. The cells were then treated with 200 ng/ml flagellin and HMGB1 (1 and 0.5 µg/ml) respectively followed by an additional 16 h incubation. The medium was then removed and the cells were gently washed with PBS for 3 times. After the final washing step, 1 ml PBS was added to each well followed by the transfer of cells into subsequent cryotubes (1.5 ml) and stored at −80°C until ready for qRT-PCR. RNA was ex tracted using the miRNeasy Mini Kit from Qiagen (Valencia, CA, USA). cDNA were synthesized using RT2 Easy First Strand cDNA Synthesis Kit per manufacturer’s instructions. TNF-a, IL-8 and GAPDH primers were purchased from SABiosceinces (Valencia, CA, USA). qPCR was performed on a CFX96 Real-Time PCR detection system (Bio-Rad, CA) using the SYBR Green method and the data were analyzed by ΔΔCt method, as previously described (Cheng et al., 2015).
ELISA
ELISA was performed using a kit from BD Biosciences (San Jose, CA) to measure TNF-a and IL-8 expression levels. THP-1 cells were seeded in 6-well plates at 1×106 cells per well with 2 ml medium [RPMI 1640 medium supplemented with 10% FBS, 1% penicillin/streptomycin and grown for 24 h at 37°C in a 5% CO2 humidified incubator. Prior to addition of ligands and compounds, cells were differentiated to macrophages using 20 ng/ml/well phorbol 12-myristate 13-acetate (PMA) for 24 h. Nonadherent cells and medium were removed and replaced with fresh unsupplemented RPMI 1640 medium followed by the treatment of various compounds and incubated for an additional 24 h. the cell culture supernatants were used for cytokine measurement.
HMGB1 protein expression and purification
The plasmids for full-length HMGB1 with N-terminal CBP and 6X His tags (Figure S5A) were the generous gifts from Prof. Kevin J. Tracey and Prof. Patrick C Swanson labs respectively. Full-length proteins were purified using previously published protocols (Li et al., 2004). Briefly, the plasmids of CBP tag 15N-HMGB1 and 6X His tag 15N-HMGB1 full-length and tailless were transformed into BL21(DE3) and BL21(DE3)PLys E. coli strains for large-scale protein expression. A single colony was picked to inoculate four 50 ml precultures of LB media with 100 µg/ml ampicillin antibiotic in each. Precultures were grown overnight at 37°C in a shaker incubator and transferred to four 1 L of LB media with the same ampicillin antibiotic (100 µg/ml). Protein overexpression was induced by 0.4 mM isopropyl-β-D-thiogalactopyranoside (IPTG). For CBP tag-HMGB1, protein overexpression was continued at 37 °C for 3 h, while for 6× His tag HMGB1 and tailless protein, overexpression was continued at 30°C for 5–6 h. The cells were harvested by centrifugation at 5,000–6,000xg for 10 min at room temperature. The cell pellet for CBP tag-15N HMGB1 (full-length) was resuspended in 20 ml of 20 mM Tris–HCl, pH 8.6, 150 mM NaCl, 2.5 mM CaCl2 (Buffer A), while His tag-15N HMGB1 full-length and HMGB1 tailless were resuspended in 40 mM Tris–HCl, pH 7.5, 500 mM NaCl and stored at −80°C.
Modified protocols of previously published procedures were used to purify CBP tag-15N isotope labeled HMGB1 (full-length), 6XHis tag- 15N isotope labeled HMGB1 (full-length) and 6X His tag- 15N HMGB1 tailless proteins (Figure S5B and S5C). 15N isotope labeling method for overexpressing protein in bacterial culture was adapted from (Das et al., 2015) (See the supplemental experimental procedure for the detailed method).
Expression and purification of Danio rerio TLR5-N14VLR (drTLR5)
The Danio rerio TLR5-N14VLR (drTLR5) was cloned into a pFastBac vector, expressed and purified as described (Yoon et al., 2012). The drTLR5 protein was expressed in Hi5 insect cells for three days at 27°C with an N-terminal gp67 signal peptide, a C-terminal thrombin cleavage site, Strep-Tactin II and His6-tags. drTLR5 was purified through a His-tag affinity purification step following by gel filtration chromatography (Superdex-200, Pharmacia) and then cleaved overnight by thrombin to remove the tags. Cleaved drTLR5 was further purified by another His-tag affinity purification step and gel filtration in 50 mM HEPES, 100 mM NaCl, pH 7.5 buffer. The drTLR5 protein eluted as a monomer, and was concentrated to 2.5 mg/ml and stored at −80°C.
Fluorescence anisotropy assay
Apparent Kd values were determined by measuring the fluorescence anisotropy of 200 nM or 5 µg/ml fluorescein-labeled HMGB1 as a function of increasing concentrations of drTLR5 ECD and MD-2 proteins. The labeled protein was incubated with varying amounts of the target protein (drTLR5 ECD and MD-2) for 30 mins at room temperature in binding buffer (40 mM HEPES, pH 7.5, 150 mM NaCl). All measurements were made on a Horiba Fluorolog-3 fluorometer. The wavelengths for monitoring excitation (Ex), emission (Em), and the emission cutoffs (Co) for fluorescein were: Ex = 494 nm, Em = 518 nm, and Co = 515 nm. Slit widths of 5 nm were used in all experiments. Data analysis was performed as described (Dewan et al., 2012) by fitting the data to a 1:1 binding model with a correction for changes in fluorophore intensity caused by protein binding (OriginPro 8 SRO). Protein concentrations were estimated using the Bradford method (Bradford, 1976).
NMR Spectroscopy
drTLR5/HMGB1 interactions were determined by solution NMR spectroscopy on the CBP- and 6X His tags containing full-length and tailless 15N uniformly labeled HMGB1 proteins ranging from 80–100 µM, 10% (v/v) 2H20 in 10 mM Tris HCl (pH 8.0) and 1 mM deuterated DTT. Titrations of 1:1 and 1:2 (mol/mol) drTLR5/HMGB1 (CBP- and 6X His tag containing) were used All spectra were collected either in a Varian 800 MHz or 900 MHz spectrometer equipped with triple resonance cryoprobe at 25°C housed at Univ. Colorado Boulder and Denver campuses.
Accession Number
Full-length HMGB1 sequence specific backbone amide resonance assignments were performed by chemical shift mapping using the BioMagResBank (BMRB) accession number 15502 deposited by (Watson et al., 2007).
Animals
Pathogen-free adult male Sprague Dawley rats (12 weeks old on arrival; Envigo Labs, Indianapolis, IN, USA) were used in all experiments. Rats were housed in temperature-controlled (23±3°C) and light-controlle d (12 h light–dark cycle; lights on at 07:00 hours) rooms with standard rodent chow and water available ad libitum. All procedures were approved by the Institutional Animal Care and Use Committee of the University of Colorado Boulder.
Subcutaneous injections
All drugs were administered into one plantar hind paw, with the needle directed between the toes with the needle tip placed subcutaneously into the plantar surface of the hindpaw to avoid backleak of the injectate. To determine whether peripheral TLR5 functionally contributed to allodynia, either 0 (vehicle control), 3, or 10 µg flagellin was administered (100 µl; saline vehicle). To determine whether HMGB1 induced TLR5-dependent allodynia, either 0 (vehicle control), 0.3, or 1 µg of TH1020 was administered (10 µl: 3% DMSO in saline), followed 30 min later by HMGB1 (10 µg in 10 µl; saline vehicle).
Mechanical allodynia
Testing was conducted blind with respect to group assignment. Rats received at least three 60-min habituations to the test environment before behavioral testing. The von Frey test (Chaplan et al., 1994) was performed at the distal region of the heel in the hindpaws, within the region of sciatic innervation as previously described (Chacur et al., 2001). Assessments were made before injection (baseline), and 2, 4, 6, and 24 h after the last injection. A logarithmic series of 10 calibrated Semmes–Weinstein monofilaments (von Frey hairs; Stoelting) was applied to the hindpaw to define the threshold stimulus intensity required to elicit a paw withdrawal response. Log stiffness of the hairs ranged from manufacturer-designated 3.61 (0.40 g) to 5.18 (15.14 g) filaments. The behavioral responses were used to calculate absolute threshold (50% probability of response) by fitting a Gaussian integral psychometric function using a maximum-likelihood fitting method (Harvey, 1986) as described (Milligan et al., 2000).
Statistical analysis
Animal study: Mechanical allodynia was analyzed as the interpolated 50% thresholds (absolute threshold). One-way ANOVAs followed by Tukey’s post hoc test were used to confirm that there were no baseline differences in absolute thresholds between treatment groups. Differences between treatment groups were determined using repeated measures two-way ANOVA, with treatment and time as main effects, followed by Sidak’s post hoc test. Differences between timepoints for HMGB1 treatment were determined using repeated measures one-way ANOVA, followed by Dunnett post hoc test. P < 0.05 was considered significant.
In vitro cellular assay: All data presented for SEAP and NO assays are mean ± SD. Differences between treatment groups were determined by unpaired student’s t-test; P < 0.05 was considered statistically significant.
Supplementary Material
In Brief.
Das et al. show that HMGB1 is an agonist of TLR5, activating proinflammatory signaling through MyD88. The C-terminal acidic tail region of HMGB1 facilitates interaction with the TLR5 extracellular domain. This interaction results in the release of various proinflammatory cytokines and causes pain hypersensitivity in an animal model.
Highlights.
HMGB1 is identified as a DAMP molecule for TLR5 along with TLR2 and TLR4
HMGB1-induced TLR5 activation causes pain
The C-terminal acidic tail of HMGB1 is involved in HMGB1-TLR5 interaction
Acknowledgments
We thank the NIH National Institute of General Medicine (GM101279 to HY) and (AI042266 to IAW) for financial support, Prof. Kevin J. Tracey and Dr. Huan Yang for generously providing rat HMGB1 protein and plasmid, Prof. Patrick Swanson for human HMGB1 plasmids, Dr. J. Isaac Godfroy for Jurkat TLR5 cells and manuscript discussions, Lei Yan for providing TH1020 and Dr. Geoff Armstrong for assistance with NMR experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
VD, ND, PG and HY designed the research; VD, ND conducted biochemical and cell-based assays. ND purified HMGB1 and conducted NMR studies. PG isolated PBMCs and conducted allodynia experiments in rat. RT synthesized peptides. RG, NT purified TLR5. VD, ND, PG, RG, LW, IW and HY analyzed data; ND, VD prepared figures. ND, VD, PG, RT, RG, LW, IW, and HY, wrote the manuscript.
REFERENCES
- Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Aravalli RN, Peterson PK, Lokensgard JR. Toll-like receptors in defense and damage of the central nervous system. J Neuroimmune Pharmacol. 2007;2:297–312. doi: 10.1007/s11481-007-9071-5. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Friggeri A, Liu G, Abraham E. The C-terminal acidic tail is responsible for the inhibitory effects of HMGB1 on efferocytosis. J Leukoc Biol. 2010;88:973–979. doi: 10.1189/jlb.0510262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brown GT, Mcintyre TM. Lipopolysaccharide signaling without a nucleus: kinase cascades stimulate platelet shedding of proinflammatory IL-1beta-rich microparticles. J Immunol. 2011;186:5489–5496. doi: 10.4049/jimmunol.1001623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacur M, Milligan ED, Gazda LS, Armstrong C, Wang H, Tracey KJ, Maier SF, Watkins LR. A new model of sciatic inflammatory neuritis (SIN): induction of unilateral and bilateral mechanical allodynia following acute unilateral peri-sciatic immune activation in rats. Pain. 2001;94:231–244. doi: 10.1016/S0304-3959(01)00354-2. [DOI] [PubMed] [Google Scholar]
- Chamberlain ND, Vila OM, Volin MV, Volkov S, Pope RM, Swedler W, Mandelin AM, 2nd, Shahrara S. TLR5, a novel and unidentified inflammatory mediator in rheumatoid arthritis that correlates with disease activity score and joint TNF-α levels. J Immunol. 2012;189:475–483. doi: 10.4049/jimmunol.1102977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods. 1994;53:55–63. doi: 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
- Cheng K, Gao M, Godfroy JI, Brown PN, Kastelowitz N, Yin H. Specific activation of the TLR1-TLR2 heterodimer by small-molecule agonists. Sci Adv. 2015;1:e1400139. doi: 10.1126/sciadv.1400139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JC, Young DW, Golenbock DT, Christ WJ, Gusovsky F. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J Biol Chem. 1999;274:10689–10692. doi: 10.1074/jbc.274.16.10689. [DOI] [PubMed] [Google Scholar]
- Csakai A, Smith C, Davis E, Martinko A, Coulup S, Yin H. Saccharin derivatives as inhibitors of interferon-mediated inflammation. J Med Chem. 2014;57:5348–5355. doi: 10.1021/jm500409k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das N, Dai J, Hung I, Rajagopalan MR, Zhou HX, Cross TA. Structure of CrgA, a cell division structural and regulatory protein from Mycobacterium tuberculosis, in lipid bilayers. Proc Natl Acad Sci U S A. 2015;112:E119–E126. doi: 10.1073/pnas.1415908112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CN, Mann E, Behrens MM, Gaidarova S, Rebek M, Rebek J, Jr, Bartfai T. MyD88-dependent and - independent signaling by IL-1 in neurons probed by bifunctional Toll/IL-1 receptor domain/BB-loop mimetics. Proc Natl Acad Sci U S A. 2006;103:2953–2958. doi: 10.1073/pnas.0510802103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewan V, Wei M, Kleiman L, Musier-Forsyth K. Dual role for motif 1 residues of human lysyl-tRNA synthetase in dimerization and packaging into HIV-1. J Biol Chem. 2012;287:41955–41962. doi: 10.1074/jbc.M112.421842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erridge C, Kennedy S, Spickett CM, Webb DJ. Oxidized phospholipid inhibition of toll-like receptor (TLR) signaling is restricted to TLR2 and TLR4: roles for CD14, LPS-binding protein, and MD2 as targets for specificity of inhibition. J Biol Chem. 2008;283:24748–24759. doi: 10.1074/jbc.M800352200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina C, Krumbholz M, Giese T, Hartmann G, Aloisi F, Meinl E. Preferential expression and function of Toll-like receptor 3 in human astrocytes. J Neuroimmunol. 2005;159:12–19. doi: 10.1016/j.jneuroim.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Gewirtz AT, Yu Y, Krishna US, Israel DA, Lyons SL, Peek RM. Helicobacter pylori Flagellin Evades Toll-Like Receptor 5-Mediated Innate Immunity. Journal of Infectious Diseases. 2004;189:1914–1920. doi: 10.1086/386289. [DOI] [PubMed] [Google Scholar]
- Goodwin GH, Sanders C, Johns EW. A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur J Biochem. 1973;38:14–19. doi: 10.1111/j.1432-1033.1973.tb03026.x. [DOI] [PubMed] [Google Scholar]
- Grace PM, Hutchinson MR, Maier SF, Watkins LR. Pathological pain and the neuroimmune interface. Nat Rev Immunol. 2014;14:217–231. doi: 10.1038/nri3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey LO. Efficient estimation of sensory thresholds. Behavior Research Methods, Instruments, & Computers. 1986;18:623–632. [Google Scholar]
- Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM, Aderem A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- Hori O, Brett J, Slattery T, Cao R, Zhang J, Chen JX, Nagashima M, Lundh ER, Vijay S, Nitecki D, et al. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin. Mediation of neurite outgrowth and co-expression of rage and amphoterin in the developing nervous system. J Biol Chem. 1995;270:25752–25761. doi: 10.1074/jbc.270.43.25752. [DOI] [PubMed] [Google Scholar]
- Ji RR, Xu ZZ, Gao YJ. Emerging targets in neuroinflammation-driven chronic pain. Nat Rev Drug Discov. 2014;13:533–548. doi: 10.1038/nrd4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim MA, Cho IH, Kim MS, Lee S, Jo EK, Choi SY, Park K, Kim JS, Akira S, Na HS, Oh SB, Lee SJ. A critical role of toll-like receptor 2 in nerve injury-induced spinal cord glial cell activation and pain hypersensitivity. J Biol Chem. 2007;282:14975–14983. doi: 10.1074/jbc.M607277200. [DOI] [PubMed] [Google Scholar]
- Kim JB, Sig Choi J, Yu YM, Nam K, Piao CS, Kim SW, Lee MH, Han PL, Park JS, Lee JK. HMGB1, a novel cytokine-like mediator linking acute neuronal death and delayed neuroinflammation in the postischemic brain. J Neurosci. 2006;26:6413–6421. doi: 10.1523/JNEUROSCI.3815-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang H, Mason JM, Levine J, Yu M, Ulloa L, Czura CJ, Tracey KJ, Yang H. Recombinant HMGB1 with cytokine-stimulating activity. J Immunol Methods. 2004;289:211–223. doi: 10.1016/j.jim.2004.04.019. [DOI] [PubMed] [Google Scholar]
- Liesz A, Dalpke A, Mracsko E, Antoine DJ, Roth S, Zhou W, Yang H, Na SY, Akhisaroglu M, Fleming T, Eigenbrod T, Nawroth PP, Tracey KJ, Veltkamp R. DAMP signaling is a key pathway inducing immune modulation after brain injury. J Neurosci. 2015;35:583–598. doi: 10.1523/JNEUROSCI.2439-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesz A, Zhou W, Mracsko E, Karcher S, Bauer H, Schwarting S, Sun L, Bruder D, Stegemann S, Cerwenka A, Sommer C, Dalpke AH, Veltkamp R. Inhibition of lymphocyte trafficking shields the brain against deleterious neuroinflammation after stroke. Brain. 2011;134:704–720. doi: 10.1093/brain/awr008. [DOI] [PubMed] [Google Scholar]
- Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- Means TK, Hayashi F, Smith KD, Aderem A, Luster AD. The Toll-like receptor 5 stimulus bacterial flagellin induces maturation and chemokine production in human dendritic cells. J Immunol. 2003;170:5165–5175. doi: 10.4049/jimmunol.170.10.5165. [DOI] [PubMed] [Google Scholar]
- Milligan ED, Mehmert KK, Hinde JL, Harvey LO, Martin D, Tracey KJ, Maier SF, Watkins LR. Thermal hyperalgesia and mechanical allodynia produced by intrathecal administration of the human immunodeficiency virus-1 (HIV-1) envelope glycoprotein, gp120. Brain Res. 2000;861:105–116. doi: 10.1016/s0006-8993(00)02050-3. [DOI] [PubMed] [Google Scholar]
- Mishra BB, Mishra PK, Teale JM. Expression and distribution of Toll-like receptors in the brain during murine neurocysticercosis. J Neuroimmunol. 2006;181:46–56. doi: 10.1016/j.jneuroim.2006.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22:240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Scaffidi P, Degryse B, Bonaldi T, Ronfani L, Agresti A, Beltrame M, Bianchi ME. New EMBO members’ review: the double life of HMGB1 chromatin protein: architectural factor and extracellular signal. Embo j. 2001;20:4337–4340. doi: 10.1093/emboj/20.16.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio L, Martino G, Furlan R. Multifaceted aspects of inflammation in multiple sclerosis: the role of microglia. J Neuroimmunol. 2007;191:39–44. doi: 10.1016/j.jneuroim.2007.09.016. [DOI] [PubMed] [Google Scholar]
- Park JS, Gamboni-Robertson F, He Q, Svetkauskaite D, Kim JY, Strassheim D, Sohn JW, Yamada S, Maruyama I, Banerjee A, Ishizaka A, Abraham E. High mobility group box 1 protein interacts with multiple Toll-like receptors. Am J Physiol Cell Physiol. 2006;290:C917–C924. doi: 10.1152/ajpcell.00401.2005. [DOI] [PubMed] [Google Scholar]
- Park JS, Svetkauskaite D, He Q, Kim JY, Strassheim D, Ishizaka A, Abraham E. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J Biol Chem. 2004;279:7370–7377. doi: 10.1074/jbc.M306793200. [DOI] [PubMed] [Google Scholar]
- Qi J, Buzas K, Fan H, Cohen JI, Wang K, Mont E, Klinman D, Oppenheim JJ, Howard OM. Painful pathways induced by TLR stimulation of dorsal root ganglion neurons. J Immunol. 2011;186:6417–6426. doi: 10.4049/jimmunol.1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Zhao G, Aoki Y, Shi L, Uyei A, Nazarian S, Ng JC, Kao PN. Immunosuppressant PG490 (triptolide) inhibits T-cell interleukin-2 expression at the level of purine-box/nuclear factor of activated T-cells and NF-κB transcriptional activation. J Biol Chem. 1999;274:13443–13450. doi: 10.1074/jbc.274.19.13443. [DOI] [PubMed] [Google Scholar]
- Ren K, Dubner R. Interactions between the immune and nervous systems in pain. Nat Med. 2010;16:1267–1276. doi: 10.1038/nm.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharwade SC, Sharma PK, Mukhopadhaya A. Vibrio cholerae porin OmpU induces pro-inflammatory responses, but down-regulates LPS-mediated effects in RAW 264.7, THP-1 and human PBMCs. PLoS One. 2013;8:e76583. doi: 10.1371/journal.pone.0076583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- Stokes JA, Corr M, Yaksh TL. Spinal toll-like receptor signaling and nociceptive processing: regulatory balance between TIRAP and TRIF cascades mediated by TNF and IFNβ. Pain. 2013;154 doi: 10.1016/j.pain.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Tanga FY, Nutile-Mcmenemy N, Deleo JA. The CNS role of Toll-like receptor 4 in innate neuroimmunity and painful neuropathy. Proc Natl Acad Sci U S A. 2005;102:5856–5861. doi: 10.1073/pnas.0501634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo SH, Grasser KD, Thomas JO. Differences in the DNA-binding properties of the HMG-box domains of HMG1 and the sex-determining factor SRY. Eur J Biochem. 1995;230:943–950. doi: 10.1111/j.1432-1033.1995.tb20640.x. [DOI] [PubMed] [Google Scholar]
- Thibault S, Imbeault M, Tardif MR, Tremblay MJ. TLR5 stimulation is sufficient to trigger reactivation of latent HIV-1 provirus in T lymphoid cells and activate virus gene expression in central memory CD4+ T cells. Virology. 2009;389:20–25. doi: 10.1016/j.virol.2009.04.019. [DOI] [PubMed] [Google Scholar]
- Van Zoelen MA, Yang H, Florquin S, Meijers JC, Akira S, Arnold B, Nawroth PP, Bierhaus A, Tracey KJ, Van Der Poll T. Role of toll-like receptors 2 and 4, and the receptor for advanced glycation end products in high-mobility group box 1-induced inflammation in vivo. Shock. 2009;31:280–284. doi: 10.1097/SHK.0b013e318186262d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins LR, Hutchinson MR, Milligan ED, Maier SF. “Listening” and “talking” to neurons: implications of immune activation for pain control and increasing the efficacy of opioids. Brain Res Rev. 2007;56:148–169. doi: 10.1016/j.brainresrev.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M, Stott K, Thomas JO. Mapping intramolecular interactions between domains in HMGB1 using a tail-truncation approach. J Mol Biol. 2007;374:1286–1297. doi: 10.1016/j.jmb.2007.09.075. [DOI] [PubMed] [Google Scholar]
- Xu ZZ, Kim YH, Bang S, Zhang Y, Berta T, Wang F, Oh SB, Ji RR. Inhibition of mechanical allodynia in neuropathic pain by TLR5-mediated A-fiber blockade. Nat Med. 2015;21:1326–1331. doi: 10.1038/nm.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Liang J, Yao C, Wu P, Zeng X, Cheng K, Yin H. Pyrimidine Triazole Thioether Derivatives as Toll-Like Receptor 5 (TLR5)/Flagellin Complex Inhibitors. ChemMedChem. 2016;11:822–826. doi: 10.1002/cmdc.201500471. [DOI] [PubMed] [Google Scholar]
- Yang H, Hreggvidsdottir HS, Palmblad K, Wang H, Ochani M, Li J, Lu B, Chavan S, Rosas-Ballina M, Al-Abed Y, Akira S, Bierhaus A, Erlandsson-Harris H, Andersson U, Tracey KJ. A critical cysteine is required for HMGB1 binding to Toll-like receptor 4 and activation of macrophage cytokine release. Proc Natl Acad Sci U S A. 2010;107:11942–11947. doi: 10.1073/pnas.1003893107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Ju Z, Ragab AA, Lundback P, Long W, Valdes-Ferrer SI, He M, Pribis JP, Li J, Lu B, Gero D, Szabo C, Antoine DJ, Harris HE, Golenbock DT, Meng J, Roth J, Chavan SS, Andersson U, Billiar TR, Tracey KJ, Al-Abed Y. MD-2 is required for disulfide HMGB1-dependent TLR4 signaling. J Exp Med. 2015;212:5–14. doi: 10.1084/jem.20141318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SI, Kurnasov O, Natarajan V, Hong M, Gudkov AV, Osterman AL, Wilson IA. Structural basis of TLR5-flagellin recognition and signaling. Science. 2012;335:859–864. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SL, Lyddon TD, Jorgenson RL, Misfeldt ML. Expression of Toll-like receptors in human endometrial epithelial cells and cell lines. Am J Reprod Immunol. 2004;52:67–73. doi: 10.1111/j.1600-0897.2004.00189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.