Abstract
Background
Plasmodium vivax causes the majority of malaria episodes outside Africa, but remains a relatively understudied pathogen. The pathology of P. vivax infection depends critically on the parasite’s ability to recognize and invade human erythrocytes. This invasion process involves an interaction between P. vivax Duffy Binding Protein (PvDBP) in merozoites and the Duffy antigen receptor for chemokines (DARC) on the erythrocyte surface. Whole-genome sequencing of clinical isolates recently established that some P. vivax genomes contain two copies of the PvDBP gene. The frequency of this duplication is particularly high in Madagascar, where there is also evidence for P. vivax infection in DARC-negative individuals. The functional significance and global prevalence of this duplication, and whether there are other copy number variations at the PvDBP locus, is unknown.
Methodology/Principal Findings
Using whole-genome sequencing and PCR to study the PvDBP locus in P. vivax clinical isolates, we found that PvDBP duplication is widespread in Cambodia. The boundaries of the Cambodian PvDBP duplication differ from those previously identified in Madagascar, meaning that current molecular assays were unable to detect it. The Cambodian PvDBP duplication did not associate with parasite density or DARC genotype, and ranged in prevalence from 20% to 38% over four annual transmission seasons in Cambodia. This duplication was also present in P. vivax isolates from Brazil and Ethiopia, but not India.
Conclusions/Significance
PvDBP duplications are much more widespread and complex than previously thought, and at least two distinct duplications are circulating globally. The same duplication boundaries were identified in parasites from three continents, and were found at high prevalence in human populations where DARC-negativity is essentially absent. It is therefore unlikely that PvDBP duplication is associated with infection of DARC-negative individuals, but functional tests will be required to confirm this hypothesis.
Author Summary
Malaria parasites must be adaptable to evade the human immune system and successfully transmit themselves to different individuals. Key to this adaptability is the fact that malaria parasite genomes are highly variable, containing mutations ranging from simple small changes in DNA sequence to complex large-scale changes in the number of copies of individual genes. Some samples of Plasmodium vivax, the parasite that causes most malaria episodes outside Africa, have recently been found to have duplicated the gene encoding Duffy Binding Protein, which enables P. vivax parasites to recognize and invade human red blood cells. By studying parasites from Cambodian patients with P. vivax malaria, we have discovered that there are actually two different types of gene duplication, with the new duplication type identified in this study present in 20% to 38% of Cambodian P. vivax isolates tested. The same gene duplication was also found in parasites from Brazilian and Ethiopian patients with P. vivax malaria, suggesting that variation in this gene is more complex and common than previously thought. The functional significance and origin of these two gene duplications requires further study.
Introduction
Plasmodium parasites must recognize and invade erythrocytes to multiply and cause clinical disease in humans. Erythrocyte invasion is a complex multi-step process involving multiple protein-protein interactions between the extracellular Plasmodium merozoite and its target erythrocyte. While P. falciparum merozoite proteins have some overlapping and partially redundant interactions with several different erythrocyte receptors [1], P. vivax merozoites appear to rely heavily on the interaction between P. vivax Duffy Binding Protein (PvDBP) and the erythrocyte Duffy antigen receptor for chemokines (DARC) to invade human erythrocytes [2–4]. There are numerous common genetic variants at the DARC locus, including the DARC-negative or FY*BES/*BES genotype, where a polymorphism in the DARC promoter eliminates expression of DARC specifically in erythroid cells, producing an Fy(a-b-) phenotype [5]. DARC-negativity is nearly ubiquitous in West and Central Africa and, given that erythrocyte invasion is essential for parasite survival, is associated with protection against P. vivax infection [6]. PvDBP is therefore a high-priority P. vivax vaccine candidate [7]. The essentiality of the PvDBP-DARC interaction for P. vivax invasion has recently been challenged by the discovery of confirmed P. vivax infections in DARC-negative individuals in Madagascar [8], a finding that is consistent with several independent reports of P. vivax infections in DARC-negative individuals in West and Central Africa [9]. The molecular basis of P. vivax invasion into DARC-negative erythrocytes is currently unknown, although it may still involve PvDBP.
Given this focus on PvDBP for both vaccine and biological studies, understanding the extent and functional consequences of PvDBP variation is important. Previous attention to PvDBP variation has focused on single-nucleotide polymorphisms (SNPs), which are common and appear to result in strain-specific immune responses [10,11]. Recent whole-genome sequencing of a P. vivax strain from Madagascar identified another source of variation, namely duplication of the entire PvDBP locus [12]. A simple PCR assay based on these whole-genome sequence data revealed that PvDBP duplication was present in more than half of Malagasy P. vivax samples, and in much lower proportions of East African, Asian, and South American samples. This finding has since been confirmed by whole-genome sequencing of >200 P. vivax isolates from around the world, where evidence for duplication based on increased sequence coverage was found in 35% of Cambodian isolates [13].
The functional consequences of PvDBP duplication are unknown, although its high prevalence in Madagascar raised the intriguing possibility that it is somehow associated with successful P. vivax infection of DARC-negative individuals. Whether the PvDBP duplication arose only once and spread globally, or arose independently more than once, which would suggest localized selection pressure, is also unknown. While such locally-selected copy number variations (CNVs) in Plasmodium genes are known to associate with drug resistance, such as CNVs at the PfMDR1 locus in association with mefloquine resistance [14,15], such CNV selection has not yet been associated with genes involved in fundamental biological processes such as erythrocyte invasion. To address the origins of CNVs at the PvDBP locus, we investigated the prevalence and nature of PvDBP duplications in 37 P. vivax samples collected in a clinical study of human genetic resistance to P. vivax malaria in Cambodia [16], which were subsequently sequenced by the Malaria Programme at the Wellcome Trust Sanger Institute (WTSI) as part of a larger study [13]. This identified a new type of duplication, which was then validated in >350 samples from all three continents where P. vivax infections are found.
Methods
Patient samples and ethics statement
Cambodia: During each annual malaria season (June-December) from 2008 to 2011, patients presenting with malaria symptoms were screened at Sampov Meas Referral Hospital in Pursat. A total of 898 patients were diagnosed with P. vivax malaria and participated in a study of human genetic resistance to this disease. The clinical protocol was approved by the National Ethics Committee for Health Research in Cambodia and the NIAID Institutional Review Board in the United States (ClinicalTrials.gov identifier NCT00663546). Following written informed consent, patients provided a venous blood sample, and DNA was extracted from 200 μl of whole blood using a QiaAmp kit (Qiagen, USA). Of these samples, 49–50 samples per year were randomly selected for analysis.
Ethiopia: During the peak malaria season (September-November) in 2014, patients presenting with malaria symptoms were screened at health centers in Jimma. Finger-prick blood samples (50 μl) were collected and screened for P. vivax infection by both microscopy and nested PCR. A total of 25 patients were diagnosed with P. vivax malaria and provided written informed consent to participate in this study. The clinical protocol was approved by the institutional review boards of Jimma University in Ethiopia and University of California, Irvine in the United States.
India: From July 2013 to December 2015, patients presenting with malaria symptoms were screened at Goa Medical College by both rapid diagnostic test (RDT) and microscopy. Patients with P. vivax monoinfection were then referred to the Malaria Evolution in South Asia (MESA) International Center for Excellence in Malaria Research (ICEMR) study team. Written informed consent was obtained from adult patients and the parents or guardians of minors; verbal assent was provided by children aged 8–17 years. Subjects provided 4–6 ml of venous blood, and parasite species was confirmed by RDT (FalciVax, Zephyr Biomedicals, India) and microscopy. A random selection of 72 P. vivax monoinfections (from a total of 838 P. vivax monoinfections diagnosed during the study period) was tested for the presence of PvDBP duplication. DNA was extracted from 50–500 μl of whole blood using a QiaAmp kit (Qiagen, USA). The clinical protocol was approved by the institutional review boards of Goa Medical College and Hospital, University of Washington, NIAID Division of Microbiology and Infectious Diseases, and Government of India Health Ministry Screening Committee.
Brazil: From 2008 to 2011, patients presenting with malaria symptoms at clinics in Acrelândia and Plácido de Castro [17] were screened by microscopy. Patients with Plasmodium infection provided 15-ml venous blood samples that were further examined for malaria parasites by a quantitative real-time PCR specific for the 18S rRNA gene. Written informed consent was obtained from adult patients and the parents or guardians of minors. A random selection of six P. vivax monoinfections was tested for the presence of PvDBP duplication. These samples were collected under protocols approved by the Institutional Review Board of the Institute of Biomedical Sciences, University of São Paulo, Brazil (936/CEP, 2010).
Brazil: From June 2014 to September 2015, patients aged 5–63 years presenting with malaria symptoms at clinics in the urban area of Mâncio Lima were screened by microscopy. Patients positive for P. vivax infection were enrolled in a clinical trial of P. vivax response to chloroquine alone vs. chloroquine plus primaquine (ClinicalTrials.gov Identifier: NCT02691910) and provided 40-ml venous blood samples. Written informed consent was obtained from adult patients and the parents or guardians of minors. A random selection of 54 P. vivax monoinfections was tested for the presence of PvDBP duplication. These samples were collected under protocols approved by the Institutional Review Board of the Institute of Biomedical Sciences, University of São Paulo, Brazil (1169/CEP, 2014).
A random selection of samples were tested from Cambodia, Ethiopia, India and Brazil, with numbers based on sample availability.
Identification of a novel PvDBP duplication in Cambodian P. vivax isolates
A subset of 37 Cambodian patients with P. vivax malaria provided an 8-ml blood sample, which was depleted of leukocytes using CF11 prior to DNA extraction [18]. These samples were sequenced at the WTSI using Illumina sequencing technology, as described [13]. As part of the genome analyses, the generated sequences were mapped individually to the P. vivax Sal1 reference genome (http://plasmodb.org/common/downloads/release-10.0/PvivaxSal1/fasta/data/PlasmoDB-10.0_PvivaxSal1_Genome.fasta) [19] using bwa version 0.5.9-r16 with default parameters [20]. In the present study, the resulting assembly bam files were reviewed in the region containing PvDBP (chromosome 6: 976329–980090, GenBank accession ID = PVX_110810) using the Artemis genome viewer [21] or Lookseq [22]. Mate pairs that were oriented tail-to-tail, and thus signified that PvDBP was duplicated, were visualized in either Artemis using a “non-proper pair” read filter or Lookseq using the “face-away” setting. To compare the breakpoints in Cambodian and Malagasy isolates, the read data for the Malagasy P. vivax M15 isolate (with a duplicated PvDBP) (SRA accession SRX266275) were mapped to the reference P. vivax Sal1 genome using bowtie2 v2.1.0 [23] with default parameters, except for counting overlapping reads as concordant pairs (--dovetail). In all sequences the PvDBP genes were reviewed for evidence of intra-isolate SNPs with a frequency of 0.3–0.7 and a minimum read coverage of 5 for both reference and alternative alleles using all SNPs called using GATK’s Haplotype caller with default settings [24,25]. Average coverage across all 14 reference P. vivax Sal1 chromosomes and the PvDBP locus (chromosome 6: 976329 to 980090) was computed using GATK’s DepthOfCoverage tool and compared to estimate PvDBP amplification [24,25].
PvDBP duplication-specific PCR assay
We used previously-published primers and new primers to identify PvDBP duplications in our samples. Previously-published primers from Menard et al. [12] are as follows: primer pair 5’-CCATAAAAGGTAGGAAATTGGAAA-3’ (AF) and 5’-GCATTTTATGAAAACGGTGCT-3’ (AR), which amplifies a 613-bp region surrounding the Malagasy isolates’ breakpoint at position 982947, and primer pair 5’-TCATCGAGCATGTTCCTTTG-3’ (BF) and 5’-TTGCACGTACTCGAAACTCAG-3’ (BR), which amplifies a 643-bp region surrounding the breakpoint at position 974770. BF+AR are predicted to amplify a 612-bp product that contains the junction between the PvDBP copies in isolates with duplications matching those in Malagasy isolates (based on GenBank accession ID KF159580). BF+BR are predicted to amplify the identical breakpoint in Cambodian isolates. Primer pair 5’-ACGCGATGTATCTTCTTTTCA-3’ (AF2) and 5’-TAGAACGCACAGTTATTGGC-3’ (AR2) are designed to amplify a 657-bp region surrounding the breakpoint at position 982100 in Cambodian samples. AF2+AR2 are expected to amplify the region in samples with or without the PvDBP duplication and were used as positive controls. In samples with the Cambodian PvDBP duplication, BF+AR2 are expected to amplify a 736-bp product that contains the junction between the PvDBP copies. In Malagasy samples, BF+AR2 are expected to amplify a 1584-bp product containing the PvDBP duplication. These primers are opposite-facing in samples without the duplication and thus are not expected to produce a product. Duplication PCR products were capillary sequenced, and chromatograms were reviewed to determine the exact breakpoint. PCR reaction volumes contained 20 μl Platinum PCR Supermix (Thermo Fisher Scientific, USA), 1 μl DNA template, and 0.5 μl each primer (10 μM working stocks). PCR conditions were: 94°C for 2 min, followed by 35 cycles of 94°C for 20 s, 55°C for 30 s, and 68°C for 60 s, followed by a 4-min extension.
Duffy genotype PCR assay
To genotype the Duffy blood group (Fy), we used the following published primers (see SI Appendix C in [8]): primer pair 5’-GTGGGGTAAGGCTTCCTGAT-3’ (Fy-F) and 5’-CAGAGCTGCGAGTGCTACCT-3’ (Fy-R), which amplifies a 1100-bp region spanning the GATA-1 transcription factor-binding site that determines DARC expression on erythrocytes [FY*B versus FY*BES in the promotor region of PvDBP (-33, TTATCT for wild-type versus TTACCT for erythrocyte silent FY*BES/FY*BES)] through the exon-two SNPs determining FY*A versus FY*B genotypes [codon 42, GGT (G) for FY*A versus GAT (D) for FY*B] and the FY*Bweak genotype (codon 89, CGC (R) for FY*B versus TGC (C) for FY*Bweak). The FY*Bweak genotype is only known to be associated with the FY*B genotype. PCR reaction volumes contained 20 μl Platinum PCR Supermix, 1 μl DNA template, and 0.5 μl each primer (10 μM working stocks). PCR conditions were: 94°C for 2 min, followed by 35 cycles of 94°C for 20 s, 58°C for 30 s, and 68°C for 60 s, followed by a 4-min extension. PCR products were capillary sequenced using the following four published primers: 5’- GTGGGGTAAGGCTTCCTGAT-3’ (Fy-F), 5’- CAAACAGCAGGGGAAATGAG-3’ (GATA-1-R), 5’-CTTCCGGTGTAACTCTGATGG-3’ (Exon2-F), and 5’- CAGAGCTGCGAGTGCTACCT-3’ (Fy-R) (see SI Appendix C in [8]). Sequences were aligned to a reference sequence for DARC (GenBank accession number S76830.1, http://www.ncbi.nlm.nih.gov/nuccore/S76830.1), and chromatograms were visually inspected to determine FY genotypes. We determined the sequence at all three SNP sites in 129 samples, and at only the FY*A and FY*B sites in another 33 samples.
PvDBP haplotypes and phylogeny
PvDBP haplotypes of the most abundant clone in each of 37 whole-genome-sequenced Cambodian samples and one Malagasy P. vivax M15 sample were reconstructed using GATK’s HaplotypeCaller v 3.4 [24,25], setting “sample ploidy” to 1 for the PvDBP locus (chromosome 6: 976329 to 980090). The resulting vcf file was used with GATK’s FastaAlternateReferenceMaker to construct FASTA files representing one PvDBP gene in each sample. The haplotype of the PvDBP region for P. vivax M15 was identical to the sequence deposited in GenBank (accession ID = KF159580), except for calling the majority “T” at position 50 instead of using the IUPAC code “Y,” representing the C/T polymorphic site. PvDBP sequences of all 37 Cambodian samples, Malagasy isolate P. vivax M15, and P. vivax Sal1 were aligned with ClustalW [26] based on default settings followed by manual editing in Sequence Alignment Editor v1.d1 [27]. Haplotype diversity was assessed using DnaSP v5.10.1 [28] with default settings. A phylogenetic tree was reconstructed using the maximum likelihood method implemented in RAxML [29] with 500 bootstrap replicates to assess clade support.
Results
A new type of PvDBP duplication is present in Cambodian isolates
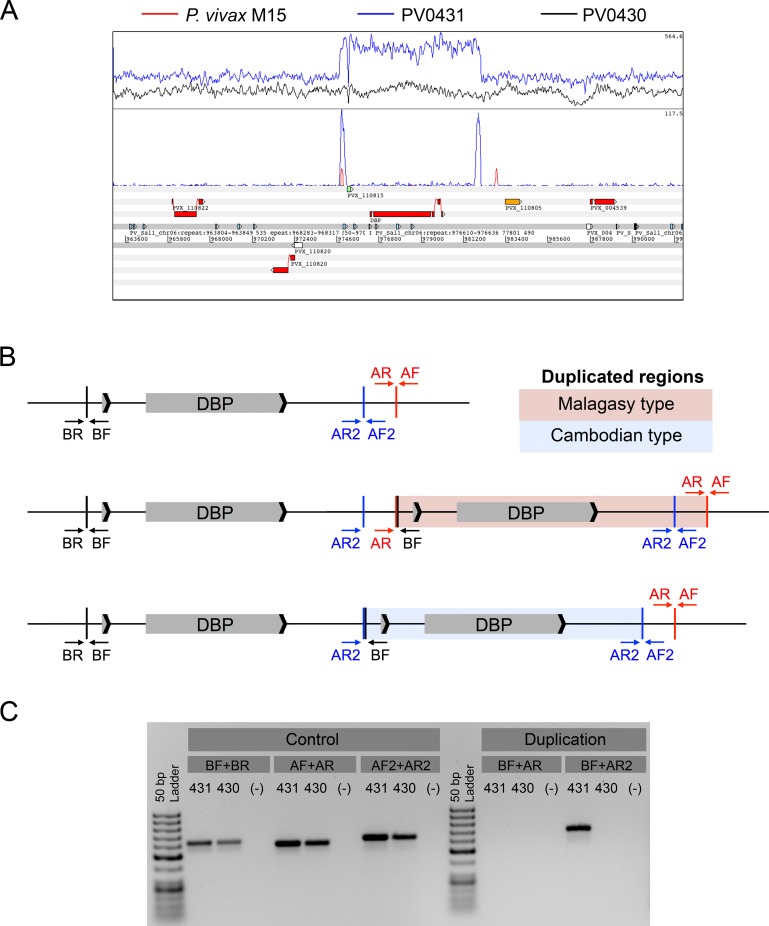
Initial attempts to detect PvDBP duplications in Cambodian P. vivax samples using previously-published PCR primers [12] failed repeatedly (example shown in Fig 1C). To establish whether this was due to technical issues or the absence of PvDBP duplications in Cambodian P. vivax isolates, we used bwa [20] to map previously-generated Illumina sequence reads from individual Cambodian P. vivax isolates to the Sal1 reference genome [13]. Visual inspection of the alignments in Artemis [21] and LookSeq [22] clearly showed increased sequence coverage at the PvDBP locus in some isolates, including some where PCR-based detection had failed. Isolate PV0431 showed increased sequence coverage at the PvDBP gene region compared to flanking regions (Fig 1A, upper panel, blue trace), while isolate PV0430 showed even sequence coverage over these same regions (black trace). A PvDBP duplication was confirmed by identifying paired-end reads that mapped in the tail-to-tail, instead of the expected head-to-head, orientation (Fig 1A, middle panel, blue trace). To investigate whether this duplication was identical to the one previously identified in a Malagasy isolate [12], we mapped tail-to-tail reads for the Malagasy M15 isolate in the same manner (Fig 1A, middle panel, red trace). While the 5’ boundary of the duplication was identical in both genomes, the 3’ boundary was not, indicating that the tandemly-duplicated PvDBP region in Cambodian isolate PV0431 is shorter than that previously observed in Malagasy P. vivax isolates. Importantly, one of the previously-published PCR primers (AR) used to detect PvDBP duplication in Malagasy samples maps to the region that is not duplicated in these genomes, explaining how this primer failed to detect PvDBP duplication in Cambodian samples.
Fig 1. PvDBP duplication in Cambodian isolates.
(A) Artemis genome view [21] showing whole-genome sequence coverage in the PvDBP region of PV0431 (blue trace) and PV0430 (black trace) for all reads (upper panel) and non-proper paired reads (middle panel). The Malagasy P. vivax M15 isolate’s non-proper paired reads (red trace, middle panel) suggest that its PvDBP duplication length is longer than that of PV0431. (B) Schematic of the PvDBP region and primer-binding locations in samples with a single PvDBP gene (top), a Malagasy PvDBP duplication region (middle), and a shorter Cambodian PvDBP duplication region (bottom). AR2+BF primers amplify both Malagasy and Cambodian breakpoints spanning the duplicated regions, while AR+BF primers only amplify Malagasy breakpoints. (C) Cambodian isolate PV0431 with a duplicated PvDBP region and PV0430 with a single PvDBP region, as determined by whole-genome sequencing. BF+AR primers failed to amplify the Malagasy PvDBP duplication in both Cambodian samples, while BF+AR2 primers successfully amplified the Cambodian PvDBP duplication in PV0431. AF2+AR2 primers detected PvDBP duplications in both Cambodian isolates, but not in DNA-negative controls (-).
To investigate how widespread this new duplication is, we designed and tested new primers to amplify the alternative 3’ breakpoint observed in Cambodian isolate PV0431, hereafter referred to as the Cambodian, as opposed to the Malagasy, duplication (Fig 1B). We tested both new (AR2) and old (AR) 3’ primers on two Cambodian isolates that sequencing had identified as carrying (PV0431) or lacking (PV0430) a PvDBP duplication. As expected, the old BF+AR primers that detect the Malagasy duplication did not amplify a PCR product from PV0431, whereas the new BF+AR2 primers that detect the Cambodian duplication did amplify a PCR product of the expected size (Fig 1C). We detected no duplication in PV0430, which Illumina sequencing confirmed lacked a PvDBP duplication, and control primers confirmed that the regions flanking the PvDBP gene were present in both isolates. Importantly, the new BF+AR2 primers are able to amplify both Malagasy and Cambodian types of PvDBP duplication, but yield differentially-sized products.
These primers were then used to confirm the presence or absence of PvDBP duplication in all 35 whole-genome-sequenced Cambodian samples for which DNA was still available (Table 1). Identically-sized PCR products validated the Cambodian-type PvDBP duplication for 14/35 isolates tested, 11/11 of which contained both elevated coverage and the presence of tail-to-tail reads (no DNA remained for a 12th sample with both elevated coverage and tail-to-tail reads). Duplication of PvDBP was supported in 2/3 additional isolates (PH0180-C, PH1113-C, PH1116-C) which lacked elevated coverage of PvDBP (1.17–1.38x) but did contain both tail-to-tail reads and intra-isolate SNPs. This suggests the presence of minor clones with the PvDBP duplication in these isolates, for which the limit of detection was reached in the case of PH0180-C. One additional isolate (PH0177-C), which lacked both tail-to-tail mates and elevated coverage of PvDBP, showed the PvDBP duplication by PCR. This may represent an additional minor clone not detected by whole-genome sequencing. Taken together, these results suggest that increased coverage, while least sensitive, is a useful marker for PvDBP duplications in this dataset. The presence of tail-to-tail read pairs and/or duplication-detecting PCRs, while not in complete agreement, have greater sensitivity and are likely both better able to detect PvDBP duplications in minor clones.
Table 1. Cambodian isolates with single and duplicated copies of PvDBP.
| PvDBP status | WTSI ID | NIH ID | Average genome coverage | Average PvDBP coverage | PvDBP copy # | Tail-to-tail mate pairs (Y) | PCR supports PvDBP dup. (N/Y) † | Intra-isolate SNPs*** |
|---|---|---|---|---|---|---|---|---|
| Single PvDBP* | PH0067-C | 11.67 | 12.75 | 1.09 | N | 0 | ||
| PH0068-C | 8.64 | 11.66 | 1.35 | N | 0 | |||
| PH0075-C | 6.92 | 9.15 | 1.32 | N | 0 | |||
| PH0086-C | 7.68 | 8.51 | 1.11 | N | 0 | |||
| PH0089-C | 10.33 | 13.47 | 1.3 | N | 0 | |||
| PH0098-C | 7.64 | 7.67 | 1 | N | 0 | |||
| PH0177-C | 177.36 | 160.49 | 0.9 | Y | 0 | |||
| PH0180-C | 57.69 | 70.6 | 1.22 | Y | N | 10 | ||
| PH0187-C | 93.72 | 120.48 | 1.29 | N | 7 | |||
| PH0189-C | PV0430 | 113.45 | 123.58 | 1.09 | N | 0 | ||
| PH0310-C | 71.63 | 74.04 | 1.03 | N | 0 | |||
| PH0320-C | 94.67 | 79.45 | 0.84 | N | 0 | |||
| PH0385-C | 35.84 | 43.12 | 1.2 | N | 0 | |||
| PH1078-C | 13.94 | 13.72 | 0.98 | na | 0 | |||
| PH1109-C | 50.66 | 50.44 | 1 | N | 0 | |||
| PH1111-C | 5.3 | 6.67 | 1.26 | N | 0 | |||
| PH1112-C | 17.18 | 19.25 | 1.12 | N | 0 | |||
| PH1113-C | 18.58 | 21.75 | 1.17 | Y | Y | 2 | ||
| PH1116-C | 11.98 | 16.52 | 1.38 | Y | Y | 15 | ||
| PH1117-C | 5.78 | 6.58 | 1.14 | N | 0 | |||
| PH1121-C | 10.29 | 12.16 | 1.18 | N | 0 | |||
| PH1124-C | 18.06 | 17.8 | 0.99 | N | 0 | |||
| PH1126-C | 10.9 | 9.95 | 0.91 | N | 6 | |||
| PH1131-C | 7.25 | 8.87 | 1.22 | N | 0 | |||
| PH1132-C | 6.26 | 6.82 | 1.09 | N | 0 | |||
| Duplicated PvDBP** | PH0178-C | 82.52 | 183.31 | 2.22 | Y | Y | 0 | |
| PH0184-C | 47.65 | 111.6 | 2.34 | Y | Y | 0 | ||
| PH0188-C | 176.17 | 372.57 | 2.11 | Y | Y | 0 | ||
| PH0190-C | PV0431 | 152.9 | 427.71 | 2.8 | Y | Y | 0 | |
| PH0309-C | 21.37 | 49 | 2.29 | Y | Y | 0 | ||
| PH0182-C | 25.68 | 46.37 | 1.81 | Y | Y | 28 | ||
| PH1084-C | 6.89 | 15.06 | 2.19 | Y | na | 0 | ||
| PH1115-C | 11.22 | 25.6 | 2.28 | Y | Y | 0 | ||
| PH1119-C | 10.46 | 21.32 | 2.04 | Y | Y | 0 | ||
| PH1125-C | 14.92 | 28.86 | 1.93 | Y | Y | 0 | ||
| PH1133-C | 15.07 | 34.33 | 2.28 | Y | Y | 6 | ||
| PH1135-C | 6.42 | 13.77 | 2.14 | Y | Y | 0 |
*Coverage of PvDBP 0.8–1.4x
**Coverage of PvDBP 1.8–2.8x
***Minimum of 5 reads supporting reference and alternative allele; allele frequencies 0.3–0.7
†All PCR results support only the shorter Cambodian PvDBP duplication
na, No remaining DNA available
Genetic diversity in PvDBP sequences
For the 12 isolates with PvDBP duplication supported by 1.8–2.8x coverage of PvDBP compared to average coverage of the entire genome, we reviewed intra-isolate SNPs in PvDBP to determine whether the two gene copies in a given isolate were identical. In 10 of the 12 isolates, there were no SNPs between the two PvDBP copies. In the other two isolates (PH1133-C, PH0182-C), there were SNPs at 30% and 70% frequency between the two PvDBP copies (Table 1), as well as SNPs at 30–70% frequency throughout the genome. These data suggest that these are polyclonal isolates, rather than clonal isolates containing two sequence-divergent PvDBP copies.
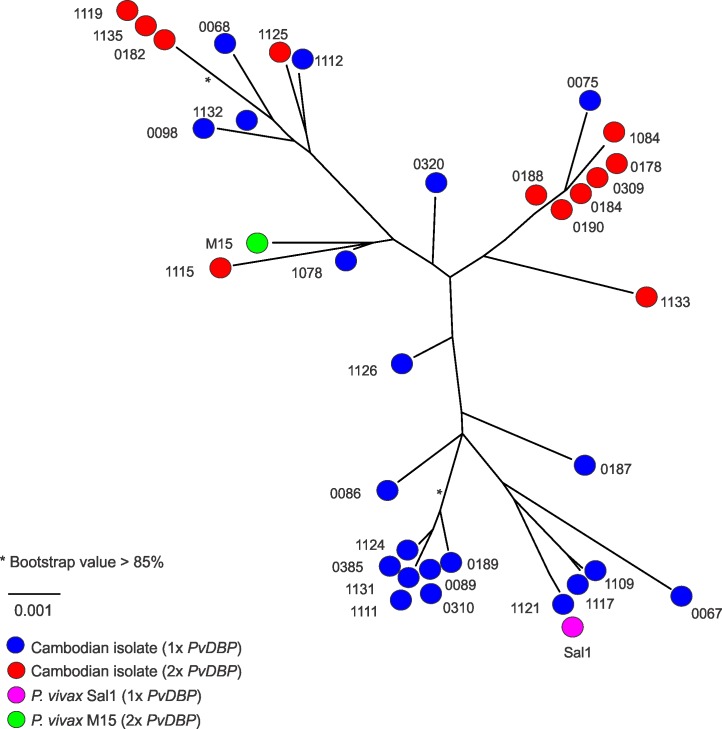
We aligned the PvDBP DNA sequences (including introns) of the reference P. vivax Sal1 line, Malagasy P. vivax M15 isolate, and all 37 whole-genome-sequenced Cambodian P. vivax isolates (including only the dominant haplotype when SNPs were present). Out of 3804 aligned bases, 51 sites were polymorphic (1.3%), excluding indels; we found unique PvDBP haplotypes in 77% (30/39) of samples, with a haplotype diversity of 0.98 ± 0.01. We categorized isolates as having either one or two PvDBP copies, excluding PH0177-C, PH0180-C, PH1113-C, and PH1116-C, which contained tail-to-tail read pairs and/or PCR support for duplication, but no increase in regional coverage. In a maximum likelihood tree analysis, isolates with either one or two PvDBP copies were intermixed and did not form distinct clades (Fig 2). This suggests that PvDBP duplication in Cambodian parasites is not the result of a single duplication event that has spread throughout the population. We found only three clades with bootstrap values >0.85, indicating that there are too few SNPs shared overall (likely due to outbreeding) to support the phylogenetic relationships between samples.
Fig 2. Maximum likelihood tree for PvDBP sequences in Cambodia.
Maximum likelihood tree reconstructed based on alignment by ClustalW with bootstrap analyses to assess clade support (500 replicates) using a single consensus sequence for PvDBP (including introns) for 33 Cambodian isolates with PvDBP duplications (red) or single copies of PvDBP (blue) with numbers corresponding to the WTSI ID numbers in Table 1. Duplications were predicted by 1.8–2.8x average coverage in PvDBP compared to genome-wide average coverage, the presence of tail-to-tail mate pairs and PCR results (Table 1). P. vivax Sal1 containing one PvDBP gene (magenta) and Malagasy P. vivax M15 containing two PvDBP genes (green) are included as references. *bootstrap values >85%.
Prevalence of PvDBP duplication in Cambodia, and its association with parasitemia and DARC genotype
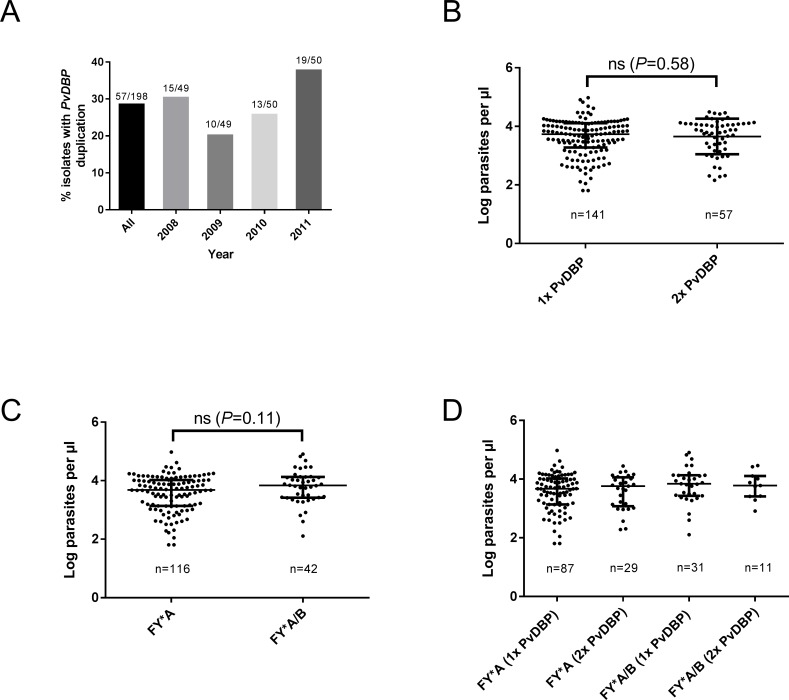
We used the new BF+AR2 primers to measure the prevalence of PvDBP duplications in 198 Cambodian P. vivax isolates collected over four years in this seasonal transmission setting. None of these samples had been previously studied for PvDBP duplication by whole-genome sequencing or any other method. Overall, 29% (57/198) of P. vivax samples contained the duplication, with an annual prevalence ranging from 20% to 38% (Fig 3A). PCR product sizes suggested that all samples contained the Cambodian but not the Malagasy PvDBP duplication. This was confirmed by capillary sequencing of PCR products in 43 isolates with the duplication; in all cases, the duplication breakpoints occurred at an identical poly-T region marking the boundary of the Cambodian PvDBP duplication (linker sequence in S1 Fig). The 29% prevalence of PvDBP duplication in 198 Cambodian samples, as measured using the new BF+AR2 primers, is similar to the 32% (6/19) prevalence previously measured by whole-genome sequencing of a much smaller number of Cambodian isolates, although as noted this analysis only scored the presence of the duplication, not its precise boundary [13].
Fig 3. Prevalence of PvDBP duplication in Cambodia, and its association with parasite density and DARC genotypes.
(A) The proportion of parasites carrying a PvDBP duplication (detected by PCR) over four years is shown. (B-D) P. vivax densities were not significantly different when stratified by (B) PvDBP copy number, (C) DARC genotype, or (D) combinations of PvDBP copy number and DARC genotype (Mann-Whitney test, P>0.05). Median (interquartile range, IQR) parasite densities are shown. ns, not significant. P values for 3D are reported in S1 Table.
To assess whether PvDBP duplication is associated with higher parasite densities (which might suggest increased invasion efficiency) in Cambodian patients with malaria, we stratified parasite densities according to PvDBP copy number across these 198 newly-genotyped isolates. Parasite densities were not significantly different between parasites carrying one or two PvDBP genes (Fig 3B). There is no reported DARC-negativity in Cambodia, making it unlikely that the high prevalence of PvDBP duplication is associated with invasion into DARC-negative erythrocytes in this country. However, different DARC genotypes are present, and it has been recently found that susceptibility to P. vivax malaria episodes is higher in the FY*B/FY*B genotype relative to the FY*A/FY*A genotype, possibly due to significantly reduced binding of PvDBP to Fy(a+b-) erythrocytes [30,31]. To establish whether there is any association between FY*A or FY*B genotypes and PvDBP duplication in Cambodia, we sequenced the DARC genotypes of 159 patient samples that were also genotyped for PvDBP duplications. As shown in Fig 3C, 73% (116/159) of patients were FY*A homozygotes and 26% (42/159) were FY*A/*B heterozygotes. A single patient (1/159) was homozygous for FY*B. As expected, we found no evidence for the DARC-negative FY*BES allele in these 159 samples. Neither FY*A nor FY*A/*B were significantly associated with PvDBP duplications in this study population (Fig 3D).
Global distribution of Cambodian and Malagasy PvDBP duplications
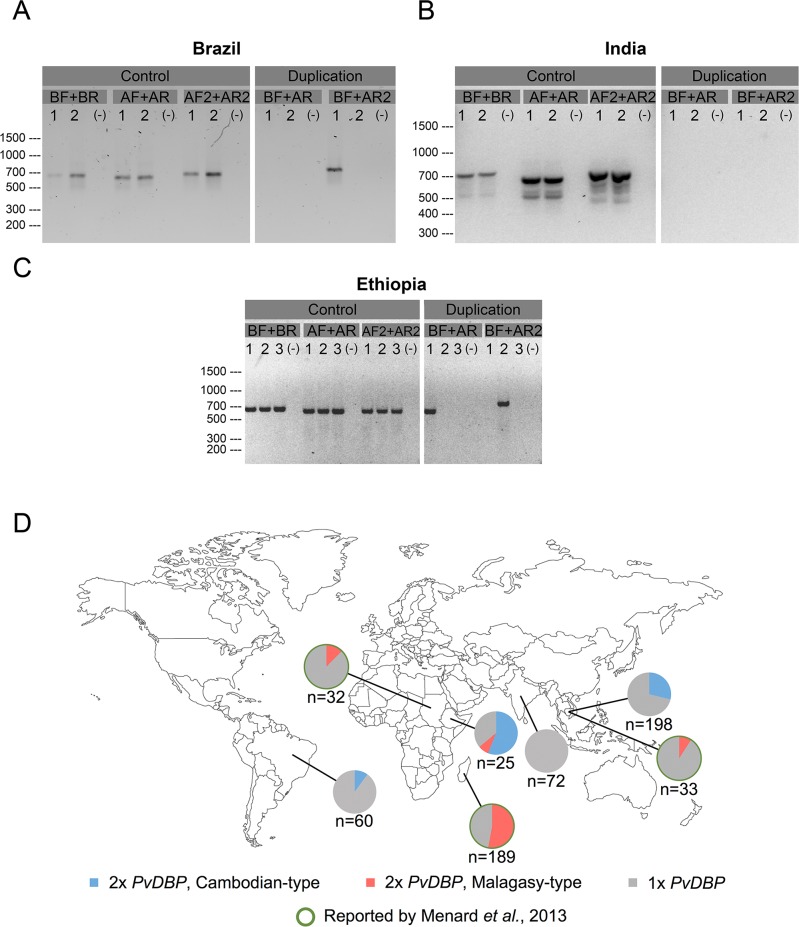
To determine whether the Cambodian PvDBP duplication is present outside Cambodia, we used the new BF+AR2 primers to amplify the PvDBP region in P. vivax clinical isolates, none of which had been previously analysed for PvDBP duplication, from three different continents. While we did not detect Cambodian or Malagasy PvDBP duplications in 72 samples from Goa, India (Fig 4B), we found Cambodian PvDBP duplications in 10% (6/60) of samples from Brazil (Fig 4A) and 56% (14/25) of samples from Ethiopia (Fig 4C). Interestingly, Malagasy duplications were also found in samples from Ethiopia (Fig 4C) but not Brazil (Fig 4A). These data, combined with those previously published for the Malagasy PvDBP duplication [12], indicate that both types of PvDBP duplication are globally distributed (Fig 4D).
Fig 4. Global distribution of Cambodian and Malagasy PvDBP duplications. PCR primers that detect Cambodian and Malagasy PvDBP duplications were tested in three P. vivax-endemic countries (Brazil, India, Ethiopia).
(A) Brazilian isolates 1 and 2 contain a Cambodian PvDBP duplication and a single PvDBP region, respectively. Note that the sizes (and sequences, S1 Fig) of the PCR products match the Cambodian PvDBP duplication detected by the BF+AR2 primers, but not the Malagasy PvDBP duplication detected by the BF+AR primers. (B) Indian isolates 1 and 2 lack Cambodian and Malagasy PvDBP duplications. (C) Ethiopian isolates 1, 2, and 3 contain a Malagasy PvDBP duplication, a Cambodian PvDBP duplication, and a single PvDBP region, respectively. (A-C) All isolates show bands for the positive control AF2+AR2 primers, and no bands for DNA-negative controls (-). (D) In the present study, the prevalence of Cambodian PvDBP duplication is 0/72 in India, 6/60 in Brazil, 14/25 in Ethiopia, and 57/198 in Cambodia, and the prevalence of Malagasy PvDBP duplication is 0/72 in India, 0/60 in Brazil, 2/25 in Ethiopia, and 0/198 in Cambodia. In a previous study [12], the prevalence of Malagasy PvDBP duplication was 4/32 in Sudan, 100/189 in Madagascar, and 3/33 in Cambodia.
Discussion
Although P. vivax causes the majority of malaria cases in Asia, South America, and the Pacific, research of the P. vivax genome lags significantly behind that of the P. falciparum genome. While thousands of P. falciparum clinical isolates have already been sequenced and analyzed, revealing insights into global epidemiology, as well as the origin and spread of drug resistance [32–34], only a handful of P. vivax genome sequences were publicly available until recently. Genomic data from four isolates suggested that P. vivax diversity may be more extensive than P. falciparum diversity [35], and sequencing additional isolates from Madagascar and Cambodia revealed CNVs and indels in genes associated with erythrocyte invasion [12,36], including a PvDBP gene duplication. Given the number of samples in these studies, however, it was not possible to infer whether any of these features are confined to individual infections or geographic locations, or are much more widespread and common forms of variation. The recent publication of more than 200 P. vivax genomes from across the world revealed that CNV at the PvDBP locus is one of the most common CNVs in the P. vivax genome [13]. In contrast, no PvDBP duplications were found in a concurrent population genomics study of 182 P. vivax isolates from 11 endemic countries [37]. This discrepancy may be due in large part to the reliance on the Malagasy duplication primers for validation in the latter study, which as noted here will miss the newly-identified Cambodian duplication [37]. In this study, we used sequence data from clinical isolates [13] to infer that PvDBP duplications in Cambodia had different boundaries than the PvDBP duplication in Madagascar [12]. Using these sequence data to define new duplication-specific PCR primers, we defined the boundaries of this new duplication type, showed that it was present at a prevalence between 20% and 38% over four consecutive transmission seasons in Cambodia, and that it was also present in P. vivax infections in Ethiopia and Brazil, although absent from a study site in India. In Cambodia, there was no association between PvDBP duplication and either parasite burden or DARC genotype. Thus, there are at least two common forms of PvDBP duplication with global distribution; the duplication-specific PCR primers defined in this study will help to estimate their prevalence in other P. vivax-endemic regions and enable genotype-phenotype correlation studies.
One key question is whether the Cambodian and Malagasy duplications arose independently in specific locations and then spread around the world, or whether PvDBP CNVs are continuously arising de novo, but consistently occur at the same defined boundaries, leading to either “Malagasy-type” or “Cambodia-type” PvDBP duplications. Phylogenetic analysis at this stage favors the latter hypothesis. PvDBP is known to be under positive selection pressure [38], with widespread diversity even within the DARC-binding region, Domain II. Of the 12 Cambodian isolates that have been whole-genome sequenced and have PvDBP duplications, ten have duplicated PvDBP genes that are identical in sequence. In phylogenetic trees, these sequences are interspersed with PvDBP sequences from samples without duplications, strongly suggesting that the Cambodian duplication arose independently multiple times on different genetic backgrounds. An alternative explanation is that the Cambodian duplication arose only once, and the two PvDBP genes subsequently diverged due to immune selection pressure. Under this hypothesis, frequent gene conversion would be required to maintain sequence identity between the two PvDBP copies within a given isolate. Whether this is likely is unclear, as little is known about the frequency of gene conversion in Plasmodium parasites, although this phenomenon of shared mutations between identical adjacent regions of the genome has previously been seen in other invasion genes in P. falciparum [39]. At this moment, however, independent origins of PvDBP duplication at defined amplification hotspots appears to be the simplest explanation.
The nature of the PvDBP duplication boundaries also favors this hypothesis and hints at a potential molecular mechanism. While there is no homology between the 3’ boundaries of the Cambodian and Malagasy duplications, both occur in homopolymeric T tracks, which are not as frequent in the P. vivax genome as they are in the AT-rich P. falciparum genome. Recent in vitro studies selecting for P. falciparum parasites with resistance to DSM1, an inhibitor of dihydroorotate dehydrogenase (DHODH), produced multiple lines with CNVs at the PfDHODH locus [40]. These CNVs had different boundaries, but notably all of them contained homopolymeric A or T tracks, suggesting that such tracks facilitate frequent localized recombination and CNV generation during DNA replication. Such CNVs may provide raw material for evolution, by providing a second copy of a given gene or enabling the accumulation of mutations in one copy while limiting the potential risk of deleterious mutations by maintaining a second wild-type copy. In Plasmodium parasites, which are faced with numerous strong selection pressures in both humans and mosquitoes, the advantages of such a mechanism are obvious. While it is not known whether this mechanism is common, the parallels between PfDHODH and PvDBP CNVs are intriguing, and a genome-wide analysis of common CNVs could be revealing.
What then is driving CNVs at the PvDBP locus? The best known examples of CNVs at Plasmodium genes involve antimalarial drug resistance; for example, selection pressure from mefloquine amplifies the MDR1 gene in P. falciparum [41] and perhaps also in P. vivax [13]. Antimalarial drug pressure appears unlikely in this context, although not inconceivable; for example, if PvDBP duplication confers an increased growth rate to P. vivax parasites, this could provide an advantage against some slow-acting drugs. However, given the critical role of the PvDBP-DARC interaction in P. vivax erythrocyte invasion, it is likely that CNVs at PvDBP are being selected by genetic variation in DARC. Is this variation DARC-negativity? The presence of PvDBP duplications at frequencies approaching 40% both globally [12,13], and over multiple transmission seasons in Cambodia where DARC-negativity is essentially absent (Fig 3), makes this unlikely. Another possibility is that PvDBP duplications increase the efficiency of parasite invasion into erythrocytes carrying other common DARC genotypes, such as FY*A or FY*B. No association between PvDBP duplication and these FY genotypes was seen in this study, but the fact that P. vivax clinical isolates were obtained only from patients with acute malaria episodes may confound this analysis. A much larger study that genotypes both the DARC and PvDBP loci, is conducted where the prevalence of DARC-negativity is high, and includes both asymptomatic and clinical cases is needed to comprehensively test for association between PvDBP duplication and DARC genotype. In addition, testing the functional consequences of PvDBP duplication, through protein binding assays or ex vivo growth assays, are clearly needed. Since Plasmodium parasites are highly adaptable, understanding how they respond to human erythrocyte variation will be particularly relevant to studies of naturally-acquired immunity to PvDBP and the development of any PvDBP-based vaccine.
Supporting Information
(DOC)
Fasta file representing the 736-bp PCR product linking the duplicated regions in samples with the Cambodian-type duplication. Primer sites for BF (red), AR2 (blue), and the poly-T break point (bold black) are shown.
(TIF)
(XLSX)
Acknowledgments
We give our deepest thanks to all study participants and field site staff, without whom this work would not have been possible.
Data Availability
All relevant data are within the paper and its Supporting Information files. All reconstructed PvDBP haplotypes and links for raw WGS data can be accessed at www.malariagen.net/resource/19.
Funding Statement
This work was funded by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) (to RF), the Wellcome Trust grant 090851 (to JCR), and NIH grant R21 AI101802 (to GY). The “Malaria Evolution in South Asia” project in the International Center of Excellence for Malaria Research (ICEMR) program was supported by US NIAID, NIH agreement U19 AI089688 (Program Director, PKR). UK was supported by a Canadian Institutes of Health Postdoctoral Fellowship. Field work in Acrelândia, Brazil was supported by NIH research grants (U19 AI089681 to Joseph M. Vinetz) and the Fundação de Amparo à Pesquisa do Estado de São Paulo (2010/51835-7). Field work in Mâncio Lima, Brazil was supported by research grants from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Ministry of Health of Brazil (404067/2012-3). MUF is supported by a senior researcher scholarship from CNPq, and a senior researcher scholarship from the Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Weiss GE, Crabb BS, Gilson PR (2016) Overlaying Molecular and Temporal Aspects of Malaria Parasite Invasion. Trends Parasitol 32: 284–295. 10.1016/j.pt.2015.12.007 [DOI] [PubMed] [Google Scholar]
- 2.Fang XD, Kaslow DC, Adams JH, Miller LH (1991) Cloning of the Plasmodium vivax Duffy receptor. Mol Biochem Parasitol 44: 125–132. [DOI] [PubMed] [Google Scholar]
- 3.Batchelor JD, Zahm JA, Tolia NH (2011) Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nat Struct Mol Biol 18: 908–914. 10.1038/nsmb.2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batchelor JD, Malpede BM, Omattage NS, DeKoster GT, Henzler-Wildman KA, et al. (2014) Red blood cell invasion by Plasmodium vivax: structural basis for DBP engagement of DARC. PLoS Pathog 10: e1003869 10.1371/journal.ppat.1003869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tournamille C, Colin Y, Cartron JP, Le Van Kim C (1995) Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet 10: 224–228. 10.1038/ng0695-224 [DOI] [PubMed] [Google Scholar]
- 6.Miller LH, Mason SJ, Clyde DF, McGinniss MH (1976) The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med 295: 302–304. 10.1056/NEJM197608052950602 [DOI] [PubMed] [Google Scholar]
- 7.Mueller I, Shakri AR, Chitnis CE (2015) Development of vaccines for Plasmodium vivax malaria. Vaccine 33: 7489–7495. 10.1016/j.vaccine.2015.09.060 [DOI] [PubMed] [Google Scholar]
- 8.Menard D, Barnadas C, Bouchier C, Henry-Halldin C, Gray LR, et al. (2010) Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci U S A 107: 5967–5971. 10.1073/pnas.0912496107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howes RE, Reiner RC Jr., Battle KE, Longbottom J, Mappin B, et al. (2015) Plasmodium vivax Transmission in Africa. PLoS Negl Trop Dis 9: e0004222 10.1371/journal.pntd.0004222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xainli J, Adams JH, King CL (2000) The erythrocyte binding motif of plasmodium vivax duffy binding protein is highly polymorphic and functionally conserved in isolates from Papua New Guinea. Mol Biochem Parasitol 111: 253–260. [DOI] [PubMed] [Google Scholar]
- 11.VanBuskirk KM, Cole-Tobian JL, Baisor M, Sevova ES, Bockarie M, et al. (2004) Antigenic drift in the ligand domain of Plasmodium vivax duffy binding protein confers resistance to inhibitory antibodies. J Infect Dis 190: 1556–1562. 10.1086/424852 [DOI] [PubMed] [Google Scholar]
- 12.Menard D, Chan ER, Benedet C, Ratsimbasoa A, Kim S, et al. (2013) Whole genome sequencing of field isolates reveals a common duplication of the Duffy binding protein gene in Malagasy Plasmodium vivax strains. PLoS Negl Trop Dis 7: e2489 10.1371/journal.pntd.0002489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearson RD, Amato R, Auburn S, Miotto O, Almagro-Garcia J, et al. (2016) Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat Genet 48: 959–964. 10.1038/ng.3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowman AF, Galatis D, Thompson JK (1994) Selection for mefloquine resistance in Plasmodium falciparum is linked to amplification of the pfmdr1 gene and cross-resistance to halofantrine and quinine. Proc Natl Acad Sci U S A 91: 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peel SA, Bright P, Yount B, Handy J, Baric RS (1994) A strong association between mefloquine and halofantrine resistance and amplification, overexpression, and mutation in the P-glycoprotein gene homolog (pfmdr) of Plasmodium falciparum in vitro. Am J Trop Med Hyg 51: 648–658. [DOI] [PubMed] [Google Scholar]
- 16.Amaratunga C, Sreng S, Mao S, Tullo GS, Anderson JM, et al. (2014) Chloroquine remains effective for treating Plasmodium vivax malaria in Pursat province, Western Cambodia. Antimicrob Agents Chemother 58: 6270–6272. 10.1128/AAC.03026-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vargas-Rodriguez Rdel C, da Silva Bastos M, Menezes MJ, Orjuela-Sanchez P, Ferreira MU (2012) Single-nucleotide polymorphism and copy number variation of the multidrug resistance-1 locus of Plasmodium vivax: local and global patterns. Am J Trop Med Hyg 87: 813–821. 10.4269/ajtmh.2012.12-0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venkatesan M, Amaratunga C, Campino S, Auburn S, Koch O, et al. (2012) Using CF11 cellulose columns to inexpensively and effectively remove human DNA from Plasmodium falciparum-infected whole blood samples. Malar J 11: 41 10.1186/1475-2875-11-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, et al. (2008) Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455: 757–763. 10.1038/nature07327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA (2012) Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28: 464–469. 10.1093/bioinformatics/btr703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manske HM, Kwiatkowski DP (2009) LookSeq: a browser-based viewer for deep sequencing data. Genome Res 19: 2125–2132. 10.1101/gr.093443.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 27.Rambaut A (2002) Se-Al: sequences alignment editor v. 2.0a11 Edinburgh: Institute of Evolutionary Biology. [Google Scholar]
- 28.Rozas J (2009) DNA sequence polymorphism analysis using DnaSP. Methods Mol Biol 537: 337–350. 10.1007/978-1-59745-251-9_17 [DOI] [PubMed] [Google Scholar]
- 29.Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King CL, Adams JH, Xianli J, Grimberg BT, McHenry AM, et al. (2011) Fy(a)/Fy(b) antigen polymorphism in human erythrocyte Duffy antigen affects susceptibility to Plasmodium vivax malaria. Proc Natl Acad Sci U S A 108: 20113–20118. 10.1073/pnas.1109621108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chittoria A, Mohanty S, Jaiswal YK, Das A (2012) Natural selection mediated association of the Duffy (FY) gene polymorphisms with Plasmodium vivax malaria in India. PLoS One 7: e45219 10.1371/journal.pone.0045219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manske M, Miotto O, Campino S, Auburn S, Almagro-Garcia J, et al. (2012) Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature 487: 375–379. 10.1038/nature11174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miotto O, Almagro-Garcia J, Manske M, Macinnis B, Campino S, et al. (2013) Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet 45: 648–655. 10.1038/ng.2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malaria GENPfCP (2016) Genomic epidemiology of artemisinin resistant malaria. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neafsey DE, Galinsky K, Jiang RH, Young L, Sykes SM, et al. (2012) The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum. Nat Genet 44: 1046–1050. 10.1038/ng.2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hester J, Chan ER, Menard D, Mercereau-Puijalon O, Barnwell J, et al. (2013) De novo assembly of a field isolate genome reveals novel Plasmodium vivax erythrocyte invasion genes. PLoS Negl Trop Dis 7: e2569 10.1371/journal.pntd.0002569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hupalo DN, Luo Z, Melnikov A, Sutton PL, Rogov P, et al. (2016) Population genomics studies identify signatures of global dispersal and drug resistance in Plasmodium vivax. Nat Genet 48: 953–958. 10.1038/ng.3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baum J, Thomas AW, Conway DJ (2003) Evidence for diversifying selection on erythrocyte-binding antigens of Plasmodium falciparum and P. vivax. Genetics 163: 1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rayner JC, Tran TM, Corredor V, Huber CS, Barnwell JW, et al. (2005) Dramatic difference in diversity between Plasmodium falciparum and Plasmodium vivax reticulocyte binding-like genes. Am J Trop Med Hyg 72: 666–674. [PubMed] [Google Scholar]
- 40.Guler JL, Freeman DL, Ahyong V, Patrapuvich R, White J, et al. (2013) Asexual populations of the human malaria parasite, Plasmodium falciparum, use a two-step genomic strategy to acquire accurate, beneficial DNA amplifications. PLoS Pathog 9: e1003375 10.1371/journal.ppat.1003375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, et al. (2004) Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364: 438–447. 10.1016/S0140-6736(04)16767-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
Fasta file representing the 736-bp PCR product linking the duplicated regions in samples with the Cambodian-type duplication. Primer sites for BF (red), AR2 (blue), and the poly-T break point (bold black) are shown.
(TIF)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. All reconstructed PvDBP haplotypes and links for raw WGS data can be accessed at www.malariagen.net/resource/19.