Abstract
Vibrio vulnificus is a bacterium responsible for severe gastroenteritis, sepsis and wound infections. Gastroenteritis and sepsis are commonly associated with the consumption of raw oysters, whereas wound infection is often associated with the handling of contaminated fish. Although classical virulence factors of this emerging pathogen are well characterised, there remains a paucity of knowledge regarding the general biology of this species. To investigate the presence of previously unreported virulence factors, we applied whole genome sequencing to a panel of ten V. vulnificus strains with varying virulence potentials. This identified two novel type 6 secretion systems (T6SSs), systems that are known to have a role in bacterial virulence and population dynamics. By utilising a range of molecular techniques and assays we have demonstrated the functionality of one of these T6SSs. Furthermore, we have shown that this system is subject to thermoregulation and is negatively regulated by increasing salinity concentrations. This secretion system was also shown to be involved in the killing of V. vulnificus strains that did not possess this system and a model is proposed as to how this interaction may contribute to population dynamics within V. vulnificus strains. In addition to this intra-species killing, this system also contributes to the killing of inter bacterial species and may have a role in the general composition of Vibrio species in the environment.
Introduction
V. vulnificus is a Gram negative opportunistic pathogen typically isolated from marine and coastal environments [1]. The bacterium is capable of causing an array of disease symptoms such as gastroenteritis, wound infection and primary septicaemia [2]. Wound infection is commonly associated with recreational activities in contaminated water or handling of infected fish and can present with edema, bullae, cellulitis and necrotising fasciitis [2]. In severe cases amputation of limbs may be required. Unlike wound infections which have on average a 25% mortality rate, primary septicaemia, which is often associated with the consumption of contaminated seafood, has a greater than 50% mortality rate rising to 100% without the prompt administration of antibiotic therapy [1]. Currently, in America, the bacterium is the leading cause of seafood related deaths, and due to climate change causing an increase in surface sea temperature, the emergence of this pathogen in colder regions such as the Baltic Sea are becoming increasingly common [3].
Although this pathogen is known to be highly pathogenic, the number of severe human infections reported each year is extremely low given the virulence potential and environmental prevalence of this bacterium [4]. This has led researchers to believe that not all strains are equal in virulence, with some strains better adapted to causing human disease than others [5]. However, a genetic basis for this difference in virulence potential is yet to be established. Furthermore, there is limited knowledge surrounding the general biology of V. vulnificus and to date only two secretion systems have been characterised in this bacterium, the type 1 secretion system (T1SS) and the type 2 secretion system (T2SS) [6]. The former is known to secrete the well-studied toxin RtxA which has been shown to be required for both gut pathogenesis and dissemination throughout the body [7], whereas the latter is required for the secretion of the haemolysin, VvhA [6], a toxin which is also required for gut pathogenesis [7]. To investigate the presence of previously unreported virulence factors in V. vulnificus, we performed whole genome sequencing which led us to identify previously unreported T6SSs.
The T6SS is found in approximately 25% of all sequenced Gram negative bacterial genomes, and is made up of 13 conserved proteins [8]. These 13 proteins are hypothesised to form a macromolecular structure similar to the contractile tail of bacteriophage [9]. The needle like structure of the T6SS is composed of Hcp, VrgG and PAAR proteins [10], with ATPase activity being provided by ClpV and the core scaffolding protein, IcmF [11, 12]. Unlike previously identified secretion systems, which usually export anti-eukaryotic effectors, the T6SS has dual functions, secreting both anti-eukaryotic and anti-prokaryotic effectors [13]. The latter function in particular has captured the attention of many researchers, as it poses the potential for development of novel antimicrobials [14]. In addition to secreting anti-bacterial toxins, it has also been shown that anti-bacterial T6SS encoding bacteria also harbour immunity proteins. These immunity proteins are able to neutralise the deleterious effects of toxins to prevent self-intoxication and provide immunity between T6SS harbouring sister cells [14, 15].
To study the role of the T6SS in affecting the population dynamics of Vibrio spp. this study employed co-culturing assays. These assays demonstrated that the T6SS present in V. vulnificus can play an important role in bacterial population composition through anti-bacterial activity. Furthermore, we present a hypothesis describing how the T6SS may provide an explanation for the limited number of serious human infections attributed to V. vulnificus given the high abundance of this bacterium in the environment.
Results
V. vulnificus strains vary in their T6SS genetic architecture
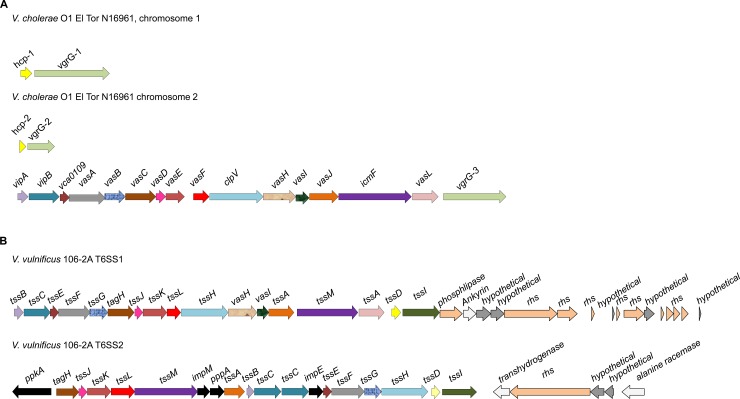
In an approach to identify genetic differences between V. vulnificus isolates that may account for species specific phenotypes, whole genome sequencing was applied to a panel of ten strains of varying virulence potential and isolation source (Table 1) [5]. Analysis of this data identified a novel T6SS, herein referred to as T6SS2, which was present in all ten sequenced isolates (S1 Fig). Furthermore, an additional T6SS, present in three out of the ten isolates, T6SS1, was also identified (S2 Fig). This T6SS1 exhibited synteny with the previously characterised T6SS from V. cholerae [16] having 76% similarity over 19.2 kb. However, several differences between T6SS1 of V. vulnificus and the T6SS of V. cholerae exist and are depicted in Fig 1. For example, V. cholerae (Fig 1A) contains two hcp genes, which are distributed across chromosomes 1 and 2 whereas the sole hcp gene of V. vulnificus T6SS1 is located within the T6SS operon on chromosome 2 (Fig 1B). Both T6SS1 and T6SS2 of V. vulnificus contain the 13 core genes required for a functional T6SS (Fig 1B) [8]. However, T6SS2 contains several accessory genes, encoding for a phosphatase and kinase, which are absent from T6SS1. Bioinformatic investigation of three V. vulnificus reference genomes, YJ016, MO6-24/O and CMCP6 revealed that all three genomes did not contain T6SS1 but all had T6SS2, although the latter were not annotated as such in their Genbank depositions (S1 Fig).
Table 1. Bacterial strains.
| Bacterial strains | Description | Virulence Grouping a | Source |
|---|---|---|---|
| V. vulnificus 106-2A | Wild-type strain, environmental isolate, T6SS1+ T6SS2+ | 3 | CEFAS |
| V. vulnificus 99–743 | Environmental isolate, T6SS1- T6SS2+ | 3 | CEFAS |
| V. vulnificus MO6-24/O | Clinical isolate, T6SS1- T6SS2+ | 4 | CEFAS |
| V. vulnificus 99–796 | Environmental isolate, T6SS1+ T6SS2+ | 4 | CEFAS |
| V. vulnificus ORL-1506 | Clinical isolate, T6SS1- T6SS2+ | 5 | CEFAS |
| V. vulnificus NSV-5830 | Clinical isolate, T6SS1- T6SS2+ | 5 | CEFAS |
| V. vulnificus DAL-79087 | Clinical isolate, T6SS1- T6SS2+ | 3 | CEFAS |
| V. vulnificus DAL-79040 | Clinical isolate, T6SS1- T6SS2+ | 2 | CEFAS |
| V. vulnificus S3-16 | Environmental isolate, T6SS1+ T6SS2+ | 1 | CEFAS |
| V. vulnificus S2-22 | Environmental isolate, T6SS1- T6SS2+ | 3 | CEFAS |
| V. vulnificus ATL-9824 | Clinical isolate, T6SS1- T6SS2+ | 2 | CEFAS |
| V. fluvialis NCTC 11327 | Clinical isolate | N/A | CEFAS |
| V. vulnificus SRC1 | 106-2A ΔicmF1 in-frame, T6SS1- T6SS2+ | N/A | This study |
| V. vulnificus SRC2 | 106-2A ΔicmF2 in-frame, T6SS1+ T6SS2- | N/A | This study |
| V. vulnificus 106-2A pSCrhaB3 | Wild-type strain containing pSCrhaB3 (Tpr) | N/A | This study |
| V. vulnificus 106-2A:icmF1 pSCrhaB3 | 106-2A ΔicmF1 containing pSCrhaB3 (Tpr) | N/A | This study |
| V. vulnificus 106-2A:icmF2 pSCrhaB3 | 106-2A ΔicmF2 containing pSCrhaB3 (Tpr) | N/A | This study |
| V. vulnificus 99–743 pBHR4-groS-RFP | V. vulnificus 99–743 containing pBHR4-groS-RFP (Cmr) | N/A | This study |
| V. fluvialis NCTC 11327 pBHR-4groS-RFP | V. fluvialis NCTC 11327 containing pBHR4-groS-RFP (Cmr) | N/A | This study |
| V. vulnificus MO6-24/O pBHR-4groS-RFP | V. vulnificus MO6-24/O containing pBHR4-groS-RFP (Cmr) | N/A | This study |
| V. cholerae V52 | Wild-type strain (Smr) | N/A | Dr. S. Pukatizki |
| Escherichia coli DH5α | Cloning host | N/A | Lab stock |
| Escherichia coli S17 λpir | Cloning host and donor strain | N/A | Lab Stock |
| Escherichia coli TOP10 | Cloning host | N/A | Invitrogen |
a Virulence grouping was determined from the in vivo study performed by Thiaville et al., 2011, virulence increases with numerical value.
Fig 1. T6SSs of Vibrio vulnificus.
(A) Schematic diagram showing the genetic organisation of T6SS of V. cholerae. (B) Schematic representation of the genetic organisation of T6SSs from V. vulnificus. Accessory proteins present in T6SS2 and absent from T6SS1 are shown in black. Homologous genes between the two species have been colour matched to represent their similarity.
V. vulnificus T6SS1 secretes Hcp-1 in a temperature and salinity dependent manner
As T6SS1 of V. vulnificus exhibits synteny with the previously described T6SS of V. cholerae, we first sought to demonstrate that T6SS1 was functional in V. vulnificus. This was assessed by confirming secretion of Hcp-1, a prototypical T6SS secreted component. Western blot analysis using an antibody against the Hcp peptide sequence; AGTSGSDDWRKPIEA [17] confirmed that Hcp-1 was secreted into the culture filtrate (Fig 2A). We were able to use the anti- Hcp-1 antibody raised against Hcp-1 from V. cholerae due to the high level of homology between V. cholerae and V. vulnificus (Fig 2B). Previous characterisation of T6SSs from V. cholerae and V. parahaemolyticus has demonstrated that salinity and temperature can affect activation of these systems [18, 19]. We therefore investigated the effect of temperature and salinity on T6SS1 of V. vulnificus in order to determine whether it was similarly regulated.
Fig 2. T6SS1 of V. vulnificus is functional.
(A) Western blot employing an antibody against Hcp-1 from V. cholerae demonstrating the presence of Hcp-1 in the culture filtrate and cell pellet of V. vulnificus 106-2A (lane 1 and 2). Lane 3, protein extract from V. cholerae V52 used as a positive control to confirm the specificity of the antibody (14 μg of protein per lane). (B) Amino acid alignment of Hcp-1 from V. cholerae and V. vulnificus. Amino acid differences are boxed in black and epitope to which the anti-Hcp-1 antibody was raised is highlighted in red.
The Western blot using the anti- Hcp-1 antibody shown in Fig 3 demonstrates that Hcp-1 is secreted at 23°C and 30°C (lanes 2 and 6), temperatures similar to those seen in marine environments but not at 37°C (lane 10). When V. vulnificus was cultured at these temperatures in the presence of 3% w/v NaCl, a similar pattern of secretion was observed but at lower levels (cf; lanes 4, 8 and 12). Qualitative analysis of the cell lysates of these experiments demonstrated that Hcp-1 was expressed under all conditions but with secretion by the T6SS1 reduced by high temperature and increased salinity.
Fig 3. Hcp-1 expression from T6SS1 of V. vulnificus 106-2A is dependent upon temperature and salinity.
Western blot of cell lysates and supernatants from V. vulnificus 106-2A cultures grown at 23°C, 30°C and 37°C, using anti-Hcp-1 antibody. Cultures were grown in LB or LB supplemented with 3% NaCl w/v (SLB) (14 μg of protein per lane).
V. vulnificus utilises T6SS1 in intra-species killing
The T6SS of V. parahaemolyticus is active against other bacteria at marine like temperatures [18]. Therefore, we hypothesised that T6SS1 of V. vulnificus may be similarly involved in bacterial killing as this system is also functional under the same conditions observed for V. parahaemolyticus [18]. To test this hypothesis we first sought to determine whether a V. vulnificus T6SS1 positive strain could kill a V. vulnificus T6SS1 negative strain, based on the rationale that the latter strain would be deficient in immunity proteins to T6SS1 effectors. This was investigated by performing co-culture assays at 30°C using a naturally occurring T6SS1 negative V. vulnificus environmental strain, (99–743) as prey and, a T6SS1 positive environmental strain, (106-2A) as the attacker. These strains were chosen as 106-2A contains both T6SS1 and T6SS2, whereas 99–743 is a naturally occurring T6SS1 mutant. To test whether T6SS1 conferred a competitive advantage in 106-2A, an in-frame deletion of icmF1, (a gene encoding a core scaffolding protein of the T6SS) was created in strain 106-2A, resulting in strain SRC1. This mutation was confirmed by whole genome sequencing (S3 Fig). To confirm that this mutation resulted in an inactive T6SS1, Western blot analysis was performed to observe secretion of Hcp-1. As can be seen in Fig 4A, SRC1 was unable to secrete Hcp to the culture filtrate at 30°C when compared to the wild type strain. Furthermore, for enumeration purposes the competitive assay was performed using the prey strain, 99–743 which has been engineered to be chloramphenicol resistant and the attacker strains, 106-2A, SRC1 to be trimethoprim resistant. In addition to SRC1, an in-frame icmF2 mutant was generated in T6SS2 (SRC2) to determine whether T6SS2 played a role in bacterial targeting.
Fig 4.
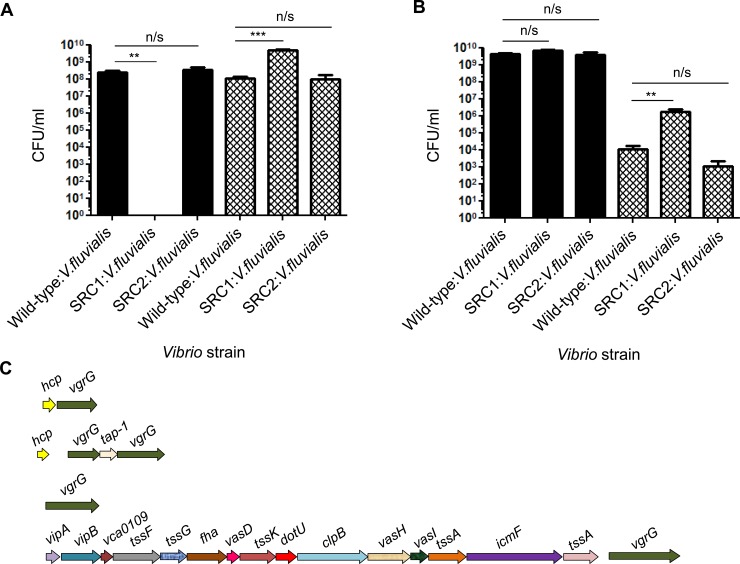
T6SS1 targets T6SS1 deficient strains of V. vulnificus (A) Western blot employing an antibody against Hcp-1 demonstrating the presence of Hcp-1 in the supernatant and cell pellet of V. vulnificus 106-2A wild-type. Whereas, Hcp-1 is only detected in the cell pellet of the icmF1 mutant, SRC1 and not in the supernatant. (B) Competitive index of V. vulnificus 99–743 (prey) following co-culture for two hours with V. vulnificus 106-2A (attacker) demonstrating an increase in the survival of the naturally occurring T6SS1 negative prey strain, 99–743 when co-cultured with SRC1 compared to 106-2A wild-type. (C) Competitive index of V. vulnificus MO6-24/O (prey) following co-culture for two hours with V. vulnificus 106-2A (attacker). Competitive index was calculated using the following equation: (input attacker/input prey)/(output attacker/output prey). Statistical analysis was performed using the Wilcoxon signed-rank test on log-transformed data. *** = p < 0.0005.
The competitive assays demonstrated that strain 106-2A could indeed target 99–743 at 30°C, resulting in a significant reduction in numbers of the prey strain (Fig 4B). In contrast to the observed inhibition of growth of the prey strain 99–743 when co-cultured with wild-type 106-2A, co-culture of 99–743 with SRC1 resulted in a significant increase in the competitive index of the prey strain (Fig 4B). Furthermore, we demonstrated that T6SS2 of 106-2A did not inhibit 99–743 as evidenced by co-culture with an icmF2 in-frame deletion mutant of 106-2A (SRC2). This latter experiment resulted in a phenotype comparable to that of the wild-type strain, Fig 4B.
As the prey strain 99–743 is an environmental isolate, we sought to determine whether T6SS1 of 106-2A could target a hyper-virulent clinical isolate. In order to test this hypothesis the hyper-virulent reference strain, MO6-24/O which was isolated from a patient with septicaemia [20] was selected as a prey strain for use in the co-culture assays. These co-culture assays demonstrated that as was the case for 99–743, the hyper-virulent clinical strain MO6-24/O was inhibited by the environmental strain 106-2A in a T6SS1 dependent manner, as evidenced by a lack of killing by SRC1 (the icmF1 mutant strain) Fig 4C.
V. vulnificus utilises T6SS1 in inter-species killing
T6SSs of several bacteria have been shown to play a role in effecting microbial population dynamics through inter-species killing [21]. In addition, it is known that several pathogenic Vibrio species exist alongside V. vulnificus within marine environments and bivalve molluscs [22]. We hypothesised that the T6SS1 of V. vulnificus may, in addition to the intra-species killing demonstrated above, play a role in effecting its proximal marine microflora through inter-species killing by T6SS1. We selected Vibrio fluvialis as a model bacterium to test this hypothesis as V. fluvialis, along with V. vulnificus, is known to be a shellfish-associated bacterium [23]. Furthermore, these strains were employed as post co- culture enumeration was performed on TCBS agar, on which V. vulnificus 106-2A forms green colonies and V. fluvialis forms yellow colonies. TCBS enumeration plates were incubated at 37°C, at which temperature T6SS1 did not secrete Hcp-1 (Fig 3, lane 10), excluding the possibility that killing due to T6SS1 was occurring on the enumeration plates.
Unlike the previous intra-species assays, the competitive index could not be calculated for either V. fluvialis or V. vulnificus. This is due to the complete removal of V. fluvialis and V. vulnificus in several of the assays resulting in an enumeration count of zero. Therefore, the number of recovered cells following co-culture is presented in place of the competitive index value.
In co-culture assays where V. fluvialis NCTC 11327 was used as a prey strain, and the T6SS1 mutant of V. vulnificus (SRC1) as the attacker, it was observed that V. fluvialis exhibited a significant increase in growth compared to when co-cultured with wild-type V. vulnificus at 30°C (Fig 5A, hatched bars). This suggests that the T6SS1 of V. vulnificus is involved in the inter-species killing of V. fluvialus. Furthermore, this observed phenotype is unlikely to be attributable to the T6SS2 of V. vulnificus, as there was no significant difference in the survival of V. fluvialis when co-cultured with the T6SS2 mutant (SRC2) compared to when co-cultured with wild-type V. vulnificus (Fig 5A, hatched bars).
Fig 5. V. vulnificus utilises T6SS1 for inter-species targeting at 30°C.
(A) The CFU/ml of recovered bacteria was recorded following a co-culture T6SS killing assay at 30°C for 5 hours when T6SS1 is active in the attacker strain. Attacker strain V. vulnificus 106-2A (solid bars) Prey strain V. fluvialis NCTC11327 (hatched bars). Co culture enumeration was performed at 37°C on TCBS where the T6SS is not active in the attacker strain. Statistical analysis was performed using a 1-way paired Student’s t-test, ** = p < 0.005. (B) Recovered CFU/ml of bacteria following co-culture at 30°C and enumeration at 37°C using antibiotic resistant strains on antibiotic plates. Hatched bars represent the prey strain, V. fluvialis NCTC11327. Solid bars represent V.vulnificus attacker strains. Statistics were performed using the 2-way unpaired Student’s t-test *** = p < 0.005. (C) Schematic diagram of the V. fluvialis T6SS gene cluster and distally encoded T6SS associated genes.
However, it was also observed that when V. fluvialis was co-cultured with the T6SS1 mutant of V. vulnificus (SRC1), this latter strain appeared to become a prey strain for V. fluvialis as evidenced by a complete lack of growth of SRC1 (Fig 5A, solid black bars). Therefore, we reasoned that this observed inhibition of SRC1 growth may be attributable to a T6SS present in V. fluvialis which is active during enumeration at 37°C. To test this hypothesis, we generated antibiotic resistant V. vulnificus and V. fluvialis strains allowing for the prey and attacker strains to be enumerated on separate antibiotic plates. This approach removed the ability for intra-strain targeting to occur during enumeration at 37°C. Enumeration of a subsequent co-culture assay on antibiotic containing enumeration plates revealed that there was no inhibition of SRC1 (Fig 5B, solid black bar), suggesting that V. fluvialis may indeed be targeting SRC1 at 37°C on TCBS agar. This experiment also revealed that during the co-culture at 30°C, V. vulnificus can target V. fluvialis in a T6SS1 dependent manner as evidenced by an increase in growth of V. fluvialis when co-cultured with the SRC1 compared to wild-type and SRC2 (Fig 5B, hatched bar).
Identification of a T6SS in V. fluvialis NCTC 11327
To determine whether V. fluvialis contained a T6SS, which may be contributing to the observed targeting of SRC1, whole genome sequencing of V. fluvialis NCTC 11327 was employed. This approach, coupled with genome annotation, did identify a putative T6SS in V. fluvialis, Fig 5C. This system contains all of the 13 conserved genes required for a functional T6SS and is similar in gene organisation to T6SS1 of V. vulnificus. However, unlike V. vulnificus, V. fluvialis encodes two hcp genes, which are located distally from the large T6SS operon. This localisation of hcp genes is similar to that seen in V. cholerae [24]. Furthermore, V. fluvialis also possesses distally encoded vgrG genes, as well as a homologue of the recently identified T6SS adapter protein-1 (tap-1) (Fig 5C) [25].
The T6SSs in V. vulnificus 106-2A are not virulent towards Galleria mellonella
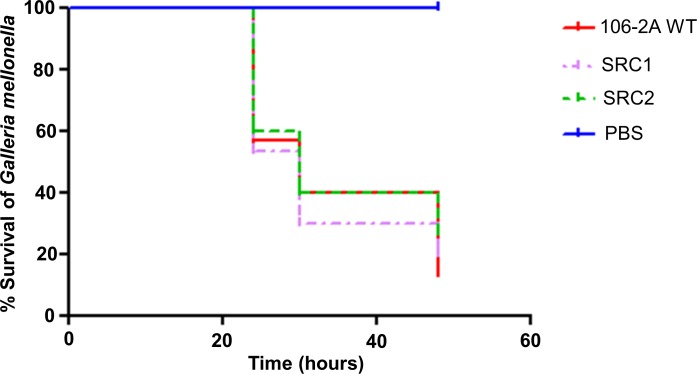
To further support our hypothesis that the T6SS1 of V. vulnificus is predominantly involved in anti-bacterial killing rather than anti-eukaryotic targeting, we investigated the role of T6SS1 in a Galleria melonella infection model. Groups of 10 larvae were injected with either, V. vulnificus 106-2A wild-type, SRC1 or SRC2 at 105 cells in the upper most pro-leg and monitored for signs of disease. The results however demonstrated that both SRC1 and SRC2 were as virulent compared to the wild-type strain, Fig 6, indicating that neither T6SS1 nor T6SS2 is required by V. vulnificus for virulence in this infection model, further supporting our hypothesis that the T6SS1 of V. vulnificus is involved in modulating the composition of environmental bacterial populations.
Fig 6. The T6SSs of V. vulnificus do not contribute towards virulence in the infection model, Galleria mellonella.
V. vulnificus is virulent in the infection model G. mellonella, however neither the icmF T6SS1 mutant, SRC1 nor the icmF T6SS2 mutant, SRC2 are attenuated in G. mellonella. Phosphate buffered saline (PBS) was used as a negative control demonstrating 100% survival. Experiments were performed three times in triplicate where n = 10 and statistics performed using GraphPad Prism version 5 using the Log-rank (Mantel-Cox) test.
Discussion
Although T6SSs have been characterised in many Vibrio species, this is the first report, to our knowledge, of a functional T6SS in V. vulnificus. Our study has shown a variation in the number of T6SSs present in ten different V. vulnificus isolates, with some having two T6SSs and others only one. This in contrast to V. cholerae, where numerous strains have been shown to possess just one conserved T6SS [26]. All V. vulnificus strains studied contained the T6SS, T6SS2 with a subset of strains containing an additional T6SS, T6SS1. Bioinformatic analysis of both T6SSs of V. vulnificus demonstrated that these systems contained the full repertoire of genes required for a functional secretion system [8]. V. vulnificus T6SS2 contained accessory genes encoding for proteins such as a phosphatase and kinase, which have previously been shown to be required for T6SS regulation in P. aeruginosa [27]. Although the accessory proteins observed in T6SS2 were not observed in T6SS1, T6SS1 does contain vasH. This gene encodes for an alternative sigma54 factor, which has previously been shown to be involved in initiation of transcription of the T6SS in V. cholerae [28, 29] and as such may have a regulatory role in the transcription of T6SS1 in V. vulnificus.
T6SS1 of V. vulnificus exhibits considerable overall synteny with the T6SS of V. cholerae [30]. However, the most notable differences between the T6SSs of these bacteria are predominantly associated with the hcp and vgrG genes. For example, V. cholerae contains two hcp and three vgrG genes, in contrast to the apparent presence of only one copy of each in the V. vulnificus T6SSs. VgrGs play a structural role in the formation of the T6SS, however several vgrGs have also been shown to encode for effector functions, such as those identified in V. cholerae, and as such are denoted as evolved VgrGs [17, 24]. Evolved vgrGs are distinguishable from structural vgrGs based on the presence of a C-terminal extension. The vgrG genes in T6SS1 and T6SS2 of V. vulnificus appear not to be evolved vgrG genes due to the lack of a C-terminal extension.
In this paper we show for the first time that T6SS1 of V. vulnificus is functional. Expression and secretion of Hcp-1 was shown to be affected by both temperature and salinity as has been observed previously in both V. cholerae and V. parahaemolyticus [18, 19]. Our initial findings demonstrated that the temperature regulation of V. vulnificus Hcp-1 was identical to those of Hcp-2 in V. parahaemolyticus [18]. In contrast to this temperature dependent similarity, salinity appeared to have a differential effect on secretion of V. vulnificus Hcp-1 relative to V. parahaemolyticus Hcp-2 [18]. Differential regulation of T6SSs in other Vibrio species is not uncommon, as V. cholerae V52 Hcp has been shown to be secreted at 37°C but Hcp of the O1 V. cholerae strain, A1552, is secreted at only 23°C [19, 30]. Our observation that Hcp-1 is not secreted at 37°C suggests that the function of V. vulnificus T6SS1 is unlikely to be associated with an anti-eukaryotic phenotype. A hypothesis supported by our observation that SRC1 displays the same virulence in a G. mellonella model of infection to that of the wild-type.
Based on T6SS1’s temperature regulation and similarity with the anti-bacterial T6SS of V. cholerae, [21] we hypothesised that V. vulnificus T6SS1 may exhibit anti-bacterial properties. Furthermore, our observation that only some strains of V. vulnificus contain this additional T6SS1 led us to propose that this system may convey a competitive advantage. Through the use of co-culturing assays this hypothesis was supported by the observation that T6SS1 positive strains do have a competitive advantage over those that are T6SS1 negative. Furthermore, mutagenesis studies demonstrated that this phenotype was attributed to T6SS1 and not T6SS2.
Given the observed antagonistic role of V. vulnificus T6SS1, it is tempting to speculate on this system’s role in the natural environment, and potentially more widely in terms of clinical impact. Taken together, we hypothesise that the limited number of serious V. vulnificus human infections compared with the high abundance of this pathogen [4] may be attributable to T6SS1 dependent killing of certain hyper virulent V. vulnificus strains. This hypothesis is based on data in this study which has shown that a lesser virulent V. vulnificus strain can target a hyper virulent strain in a T6SS1 dependent manner, leading us to suggest that the same scenario may also occur naturally in the environment. This proposed scenario would potentially lead to a decrease in the number of hyper virulent strains in the environment, which may lessen the chances of humans being exposed to hyper virulent strains, and as such only a limited number of serious cases reported each year. This scenario is appealing as our study identified a predominance of T6SS1 among lesser virulent environmental strains [5]. In accordance with our hypothesis, other researchers have also suggested that the main role of the T6SS is to attack competing cells and enhance the fitness of the secreting cell in poly-microbial communities [14, 31]. However, it should be noted that although this study identified a predominance of T6SS1 among lesser virulent strains, T6SS1 was also identified in a hyper virulent environmental strain, 99–796. Furthermore, given the increase in knowledge regarding different effector/immunity proteins in V. cholerae allowing for differing strains of V. cholerae to outcompete each other based on their effector repertoire [26] this hypothesis may be too simplistic. Initial in silico investigations into the V. vulnificus T6SS1 and T6SS2 effectors has demonstrated that V. vulnificus contains mainly rhs effectors with DNase activity. However, to fully answer our proposed hypothesis the identification of V. vulnificus T6SS1 and T6SS2 effectors and immunity proteins needs to be elucidated, a study which our laboratory is currently undertaking.
As well as intra-species targeting, bacterial species such as V. cholerae and S. marcescens utilise their T6SS for inter-species targeting [21, 32]. Therefore, we sought to determine whether V. vulnificus T6SS1 could target other bacterial species. To test this hypothesis, co-culture assays were performed using the prey strain, V. fluvialis NCTC 11327. Both the anti-bacterial properties of T6SS1 and T6SS2 were investigated as it was hypothesised that T6SS1 may be specific to intra-species targeting and T6SS2 inter-species targeting. However, the results demonstrated that T6SS2 played no role in inter-species targeting. The finding that T6SS2 plays no role in anti-bacterial killing is not unusual as bacteria containing multiple T6SSs can often utilise these system for distinct roles. For example, although B. thailandensis contains a total of five T6SS, only T6SS-1 has been shown to be anti-bacterial, while T6SS-5 has been shown to possess an anti-eukaryotic phenotype [33]. Although our studies did not demonstrate any role for V. vulnificus T6SS2, it is likely that this system is tightly regulated. This hypothesis is based on the finding that T6SS2 contains the accessory proteins, phosphatase and kinases, two proteins which have been shown to be involved with tight regulation of the T6SS, HSI-I in P. aeruginosa [27].
During co-enumeration on TCBS agar, the V. vulnificus T6SS1 mutant, SRC1 was unable to survive in the presence of V. fluvialis NCTC 11327 at 37°C. Suggesting that V. vulnificus requires T6SS1 for survival in mixed bacterial populations. Based on these findings, WGS and annotation was performed to determine whether V. fluvialis NCTC 11327 contained a T6SS which may account for the antagonistic manner of this strain. These investigations identified the presence of a T6SS locus in V. fluvialis NCTC 11327. Although the functionality of this system was not investigated, the T6SS locus contains all of the 13 genes required for a functional T6SS [8] and is similar in gene organisation to the T6SS of V. cholerae V52 [24]. Similarly, a recent bioinformatic study using the V. fluvialis strain, 85003 also identified the presence of a T6SS cluster, suggesting that the T6SS is common among V. fluvialis strains [34]. The widespread identification of the T6SS in the Vibrio family is not surprising as many Vibrio spp inhabit similar niches, therefore containing a mechanism such as the T6SS which can displace competing bacteria when resources such as space and nutrients are limited would be beneficial [14].
In addition to environmental attacks, a recent publication demonstrated the antagonistic behavior of the T6SS in vivo [31]. The authors extrapolated this information to suggest the pivotal role played by the T6SS in affecting the composition of many poly-microbial communities and the effects of this system on health and disease outcomes [31]. Similarly, we hypothesise that the T6SS may play an important role in determining the severity of V. vulnificus disease. For example, V. vulnificus and V. fluvialis share the oyster as a common niche and human infection route [4, 23] and in this study we have demonstrated that at 37°C in the absence of T6SS1, V. fluvialis can target V. vulnificus. Furthermore, we have identified that the majority of V. vulnificus strains are T6SS1 negative. Therefore, we propose that upon human ingestion of an oyster containing V. fluvialis and T6SS1 negative V. vulnificus cells, V. fluvialis may target and remove V. vulnificus. In comparison to V. vulnificus which can exhibit varying degrees of pathogenicity [5], V. fluvilais is commonly associated with mild gastroenteritis [23]. Therefore, we speculate that in this instance, V. fluvialis could acts as a “probiotic”, by targeting and removing hyper virulent V. vulnificus cells, leaving only the lesser virulent V. fluvialis for the host’s immune system to eliminate. Based on this hypothesis, it is tempting to conclude that only a limited number of serious V. vulnificus infections would be reported each year. Further work is required to understand whether the removal of V. vulnificus by V. fluvialis is due to the V. fluvialis T6SS or whether there are potentially other diffusible anti-microbial molecules secreted by V. fluvilais. This is based on the finding that V. fluvilais was able to target V. vulnificus on enumeration plates where colonies were sufficiently diluted to not be in contact. It is highly likely that V. fluvialis may secrete diffusible anti-microbial molecules as other Vibrios have previously been shown to secrete anti-microbial molecules other than those associated with the T6SS [35].
In conclusion, this study has identified for the first time two T6SSs in V. vulnificus. Furthermore, we have identified the conditions under which T6SS1 is active, and demonstrated that T6SS1 contains anti-bacterial properties. These findings have been used to hypothesise how V. vulnificus T6SS1 may be used to shape Vibrio microbial populations and how inter-bacterial interactions may explain the limited number of serious human infections attributed to V. vulnificus given the natural prevalence and virulence potential of this bacterium.
Materials and Methods
Bacterial strains, plasmids and culture conditions
V. vulnificus strains and V. fluvialis NCTC 11327, were routinely cultured at 30°C and V. choerlae V52 at 37°C in Luria-Bertani (LB) broth (Oxoid) unless otherwise stated. Where appropriate, antibiotics (Sigma) were added at the following concentrations; ampicillin (Amp) 100 μg/ml, Kanamycin (Km) 50 μg/ml, chloramphenicol (Cm) 35 μg/ml for Escherichia coli and 10 μg/ml for Vibrio vulnificus, Streptomycin 100 μg/ml and Trimethoprim (Tp) 100 μg/ml. Strains, plasmids and primers used in this study are listed in Tables 1, 2 and 3.
Table 2. Plasmids.
| Plasmids | Description | Source |
|---|---|---|
| pDM4 | Suicide vector (Cmr, R6K origin, sacBR) | Milton, et al 1996 |
| pGEM®-T Easy | High copy number cloning vector (Ampr) | Promega |
| pRK2013 | Conjugation, helper strain (Kmr) | Lab stock |
| pSCrhaB3 | Vector containing Tpr cassette | Dr. C. Hemsley |
| pBHR4-groS-RFP | Vector containing Cmr cassette | Dr. C. Hemsley |
| pSRC6a | pGEM®-T Easy containing 106-2A icmF2 left flanking region | This Study |
| pSRC7a | pGEM®-T Easy containing 106-2A icmF2 right flanking region | This Study |
| pSRC8a | pGEM®-T Easy containing 106-2A icmF1 left flanking region | This Study |
| pSRC9b | pGEM®-T Easy containing 106-2A icmF1 right flanking region | This Study |
| pSRC10 | pDM4 containing containing NdeI ligated106-2A icmF2 left flank and right flank | This Study |
| pSRC11 | pDM4 containing containing NdeI ligated106-2A icmF1 left flank and right flank | This Study |
Table 3. Primer sequences.
| Primer Name | Description | Primer Sequence 5’–3’ |
|---|---|---|
| M13 Forward | Sequencing primer | TGTAAAACGACGGCCAGT |
| M13 Reverse | Sequencing primer | CAGGAAACAGCTATGAC |
| LFF 106-2A icmF1 SmaI | 106-2A left flanking region IcmF forward | TGCCCGGGAGTGATTGGTCTGAGGCCATTG |
| LFR 106-2A icmF1 NdeI | 106-2A left flanking region IcmF reverse | TGCATATGTATCGGGTCTTGACGTAACTGG |
| RFF 106-2A icmF1 NdeI | 106-2A right flanking region IcmF forward | TGCATATGTGGGCATTCTTTCGATTGTTAG |
| RFR 106-2A icmF1 ApaI | 106-2A right flanking region IcmF reverse | TTGGGCCCCTCGGCACGATAAAGCTCTCTC |
| LFF 106-2A icmF2 SmaI | 106-2A right flanking region IcmF2 forward | TGCCCGGGACCAGAGTGCGGATTATATTTC |
| LFR 106-2A icmF2 NdeI | 106-2A left flanking region IcmF2 reverse | TGCATATGAATAACCAACAAACGGTTAAAG |
| RFF 106-2A icmF2 NdeI | 106-2A right flanking region IcmF2 forward | TACATATGGCAAGGCCGCAATCTCGCAAAG |
| RFR 106-2A I icmF2 ApaI | 106-2A right flanking region IcmF2 reverse | TCGGGCCCTGCCAATTTGAGGTAAACCATC |
| CmpDM4_F | first crossover integrant primer forward | ATGGAGAAAAAAATCACTGGATATACCACC |
| CmpDM4_R | first crossover integrant primer reverse | TTACGCCCCGCCCTGCCACTCATCGCAGTA |
| LFF_ icmF1_MutScr | Second cross over confirmation primer | GGCGGAAAGTGGAACAAAGC |
| RFR_ icmF1_MutScr | Second cross over confirmation primer | TACTCGGTTTGCTGGTACTC |
| LFF_ icmF2_MutScr | Second cross over confirmation primer | CGGCTGTGTTAGTCAGTGTG |
| RFR_ icmF2_MutScr | Second cross over confirmation primer | CTCATTCGATCACCGTTACC |
Identification of putative T6SS(s) in V. vulnificus 106-2A
WGS of V. vulnificus isolates was carried out by the Exeter Sequencing Service, University of Exeter, Exeter, UK. De-novo assembly of raw reads was performed using the A5-piepeline [36] followed by annotation of contigs using the RAST server [37]. Mapping of the putative T6SS clusters identified using RAST were performed using Clone Manager Professional Suite 8. Prior to GenBank submission assembled scaffolds were screened for contaminants against the UniVec database. Genomic features were predicted and annotated using prokka software and after manual curation submitted through the NCBI wgs portal. The genomes are accessible through the NCBI database with the following accession numbers: Vibrio vulnificus S2-22 (LKUU00000000); Vibrio vulnificus 106-2A (LMTD00000000); Vibrio vulnificus ORL-1506 (LMXV00000000); Vibrio vulnificus NSV-5830 (LMXW00000000); Vibrio vulnificus 99–796 (LMXX00000000); Vibrio vulnificus 99–743 (LMXY00000000); Vibrio vulnificus DAL-79087 (LMXZ00000000); Vibrio vulnificus DAL-79040 (LMYA00000000); Vibrio vulnificus ATL-9824 (LMYB00000000); Vibrio vulnificus S3-16 (LMTC00000000); Vibrio fluvialis NCTC11327 (LMTE00000000).
Generation of V. vulnificus SRC1 and SRC2 mutants
SRC1 and SRC2 mutants of V. vulnificus 106-2A were produced using pDM4 [38]. Flanking regions upstream and downstream of the target gene were PCR amplified using the primers described in Table 3. Correctly sequenced flanking regions were digested, purified and ligated into pDM4, resulting in pSRC10 and pSRC11, described in Table 2. pSRC10 and pSRC11 were subsequently transformed into E.coli DH5α and chromosomally introduced to V. vulnificus 106-2A using the conjugal donor pRK2013 and plated onto Cholera Medium TCBS (Oxoid) supplemented with choloramphenicol at 10 μg/ml. Meridiploid strains were confirmed using LFF or RFR flanking primers with cmpDM4_F or cmpDM4_R primers. Chromosomally confirmed first cross-over integrants were then plated onto LB supplemented with 10% sucrose (Sigma) incubated at 24°C to exercise the plasmid in a second cross-over event replacing the wildtype gene with the mutated gene. Mutants were confirmed using MutScr primers detailed in Table 3, DNA sequencing and WGS.
Generation of antibiotic resistant Vibrio strains
Antibiotic resistant strains were generated using tri-parental conjugal mating using the helper strain pRK2013 and plated onto TCBS agar containing appropriate antibiotic.
Hcp secretion analysis
Overnight V. vulnificus cultures were back-diluted to an OD590 0.03 in 25 ml volumes and grown to mid-log OD590 1.5. Cellular fractions were normalised to OD590 1 in 1ml LB, centrifuged at 2000 rpm for 3 min, supernatant removed and pellets re-suspended in 50 μl bug buster, 25 μl PBS and 25 μl 4x loading buffer and heated at 100°C for 10min. Supernatants were filter sterilised using a 0.2 μm filter and proteins precipitated with 100% TCA on ice for 45mins and centrifuged at 20,000 g for 5mins at 4°C. Pellets were washed with acetone, heat dried at 95°C and re-suspended in sterile PBS. Precipitated proteins and cell pellets were quantified using the BCA assay (Thermo Fisher) and equal protein loadings were subjected to SDS-PAGE and Western blotting using primary Hcp-1 antibody [17] (1:500) and secondary anti-rabbit antibody. All were performed in conjunction with the strain SRC1 as a lysis control. Blots were visualised using Odyssey CLx Infared imaging system.
Co-culture assays using TCBS enumeration plates
Overnight cultures of Vibrio strains were adjusted to OD590 0.03 and grown to OD590 ~1.00, cultures were then re-adjusted to OD590 0.8. Attacker and prey strains were then mixed at a ratio of 3 mls: 1 mls respectively. 200 ml of these mixed cultures were then incubated at 30°C for 5 hours on LB plates to allow contact dependent-killing in triplicate. Following incubation, co-cultures were harvested from the plates and re-suspended in 1 ml of sterile PBS for enumeration. 10-fold serial dilutions of these suspensions were prepared and 10 μl of each dilution was spotted onto TCBS agar plates in triplicate. These enumeration plates were then incubated at 37°C overnight. The average cfu/ml was recorded for each triplicate dilution.
Co-culture assays using selective antibiotic enumeration plates
Assays were performed as described above with the following modifications: prey and attacker strains were grown overnight in LB containing chloramphenicol (10mg/ml) and trimethoprim (100 mg/ml) respectively. Prior to mixing of the prey and attacker strains, overnight cultures were centrifuged and re-suspended in fresh LB media containing no antibiotic to prevent killing due to the antibiotics. Co-cultures using V. fluvialis NCTC 11327 as prey were incubated on LB plates at 30°C for five hours and co-cultures using V. vulnificus MO6-24/O as prey were incubated on LB plates at 30°C for two hours. Following incubation to determine T6SS dependent killing, co-cultures were harvested from the plates and re-suspended in 1 ml of sterile PBS for enumeration. 10-fold serial dilutions of these suspensions were prepared and 10 μl of each dilution was spotted in triplicate onto LB agar plates that contained trimethoprim and onto another set of LB plates containing chloramphenicol. These enumeration plates were then incubated at 37°C overnight. The average cfu/ml was recorded for each triplicate dilution.
G. mellonella infection assay
G. mellonella weighing between 2.0 and 3.0 g were injected in the upper most right proleg with V. vulnificus cells that had been grown overnight at 37°C in LB using a 25 μl Hamilton Microlitre™ syringe 800 series with removable needle (Sigma). Overnight bacterial cultures were adjusted to ~ 1x107 cfu/ ml (equating to ~ 1x105 cfu/10 μl). Enuermation of the infective dose was also performed to ensure the correct bacterial input. Infected larvae were then incubated at 37°C and monitored for signs of infection, death was recorded when larvae failed to respond to touch.
Supporting Information
Boxed maps are those of previously sequenced reference strains.
(PDF)
(PDF)
(PDF)
Acknowledgments
We would like to thank Stefan Pukatzki for the gift of the Hcp antibody, Claudia Hemsley for help with figure preparations and Ken Haynes and Tom Richards for critical review of the manuscript.
Data Availability
The genomes are accessible through the NCBI database with the following accession numbers: Vibrio vulnificus S2-22 (LKUU00000000); Vibrio vulnificus 106-2A (LMTD00000000); Vibrio vulnificus ORL-1506 (LMXV00000000); Vibrio vulnificus NSV-5830 (LMXW00000000); Vibrio vulnificus 99-796 (LMXX00000000); Vibrio vulnificus 99-743 (LMXY00000000); Vibrio vulnificus DAL-79087 (LMXZ00000000); Vibrio vulnificus DAL-79040 (LMYA00000000); Vibrio vulnificus ATL-9824 (LMYB00000000); Vibrio vulnificus S3-16 (LMTC00000000); Vibrio fluvialis NCTC11327 (LMTE00000000).
Funding Statement
This work was funded by a CEFAS-Exeter University Alliance PhD Studentship awarded to SRC and SLM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Baker-Austin C, Lemm E, Hartnell R, Lowther J, Onley R, Amaro C, et al. pilF polymorphism-based real-time PCR to distinguish Vibrio vulnificus strains of human health relevance. Food Microbiol. 2012;30(1):17–23. Epub 2012/01/24. doi: S0740-0020(11)00222-X [pii] 10.1016/j.fm.2011.09.002 . [DOI] [PubMed] [Google Scholar]
- 2.Kwak JS, Jeong HG, Satchell KJ. Vibrio vulnificus rtxA1 gene recombination generates toxin variants with altered potency during intestinal infection. Proc Natl Acad Sci U S A. 2011;108(4):1645–50. Epub 2011/01/12. doi: 1014339108 [pii] 10.1073/pnas.1014339108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bier N, Bechlars S, Diescher S, Klein F, Hauk G, Duty O, et al. Genotypic diversity and virulence characteristics of clinical and environmental Vibrio vulnificus isolates from the Baltic Sea region. Appl Environ Microbiol. 2013;79(12):3570–81. 10.1128/AEM.00477-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strom MS, Paranjpye RN. Epidemiology and pathogenesis of Vibrio vulnificus. Microbes Infect. 2000;2(2):177–88. Epub 2000/04/01. doi: S1286-4579(00)00270-7 [pii]. . [DOI] [PubMed] [Google Scholar]
- 5.Thiaville PC, Bourdage KL, Wright AC, Farrell-Evans M, Garvan CW, Gulig PA. Genotype is correlated with but does not predict virulence of Vibrio vulnificus biotype 1 in subcutaneously inoculated, iron dextran-treated mice. Infect Immun. 2011;79(3):1194–207. Epub 2011/01/05. doi: IAI.01031-10 [pii] 10.1128/IAI.01031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang W, Lee NY, Kim J, Lee MA, Kim KS, Lee KH, et al. Functional characterization of EpsC, a component of the type II secretion system, in the pathogenicity of Vibrio vulnificus. Infect Immun. 2011;79(10):4068–80. Epub 2011/07/27. doi: IAI.05351-11 [pii] 10.1128/IAI.05351-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeong HG, Satchell KJ. Additive Function of Vibrio vulnificus MARTX(Vv) and VvhA Cytolysins Promotes Rapid Growth and Epithelial Tissue Necrosis During Intestinal Infection. PLoS Pathog. 2012;8(3):e1002581 Epub 2012/03/30. 10.1371/journal.ppat.1002581 PPATHOGENS-D-11-01887 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: a beginner's guide. Curr Opin Microbiol. 2008;11(1):3–8. Epub 2008/02/22. doi: S1369-5274(08)00007-6 [pii] 10.1016/j.mib.2008.01.006 . [DOI] [PubMed] [Google Scholar]
- 9.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483(7388):182–6. 10.1038/nature10846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500(7462):350–3. 10.1038/nature12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma LS, Narberhaus F, Lai EM. IcmF family protein TssM exhibits ATPase activity and energizes type VI secretion. J Biol Chem. 2012;287(19):15610–21. 10.1074/jbc.M111.301630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cascales E, Cambillau C. Structural biology of type VI secretion systems. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2012;367(1592):1102–11. 10.1098/rstb.2011.0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang F, Waterfield NR, Yang J, Yang G, Jin Q. A Pseudomonas aeruginosa type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host Microbe. 2014;15(5):600–10. 10.1016/j.chom.2014.04.010 . [DOI] [PubMed] [Google Scholar]
- 14.Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol. 2014;12(2):137–48. Epub 2014/01/05. doi: nrmicro3185 [pii] 10.1038/nrmicro3185 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang X, Moore R, Wilton M, Wong MJ, Lam L, Dong TG. Identification of divergent type VI secretion effectors using a conserved chaperone domain. Proc Natl Acad Sci U S A. 2015. 10.1073/pnas.1505317112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406(6795):477–83. 10.1038/35020000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma AT, McAuley S, Pukatzki S, Mekalanos JJ. Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe. 2009;5(3):234–43. Epub 2009/03/17. doi: S1931-3128(09)00068-7 [pii] 10.1016/j.chom.2009.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salomon D, Gonzalez H, Updegraff BL, Orth K. Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS One. 2013;8(4):e61086 10.1371/journal.pone.0061086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishikawa T, Sabharwal D, Broms J, Milton DL, Sjostedt A, Uhlin BE, et al. Pathoadaptive conditional regulation of the type VI secretion system in Vibrio cholerae O1 strains. Infect Immun. 2012;80(2):575–84. Epub 2011/11/16. doi: IAI.05510-11 [pii] 10.1128/IAI.05510-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wright AC, Simpson LM, Oliver JD, Morris JG Jr. Phenotypic evaluation of acapsular transposon mutants of Vibrio vulnificus. Infect Immun. 1990;58(6):1769–73. Epub 1990/06/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci U S A. 2010;107(45):19520–4. Epub 2010/10/27. doi: 1012931107 [pii] 10.1073/pnas.1012931107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maugeri TL, Caccamo D, Gugliandolo C. Potentially pathogenic vibrios in brackish waters and mussels. J Appl Microbiol. 2000;89(2):261–6. . [DOI] [PubMed] [Google Scholar]
- 23.Ramamurthy T, Chowdhury G, Pazhani GP, Shinoda S. Vibrio fluvialis: an emerging human pathogen. Front Microbiol. 2014;5:91 10.3389/fmicb.2014.00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A. 2007;104(39):15508–13. Epub 2007/09/18. doi: 0706532104 [pii] 10.1073/pnas.0706532104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tietze R, Zaloga J, Unterweger H, Lyer S, Friedrich RP, Janko C, et al. Magnetic nanoparticle-based drug delivery for cancer therapy. Biochem Biophys Res Commun. 2015. 10.1016/j.bbrc.2015.08.022 . [DOI] [PubMed] [Google Scholar]
- 26.Unterweger D, Miyata ST, Bachmann V, Brooks TM, Mullins T, Kostiuk B, et al. The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nature communications. 2014;5:3549 10.1038/ncomms4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol. 2007;9(7):797–803. Epub 2007/06/15. doi: ncb1605 [pii] 10.1038/ncb1605 . [DOI] [PubMed] [Google Scholar]
- 28.Bernard CS, Brunet YR, Gavioli M, Lloubes R, Cascales E. Regulation of type VI secretion gene clusters by sigma54 and cognate enhancer binding proteins. J Bacteriol. 2011;193(9):2158–67. 10.1128/JB.00029-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitaoka M, Miyata ST, Brooks TM, Unterweger D, Pukatzki S. VasH is a transcriptional regulator of the type VI secretion system functional in endemic and pandemic Vibrio cholerae. J Bacteriol. 2011;193(23):6471–82. 10.1128/JB.05414-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A. 2006;103(5):1528–33. Epub 2006/01/25. doi: 0510322103 [pii] 10.1073/pnas.0510322103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerc AJ, Diepold A, Trunk K, Porter M, Rickman C, Armitage JP, et al. Visualization of the Serratia Type VI Secretion System Reveals Unprovoked Attacks and Dynamic Assembly. Cell Rep. 2015;12(12):2131–42. 10.1016/j.celrep.2015.08.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murdoch SL, Trunk K, English G, Fritsch MJ, Pourkarimi E, Coulthurst SJ. The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J Bacteriol. 2011;193(21):6057–69. 10.1128/JB.05671-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, Hood RD, et al. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 2010;6(8):e1001068 Epub 2010/09/25. 10.1371/journal.ppat.1001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu X, Liang W, Wang Y, Xu J, Zhu J, Kan B. Identification of genetic bases of vibrio fluvialis species-specific biochemical pathways and potential virulence factors by comparative genomic analysis. Appl Environ Microbiol. 2014;80(6):2029–37. 10.1128/AEM.03588-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wietz M, Mansson M, Gotfredsen CH, Larsen TO, Gram L. Antibacterial compounds from marine Vibrionaceae isolated on a global expedition. Marine drugs. 2010;8(12):2946–60. 10.3390/md8122946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tritt A, Eisen JA, Facciotti MT, Darling AE. An integrated pipeline for de novo assembly of microbial genomes. PLoS One. 2012;7(9):e42304 Epub 2012/10/03. 10.1371/journal.pone.0042304 PONE-D-11-23103 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. Epub 2008/02/12. doi: 1471-2164-9-75 [pii] 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milton DL, O'Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol. 1996;178(5):1310–9. Epub 1996/03/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Boxed maps are those of previously sequenced reference strains.
(PDF)
(PDF)
(PDF)
Data Availability Statement
The genomes are accessible through the NCBI database with the following accession numbers: Vibrio vulnificus S2-22 (LKUU00000000); Vibrio vulnificus 106-2A (LMTD00000000); Vibrio vulnificus ORL-1506 (LMXV00000000); Vibrio vulnificus NSV-5830 (LMXW00000000); Vibrio vulnificus 99-796 (LMXX00000000); Vibrio vulnificus 99-743 (LMXY00000000); Vibrio vulnificus DAL-79087 (LMXZ00000000); Vibrio vulnificus DAL-79040 (LMYA00000000); Vibrio vulnificus ATL-9824 (LMYB00000000); Vibrio vulnificus S3-16 (LMTC00000000); Vibrio fluvialis NCTC11327 (LMTE00000000).