Abstract
In mammals, because they share a single synthetic pathway, n-6/n-3 ratios of dietary PUFAs impact tissue arachidonic acid (ARA) and DHA content. Likewise, SNPs in the human fatty acid desaturase (FADS) gene cluster impact tissue ARA and DHA. Here we tested the feasibility of using heterozygous Fads2-null-mice (HET) as an animal model of human FADS polymorphisms. WT and HET mice were fed diets with linoleate/α-linolenate ratios of 1:1, 7:1, and 44:1 at 7% of diet. In WT liver, ARA and DHA in phospholipids varied >2× among dietary groups, reflecting precursor ratios. Unexpectedly, ARA content was only <10% lower in HET than in WT livers, when fed the 44:1 diet, likely due to increased Fads1 mRNA in response to reduced Fads2 mRNA in HET. Consistent with the RNA data, C20:3n-6, which is elevated in minor FADS haplotypes in humans, was lower in HET than WT. Diet and genotype had little effect on brain PUFAs even though brain Fads2 mRNA was low in HET. No differences in cytokine mRNA were found among groups under unstimulated conditions. In conclusion, differential PUFA profiles between HET mice and human FADS SNPs suggest low expression of both FADS1 and 2 genes in human minor haplotypes.
Keywords: brain lipids, cytokines, diet and dietary lipids, fatty acid/desaturases, fatty acid/elongases, inflammation, liver, muscle, omega-3 fatty acids, fatty acid desaturase 1, fatty acid desaturase 2, polyunsaturated fatty acid
Linoleic acid (C18:2 n-6; LA) and α-linolenic acid (C18:3 n-3; ALA) are essential FAs because they cannot be synthesized in mammals and must be obtained from diet (1, 2). They are precursors of the physiologically essential highly unsaturated FAs (HUFAs) arachidonic acid (C20:4 n-6; ARA) and DHA (C22:6 n-3), respectively. The n-6 and n-3 FA families share the same synthetic pathway that involves a series of desaturation and elongation (2, 3). Therefore, competition exists between n-6 and n-3 FAs for utilizing the enzymes, and overconsumption of one results in a significant decrease in products of the other (4, 5). In general, the n-6 and n-3 families have an opposite effect in the progress of inflammation through their derivatives (6). Upon activation of phospholipase A2, ARA and its n-3 counterpart EPA are released from cell membrane phospholipids (PLs) and compete for synthesis of eicosanoids such as prostaglandins and leukotrienes (7). Generally, 2-series prostaglandins and 4-series leukotrienes derived from ARA promote inflammation, whereas EPA-originated 3-series prostaglandins and 5-series leukotrienes are less inflammatory or anti-inflammatory (6, 8, 9). Also, docosanoids such as resolvins and neuroprotectins derived from DHA were reported to have anti-inflammatory effects (10). Chronic inflammation contributes to the development of several diseases including atherosclerosis, cancer, and neurological disorders (11–13).
Recently, large-scale, genome-wide association studies identified strong associations of SNPs in the region of the fatty acid desaturase (FADS) gene cluster with disease or its risk factors such as blood lipids (14–16), blood glucose (16), and asthma (17). Minor alleles (designated by the NCBI dbSNP database) in the FADS gene cluster, where two desaturase genes, FADS1 and FADS2, for the ARA and DHA synthesis are located, were generally associated with a lower product/substrate ratio of n-6 and n-3 PUFAs in serum or red blood cells (RBCs), indicating lower capacity of ARA, DHA synthesis (18–22). Furthermore, the allele frequency of the FADS cluster differs among populations; some minor alleles in most populations occur predominantly in certain populations (23, 24), suggesting environmental effects on the allele frequency. Taken together, the genetic capacity of endogenous synthesis of ARA and DHA likely interacts with environmental factors and affects disease susceptibility and survival. Although the environmental factors that affect the allele frequency in the FADS gene cluster are yet to be identified, a dietary n-6/n-3 ratio is likely one of them. For example, a reduced capacity in synthesis when combined with a diet with a high LA/ALA ratio may attenuate elevation of tissue ARA and subsequent overreaction of inflammatory response, whereas it may compromise adequate supply of DHA that can be compensated if a diet contains sufficient preformed DHA.
Our laboratory created Fads2-null mice, in which synthesis of ARA and DHA was disabled. Because heterozygous Fads2-null mice (HET) have reduced delta-6-desaturase expression, we conducted this study to address if the HET mouse is suitable as a model of human polymorphism in the FADS gene cluster. To achieve this objective, we investigated the effects of Fads2 genotypes on tissue ARA and DHA abundance in liver, brain, gastrocnemius muscle, and RBCs and on the gene expression of synthetic pathway of these HUFAs under diets with a varying ratio of dietary precursors, LA and ALA.
MATERIALS AND METHODS
Animals and diets
All the animal experiments were approved by the University of Illinois Institutional Animal Care and Use Committee and were conducted in conformity with the Public Health Service Policy on Humane Care and Use of Laboratory Animals. Production of the Fads2-null mouse was described previously (3). The mouse was subsequently backcrossed to C57BL/6J background. Heterozygous and WT mice from our breeding colony were mated to produce WT and HET animals for the study. Until 2 months of age, the animals were fed a standard purified diet, AIN93G diet (Dyets Inc., Bethlehem, PA). At 2 months, 18 WT and 18 HET male mice were evenly divided into three dietary groups and were singly housed in a 12 h light/dark cycle. They had free access to food and water. Body weight was recorded every week. Three experimental diets were prepared by adding 7% (w/w) soybean oil (SO), corn oil (CO), and a mixture of flaxseed and canola oil (FCO) to a fat-free AIN93G diet (Dyets Inc.) to achieve the ratio of LA/ALA 7:1, 44:1, and 1:1, respectively, as shown in Table 1. After being fed the respective diets for 14 weeks, mice were fasted overnight before euthanization. Final body weight was recorded. Blood was drawn by cardiac puncture after mice were anesthetized under isoflurane gas. Plasma and erythrocytes (RBCs) were separated by centrifuging at 800 g for 10 min at 4°C. Liver, brain, and gastrocnemius were removed and snap frozen in liquid N2. All samples were stored in a −80°C freezer until further analysis.
TABLE 1.
FA composition of experimental diets
| FCO | SO | CO | |
| 16:0 | 4.4 | 10.2 | 10.6 |
| 18:0 | 2.5 | 4.5 | 1.8 |
| 18:1 | 53.2 | 22.7 | 27.3 |
| 18:2 n-6 | 18.4 | 54.8 | 53.2 |
| 18:3 n-3 | 18.4 | 7.8 | 1.2 |
| n-6:n-3 | 1:1 | 7:1 | 44:1 |
Values are wt. % of total FAs. Each diet contains 7% fat. FCO was prepared by mixing flaxseed oil and canola oil in a ratio of 1:4.
Plasma lipid analysis
Plasma total cholesterol and TGs were measured by enzymatic kits (Cholesterol E and L-Type TG M; Wako Life Sciences Inc., Mountain View, CA) according to the manufacturer’s instructions.
Total RNA extraction and quantitative PCR
Total RNA was isolated from liver, brain, and gastrocnemius muscle with a Trizol reagent and a purification kit Direct-ZolTM RNA Miniprep (Zymo Research Corp., Irvine, CA). About 25 mg of liver and 60 mg of brain and gastrocnemius muscle were used. After adjusting the RNA concentration to 100 ng/µl, 10 µl from each sample was prepared for the synthesis of cDNA by a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA). Then, 25ng cDNA template was amplified by a 7300 real-time PCR system (Applied Biosystems) under the following conditions: 95°C for 10 min, then 15 s at 95°C and 1 min at 60°C for 35 cycles. The primers were purchased from Integrated DNA Technologies (Coralville, IA). The sequences were previously published for the primers of Fads2 (25), elongase of very long chain fatty acid (Elovl) 5 (26), interleukin 1 β (Il1b) (27), interleukin 6 (Il6) (27), and tumor necrosis factor (Tnf) (28). These three cytokines are chosen as markers of proinflammatory condition (29). Additional genes were analyzed using the following primer pairs: Fads1, 5′ primer, 5′-CCAGCTTTGAACCCACCAA-3′ and 3′ primer, 5′-CATGAGGCCCATTCGCTCTA-3′ Elovl2, 5′ primer, 5′-GCTAATGGCATGACGGACA-3′ and 3′ primer, 5′-GTTCCCCGGCACTTCATTT-3′ ribosomal protein L7α (Rpl7a as a housekeeping gene) 5′ primer, 5′-AACTTCGGCATTGGACAGG ACA-3′, and 3′ primer, 5′-TTTGAGCCGCTTGTAGAGGATAGC-3′. A standard curve was constructed in every run to account for the efficiency of each primer set. A pooled cDNA was sequentially diluted to half each time from 100 to 3.1 ng and was used for the standard curve construction. All gene expression data were normalized with Rpl7a. In addition, to compare gene expression between different genes and organs, a normalizing factor 2−ΔΔCt was back-calculated from the standard curves using the following formula:
ΔΔCt = [(Ct of Gene X in Tissue Y at 25 ng − Ct of Rp17a in Tissue Y at 25 ng) − (Ct of Fads2 in liver at 25 ng − Ct of Rp17a in liver at 25 ng)]
Thus, all RNA expression data were expressed relative to the mean of liver Fads2, which was set as 100.
Total lipid extraction, thin-layer chromatography, and methylation
A one-step extraction-methylation method was used for brain, gastrocnemius, and RBC samples (30). Briefly, ∼100 mg tissue was added to a glass tube containing 2 ml of 4:1 methanol and hexane and 100 µg butylated hydroxytoluene. After homogenization on ice and flushing a tube with nitrogen for 20 s, 200 µl of acetyl chloride was slowly added. The tube was then flushed with nitrogen, capped tightly, and incubated for 1 h in a water bath at 100°C. Five milliliters of 6% K2CO3 was added to the tube after cooling on ice. After centrifugation at 3,200 g for 3 min, the upper layer containing FA methyl esters was transferred to a 2.0 ml vial. For liver samples, total lipid was first extracted from 150 mg of frozen samples by the method of Folch (31). The lipid extracts were separated with thin-layer chromatography to cholesteryl ester (CE), TG, and PL fractions, which were then methylated individually (3).
FA analysis
Methylated FAs in hexane were analyzed using a GC-2010 Plus gas chromatograph (Shimadzu, Columbia, MD) equipped with a flame ionization detector and a DB-FFAP capillary column (15 m × 0.1 mm inner diameter, 0.1 μm film thickness; Agilent Technologies, Santa Clara, CA). A program was based on the method of Masood (32). FA data were analyzed by GC Solution (Shimadzu) and presented as weight percent of total FAs.
Statistical analyses
Data are presented as mean ± SD (n = 6 for each group). Six animals per group was sufficient to reach a power of 0.8 with a type I error rate of 0.05. All data were analyzed using two-way ANOVA (2 genotypes × 3 diets), and when significance was found, data sets were further analyzed using one-way ANOVA followed by a post hoc test using the least significant difference (LSD) multiple comparison. Analysis by another post hoc test, Duncan’s multiple range test, yielded essentially identical results to LSD. Difference was considered significant at P < 0.05.
RESULTS
Both diet and genotype affected mRNA expression of desaturases and elongases in liver
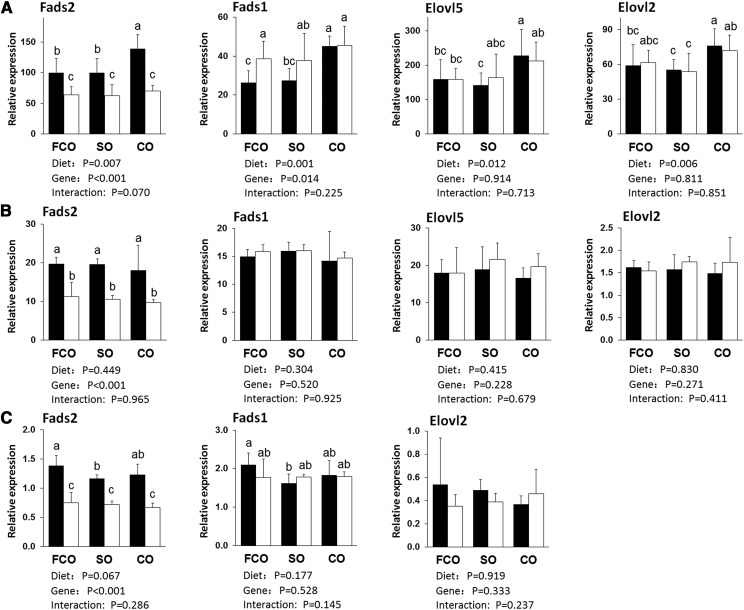
No significant dietary or genotype effect was observed in body weight, tissue weight, plasma lipids, or liver TG, although brain was heavier in SO-fed HET than in CO-fed WT, and plasma cholesterol was lower in SO-fed HET than in CO-fed HET by LSD test (supplemental Table S1). In the liver of WT, the CO group with a high ratio of dietary n-6/n-3 precursors increased the mRNA of all 4 synthetic enzymes, Fads2, Fads1, Elovl5, and Elovl2, compared with SO and FCO groups, whereas none of four genes was different between SO and FCO (Fig. 1A). As expected, the expression of Fads2 gene in HET was about half of that in WT. Fads1 gene expression in HET liver was higher than that in WT in the FCO and SO groups, suggesting compensatory increase of Fads1 expression when Fads2 expression is limited. However, in the CO group, Fads1 in HET did not increase over WT, suggesting the Fads1 expression may have reached maximum in the CO group. On the other hand, the genotype had no effect on Elovl2 or Elovl5.
Fig. 1.
Expression of genes for HUFA synthesis in liver (A), brain (B), and gastrocnemius muscle (C). A: In liver, CO diet with high LA/ALA ratio increased Fads2, Fads1, Elovl2, and Elovl5 mRNA in WT mice liver. HET showed elevated Fads-1 expression compared with WT. In HET mice, Fads2 mRNA was about half of that in WT. B: Except for decreased Fads2 expression in HET, no other dietary or genotype effect was found in brain. The expression of Elovl2 was an order of magnitude lower than other three genes. C: In gastrocnemius muscle, Fads2 in HET was about half in WT. Expression of Fads genes were much lower than in liver and brain. Black bars, WT; white bars, HET. Values were normalized with the housekeeping gene (Rpl7a) and expressed as relative to mean liver Fads2, which was set as 100. Mean ± SD, n = 6. Bars without same letter were significantly different, P < 0.05 by LSD multiple comparison. P values for the effect of diet, gene, and their interaction were calculated by two-way ANOVA.
In the brain of WT, no dietary effect was observed in the expression of Fads2, Fads1, Elovl5, or Elovl2 (Fig. 1B). Fads2 expression in HET was about half of WT, but no compensatory change was observed in other genes, indicating constitutive expression in brain. Surprisingly, Elovl2 expression was an order of magnitude lower than other three genes. This expression pattern suggests that the role of the pathway in brain may be primarily supplying ARA, and that brain may acquire DHA mostly from circulation.
In the gastrocnemius muscle, mRNA of Fads2 and Fads1 was much lower than in liver and brain, suggesting a limited synthetic capacity. In the muscle of WT, unlike liver, the FCO group showed small but significantly higher Fads2 and Fads1 mRNA compared with the SO group, whereas the expression of these genes in the CO group fell in between (Fig. 1C). Like brain, no genotype effect was observed except Fads2 expression in HET was about half of WT.
Diet showed stronger effects than genotype on PUFA composition in liver
As shown in Table 2, two-way ANOVA revealed that the n-6/n-3 ratio of precursor FAs in diet had strong effects on the abundance of product HUFAs in liver PLs in both genotypes. ARA in the SO and CO groups was nearly 2× and 3×, respectively, higher than that of FCO, whereas DHA in FCO and SO was more than twice as that in the CO group as dietary ALA was increased. C22:5n-6 significantly increased only in the CO group, whereas EPA decreased from 4% in FCO to <1% in SO and CO groups. In liver TGs, as the dietary LA to ALA ratio increased, all products of LA were significantly increased, whereas all ALA products were decreased significantly although the abundance of each HUFA is 2% or less in TGs (supplemental Table S2). PUFAs in the liver CE fraction also showed dietary effects similar to FAs in TGs (supplemental Table S3). In WT animals, LA was significantly lower, and ARA was higher in CO than in SO in all PL, TG, and CE fractions, although ARA in TGs did not reach statistical significance (Table 2; supplemental Tables S2 and S3), reflecting induction of genes of the synthetic pathway (Fig. 1A).
TABLE 2.
PUFA composition of liver PLs (wt. %)
| FCO | SO | CO | P | ||||||
| Fatty Acid | WT | HET | WT | HET | WT | HET | Diet | Gene | Gene × Diet |
| 18:2 n-6 | 13.65 ± 0.81b | 14.11 ± 1.2b | 16.16 ± 1.86a | 17.49 ± 1.96a | 13.24 ± 1.68b | 16.75 ± 0.86a | *** | ** | * |
| 18:3 n-6 | n. d. | n. d. | n. d. | n. d. | n. d. | n. d. | |||
| 20:2 n-6 | 0.34 ± 0.03bc | 0.41 ± 0.08ab | 0.29 ± 0.07c | 0.46 ± 0.07a | 0.31 ± 0.04c | 0.44 ± 0.04a | *** | ||
| 20:3 n-6 | 2 ± 0.28a | 1.45 ± 0.2b | 2.07 ± 0.34a | 1.33 ± 0.08bc | 1.84 ± 0.28a | 1.15 ± 0.15c | * | *** | |
| 20:4 n-6 | 8.77 ± 1.01d | 8.5 ± 1.3d | 17.76 ± 1.1c | 16.49 ± 0.97c | 24.15 ± 1.21a | 22.16 ± 0.9b | *** | ** | |
| 22:4 n-6 | 0.05 ± 0.01c | 0.05 ± 0.04c | 0.21 ± 0.11b | 0.27 ± 0.13b | 0.39 ± 0.02a | 0.37 ± 0.02a | *** | ||
| 22:5 n-6 | 0.04 ± 0.01b | 0.05 ± 0.02b | 0.19 ± 0.15b | 0.25 ± 0.17b | 2.37 ± 0.29a | 2.25 ± 0.29a | *** | ||
| 18:3 n-3 | 0.64 ± 0.13a | 0.67 ± 0.1a | 0.19 ± 0.03b | 0.22 ± 0.05b | 0.04 ± 0.04c | 0.03 ± 0.02c | *** | ||
| 20:3 n-3 | n. d. | n. d. | n. d. | n. d. | n. d. | n. d. | |||
| 20:5 n-3 | 4.36 ± 0.29a | 3.87 ± 0.87b | 0.66 ± 0.13c | 0.55 ± 0.21cd | 0.11 ± 0.03d | 0.09 ± 0.04d | *** | ||
| 22:5 n-3 | 0.81 ± 0.02a | 0.79 ± 0.1a | 0.41 ± 0.03b | 0.35 ± 0.07c | 0.12 ± 0.01d | 0.1 ± 0.02d | *** | ||
| 22:6 n-3 | 13.62 ± 0.72a | 13.36 ± 0.54a | 10.82 ± 0.49b | 10.54 ± 0.59b | 5.55 ± 1.36c | 4.98 ± 0.18c | *** | ||
| Total n-6 | 24.85 ± 0.77c | 24.56 ± 1.28c | 36.68 ± 1.24b | 36.29 ± 1.57b | 42.3 ± 1.33a | 43.12 ± 0.85a | *** | ||
| Total n-3 | 19.43 ± 0.65a | 18.68 ± 1.06a | 12.09 ± 0.62b | 11.66 ± 0.87b | 5.81 ± 1.37c | 5.2 ± 0.17c | *** | ||
| n-6/n-3 | 1.28 ± 0.08c | 1.32 ± 0.13c | 3.04 ± 0.17b | 3.13 ± 0.23b | 7.64 ± 1.83a | 8.3 ± 0.38a | *** | ||
| AA/DHA | 0.65 ± 0.10c | 0.64 ± 0.09c | 1.65 ± 0.16b | 1.57 ± 0.14b | 4.53 ± 0.89a | 4.46 ± 0.19a | *** | ||
n. d., not detected. Values are means ± SD (n = 6); data that do not share same superscript letter(s) within a row are significantly different, P < 0.05 by LSD multiple comparison. Effects of diet, gene, and their interaction were calculated by two-way ANOVA; * P < 0.05, ** P < 0.01, and *** P < 0.001.
Contrary to our expectation, the magnitude of the genotype effect was weaker than the dietary effect. The HET animals exhibited significantly lower ARA than WT in liver PLs only in the CO group in which dietary LA/ALA was high, but the difference was small, a decrease of only 9% (24% and 22% in WT and HET, respectively) (Table 2). A decline of ARA and DHA with a larger magnitude was found in liver CE (supplemental Table S3), confirming a previous observation (3). The most consistently found genotype effects across dietary treatment were a decrease in dihomo-γ-linolenic acid (C20:3 n-6; DGLA) and an increase in C20:2n-6 in liver PLs from HET compared with WT (Table 2). Also, DGLA in HET was significantly lower in TG and CE fractions of liver (supplemental Tables S2 and S3). Even though decreased expression of Fads2 in HET mice, neither DHA nor C22:5 n-6, the products of the second desaturation by Δ6 desaturase, decreased in any lipid fractions of HET except a small decline of DHA in the CE fraction (Table 2; supplemental Tables S2 and S3). This lack of genotype effects suggests that the second desaturation by Δ6 desaturase is not rate limiting.
Brain PUFA composition was less affected by diet and genotype than in liver
As presented in Table 3, the diet had a significant effect on all PUFAs except the LA in brain. However, as expected, the changes in ARA and DHA, the two major HUFAs in brain, were much smaller compared with the changes in liver PLs. In HET, the level of DGLA was significantly lower compared with WT. In general, the brain PUFA composition was maintained in a narrow range, indicating the presence of a mechanism to resist dietary and genetic influence.
TABLE 3.
PUFA composition of brain (wt. %)
| FCO | SO | CO | P | ||||||
| Fatty Acid | WT | HET | WT | HET | WT | HET | Diet | Gene | Gene × Diet |
| 18:2 n-6 | 0.41 ± 0.06b | 0.45 ± 0.04ab | 0.65 ± 0.24ab | 0.65 ± 0.05ab | 0.42 ± 0.04b | 1.03 ± 1.22a | |||
| 18:3 n-6 | n. d. | n. d. | n. d. | n. d. | n. d. | n. d. | |||
| 20:2 n-6 | n. d. | n. d. | n. d. | n. d. | n. d. | n. d. | |||
| 20:3 n-6 | 0.44 ± 0.01a | 0.39 ± 0.04b | 0.35 ± 0.06b | 0.27 ± 0.06c | 0.25 ± 0.02c | 0.20 ± 0.02d | *** | *** | |
| 20:4 n-6 | 7.54 ± 0.38b | 7.51 ± 0.58b | 8.63 ± 0.41a | 8.29 ± 0.39a | 8.80 ± 0.52a | 8.68 ± 0.37a | *** | ||
| 22:4 n-6 | 1.71 ± 0.06c | 1.70 ± 0.10c | 2.06 ± 0.15b | 1.97 ± 0.04b | 2.33 ± 0.15a | 2.31 ± 0.08a | *** | ||
| 22:5 n-6 | 0.08 ± 0.02b | 0.09 ± 0.02b | 0.15 ± 0.03b | 0.15 ± 0.04b | 0.84 ± 0.15a | 0.81 ± 0.08a | *** | ||
| 18:3 n-3 | n. d. | n. d. | n. d. | n. d. | n. d. | n. d. | |||
| 20:3 n-3 | n. d. | n. d. | n. d. | n. d. | n. d. | n. d. | |||
| 20:5 n-3 | 0.12 ± 0.01a | 0.14 ± 0.05a | 0.05 ± 0.03b | 0.06 ± 0.02b | 0.04 ± 0.01b | 0.05 ± 0.03b | *** | ||
| 22:5 n-3 | 0.29 ± 0.02a | 0.31 ± 0.03a | 0.13 ± 0.02b | 0.14 ± 0.01b | 0.07 ± 0.02c | 0.08 ± 0.01c | *** | * | |
| 22:6 n-3 | 13.12 ± 1.56a | 13.03 ± 1.20a | 12.76 ± 1.12a | 12.05 ± 0.93ab | 11.28 ± 1.25b | 11.25 ± 0.89b | ** | ||
| Total N-6 | 10.18 ± 0.40c | 10.13 ± 0.66c | 11.85 ± 0.52b | 11.32 ± 0.43b | 12.64 ± 0.77a | 13.03 ± 0.91a | *** | ||
| Total N-3 | 13.53 ± 1.58a | 13.48 ± 1.24a | 12.94 ± 1.14a | 12.25 ± 0.95ab | 11.38 ± 1.27b | 11.38 ± 0.89b | *** | ||
| N-6/N-3 | 0.76 ± 0.06c | 0.75 ± 0.03c | 0.92 ± 0.06b | 0.93 ± 0.06b | 1.12 ± 0.06a | 1.15 ± 0.14a | *** | ||
| AA/DHA | 0.58 ± 0.04c | 0.58 ± 0.02c | 0.68 ± 0.05b | 0.69 ± 0.04b | 0.78 ± 0.05a | 0.77 ± 0.04a | *** | ||
n. d., not detected. Values are means ± SD (n = 6); data that do not share same superscript letter(s) within a row are significantly different, P < 0.05 by LSD multiple comparison. Effects of diet, gene, and their interaction were calculated by two-way ANOVA; * P < 0.05, ** P < 0.01, and *** P < 0.001.
PUFA composition in gastrocnemius muscle was strongly affected by diet but much less by genotype
As shown in Table 4, high dietary LA/ALA ratio significantly elevated proportions of all n-6 PUFAs. The LA level in the SO and CO groups was more than twice of that in FCO. The ARA in the SO and CO groups doubled and tripled, respectively, compared with the FCO group. The n-3 family exhibited an opposite trend. Particularly, the ALA in the FCO groups was 20-fold higher than that in the CO group, and the DHA level in FCO was twice as that in CO (Table 4). The genotype effect was less pronounced; the DGLA level was lower in HET compared with WT, whereas C20:2 n-6 was increased in HET (Table 4).
TABLE 4.
PUFA composition of gastrocnemius muscle (wt. %)
| FCO | SO | CO | P | ||||||
| Fatty Acid | WT | HET | WT | HET | WT | HET | Diet | Gene | Gene × Diet |
| 18:2 n-6 | 8.69 ± 1.34b | 8.47 ± 1.24b | 19.59 ± 2.73a | 19.10 ± 4.61a | 20.11 ± 2.81a | 19.16 ± 4.59a | *** | ||
| 18:3 n-6 | n. d. | n. d. | n. d. | n. d. | n. d. | n. d. | |||
| 20:2 n-6 | 0.07 ± 0.03c | 0.09 ± 0.03c | 0.17 ± 0.04b | 0.22 ± 0.05ab | 0.20 ± 0.03b | 0.26 ± 0.07a | *** | * | |
| 20:3 n-6 | 0.33 ± 0.12bc | 0.26 ± 0.11c | 0.41 ± 0.09ab | 0.28 ± 0.12bc | 0.50 ± 0.14a | 0.39 ± 0.16abc | * | * | |
| 20:4 n-6 | 1.53 ± 0.74c | 1.68 ± 0.61c | 3.30 ± 0.64bc | 3.68 ± 1.61ab | 5.22 ± 1.91a | 5.36 ± 2.58a | *** | ||
| 22:4 n-6 | 0.06 ± 0.03c | 0.07 ± 0.02c | 0.33 ± 0.05bc | 0.36 ± 0.16b | 0.94 ± 0.33a | 0.89 ± 0.44a | *** | ||
| 22:5 n-6 | 0.06 ± 0.03b | 0.06 ± 0.03b | 0.43 ± 0.15b | 0.61 ± 0.44b | 3.04 ± 0.99a | 3.16 ± 1.86a | *** | ||
| 18:3 n-3 | 3.43 ± 1.20a | 3.22 ± 0.85a | 1.60 ± 0.34b | 1.52 ± 0.46b | 0.17 ± 0.09c | 0.16 ± 0.07c | *** | ||
| 20:3 n-3 | 0.03 ± 0.01a | 0.03 ± 0.01a | 0.01 ± 0.00b | 0.02 ± 0.02ab | 0.02 ± 0.01ab | 0.03 ± 0.01a | |||
| 20:5 n-3 | 0.46 ± 0.39a | 0.23 ± 0.07b | 0.09 ± 0.02b | 0.07 ± 0.02b | 0.06 ± 0.06b | 0.05 ± 0.03b | *** | ||
| 22:5 n-3 | 1.64 ± 0.70ab | 1.78 ± 0.75a | 1.12 ± 0.35b | 1.07 ± 0.63b | 0.35 ± 0.14c | 0.43 ± 0.31c | *** | ||
| 22:6 n-3 | 8.68 ± 4.93a | 9.12 ± 3.98a | 7.18 ± 1.70ab | 8.63 ± 4.00a | 3.50 ± 1.17b | 4.05 ± 2.68b | ** | ||
| Total n-6 | 10.74 ± 0.48c | 10.62 ± 0.63c | 24.24 ± 1.94b | 24.25 ± 2.74b | 30.01 ± 1.24a | 29.22 ± 1.36a | *** | ||
| Total n-3 | 14.23 ± 4.68a | 14.38 ± 3.89a | 10.00 ± 1.71b | 11.31 ± 4.04ab | 4.10 ± 1.23c | 4.72 ± 2.95c | *** | ||
| n-6:n-3 | 0.84 ± 0.33b | 0.79 ± 0.23b | 2.49 ± 0.49b | 2.54 ± 1.40b | 7.74 ± 1.76a | 7.80 ± 3.53a | *** | ||
| AA/DHA | 0.18 ± 0.77c | 0.18 ± 0.52c | 0.46 ± 0.16b | 0.43 ± 0.38b | 1.49 ± 0.06a | 1.32 ± 0.12a | *** | ||
n. d., not detected. Values are means ± SD (n = 6); data that do not share same superscript letter(s) within a row are significantly different, P < 0.05 by LSD multiple comparison. Effects of diet, gene, and their interaction were calculated by two-way ANOVA; * P < 0.05, ** P < 0.01, and *** P < 0.001.
PUFA composition in RBCs was similar to that in liver PLs
As shown in Table 5, the diet had a very strong effect on all n-6 PUFAs except on DGLA. The CO and SO groups showed similar ARA level, which was twice as much as that in the FCO group. The n-3 PUFAs showed an opposite trend of n-6 PUFAs. As seen in the other tissues, DGLA showed lower percentage in HET than in WT, whereas the 20:2 n-6 increased in HET compared with WT. No genotype effect was found in n-3 PUFA composition.
TABLE 5.
PUFA composition of RBCs (wt. %)
| FCO | SO | CO | P | ||||||
| Fatty Acid | WT | HET | WT | HET | WT | HET | Diet | Gene | Gene × Diet |
| 18:2 n-6 | 8.1 ± 0.38c | 8.82 ± 0.79bc | 11.25 ± 1.38a | 12.17 ± 1.19a | 10.44 ± 0.94ab | 11.55 ± 3.51a | *** | ||
| 18:3 n-6 | n. d. | n. d. | n. d. | n. d. | n. d. | n. d. | |||
| 20:2 n-6 | 0.14 ± 0.05c | 0.19 ± 0.09bc | 0.24 ± 0.05b | 0.35 ± 0.1a | 0.24 ± 0.03b | 0.26 ± 0.05b | *** | * | |
| 20:3 n-6 | 1.38 ± 0.21a | 1.02 ± 0.2b | 1.61 ± 0.33a | 0.98 ± 0.23b | 1.42 ± 0.24a | 1.03 ± 0.29b | *** | ||
| 20:4 n-6 | 9.84 ± 1.49b | 9.89 ± 1.18b | 17.68 ± 2.23a | 16.87 ± 1.65a | 18.67 ± 2.46a | 18.21 ± 5.23a | *** | ||
| 22:4 n-6 | 0.26 ± 0.12c | 0.25 ± 0.14c | 1.42 ± 0.31b | 1.42 ± 0.25b | 2.14 ± 0.39a | 2.14 ± 0.64a | *** | ||
| 22:5 n-6 | 0.15 ± 0.17b | 0.15 ± 0.16b | 0.45 ± 0.15b | 0.47 ± 0.17b | 2.46 ± 0.59a | 2.39 ± 0.8a | *** | ||
| 18:3 n-3 | 0.52 ± 0.34a | 0.62 ± 0.26a | 0.18 ± 0.13b | 0.18 ± 0.12b | 0.03 ± 0.01b | 0.02 ± 0.03b | *** | ||
| 20:3 n-3 | n. d. | n. d. | n. d. | n. d. | n. d. | n. d. | |||
| 20:5 n-3 | 2.91 ± 0.48a | 2.82 ± 0.57a | 0.45 ± 0.15b | 0.32 ± 0.17bc | 0.03 ± 0.02c | 0.03 ± 0.01c | *** | ||
| 22:5 n-3 | 1.85 ± 0.31a | 1.84 ± 0.29a | 0.86 ± 0.16b | 0.81 ± 0.26b | 0.18 ± 0.04c | 0.17 ± 0.05c | *** | ||
| 22:6 n-3 | 7.9 ± 1.01a | 7.7 ± 0.87a | 6.08 ± 0.7b | 5.92 ± 1.13b | 2.65 ± 0.45c | 2.54 ± 0.74c | *** | ||
| Total n-6 | 19.73 ± 1.54c | 20.13 ± 0.81c | 32.41 ± 1.83b | 31.92 ± 1.61b | 35.13 ± 2.63a | 35.33 ± 3.4a | *** | ||
| Total n-3 | 13.18 ± 1.44a | 12.98 ± 1.49a | 7.58 ± 0.81b | 7.24 ± 1.45b | 2.88 ± 0.48c | 2.77 ± 0.76c | *** | ||
| n-6:n-3 | 1.5 ± 0.06c | 1.57 ± 0.24c | 4.32 ± 0.55b | 4.58 ± 1.05b | 12.41 ± 1.64a | 13.86 ± 4.72a | *** | ||
| AA/DHA | 1.24 ± 0.05c | 1.3 ± 0.24c | 2.92 ± 0.3b | 2.93 ± 0.57b | 7.12 ± 0.59a | 7.18 ± 0.15a | *** | ||
n. d., not detected. Values are means ± SD (n = 6); data that do not share same superscript letter(s) within a row are significantly different, P < 0.05 by LSD multiple comparison. Effects of diet, gene, and their interaction were calculated by two-way ANOVA; * P < 0.05, ** P < 0.01, and *** P < 0.001.
Proinflammatory cytokine gene expression
As low-grade inflammation is implicated to be involved with the development of chronic diseases in tissues including liver and brain, we investigated if abundance of tissue ARA affected cytokine expression. Shown in Table 6 is the mRNA of proinflammatory cytokines in liver and brain. No significant difference was found in any cytokines tested, indicating neither the diet nor the genotype had an influence on these cytokines gene expression in an unstimulated condition.
TABLE 6.
Cytokine gene expression in liver and brain
| FCO | SO | CO | ||||
| WT | HET | WT | HET | WT | HET | |
| Liver | ||||||
| Il1b | 0.66 ± 0.28 | 0.62 ± 0.51 | 0.75 ± 0.42 | 0.89 ± 0.68 | 0.54 ± 0.10 | 0.66 ± 0.24 |
| Il6 | 0.21 ± 0.16 | 0.15 ± 0.11 | 0.15 ± 0.08 | 0.20 ± 0.13 | 0.14 ± 0.06 | 0.11 ± 0.05 |
| Tnf | 0.21 ± 0.13 | 0.29 ± 0.30 | 0.23 ± 0.23 | 0.42 ± 0.35. | 0.14 ± 0.05 | 0.16 ± 0.08 |
| Brain | ||||||
| Il1b | 0.16 ± 0.02 | 0.15 ± 0.03 | 0.16 ± 0.05 | 0.13 ± 0.03 | 0.12 ± 0.05 | 0.14 ± 0.06 |
| Il6 | 0.12 ± 0.07 | 0.17 ± 0.11 | 0.21 ± 0.12 | 0.15 ± 0.08 | 0.14 ± 0.06 | 0.16 ± 0.13 |
Means ± SD (n = 6); values were normalized with the housekeeping gene (Rpl7a) and expressed as relative to mean liver Fads2, which was set as 100.
DISCUSSION
Impact of dietary LA/ALA ratio on HUFA synthesis
The enzyme activity (33) and mRNA (34) of Δ6 desaturase increase when animals are fed a diet free of n-6 and n-3 essential FAs. The activity of Δ6 desaturase, the first step of ARA and DHA synthesis is primarily regulated at the transcriptional level by transcription factors such as sterol regulatory element-binding protein (SREBP) 1c (35), PPARα (36), and Elk (37). Elk is likely to mediate the effect of growth hormone on Fads2 (38, 39). Strong associations of SNPs in the FADS gene cluster with HUFA profiles and chronic diseases further suggest physiological importance of transcriptional regulation of the pathway. In this study, we found that gene expression of Fads2 as well as Fads1, Elovl 5, and Elovl 2 were also regulated by a ratio of dietary LA and ALA. The CO diet with LA/ALA ratio of 44:1 increased the expression of all four genes required for synthesis of DHA and ARA in the liver of WT compared with the SO diet with the ratio of 7:1. This upregulation of synthetic pathway in the CO group is likely a mechanism to compensate the decreased conversion of dietary ALA to DHA, but the upregulation also increased the conversion of LA to ARA, resulting in a large increase in ARA in liver PL and CE fractions and a decrease in LA in liver PL, TG, and CE fractions. In contrast, changing the LA/ALA ratio to 1:1 from 7:1 did not alter the expression of the desaturase and elongase genes in liver even though ARA in the liver PLs decreased to half. This lack of upregulation in response to decreased ARA suggests that the pathway of DHA and ARA synthesis is primarily regulated to meet DHA requirement rather than ARA requirement in liver. The mechanism underlying this differential regulation by DHA and ARA is yet to be elucidated.
In contrast, the brain gene expression was unaffected by diet. Our results are consistent with studies by Igarashi et al. (40–42) that reported upregulation of synthetic genes in liver but not in brain when rats were fed diets devoid of n-3 FAs for 15 weeks even though brain DHA decreased by nearly 40%. Skeletal and heart muscle in mice contains high DHA (43, 44) compared with other species such as rats (45), pigs (46), and humans (47, 48). Our study showed that unlike brain, the DHA content in gastrocnemius muscle markedly decreased and partly replaced with 22:5 n-6 when the dietary LA/ALA ratio was increased, although it is currently unknown if this change affects physiological function of skeletal muscle. The mechanism underlying the higher expression of both Fads2 and Fads1 mRNA in the WT FCO than in WT SO in muscle is unknown, but it is possible that the pathway may be under feedback regulation by ARA in the muscle.
Differences in FA profiles between HET mice and human FADS SNPs
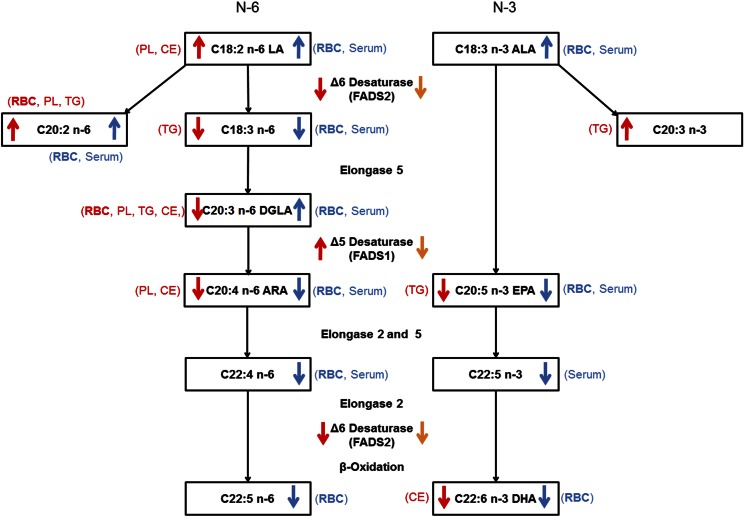
Our study revealed critical differences in FA profiles between HET mouse model and human SNPs in the FADS gene cluster. Figure 2 summarizes the changes in the gene expression and FA compositions in HET animals compared with WT (shown in red). Also, changes in FA profile in minor alleles of human SNPs compared with major alleles are overlaid in Fig. 2 (shown in blue). Although HET mice and human minor SNPs share similar changes in FA profiles, human SNPs showed more extensive changes (Fig. 2). There is a notable difference in the abundance of DGLA between HET mice and human FADS SNPs. Decreased DGLA was the most consistent change of FA profile in HET mice, which can be explained by decreased Fads2 mRNA and increased Fads1 mRNA in liver (Fig. 2). In contrast, DGLA is increased in the human FADS minor alleles, suggesting the expression of both FADS2 and FADS1 is decreased (Fig. 2, shown in orange). Our study and human studies suggest that control of FADS2, the first step of ARA and DHA synthesis, alone is insufficient for regulating the synthesis rate, and requires coordinated regulation of both FADS2 and FADS1.
Fig. 2.
Summary of genotype effects of HET mice and comparison with minor alleles in human FADS gene cluster. Red arrows: changes in gene expression in liver and FA abundance in RBC and liver in HET mice compared with WT; lipid fractions in liver and RBC total lipid that showed a significant change are shown in parentheses in red. Blue arrows: changes in FA abundance in RBC (21) and serum (22) in minor alleles of SNPs compared with major alleles in the human FADS cluster; specimens that showed a significant change are shown in parentheses in blue. Orange arrows: likely changes in human FADS2 and FADS1 expression based on the comparison with the HET mouse gene expression and FA profiles; increased LA, ALA, and C20:2 n-6, as well as decreased C18:3 n-6, indicate decreased expression of FADS2, whereas increased DGLA in contrast to HET mouse and a more extensive decrease in downstream products suggest a decrease in FADS1 expression in human minor alleles. RBC, the common data from both human and mice, is shown in bold. Although no data of gene expression or FA profiles in human liver are available, the serum FA profile likely reflects the hepatic synthesis because the FAs in fasting serum are largely originated from liver.
Because the SNPs occurring in FADS2 and FADS1 genes exhibit a strong linkage disequilibrium (18, 22), it is likely that the expression of FADS2 and FADS1 is regulated together in most haplotypes, and that expression of both FADS1 and FADS2 are lower in the minor alleles than the major alleles in the FADS gene cluster. In addition, because the transcription start sites of FADS2 and FADS1 genes are located only 20 kb apart in diverging orientation in human genome (2), these two genes may share the same regulatory elements, resulting in functional alleles affecting expression of both genes. Indeed, studies with rodents showed that both Fads2 and Fads1 genes were induced, albeit Fads1 in lesser extent, in response to fat-free diet, triolein diet and administration of a PPARα agonist (36, 49). Taken together, as seen in other pathways, coordination between the upstream and downstream of a pathway seems to be important for the regulation of the DHA and ARA synthesis in haplotypes as well as in the physiological responses.
A study reported that the major haplotype in the FADS cluster emerged in humans after divergence from Neanderthals and before exodus from Africa during the human evolution, suggesting upregulation of the synthetic capacity to supply DHA for larger brain as an important event in human evolution (50). In addition to strong associations of haplotypes in the FADS gene cluster with tissue ARA and disease risks, allele frequency differs among populations. As mentioned in the introduction, a “minor allele” becomes major in some populations, indicating that both haplotypes have a survival advantage depending on the environment they live in (23, 24). The conservation of haplotypes and differing occurrences of the FADS cluster underscore the physiological importance of the endogenous synthesis of ARA and DHA. Further study is warranted to investigate the interactive effects of environment and SNPs in the FADS gene cluster on health.
Lack of effects of tissue HUFAs on baseline cytokine expression
The n-3 FAs have been shown to decrease inflammatory cytokines both in vivo (51, 52) and in vitro (53, 54). However, we observed no difference in cytokine expression between groups under an unstimulated condition despite a large difference in the tissue ARA/(EPA + DHA) ratio among the dietary treatments. Consistent with our results, Moranis et al. (55) reported that mice with an n-3-deficient diet showed no difference in Il6 and Il10 concentrations in plasma and brain cortex as compared with the ones with normal diet. Also, Huang et al. (56) found that compared with an SO-based diet, fish oil significantly decreased dextran sulfate sodium-induced Tnf, Il1b, and Il6 secretion in colon but not in untreated groups. Because regulatory enzymes for eicosanoids synthesis such as phospholipases A2 and cyclooxygenase-2 are activated and induced, respectively, in response to inflammation (57, 58), a high tissue ARA/(EPA + DHA) is likely to have a minimal effect on cytokine production in basal condition, while it could exaggerate a response against an inflammatory stimulus by increasing proinflammatory eicosanoids such as prostaglandin E2. The overproduction of prostaglandins may further promote the inflammation by amplifying the cytokine signaling and may form a positive feedback on its own production (8).
CONCLUSIONS
In conclusion, a diet with a high ratio of LA/ALA (44:1) induced all four genes for ARA and DHA synthesis in WT liver compared with a diet with LA/ALA of 7:1 in response to decreased tissue DHA, whereas a diet with LA/ALA of 1:1 did not change expression of synthetic genes from the 7:1 diet despite marked decrease in ARA, suggesting that DHA exerts stronger regulation to the synthetic pathway than ARA. ARA in liver PL was small but significantly lower in HET than WT when fed a high LA/ALA diet, whereas DHA was unchanged in HET in all tissues examined. Fads1 was induced in response to decreased Fads2 expression in HET liver, resulting in decreased DGLA in all tissues examined. This Fads1 induction in HET is likely to have attenuated the decline of HUFA supply. In contrast to the HET mouse, minor alleles of SNPs in the human FADS cluster show increased DGLA in serum and RBC, as well as a strong linkage disequilibrium, suggesting reduced expression of both FADS2 and FADS1 genes.
Supplementary Material
Footnotes
Abbreviations:
- ALA
- alpha-linolenic acid
- ARA
- arachidonic acid
- CE
- cholesteryl ester
- CO
- corn oil
- DGLA
- dihomo-γ-linolenic acid
- Elovl
- elongation of very long chain fatty acids
- FADS/Fads
- fatty acid desaturase
- FCO
- flaxseed plus canola oil
- HUFA
- highly unsaturated fatty acid
- HET
- heterozygous Fads2-null mouse
- Il1b
- interleukin 1 beta
- Il6
- interleukin 6
- LA
- linoleic acid
- LSD
- least significant difference
- PL
- phospholipid
- RBC
- red blood cell, erythrocyte
- Rpl7a
- ribosomal protein L7alpha
- SO
- soybean oil
This work was supported in part by the U.S. Department of Agriculture, National Institute of Food and Agriculture Grant ILLU-971-365 (M.T.N.) and the China Scholarship Council (H.S.).
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Holman R. T. 1998. The slow discovery of the importance of ω3 essential fatty acids in human health. J. Nutr. 128: 427S–433S. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura M. T., and Nara T. Y.. 2004. Structure, function, and dietary regulation of Δ6, Δ5, and Δ9 desaturases. Annu. Rev. Nutr. 24: 345–376. [DOI] [PubMed] [Google Scholar]
- 3.Stroud C. K., Nara T. Y., Roqueta-Rivera M., Radlowski E. C., Lawrence P., Zhang Y., Cho B. H., Segre M., Hess R. A., Brenna J. T., et al. 2009. Disruption of FADS2 gene in mice impairs male reproduction and causes dermal and intestinal ulceration. J. Lipid Res. 50: 1870–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Umesha S. S., and Naidu K. A.. 2012. Vegetable oil blends with α-linolenic acid rich Garden cress oil modulate lipid metabolism in experimental rats. Food Chem. 135: 2845–2851. [DOI] [PubMed] [Google Scholar]
- 5.Liou Y. A., King D. J., Zibrik D., and Innis S. M.. 2007. Decreasing linoleic acid with constant α-linolenic acid in dietary fats increases (n-3) eicosapentaenoic acid in plasma phospholipids in healthy men. J. Nutr. 137: 945–952. [DOI] [PubMed] [Google Scholar]
- 6.Schmitz G., and Ecker J.. 2008. The opposing effects of n-3 and n-6 fatty acids. Prog. Lipid Res. 47: 147–155. [DOI] [PubMed] [Google Scholar]
- 7.Funk C. D. 2001. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 294: 1871–1875. [DOI] [PubMed] [Google Scholar]
- 8.Aoki T., and Narumiya S.. 2012. Prostaglandins and chronic inflammation. Trends Pharmacol. Sci. 33: 304–311. [DOI] [PubMed] [Google Scholar]
- 9.Calder P. C. 2006. n-3 Polyunsaturated fatty acids, inflammation, and inflammatory diseases. Am. J. Clin. Nutr. 83: 1505S–1519S. [DOI] [PubMed] [Google Scholar]
- 10.Kohli P., and Levy B. D.. 2009. Resolvins and protectins: mediating solutions to inflammation. Br. J. Pharmacol. 158: 960–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Libby P. 2006. Inflammation and cardiovascular disease mechanisms. Am. J. Clin. Nutr. 83: 456S–460S. [DOI] [PubMed] [Google Scholar]
- 12.Balkwill F. R., and Mantovani A.. 2012. Cancer-related inflammation: common themes and therapeutic opportunities. Semin. Cancer Biol. 22: 33–40. [DOI] [PubMed] [Google Scholar]
- 13.Amor S., Puentes F., Baker D., and Van Der Valk P.. 2010. Inflammation in neurodegenerative diseases. Immunology. 129: 154–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabatti C., Service S. K., Hartikainen A. L., Pouta A., Ripatti S., Brodsky J., Jones C. G., Zaitlen N. A., Varilo T., Kaakinen M., et al. 2009. Genomewide association analysis of metabolic phenotypes in a birth cohort from a founder population. Nat. Genet. 41: 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kathiresan S., Willer C. J., Peloso G., Demissie S., Musunuru K., Schadt E., Lee K., Bennett D., Li Y., Tanaka T., et al. 2009. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 41: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A. U., Wheeler E., Glazer N. L., Bouatia-Naji N., Gloyn A. L., et al. 2010. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 42: 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma S., Zhou X., Thibault D. M., Himes B. E., Liu A., Szefler S. J., Strunk R., Castro M., Hansel N. N., Diette G. B., et al. 2014. A genome-wide survey of CD4+ lymphocyte regulatory genetic variants identifies novel asthma genes. J. Allergy Clin. Immunol. 134: 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bokor S., Dumont J., Spinneker A., Gonzalez-Gross M., Nova E., Widhalm K., Moschonis G., Stehle P., Amouyel P., De Henauw S., et al. 2010. Single nucleotide polymorphisms in the FADS gene cluster are associated with delta-5 and delta-6 desaturase activities estimated by serum fatty acid ratios. J. Lipid Res. 51: 2325–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steer C. D., Hibbeln J. R., Golding J., and Smith G. D.. 2012. Polyunsaturated fatty acid levels in blood during pregnancy, at birth and at 7 years: their associations with two common FADS2 polymorphisms. Hum. Mol. Genet. 21: 1504–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathias R. A., Vergara C., Gao L., Rafaels N., Hand T., Campbell M., Bickel C., Ivester P., Sergeant S., Barnes K. C., et al. 2010. FADS genetic variants and ω-6 polyunsaturated fatty acid metabolism in a homogeneous island population. J. Lipid Res. 51: 2766–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koletzko B., Lattka E., Zeilinger S., Illig T., and Steer C.. 2011. Genetic variants of the fatty acid desaturase gene cluster predict amounts of red blood cell docosahexaenoic and other polyunsaturated fatty acids in pregnant women : findings from the Avon Longitudinal Study of Parents and Children. Am. J. Clin. Nutr. 93: 211–219. [DOI] [PubMed] [Google Scholar]
- 22.Schaeffer L., Gohlke H., Müller M., Heid I. M., Palmer L. J., Kompauer I., Demmelmair H., Illig T., Koletzko B., and Heinrich J.. 2006. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum. Mol. Genet. 15: 1745–1756. [DOI] [PubMed] [Google Scholar]
- 23.Mathias R. A., Sergeant S., Ruczinski I., Torgerson D. G., Hugenschmidt C. E., Kubala M., Vaidya D., Suktitipat B., Ziegler J. T., Ivester P., et al. 2011. The impact of FADS genetic variants on ω6 polyunsaturated fatty acid metabolism in African Americans. BMC Genet. 12: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kothapalli K. S. D., Ye K., Gadgil M. S., Carlson S. E., O’Brien K. O., Zhang J. Y., Park H. G., Ojukwu K., Zou J., Hyon S. S., et al. 2016. Positive selection on a regulatory insertion-deletion polymorphism in FADS2 influences apparent endogenous synthesis of arachidonic acid. Mol. Biol. Evol. 33: 1726–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenblat M., Volkova N., Roqueta-Rivera M., Nakamura M. T., and Aviram M.. 2010. Increased macrophage cholesterol biosynthesis and decreased cellular paraoxonase 2 (PON2) expression in Δ6-desaturase knockout (6-DS KO) mice: beneficial effects of arachidonic acid. Atherosclerosis. 210: 414–421. [DOI] [PubMed] [Google Scholar]
- 26.Moon Y. A., Hammer R. E., and Horton J. D.. 2009. Deletion of ELOVL5 leads to fatty liver through activation of SREBP-1c in mice. J. Lipid Res. 50: 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bodeman C. E., Dzierlenga A. L., Tally C. M., Mulligan R. M., Lake A. D., Cherrington N. J., and McKarns S. C.. 2013. Differential regulation of hepatic organic cation transporter 1, organic anion-transporting polypeptide 1a4, bile-salt export pump, and multidrug resistance-associated protein 2 transporter expression in lymphocyte-deficient mice associates with interleukin-6 production. J. Pharmacol. Exp. Ther. 347: 136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi H., Kokoeva M. V., Inouye K., Tzameli I., Yin H., and Flier J. S.. 2006. TLR4 links innate immunity and fatty acid–induced insulin resistance. J. Clin. Invest. 116: 3015–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Godbout J. P., Berg B. M., Krzyszton C., and Johnson R. W.. 2005. α-Tocopherol attenuates NFκB activation and pro-inflammatory cytokine production in brain and improves recovery from lipopolysaccharide-induced sickness behavior. J. Neuroimmunol. 169: 97–105. [DOI] [PubMed] [Google Scholar]
- 30.Lepage G., and Roy C. C.. 1986. Direct transesterification of all classes of lipids in a one-step reaction. J. Lipid Res. 27: 114–120. [PubMed] [Google Scholar]
- 31.Folch J., Lees M., and Stanley G. H. S.. 1957. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 32.Masood A., Stark K. D., and Salem N. Jr. 2005. A simplified and efficient method for the analysis of fatty acid methyl esters suitable for large clinical studies. J. Lipid Res. 46: 2299–2305. [DOI] [PubMed] [Google Scholar]
- 33.Castuma J. C., Catala A., and Brenner R. R.. 1972. Oxidative desaturation of eicosa-8,11-dienoic acid to eicosa-5,8,11-trienoic acid: comparison of different diets on oxidative desaturation at the 5,6 and 6,7 positions. J. Lipid Res. 13: 783–789. [PubMed] [Google Scholar]
- 34.Cho H. P., Nakamura M. T., and Clarke S. D.. 1999. Cloning, expression, and nutritional regulation of the mammalian Δ-6 desaturase. J. Biol. Chem. 274: 471–477. [DOI] [PubMed] [Google Scholar]
- 35.Nara T. Y., He W. S., Tang C., Clarke S. D., and Nakamura M. T.. 2002. The E-box like sterol regulatory element mediates the suppression of human Δ-6 desaturase gene by highly unsaturated fatty acids. Biochem. Biophys. Res. Commun. 296: 111–117. [DOI] [PubMed] [Google Scholar]
- 36.Li Y., Nara T. Y., and Nakamura M. T.. 2005. Peroxisome proliferator-activated receptor α is required for feedback regulation of highly unsaturated fatty acid synthesis. J. Lipid Res. 46: 2432–2440. [DOI] [PubMed] [Google Scholar]
- 37.Lattka E., Eggers S., Moeller G., Heim K., Weber M., Mehta D., Prokisch H., Illig T., and Adamski J.. 2010. A common FADS2 promoter polymorphism increases promoter activity and facilitates binding of transcription factor ELK1. J. Lipid Res. 51: 182–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clarkson R. W. E., Shang C. A., Levitt L. K., Howard T., and Waters M. J.. 1999. Ternary complex factors Elk-1 and Sap-1a mediate growth hormone- induced transcription of Egr-1 (early growth response factor-1) in 3T3–F442A preadipocytes. Mol. Endocrinol. 13: 619–631. [DOI] [PubMed] [Google Scholar]
- 39.Nakamura M. T., Phinney S. D., Tang A. B., Oberbauer A. M., German J. B., and Murray J. D.. 1996. Increased hepatic Δ6-desaturase activity with growth hormone expression in the MG101 transgenic mouse. Lipids. 31: 139–143. [DOI] [PubMed] [Google Scholar]
- 40.Igarashi M., Ma K., Chang L., Bell J. M., and Rapoport S. I.. 2007. Dietary n-3 PUFA deprivation for 15 weeks upregulates elongase and desaturase expression in rat liver but not brain. J. Lipid Res. 48: 2463–2470. [DOI] [PubMed] [Google Scholar]
- 41.Igarashi M., DeMar J. C., Ma K., Chang L., Bell J. M., and Rapoport S. I.. 2007. Docosahexaenoic acid synthesis from α-linolenic acid by rat brain is unaffected by dietary n-3 PUFA deprivation. J. Lipid Res. 48: 1150–1158. [DOI] [PubMed] [Google Scholar]
- 42.Igarashi M., DeMar J. C., Ma K., Chang L., Bell J. M., and Rapoport S. I.. 2007. Upregulated liver conversion of α-linolenic acid to docosahexaenoic acid in rats on a 15 week n-3 PUFA-deficient diet. J. Lipid Res. 48: 152–164. [DOI] [PubMed] [Google Scholar]
- 43.Kelley D. S., Bartolini G. L., Newman J. W., Vemuri M., and Mackey B. E.. 2006. Fatty acid composition of liver, adipose tissue, spleen, and heart of mice fed diets containing t10, c12-, and c9, t11-conjugated linoleic acid. Prostaglandins Leukot. Essent. Fatty Acids. 74: 331–338. [DOI] [PubMed] [Google Scholar]
- 44.Fedor D. M., Adkins Y., Newman J. W., Mackey B. E., and Kelley D. S.. 2013. The effect of docosahexaenoic acid on t10, c12-conjugated linoleic acid-induced changes in fatty acid composition of mouse liver, adipose, and muscle. Metab. Syndr. Relat. Disord. 11: 63–70. [DOI] [PubMed] [Google Scholar]
- 45.Stark K. D., Lim S., and Salem N. Jr. 2007. Artificial rearing with docosahexaenoic acid and n-6 docosapentaenoic acid alters rat tissue fatty acid composition. J. Lipid Res. 48: 2471–2477. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura M. T., Tang A. B., Villanueva J., Halsted C. H., and Phinney S. D.. 1993. The body composition and lipid metabolic effects of long-term ethanol feeding during a high ω6 polyunsaturated fatty acid diet in micropigs. Metabolism. 42: 1340–1350. [DOI] [PubMed] [Google Scholar]
- 47.Rocquelin G., Guenot L., Astorg P. O., and David M.. 1989. Phospholipid content and fatty acid composition of human heart. Lipids. 24: 775–780. [DOI] [PubMed] [Google Scholar]
- 48.Baur L. A., O’Connor J., Pan D. A., Kriketos A. D., and Storlien L. H.. 1998. The fatty acid composition of skeletal muscle membrane phospholipid: its relationship with the type of feeding and plasma glucose levels in young children. Metabolism. 47: 106–112. [DOI] [PubMed] [Google Scholar]
- 49.He W. S., Nara T. Y., and Nakamura M. T.. 2002. Delayed induction of Δ-6 and Δ-5 desaturases by a peroxisome proliferator. Biochem. Biophys. Res. Commun. 299: 832–838. [DOI] [PubMed] [Google Scholar]
- 50.Ameur A., Enroth S., Johansson Å., Zaboli G., Igl W., Johansson A. C. V., Rivas M. A., Daly M. J., Schmitz G., Hicks A. A., et al. 2012. Genetic adaptation of fatty-acid metabolism: a human-specific haplotype increasing the biosynthesis of long-chain omega-3 and omega-6 fatty acids. Am. J. Hum. Genet. 90: 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hassan A., Ibrahim A., Mbodji K., Coeffier M., Ziegler F., Bounoure F., Chardigny J. M., Skiba M., Savoye G., Dechelotte P., et al. 2010. An α-linolenic acid-rich formula reduces oxidative stress and inflammation by regulating NF-κB in rats with TNBS-induced colitis. J. Nutr. 140: 1714–1721. [DOI] [PubMed] [Google Scholar]
- 52.Sadeghi S., Wallace F. A., and Calder P. C.. 1999. Dietary lipids modify the cytokine response to bacterial lipopolysaccharide in mice. Immunology. 96: 404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mullen A., Loscher C. E., and Roche H. M.. 2010. Anti-inflammatory effects of EPA and DHA are dependent upon time and dose-response elements associated with LPS stimulation in THP-1-derived macrophages. J. Nutr. Biochem. 21: 444–450. [DOI] [PubMed] [Google Scholar]
- 54.Zhao G., Etherton T. D., Martin K. R., Gillies P. J., West S. G., and Kris-Etherton P. M.. 2007. Dietary alpha-linolenic acid inhibits proinflammatory cytokine production by peripheral blood mononuclear cells in hypercholesterolemic subjects. Am. J. Clin. Nutr. 85: 385–391. [DOI] [PubMed] [Google Scholar]
- 55.Moranis A., Delpech J. C., De Smedt-Peyrusse V., Aubert A., Guesnet P., Lavialle M., Joffre C., and Layé S.. 2012. Long term adequate n-3 polyunsaturated fatty acid diet protects from depressive-like behavior but not from working memory disruption and brain cytokine expression in aged mice. Brain Behav. Immun. 26: 721–731. [DOI] [PubMed] [Google Scholar]
- 56.Huang C. H., Hou Y. C., Yeh C. L., and Yeh S. L.. 2015. A soybean and fish oil mixture with different n-6/n-3 PUFA ratios modulates the inflammatory reaction in mice with dextran sulfate sodium-induced acute colitis. Clin. Nutr. 34: 1018–1024. [DOI] [PubMed] [Google Scholar]
- 57.Kudo I., and Murakami M.. 2002. Phospholipase A2 enzymes. Prostaglandins Other Lipid Mediat. 68–69: 3–58. [DOI] [PubMed] [Google Scholar]
- 58.Clària J. 2003. Cyclooxygenase-2 biology. Curr. Pharm. Des. 9: 2177–2190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.