Abstract
Septins are highly conserved filamentous proteins first characterized in budding yeast and subsequently identified in all eukaryotes. Septins can bind and hydrolyze GTP which is intrinsically related to their formation of septin hexamers and functional protein interactions. The human septin family is composed of 14 loci, SEPT1-SEPT14, which encodes dozens of different septin proteins. Their central GTPase and polybasic domain regions are highly conserved but they diverge in their N-terminus and/or C-terminus. The mechanism by which the different isoforms are generated is not yet well understood, but one can hypothesize that the use of different promoters and/or alternative splicing could give rise to these variants.
Septins perform diverse cellular functions according to tissue expression and their interacting partners. Functions identified to date include cell division, chromosome segregation, protein scaffolding, cellular polarity, motility, membrane dynamics, vesicle trafficking, exocytosis, apoptosis, and DNA damage response. Their expression is tightly regulated to maintain proper filament assembly and normal cellular functions. Alteration of these proteins, by mutation or expression changes, has been associated with a variety of cancers and neurological diseases. The association of septins with cancer results from expression alterations in solid tumorsor translocations in leukemias (MLL). Expression changes in septins have also been associated with neurological conditions such as Alzheimer’s and Parkinson’s disease, as well as retinopathies, Hepatitis C, spermatogenesis and Listeria infection. Pathogenic mutations of SEPT9 were identified in the autosomal dominant neurological disorder Hereditary Neuralgic Amyotrophy (HNA).
Human septin research over the past decade has established their importance in cell biology and human disease. Further functional characterization of septins is crucial to our understanding of their possible diagnostic, prognostic, and therapeutic applications.
Keywords: cancer, cell division, GTPases, human septins, neurological disorders, SEPT9
Introduction
Forty years ago, septin’s pioneer researchers may have never imagined that their discovery of four novel yeast genes would open the door to a biologically complex world of cytoskeletal GTPases that were highly relevant to human disease. Septins were first characterized in Saccharomyces cerevisiae as a group of cytoskeletal GTP-binding proteins essential for proper cell division. Genetic screening performed by Hartwell and colleagues for budding yeast mutants lead to the discovery of the first septin genes, CDC3, CDC10, CDC11 and CDC12, in which temperature sensitive mutations caused defects in budding morphology and cell cycle arrest (1). These genes were further characterized in Saccharomyces pombe in which mutations led to cytokinesis defects during mitotis. Subsequently, septins have been found to be highly conserved in all eukaryotes except plants, and many of the loci encode for different variants and/or protein isoforms (Table 1).
Table 1.
Human septins variants and isoforms per loci**
| Human septin loci | Number of recognized variants/isoforms | Variable region(s) |
|---|---|---|
| SEPT1 | - | - |
| SEPT2 | 4 transcripts/1 isoform | 5′UTR and 3′UTR |
| SEPT3 | 2 isoforms | 3′UTR and C-termini |
| SEPT4 | 3 isoforms | 5′UTR and 3′UTR N-and C-termini |
| SEPT5 | - | - |
| SEPT6 | 4 isoforms | 3′UTR C-termini |
| SEPT7 | 2 isoforms | alternative splicing exon2 |
| SEPT8 | 4 isoforms | 5′UTR and 3′UTR N-and C-termini |
| SEPT9 | 7 transcripts/6 isoforms | 5′UTR and 3′UTR alternative splicing of 5′ exons |
| SEPT10 | 2 isoforms | alternative splicing exon 2 |
| SEPT11 | - | - |
| SEPT12 | 2 isoforms | alternative splicing central exon |
| SEPT13 | - | - |
| SEPT14 | - | - |
Data gathered from NCBI Reference Sequence (Refseq)
Recently, phylogenetic and evolutionary analysis for septins of metazoans demonstrated that all septin proteins could be clustered into four major subgroups (Fig 1). It has been proposed that the emergence of these four subgroups occurred prior to the divergence of vertebrates and invertebrates and that septin expansion in number was due to duplication of pre-existing genes (2). Septin research has grown impressively, from four genes identified in budding yeast to 14 core loci identified in humans to date. The variety of existing septin isoforms and the diversity of their biological functions in eukaryotic cells are both intriguing and exciting. Contributions to septin biology over the last few years include the first crystal structure of septin oligomers, septin protein interactions, expression profiles, associations with well known signaling pathways and involvement in human disease. These observations have set the stage for more exciting experimental strategies to further understand the role of septins in health and disease. Many questions remain related to their multiple physiological functions in cells as related to normal development and health, and their implicated roles in the development of human diseases.
Figure 1. Phylogenetic tree of the human septin family.
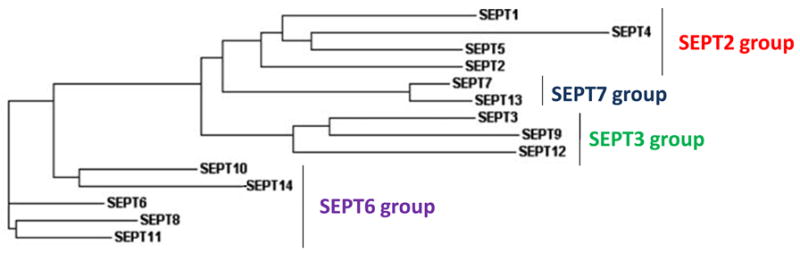
A dendrogram tree illustrates the phylogenetic relationship of the human septin family members as determined by Clustal W analysis (modified from Peterson E.A., et al. (2008) Mamm Genome).
Human septin family conserved domains
The four human septin subgroups are characterized by sequence homology and domain composition (Fig 1 and Fig 2). All human septins share a highly conserved central region that contains a polybasic domain and the GTP-binding domain. The N- and C-terminal regions vary in length and amino acid composition. They contain a proline-rich domain and an α-helical coiled-coil domain, respectively. The relevance of the differences in the N- and C- termini of some septins and their isoforms is not completely understood, but these variations might play a role in their unique cellular localizations, protein interactions, and biological functions. While the specific functions of these domains remain to be fully elucidated, recent studies have begun to reveal information about the possible roles of these domains, as described later on. Several members of the human septin family also contain the less well studied proline-rich domain near the N terminal region. Even though there are no specific studies dedicated to understanding the functional role of the proline-rich domain of septins, one can hypothesize that this domain may be essential for protein-protein interactions, including interaction with proteins containing SH3 domains (3, 4). Preceding the GTPase domain, all mammalian septins, including human septins, contain a highly conserved polybasic domain responsible for the association of septins with cellular membranes. Studies of the polybasic domain of SEPT4 have shown that this domain binds to phosphoinositol phosphates (5). The polybasic domain might mediate the targeting of septins to membrane domains relevant for their role as diffusion barriers in yeast and mammalian cells. The Wittinghofer team studied the human septin complex composed of SEPT2, SEPT6 and SEPT7 and found that these three septins form a hexameric complex of two copies of each septin through a GTP/GDP bound interface, which is also crucial for the GTPase activity (6, 7). Future studies of these domains are necessary for comprehensive insight into septins’ intricate functional roles in mammalian cells.
Figure 2. General structure of septins.
N- and C- termini vary between Septin family members and their isoforms. Some septins contain a proline rich domain in their amino terminus that is involved in protein interaction. The C- terminus of some septins consists of a coiled-coil domain predicted to contain a α-helix. The polybasic domain is responsible for the association of septins with the plasma membrane. The central GTP-binding domain contains conserved motifs G1 (GxxxxGK[S/T]), G3 (DxxG) and G4 (xKxD) and is implicated in the formation of septin heteroligomers and protein interaction.
Septins are widely subjected to functionally significant post-translational modifications. For example, sumoylation of yeast septins in G2/M-arrested cells is well described, and the ubiquitin-protein ligase (E3) Siz1p is required for yeast septin sumoylation (8). Sumoylation of mammalian septins has not yet been reported. However, mammalian septins contain multiple phosphorylation sites and they can be phosphorylated by Ser/Thr kinases in post-mitotic neurons (9–11) and by Aurora-B kinase in mitotic cells (12). This suggests that sumoylation and/or phosphorylation of septins might modulate conformational changes that can alter their functional role.
Functional role of septins
Despite the mechanistic differences between budding yeast and dividing animal cells (13), several orthologs of yeast septins were first identified in Caenorhabditis elegans (unc-59 and unc-61), Drosophila melanogaster (pnut) and mammals (SEPT2 (Nedd5) and SEPT4(H5)) (14–17). Similar to yeast septins, animal septins can form polymeric actin-associated filaments, hydrolyze GTP and produce multinucleated cells when mutated (18, 19). Septins can form microfilaments by interacting with each other or with cytoskeletal and filamentous proteins such as actin, myosin and tubulin, indicative of their functional roles in cytokinesis during contractile ring formation, cell morphology changes and dynamic scaffolds (14, 20–24). Several mammalian septins have many interacting partners (Table 2) and form complexes between family members to create filaments important in different cellular functions. One can speculate that some of the diverse functional roles of mammalian septins are intrinsically related to their physical interactions, which might explain how one septin can have multiple functions in different tissues and at different times during cell cycle and development.
Table 2.
Representative interacting partners and functional roles of mammalian septins
| Complexes | Putative Functions | Implicated Septin Domain | Reference |
|---|---|---|---|
| SEPT1/Aurora B | Chromosome segregation, Cytokinesis | not determined | Qi et al., 2005 |
| SEPT2/F-Actin | Cytokinesis | GTP binding | Kinoshita et al., 1997 |
| SEPT2/5/6/Anillin | Cytokinesis | not determine | Kinoshita et al., 2002 |
| SEPT2/6/7 | Filament formation | C- & N- termini | Low et al., 2006 |
| SEPT2/GLAST | Neurotransmitter release | not determined | Kinoshita et al., 2004 |
| SEPT2/myosin II | Cytokinesis | not determined | Joo et al., 2007 |
| SEPT2/6/CENP-E | Chromosome segregation | not determined | Spiliotis et at., 2005 |
| SEPT2/6/7/MAP4 | Microtubule stability | not determined | Kremer et al., 2005 |
| SEPT2/4/7/Sec6/8 | Vesicle trafficking | not determined | Hsu et al., 1998 |
| SEPT3/5/7 | Neuronal biology | not determined | Fujishuma et al., 2007 |
| SEPT4/5/8 | Vesicle Targeting/Exocytosis | N- & C- termini | Martinez et al., 2006 |
| SEPT4/8 | Platelet biology | not determined | Blaser et al., 2004 |
| SEPT5/parkin | Parkinson’s pathogenesis | not determined | Choi et al., 2003 |
| SEPT5/11 | Exocytosis in endothelial cells | GTP binding & C- termini | Blaser et al., 2006 |
| SEPT5/SNARE complex | Vesicle Targeting/Exocytosis | GTP binding & C- terminal | Beites et al., 2005 |
| SEPT6/12 | Filament formation | not determined | Ding et al., 2007 |
| SEPT7/9/11/Actin | Filament formation | N-terminal | Nagata et al., 2004 |
| SEPT7/CENP-E | Chromosome segregation | not determined | Zhu et al., 2008 |
| SEPT8/Vamp2 | Snare complex formation Neurotransmitter release | not determined | Ito et al., 2009 |
| SEPT9/Actin | Stress Fiber | not determined |
Nagata et al., 2005 González et al., 2007 |
| SEPT9/SA-RhoGEF/Actin | Rho Signaling | N-terminal | Nagata et al., 2005 |
| SEPT9/HIF-1 | Cell proliferation, angiogenesis, prostate Cancer | N-terminus GTP binding |
Amir et al., 2006 Amir et al., 2009 |
| SEPT9/JNK | Cell proliferation, breast cancer | GTP binding | Gonzalez et al., 2009 |
| SEPT9/Tubulins | Filament formation, microtubules, spindle formation, | GTP binding |
Surka et al., 2002 Nagata et al., 2003 González et al., 2007 |
| SEPT11/12 | Filament formation | not determined | Ding et al., 2008 |
| SEPT14/SEPT 1-7/9/11-12 | Testicular biology | not determined | Peterson et al., 2008 |
In addition to their roles in cytokinesis, mammalian septins have been associated with other distinct cellular processes including vessicle trafficking, cell polarity, cytoskeletal dynamics, apoptosis, neurodegeneration and oncogenesis. The diversity and complexity of these septin-associated functions is not well understood, but it is believed that a wide range of hetero-oligomeric septin complexes are major players in these cellular processes. For example, SEPT4/ARTS, an alternative transcript of SEPT4, is translocated from the mitochondrion to the nucleus upon exposure to the apoptotic agent TGF-β, possibly providing a mechanistic link between cell division and cell death (25–27). Recent data indicate that several septins, including SEPT2, 4, 6 and 7, co-precipitate with the mammalian sec6/8 complex, suggesting an association of septins with membrane dynamics and vesicle trafficking (28–31). In addition, septins have been implicated in platelet function (SEPT4, 5 and 8) (32–35), cardiac myocyte development (36), chromosome dynamics (SEPT2) (37), cell polarity, motility and microtubule dynamics (SEPT9 and others) (20, 22–24, 38).
In a very elegant study, Kremer et al. showed that a complex of septin proteins (SEPT2, 6 and 7) interacts with SOCS7, which is necessary to retain SOCS7 and NCK in the cytoplasm at a steady state to regulate actin organization and the DNA damage response. The SOCS7/NCK complex accumulates in the nucleus to activate CHK2 and p53 mediated cell cycle arrest upon DNA damage. This action is potentiated by the depletion of the septin complex by siRNA. The spatial distribution of NCK as mediated by the septin complex is important in the reorganization of the actin cytoskeleton, cell polarity and the DNA damage response. One can predict that in vivo the distribution of septins in the cytoplasm is an important mechanism that regulates the localization of many proteins and their site of action in the cell (39).
SEPT2 and SEPT11 are required for phagosome formation which is important for cell membrane dynamics (31). SEPT2 is also required for epithelial cell polarity by associating with tubulin networks and facilitating vesicle transport by preventing polyGlu microtubule tracts from binding to MAP4 (38). SEPT12 has been implicated in mammalian spermatogenesis (40, 41). In addition, SEPT4 and SEPT7 serve as diagnostic markers for human male asthonozoospermia, since healthy individuals show septin expression in the annuli of spermatozoa while infertile patients do not (42). Ihara et al. demonstrated that expression of murine Sept4 is important for the cortical organization required for morphology and motility of the sperm flagellum (43). SEPT2 and SEPT11 are modulators of InlB-mediated invasion by Listeria monocytogenes. SEPT2 is essential for the entry of Listeria to the cells, but SEPT11 expression restricts the efficacy of Listeria invasion. Overall, these findings demonstrate the diverse roles of septins despite their high structural conservation (44–46).
Most recently SEPT14, a novel testis-specific septin was identified and characterized as the newest member of the human septin family. This septin was identified in an effort to find novel interactors of SEPT9 isoforms using a yeast-two hybrid system. SEPT14 maps to 7p11.2 in humans and includes a conserved GTPase domain and a carboxy-terminus coil-coiled domain characteristic of other septins. SEPT14 was found to interact with 10 other members of the human septins and localized with actin stress fibers, a general feature of septin filaments. Expression analyses in many fetal, adult and cancer tissues showed that SEPT14 expression is limited to the normal testes by Northern blot and RT-PCR. Interestingly, SEPT14 expression was negative when tested in cancer cell lines isolated from testis (47).
Septins as microtubule associated proteins
Several studies suggest that septins might be involved in microtubule dynamics and chromosome segregation. Two groups showed that mammalian SEPT2 and SEPT7 may form a mitotic scaffold for CENP-E and other effectors to coordinate chromosome segregation and cytokinesis (37, 48). Kremer et al. proposed a novel molecular function for septins in mammalian cells through the modulation of microtubule dynamics via an interaction with MAP4 (22). Looking specifically at SEPT9 isoforms, SEPT9_v1 (MSFA) was found to localize specifically with microtubules. This localization was required for the completion of cytokinesis and it could be disrupted by nocodazole treatment (24). Interaction of SEPT9_v1 with microtubule networks was found to be essential for septin filament formation (23). We propose that there is a tight regulation between tubulin filaments, microtubules and septins filaments. How this regulation is mediated, and what other proteins are involved remains unclear. Fluorescence microscopy and further characterization of protein interactions might give better insight into septin-tubulin interaction and its regulation of chromosome segregation during mitosis.
Septins and neurological disorders
Possible roles for septins in neurological disorders have emerged based on the brain-specific expression of some septins (Table 3). SEPT2/NEDD5, SEPT1/DIFF6 and SEPT4/H5 have been found to associate with Alzheimer-specific neurofibrillary tangles (5, 49). The SEPT5/CDCREL-1 interaction with Parkin, a pathogenic protein in Parkinson’s disease (50, 51), provides evidence for the involvement of another subset of septins in neuronal development and disease. SEPT4 has also been implicated in brain pathogenesis by its association with cytoplasmic inclusions in Parkinson’s disease and other synocleinopathies (52). The SEPT3/5/7 complex was identified in the mammalian brain and SEPT3 specifically is developmentally regulated and enriched in presynaptic nerve terminals, suggesting a role for these septins in neuronal biology (53, 54). SEPT2 and SEPT8 are associated with neurotransmitter release due to their interaction with the glutamate transporter (Glast) and the synaptic vesicle protein synaptobrevin 2 (Vamp2), respectively (55, 56). Other septin complexes have been associated with myelin formation in the peripheral nervous system (57). Recently, SEPT9 point mutations and a duplication within the gene have been identified in patients with the autosomal dominant neuropathy Hereditary Neuralgic Amyotrophy (HNA) (58–60), further supporting a role for septins in neurological disease. The mechanism by which these mutations are pathogenic in HNA is still not fully elucidated, but McDade et al. showed that isoforms v4 and v4* have distinct 5′ ends encoded by exons where germline mutations of HNA are found. The two mRNAs are translated with different efficiencies and cellular stress can alter this pattern (61). These mutations dramatically enhance the translational efficiency of the v4 5′-UTR, leading to elevated SEPT9_v4 protein under hypoxic conditions (61). These data provide mechanistic insight into the effect of HNA mutations on fine control of SEPT9_v4 protein and its regulation under physiologically relevant conditions. The data are consistent with the episodic and stress-induced nature of the clinical features of HNA (61).
Table 3.
Characteristics of human septin loci and disease associations
| Human Septin | Alternative Names | Human Chromosome Location | Expression | Disease Associations |
|---|---|---|---|---|
| SEPT1 | DIFF6 | 16p11.1 | Brain, Lymphocytes, Others | Alzheimer’s disease, Leukemia, Lymphoma |
| SEPT2 | NEDD5, DIFF6 | 2q37 | Brain, Lymphocytes, Others | Brain, Liver and Renal cancer, Von Hippel-Lindau syndrome, Alzheimer’s disease Shigella and Listeria infections |
| SEPT3 | SEP3, G-SEPTIN | 22q13.2 | Brain-specific | Brain cancer, Alzheimer’s disease |
| SEPT4 | ARTS, H5, PNUTL2, BRADEION, SEP4 | 17q22 | Brain, Testes, Eye, Lymphocytes | Alzheimer’s disease, Skin, Urogenital and Colon cancer, Leukemia, Male infertility (spermatogenesis defect) |
| SEPT5 | CDCREL-1, PNUTL1 | 22q11.21 | Brain, Eye, Platelets | Leukemia, Parkinson’s disease, Schizophrenia, Pancreatic Cancer |
| SEPT6 | SEP2, KIAA0128 | Xq24 | Ubiquitous | Schizophrenia, Leukemia, Hepatitis C |
| SEPT7 | CDC10 | 7q36.1 | Brain, Testes | Nervous system cancers, Male infertility (spermatogenesis defect) |
| SEPT8 | KIAA0202 | 5q31 | Brain, Retina and others | Retinal Degeneration |
| SEPT9 | MSF, PNUTL4 AF17Q25, OvBrSEPT, SINT1, KIAA0991 | 17q25 | Ubiquitous | Hereditary Neuralgic Amyotrophy, Leukemia, Breast, Ovarian and Colorectal cancers, Head and Neck cancers, classical Hodgkin lymphoma, Shigella and Listeria infections |
| SEPT10 | SEPT1-like | 2q13 | Ubiquitous | - |
| SEPT11 | FLJ10849 | 4q21.1 | Ubiquitous | Schizophrenia, Bipolar Disorder, Leukemia, Shigella and Listeria infections |
| SEPT12 | FLJ25410 | 16p13.3 | Lymphocytes, Testes | Male Infertility (spermatogenesis defect) |
| SEPT13 | - | 7p13 | Ubiquitous | - |
| SEPT14 | - | 7q11 | Testes | - |
Septins in cancer
The first evidence that septins may contribute to neoplasia occurred with the discovery of septin SEPT5/CDCREL-1 as a carboxy-terminus fusion partner with mixed lineage leukemia (MLL) in an acute myeloid leukemia (AML) patient with a t(11;22)(q23;q11.2) translocation (62). Subsequently, SEPT9/MSF, SEPT6, SEPT2, and SEPT11 were also identified as fusion partners with MLL in human leukemic cells (63–66). MLL, the human homolog of Drosophila trithorax, is a common translocation partner in leukemias with more than 80 rearrangements with 50 fusion partners identified (67). The well-characterized MLL fusion products produce in-frame translated chimeric proteins associated with phenotypic disease variability such as leukemia-type and prognostic outcome. These carboxy partners, in addition to MLL, appear to be essential contributors to the pathogenesis of leukemia (68).
Five human septins have also been implicated in other types of cancer including SEPT2, SEPT3, SEPT4, SEPT5 and SEPT9 (25, 69, 70). In fact, SEPT4 acts as a tumor suppressor in leukemias and solid tumors. The SEPT4 gene promotes apoptosis, and is lost in 70% of leukemia patients (26), in addition to being associated with inhibition of colorectal cancer tumorigenesis (71). SEPT9 seems to be important not only in leukemias but also in solid tumor cancers (72–77). SEPT2 and SEPT3 were abundantly expressed in several brain tumors and brain tumor cell lines, suggesting that these family members are potential oncogenes (70). SEPT2 phosphorylation by casein kinase 2 was found to be important for hepatoma carcinoma cell proliferation (78). In addition, SEPT2 is the only human septin that has been associated with a specific human cancer predisposition syndrome, Von Hippel-Lindau (VHL). VHL is anautosomal dominant genetic condition characterized by hemangioblastomas in different tissues and renal cell carcinoma. Increased SEPT2 expression is a common event in renal cell carcinomas (78, 79). A proteomic study by Craven et al. showed that SEPT2 was downregulated upon expression of VHL in the UMRC2 renal cell carcinoma line. This suggests that SEPT2 is subject to either direct or indirect VHL regulation(78, 79).
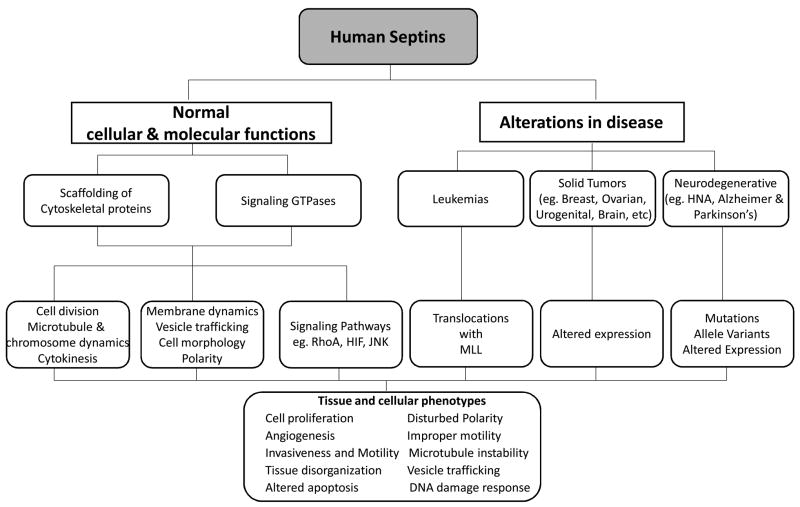
Overall, these studies show that human septins are important in many cellular and molecular functions, as summarized in Fig 3. It seems that their expression needs to be highly regulated and the dynamics with their interaction partners are crucial for their function as either scaffolds for cytoskeletal proteins or signaling GTPases. Mutations or altered expression of these genes can arise by multiple mechanisms that are not well understood. Many septins are strikingly associated with multiple human diseases, primarily cancers and neurological disorders (Table 3 and Fig 3).
Figure 3. Human septins in health and disease.
Human septins are involved in multiple tissue and cellular phenotypes. Alterations in their expression or gene mutations affect these phenotypes, which are mechanistically important in the pathogenesis of many diseases such neurological diseases and cancer.
Human SEPT9 locus
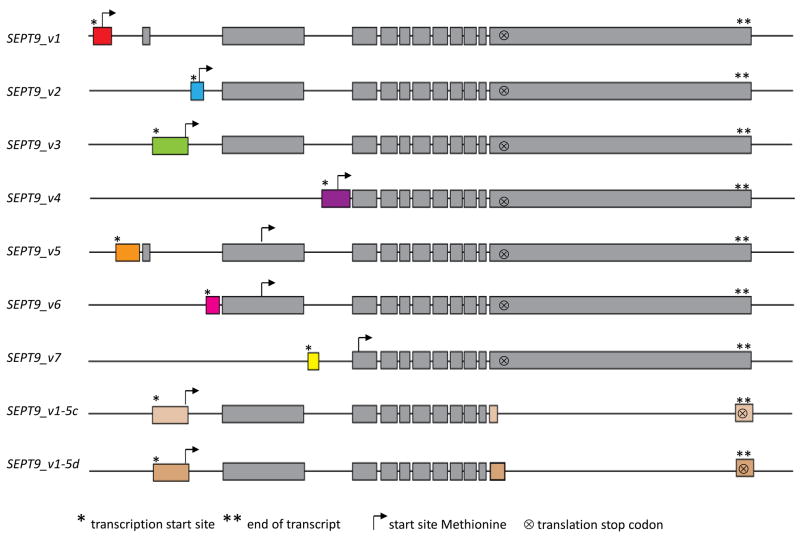
SEPT9, with demonstrated roles in both cancer and neurological disease, provides an illustration of the diversity of human septin loci. Human SEPT9 became of great interest to the septin scientific community after it was found to be an MLL-translocation partner in leukemia (66). Subsequently, SEPT9 was mapped to chromosome 17q25, a region linked to breast and ovarian cancer, by positional cloning to a region of allelic imbalance in breast and ovarian cancers (74, 80–82). This suggested that alterations of this novel septin gene may be important not only leukemia, but also in breast and ovarian cancer. Afterward, it was shown that the SEPT9 locus gives rise to multiple alternative transcripts encoding at least seven annotated isoforms (SEPT9_v1-SEPT9_v7) (74). The SEPT9 locus contains 13 exons and shows high variability at the 5′ and 3′ ends by either alternative splicing of exons or different transcription start sties. The variable exons 1–3 and 12–13 encode alternative translational start and stop sequences and are spliced onto a core of 8 coding exons (Fig 4) (74). These isoforms maintain the general structure of septins with a highly conserved central region containing the polybasic domain and GTP-binding domain (Fig 2). They vary in the 5′-and 3′-untranslated regions (UTR) and at the N- and C- terminus of the protein. Interestingly, the SEPT9_v4 and SEPT9_v4* isoforms encode for the exact same protein, but they differ at their 5′ UTRs, suggesting that these transcripts might be differentially regulated in time or tissue of expression (73, 76, 77). SEPT9_v1 and SEPT9_v3, the longest transcripts, are highly similar except for 25 distinct amino acids at the N- terminus of SEPT9_v1. The biological reason for the presence of multiple SEPT9 transcripts is still not elucidated, but many studies, including expression analyses, show that these isoforms are differentially expressed and may have distinct functional roles in mammalian cells.
Figure 4. SEPT9 genomic structure.
Genomic structure of SEPT9 alternatively spliced transcripts and variants. SEPT9 exhibits 5′ and 3′ alternative spliced and variable exons and is composed of 13 exons in total. Gray boxes indicate common exons and colored boxes indicate variant-specific exons, all drawn to scale. Transcriptional start sites are indicated by (*) and the end of each transcript by (**). The variable exons (colored boxes) encode different translational start (arrow) and stop (⊗) and are spliced into a core of common coding exons shared between variants (gray boxes).
SEPT9 is widely expressed based on ubiquitous adult and fetal transcript expression, although individual isoforms may have tissue specific expression (74). The cellular localization of SEPT9 isoforms is largely cytoplasmic in interphase cells, but the SEPT9_v1 isoform has a bipartite nuclear localization signal that may direct the shuttling of SEPT9_v1 between the nucleus and the cytoplasm. In mitotic cells SEPT9 exhibits a punctate staining pattern located between the separating chromosomes and at the cleavage furrow during telophase (24). SEPT9_v1 is localized to microtubule networks in interphase cells and in mitotic cells it is localized at the mitotic spindle and the bundle of microtubules at the midzone (23). Both studies showed that when SEPT9 is ablated, cells become binucleated, suggesting a role for SEPT9 in cell division via interaction with components of the cytoskeleton such as tubulins.
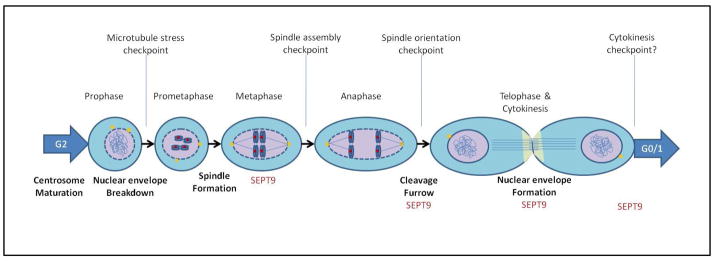
Since they were discovered, septins had been characterized as cytoskeletal GTPases important in cell division. Mutations of this family of genes in yeast lead to budding defects, cell cycle arrest and cytokinesis defects. In mammals, several septins(eg. SEPT2, SEPT5, SEPT7) also have been implicated in mitosis, specifically in chromosome segregation dynamics and cytokinesis (Table 2). SEPT9 is not an exception. Its localization to microtubules, the cleavage furrow and midbody at different stages in mitosis suggests a possible specific role of SEPT9 isoforms in chromosome segregation and/or completion of cell division by affecting the mitotic spindle assembly, disassembly or dynamics and the cytokinesis process (Fig 5).
Figure 5. SEPT9 functional role in mitosis.
Diagram showing the phases in mitosis in which SEPT9 expression is present. SEPT9 is localized to the microtubules at the midzone during anaphase and at the cleavage furrow in telophase. Its particular localization during mitosis suggests a role in chromosome segregation and cytokinesis.
Intricate human SEPT9 nomenclature
Human septin nomenclature has evolved on the past decade as illustrated by many alternative names as shown in Table 3, making the septin literature difficult to reconcile. SEPT9 is no exception, as it was originally called MLL septin-like fusion (MSF) due to the fact that it was identified as a fusion partner of MLL in leukemias (66). In 2002, a committee of septin researchers developed an official nomenclature system for SEPT9 transcripts (83). This new name was derived from the functional role of these genes in septae formation in yeast. Some research groups published with the old nomenclature while others used their own versions, making progress in SEPT9 research hard to reconcile. New nomenclature was established by the National Center for Biotechnology Information (NCBI) and HUGO Gene Nomenclature Committee (HGNC), but it has yet to be universally accepted by the septin community. These nomenclature changes are depicted in Table 4 by group and year. Throughout this review the nomenclature utilized was established in 2005 by Scott et al. and endorsed at the 2009 International Septin Meeting(76).
Table 4.
Human SEPT9 nomenclature
| Kalikin et al. | McIlhatton et al. | Macara et al. | Inagaki et al. | Scott et al. | NCBI | HGNC | NCBI Accession Numbers |
|---|---|---|---|---|---|---|---|
| 2000 | 2001 | 2002 | 2004 | 2005β | 2008 | 2008 | |
| variant/isoform | isoform | transcript/protein | |||||
| MSF-A | epsilon | SEPT9_v1 | SEPT9a | SEPT9_v1a | SEPT9_v1/SEPT9a | SEPT9_i1 | NM_001113491/NP_001106963 |
| - | gamma | - | - | SEPT9_v2a | SEPT9_v2/SEPT9b | SEPT9_i2 | NM_001113493/NP_001106965 |
| MSF | alpha | SEPT9 | SEPT9b | SEPT9_v3a | SEPT9_v3/SEPT9c | SEPT9_i3 | NM_006640/NP_006631 |
| - | - | - | - | - | SEPT9_v4/SEPT9d | SEPT9_i4 | NM_001113495/NP_001106967 |
| MSF-B | zeta | SEPT9_v2 | SEPT9c | SEPT9_v4*a | SEPT9_v5/SEPT9ea | SEPT9_i5 | NM_001113492/NP_001106966 |
| MSF-C | beta | SEPT9_v3 | SEPT9c | SEPT9_v4a | SEPT9_v6/SEPT9ea | SEPT9_i6 | NM_001113494/NP_001106966 |
| - | delta | SEPT9_v4 | - | SEPT9_v5a | SEPT9_v7/SEPT9f | SEPT9_i7 | NM_001113496/NP_001106968 |
| - | - | SEPT9_v5 | - | SEPT9_v1-5b-c* | - | - | - |
SEPT9 variant 5 and 6 differ in their 5′ UTR but encode the same protein isoform.
SEPT9_v1-5b-c variants differ at the 3′ UTR. No well characterized.
Current nomenclature in peer reviewed publications
SEPT9 as a translocation partner of MLL in leukemia
Originally, SEPT9 was identified as part of a fusion protein with the myeloid/lymphoid leukemia gene (MLL) in a therapy-induced acute myeloid leukemia (t-AML) patient with a t(11;17)(q23;q25) rearrangement and was named MSF (MLL septin-like fusion; (66)). The translocation codes for an in-frame transcript joining the 5′ of MLL through exon 5 to SEPT9 at the start of exon 3 through the 3′ terminus. A reciprocal transcript was amplified in one patient joining the 5′ of MSF through exon 1 to exon 7 of MLL but was out of frame (66, 84). The fusion protein was composed of the amino protein terminus of MLL, including the nuclear localization signal, the A-T hook DNA binding domain, and the DNA methyltransferase-like DNA binding domain, and a carboxy terminus of SEPT9_v1, including the proline rich domain, the polybasic and GTPase domains. Lost from the carboxy terminus of MLL is the PHD zinc finger protein-protein interaction domain and the SET domain thought to regulate gene expression through chromatin remodeling (66, 84). It has been suggested that MLL is the sole clinical cause in leukemias with 11q23 rearrangements as it is fused to a wide variety of other genes. However, all of the well-characterized MLL fusion products produce in-frame translated chimeric proteins associated with phenotypic disease variability such as leukemia type and prognostic outcome which provide further evidence that proteins at the varying reciprocally translocated chromosomes are essential contributors to the pathogenesis of leukemia (67, 68, 85). Thus, speculation on the contributions of MLL-SEPT9 fusion protein expression to haematopoetic cellular transformation would include potential mislocalization of MSF from the cytoplasm to the nucleus, aberrant expression of MLL target proteins and altered activation of MSF GTPase signaling pathways. This suggests that these carboxy partners, in addition to MLL, are essential contributors to the pathogenesis of leukemia (68) and implicates septin family members in oncogenesis(85).
SEPT9 model of oncogenesis in solid tumors
A compelling example of the role of septins in the oncogenesis of solid tumors was the identification of the MSF/SEPT9 murine ortholog Sint1/Sept9 at a provirus insertion site in SL3-3 MLV-induced lymphomas (86), which demonstrated that this septin locus is a site for oncogenic integration. SEPT9 also maps to a region of allelic imbalance at chromosome 17q25.3 in breast cancer (74) and sporadic ovarian cancer (82). Additonally, there is differential expression of SEPT9 isoforms in tumors of various tissues (21, 77, 87, 88). SEPT9 has been characterized as a candidate gene in head and neck squamous cell carcinomas by a genome-wide screen for methylated genes (89, 90). SEPT9 was also identified as a possible candidate gene for classical Hodgkin lymhoma (cHL) (91). Circulating methylated SEPT9 DNA in plasma was recently identified as a valuable biomarker for minimally invasive detection of colorectal cancer. This led to the development of a new methylation (m)SEPT9 assay which may prove useful in clinical research for the detection of invasive colorectal cancer with a simple blood test from patients (92–94).
Finally, an isoform of SEPT9, SEPT9_v1, might be important as a biomarker for therapeutic resistance of many cancers to microtubule disrupting agents. SEPT9_v1 expression was strongly correlated with susceptibility of a wide range of cancer cells to drugs such as paclitaxel (95). In general, cancer cells with high SEPT9_v1 expression were more resistant to these drugs (95).
SEPT9 isoforms have been associated with signaling pathways relevant to oncogenesis. For example, SEPT9_v1 interacts with HIF1-α preventing its ubiquitination and degradation, thus activating HIF downstream survival genes to promote tumor progression and angiogenesis in prostate cancer cells (72). Also, a new study showed that SEPT9 up-regulates HIF-1 by preventing Rack1-mediated degradation in an oxygen independent manner (96). In addition, a gene expression profile of prostate cancer showed that SEPT9 was over-expressed in tumors with chromosomal fusion between androgen-regulated transmembrane protease serine 2 (TMPRSS2) and v-ets erythroblastocis virus E26 oncogne (ERG). This fusion is associated with a more aggressive clinical phenotype and tumors with TMPRSS2-ERG are regulated by a estrogen-dependent signaling and are considered a distinct molecular subclass (97). The SEPT9_v3 isoform binds SA-RhoGEF, functioning as a scaffold to keep it in an inactive state, thereby inhibiting SA-RhoGEF mediated Rho activation (98, 99). This finding was crucial for the study of septins in cancer because it provided a direct link between septins and signaling proteins involved in cellular functions such as actin cytoskeletal organization, transcriptional activation, tumor invasion, cell morphology, cell motility and cytokinesis.
SEPT9 expression is also altered in many types of cancers including head and neck tumors, ovarian, prostate, and breast cancers and it is over-expressed in a MMTV mouse model of mammary tumorigenesis (20, 72–76, 86, 92). Most recently, Gonzalez et al., showed that preferential expression of SEPT9_v1 in human mammary epithelial cells promotes pro-oncogenic phenotypes. Briefly, ectopic expression of SEPT9_v1 using retroviral constructs in two immortalized human mammary epithelial cells, MCF10A and HPV 4-12, increases cell proliferation, decreases apoptotic response and enhances motility and invasiveness, in addition to present a reminiscent of epithelial to mesenchymal transition (EMT), all hallmarks of oncogenic transformation in epithelial tissues. In addition, knockdown of SEPT9, and specifically SEPT9_v1, via RNA interference assays in two cancer cell lines wth high endogenous levels of SEPT9_v1 (MDA-MB-231 and BT-549) complemented the observations made in the expression model by rescuing tumorigenic phenotypes. Another striking phenotype found was that SEPT9_v1 high expression disrupts proper formation of tubulin microfilaments in interphase cells and increases aneuploidy. This study provides compelling evidence that SEPT9_v1 could drive malignant progression in mammary epithelial cells (20). Another study also by Gonzalez et al., demonstrated that up-regulation of SEPT9_v1 might increases proliferation rates of mammary epithelial cells by stabilizing c-Jun-N-terminal kinase (JNK) proteins, which are involved in cell proliferation and cell cycle progression. High SEPT9_v1 expression also showed to increase JNK kinase activiy and the transcriptional activation of JNK target genes important in cell cycle progression, including cyclin D1(100).
As a result of these studies, SEPT9 is now accepted as a novel cancer associated protein and is emerging as a potential biomarker for diagnosis and chemotherapeutic response in some epithelial cancers.
Conclusion
Since septins were discovered almost 40 years ago, septin research has grown impressively, from four genes identified in budding yeast in the mid 1970’s to 14 members identified in humans today. In yeast, septins were primarily described as filamentous proteins necessary for budding morphology and cell cycle progression by possibly interacting with cytoskeleton components. The variety of septin isoforms and the significant diversity of their functions in eukaryotic cells have proven to be intriguing and exciting. Studies of mammalian septins have been primarily focused on their association with cancer and neurological diseases. However, many other interesting functions are coming to light such as the role of certain septins in bacterial infection (44–46), angiogenesis (72), spermatogenesis (40–43, 101), retinal degeneration (102, 103), platelet-release reaction (32–35, 104), and beyond. Still, questions related to their normal function in cells and to their implication in human diseases are still puzzling. Major fundamental questions still need to be studied including but not limited to: which factors regulate the tissue-specific and temporal-specific expression of these proteins?, why do human cells needs the presence of 14 distinct septins?, or which molecular and/or cellular alterations dictate the involvement of each specific septin in human diseases? Still, many of the contributions in septin biology in the last years including the first crystal structure of septin oligomers, additional protein interactions, expression profiles and associations with signaling pathways and human diseases have set the path to more exciting experimental strategies to understand further their role in sickness and in health.
Acknowledgments
We thank Lisa M. Privette Vinnedge, Janice Loffreda-Wren, Marta J. Gonzalez-Hernandez and Jennifer A. Keller for critical review and helpful editing of this manuscript. We also thank Megan Landsverk for the helpful review of the SEPT9 nomenclature table.
Funding
This work was supported by the National Institute of Health [RO1CA072877 to E.M.P., 5F31CA12363902 to E.A.P].
Abbreviations
- MLL
myeloid/lymphoid leukemia gene
- HNA
Hereditary Neuralgic Amyotrophy
- MSF
MLL septin-like fusion
- VHL
von Hippel Lindau
- NCBI
National Center for Biotechnology
- HGNC
HUGO Gene Nomenclature Committee
- MMTV
mouse mammary tumor virus
- SEPT
septin
- NEDD5
Neural precursor cell expressed, developmentally down regulated 5
- CDCREL
cell division control-related protein
- DIFF
differentiation protein
- PNUTL
peanut-like protein
Footnotes
Conflict of interest: The authors declare that no conflict of interest exists.
References
- 1.Hartwell LH. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- 2.Cao L, Ding X, Yu W, et al. Phylogenetic and evolutionary analysis of the septin protein family in metazoan. FEBS Lett. 2007;581:5526–5532. doi: 10.1016/j.febslet.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 3.Feng S, Chen JK, Yu H, et al. Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 4.Lim WA, Richards FM. Critical residues in an SH3 domain from Sem-5 suggest a mechanism for proline-rich peptide recognition. Nat Struct Biol. 1994;1:221–225. doi: 10.1038/nsb0494-221. [DOI] [PubMed] [Google Scholar]
- 5.Garcia W, de Araujo AP, Neto Mde O, et al. Dissection of a human septin: definition and characterization of distinct domains within human SEPT4. Biochemistry. 2006;45:13918–13931. doi: 10.1021/bi061549z. [DOI] [PubMed] [Google Scholar]
- 6.Low C, Macara IG. Structural analysis of septin 2, 6, and 7 complexes. J Biol Chem. 2006;281:30697–30706. doi: 10.1074/jbc.M605179200. [DOI] [PubMed] [Google Scholar]
- 7.Sirajuddin M, Farkasovsky M, Hauer F, et al. Structural insight into filament formation by mammalian septins. Nature. 2007;449:311–315. doi: 10.1038/nature06052. [DOI] [PubMed] [Google Scholar]
- 8.Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 9.She YM, Huang YW, Zhang L, et al. Septin 2 phosphorylation: theoretical and mass spectrometric evidence for the existence of a single phosphorylation site in vivo. Rapid Commun Mass Spectrom. 2004;18:1123–1130. doi: 10.1002/rcm.1453. [DOI] [PubMed] [Google Scholar]
- 10.Xue J, Milburn PJ, Hanna BT, et al. Phosphorylation of septin 3 on Ser-91 by cGMP-dependent protein kinase-I in nerve terminals. Biochem J. 2004;381:753–760. doi: 10.1042/BJ20040455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xue J, Wang X, Malladi CS, et al. Phosphorylation of a new brain-specific septin, G-septin, by cGMP-dependent protein kinase. J Biol Chem. 2000;275:10047–10056. doi: 10.1074/jbc.275.14.10047. [DOI] [PubMed] [Google Scholar]
- 12.Qi M, Yu W, Liu S, et al. Septin1, anew interaction partner for human serine/threonine kinase aurora-B. Biochem Biophys Res Commun. 2005;336:994–1000. doi: 10.1016/j.bbrc.2005.06.212. [DOI] [PubMed] [Google Scholar]
- 13.Sanders SL, Field CM. Cell division. Septins in common? Curr Biol. 1994;4:907–910. doi: 10.1016/s0960-9822(00)00201-3. [DOI] [PubMed] [Google Scholar]
- 14.Kinoshita M, Kumar S, Mizoguchi A, et al. Nedd5, a mammalian septin, is a novel cytoskeletal component interacting with actin-based structures. Genes Dev. 1997;11:1535–1547. doi: 10.1101/gad.11.12.1535. [DOI] [PubMed] [Google Scholar]
- 15.Neufeld TP, Rubin GM. The Drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell. 1994;77:371–379. doi: 10.1016/0092-8674(94)90152-x. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen TQ, Sawa H, Okano H, et al. The C. elegans septin genes, unc-59 and unc-61, are required for normal postembryonic cytokineses and morphogenesis but have no essential function in embryogenesis. J Cell Sci. 2000;113(Pt 21):3825–3837. doi: 10.1242/jcs.113.21.3825. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Kong C, Xie H, et al. Phosphatidylinositol polyphosphate binding to the mammalian septin H5 is modulated by GTP. Curr Biol. 1999;9:1458–1467. doi: 10.1016/s0960-9822(00)80115-3. [DOI] [PubMed] [Google Scholar]
- 18.Field CM, Kellogg D. Septins: cytoskeletal polymers or signalling GTPases? Trends Cell Biol. 1999;9:387–394. doi: 10.1016/s0962-8924(99)01632-3. [DOI] [PubMed] [Google Scholar]
- 19.Kartmann B, Roth D. Novel roles for mammalian septins: from vesicle trafficking to oncogenesis. J Cell Sci. 2001;114:839–844. doi: 10.1242/jcs.114.5.839. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez ME, Peterson EA, Privette LM, et al. High SEPT9_v1 expression in human breast cancer cells is associated with oncogenic phenotypes. Cancer Res. 2007;67:8554–8564. doi: 10.1158/0008-5472.CAN-07-1474. [DOI] [PubMed] [Google Scholar]
- 21.Hall PA, Jung K, Hillan KJ, et al. Expression profiling the human septin gene family. J Pathol. 2005;206:269–278. doi: 10.1002/path.1789. [DOI] [PubMed] [Google Scholar]
- 22.Kremer BE, Haystead T, Macara IG. Mammalian septins regulate microtubule stability through interaction with the microtubule-binding protein MAP4. Mol Biol Cell. 2005;16:4648–4659. doi: 10.1091/mbc.E05-03-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagata K, Kawajiri A, Matsui S, et al. Filament formation of MSF-A, a mammalian septin, in human mammary epithelial cells depends on interactions with microtubules. J Biol Chem. 2003;278:18538–18543. doi: 10.1074/jbc.M205246200. [DOI] [PubMed] [Google Scholar]
- 24.Surka MC, Tsang CW, Trimble WS. The mammalian septin MSF localizes with microtubules and is required for completion of cytokinesis. Mol Biol Cell. 2002;13:3532–3545. doi: 10.1091/mbc.E02-01-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottfried Y, Rotem A, Lotan R, et al. The mitochondrial ARTS protein promotes apoptosis through targeting XIAP. Embo J. 2004;23:1627–1635. doi: 10.1038/sj.emboj.7600155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larisch S. The ARTS connection: role of ARTS in apoptosis and cancer. Cell Cycle. 2004;3:1021–1023. [PubMed] [Google Scholar]
- 27.Larisch S, Yi Y, Lotan R, et al. A novel mitochondrial septin-like protein, ARTS, mediates apoptosis dependent on its P-loop motif. Nat Cell Biol. 2000;2:915–921. doi: 10.1038/35046566. [DOI] [PubMed] [Google Scholar]
- 28.Beites CL, Campbell KA, Trimble WS. The septin Sept5/CDCrel-1 competes with alpha-SNAP for binding to the SNARE complex. Biochem J. 2005;385:347–353. doi: 10.1042/BJ20041090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beites CL, Xie H, Bowser R, et al. The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat Neurosci. 1999;2:434–439. doi: 10.1038/8100. [DOI] [PubMed] [Google Scholar]
- 30.Hsu SC, Hazuka CD, Roth R, et al. Subunit composition, protein interactions, and structures of the mammalian brain sec6/8 complex and septin filaments. Neuron. 1998;20:1111–1122. doi: 10.1016/s0896-6273(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 31.Huang YW, Yan M, Collins RF, et al. Mammalian septins are required for phagosome formation. Mol Biol Cell. 2008;19:1717–1726. doi: 10.1091/mbc.E07-07-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blaser S, Horn J, Wurmell P, et al. The novel human platelet septin SEPT8 is an interaction partner of SEPT4. Thromb Haemost. 2004;91:959–966. doi: 10.1160/TH03-09-0578. [DOI] [PubMed] [Google Scholar]
- 33.Blaser S, Roseler S, Rempp H, et al. Human endothelial cell septins: SEPT11 is an interaction partner of SEPT5. J Pathol. 2006;210:103–110. doi: 10.1002/path.2013. [DOI] [PubMed] [Google Scholar]
- 34.Dent J, Kato K, Peng XR, et al. A prototypic platelet septin and its participation in secretion. Proc Natl Acad Sci U S A. 2002;99:3064–3069. doi: 10.1073/pnas.052715199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez C, Corral J, Dent JA, et al. Platelet septin complexes form rings and associate with the microtubular network. J Thromb Haemost. 2006;4:1388–1395. doi: 10.1111/j.1538-7836.2006.01952.x. [DOI] [PubMed] [Google Scholar]
- 36.Ahuja P, Perriard E, Trimble W, et al. Probing the role of septins in cardiomyocytes. Exp Cell Res. 2006;312:1598–1609. doi: 10.1016/j.yexcr.2006.01.029. [DOI] [PubMed] [Google Scholar]
- 37.Spiliotis ET, Kinoshita M, Nelson WJ. A mitotic septin scaffold required for Mammalian chromosome congression and segregation. Science. 2005;307:1781–1785. doi: 10.1126/science.1106823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spiliotis ET, Hunt SJ, Hu Q, et al. Epithelial polarity requires septin coupling of vesicle transport to polyglutamylated microtubules. J Cell Biol. 2008;180:295–303. doi: 10.1083/jcb.200710039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kremer BE, Adang LA, Macara IG. Septins regulate actin organization and cell-cycle arrest through nuclear accumulation of NCK mediated by SOCS7. Cell. 2007;130:837–850. doi: 10.1016/j.cell.2007.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin YH, Lin YM, Wang YY, et al. The expression level of septin12 is critical for spermiogenesis. Am J Pathol. 2009;174:1857–1868. doi: 10.2353/ajpath.2009.080955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steels JD, Estey MP, Froese CD, et al. Sept12 is a component of the mammalian sperm tail annulus. Cell Motil Cytoskeleton. 2007;64:794–807. doi: 10.1002/cm.20224. [DOI] [PubMed] [Google Scholar]
- 42.Sugino Y, Ichioka K, Soda T, et al. Septins as diagnostic markers for a subset of human asthenozoospermia. J Urol. 2008;180:2706–2709. doi: 10.1016/j.juro.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 43.Ihara M, Kinoshita A, Yamada S, et al. Cortical organization by the septin cytoskeleton is essential for structural and mechanical integrity of mammalian spermatozoa. Dev Cell. 2005;8:343–352. doi: 10.1016/j.devcel.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 44.Mostowy S, Cossart P. Cytoskeleton rearrangements during Listeria infection: Clathrin and septins as new players in the game. Cell Motil Cytoskeleton. 2009 doi: 10.1002/cm.20353. [DOI] [PubMed] [Google Scholar]
- 45.Mostowy S, Danckaert A, Tham TN, et al. Septin 11 restricts InlB-mediated invasion by Listeria. J Biol Chem. 2009;284:11613–11621. doi: 10.1074/jbc.M900231200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mostowy S, Nam Tham T, Danckaert A, et al. Septins regulate bacterial entry into host cells. PLoS ONE. 2009;4:e4196. doi: 10.1371/journal.pone.0004196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peterson EA, Kalikin LM, Steels JD, et al. Characterization of a SEPT9 interacting protein, SEPT14, a novel testis-specific septin. Mamm Genome. 2007;18:796–807. doi: 10.1007/s00335-007-9065-x. [DOI] [PubMed] [Google Scholar]
- 48.Zhu M, Wang F, Yan F, et al. Septin 7 interacts with centromere-associated protein E and is required for its kinetochore localization. J Biol Chem. 2008;283:18916–18925. doi: 10.1074/jbc.M710591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kinoshita A, Kinoshita M, Akiyama H, et al. Identification of septins in neurofibrillary tangles in Alzheimer’s disease. Am J Pathol. 1998;153:1551–1560. doi: 10.1016/S0002-9440(10)65743-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi P, Snyder H, Petrucelli L, et al. SEPT5_v2 is a parkin-binding protein. Brain Res Mol Brain Res. 2003;117:179–189. doi: 10.1016/s0169-328x(03)00318-8. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, Gao J, Chung KK, et al. Parkin functions as an E2-dependent ubiquitin-protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci U S A. 2000;97:13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ihara M, Tomimoto H, Kitayama H, et al. Association of the cytoskeletal GTP-binding protein Sept4/H5 with cytoplasmic inclusions found in Parkinson’s disease and other synucleinopathies. J Biol Chem. 2003;278:24095–24102. doi: 10.1074/jbc.M301352200. [DOI] [PubMed] [Google Scholar]
- 53.Fujishima K, Kiyonari H, Kurisu J, et al. Targeted disruption of Sept3, a heteromeric assembly partner of Sept5 and Sept7 in axons, has no effect on developing CNS neurons. J Neurochem. 2007;102:77–92. doi: 10.1111/j.1471-4159.2007.04478.x. [DOI] [PubMed] [Google Scholar]
- 54.Xue J, Tsang CW, Gai WP, et al. Septin 3 (G-septin) is a developmentally regulated phosphoprotein enriched in presynaptic nerve terminals. J Neurochem. 2004;91:579–590. doi: 10.1111/j.1471-4159.2004.02755.x. [DOI] [PubMed] [Google Scholar]
- 55.Ito H, Atsuzawa K, Morishita R, et al. Sept8 controls the binding of vesicle-associated membrane protein 2 to synaptophysin. J Neurochem. 2009;108:867–880. doi: 10.1111/j.1471-4159.2008.05849.x. [DOI] [PubMed] [Google Scholar]
- 56.Kinoshita N, Kimura K, Matsumoto N, et al. Mammalian septin Sept2 modulates the activity of GLAST, a glutamate transporter in astrocytes. Genes Cells. 2004;9:1–14. doi: 10.1111/j.1356-9597.2004.00696.x. [DOI] [PubMed] [Google Scholar]
- 57.Buser AM, Erne B, Werner HB, et al. The septin cytoskeleton in myelinating glia. Mol Cell Neurosci. 2009;40:156–166. doi: 10.1016/j.mcn.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 58.Hannibal MC, Ruzzo EK, Miller LR, et al. SEPT9 gene sequencing analysis reveals recurrent mutations in hereditary neuralgic amyotrophy. Neurology. 2009;72:1755–1759. doi: 10.1212/WNL.0b013e3181a609e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuhlenbaumer G, Hannibal MC, Nelis E, et al. Mutations in SEPT9 cause hereditary neuralgic amyotrophy. Nat Genet. 2005;37:1044–1046. doi: 10.1038/ng1649. [DOI] [PubMed] [Google Scholar]
- 60.Landsverk ML, Ruzzo EK, Mefford HC, et al. Duplication within the SEPT9 gene associated with a founder effect in North American families with hereditary neuralgic amyotrophy. Hum Mol Genet. 2009;18:1200–1208. doi: 10.1093/hmg/ddp014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McDade SS, Hall PA, Russell SE. Translational control of SEPT9 isoforms is perturbed in disease. Hum Mol Genet. 2007;16:742–752. doi: 10.1093/hmg/ddm003. [DOI] [PubMed] [Google Scholar]
- 62.Megonigal MD, Rappaport EF, Jones DH, et al. t(11;22)(q23;q11.2) In acute myeloid leukemia of infant twins fuses MLL with hCDCrel, a cell division cycle gene in the genomic region of deletion in DiGeorge and velocardiofacial syndromes. Proc Natl Acad Sci U S A. 1998;95:6413–6418. doi: 10.1073/pnas.95.11.6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Borkhardt A, Teigler-Schlegel A, Fuchs U, et al. An ins(X;11)(q24;q23) fuses the MLL and the Septin 6/KIAA0128 gene in an infant with AML-M2. Genes Chromosomes Cancer. 2001;32:82–88. doi: 10.1002/gcc.1169. [DOI] [PubMed] [Google Scholar]
- 64.Cerveira N, Correia C, Bizarro S, et al. SEPT2 is a new fusion partner of MLL in acute myeloid leukemia with t(2;11)(q37;q23) Oncogene. 2006 doi: 10.1038/sj.onc.1209626. [DOI] [PubMed] [Google Scholar]
- 65.Kojima K, Sakai I, Hasegawa A, et al. FLJ10849, a septin family gene, fuses MLL in a novel leukemia cell line CNLBC1 derived from chronic neutrophilic leukemia in transformation with t(4;11)(q21;q23) Leukemia. 2004;18:998–1005. doi: 10.1038/sj.leu.2403334. [DOI] [PubMed] [Google Scholar]
- 66.Osaka M, Rowley JD, Zeleznik-Le NJ. MSF (MLL septin-like fusion), a fusion partner gene of MLL, in a therapy-related acute myeloid leukemia with a t(11;17)(q23;q25) Proc Natl Acad Sci U S A. 1999;96:6428–6433. doi: 10.1073/pnas.96.11.6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meyer C, Schneider B, Jakob S, et al. The MLL recombinome of acute leukemias. Leukemia. 2006;20:777–784. doi: 10.1038/sj.leu.2404150. [DOI] [PubMed] [Google Scholar]
- 68.Ernst P, Wang J, Korsmeyer SJ. The role of MLL in hematopoiesis and leukemia. Curr Opin Hematol. 2002;9:282–287. doi: 10.1097/00062752-200207000-00004. [DOI] [PubMed] [Google Scholar]
- 69.Capurso G, Crnogorac-Jurcevic T, Milione M, et al. Peanut-like 1 (septin 5) gene expression in normal and neoplastic human endocrine pancreas. Neuroendocrinology. 2005;81:311–321. doi: 10.1159/000088449. [DOI] [PubMed] [Google Scholar]
- 70.Kim DS, Hubbard SL, Peraud A, et al. Analysis of mammalian septin expression in human malignant brain tumors. Neoplasia. 2004;6:168–178. doi: 10.1593/neo.03310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanaka M, Kijima H, Itoh J, et al. Impaired expression of a human septin family gene Bradeion inhibits the growth and tumorigenesis of colorectal cancer in vitro and in vivo. Cancer Gene Ther. 2002;9:483–488. doi: 10.1038/sj.cgt.7700460. [DOI] [PubMed] [Google Scholar]
- 72.Amir S, Wang R, Matzkin H, et al. MSF-A interacts with hypoxia-inducible factor-1alpha and augments hypoxia-inducible factor transcriptional activation to affect tumorigenicity and angiogenesis. Cancer Res. 2006;66:856–866. doi: 10.1158/0008-5472.CAN-05-2738. [DOI] [PubMed] [Google Scholar]
- 73.Burrows JF, Chanduloy S, McIlhatton MA, et al. Altered expression of the septin gene, SEPT9, in ovarian neoplasia. J Pathol. 2003;201:581–588. doi: 10.1002/path.1484. [DOI] [PubMed] [Google Scholar]
- 74.Kalikin LM, Sims HL, Petty EM. Genomic and expression analyses of alternatively spliced transcripts of the MLL septin-like fusion gene (MSF) that map to a 17q25 region of loss in breast and ovarian tumors. Genomics. 2000;63:165–172. doi: 10.1006/geno.1999.6077. [DOI] [PubMed] [Google Scholar]
- 75.Montagna C, Lyu MS, Hunter K, et al. The Septin 9 (MSF) gene is amplified and overexpressed in mouse mammary gland adenocarcinomas and human breast cancer cell lines. Cancer Res. 2003;63:2179–2187. [PubMed] [Google Scholar]
- 76.Scott M, Hyland PL, McGregor G, et al. Multimodality expression profiling shows SEPT9 to be overexpressed in a wide range of human tumours. Oncogene. 2005;24:4688–4700. doi: 10.1038/sj.onc.1208574. [DOI] [PubMed] [Google Scholar]
- 77.Scott M, McCluggage WG, Hillan KJ, et al. Altered patterns of transcription of the septin gene, SEPT9, in ovarian tumorigenesis. Int J Cancer. 2006;118:1325–1329. doi: 10.1002/ijc.21486. [DOI] [PubMed] [Google Scholar]
- 78.Yu W, Ding X, Chen F, et al. The phosphorylation of SEPT2 on Ser218 by casein kinase 2 is important to hepatoma carcinoma cell proliferation. Mol Cell Biochem. 2009;325:61–67. doi: 10.1007/s11010-008-0020-2. [DOI] [PubMed] [Google Scholar]
- 79.Craven RA, Stanley AJ, Hanrahan S, et al. Proteomic analysis of primary cell lines identifies protein changes present in renal cell carcinoma. Proteomics. 2006;6:2853–2864. doi: 10.1002/pmic.200500549. [DOI] [PubMed] [Google Scholar]
- 80.Kalikin LM, Frank TS, Svoboda-Newman SM, et al. A region of interstitial 17q25 allelic loss in ovarian tumors coincides with a defined region of loss in breast tumors. Oncogene. 1997;14:1991–1994. doi: 10.1038/sj.onc.1201013. [DOI] [PubMed] [Google Scholar]
- 81.Kalikin LM, Qu X, Frank TS, et al. Detailed deletion analysis of sporadic breast tumors defines an interstitial region of allelic loss on 17q25. Genes Chromosomes Cancer. 1996;17:64–68. doi: 10.1002/(SICI)1098-2264(199609)17:1<64::AID-GCC10>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 82.Russell SE, McIlhatton MA, Burrows JF, et al. Isolation and mapping of a human septin gene to a region on chromosome 17q, commonly deleted in sporadic epithelial ovarian tumors. Cancer Res. 2000;60:4729–4734. [PubMed] [Google Scholar]
- 83.Macara IG, Baldarelli R, Field CM, et al. Mammalian septins nomenclature. Mol Biol Cell. 2002;13:4111–4113. doi: 10.1091/mbc.E02-07-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Taki T, Ohnishi H, Shinohara K, et al. AF17q25, a putative septin family gene, fuses the MLL gene in acute myeloid leukemia with t(11;17)(q23;q25) Cancer Res. 1999;59:4261–4265. [PubMed] [Google Scholar]
- 85.Corral J, Lavenir I, Impey H, et al. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell. 1996;85:853–861. doi: 10.1016/s0092-8674(00)81269-6. [DOI] [PubMed] [Google Scholar]
- 86.Sorensen AB, Lund AH, Ethelberg S, et al. Sint1, a common integration site in SL3-3-induced T-cell lymphomas, harbors a putative proto-oncogene with homology to the septin gene family. J Virol. 2000;74:2161–2168. doi: 10.1128/jvi.74.5.2161-2168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hall PA, Russell SE. The pathobiology of the septin gene family. J Pathol. 2004;204:489–505. doi: 10.1002/path.1654. [DOI] [PubMed] [Google Scholar]
- 88.McIlhatton MA, Burrows JF, Donaghy PG, et al. Genomic organization, complex splicing pattern and expression of a human septin gene on chromosome 17q25.3. Oncogene. 2001;20:5930–5939. doi: 10.1038/sj.onc.1204752. [DOI] [PubMed] [Google Scholar]
- 89.Bennett KL, Karpenko M, Lin MT, et al. Frequently methylated tumor suppressor genes in head and neck squamous cell carcinoma. Cancer Res. 2008;68:4494–4499. doi: 10.1158/0008-5472.CAN-07-6509. [DOI] [PubMed] [Google Scholar]
- 90.Bennett KL, Lee W, Lamarre E, et al. HPV status-independent association of alcohol and tobacco exposure or prior radiation therapy with promoter methylation of FUSSEL18, EBF3, IRX1, and SEPT9, but not SLC5A8, in head and neck squamous cell carcinomas. Genes Chromosomes Cancer. 2009 doi: 10.1002/gcc.20742. [DOI] [PubMed] [Google Scholar]
- 91.Giefing M, Arnemann J, Martin-Subero JI, et al. Identification of candidate tumour suppressor gene loci for Hodgkin and Reed-Sternberg cells by characterisation of homozygous deletions in classical Hodgkin lymphoma cell lines. Br J Haematol. 2008;142:916–924. doi: 10.1111/j.1365-2141.2008.07262.x. [DOI] [PubMed] [Google Scholar]
- 92.Devos T, Tetzner R, Model F, et al. Circulating Methylated SEPT9 DNA in Plasma Is a Biomarker for Colorectal Cancer. Clin Chem. 2009 doi: 10.1373/clinchem.2008.115808. [DOI] [PubMed] [Google Scholar]
- 93.Grutzmann R, Molnar B, Pilarsky C, et al. Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PLoS ONE. 2008;3:e3759. doi: 10.1371/journal.pone.0003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lofton-Day C, Model F, Devos T, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 2008;54:414–423. doi: 10.1373/clinchem.2007.095992. [DOI] [PubMed] [Google Scholar]
- 95.Amir S, Mabjeesh NJ. SEPT9_V1 protein expression is associated with human cancer cell resistance to microtubule-disrupting agents. Cancer Biol Ther. 2007;6:1926–1931. doi: 10.4161/cbt.6.12.4971. [DOI] [PubMed] [Google Scholar]
- 96.Amir S, Wang R, Simons JW, et al. SEPT9_v1 up-regulates hypoxia-inducible factor 1 by preventing its RACK1-mediated degradation. J Biol Chem. 2009;284:11142–11151. doi: 10.1074/jbc.M808348200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Setlur SR, Mertz KD, Hoshida Y, et al. Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. J Natl Cancer Inst. 2008;100:815–825. doi: 10.1093/jnci/djn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ito H, Iwamoto I, Morishita R, et al. Possible role of Rho/Rhotekin signaling in mammalian septin organization. Oncogene. 2005;24:7064–7072. doi: 10.1038/sj.onc.1208862. [DOI] [PubMed] [Google Scholar]
- 99.Nagata K, Inagaki M. Cytoskeletal modification of Rho guanine nucleotide exchange factor activity: identification of a Rho guanine nucleotide exchange factor as a binding partner for Sept9b, a mammalian septin. Oncogene. 2005;24:65–76. doi: 10.1038/sj.onc.1208101. [DOI] [PubMed] [Google Scholar]
- 100.Gonzalez ME, Makarova O, Peterson EA, et al. Up-regulation of SEPT9_v1 stabilizes c-Jun-N-terminal kinase and contributes to its pro-proliferative activity in mammary epithelial cells. Cell Signal. 2009;21:477–487. doi: 10.1016/j.cellsig.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kissel H, Georgescu MM, Larisch S, et al. The Sept4 septin locus is required for sperm terminal differentiation in mice. Dev Cell. 2005;8:353–364. doi: 10.1016/j.devcel.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 102.Xin X, Pache M, Zieger B, et al. Septin expression in proliferative retinal membranes. J Histochem Cytochem. 2007;55:1089–1094. doi: 10.1369/jhc.7A7188.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pache M, Zieger B, Blaser S, et al. Immunoreactivity of the septins SEPT4, SEPT5, and SEPT8 in the human eye. J Histochem Cytochem. 2005;53:1139–1147. doi: 10.1369/jhc.4A6588.2005. [DOI] [PubMed] [Google Scholar]
- 104.Baretton G, Vogt T, Valina C, et al. Prostate cancers and potential precancerous conditions: DNA cytometric investigations and interphase cytogenetics. Verh Dtsch Ges Pathol. 1993;77:86–92. [PubMed] [Google Scholar]