Abstract
The α,β polyunsaturated lipid aldehydes are potent lipid electrophiles that covalently modify lipids, proteins, and nucleic acids. Recent work highlights the critical role these lipids play under both physiological and pathological conditions. Protein carbonylation resulting from nucleophilic attack of lysine, histidine, and cysteine residues is a major outcome of oxidative stress and functions as a redox-sensitive signaling mechanism with roles in autophagy, cell proliferation, transcriptional control, and apoptosis. In addition, protein carbonylation is implicated as an initiating factor in mitochondrial dysfunction and endoplasmic reticulum stress, providing a mechanistic connection between oxidative stress and metabolic disease. In this review, we discuss the generation and metabolism of reactive lipid aldehydes, as well as their signaling roles.
Keywords: cardiolipin, oxidized lipids, lipids/peroxidation
Lipotoxicity refers to a state in which lipids accumulate in cells and tissues that are not equipped to adequately metabolize or store them. This definition includes the biology of cytotoxic lipids and their resultant impact on cellular homeostasis. Importantly, small changes in abundance, composition, or location of such lipids can have profound effects on cellular viability and function (1). This review is focused on one class of cytotoxic lipids: the α,β polyunsaturated lipid aldehydes. These lipids are strong electrophiles whose production is induced by oxidative stress and that modify proteins, RNA, and DNA (2, 3). Although historically it has been accepted that the generation of these lipids is simply an endpoint of oxidative damage, it has become clear that they play an important role in many cellular processes, including autophagy, the unfolded protein response (UPR), endoplasmic reticulum (ER) stress, mitochondrial function, DNA-damage response, and apoptosis. Furthermore, despite the highly regulated antioxidant system in place to manage the generation of such lipids, under conditions of chronic and severe oxidative stress the α,β polyunsaturated lipid aldehydes can initiate mitochondrial dysfunction eventually culminating in apoptosis. Here, we review the mechanisms that lead to formation of these lipids and the signaling pathways impacted by their presence.
LIPID PEROXIDATION AND FORMATION OF REACTIVE LIPID ALDEHYDES
Oxidative stress is an imbalance in the tightly regulated production and metabolism of reactive oxygen species (ROS) such that the equilibrium is skewed toward an increasingly oxidative environment (4, 5). Lipid peroxidation is the result of hydroxyl radical attack of fatty acyl chains of phospholipids and triglycerides, and has garnered much attention due to the resultant widespread effects on cellular function. Although all organelles and compartments of the cell produce ROS, mitochondrial generation of hydrogen peroxide (H2O2) is generally considered to be the major source of oxidants (6, 7). Mitochondrial generation of superoxide anion via complexes I and III leads to elevated H2O2 through the action of superoxide dismutase (8). In the presence of ferrous iron, H2O2 undergoes Fenton chemistry to yield hydroxyl radicals (9, 10). The hydroxyl radical is highly reactive and rapidly abstracts hydrogen atoms from biomolecules in its immediate environment (10). Membrane phospholipids and triglycerides are primary targets for hydroxyl mediated attack and formation of lipid radicals (9, 11). Such lipid radicals are quickly oxidized leading to lipid peroxidation of acyl chains. Following peroxidation, membrane phospholipids undergo bond rearrangement allowing for further capture of radicals and chain propagation. Peroxidized acyl chains subsequently undergo nonenzymatic Hock cleavage, producing a family of aldehydes of various carbon lengths [e.g., malondialdehyde (n3), hydroxy hexenal (n6), hydroxy nonenal (n9)] depending upon the level of unsaturation and the lipid species oxidized (12, 13). Importantly, the second product of Hock cleavage, the parent phospholipid or triglyceride, now contains a shortened acyl chain that affects lipid packing and, therefore, membrane permeability (14). Broadly, the consequences of lipid peroxidation are twofold: generation of downstream reactive molecules such as the α,β polyunsaturated lipid aldehydes, as well as the concomitant changes in membrane organization and architecture (Fig. 1).
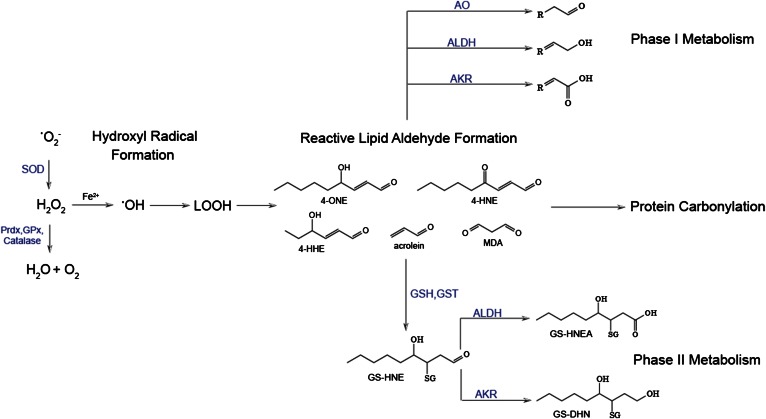
Fig. 1.
Generation and metabolism of reactive lipid aldehydes. Increased production of superoxide anion (•O2−) leads to the production of the hydroxyl radical (•OH) and subsequent lipid peroxidation (LOOH). This eventually results in the generation of a variety of reactive lipid aldehydes that can covalently modify proteins in a process called protein carbonylation. Under normal conditions, these lipids are detoxified by phase I and phase II antioxidant enzymes. In metabolic disease, the antioxidant milieu is depressed leading to accumulation of reactive aldehydes and protein carbonylation. SOD, superoxide dismutase; AO, alkenal/one oxidoreductase; AKR, aldo-keto reductase.
The formation of α,β unsaturated lipid aldehydes is largely dependent on the specific n-3 and n-6 PUFA species modified by the hydroxyl radical. Depending upon the lipid, aldehyde-containing secondary oxidation products, such as 4-hydroxy nonenal (4-HNE), 4-hydroxy hexenal (4-HHE), malondialdehyde, and acrolein, are commonly generated (Fig. 1) (13). The 4-HNE is the most well-studied lipid peroxidation product and it remains a major focus in the field due to its role in the etiology of many metabolic disease states, including cardiovascular disease, obesity, type II diabetes, neurodegenerative disease, atherosclerosis, asthma, chronic obstructive pulmonary disease, and cancer (15–23). Notwithstanding, a wide variety of other reactive lipid aldehyde species are formed at appreciable levels in mammalian cells, though the specific effect of these lipids on cellular metabolism is less defined.
While the chemical mechanisms that generate hydroxy-alkenals from PUFAs have been relatively well-characterized, the localization in vivo of these reactions remains largely a matter of assumption and conjecture. Interestingly, lipidomic profiling of wild-type C56Bl/6J mice indicated that individual tissues have unique lipid aldehyde profiles, suggesting specificity for the generation of particular lipids depending on the cell type. For example, in subcutaneous and visceral adipose depots, both 4-HNE and 4-oxononenal (4-ONE) are produced at high levels, while 4-HHE is undetectable (24). In contrast, in the liver, 4-HHE was observed to be the most abundant reactive lipid, with levels over an order of magnitude greater than 4-HNE (J. S. Burrill and D. A. Bernlohr, unpublished observations). Adipose tissue contains high relative concentrations of n-6 PUFAs, such as arachidonic acid and linoleic acid, both of which are the precursors to 4-HNE and 4-ONE. Conversely, the brain contains high levels of n-6 PUFAs and, therefore, may be ideally primed for the production of 4-HHE from docosahexaenoic acid or α-linoleic acid (25–27). Together these observations suggest that differences in reactive lipid aldehyde profiles are principally due to the PUFA composition of individual cell types.
LIPID PEROXIDATION AND APOPTOSIS
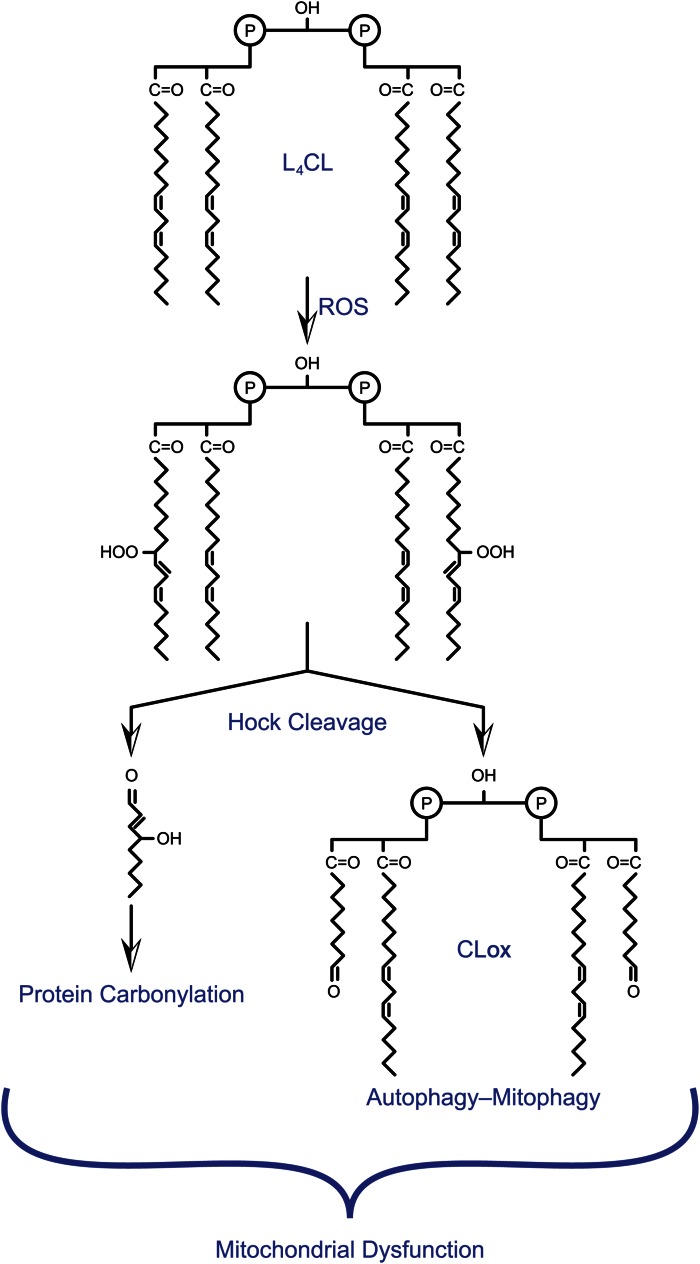
A variety of studies indicate that damage to membranes as a result of oxidation of PUFAs is associated with ageing and metabolic disease. Specifically, oxidation of lipid membranes can cause changes in membrane fluidity and permeability. This, in turn, may lead to altered enzymatic activity of membrane bound complexes. Recent work focuses on the finding that the bisphosphatidyl glycerol phospholipid, cardiolipin (CL), which is localized to the inner mitochondrial membrane, is particularly susceptible to lipid peroxidation, resulting in the generation of a variety of hydroperoxides. CL, which is distinct from other phospholipids in that it contains three glycerol moieties and four fatty acyl chains in the same molecule, is critical for maintenance of cristae structure as well as stabilizing mitochondrial electron transport complexes (28, 29). Lipidomic analysis demonstrated that oxidation of tetralinoleoyl CL (L4CL), the most common mitochondrial CL in mammalian tissues, not only generates lipid hydroperoxides that remain in the membrane, but also contributes to the production of mitochondrial 4-HNE (14, 30, 31) (Fig. 2). Furthermore, oxidation of CL is known to be accompanied by decreased activity of complexes I and IV, leading to impaired mitochondrial respiration (32–34). Accordingly, decreases in membrane CL content as well as oxidation of CL are strongly correlated with a variety of disease models in which mitochondrial dysfunction plays a major role, including atherosclerosis, obesity, and cardiac injury during ischemia/reperfusion (35, 36).
Fig. 2.
Oxidative modification of CL. Representation of L4CL oxidation by hydroxyl radicals (ROS) and subsequent production of α,β unsaturated lipid aldehydes as well as an CLox remnant. The head group of CL is drawn schematically to emphasize chemistry within the acyl chains. Unsaturated aldehydes lead to protein carbonylation, while CLox potentiates apoptosis, autophagy, or mitophagy leading to generalized mitochondrial dysfunction.
Critical work from Kagan et al. (37) showed that oxidation of CL is a required step for the release of pro-apoptotic factors during mitochondrial-driven (intrinsic) apoptosis. Pro-apoptotic stimuli induced the H2O2-dependent peroxidation of CL by cytochrome C (37). Because oxidized CL (CLox) (shown as CLox in Fig. 3) exhibits a reduced binding affinity for cytochrome C over CL (38, 39), peroxidation of CL results in the release of cytochrome C and mobilization to the outer mitochondrial membrane, an event that is an early required step that precedes the release of cytochrome C into the cytosol. Moreover, initial studies showed that CL specifically is essential for Bcl2-mediated membrane permeabilization through Bax and Bid (40). More recent work has indicated that oxidation of CL results not only in cytochrome C mobilization in the membrane, but also in migration of CL from the inner mitochondrial membrane to the outer mitochondrial membrane (41, 42). Here, CLox recruits and interacts with Bax to initiate formation of the mitochondrial transition pore. Furthermore, activation of Bax by pro-apoptotic stimuli has been shown to increase ROS production (43, 44), indicating that recruitment of Bax to the outer mitochondrial membrane engages the mitochondria into a feed-forward loop in which increased ROS perpetuate that apoptotic signaling cascade. Supporting this hypothesis is the observation by Jiang et al. (45) that ablation of Bax/Bak protects against actinomycin D-induced ROS production and CL peroxidation, while expression of recombinant Bax induces CL peroxidation. Together these data demonstrate that lipid peroxidation of membrane phospholipids can have widespread effects through alterations in membrane structure and function, as well as through protein-lipid interactions. These interactions are particularly important for the regulation of the intrinsic apoptotic pathway (Fig. 3).
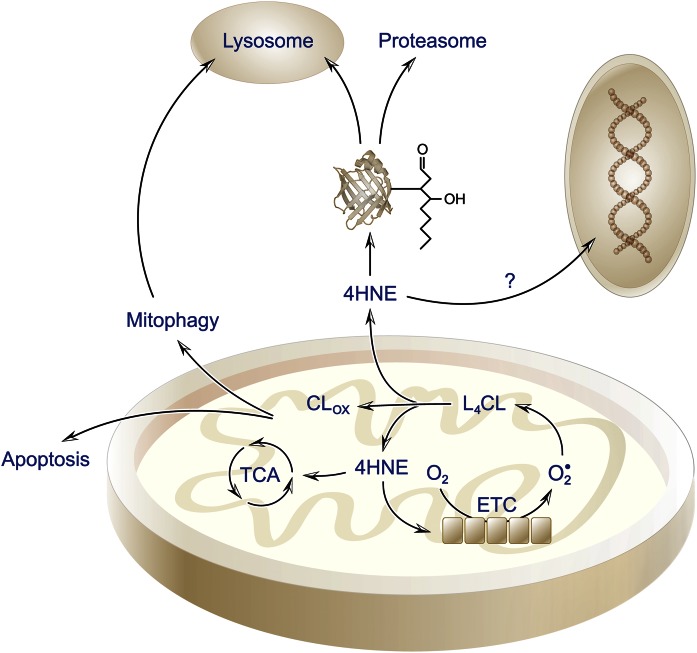
Fig. 3.
Cartoon representation of lipotoxic lipids and cellular responses. Peroxidation of CL (L4CL) within the mitochondrial inner membrane leads to oxidized CL (CLox) with shortened acyl chains that may initiate an apoptotic response. In parallel, resultant lipid aldehydes modify mitochondrial proteins leading to dysfunction and potential induction of mitophagy. Aldehydes may diffuse from the mitochondrion and modify proteins within the cytoplasm, nuclear region, or ER affecting cellular homeostasis and signaling response.
PROTEIN CARBONYLATION
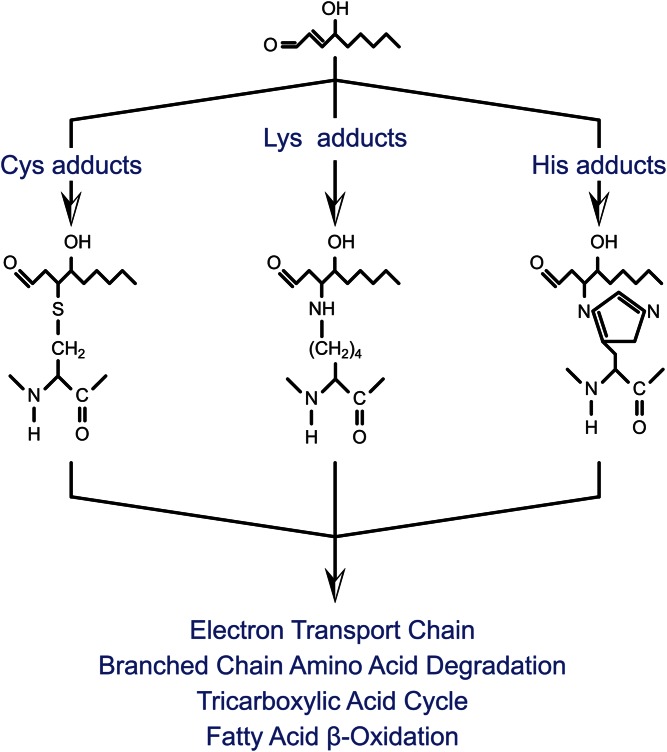
Under normal conditions, there are several classes of aldehyde-metabolizing enzymes that function to detoxify reactive lipid aldehydes prior to oxidative cellular damage (Fig. 1). However, under conditions of prolonged or elevated oxidative stress, α,β unsaturated lipid aldehydes accumulate. Due to the electron-withdrawing property of the carbonyl and hydroxyl oxygen atoms, α,β unsaturated aldehydes are strong electrophiles and highly reactive toward attack by proteins and nucleic acids. Furthermore, unlike many other oxidative species, the lipids are relatively stable and highly diffusible, allowing for a large functional boundary within the cellular environment. In addition, such lipid aldehydes may diffuse through membranes and can be found extracellularly (46). Protein carbonylation typically refers to the posttranslational modification of cysteine, histidine, and lysine residues by reactive lipid aldehydes. This class of posttranslational modification is the result of nucleophilic attack by sulfhydryl (cysteine), imidazole (histidine), or ε-amine (lysine) moieties on the C3 carbon of the lipid, resulting in the formation of a stable protein-lipid Michael adduct (3, 47) (Fig. 4). Schiff base formation can also occur between the ε-amine of lysine and lipid, though these adducts are generally believed to be less stable and do not retain the free carbonyl (3).
Fig. 4.
Protein carbonylation and formation of protein-lipid adducts. Alkylation of the side chains of lysine, histidine, and cysteine are shown and the major affected mitochondrial pathways. For diagrammatic purposes, only the modification of amino acid side chains by 4-HNE is shown. Additional parallel modification reactions with other electrophiles occur.
The effect of protein carbonylation on function is largely considered to affect two major properties: protein catalytic activity and/or protein-protein interactions. Due to the centrality of cysteine, lysine, and histidine to enzymatic catalysis, protein carbonylation typically represents a loss-of-function modification. In general, however, the stoichiometry of modification is measured to be quite low, typically less than 5% of any protein being carbonylated. Despite this, when multiple proteins of a biochemical pathway are concurrently carbonylated, flux through major components of metabolism can be affected. For example, a number of studies have reported that multiple enzymes of glycolysis are carbonylated (glyceraldehyde 3-phosphate dehydrogenase, enolase, fructose 1,6 bisphosphate aldolase) (48–51). Reduction in activity of multiple steps, even if relatively modest for each step, can have significant influence on overall pathway flux. Secondarily, carbonylation of lysine residues would block subsequent ubiquitination, thereby affecting cellular degradation and increasing the half-life of target proteins. In addition, cysteine, lysine, and histidine residues are frequently found in solvent-accessible regions of proteins, suggesting that carbonylation may lead to the introduction of a hydrophobic region on the surface of a polypeptide. If a modification occurs in a relatively nonpolar region, carbonylation may enable protein-protein interactions or complex formation. If such an assembly introduces functionality, carbonylation may lead to a gain-of-function modification for a system.
Protein carbonylation has traditionally been used as a biomarker for oxidative stress and is widely observed in a variety of metabolic tissues under both basal and diseased conditions. Carbonylation targets have been studied at length in metabolically active tissues, such as brain, skeletal muscle, liver, adipose, and cancer cells (14, 52–55). As detection techniques improve, identification of specific proteins that become modified has facilitated the discovery that protein carbonylation plays an active role in a wide array of cellular mechanisms, including the oxidative stress response, autophagy, ER stress response, proliferation, mitochondrial function, and apoptosis. In obesity, for example, total protein carbonylation is significantly increased in visceral adipose tissue (16, 56) and in the heart (34). Although the specific role of protein carbonylation remains complex, a number of studies suggest that protein carbonylation is a mechanistic connection between obesity and the development of metabolic dysfunction, particularly in the adipose tissue (48). In addition to obesity, elevated protein carbonylation has been observed in human and mouse models of neurodegenerative disease, cardiovascular disease, diabetes, cancer, hepatosteatosis, asthma, and pulmonary disease.
It remains unclear, in many cases, whether protein carbonylation plays an initiating/causal role or rather is the endpoint of another underlying dysfunction. As such, identification of specific modified proteins and a better understanding of how the modification affects protein function is a necessary step. To date, several large proteomics studies have aided our understanding of both the specificity and localization of protein carbonyls in tissue, such as adipose, liver, skeletal muscle, plasma, lung, and cardiac tissue (46, 48, 49, 57–61). Mass-spectrometry-based approaches have allowed for the identification of hundreds of direct targets of 4-HNE, 4-HHE, and acrolein carbonylation targets with cytosolic, mitochondrial, ER, and nuclear localization.
The mitochondrion, in particular, is a major source of ROS due to electron leak from complexes I and III of the electron transport chain (5). In addition, the mitochondrial inner membrane is rich in CL, making the mitochondrion a likely source of reactive lipid aldehydes and site of protein carbonylation. These observations, combined with the dogma that oxidative stress is often coincident with mitochondrial dysfunction and impaired respiration, spurred a number of studies focused on the role of mitochondrial carbonylation and mitochondrial dysfunction. Using skeletal muscle fibers from rats, Meany et al. (49) utilized an iTRAQ-based proteomic approach to identify proteins susceptible to protein carbonylation in mitochondrial lysates. Ninety-four protein targets were identified, nearly 40% of which were involved in oxidative phosphorylation. Moreover, quantitative analysis of mitochondrial carbonylation indicated that carbonylation of many targets increased with age, specifically in fast twitch fibers (62). In 2012, Curtis et al. (52) undertook a proteomics study in which they identified over 200 mitochondrial targets of protein carbonylation in glutathione-S-transferase (GST)A4-silenced 3T3-L1 adipocytes. KEGG analysis of the top pathways represented in the dataset revealed that 19% of the proteins involved in oxidative phosphorylation, 33% of the TCA cycle pathway, and 28% of the branched-chain amino acid (BCAA) catabolic pathway were carbonylated (48). Notably, in this model system, mitochondrial function was significantly impaired, as measured by decreased complex I activity, decreased membrane potential, reduced mitochondrial respiration, and increased ROS production. Other key targets identified included several proteins critical for efficient respiration, such as complex I proteins, NADH dehydrogenase (ubiquinone) 1α subunits 2 and 3, and the mitochondrial phosphate carrier (MPCP). Consistent with these observations, an emerging hypothesis is that protein carbonylation is a mechanism that links oxidative stress to impaired mitochondrial function through direct modification of electron transport machinery (Fig. 3).
The observation that the BCAA catabolic pathway is enriched for carbonylation is intriguing because serum levels of BCAAs are positively associated with obesity and insulin resistance (63–68). The BCAAs (leucine, isoleucine, and valine) are important sources of succinyl-CoA and acetyl-CoA to replenish the TCA cycle in adipocytes in order to fuel lipogenesis. Interestingly, treatment of cultured 3T3-L1 adipocytes with the inflammatory cytokine, TNF-α, led to decreased [14C]leucine uptake and conversion to triglyceride (69). While investigation of how carbonylation specifically affects the function of the enzymes involved in BCAA is ongoing, these data suggest a correlation between increased carbonylation and decreased lipogenesis, resulting in accumulation of BCAAs in the serum. As such, increased serum BCAA may be a biomarker for adipose inflammation and mitochondrial dysfunction.
Recent work by Zhao et al. (70) led to the identification of specific 4-HNE adducts in cardiac tissue from mice treated with doxorubicin. Proteomic analysis of the mitochondrial homogenates identified several carbonylation targets involved in energy metabolism, such as ATP synthase subunit β, succinate dehydrogenase (ubiquinone) flavoprotein subunit, creatine kinase S-type, and succinyl-CoA:3 ketoacid-CoA transferase. The observed changes in protein carbonylation were paralleled by a decrease in oxygen consumption rate. Interestingly, complex I subunit, NADH dehydrogenase 1α subunit 2, was among the targets identified, indicating that complex I may be a hotspot for protein carbonylation across multiple tissues.
Alcoholic liver disease (ALD) is characterized by impaired mitochondrial function and oxidative stress. Similar to the case in obese adipose tissue, oxidative stress in the liver is accompanied by increases in lipid peroxidation and production of reactive lipid aldehydes (54). Consistent with this, in mouse models of chronic alcohol consumption, protein carbonylation is significantly increased in the liver. As such, several groups have pursued the hypothesis that protein carbonylation contributes to impaired hepatocyte function and metabolism in ALD. Proteomic analysis of liver extracts from ethanol-treated mice led to the identification of hundreds of carbonylation targets, including 4-HNE, 4-ONE, and acrolein modification sites (71). Consistent with the observed mitochondrial dysfunction and lipid accumulation characteristics of this disease model, bioinformatic pathway analysis revealed that fatty acid metabolism, oxidative phosphorylation, glycolysis, and drug metabolism were among the top modified pathways. More recent work has focused on the identification of mitochondrial-specific targets in both mouse and rat models of ALD (57, 72). Although the functional consequences of these modifications on mitochondrial dysfunction and the progression of ALD have yet to be determined, it is tempting to speculate that carbonylation of specific mitochondrial proteins plays a central role in regulating hepatocyte metabolic flux in ALD.
Antioxidant enzymes that detoxify hydroperoxides or lipid aldehydes are critically important for the regulation of carbonylation and prevention of uncontrolled oxidative damage to cellular machinery. This point is exemplified by the many human diseases that are characterized by decreased activity or expression of specific families of antioxidant enzymes, such as the aldehyde dehydrogenases (ALDHs), glutathione peroxidases (GPXs), and GSTs (16, 24, 73–76). The relative specificity of these enzymes toward the detoxification of lipid peroxidation products makes a strong case for the lipotoxicity of reactive lipid aldehydes and protein carbonylation. For example, GSTA4 is a key phase II enzyme that glutathionylates lipid peroxidation products, allowing for their eventual export from the cell (Fig. 1). In 2007, Grimsrud et al. (16) observed that Gsta4 mRNA expression was decreased 3- to 4-fold in obese adipose tissue from mice, an observation that correlated with increased protein carbonylation in whole cell extracts from obese adipose depots. Strikingly, Gsta4-null mice on a 129/sv background exhibit high levels of 4-HNE and become obese when fed a chow diet (77). Conversely, Gsta4-null mice on a C57BL/6 background do not display elevated 4-HNE levels and do not become obese, suggesting that accumulation of 4-HNE is linked to metabolic dysfunction (77). Following this work, Curtis et al. (78) showed that silencing of GSTA4 in 3T3-L1 adipocytes led to increased mitochondrial protein carbonylation and impaired mitochondrial function. Recent work by Shearn et al. (57) highlights the role of GSTA4 in the liver. In this study, mitochondria were isolated from the livers of GSTA4−/− mice treated chronically with ethanol. Using mass spectrometry-based proteomics, they were able to identify over 800 targets of protein carbonylation, 417 of which were new carbonylation targets in chronic ethanol models of hepatic steatosis.
In addition to Gsta4, mRNA expression of GPx4, peroxiredoxin (Prdx)3, and Aldh2 is significantly downregulated in visceral adipose depots of obese mice compared with lean controls (24). PRDX3 is a mitochondrially localized peroxide reductase that catalyzes the conversion of H2O2 to O2 and water, thereby protecting the cell from H2O2-mediated oxidative damage. GPX4 and ALDH both function to reduce lipid hydroperoxides. As such, the end result of the concerted downregulation of these key enzymes is the unregulated production of lipid peroxidation end products (Fig. 1). Consistent with this, Long, Olson, and Bernlohr (24) observed a 5-fold increase in free 4-HNE and 4-ONE levels in visceral adipose depots of high-fat-fed C57Bl/6J mice. The downregulation of Gsta4, GPx4, and Prdx3, as well as the concomitant elevation in 4-HNE and 4-HHE, has been observed in both diet-induced and genetic (ob/ob mouse) models of obesity. Recent work by Katunga et al. (34) highlighted the importance of GPx4 in the context of obesity. While whole body knockout of this enzyme is embryonic lethal, Gpx4 haplo-insufficient mice maintained on a high-fat high-sucrose diet exhibit increased carbonyl stress, exemplified by increased 4-HNE adduct in the liver and heart accompanied by elevated ROS and mitochondrial dysfunction in the heart (34). Strikingly, decreased GPx4 mRNA expression was observed in human heart tissue from subjects with diabetes compared with nondiabetic controls. This expression pattern was accompanied by an increase in 4-HNE adducts in the diabetic samples, further highlighting the importance of this enzyme in mediating the effect of lipid peroxidation (34, 79).
LIPOTOXICITY AND ER STRESS
Protein carbonylation within the ER is emerging as a critical signaling mechanism within many cell types. ER stress and the accumulation of unfolded proteins are correlated with increased protein carbonyls in several mammalian cell types (80, 81). Similarly, amelioration of ER stress is concomitant with reduction in lipid peroxidation and protein carbonylation (80–84). Recent work in a variety of models suggests that lipid electrophiles play a primary role in ER stress and can directly activate all three arms of the UPR. In human stable and advanced atherosclerotic lesions, 4-HNE adducts colocalize with ER stress marker, Ire1α, and treatment of cultured human endothelial cells with 4-HNE induces phosphorylation of Ire1α and eIF2α (85). Similarly, treatment of human umbilical vein endothelial cells with 4-HNE (86) or acrolein (87) led to a robust activation of each major component of the UPR. In the 4-HNE-treated cells, HNE-protein adducts colocalized specifically with the ER and proteomic analysis led to the identification of several ER-specific chaperone proteins. The observation that protein chaperones are particularly susceptible to oxidation has been observed in several other contexts. In human leukemia cell line HL60, apoptosis-inducing photodynamic therapy selectively led to the carbonylation of key molecular chaperones: glucose-regulated protein (GRP)78, heat-shock protein (HSP)60, heat-shock protein cognate 71 (HSC71), phosphate disulphide isomerase, and calreticulin (88). In addition, proteomic analysis of lysates from aged murine liver (89), Alzheimer’s disease brain (19), and mouse hippocampal neuronal cells (90) suggest targeted carbonylation of critical ER proteins, such as phosphate disulphide isomerase and calreticulin. Proteomic analysis of lysates from a rodent model of ALD confirms the susceptibility of chaperones HSP72, HSP90, and GRP78 to adduction by lipid aldehydes (91–93). The 4-HNE modification of cysteine residues on HSP72 and Hsp90 correlated with impaired chaperone activity, while modification of GRP78 was localized to the ATPase domain and had a minimal effect on chaperone activity. Collectively, these results are consistent with the hypothesis that oxidized lipids function as a trigger for ER stress. Furthermore, although the mechanisms connecting elevated levels of 4-HNE and other electrophilic lipids to specific facets of the UPR remain unclear, these data suggest that modification of ER proteins, particularly chaperones involved in protein folding, plays a role in initiating this response.
Interestingly, recent work by Haberzettl and colleagues suggests a role for reactive lipid aldehydes in promoting autophagy through activation of the UPR. These studies stemmed from the observation that treatment of rat aortic smooth muscle cells with 4-HNE or acrolein resulted in protein carbonylation and activation of the autophagic machinery (94). Pretreatment of cells with rapamycin accelerated the rate of removal of 4-HNE-modified proteins, indicating that autophagy is one mechanism by which cells remove oxidatively damaged proteins. Consistent with this, cotreatment of 4-HNE with the autophagic inhibitor, 3-methyladenine, or the proteasome inhibitor, MG-132, resulted in cell death. Interestingly, proteomic analysis of 4-HNE-modified proteins in 4-HNE-treated rat aortic smooth muscle cells indicated that modified proteins were primarily localized to the ER. Several ER-specific chaperone proteins, including protein-disulfide isomerase and glucose-regulated proteins 58 and 78, were identified (GRP58, GRP78) (95). Moreover, 4-HNE treatment resulted in the selective activation of the PERK-dependent arm of the UPR, culminating in the activation of stress kinases, including JNK. A key finding in this study was that 4-HNE-induced increases in autophagy were entirely dependent on activation of the UPR with JNK activation as a critical signaling event that connected protein carbonylation in the ER to the induction of autophagy (95).
PROTEIN CARBONYLATION AND PROTEOSTASIS
Oxidized proteins are proteolytically degraded via a variety of mechanisms; however, under conditions of oxidative stress, either the increased production of unfolded damaged proteins or the decreased capacity to degrade them can lead to large protein aggregates that negatively impact cellular function. Such protein aggregates are known to increase with age as well as play a major functional role in many diseases of ageing (96, 97). Because a variety of proteins are modified by carbonylation, the definition of how carbonylation affects protein quality control mechanisms has been intensely pursued.
Protein proteolysis is carried out by three broad classes of proteases: the lysosomal proteases (e.g., cathepsins), calcium-dependent proteases (e.g., calpains), and the multicatalytic proteases (proteasomes). There are a number of hypotheses regarding the primary mechanism responsible for targeted degradation of carbonylated peptides, though much of the data indicates that the multicatalytic proteases, including the 20S and 26S proteasomes, play a major role in this process. The 20S proteasome, which exhibits chymotrypsin-like, trypsin-like, and peptidyl glutamyl peptide hydrolase-like activity, is the catalytic core of the proteasome. The 19S (PA700) subunit associates with the 20S catalytic core, yielding the 26S proteasome that efficiently carries out ATP-dependent proteolysis of ubiquitinated proteins. It is generally accepted that the 20S proteasome specifically degrades oxidized proteins, an observation that suggests that this may also be the case for carbonylated peptides (98) (Fig. 3). Initial studies in which liver cells were treated with 4-HNE indicated that 4-HNE-modified proteins were selectively removed by the proteasome and, further, proteasome activity was stimulated upon treatment with low concentrations of 4-HNE (32, 33, 99). In vitro carbonylated histones were rapidly proteolyzed by the 20S proteasome relative to the unmodified proteins, suggesting that modified histones were preferentially degraded (35, 100). Conversely, covalent modification of alcohol dehydrogenase with 4-HNE led to polyubiquitination and a 1.5-fold increase in its 26S-dependent degradation (101). These studies, along with several others, collectively indicate that 4-HNE-modified proteins can be degraded by the proteasome, though whether or not this is the sole, or even the primary, mechanism for removal of carbonylated protein remains controversial. Furthermore, reports on the mechanism by which proteasomal degradation may occur are inconsistent. Namely, it is unclear whether ubiquitination is required for recognition and targeting of carbonylated proteins to the proteasome. In addition to these inconsistencies, several studies challenge the proteasome-dependent degradation hypothesis itself. In a 2004 study, Marques et al. (102) demonstrated that 4-HNE-modified proteins were ubiquitinated preferentially in a cell free system. In lens epithelial cells, inhibition of the catalytic activity of the 26S proteasome had no effect on the removal of 4-HNE-modified proteins, whereas inhibition of lysosomal activity resulted in the accumulation of 4-HNE-modified proteins (102), suggesting a ubiquitin-dependent lysosomal degradation pathway.
In addition to the role the 20S and 26S proteasomes likely play in degrading 4-HNE-modified proteins, these complexes are also substrates for protein carbonylation. In several systems, elevated levels of reactive lipid aldehydes are coincident with decreased proteasomal activity both in vitro and in vivo (100, 103–106). It has been observed that 4-HNE-modified proteins make poor substrates for proteolysis (103, 107). Furthermore, the catalytic activity of the proteasome can be inhibited by site-specific carbonylation of various subunits leading to impaired protein turnover. In renal homogenates from rats treated with ferric nitrilotriacetate to induce oxidative stress, Okada et al. (108) observed 4-HNE-proteasome adducts. Appearance of these adducts correlated with decreased trypsin-like and peptidylglutamyl peptide hydrolase activity of the proteasome (70, 108). Proteasome-specific 4-HNE adducts coincident with inhibition of proteasome activity have been observed in many pathological states/tissues, including in neurodegenerative disease (72, 77, 104, 109), cerebral ischemia-reperfusion injury (77, 110), cardiac ischemia-reperfusion injury (111–113), and in lymphocytes from aged humans (57, 106). In-depth study of proteasome oxidation revealed that specific subunits are preferentially modified by reactive lipids and that, depending on which subunit is modified, catalytic activity of some, or all three, of the catalytic peptidase activities of the 20S proteasome can be inhibited (111, 114, 115). Although the precise mechanisms behind such inhibition of proteolysis remain complex, it is clear that protein carbonylation is a dynamic regulatory process in proteolysis.
CONCLUDING REMARKS
Reactive lipid aldehydes are critical mediators of the cellular response to oxidative stress under both physiological and pathological conditions. Significant advances in the technologies to detect both free aldehydes and lipid-protein, lipid-lipid, and lipid-nucleic acid adducts have widened our understanding of the functional scope that such lipids possess. Work in the last three decades has largely focused on identification of carbonylated proteins in oxidative stress models, and these efforts are exemplified by the hundreds of known carbonylation targets reported in the literature. Despite these advances, clear and specific roles for protein carbonylation in the pathophysiology of metabolic disease remain unclear. Analysis of the Gsta4-null mouse has given us important clues as to the critical role that protein carbonylation plays in metabolic disease. The metabolic dysfunction observed upon ablation of Gsta4 strongly argues for a critical role of protein carbonylation in cellular homeostasis. In moving forward, studies aimed at identifying mechanistically how modification of proteins affects not only their function, but also how these events initiate signaling changes, will be a particularly important goal. Furthermore, a better understanding of the biologically relevant differences between distinct lipid peroxidation products will be informative. Finally, a number of studies demonstrate that targeting enzymes or reactions that produce reactive lipids leads to metabolic benefits at the level of cultured cells, organs, and even whole organisms. As such, a clear understanding of the balance between production and metabolism of reactive lipids holds promise for therapeutic intervention for metabolic disease.
Acknowledgments
The authors would like to thank the Bernlohr laboratory for helpful discussions and Mr. Anthony Hertzel for assistance with the graphics.
Footnotes
Abbreviations:
- ALD
- alcoholic liver disease
- Aldh
- aldehyde dehydrogenase
- BCAA
- branched-chain amino acid
- CL
- cardiolipin
- CLox
- oxidized cardiolipin
- ER
- endoplasmic reticulum
- Gpx
- glutathione peroxidase
- Grp
- glucose-regulated protein
- GST
- glutathione-S-transferase
- 4-HHE
- 4-hydroxy hexenal
- 4-HNE
- 4-hydroxy nonenal
- H2O2
- hydrogen peroxide
- Hsp
- heat-shock protein
- JNK
- jun amino-terminal kinase
- L4CL
- tetralinoleoyl cardiolipin
- 4-ONE
- 4-oxononenal
- Prdx
- peroxiredoxin
- ROS
- reactive oxygen species
- UPR
- unfolded protein response
This work was supported by Office of Extramural Research, National Institutes of Health Grants DK084669 to D.A.B. and T32 GM008347 to A.K.H. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Ayala A., Muñoz M. F., and Argüelles S.. 2014. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev. 2014: 360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uchida K. 2003. 4-Hydroxy-2-nonenal: a product and mediator of oxidative stress. Prog. Lipid Res. 42: 318–343. [DOI] [PubMed] [Google Scholar]
- 3.Esterbauer H., Schaur R. J., and Zollner H.. 1991. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 11: 81–128. [DOI] [PubMed] [Google Scholar]
- 4.Pisoschi A. M., and Pop A.. 2015. The role of antioxidants in the chemistry of oxidative stress: a review. Eur. J. Med. Chem. 97: 55–74. [DOI] [PubMed] [Google Scholar]
- 5.Hamanaka R. B., and Chandel N. S.. 2010. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem. Sci. 35: 505–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sena L. A., and Chandel N. S.. 2012. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell. 48: 158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brand M. D. 2010. The sites and topology of mitochondrial superoxide production. Exp. Gerontal. 45: 466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fridovich I. 1995. Superoxide radical and superoxide dismutases. Annu. Rev. Biochem. 64: 97–112. [DOI] [PubMed] [Google Scholar]
- 9.Gardner H. W. 1989. Oxygen radical chemistry of polyunsaturated fatty acids. Free Radic. Biol. Med. 7: 65–86. [DOI] [PubMed] [Google Scholar]
- 10.Gutteridge J. M. 1984. Lipid peroxidation initiated by superoxide-dependent hydroxyl radicals using complexed iron and hydrogen peroxide. FEBS Lett. 172: 245–249. [DOI] [PubMed] [Google Scholar]
- 11.Bielski B. H., Arudi R. L., and Sutherland M. W.. 1983. A study of the reactivity of HO2/O2- with unsaturated fatty acids. J. Biol. Chem. 258: 4759–4761. [PubMed] [Google Scholar]
- 12.Vigo-Pelfrey C. 1990. Membrane lipid oxidation. CRC Press, Boca Raton, FL. [Google Scholar]
- 13.Benedetti A., Comporti M., and Esterbauer H.. 1980. Identification of 4-hydroxynonenal as a cytotoxic product originating from the peroxidation of liver microsomal lipids. Biochim. Biophys. Acta. 620: 281–296. [DOI] [PubMed] [Google Scholar]
- 14.Zhong H., and Yin H.. 2015. Role of lipid peroxidation derived 4-hydroxynonenal (4-HNE) in cancer: focusing on mitochondria. Redox Biol. 4: 193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrera G., Pizzimenti S., Ciamporcero E. S., Daga M., Ullio C., Arcaro A., Cetrangolo G. P., Ferretti C., Dianzani C., Lepore A., et al. 2015. Role of 4-hydroxynonenal-protein adducts in human diseases. Antioxid. Redox Signal. 22: 1681–1702. [DOI] [PubMed] [Google Scholar]
- 16.Grimsrud P. A., Picklo M. J., Griffin T. J., and Bernlohr D. A.. 2007. Carbonylation of adipose proteins in obesity and insulin resistance: identification of adipocyte fatty acid-binding protein as a cellular target of 4-hydroxynonenal. Mol. Cell. Proteomics. 6: 624–637. [DOI] [PubMed] [Google Scholar]
- 17.Wagner T. M., Mullally J. E., and Fitzpatrick F. A.. 2006. Reactive lipid species from cyclooxygenase-2 inactivate tumor suppressor LKB1/STK11. J. Biol. Chem. 281: 2598–2604. [DOI] [PubMed] [Google Scholar]
- 18.Barreiro E., del Puerto-Nevado L., Puig-Vilanova E., Pérez-Rial S., Sanchez F., Martínez-Galán L., Rivera S., Gea J., González-Mangado N., and Peces-Barba G.. 2012. Cigarette smoke-induced oxidative stress in skeletal muscles of mice. Respir. Physiol. Neurobiol. 182: 9–17. [DOI] [PubMed] [Google Scholar]
- 19.Castegna A., Aksenov M., Aksenova M., Thongboonkerd V., Klein J. B., Pierce W. M., Booze R., Markesbery W. R., and Butterfield D. A.. 2002. Proteomic identification of oxidatively modified proteins in Alzheimer’s disease brain. Part I: creatine kinase BB, glutamine synthase, and ubiquitin carboxy-terminal hydrolase L-1. Free Radic. Biol. Med. 33: 562–571. [DOI] [PubMed] [Google Scholar]
- 20.Dalle-Donne I., Rossi R., Giustarini D., Milzani A., and Colombo R.. 2003. Protein carbonyl groups as biomarkers of oxidative stress. Clin. Chim. Acta. 329: 23–38. [DOI] [PubMed] [Google Scholar]
- 21.Pirinccioglu A. G., Gökalp D., Pirinccioglu M., Kizil G., and Kizil M.. 2010. Malondialdehyde (MDA) and protein carbonyl (PCO) levels as biomarkers of oxidative stress in subjects with familial hypercholesterolemia. Clin. Biochem. 43: 1220–1224. [DOI] [PubMed] [Google Scholar]
- 22.Leonarduzzi G., Chiarpotto E., Biasi F., and Poli G.. 2005. 4-Hydroxynonenal and cholesterol oxidation products in atherosclerosis. Mol. Nutr. Food Res. 49: 1044–1049. [DOI] [PubMed] [Google Scholar]
- 23.Zarkovic K. 2003. 4-hydroxynonenal and neurodegenerative diseases. Mol. Aspects Med. 24: 293–303. [DOI] [PubMed] [Google Scholar]
- 24.Long E. K., Olson D. M., and Bernlohr D. A.. 2013. High-fat diet induces changes in adipose tissue trans-4-oxo-2-nonenal and trans-4-hydroxy-2-nonenal levels in a depot-specific manner. Free Radic. Biol. Med. 63: 390–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pawlosky R. J., Bacher J., and Salem N.. 2001. Ethanol consumption alters electroretinograms and depletes neural tissues of docosahexaenoic acid in rhesus monkeys: nutritional consequences of a low n-3 fatty acid diet. Alcohol. Clin. Exp. Res. 25: 1758–1765. [PubMed] [Google Scholar]
- 26.Salem N., Litman B., Kim H. Y., and Gawrisch K.. 2001. Mechanisms of action of docosahexaenoic acid in the nervous system. Lipids. 36: 945–959. [DOI] [PubMed] [Google Scholar]
- 27.Lim S-Y., Doherty J. D., and Salem N.. 2005. Lead exposure and (n-3) fatty acid deficiency during rat neonatal development alter liver, plasma, and brain polyunsaturated fatty acid composition. J. Nutr. 135: 1027–1033. [DOI] [PubMed] [Google Scholar]
- 28.Mileykovskaya E., and Dowhan W.. 2014. Cardiolipin-dependent formation of mitochondrial respiratory supercomplexes. Chem. Phys. Lipids. 179: 42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paradies G., Paradies V., De Benedictis V., Ruggiero F. M., and Petrosillo G.. 2014. Functional role of cardiolipin in mitochondrial bioenergetics. Biochim. Biophys. Acta. 1837: 408–417. [DOI] [PubMed] [Google Scholar]
- 30.Zhong H., Lu J., Xia L., Zhu M., and Yin H.. 2014. Formation of electrophilic oxidation products from mitochondrial cardiolipin in vitro and in vivo in the context of apoptosis and atherosclerosis. Redox Biol. 2: 878–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu W., Porter N. A., Schneider C., Brash A. R., and Yin H.. 2011. Formation of 4-hydroxynonenal from cardiolipin oxidation: intramolecular peroxyl radical addition and decomposition. Free Radic. Biol. Med. 50: 166–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paradies G., Petrosillo G., Pistolese M., and Ruggiero F. M.. 2000. The effect of reactive oxygen species generated from the mitochondrial electron transport chain on the cytochrome c oxidase activity and on the cardiolipin content in bovine heart submitochondrial particles. FEBS Lett. 466: 323–326. [DOI] [PubMed] [Google Scholar]
- 33.Paradies G., Petrosillo G., Pistolese M., and Ruggiero F. M.. 2002. Reactive oxygen species affect mitochondrial electron transport complex I activity through oxidative cardiolipin damage. Gene. 286: 135–141. [DOI] [PubMed] [Google Scholar]
- 34.Katunga L. A., Gudimella P., Efird J. T., Abernathy S., Mattox T. A., Beatty C., Darden T. M., Thayne K. A., Alwair H., Kypson A. P., et al. 2015. Obesity in a model of gpx4 haploinsufficiency uncovers a causal role for lipid-derived aldehydes in human metabolic disease and cardiomyopathy. Mol. Metab. 4: 493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martens J-C., Keilhoff G., Halangk W., Wartmann T., Gardemann A., Päge I., and Schild L.. 2015. Lipidomic analysis of molecular cardiolipin species in livers exposed to ischemia/reperfusion. Mol. Cell. Biochem. 400: 253–263. [DOI] [PubMed] [Google Scholar]
- 36.Paradies G., Petrosillo G., Paradies V., and Ruggiero F. M.. 2009. Role of cardiolipin peroxidation and Ca2+ in mitochondrial dysfunction and disease. Cell Calcium. 45: 643–650. [DOI] [PubMed] [Google Scholar]
- 37.Kagan V. E., Tyurin V. A., Jiang J., Tyurina Y. Y., Ritov V. B., Amoscato A. A., Osipov A. N., Belikova N. A., Kapralov A. A., Kini V., et al. 2005. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat. Chem. Biol. 1: 223–232. [DOI] [PubMed] [Google Scholar]
- 38.Shidoji Y., Komura S., Ohishi N., and Yagi K.. 2002. Interaction between cytochrome c and oxidized mitochondrial lipids. Subcell. Biochem. 36: 19–37. [PubMed] [Google Scholar]
- 39.Shidoji Y., Hayashi K., Komura S., Ohishi N., and Yagi K.. 1999. Loss of molecular interaction between cytochrome c and cardiolipin due to lipid peroxidation. Biochem. Biophys. Res. Commun. 264: 343–347. [DOI] [PubMed] [Google Scholar]
- 40.Kuwana T., Mackey M. R., Perkins G., Ellisman M. H., Latterich M., Schneiter R., Green D. R., and Newmeyer D. D.. 2002. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 111: 331–342. [DOI] [PubMed] [Google Scholar]
- 41.Korytowski W., Basova L. V., Pilat A., Kernstock R. M., and Girotti A. W.. 2011. Permeabilization of the mitochondrial outer membrane by Bax/truncated Bid (tBid) proteins as sensitized by cardiolipin hydroperoxide translocation. J. Biol. Chem. 286: 26334–26343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sani M-A., Dufourc E. J., and Gröbner G.. 2009. How does the Bax-alpha1 targeting sequence interact with mitochondrial membranes? The role of cardiolipin. Biochim. Biophys. Acta. 1788: 623–631. [DOI] [PubMed] [Google Scholar]
- 43.Kirkland R. A., Windelborn J. A., Kasprzak J. M., and Franklin J. L.. 2002. A Bax-induced pro-oxidant state is critical for cytochrome c release during programmed neuronal death. J. Neurosci. 22: 6480–6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manon S. 2004. Utilization of yeast to investigate the role of lipid oxidation in cell death. Antioxid. Redox Signal. 6: 259–267. [DOI] [PubMed] [Google Scholar]
- 45.Jiang J., Huang Z., Zhao Q., Feng W., Belikova N. A., and Kagan V. E.. 2008. Interplay between bax, reactive oxygen species production, and cardiolipin oxidation during apoptosis. Biochem. Biophys. Res. Commun. 368: 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madian A. G., and Regnier F. E.. 2010. Profiling carbonylated proteins in human plasma. J. Proteome Res. 9: 1330–1343. [DOI] [PubMed] [Google Scholar]
- 47.Witz G. 1989. Biological interactions of alpha,beta-unsaturated aldehydes. Free Radic. Biol. Med. 7: 333–349. [DOI] [PubMed] [Google Scholar]
- 48.Curtis J. M., Hahn W. S., Stone M. D., Inda J. J., Droullard D. J., Kuzmicic J. P., Donoghue M. A., Long E. K., Armien A. G., Lavandero S., et al. 2012. Protein carbonylation and adipocyte mitochondrial function. J. Biol. Chem. 287: 32967–32980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meany D. L., Xie H., Thompson L. V., Arriaga E. A., and Griffin T. J.. 2007. Identification of carbonylated proteins from enriched rat skeletal muscle mitochondria using affinity chromatography-stable isotope labeling and tandem mass spectrometry. Proteomics. 7: 1150–1163. [DOI] [PubMed] [Google Scholar]
- 50.Barreiro E., Gea J., Di Falco M., Kriazhev L., James S., and Hussain S. N. A.. 2005. Protein carbonyl formation in the diaphragm. Am. J. Respir. Cell Mol. Biol. 32: 9–17. [DOI] [PubMed] [Google Scholar]
- 51.England K., O’Driscoll C., and Cotter T. G.. 2004. Carbonylation of glycolytic proteins is a key response to drug-induced oxidative stress and apoptosis. Cell Death Differ. 11: 252–260. [DOI] [PubMed] [Google Scholar]
- 52.Curtis J. M., Hahn W. S., Long E. K., Burrill J. S., Arriaga E. A., and Bernlohr D. A.. 2012. Protein carbonylation and metabolic control systems. Trends Endocrinol. Metab. 23: 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Picklo M. J., Montine T. J., Amarnath V., and Neely M. D.. 2002. Carbonyl toxicology and Alzheimer’s disease. Toxicol. Appl. Pharmacol. 184: 187–197. [DOI] [PubMed] [Google Scholar]
- 54.Smathers R. L., Galligan J. J., Stewart B. J., and Petersen D. R.. 2011. Overview of lipid peroxidation products and hepatic protein modification in alcoholic liver disease. Chem. Biol. Interact. 192: 107–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lourenço Dos Santos S., Baraibar M. A., Lundberg S., Eeg-Olofsson O., Larsson L., and Friguet B.. 2015. Oxidative proteome alterations during skeletal muscle ageing. Redox Biol. 5: 267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frohnert B. I., Sinaiko A. R., Serrot F. J., Foncea R. E., Moran A., Ikramuddin S., Choudry U., and Bernlohr D. A.. 2011. Increased adipose protein carbonylation in human obesity. Obesity (Silver Spring). 19: 1735–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shearn C. T., Fritz K. S., Shearn A. H., Saba L. M., Mercer K. E., Engi B., Galligan J. J., Zimniak P., Orlicky D. J., Ronis M. J., et al. 2016. Deletion of GSTA4-4 results in increased mitochondrial post-translational modification of proteins by reactive aldehydes following chronic ethanol consumption in mice. Redox Biol. 7: 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chavez J. D., Wu J., Bisson W., and Maier C. S.. 2011. Site-specific proteomic analysis of lipoxidation adducts in cardiac mitochondria reveals chemical diversity of 2-alkenal adduction. J. Proteomics. 74: 2417–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo J., Prokai-Tatrai K., Nguyen V., Rauniyar N., Ughy B., and Prokai L.. 2011. Protein targets for carbonylation by 4-hydroxy-2-nonenal in rat liver mitochondria. J. Proteomics. 74: 2370–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coughlan C., Walker D. I., Lohr K. M., Richardson J. R., Saba L. M., Caudle W. M., Fritz K. S., and Roede J. R.. 2015. Comparative proteomic analysis of carbonylated proteins from the striatum and cortex of pesticide-treated mice. Parkinsons Dis. 2015: 812532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spiess P. C., Deng B., Hondal R. J., Matthews D. E., and van der Vliet A.. 2011. Proteomic profiling of acrolein adducts in human lung epithelial cells. J. Proteomics. 74: 2380–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feng J., Xie H., Meany D. L., Thompson L. V., Arriaga E. A., and Griffin T. J.. 2008. Quantitative proteomic profiling of muscle type-dependent and age-dependent protein carbonylation in rat skeletal muscle mitochondria. J. Gerontol. A Biol. Sci. Med. Sci. 63: 1137–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang T. J., Larson M. G., Vasan R. S., Cheng S., Rhee E. P., McCabe E., Lewis G. D., Fox C. S., Jacques P. F., Fernandez C., et al. 2011. Metabolite profiles and the risk of developing diabetes. Nat. Med. 17: 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Newgard C. B., An J., Bain J. R., Muehlbauer M. J., Stevens R. D., Lien L. F., Haqq A. M., Shah S. H., Arlotto M., Slentz C. A., et al. 2009. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 9: 311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Batch B. C., Shah S. H., Newgard C. B., Turer C. B., Haynes C., Bain J. R., Muehlbauer M., Patel M. J., Stevens R. D., Appel L. J., et al. 2013. Branched chain amino acids are novel biomarkers for discrimination of metabolic wellness. Metabolism. 62: 961–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Magkos F., Bradley D., Schweitzer G. G., Finck B. N., Eagon J. C., Ilkayeva O., Newgard C. B., and Klein S.. 2013. Effect of Roux-en-Y gastric bypass and laparoscopic adjustable gastric banding on branched-chain amino acid metabolism. Diabetes. 62: 2757–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCormack S. E., Shaham O., McCarthy M. A., Deik A. A., Wang T. J., Gerszten R. E., Clish C. B., Mootha V. K., Grinspoon S. K., and Fleischman A.. 2013. Circulating branched-chain amino acid concentrations are associated with obesity and future insulin resistance in children and adolescents. Pediatr. Obes. 8: 52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lackey D. E., Lynch C. J., Olson K. C., Mostaedi R., Ali M., Smith W. H., Karpe F., Humphreys S., Bedinger D. H., Dunn T. N., et al. 2013. Regulation of adipose branched-chain amino acid catabolism enzyme expression and cross-adipose amino acid flux in human obesity. Am. J. Physiol. Endocrinol. Metab. 304: E1175–E1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burrill J. S., Long E. K., Reilly B., Deng Y., Armitage I. M., Scherer P. E., and Bernlohr D. A.. 2015. Inflammation and ER stress regulate branched-chain amino acid uptake and metabolism in adipocytes. Mol. Endocrinol. 29: 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao Y., Miriyala S., Miao L., Mitov M., Schnell D., Dhar S. K., Cai J., Klein J. B., Sultana R., Butterfield D. A., et al. 2014. Redox proteomic identification of HNE-bound mitochondrial proteins in cardiac tissues reveals a systemic effect on energy metabolism after doxorubicin treatment. Free Radic. Biol. Med. 72: 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Galligan J. J., Smathers R. L., Fritz K. S., Epperson L. E., Hunter L. E., and Petersen D. R.. 2012. Protein carbonylation in a murine model for early alcoholic liver disease. Chem. Res. Toxicol. 25: 1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Andringa K. K., Udoh U. S., Landar A., and Bailey S. M.. 2014. Proteomic analysis of 4-hydroxynonenal (4-HNE) modified proteins in liver mitochondria from chronic ethanol-fed rats. Redox Biol. 2C: 1038–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo J-M., Liu A-J., Zang P., Dong W-Z., Ying L., Wang W., Xu P., Song X-R., Cai J., Zhang S-Q., et al. 2013. ALDH2 protects against stroke by clearing 4-HNE. Cell Res. 23: 915–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Venkateshappa C., Harish G., Mythri R. B., Mahadevan A., Bharath M. M. S., and Shankar S. K.. 2012. Increased oxidative damage and decreased antioxidant function in aging human substantia nigra compared to striatum: implications for Parkinson’s disease. Neurochem. Res. 37: 358–369. [DOI] [PubMed] [Google Scholar]
- 75.Takagi S., Iwai N., Yamauchi R., Kojima S., Yasuno S., Baba T., Terashima M., Tsutsumi Y., Suzuki S., Morii I., et al. 2002. Aldehyde dehydrogenase 2 gene is a risk factor for myocardial infarction in Japanese men. Hypertens. Res. 25: 677–681. [DOI] [PubMed] [Google Scholar]
- 76.Jo S. A., Kim E-K., Park M. H., Han C., Park H-Y., Jang Y., Song B. J., and Jo I.. 2007. A Glu487Lys polymorphism in the gene for mitochondrial aldehyde dehydrogenase 2 is associated with myocardial infarction in elderly Korean men. Clin. Chim. Acta. 382: 43–47. [DOI] [PubMed] [Google Scholar]
- 77.Singh S. P., Niemczyk M., Saini D., Awasthi Y. C., Zimniak L., and Zimniak P.. 2008. Role of the electrophilic lipid peroxidation product 4-hydroxynonenal in the development and maintenance of obesity in mice. Biochemistry. 47: 3900–3911. [DOI] [PubMed] [Google Scholar]
- 78.Curtis J. M., Grimsrud P. A., Wright W. S., Xu X., Foncea R. E., Graham D. W., Brestoff J. R., Wiczer B. M., Ilkayeva O., Cianflone K., et al. 2010. Downregulation of adipose glutathione S-transferase A4 leads to increased protein carbonylation, oxidative stress, and mitochondrial dysfunction. Diabetes. 59: 1132–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anderson E. J., Kypson A. P., Rodriguez E., Anderson C. A., Lehr E. J., and Neufer P. D.. 2009. Substrate-specific derangements in mitochondrial metabolism and redox balance in the atrium of the type 2 diabetic human heart. J. Am. Coll. Cardiol. 54: 1891–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Back S. H., Scheuner D., Han J., Song B., Ribick M., Wang J., Gildersleeve R. D., Pennathur S., and Kaufman R. J.. 2009. Translation attenuation through eIF2alpha phosphorylation prevents oxidative stress and maintains the differentiated state in beta cells. Cell Metab. 10: 13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Malhotra J. D., Miao H., Zhang K., Wolfson A., Pennathur S., Pipe S. W., and Kaufman R. J.. 2008. Antioxidants reduce endoplasmic reticulum stress and improve protein secretion. Proc. Natl. Acad. Sci. USA. 105: 18525–18530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim H-R., Lee G-H., Cho E. Y., Chae S-W., Ahn T., and Chae H-J.. 2009. Bax inhibitor 1 regulates ER-stress-induced ROS accumulation through the regulation of cytochrome P450 2E1. J. Cell Sci. 122: 1126–1133. [DOI] [PubMed] [Google Scholar]
- 83.Liu H., Bowes R. C., van de Water B., Sillence C., Nagelkerke J. F., and Stevens J. L.. 1997. Endoplasmic reticulum chaperones GRP78 and calreticulin prevent oxidative stress, Ca2+ disturbances, and cell death in renal epithelial cells. J. Biol. Chem. 272: 21751–21759. [DOI] [PubMed] [Google Scholar]
- 84.Song B., Scheuner D., Ron D., Pennathur S., and Kaufman R. J.. 2008. Chop deletion reduces oxidative stress, improves beta cell function, and promotes cell survival in multiple mouse models of diabetes. J. Clin. Invest. 118: 3378–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sanson M., Augé N., Vindis C., Muller C., Bando Y., Thiers J-C., Marachet M-A., Zarkovic K., Sawa Y., Salvayre R., et al. 2009. Oxidized low-density lipoproteins trigger endoplasmic reticulum stress in vascular cells: prevention by oxygen-regulated protein 150 expression. Circ. Res. 104: 328–336. [DOI] [PubMed] [Google Scholar]
- 86.Vladykovskaya E., Sithu S. D., Haberzettl P., Wickramasinghe N. S., Merchant M. L., Hill B. G., McCracken J., Agarwal A., Dougherty S., Gordon S. A., et al. 2012. Lipid peroxidation product 4-hydroxy-trans-2-nonenal causes endothelial activation by inducing endoplasmic reticulum stress. J. Biol. Chem. 287: 11398–11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Haberzettl P., Vladykovskaya E., Srivastava S., and Bhatnagar A.. 2009. Role of endoplasmic reticulum stress in acrolein-induced endothelial activation. Toxicol. Appl. Pharmacol. 234: 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Magi B., Ettorre A., Liberatori S., Bini L., Andreassi M., Frosali S., Neri P., Pallini V., and Di Stefano A.. 2004. Selectivity of protein carbonylation in the apoptotic response to oxidative stress associated with photodynamic therapy: a cell biochemical and proteomic investigation. Cell Death Differ. 11: 842–852. [DOI] [PubMed] [Google Scholar]
- 89.Rabek J. P., Boylston W. H., and Papaconstantinou J.. 2003. Carbonylation of ER chaperone proteins in aged mouse liver. Biochem. Biophys. Res. Commun. 305: 566–572. [DOI] [PubMed] [Google Scholar]
- 90.Choi J., Conrad C. C., Dai R., Malakowsky C. A., Talent J. M., Carroll C. A., Weintraub S. T., and Gracy R. W.. 2003. Vitamin E prevents oxidation of antiapoptotic proteins in neuronal cells. Proteomics. 3: 73–77. [DOI] [PubMed] [Google Scholar]
- 91.Carbone D. L., Doorn J. A., Kiebler Z., Sampey B. P., and Petersen D. R.. 2004. Inhibition of Hsp72-mediated protein refolding by 4-hydroxy-2-nonenal. Chem. Res. Toxicol. 17: 1459–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Carbone D. L. 2005. Modification of heat shock protein 90 by 4-hydroxynonenal in a rat model of chronic alcoholic liver disease. J. Pharmacol. Exp. Ther. 315: 8–15. [DOI] [PubMed] [Google Scholar]
- 93.Galligan J. J., Fritz K. S., Backos D. S., Shearn C. T., Smathers R. L., Jiang H., MacLean K. N., Reigan P. R., and Petersen D. R.. 2014. Oxidative stress-mediated aldehyde adduction of GRP78 in a mouse model of alcoholic liver disease: functional independence of ATPase activity and chaperone function. Free Radic. Biol. Med. 73: 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hill B. G., Haberzettl P., Ahmed Y., Srivastava S., and Bhatnagar A.. 2008. Unsaturated lipid peroxidation-derived aldehydes activate autophagy in vascular smooth-muscle cells. Biochem. J. 410: 525–534. [DOI] [PubMed] [Google Scholar]
- 95.Haberzettl P., and Hill B. G.. 2013. Oxidized lipids activate autophagy in a JNK-dependent manner by stimulating the endoplasmic reticulum stress response. Redox Biol. 1: 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jung T., Höhn A., and Grune T.. 2013. The proteasome and the degradation of oxidized proteins: Part II - protein oxidation and proteasomal degradation. Redox Biol. 2C: 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Grune T., Jung T., Merker K., and Davies K. J. A.. 2004. Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and “aggresomes” during oxidative stress, aging, and disease. Int. J. Biochem. Cell Biol. 36: 2519–2530. [DOI] [PubMed] [Google Scholar]
- 98.Davies K. J. 2001. Degradation of oxidized proteins by the 20S proteasome. Biochimie. 83: 301–310. [DOI] [PubMed] [Google Scholar]
- 99.Grune T., Reinheckel T., Joshi M., and Davies K. J.. 1995. Proteolysis in cultured liver epithelial cells during oxidative stress. Role of the multicatalytic proteinase complex, proteasome. J. Biol. Chem. 270: 2344–2351. [DOI] [PubMed] [Google Scholar]
- 100.Grune T., and Davies K. J. A.. 2003. The proteasomal system and HNE-modified proteins. Mol. Aspects Med. 24: 195–204. [DOI] [PubMed] [Google Scholar]
- 101.Carbone D. L., Doorn J. A., and Petersen D. R.. 2004. 4-Hydroxynonenal regulates 26S proteasomal degradation of alcohol dehydrogenase. Free Radic. Biol. Med. 37: 1430–1439. [DOI] [PubMed] [Google Scholar]
- 102.Marques C., Pereira P., Taylor A., Liang J. N., Reddy V. N., Szweda L. I., and Shang F.. 2004. Ubiquitin-dependent lysosomal degradation of the HNE-modified proteins in lens epithelial cells. FASEB J. 18: 1424–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Friguet B., and Szweda L. I.. 1997. Inhibition of the multicatalytic proteinase (proteasome) by 4-hydroxy-2-nonenal cross-linked protein. FEBS Lett. 405: 21–25. [DOI] [PubMed] [Google Scholar]
- 104.Cecarini V., Ding Q., and Keller J. N.. 2007. Oxidative inactivation of the proteasome in Alzheimer’s disease. Free Radic. Res. 41: 673–680. [DOI] [PubMed] [Google Scholar]
- 105.Viteri G., Carrard G., Birlouez-Aragón I., Silva E., and Friguet B.. 2004. Age-dependent protein modifications and declining proteasome activity in the human lens. Arch. Biochem. Biophys. 427: 197–203. [DOI] [PubMed] [Google Scholar]
- 106.Carrard G., Dieu M., Raes M., Toussaint O., and Friguet B.. 2003. Impact of ageing on proteasome structure and function in human lymphocytes. Int. J. Biochem. Cell Biol. 35: 728–739. [DOI] [PubMed] [Google Scholar]
- 107.Friguet B., Stadtman E. R., and Szweda L. I.. 1994. Modification of glucose-6-phosphate dehydrogenase by 4-hydroxy-2-nonenal. Formation of cross-linked protein that inhibits the multicatalytic protease. J. Biol. Chem. 269: 21639–21643. [PubMed] [Google Scholar]
- 108.Okada K., Wangpoengtrakul C., Osawa T., Toyokuni S., Tanaka K., and Uchida K.. 1999. 4-Hydroxy-2-nonenal-mediated impairment of intracellular proteolysis during oxidative stress. Identification of proteasomes as target molecules. J. Biol. Chem. 274: 23787–23793. [DOI] [PubMed] [Google Scholar]
- 109.Hyun D-H., Lee M-H., Halliwell B., and Jenner P.. 2002. Proteasomal dysfunction induced by 4-hydroxy-2,3-trans-nonenal, an end-product of lipid peroxidation: a mechanism contributing to neurodegeneration? J. Neurochem. 83: 360–370. [DOI] [PubMed] [Google Scholar]
- 110.Keller J. N., Huang F. F., Zhu H., Yu J., Ho Y. S., and Kindy T. S.. 2000. Oxidative stress-associated impairment of proteasome activity during ischemia-reperfusion injury. J. Cereb. Blood Flow Metab. 20: 1467–1473. [DOI] [PubMed] [Google Scholar]
- 111.Farout L., Mary J., Vinh J., Szweda L. I., and Friguet B.. 2006. Inactivation of the proteasome by 4-hydroxy-2-nonenal is site specific and dependant on 20S proteasome subtypes. Arch. Biochem. Biophys. 453: 135–142. [DOI] [PubMed] [Google Scholar]
- 112.Bulteau A. L., Lundberg K. C., Humphries K. M., Sadek H. A., Szweda P. A., Friguet B., and Szweda L. I.. 2001. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J. Biol. Chem. 276: 30057–30063. [DOI] [PubMed] [Google Scholar]
- 113.Campos J. C., Queliconi B. B., Dourado P. M. M., Cunha T. F., Zambelli V. O., Bechara L. R. G., Kowaltowski A. J., Brum P. C., Mochly-Rosen D., and Ferreira J. C. B.. 2012. Exercise training restores cardiac protein quality control in heart failure. PLoS One. 7: e52764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ferrington D. A., and Kapphahn R. J.. 2004. Catalytic site-specific inhibition of the 20S proteasome by 4-hydroxynonenal. FEBS Lett. 578: 217–223. [DOI] [PubMed] [Google Scholar]
- 115.Just J., Jung T., Friis N. A., Lykkemark S., Drasbek K., Siboska G., Grune T., and Kristensen P.. 2015. Identification of an unstable 4-hydroxynoneal modification on the 20S proteasome subunit α7 by recombinant antibody technology. Free Radic. Biol. Med. 89: 786–792. [DOI] [PubMed] [Google Scholar]