Summary
Identifying the spatial organization of tissues at cellular resolution from single cell gene expression profiles is essential to understanding biological systems. Using an in situ 3D multiplexed imaging method, seqFISH, we identify unique transcriptional states by quantifying and clustering up to 249 genes in 16,958 cells to examine whether the hippocampus is organized into transcriptionally distinct subregions. We identified distinct layers in the dentate gyrus corresponding to the granule cell layer and the subgranular zone and contrary to previous reports, discovered that distinct subregions within the CA1 and CA3 are composed of unique combinations of cells in different transcriptional states. In addition, we found that the dorsal CA1 is relatively homogenous at the single cell level, while ventral CA1 is highly heterogeneous. These structures and patterns are observed using different mice and different sets of genes. Together, these results demonstrate the power of seqFISH in transcriptional profiling of complex tissues.
TOC blurb
Shah et al. demonstrated multiplexing of 250-genes in-situ in mouse brain slices using amplified seqFISH. They found that there are distinct subregions of the hippocampus consisting of different combinations of cell classes, defined by the expression patterns in single cells.
Introduction
The mouse brain contains ~108 cells arranged into distinct anatomical structures. While cells in these complex structures have been traditionally classified by morphology and electrophysiology, their characterization has been recently aided by gene expression studies. In particular, the Allen Brain Atlas (ABA) provides a systematic gene expression database using in situ hybridization (ISH) of the entire mouse brain one gene at a time (Dong et al., 2009; Fanselow and Dong, 2010; Thompson et al., 2008). This comprehensive reference provides regional gene expression information, but lacks the ability to correlate the expression of different genes in the same cell. More recently, single cell RNA sequencing (RNA-seq) has identified many cell types based on gene expression profiles (Darmanis et al., 2015; Tasic et al., 2016; Zeisel et al., 2015). However, while single cell RNA-seq provides useful information on multiple genes in individual cells, it has relatively low detection efficiencies and requires cells to be removed from their native environment resulting in the loss of spatial information. These different approaches can lead to contradictory descriptions of cellular organization in the brain and other biological systems.
In the hippocampus, recent RNA-seq data suggests that the CA1 region is composed of cells with a continuum of expression states (Cembrowski et al., 2016, Zeisel et al 2015), while ABA analysis indicates that sub-regions within the CA1 have distinct expression profiles (Thompson et al, 2008). To resolve the two conflicting descriptions of hippocampal organization, a method to profile transcription in situ in the hippocampus with single cell resolution is needed. Here, we demonstrate a general technique that enables the mapping of cells and their transcription profiles with single molecule resolution in tissue, allowing an unprecedented resolution of cellular transcription states for molecular neuroscience (Fig 1A).
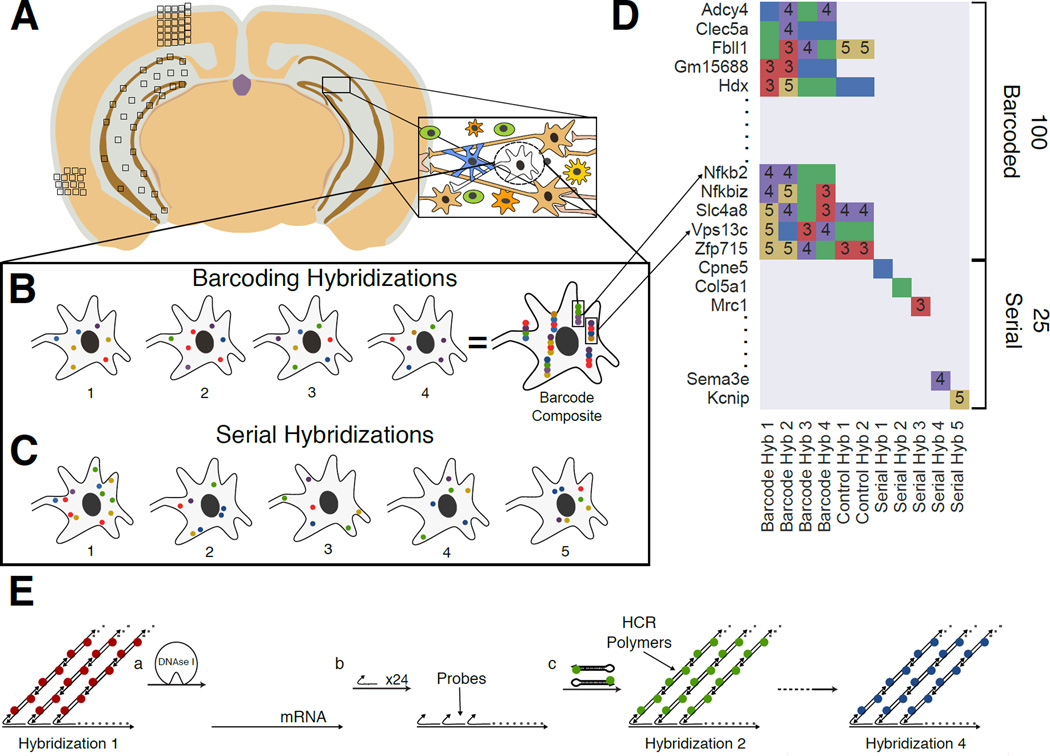
Fig. 1. Overview of the Sequential barcode FISH (seqFISH) in brain slices.
A. A coronal section from a mouse brain was mounted on a slide and imaged in all boxed areas. Each image was taken at 60x magnification. B. Example of barcoding hybridizations from one cell in field from A. The same points are re-probed through a sequence of 4 hybridizations (numbered). The sequence of colors at a given location provides a barcode readout for that mRNA (“barcode composite”). These barcodes are identified through referencing a lookup table abbreviated in D and quantified to obtain single cell expression. In principle, the maximum number of transcripts that can be identified with this approach scales to FN, where F is the number of fluorophores and N is the number of hybridizations. Error correction adds another round of hybridization. C. Serial smHCR is an alternative detection method where 5 genes are quantified in each hybridization and repeated N times. Serial hybridization scales as F*N. D. Schematic for multiplexing 125 genes in single cells. 100 genes are multiplexed in 4 hybridizations by seqFISH barcoding. This barcode scheme is tolerant to loss of any round of hybridization in the experiment. 25 genes are serially hybridized 5 genes at a time by 5 rounds of hybridization. Each number represents a color channel in single molecule HCR. As a control, 5 genes are measured both by double rounds of smHCR as well as barcoding in the same cell. E. SmHCR amplifies signal from individual mRNAs. After imaging, DNAse strips the smHCR probes from the mRNA, enabling rehybridization on the same mRNA (step a). The “color” of an mRNA can be modulated by hybridizing probes that trigger HCR polymers labeled with different dyes (step b). mRNA are amplified following hybridization by adding the complementary hairpin pair (step c). The DNAse smHCR cycle is repeated on the same mRNAs to construct a predefined barcode over time.
A great deal of progress has been made recently in developing highly quantitative methods to profile the transcriptome of single cells. Building upon single molecule fluorescence in situ hybridization (smFISH) (Femino et al., 1998; Raj et al., 2006;), Lubeck et al. devised a general method to highly multiplex single molecule in situ mRNA imaging irrespective of transcript density using super-resolution microscopy (Betzig et al., 2006; Rust et al., 2006; Lubeck and Cai, 2012;). However, the spectral barcoding methods used in these previous works is difficult to scale up beyond 20–30 genes because of the limited number of fluorophores (Fan et al., 2001; Lubeck and Cai, 2012).
To overcome the scalability problem, a temporal barcoding scheme was developed that uses a limited set of fluorophores and scales exponentially with time (Lubeck et al., 2014). Specifically, sequential probe hybridizations on the mRNAs in fixed cells impart a unique pre-defined temporal sequence of colors, generating an in situ mRNA barcodes. The multiplex capacity scales as FN, where F is the number of fluorophores and N is the number of rounds of hybridization. Thus, one can increase the multiplex capacity by increasing the number of rounds of hybridization with a limited pool of fluorophores. We called this approach Sequential barcoded Fluorescence in situ Hybridization (seqFISH) (Lubeck et al., 2014). In parallel, in situ sequencing methods were developed to directly sequence transcripts in tissue sections, but these methods suffer from low detection efficiency (<1%) (Ke et al., 2013; Lee et al., 2014). Recently, Chen et al. expanded the error correction method in the original seqFISH demonstration by using a Hamming distance 2 based error correcting barcode system, called merFISH. However, this implementation requires larger transcripts (>6kb) and many more rounds of hybridization than the method described here (Chen et al., 2015b). Furthermore, seqFISH and its variants have only been applied in cell culture systems due to the difficulty of smFISH detection in tissue. Here, we demonstrate an improved version of seqFISH in complex tissues by including signal amplification and a time-efficient error correction scheme (Fig 1A–D, Table S1), allowing us to resolve the structural organization of the hippocampus with single cell resolution.
Results
Signal amplification and error correction enable robust detection of mRNAs in tissues
To overcome the autofluorescence and scattering inherent to brain tissues, we used an amplified version of smFISH, called single molecule Hybridization Chain Reaction (smHCR) (Fig 1E) (Choi et al., 2014, Shah et al., 2016). Single molecule HCR amplified signal 22.1 ± 11.5 (mean ± s.d., n=1338, Fig S1B) fold compared to smFISH, enabling robust and rapid detection of individual mRNA molecules in tissues and facile alignment of spots between hybridizations (Fig 2A). Single transcripts can be detected and localized in 3D with just 24 probes in tissues, enabling detection of transcripts <1kb in size, with a fidelity comparable to the smFISH gold standard (Fig S1C–D) but with signals 20-fold brighter (Shah et al., 2016). Single molecule HCR DNA polymers can also be digested by DNAse and re-hybridized in brain slices, allowing HCR-seqFISH to be robustly implemented (Fig 2A). We note the smHCR enables true 3D imaging in tissues, whereas the previous sequential FISH demonstrations (Lubeck et al., 2014, Chen et al., 2015) were performed only in flat cell cultures.
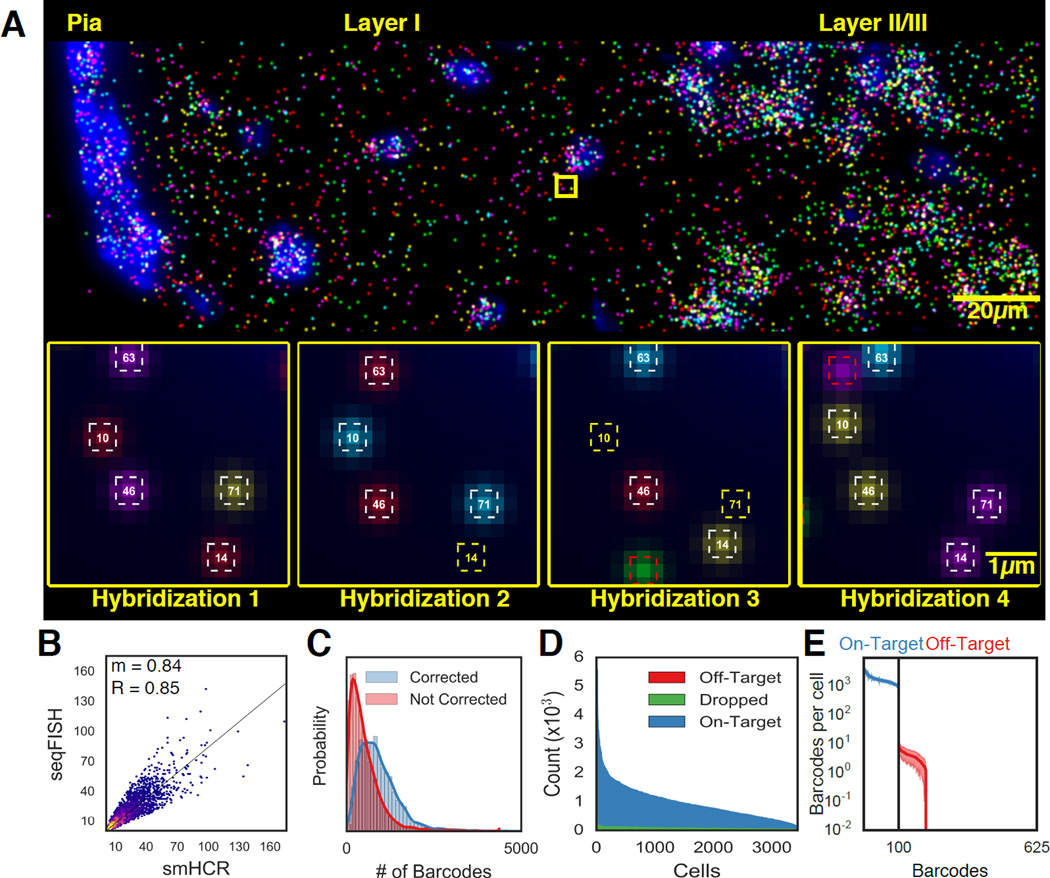
Fig. 2. seqFISH generate accurate in situ quantification of mRNA levels.
A. Image of seqFISH barcoding 100 genes in the outer layer of the mouse cortex. RNA dots in the image are z projected over 15µm. Individual mRNA points are shown across 4 hybridizations in the inset images. White squares correspond to identified barcodes, yellow squares correspond to missing transcripts in a particular hybridization, red squares correspond to spurious false positives and are not counted in any barcode measurements. Numbers in the squares correspond to barcode indices. B. seqFISH correlates with smHCR counts. After barcoding, 5 target mRNAs were measured twice by smHCR in the same cells, providing absolute counts of the transcripts. The two techniques correlate with an R=0.85 and a slope (m) of 0.84 (n=3851 measurements). The 2D histogram intensity shows the distribution of points around the regression line. A high density of points is seen along the regression line. The density falls off steeply around the regression line. C. Error correction results in a median gain of 373 (25%) counts per cell (n=3497). Red and blue curves correspond to the total barcode counts per cell before and after error correction. D. Dropped and off-target barcodes represent a small source of error in seqFISH. 100 on-target barcodes and 525 off-target barcodes are measured per cell. Dropped barcodes are due to at least two overlapping dots appearing within the same region. E. Off-target barcodes are rarely observed and contribute minimally to the expression profile in single cells. Each of the 100 on-target barcodes (blue) and 525 off-target barcodes (red) are quantified per cell. The mean is shown with shaded regions corresponding to 1 SD (N=41 imaged regions).
Furthermore, we improved upon our existing barcode system by implementing a time-efficient error correction scheme. The major source of error in seqFISH is the loss of signal due to mis-hybridization, which increases with the numbers of hybridization. We introduced an extra round of hybridization to correct loss of signal during any round of hybridization (Fig 1D) (Supplementary Text). By minimizing the number of hybridizations, this error correction scheme is efficient to implement. For example, using 5 fluorophores and 4 rounds (instead of 3 rounds) of hybridization to code for 125 genes, we can still uniquely assign barcodes to genes even when signal from any single round of hybridization is missing. Although merFISH can tolerate 2 errors in the barcodes, it requires 16 rounds of hybridization to code 140 genes (Chen et al. 2015). As increasing the number of hybridizations can potentially lead to more experimental error and analysis complexity, our simple error correction method corrects for the most common error, dropped signal. Also, the fewer rounds of hybridizations decrease the total imaging and experimental time, which is rate-limiting for tissue experiments. HCR-seqFISH with simpler error-correction scheme allows efficient and accurate quantification of transcription profiles in tissues.
Using this HCR-seqFISH method, we surveyed the regional and sub-regional transcriptional heterogeneity within the temporal and parietal cortex and hippocampus of the mouse brain by imaging similar coronal sections collected from 3 different animals. Two similar sections from separate mice were profiled with probes for 125 genes, while one additional brain slice was imaged for 249 genes. In each of the coronal slices, between 60–80 fields of view were imaged, each 216µm × 216µm × 15µm, in the cortex and hippocampus (Fig 1A and S1E). For the 125 gene set, 56 of the genes (Fig 1D, Table S1) were selected because they showed spatially heterogeneous expression based on the ABA (Lein et al., 2007), another 44 were selected from a list of transcription factors, and 25 marker genes were selected from single cell RNA-seq datasets (Zeisel et al., 2015). One hundred of these genes were barcoded by 4 rounds of hybridization (Fig 1B). The remaining 25 high abundance genes were measured individually using 5-color smHCR in 5 serial rounds of hybridizations (Fig 1C). This hybrid approach of measuring medium expression genes with barcoding seqFISH and high copy number genes serially in subsequent hybridizations allows a large dynamic range of transcripts to be profiled in the same cell.
seqFISH is an accurate and efficient method to multiplex RNA in situ
To determine the accuracy of the seqFISH method in quantifying mRNA levels in single cells in tissue, we compared the copy number of 5 of the 100 target genes measured by barcoding to the copy number found by smHCR detection in the same cell (Fig 2B, S2A) in 15µm brain sections. We found that the copy number of the RNAs per cell as measured by barcoding and smHCR agreed with an R-value of 0.85 and a slope of 0.84 (N=3851). As smHCR matches smFISH transcript quantitation (Shah et al., 2016), the barcoded seqFISH method can quantify mRNA molecules in single cells with 84% efficiency compared to the gold standard of smFISH. In comparison, single cell RNA-seq measurements are 5–20% efficient based on spike-in controls and in situ sequencing is less than 1% efficient (Darmanis et al., 2015; Klein et al., 2015; Lee et al., 2014; Macosko et al., 2015; Tasic et al., 2016; Zeisel et al., 2015; Ståhl et al., 2016). This high efficiency of detection results from a low transcript drop rate and a high barcode recovery rate due to the error correction round of hybridization. In our experiment, 78.9% of barcodes (N=2,115,477 barcodes) were found in all 4 hybridization rounds and 21.1% were identified in 3 out of the 4 hybridizations (Fig 2C), indicating that the probability of detecting a given mRNA molecule is 94% in each round of hybridization (Fig S2B).
To quantify the amount of false positive signal due to misalignment of barcodes and nonspecific binding of probes, we measured the amount of off-target barcodes that were detected. With four rounds of hybridizations and 5 fluorophores, there were 54=625 unique codes. We assigned 100 of these barcodes to measure mRNAs detected at 914.8 ± 570.5 counts per cell (mean ± s.d., N=3439). In comparison, the 525 remaining off-target barcodes that were not used were detected at 4.6 ± 4.7 (mean ± s.d., N =3439) counts per cell (Fig 2D). False positives, due to chance alignment of nonspecifically bound spots, contributed minimally to the barcode readouts because of this three order of magnitude difference in detected barcodes (on target vs. off target). The false positives we observe fall only on barcodes hamming distance one away from on-target barcodes, yet minimally contribute to undercounting on-target barcodes (Fig 2E). Furthermore, even the most frequent off-target barcode was observed 65.57 times less frequently than the most infrequent mRNA coding barcode (Fig 2E, S2). Even though during each round of hybridization, 24.8 ± 0.4% (mean ± s.e., N=4 rounds of hybridization) of the spots were nonspecifically bound probes, barcode miss-assignments did not occur frequently because non-specifically bound probes do not reappear in the same location after digestion with DNAse and re-hybridization (Fig 2A). Together the quantifications of false positive and false negative barcodes demonstrate that this method is highly efficient and accurate at detecting RNAs in situ in single cells within tissues.
Cell clusters are based on combinatorial expression profiles
We imaged the expression of 125 genes in coronal sections from two mice for a total of 14,908 cells (Fig S1E). Cortical and hippocampal cells were segmented based on DAPI and Nissl staining. A tessellation algorithm was developed to accurately segment densely packed cells in the hippocampus. To avoid capturing mRNA from neighboring cells, we contracted by 10% the borders of cells determined by the segmentation algorithm.
To group the single cell data into distinct transcriptional states, we Z-score normalized the copy number of each transcript in every cell (Fig 3A) and hierarchically clustered the cells to identify cells with similar expression patterns (Table S2, Fig S3). While these clusters do not necessarily represent canonical cell types, many of these clusters contain clear transcriptional markers of known cell types previously identified by single cell RNA-seq (Fig 3B) (Zeisel et al., 2015, Tasic et al 2016). Cell clusters 12 and 13 contained clear expression of Gja1 which marks out astrocytes (Zeisel et al., 2015, Tasic et al 2016). Cluster 12 also expresses Mfge8 while cluster 13 did not, indicating two distinct subpopulations of astrocytes (Fig 3B). There are further subclusters within each of the astrocyte populations with different spatial localization patterns (Fig S3C–E). Cluster 11 cells expressed Laptm5, a known microglia marker (Zeisel et al., 2015, Tasic et al 2016). Cluster 3 expressed interneuron genes while cluster 1–2 and 4–5 expressed genes associated with pyramidal neurons (Zeisel et al., 2015, Tasic et al 2016). The major clusters were robust to down-sampling the number of cells used in clustering (Fig S4), with some of the hippocampal pyramidal and glial clusters robustly defined even with 400 cells. Similarly, principal component analysis (PCA) visualization of the data (Fig S3H) recapitulated the major clusters that correspond to astrocyte, microglia, cortical pyramidal, hippocampal pyramidal, dentate gyrus (DG) granule, and interneuron cells.
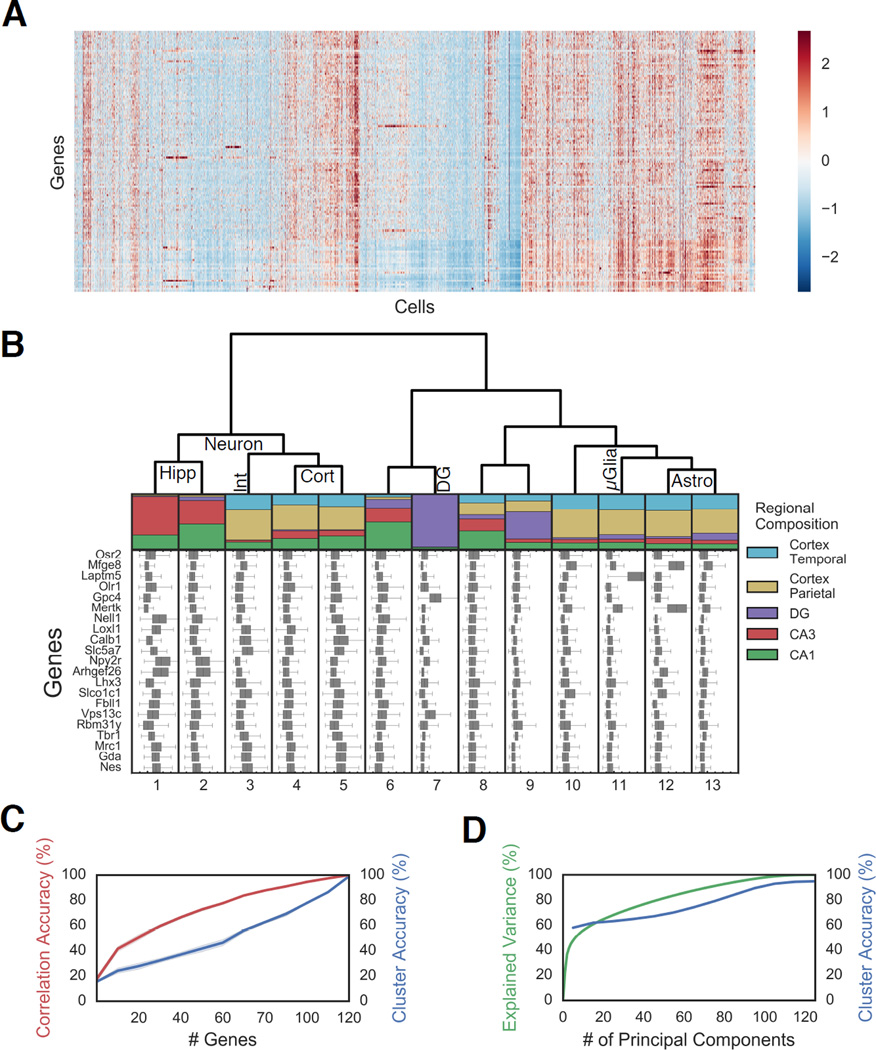
Fig. 3. Distinct clusters of cells exhibit different regional localization in the brain.
A. Gene expression of 14,908 cells presented as a Z-score normalized heatmap. B. Regional compositions of 13 cell clusters are visualized as stacked bar plots with the area corresponding the to number of cells in each region. Hippocampal regions are: CA3, CA1, Dentate Gyrus (DG). Cortical regions: parietal and temporal. Box plot of the Z scores of 21 representative genes are plotted for each cell class. The major tick marks correspond to Z score 0 while every minor tick is a z score interval of 1. Cell type assignments are shown on the dendrogram.
Abbreviations: Hippocampus pyramidal (Hipp), cortex (Cort), Dentate Gyrus (DG), Interneurons (Int), Astrocyes(Astro), Microglia (µGlia). C. Any random subset of 25 genes can recapitulate approximately 50% of the information in the correlation amongst cells (red), but a larger number of genes are required to accurately assign cells to cluster using a random forest algorithm (blue) (n=10 bootstrap replicates; shading is 95% CI), indicating that fine structures in the data require quantitative measurements of combinatorial expression of many genes. D. Similar to C, while the first ten PCs explain the coarse structure, a larger number of principal components (PCs) are required to describe the full data. Expected variation (green) and accuracy in predicting cell identity using a random forest model (blue).
As the cluster distance between different cells is proportional to the number of differentially expressed genes in the target list, an unbiased clustering of the 125 gene data without weighting specific genes should not be interpreted directly as canonical “cell types,”, but rather as grouping cells with different patterns of genes expression based on the current target list. We will refer to some of these clusters as pyramidal neurons or astrocytes for ease of notation, but strictly speaking, they are cells clusters with similar expression patterns as neurons or astrocytes.
Cell clusters show distinct regional localization
Many neuronal clusters mapped to distinct regions in the brain (Fig 3B). Several classes of pyramidal cells (cluster 1–2) showed exclusive localization to the hippocampus, while other classes (4–5) showed predominantly cortical localization. There were also a class of cells (cluster 7) that were almost exclusively present in the DG. Interestingly, these clusters segregated based solely on gene expression profiles without adding any spatial information into the clustering algorithm. These differences in transcriptional states of neurons could be due to intrinsic differences in the cells or due to different local environment and activity patterns.
In contrast, astrocyte, microglia and other non-neuronal cell clusters were generally uniformly present in all areas of the brain (Fig 3B). However, subclusters of astrocytes did localize to different regions of the brain preferentially (Fig S3E), with subcluster 12.3 localized preferentially to the cortex, while 12.1 subcluster was uniformly distributed. Similarly, cluster 9 cells contain subclusters (9.3, 9.5 and 9.6) that localize exclusively to the DG, while other subcluster (9.1) localize almost exclusively to the cortex. The regional localization of neurons are especially pronounced with cluster 1 and 2 localized almost exclusively to the hippocampus, with some of the subclusters localized predominantly to the CA3. Furthermore, while pyramidal cell clusters 4 and 5 are preferentially cortically localized, the few hippocampal cells in these clusters form their own subclusters (4.4 and 5.4) (Fig S3E). In cluster 6 cells, many subclusters with distinct expression profiles are localized almost exclusively in the CA1, CA3 or the DG (Fig S3C). In contrast, cluster 7 cells show a relatively homogenous regionalization pattern, but further subdivide based on combinatorial expression patterns (Fig S3D). Subclusters of cluster 9 also show significant regionalization where subclusters 9.1, 9.3, 9.5, and 9.6 show localization to the SGZ (Fig S3E). Overall, cell clusters with similar expression profiles exhibited similar spatial localizations across the brain with a correlation coefficient of 0.67 (Fig S3G), indicating the existence of archetypal regional expression patterns and potential spatial markers in the brain. These results show that the tissue-optimized HCR seqFISH approach can directly identify a variety of transcriptional states and quantify broad spatial patterns of expression.
Combinatorial expression patterns define fine clusters
While certain cell clusters contain strong expression of marker genes, not all clusters are defined based on a few genes. How much power do individual genes or groups of genes have in explaining the observed cell clusters? To understand this, we examined whether subsets of genes can recapitulate the observed clusters (Fig 3C–D). We found that any set of 25 genes recovers about half of the correlation structure in the cell-to-cell correlation map (Fig 3C, S3I, S4, N=10 bootstrap replicates). The fact that the selection of any 25 genes can explain the gross patterns in the data is likely due to the high correlations amongst the expression patterns of genes, as shown in the gene-to-gene correlation map (Fig S3J). Thus, a small subset of the measured genes can provide sufficient information to infer the gross transcriptional states of the cells. Interestingly, this may be the same reason why low-coverage single cell sequencing methods such as drop-seq and inDrop (Klein et al., 2015; Macosko et al., 2015) can capture the large distinction of cell types, because many highly expressed genes are correlated to other genes that collectively define cell types.
At the same time, the finer correlation structure in the data, required to define the cell clusters accurately, can only be captured with accurate quantitation of many genes (Fig 3C–D). Consistent with this, using a “random-forest” machine learning algorithm (Breiman, 2001) to classify cell clusters, we found that 75 genes are needed to classify cells with 50% accuracy, indicating that correct cluster assignment requires more detailed information from many genes (Fig 3C). Supporting this view, the first 10 principal components (PC) explained 59.5% of the variation in the data, while the rest of the variation required the remaining 115 PCs (Fig 3D, S3F). The “random forest” algorithm required 10 PCs to predict the cell cluster assignments with 50% accuracy (Fig 3D), but accuracy steadily increased with more PCs. These observations indicated two levels of information in the data: a coarse level, where large distinctions in cell clusters are observable by a few genes, and a fine level, where subtle distinctions require many more genes.
These results suggest two points experimentally. First, multiplexing at the level of 20 genes by seqFISH can give broad cell cluster identification that is not available with 2–3 gene smFISH experiments. Although single marker genes are useful for inference, we find that they frequently are not sufficient for cell classification. For example, all DG specific granule cells (clusters 7) have Gpc4 and Vps13c as their enriched marker genes (Fig 3B); yet, Gpc4 and Vps13c are also strongly expressed in other hippocampal cells outside of the DG, as seen in both our experiments and the ABA. Thus, smFISH against Gpc4 and Vps13c alone would not be sufficient to uniquely identify the DG granule cells. Furthermore, even the strongly bimodal markers that are known to define cell types (i.e. Mgfe8, Gja1, etc.) are correlated enough to overall expression profiles that cells fall into the appropriate cluster even when these genes are excluded. This point suggests that while marker genes can be essential in assigning a cell to a known cell type, they are not necessary to identify unique clusters in the dataset provided enough measurements are made.
Second, accurate measurement of combinatorial expression of many genes enabled by seqFISH can allow for more specific cell cluster identification. As a comparison, in single cell RNAseq data, CA1 pyramidal cells are clustered into a single cluster (Zeisel et. al, 2015; Habib et. al 2016) potentially because of the relatively lower detection efficiency. In our seqFISH experiments, measuring hundreds of genes quantitatively, we can resolve several clusters and subclusters with robust regionalization within the CA1 (Fig 3B, S3C–E).
Cells are patterned in the dentate gyrus
To further visualize the spatial organization of cells, we mapped cluster definitions of cells back into the images. In the DG, we observed a striking lamina layering of cell classes. The two blades of the DG (Fig 4A–B) showed mirror arrangements of cells, with cluster 9 cells, forming the subgranular zone (SGZ), leading into a granule cell layer (GCL) dominated by a single cluster of granule cells (cluster 7) (Fig 3B). In the 125 gene data set, the cells of the GCL were found to be dominated by expression of Gpc4 and Vps13c matching ISH data from the ABA (Fig S8B). Cluster 7 was found to be further subdivided into 6 subclusters (Fig S3D). These subclusters were found to have varying levels of calbindin D-28K (Calb1) expression which is known to increase with granule cell maturation (Yang et al., 2015). On the other hand, the cells of the SGZ were found to be significantly enriched in astrocyte markers such as Mfge8 and Mertk, which has been also been observed previously (Miller et al, 2013) and in the ABA data (Fig S8A). However, these cells do not cluster with typical astrocytes (cluster 12 and 13) because their combinatorial expression patterns are different from astrocytes, consistent with their classification as a completely different population of cells.
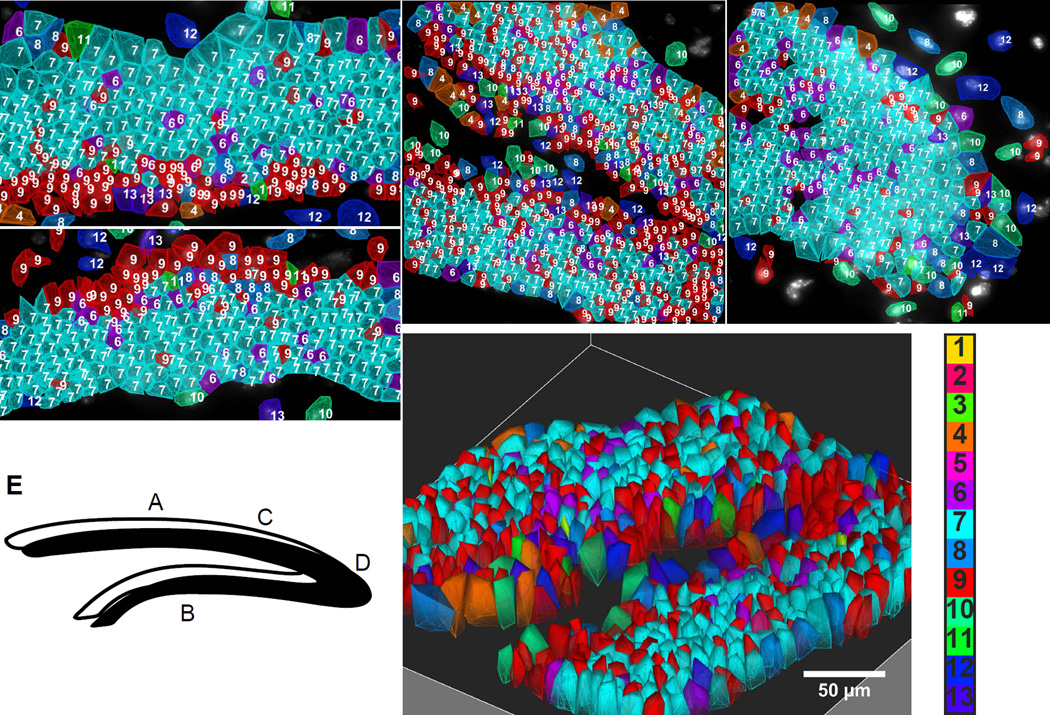
Fig. 4. Spatial layering of cell classes in the Dentate Gyrus (DG).
A–B. Suprapyramidal and infrapyramidal blades of DG. Cells of the subgranular zone and granule cells are arranged in lamina layers in mirror symmetric patterns on the upper and lower blades. C. The SGZ stays on the inner layer of the DG fork. D. Cells are patterned in the crest. Numbered color key corresponds to cluster numbers in Fig 3b. E. Letters in the cartoon of DG correspond to images. F. 3D image of the fork region shown in C.
In the fork region of the DG, the layer of cluster 9 cells appeared on the interior surface of the fork, followed by a layer of granule cells (cluster 7) (Fig 4C). A different layering pattern is seen at the crest of the DG, where astrocytes, microglia, and some other glial cells line the exterior of the crest ensheathing the GCL (Fig 4D). In both brains of the 125 gene experiments, the same cell clusters and spatial arrangements are observed. Furthermore, because the mRNAs are imaged in 3D in the 10–15um brain slices, we can obtain a 3D view of the expression profiles, shown in the fork regions of the DG (Fig 4F).
Distinct regions of CA1 and CA3 are composed of different combination of cell clusters
While each region of the DG contains similar compositions of cells, distinct subregions within the CA1 and CA3 contained different combinations of cell classes (Fig 5, S6F). In the CA1, there were 3 distinct regions defined by their individual cellular compositions. In the dorsal region of CA1 (CA1d), neuron cluster 6 (enriched in Nell1, a protein kinase C binding protein) (Table S3) was the major cell type in the pyramidal layer, with astrocyte, microglia and other cells (clusters 10–13) intercalating into the stratum pyramidale (SP) (Fig 5A–C). Transitioning into the CA1 intermediate region (CA1i) (Fig 5D), pyramidal cell cluster 4 displaced cell cluster 6 as the dominant cell, with the co-appearance of cluster 1 and 2 pyramidal cells.
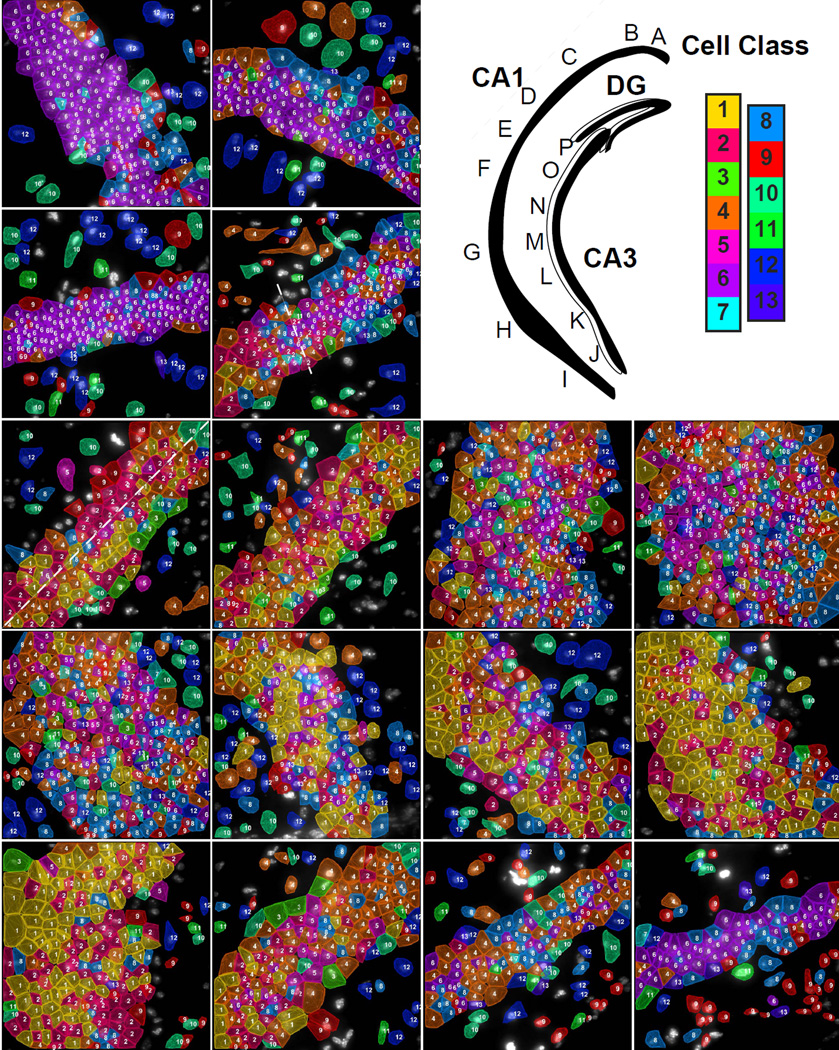
Fig. 5. Subregions of the hippocampus are composed of distinct compositions of cell classes based on the first 125 gene experiment.
Upper right panel. Cartoon of hippocampus with imaged regions labeled. Color key corresponds to the classes in Fig 3b. A–D. These images are regions from the CA1d. Astrocytes (Astro) are marked in image A and a microglia cell (µGlia) is marked in image B. Moving along the hippocampus from CA1 dorsal to ventral, cell classes transition from a homogenous dorsal population (C to D) to a mixed population in the CA1 intermediate (E–F) to regions of even larger cellular diversity in the CA1 ventral region (G–I). The dotted line in D marks the transition point of the CA1d to the CA1i. E shows two laterally segregated cell classes (marked by a dotted line) in the CA1i along with cholinergic interneurons (Int) on the interior surface of the CA1i. The ventral (J–K) and intermediate CA3 (L–M) have similar cell classes compositions to the CA1v and CA1i. The two last regions (O–P) of the dorsal CA3 shows distinct cell classes compositions that are relatively homogeneous within a field but are different than other fields of CA3. The cell class composition of field P is similar to that of the CA1d, but these cluster 6 cells are grouped into a distinct subcluster.
As the middle of the CA1i region was reached, a small amount of cluster 4 pyramidal cells remain, while cluster 1 and 2 pyramidal cells dominate (Fig 5E–F). Cluster 1 and 2 are enriched in Nell1 (EGF like protein), Npy2r (neuropeptide Y receptor), Slc4a8 (sodium bicarbonate transporter) and B3gat2 (glucuronosyltransferase) (Table S2). The CA1i region displayed a characteristic spatial organization where glial cells line the outermost regions, while pyramidal cell cluster 1 and 2 longitudinally partitioned the pyramidal layer. This separation of the inner versus the outer layers of CA1 matches those observed in previously (Dong et al., 2008). Furthermore, interneurons (cluster 3) were found to preferentially line the inner edge of the pyramidal layer in the CA1i region (Fig 5E–F). This patterning of interneurons, particularly subcluster 3.1 cells which were enriched in Slc5a7, a choline transporter, was consistent with the patterning of cholinergic interneurons observed with ChAT-GFP labeling (Yi et al., 2015). Finally, the largest amount of heterogeneity in the CA1 was seen in the ventral CA1 region (CA1v), where cell clusters 3, 5, and 10 began to mix in with clusters 1 and 2 (Fig 5G–I).
Similarly, the CA3 was found to have four transcriptionally distinct regions with different pyramidal cell compositions and abrupt transitions. The ventral most region of CA3 contained a high level of heterogeneity of pyramidal cell clusters (Fig 5J–K), while the intermediate region of CA3 contain a mixture of cell clusters 1 and 2 (Fig 5L–M). As the CA3 progressed towards the hilus of the DG, the cell types transitioned first to primarily cluster 4 neurons (enriched in dcx, doublecortin, and Col5a1, a collagen), and then to almost exclusively cluster 6 neurons in the region most proximal to the DG hilus (Fig 5O–P). It is interesting to note that while cluster 6 cells appear in both the CA1 (subcluster 6.8) and CA3 (subclusters 6.1 and 6.4), sub-clusters of 6 show distant regional localization (Fig S3E), suggesting that the gene expression differences in CA1 and CA3 cells are captured in the seqFISH data. We note that similar patterns of homogeneous dorsal and heterogeneous ventral cell populations are observed when only hippocampal cells are clustered (Figure S5).
The regionalized expression patterns we observed in the hippocampus match closely to those observed in previous literature (Thompson et al Neuron 2008 and Dong et al PNAS 2009). For example, CA1d, CA1i, CA1v boundaries correspond to the boundaries shown in Dong et al Fig 2B. In CA3, the subregions observed in our experiment match the CA3 subregion 4–7 in Thompson et al. (Thompson et al., 2008).
Lastly, we note that the two slices from two different mice in the 125 gene experiment show not only the same subregional structure (Fig 4–6), but also the same clusters of cells (Fig 5 and 6) in the different subregions of the hippocampus (Fig S6). In both brains, the CA1d consists of relatively homogenous population of cluster 6 cells, which transition to a mixture of 1 and 2 cells in CA1i, and finally to a mixture of 1–6 and 10 cells in the CA1v (Fig S6F). These results together show that the sub-regions of the hippocampus are a robust feature in the organization of CA1 and CA3, consisting of cells classes with distinct expression profiles. The stereotypical nature of the spatial arrangement of these structures suggest further experiments with seqFISH and other functional assays to probe the distinct functions of the different cell clusters in the CA1 and CA3.
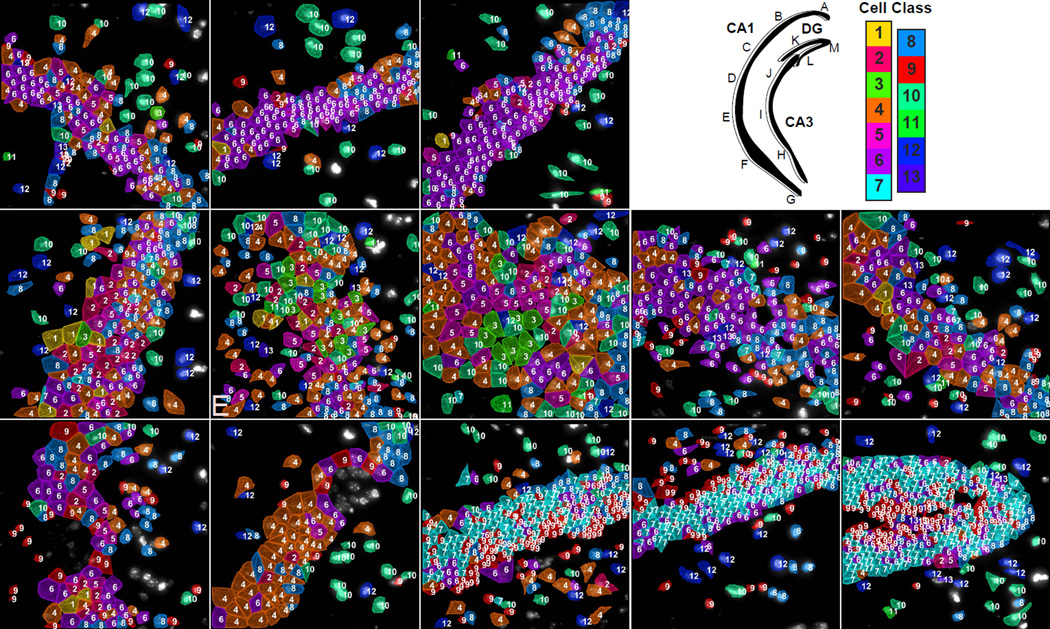
Fig. 6. Mapping of cell types to a second brain slice with 125 genes.
Upper right panel. Cartoon of hippocampus with imaged regions labeled. Color key corresponds to the classes in Fig 3b. A–D. Similar to the cell class compositions shown for the hippocampus in Fig 5, CA1d in this second coronal section from a second mouse is composed of mostly cluster 6 cells. (E) CA1i region and (F–G) the CA1 ventral regions are again composed of similar cell classes to that shown in figure 5 with increasing diversity of cell class compositions from the CA1d to the CA1i to finally the CA1v. (H–J) CA3 regions. (K–M) DG regions showing the same cell classes and layer pattern of the GCL and SGZ shown in Figure 4.
249 gene multiplex experiments show the same hippocampal subregions
To further show that the sub-regional structure of the hippocampus is independent of the target genes, we performed a 249 gene seqFISH experiment on a third coronal section. Of these 249 genes, only 22 genes overlapped with the 125 gene experiment set. For this set of genes, 214 were selected from a list of transcription factors and signaling pathway components and the remaining 35 were selected from cell identity markers from another single cell RNAseq dataset (Tasic et al, 2016). The 214 genes were barcoded by 5 rounds of hybridization, while the remaining genes were imaged in 7 rounds of non-barcoding serial hybridization. To quantify the efficiency of this experiment, 4 genes in the barcoding set (Smarca4, Sin3a, Npas3, and Neurod4) were re-probed with smHCR. The barcoding efficiency of the 249 gene probe set was found to be 71% with and R value of 0.80 (Fig S6D). In single cells, we detect on average 2807±1660 (mean±s.d., N=2050 cells) total barcoded barcodes.
The same arrangement in the DG was observed in the 249 gene experiment, despite different genes used, indicating robust identification of the layering in the DG by seqFISH (Fig 7S–T). In particular, the cells in the SGZ are clustered independently from cells in the GCL, similar to the layers observed in the 125 gene experiment. In the SGZ cells, we observed enrichment of Sox11, a key transcription factor in neurogenesis (Miller et al, 2013). Other transcription factors involved in neurogenesis, NFIA and Tbr1 are also enriched in the SGZ cells as seen in our data and the ABA images (Fig S8A). The observations of this distinct layer in both the 249 and 125 gene experiment and the combined gene enrichment pattern (increased Sox11, Sox9, NFIA, and Tbr1 in the 249 gene experiment and increased Mertk and Mfge8 in the 125 gene experiment) suggests that many cells in this layer are involved in adult neurogenesis in the SGZ. Supplementary figure 7B shows distinctive marker gene expression in the GCL of the dentate gyrus.
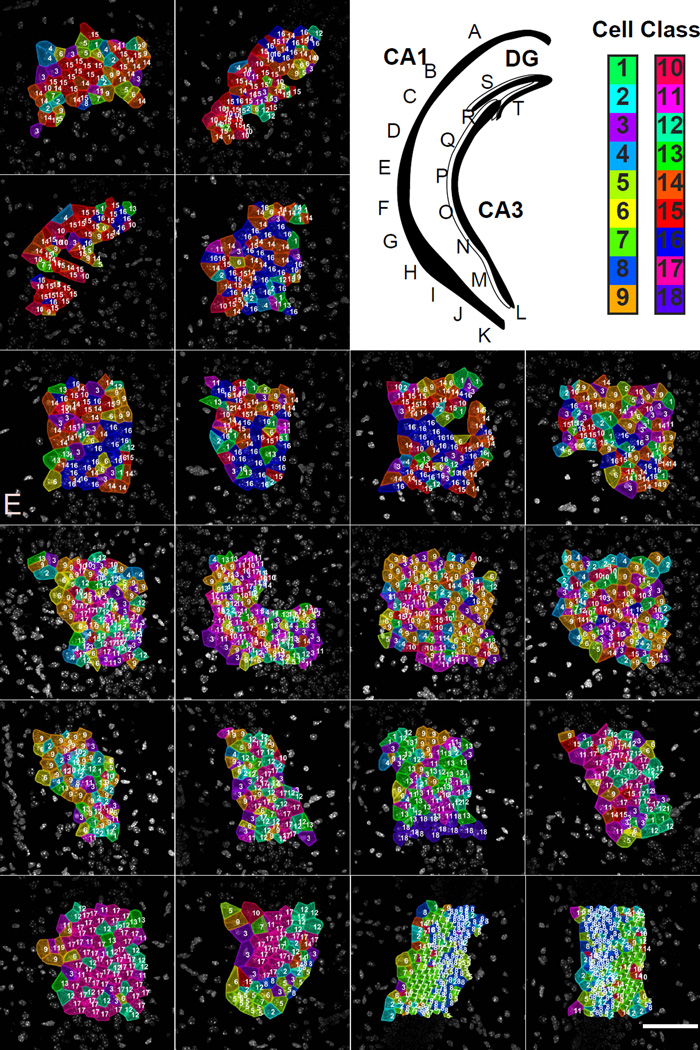
Fig. 7. Mapping of cell types to a third brain slice with 249 genes.
Upper right panel. Cartoon of hippocampus with imaged regions labeled. Color key corresponds to the classes in Fig S6C. A–C. Similar to the slice shown in Fig 5 and 6, CA1d is relatively homogenous in cell cluster composition. D–G. Images from the CA1i region show that the cell class composition is different from that of the CA1d. H–K. Again, similar to Fig 5 and 6, images from the CA1 ventral regions shows a much more complicated cellular composition and a high degree of cellular heterogeneity. L–R. Images from the CA3 region show that the cellular compositions also creates 3–4 subregions within the CA3. The cellular heterogeneity of the CA3 subregions mirrors that of the CA1, where the ventral region of the CA3 is very heterogenous while the dorsal region of the CA3 is relatively homogenous. S–T. The DG regions show the distinct SGZ versus GCL layering pattern seen in the previous two brains.
In addition, the same regionalized cellular patterns are observed in CA1d, CA1i, and CA1v, where different subregions utilize different cell classes in characteristic ratios (Fig S6F). As seen with the 125 gene experiment, while the CA1d uses only a few cell classes and is relatively homogeneous, while the CA1v region is made up of many different cell classes resulting in a high level of cellular heterogeneity. Furthermore, the distinction between CA1 and CA3 cell clusters are more clear in the 249 gene experiment suggesting more resolving power of spatial patterns (Fig 7A–K). The 249 gene experiment also suggests that the CA3 may be composed of 3–4 subregions based on cell cluster composition (Fig 7L–R). The cellular heterogeneity of the CA3 is again shown to mirror that of the CA1, where the cellular heterogeneity increases along the dorsal to ventral axis. Cells with distinctive marker gene expression in the hippocampus are shown in Supplementary figure 7A.
Discussion
Single cell data resolves cellular organizations in the sub-regions of the CA1 and CA3
Two conflicting views of the cell types in the hippocampus have been proposed based on the analysis of the Allen Brain Atlas data (Thompson 2008) as well as recent RNA-seq data (Cembrowski et al., 2016, Zeisel et al 2015). Analysis of the ABA in situ data showed that distinct subregions of the hippocampus expressed different molecular markers, indicating that the CA1 and CA3 are “regionalized” into distinct sub-structures (Fanselow and Dong, 2010; Thompson et al., 2008). However, recent bulk RNA-seq experiments on the CA1 found that gene expression patterns changed gradually along the dorsal to ventral axis, contradicting the sharp boundaries observed in the ABA analysis (Cembrowski et al., 2016). Further supporting this “continuous” cell type view of the hippocampus, analysis of the single cell RNA-seq data (Zeisel et al, 2015) identified a single continuous population of cells in the CA1 region.
Our data provides a single cell resolution picture of the spatial organization of cells in the hippocampus and reconciles both the RNA-seq and the ABA data. While our data mostly supports a regionalized view of the hippocampus, we observe that a single cell class does not in general define CA1 and CA3 sub-regions. Instead, we observed that different subregions of CA1 and CA3 are composed of distinct combinations of cell clusters (Fig 5–7). For example, CA1d consists primarily of cluster 6 pyramidal cells (Fig 5A–C), in addition to the cluster 1,2, 10, and 12 cells, while CA1v consists of a large set of cell classes including cluster 1–6 and 10 cells, but at different relative abundances (Fig 5–6, Fig S6 F–G). Due to this intermixing of cell classes in each sub-region, a bulk measurement of transcription profiles would find a lack of regionalization, but single cell analysis with spatial resolution would identify these distinct regions based on their unique cell class compositions. Indeed, when we averaged the single cell expression profile within each sub-region of the CA1, we can reproduce the continuous correlation profiles found by bulk RNA-seq between CA1v, CA1i, and CA1d (Fig 8) (Cembrowski et al., 2016). The bulk RNA-seq observation that CA1i lacked specific marker genes can also be explained. This is in fact consistent with our findings that CA1i contained cell classes present in both CA1d and CA1v (Fig 5–7). This organization of cell classes is observed in both the 125 gene experiments as well as in the 249 gene experiment.
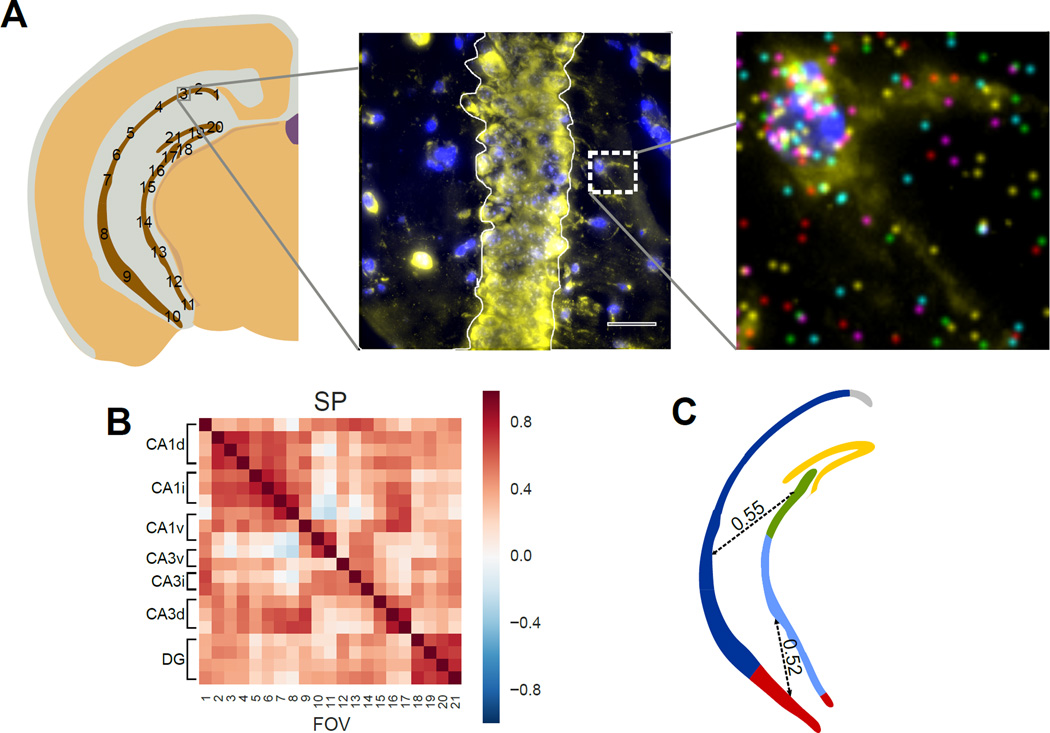
Fig 8. Correlations of the transcription profile across the pyramidal layer.
A. mRNA counts in the cell bodies in the Stratum Pyramidale (SP) are grouped within each field of view. A single cell in the Stratum Radiatum (SR) is shown to illustrate individual mRNA localization. Stratum Oriens (SO) is labeled for orientation. B. mRNAs in different subregions of pyramidal layer show both long-distance spatial correlations as well as local correlations between neighboring fields. Both CA1 and Dentate Gyrus (DG) show high regional correlations. Correlation is calculated based on the 125 gene experiment. C. Illustration of regional and long distance correlation patterns observed in B. Correlated regions are colored and long distance correlations are shown as dotted lines with their median correlation coefficient written over the dotted line.
It is worth noting that the complexity of cell populations observed in the CA1d versus the CA1v matches the functional differences in CA1. CA1d is responsible for spatial learning and navigation and contains a higher concentration of place cells and send projections to dorsal subiculum and cortical retrosplenial area (Cenquizca and Swanson, 2007; Jung et al., 1994; Risold et al, 1997; O’Keefe and Dostrovsky, 1971). We observed that CA1d is composed of a relatively homogeneous population of cells, predominantly of cluster 6 cells. In contrast, the ventral region is involved in a variety of cognitive tasks, such as stress response, emotional and social behavior (Cenquizca and Swanson, 2007; Jung et al., 1994; Fanselow and Dong, 2010; Kishi et al., 2006; Muller et al., 1996; Petrovich et al., 2001; Pitkänen et al., 2000; Saunders et al., 1988; Witter and Amaral, 1991; Yi et al., 2015). Correspondingly, we observed a large set of cell classes in the CA1v regions. It is intriguing to hypothesize that the different cell classes identified based on molecular profiles may correspond to neurons with distinct connectivity and functional patterns. This hypothesis can be investigated in future experiments combining anterograde tracing as well as electrophysiological recording followed by seqFISH.
SeqFISH cell classes versus single cell RNA-seq cell types
While the accurate measurement of 100–200 genes can provide distinctions between the large functional classes found by RNA-seq, the clusters found by seqFISH, in general, should not be interpreted as cell types. RNA-seq measurements at the whole transcriptome level defines cell types based on highly variable genes. On the other hand, seqFISH provides highly accurate measurements of fewer genes, but uses the combinatorial expression patterns to group cells into clusters. However, because only 100–200 genes are targeted in the seqFISH experiments, not all of the “cell types” are equally represented in the gene list and seqFISH cannot catalogue “cell types” in the same fashion that single cell RNAseq can. For example, in our 125 gene experiments, we cannot resolve the distinct subpopulation of interneurons because we lacked marker genes such as Vip and Sst. seqFISH and RNA-seq provide two different, yet complementary, levels of resolution into the transcriptional profiles of cells. RNA-seq measures the transcription levels of thousands of genes but at a lower quantitative accuracy, while seqFISH measures only 100’s of genes but with much greater quantitative power. The differing nature of the two sets of data informs how the data should be analyzed and interpreted. Thus, seqFISH and single cell RNAseq have complementary roles in elucidating distinct cell subpopulations in tissues. SeqFISH could be applied to find finer distinctions within cell types found by RNA-seq or to look at the spatial patterning of cell types found by RNA-seq.
seqFISH provides a generalized method to multiplex mRNA imaging in tissues
seqFISH with amplification and error correction provides a highly quantitative method to profile hundreds of mRNA species directly in single cells within their native anatomical context. Our method of stripping the probes from the RNA has many advantages. DNAse digestion of probes allows false positives to be rejected as nonspecifically bound probes do not colocalize between different rounds of hybridization (Fig 2A). In addition, the same region of the transcript can be hybridized in every round, allowing seqFISH to efficiently target mRNAs shorter than 1kb, enabling targeting of most genes. Lastly, seqFISH allows exponential scaling of barcode numbers, thus 4–5 rounds of hybridization can code for hundreds of transcripts with a simple error correction scheme. Theoretically, the entire transcriptome can be coded for with error correction by using 8–9 rounds of hybridization with seqFISH. These advantages of HCR seqFISH allows robust multiplexed RNA detection in tissues, shown here in the mouse brain.
Ultimately, the multiplexing capability of seqFISH is limited by the amount of optical space within a cell, and not by the coding capacity of the method (supplementary text). We showed previously that super-resolution microscopy can significantly increase the optical space available in the cell for transcription profile imaging, but super-resolution microscopy experiments proved difficult to image in samples thicker than 1µm, and were experimentally cumbersome and time consuming to image (Lubeck and Cai, 2012). A recent development in expansion microscopy as well as correlation methods (Coskun et al., 2016) however offers promise for multiplexing to levels of high transcript density (Chen et al., 2015a; Treweek et al., 2015, Chen et al., 2016). In addition, by labeling subcellular components (i.e. dendrites and axons) with antibodies, the local transcriptome in compartments of the cell can be measured.
We observed that, because expression patterns amongst genes are highly correlated, the distinction between large classes of cells can be determined from 10–20 genes, while a finer classification of cell clusters depends on the quantitative measurement of the combinatorial expression patterns of many genes (Fig 3C–D). This correlation amongst genes can be used to “stitch” our seqFISH data with single cell RNAseq data, similar to the approach explored with single cell RNAseq and ISH in Satija et al (Satija et al., 2015). By correlating seqFISH data to single cell RNA-seq expression data, cells types identified based on RNA-seq can be “mapped” back into our seqFISH data.
As shown here, seqFISH with hundreds of genes in tissues can become a general and widely used tool to answer a wide range of fundamental questions in biology and medicine. For neuroscience, by combining the insights into the spatial organization of transcription provided by seqFISH with connectomics and electrophysiological measurements, we can obtain a comprehensive understanding of the molecular basis of the neuroanatomy of the brain.
Experimental Procedure
Probe Design
Genes were selected from the Allen Brain Atlas database. We identified genes that are heterogeneously expressed in coronal sections containing the hippocampus at Bregma coordinates −2.68 mm anterior. We selected 100 genes that had high variances across these distinct regions and that also had low-medium expression levels. Probe sequences were designed using software developed in house. Full details are described in Supplemental Experimental Procedures.
Probe Generation
All oligoarray pools were purchased as 92k synthesis from Customarray Inc. Probes were amplified from array-synthesized oligo pool as previously described (Chen et al., 2015b). Full details are described in Supplemental Experimental Procedures.
Brain extraction and sample mounting
C57BL/6 with Ai6 Cre-reporter (uncrossed) (Jackson Labs, SN: 007906) female mice aged 50–80 days were anesthetized with isoflurane according to institute protocols (protocol #1701-14) (Madisen et al., 2012). Mice were perfused with 4% PFA and the brain was dissected out and placed in a 4% PFA buffer for 2 hours at room temperature. The brain was then immersed in 4C 30% RNAse-free Sucrose\1× PBS until the brain sank. Once sunk, the brain was embedded in OCT and sectioned. Full details are described in Supplemental Experimental Procedures.
Sample permeabilization, hybridization, and Imaging
Sections were permeabilized in 4C 70% EtOH for 12–18 hours. Brains were further permeabilized by the addition of rnase-free 8% SDS. A hybridization chamber was adhered around the brain section. RNA integrity test probes were hybridized overnight at 37 in hybridization buffer (Table S3). Samples were washed in 30% wash buffer (WB) for 30 minutes. Probes were amplified. Following amplification, samples were washed in the same 30% WB for at least 10 minutes to remove excess hairpins. Samples were stained with DAPI and submerged in pyranose oxidase antibleaching buffer (Lubeck et al., 2014). If the RNA was deemed to be intact, DAPI data was collected in this hybridization. Samples were digested with DNAse I for 4 hours at room temperature on the scope. Following DNAse I the sample was washed several times with 30% WB and the probes were hybridized overnight (Table S4 and S5). Samples were again washed and amplified. Repeating this cycle with the appropriate probes for each hybridization developed barcode digits. Fluorescent Nissl stain was collected at the end of the experiment along with images of multispectral beads to aid chromatic aberration corrections. Full details are described in Supplemental Experimental Procedures.
Image Processing
The images were first corrected for to remove the uneven illumination profiles in each channel and to remove the effects of chromatic aberration. The background intensity in the images was then subtracted. A 150-pixel border region around the image was ignored in all analysis to avoid errors from edge effects of illumination. Full details are described in Supplemental Experimental Procedures.
Image Registration
The processed images were then registered by first taking a maximum intensity projection along the z direction in each channel. All of the maximum projections of the channels of a single hybridization were then collapsed resulting in 4 composite images containing all the points in a particular round of hybridization. Each of these composite images of hybridization 1–3 was then cross-correlated individually with the composite image of hybridization 4 and the position of the maxima of the cross-correlation was used as the translation factor to align hybridizations 1–3 to hybridization 4.
Cell Segmentation
For cells in the cortex, the cells were segmented manually using the DAPI images taken in the first round of hybridization and the fluorescent nissl stain taken at the end of the experiment. Furthermore, the density of the point cloud surrounding a cell was taken into account when forming cell boundaries, especially in cells that did not stain with the nissl stain. For the hippocampus, the cells were segmented by first manually selecting the centroid in 3D of each DAPI signal of every cell. Transcripts were first assigned based on nearest centroids. These point clouds were then used to refine the centroid estimate and create a 3D voronoi tessellation with a 10% boundary-shrinking factor to eliminate ambiguous mRNA assignments from neighboring cells. Regional segmentation was performed manually using the ImageJ ROI tool.
Barcode calling
The potential mRNA signals were then found by LOG filtering the registered images and finding points of local maxima above a specified threshold value. Once all potential points in all channels of all hybridizations were obtained, dots were matched to potential barcode partners in all other channels of all other hybridizations using a 1-pixel search radius to find symmetric nearest neighbors. This procedure was repeated using each hybridization as a seed for barcode finding and only barcodes that were called similarly in at least 3 out of 4 rounds were used in the analysis. The number of each barcode was then counted in each of the assigned cell volumes and transcript numbers were assigned based on the number of on-target barcodes present in the cell volume. All image processing and image analysis code can be obtained upon request. Full details are described in Supplemental Experimental Procedures.
Clustering
To cluster the dataset with two brain measured with 125 genes, we first Z-score normalized each of the slices based on gene expression (Table S6). Once the single cell gene expression data is converted into z-scores, we compute a matrix of cell-to-cell correlations using Pearson correlation coefficients for all of the cells in the two brains. Then hierarchical clustering with Ward linkage is performed on the cell-to-cell correlation data using cells taken from the center of the field of view. To analyze the robustness of individual clusters, a random forest model was trained using varying subsets of the data and used to predict the cluster assignment of the remaining cells (Breiman, 2001). For Figure 4–6, the entire field of cells was classified using the clustered cells as the training set. A bootstrap analysis by dropping different sets of cells was performed in increments (Fig S5). To determine the effect of dropping out genes on the accuracy of the clustering analysis, we used a random forest decision tree to learn the cluster definition based on the 125 gene data. Then we ask the decision tree to re-compute the cluster assignment on cell-to-cell correlation matrices with fewer and fewer genes (Fig 3C–D, green line). Bootstrap resampling was also performed with this analysis (Fig 3C–D, blue lines). The PCA and stone analysis were performed using the same cell-to-cell z-scored Pearson correlation matrix. The cell-to-cell correlation in Fig S3EI was calculated with increasing number of principal components dropped (have their eigenvalues set to zero). The cluster assignment accuracy is again computed through the random forest decision tree. The 249 gene experiment was clustered independently with Z-score normalized data.
Supplementary Material
Highlights.
Amplified seqFISH enables in situ detection of 100’s genes in single cells in tissues.
Combinatorial expression patterns of genes define cell classes in the mouse brain.
Subregions of the hippocampus are composed of distinct combinations of cell classes.
Heterogeneity in cell class compositions increases along the dorsal to ventral axis
Acknowledgments
We thank Michael Elowitz, Henry Lester, Hongwei Dong, and Bosiljka Tasic for advice on the manuscript. We also would like to thank Niles Pierce, Viviana Gradinaru, Thanos Siapas, Mary Kennedy, and Carlos Lois for productive discussions. Raw data can be found in the Supplementary Materials of this paper. The National Institute of Health, the McKnight Foundation, and the Allen Foundation supported this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: L.C., S.S., E.L. designed the experiments. W.Z., S.S., E.L. performed the experiments. L.C., S.S., E.L. analyzed the data and wrote the manuscript.
References and Notes
- Beliveau BJ, Joyce EF, Apostolopoulos N, Yilmaz F, Fonseka CY, McCole RB, Chang Y, Li JB, Senaratne TN, Williams BR, et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc. Natl. Acad. Sci. U.S.A. 2012;109:21301–21306. doi: 10.1073/pnas.1213818110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging Intracellular Fluorescent Proteins at Nanometer Resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Breiman L. Random Forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- Cajigas IJ, Tushev G, Will TJ, Dieck S. tom, Fuerst N, Schuman EM. The Local Transcriptome in the Synaptic Neuropil Revealed by Deep Sequencing and High-Resolution Imaging. Neuron. 2012;74:453–466. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cembrowski MS, Bachman JL, Wang L, Sugino K, Shields BC, Spruston N. Spatial Gene-Expression Gradients Underlie Prominent Heterogeneity of CA1 Pyramidal Neurons. Neuron. 2016;89:351–368. doi: 10.1016/j.neuron.2015.12.013. [DOI] [PubMed] [Google Scholar]
- Cenquizca LA, Swanson LW. Spatial organization of direct hippocampal field CA1 axonal projections to the rest of the cerebral cortex. Brain Res. Rev. 2007;56:1–26. doi: 10.1016/j.brainresrev.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tillberg PW, Boyden ES. Expansion microscopy. Science. 2015a;347:543–548. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Wassie AT, Cote AJ, Sinha A, Alon S, Asano S, Daugharthy ER, Chang J-B, Marblestone A, Church GM, Raj A, Boyden ES. Nanoscale imaging of RNA with expansion microscopy. Nat Meth advance online publication. 2016 doi: 10.1038/nmeth.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015b;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HMT, Beck VA, Pierce NA. Next-Generation in Situ Hybridization Chain Reaction: Higher Gain, Lower Cost, Greater Durability. ACS Nano. 2014;8:4284–4294. doi: 10.1021/nn405717p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Gephart MGH, Barres BA, Quake SR. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H-W, Swanson LW, Chen L, Fanselow MS, Toga AW. Genomic–anatomic evidence for distinct functional domains in hippocampal field CA1. Proc. Natl. Acad. Sci. 2009;106:11794–11799. doi: 10.1073/pnas.0812608106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Braut SA, Lin Q, Singer RH, Skoultchi AI. Determination of transgenic loci by expression FISH. Genomics. 2001;71:66–69. doi: 10.1006/geno.2000.6403. [DOI] [PubMed] [Google Scholar]
- Fanselow MS, Dong H-W. Are the dorsal and ventral hippocampus functionally distinct structures? Neuron. 2010;65:7–19. doi: 10.1016/j.neuron.2009.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of Single RNA Transcripts in Situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- Habib N, Li Y, Heidenreich M, Swiech L, Trombetta JJ, Zhang F, Regev A. Div-Seq: A single nucleus RNA-Seq method reveals dynamics of rare adult newborn neurons in the CNS. bioRxiv. 2016:045989. doi: 10.1126/science.aad7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung MW, Wiener SI, McNaughton BL. Comparison of spatial firing characteristics of units in dorsal and ventral hippocampus of the rat. J. Neurosci. 1994;14:7347–7356. doi: 10.1523/JNEUROSCI.14-12-07347.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke R, Mignardi M, Pacureanu A, Svedlund J, Botling J, Wählby C, Nilsson M. In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods. 2013;10:857–860. doi: 10.1038/nmeth.2563. [DOI] [PubMed] [Google Scholar]
- Kishi T, Tsumori T, Yokota S, Yasui Y. Topographical projection from the hippocampal formation to the amygdala: A combined anterograde and retrograde tracing study in the rat. J. Comp. Neurol. 2006;496:349–368. doi: 10.1002/cne.20919. [DOI] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Droplet Barcoding for Single-Cell Transcriptomics Applied to Embryonic Stem Cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, Ferrante TC, Terry R, Jeanty SSF, Li C, Amamoto R, et al. Highly Multiplexed Subcellular RNA Sequencing in Situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lubeck E, Cai L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat. Methods. 2012;9:743–748. doi: 10.1038/nmeth.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Mao T, Koch H, Zhuo J, Berenyi A, Fujisawa S, Hsu Y-WA, Iii AJG, Gu X, Zanella S, et al. A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat. Neurosci. 2012;15:793–802. doi: 10.1038/nn.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Nathanson Jason, Franjic Daniel, Shim Sungbo, Dalley Rachel A, Shapouri Sheila, Smith Kimberly A, Sunkin Susan M., Bernard Amy, Bennett Jeffrey L, Lee Chang-Kyu, Hawrylycz Michael J, Jones Allan R, Amaral David G, Sestan Nenad, Gage Fred H, Lein Ed S. Conserved molecular signatures of neurogenesis in the hippocampal subgranular zone of rodents and primates. Development. 2013;140(22):4633–4644. doi: 10.1242/dev.097212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller R, Stead M, Pach J. The hippocampus as a cognitive graph. J. Gen. Physiol. 1996;107:663–694. doi: 10.1085/jgp.107.6.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe J, Dostrovsky J. The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res. 1971;34:171–175. doi: 10.1016/0006-8993(71)90358-1. [DOI] [PubMed] [Google Scholar]
- Petrovich GD, Canteras NS, Swanson LW. Combinatorial amygdalar inputs to hippocampal domains and hypothalamic behavior systems. Brain Res. Brain Res. Rev. 2001;38:247–289. doi: 10.1016/s0165-0173(01)00080-7. [DOI] [PubMed] [Google Scholar]
- Pitkänen A, Pikkarainen M, Nurminen N, Ylinen A. Reciprocal Connections between the Amygdala and the Hippocampal Formation, Perirhinal Cortex, and Postrhinal Cortex in Rat: A Review. Ann. N. Y. Acad. Sci. 2000;911:369–391. doi: 10.1111/j.1749-6632.2000.tb06738.x. [DOI] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA Synthesis in Mammalian Cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risold PY, Swanson LW. Structural evidence for functional domains in the rat hippocampus. Science. 1996;272:1484–1486. doi: 10.1126/science.272.5267.1484. [DOI] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Meth. 2006;3:793–796. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotech. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders RC, Rosene DL, Van Hoesen GW. Comparison of the efferents of the amygdala and the hippocampal formation in the rhesus monkey: II. Reciprocal and nonreciprocal connections. J. Comp. Neurol. 1988;271:185–207. doi: 10.1002/cne.902710203. [DOI] [PubMed] [Google Scholar]
- Shah S, Lubeck E, Schwarzkopf M, He T, Greenbaum A, Sohn C. ho, Lignell A, Choi HMT, Gradinaru V, Pierce NA, Cai L. Single-molecule RNA detection at depth via hybridization chain reaction and tissue hydrogel embedding and clearing. Development dev.138560. 2016 doi: 10.1242/dev.138560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, Mollbrink A, Linnarsson S, Codeluppi S, Borg Å, Pontén F, Costea PI, Sahlén P, Mulder J, Bergmann O, Lundeberg J, Frisén J. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 2016 doi: 10.1038/nn.4216. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CL, Pathak SD, Jeromin A, Ng LL, MacPherson CR, Mortrud MT, Cusick A, Riley ZL, Sunkin SM, Bernard A, et al. Genomic Anatomy of the Hippocampus. Neuron. 2008;60:1010–1021. doi: 10.1016/j.neuron.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Treweek JB, Chan KY, Flytzanis NC, Yang B, Deverman BE, Greenbaum A, Lignell A, Xiao C, Cai L, Ladinsky MS, et al. Whole-body tissue stabilization and selective extractions via tissue-hydrogel hybrids for high-resolution intact circuit mapping and phenotyping. Nat. Protoc. 2015;10:1860–1896. doi: 10.1038/nprot.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Maaten L, Hinton G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008;9:85. [Google Scholar]
- Witter MP. Organization of the entorhinal—hippocampal system: A review of current anatomical data. Hippocampus. 1993;3:33–44. [PubMed] [Google Scholar]
- Witter MP, Amaral DG. Entorhinal cortex of the monkey: V. Projections to the dentate gyrus, hippocampus, and subicular complex. J. Comp. Neurol. 1991;307:437–459. doi: 10.1002/cne.903070308. [DOI] [PubMed] [Google Scholar]
- Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen C-K, Lubeck E, Shah S, Cai L, Gradinaru V. Single-Cell Phenotyping within Transparent Intact Tissue through Whole-Body Clearing. Cell. 2014 doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SM, Alvarez DD, Schinder AF. Reliable Genetic Labeling of Adult-Born Dentate Granule Cells Using Ascl1 CreERT2 and Glast CreERT2 Murine Lines. J Neurosci. 2015;35(46):15379–15390. doi: 10.1523/JNEUROSCI.2345-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F, Catudio-Garrett E, Gábriel R, Wilhelm M, Erdelyi F, Szabo G, Deisseroth K, Lawrence J. Hippocampal “cholinergic interneurons” visualized with the choline acetyltransferase promoter: anatomical distribution, intrinsic membrane properties, neurochemical characteristics, and capacity for cholinergic modulation. Front. Synaptic Neurosci. 2015;7 doi: 10.3389/fnsyn.2015.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Manchado ABM, Codeluppi S, Lönnerberg P, Manno GL, Juréus A, Marques S, Munguba H, He L, Betsholtz C, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015 doi: 10.1126/science.aaa1934. aaa1934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.