Abstract
In England in 2015, Shigella sonnei isolates from men who have sex with men produced extended-spectrum β-lactamases and exhibited macrolide resistance. Whole-genome sequencing showed a close relationship among the isolates, which harbored a plasmid that was previously identified in a shigellosis outbreak among this population but has acquired a mobile element.
Keywords: Shigella, Shigella sonnei, bacteria, bacterial infections, sexually transmitted infections, STIs, men who have sex with men, MSM, outbreaks, antimicrobial resistance, England, ESBL producers, extended-spectrum beta-lactamase, ESBL
Historically, shigellosis is an infection associated with travel to countries in which this infection is endemic. In England in 2009, Shigella flexneri 3a infections dramatically increased among men who have sex with men (MSM) and were thought to be associated with sexual transmission (1). Since 2011, an emerging epidemic of S. sonnei infections in England among men has occurred (2). We describe an investigation of a cluster of multidrug-resistant S. sonnei infections among MSM in England during 2015.
The Study
In November 2015, Public Health England (PHE) identified a cluster of S. sonnei isolates by analyzing whole-genome sequencing data from specimens collected during September and October from 4 adult men residing in London and having no known travel history. Clinical reporting data indicated that 2 of the men were MSM. The isolates were later found to exhibit high levels of antimicrobial drug resistance to amoxicillin, ceftriaxone, tetracycline, sulfonamides, trimethoprim, and azithromycin and to produce extended-spectrum β-lactamase (ESBL) (Table 1). The isolates were sensitive to quinolones. PHE convened an outbreak control team to identify additional cases, characterize cases, capture exposure information, rule out a point source for the outbreak, and provide evidence for additional targeted public health efforts.
Table 1. Results of the phenotypic antimicrobial drug resistance profile of 4 isolates of Shigella sonnei, England, 2015*.
| Isolate ID | MIC for antimicrobial drugs tested, mg/mL† |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | AMC | CTX | CRO | CAZ | CFP | GEN | TOB | TMP | SEP | ATM | CIP | COL | ERT | MER | FOS | TEM | AZT | |
| 182834 | >1,052 | 1 | >32 | >32 | 2 | 0.38 | 0.5 | 0.38 | >32 | 64 | 0.38 | <0.032 | 0.19 | 0.008 | 0.06 | 1 | <1 | >256 |
| 184494 | >1,052 | 1 | >32 | >32 | 2 | 0.5 | 0.38 | 0.38 | >32 | 128 | 0.5 | <0.032 | 0.125 | 0.008 | 0.06 | 1.5 | <1 | >256 |
| 183660 | >1,052 | 1 | >32 | >32 | 1 | 0.38 | 0.38 | 0.38 | >32 | 128 | 0.38 | <0.032 | 0.125 | 0.008 | 0.06 | 1 | <1 | >256 |
| 164679 | >1,052 | 1 | >32 | >32 | 1 | 0.38 | 0.38 | 0.38 | >32 | 64 | 0.38 | <0.032 | 0.19 | 0.008 | 0.06 | 1 | <1 | >256 |
*TEM was included in the panel to aid detection of OXA-48–like carbapenemases. AMC, amoxicillin/clavulanate; AMP, ampicillin; ATM, aztreonam; AZT, azithromycin; CAZ, ceftazidime; CFP, cefepime; CIP, ciprofloxacin; COL, colistin; CRO, ceftriaxone; CTX, cefotaxime; ERT, ertapenem; FOS, fosfomycin; GEN, gentamicin; ID, identification; MER, meropenem; SEP, trimethoprim/sulfamethoxazole; TEM, temocillin; TMP, trimethoprim; TOB, tobramycin. †Phenotypic antimicrobial drug susceptibility testing was performed by using Etest (bioMérieux, Marcy l’Etoile, France) and interpreted by using breakpoints for Enterobacteriaceae established by the European Committee on Antimicrobial Susceptibility Testing (http://www.eucast.org/). Production of extended-spectrum β-lactamase was confirmed by using CTX + clavulunic acid and CAZ + clavalunic acid in combination (bioMérieux).
Since August 2015, PHE has conducted whole-genome sequencing of ≈70% of S. sonnei isolates (those voluntarily referred from hospital laboratories) (Technical Appendix). Single nucleotide polymorphism (SNP) clustering is performed, as previously described (3). Readings (in FASTQ format) from all sequences in this study can be found at the PHE Pathogens BioProject (National Center for Biotechnology Information project no. PRJNA315192).
DNA from isolate 183660 was used to elucidate plasmid sequencing (Technical Appendix). The multidrug-resistance phenotype was explained by the presence of an 89,000-bp IncFII plasmid designated p183660 (GenBank accession no. KX008967). This plasmid had >95% nt identity to pKSR100, a multidrug-resistance plasmid from an MSM case of S. flexneri 3a infection (GenBank accession no. LN624486) that had acquired the previously described pKSR100 integron (blaTEM-1, dfrA17, dfrA1, sul1, aadA5) (4) and a novel mobile element harboring blaCTX-M-27 (Figure 1). Macrolide resistance was conferred by ermB and mphA.
Figure 1.
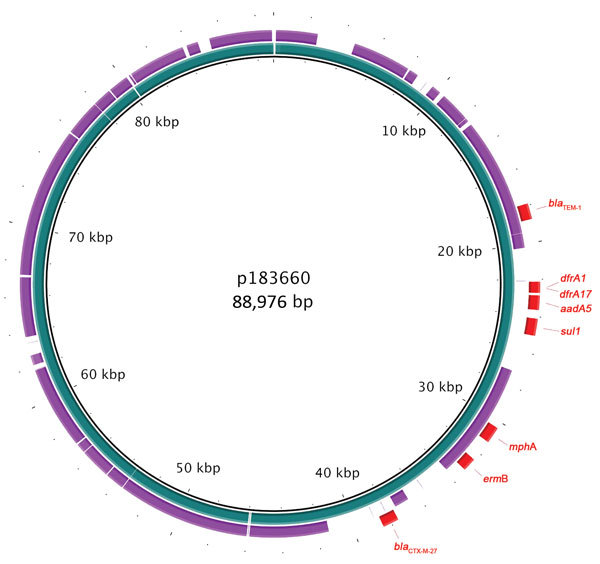
Genomic plot of multidrug-resistance plasmid p183660 (inner ring, blue) from a man in England infected with Shigella sonnei compared with pKSR100 (outer ring, purple), a multidrug-resistance plasmid from a case of S. flexneri 3a infection occurring among men who have sex in men (4). Drug-resistant elements from p183660 are shown in red. Plot produced by using BLAST Ring Image Generator (5).
For this investigation, we defined a case-patient as a resident of England with laboratory-confirmed S. sonnei infection belonging to a 5 SNP cluster with the SNP address 1.3.3.9.207.212.%, a resistance profile characterized by the presence of the plasmid p183660, and a specimen collected during September 1, 2015–February 29, 2016. We identified 9 case-patients (Figure 2) from fecal specimens; all had no known travel history in the 2 weeks before illness. Median age was 33 (range 28–83) years. Gastrointestinal symptom onset ranged from early September 2015 to late December 2015. No patients were bacteremic. Treatment failure was reported for 2 patients; both were hospitalized and 1 died , although Shigella was not recorded as a cause of death. Duration of illness was not systematically collected. Diagnoses occurred in general practice (n = 5), sexual health clinics (n = 2), and hospitals (n = 2). Seven patients resided in London; 2 resided elsewhere in England, 1 of whom reported recent travel to London. Of 8 patients with reported ethnicity, 6 self-identified as white.
Figure 2.
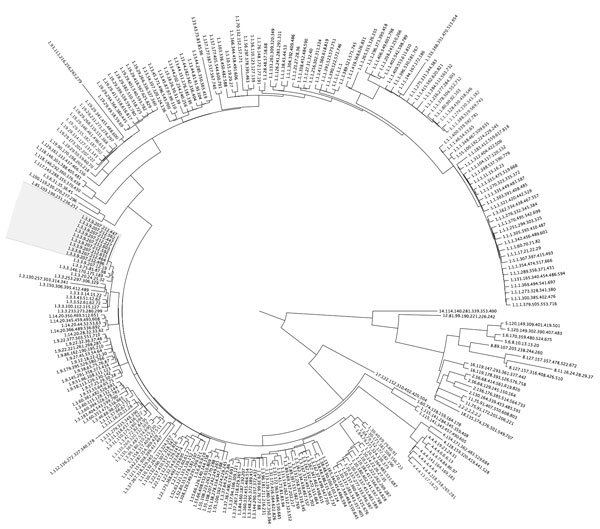
Maximum-likelihood phylogeny showing isolates from a cluster of 9 cases (gray shading) of Shigella sonnei infection among men who have sex with men in England, 2015. For context, 246 S. sonnei isolates that are representatives from each t25 cluster were included in the comparison. Isolates are labeled by single nucleotide polymorphism address.
In-depth questionnaires administered by telephone interviews collected information on sexual history, food exposures, and travel history; 8 patients completed the interviews. Seven answered detailed sexual history questions (including the 2 patients identified as MSM from clinical reporting data); all 7 reported having male partners. Six reported exposures considered to be high risk for Shigella transmission during the incubation period (e.g., fisting, sex under the influence of recreational drugs [6], oral-anal contact, attendance at sex parties, or use of sex toys with a partner) (Table 2). Reported number of sexual partners during the 2 weeks before illness was 0–5. The 7 patients who completed the sexual history questions reported previous high-risk sexual exposures; 6 of these patients and 1 patient who partially completed the questionnaire reported a history of other sexually transmitted infections, including gonorrhea, chlamydia, syphilis, nonspecific urethritis, hepatitis C, and pubic lice. Five reported using mobile apps to meet partners at some time in the past; none reported a prior shigellosis diagnosis. All had visited sexual health services (Table 2). Some patients were HIV positive (frequency is omitted to prevent deductive disclosure).
Table 2. Reported exposure history for 9 case-patients with Shigella sonnei infection in the cluster with ESBL production and macrolide resistance, England, 2015* .
| Risk factor | No. (%) exposed | No. (%) unexposed | No. unknown† | |
|---|---|---|---|---|
| Exposure ever before onset | ||||
| Self-identified as a man who has sex with men | 7 (100) | 0 | 2 | |
| Fisting | 2 (29) | 5 (71) | 2 | |
| Sex under the influence of recreational drugs | 4 (57) | 3 (43) | 2 | |
| Scat play | 0 | 7 (100) | 2 | |
| History of other sexually transmitted infections | 7 (88) | 1 (13) | 1 | |
| Use of apps to meet partners | 5 (71) | 2 (29) | 2 | |
| Awareness of Shigella infections | 0 | 7 (100) | 2 | |
| Previous Shigella diagnosis | 0 | 8 (100) | 1 | |
| Known to sexual health services |
7 (100) |
0 |
2 |
|
| Exposure during the 2 weeks before onset | ||||
| Fisting | 1 (14) | 6 (86) | 2 | |
| Sex under the influence of recreational drugs | 3 (43) | 4 (57) | 2 | |
| Oral–anal contact | 3 (43) | 4 (57) | 2 | |
| Condomless sex‡ | 5 (71) | 2 (29) | 2 | |
| Attended sex parties or live sex premises | 3 (43) | 4 (57) | 2 | |
| Use of sex toys with partner | 1 (14) | 6 (86) | 2 | |
| Scat play | 0 | 7 (100) | 2 | |
*ESBL, extended-spectrum β-lactamase; scat play, sexual arousal or activity linked to feces. †Unknowns were not included in percentage calculations; 7 of 9 patients completed and 1 partially completed interviews about sexual behavior. ‡Any condomless sexual contact.
No epidemiologic links were identified among patients, and no point sources were identified, although food exposure data was collected for 5 patients only. Two reported attending the same sex-on-premises venue in London 3 months apart (1 reported no sexual contact at the venue). None reported awareness of possible infection with Shigella spp. or of contact with others with gastrointestinal symptoms. Three thought they had acquired their infections from food. Patients reported that they accessed health promotion messages at sexual health clinics and on the internet.
Conclusions
We describe a phylogenetic cluster of ESBL-producing S. sonnei infections in MSM in England that raises concerns about the ability to manage the spread of resistant Shigella infection in this population. Whole-genome sequencing revealed that the multidrug-resistance phenotype is conferred by acquisition of an IncFII plasmid (p183660) known to be circulating in S. flexneri–affected MSM (4); this plasmid has acquired blaCTX-M-27.
Patient-reported behaviors were similar to those reported in an earlier S. flexneri epidemic among MSM: high numbers of sexual partners, high levels of condomless sex, attendance at sex parties, sex under the influence of recreational drugs, and prior HIV infection (6). We identified no point source, further indicating that sexual contact was the dominant mode of transmission.
Although S. sonnei causes self-limiting diarrhea in most patients, life-threatening invasive infections in patients co-infected with HIV have occurred (7). Quinolone and azithromycin resistance have been observed in recent outbreaks of S. sonnei infections in MSM; cepaholsporins are recognized as a suitable therapeutic choice for invasive or prolonged infections (8,9).
Primary public health concerns include possible treatment failure for severe shigellosis in immunocompromised patients; further spread of the multidrug-resistant IncFII plasmid to other enteric pathogens in MSM, with possible implications for treatment among the immunocompromised; rapid spread of drug-resistant S. sonnei infections among MSM, including outside the United Kingdom, as happened with S. flexneri 3a (4), especially among the HIV infected; and transmission through food handling and childcare centers, with potential to cause outbreaks of drug-resistant S. sonnei infections in other populations. Low awareness of Shigella infections among patients in our study suggests that prior awareness campaigns targeting high-risk MSM in England (10) have not been fully successful. Future campaigns being planned to improve awareness of Shigella infection and transmission routes will target social media, sexual health clinics, and primary care workers to increase awareness among MSM and healthcare staff. Frontline sexual health clinicians and microbiologists in England have been made aware of emerging drug resistance in Shigella spp. among this vulnerable group and of the need to perform antimicrobial drug susceptibility testing if treatment is considered necessary.
Detailed methods for whole genome sequencing of Shigella sonnei isolates and plasmid elucidation.
Acknowledgments
We thank all persons involved in the outbreak investigation, including PHE Health Protection Team staff, with whom we liaised during patient follow-up.
This work was supported by the National Institute for Health Research Health Protection Research Unit in Gastrointestinal Infections at the University of Liverpool.
Biography
Dr. Mook is an epidemiologist with the Field Epidemiology Service, Public Health England. His research interests include gastrointestinal disease surveillance and outbreak response.
Footnotes
Suggested citation for this article: Mook P, McCormick J, Bains M, Cowley LA, Chattaway MA, Jenkins C, et al. ESBL-producing and macrolide-resistant Shigella sonnei infections among men who have sex with men, England, 2015. Emerg Infect Dis. 2016 Nov [date cited]. http://dx.doi.org/10.3201/eid2211.160653
These authors contributed equally to this article.
These senior authors contributed equally to this article.
References
- 1.Borg ML, Modi A, Tostmann A, Gobin M, Cartwright J, Quigley C, et al. Ongoing outbreak of Shigella flexneri serotype 3a in men who have sex with men in England and Wales, data from 2009-2011. Euro Surveill. 2012;17:20137. [PubMed] [Google Scholar]
- 2.Simms I, Field N, Jenkins C, Childs T, Gilbart VL, Dallman TJ, et al. Intensified shigellosis epidemic associated with sexual transmission in men who have sex with men—Shigella flexneri and S. sonnei in England, 2004 to end of February 2015. Euro Surveill. 2015;20:21097. 10.2807/1560-7917.ES2015.20.15.21097 [DOI] [PubMed] [Google Scholar]
- 3.Ashton P, Nair S, Peters T, Tewolde R, Day M, Doumith M, et al. Revolutionising public health reference microbiology using whole genome sequencing: salmonella as an exemplar. bioRxiv. 2015;•••:033225. [Google Scholar]
- 4.Baker KS, Dallman TJ, Ashton PM, Day M, Hughes G, Crook PD, et al. Intercontinental dissemination of azithromycin-resistant shigellosis through sexual transmission: a cross-sectional study. Lancet Infect Dis. 2015;15:913–21. 10.1016/S1473-3099(15)00002-X [DOI] [PubMed] [Google Scholar]
- 5.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbart VL, Simms I, Jenkins C, Furegato M, Gobin M, Oliver I, et al. Sex, drugs and smart phone applications: findings from semistructured interviews with men who have sex with men diagnosed with Shigella flexneri 3a in England and Wales. Sex Transm Infect. 2015;91:598–602. 10.1136/sextrans-2015-052014 [DOI] [PubMed] [Google Scholar]
- 7.Baer JT, Vugia DJ, Reingold AL, Aragon T, Angulo FJ, Bradford WZ. HIV infection as a risk factor for shigellosis. Emerg Infect Dis. 1999;5:820–3. 10.3201/eid0506.990614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann C, Sahly H, Jessen A, Ingiliz P, Stellbrink H-J, Neifer S, et al. High rates of quinolone-resistant strains of Shigella sonnei in HIV-infected MSM. Infection. 2013;41:999–1003. 10.1007/s15010-013-0501-4 [DOI] [PubMed] [Google Scholar]
- 9.Bowen A, Eikmeier D, Talley P, Siston A, Smith S, Hurd J, et al. ; Centers for Disease Control and Prevention (CDC). Notes from the field: outbreaks of Shigella sonnei infection with decreased susceptibility to azithromycin among men who have Sex with men—Chicago and Metropolitan Minneapolis–St. Paul, 2014. MMWR Morb Mortal Wkly Rep. 2015;64:597–8. [PMC free article] [PubMed] [Google Scholar]
- 10.Public Health England. Shigella: leaflet and poster for men who have sex with men. 2013. Dec 1 [cited 2016 Feb 10]. https://www.gov.uk/government/publications/shigella-leaflet-and-poster
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed methods for whole genome sequencing of Shigella sonnei isolates and plasmid elucidation.


