Abstract
Objective
Exemestane is a potent third-generation aromatase inhibitor used as endocrine therapy in breast cancer treatment and prevention. Characterization of its metabolic pathway is incomplete with ambiguity existing in the identity of enzymes driving production of its key metabolite, 17β-dihydroexemestane. The impact of genetic variation on exemestane metabolism is also unknown. This study seeks to describe cytosolic reductase involvement in hepatic exemestane metabolism and to assess the impact of functional polymorphisms on metabolite production.
Methods
Phase I metabolites were identified in incubations of exemestane with pooled human liver cytosol or recombinant protein for AKR1Cs and CBR1. Kinetic parameters characterizing exemestane reduction were measured for purified wildtype enzymes, and nonsynonymous variants occurring at >1% minor allele frequency using UPLC/MS/MS.
Results
Human liver cytosol, CBR1, AKR1C1, AKR1C2, AKR1C3, and AKR1C4 reduce exemestane to active primary metabolite 17β-dihydroexemestane. Formation of a novel metabolite, 17α-dihydroexemestane, was catalyzed by recombinant AKR1C4 and CBR1 in addition to hepatic cytosol. Variants AKR1C3 Arg258Cys and AKR1C4 Gly135Glu had significantly decreased affinity for EXE relative to their respective wildtypes. Five common AKR1C3 polymorphisms were associated with decreased rates of catalysis while AKR1C4 Gly135Glu increased the velocity of EXE reduction.
Conclusions
AKR1Cs and CBR1 catalyze exemestane reduction in vitro. These results imply that cytosolic ketosteroid reductases may participate in the exemestane metabolic pathway in vivo. In addition, several common variants were associated with altered enzymatic activity, suggesting that functional polymorphisms could play an important role in overall exemestane metabolism and activity by altering the extent and duration of 17β-dihydroexemestane exposure.
Keywords: Exemestane, dihydroexemestane, aromatase inhibitor, hydroxysteroid dehydrogenase, aldo-keto reductase, pharmacogenetics, breast cancer
Introduction
Breast cancer was the second leading cause of cancer-related death as well as the most commonly diagnosed invasive malignancy in American women in 2012 with incidence rates almost 2.5-fold higher than lung cancer [1]. In response to an ongoing need for new endocrine therapies, third-generation aromatase inhibitors (AIs), including anastrazole, letrozole, and exemestane (6-methylenandrosa-1, 4-diene-3-17-dione), have been adopted into the armory of clinicians [2]. Since its inception, exemestane (EXE) has proven a versatile tool sans the life-threatening complications of tamoxifen (TAM), which include thromboembolism and increased risk of gynecological abnormalities, including endometrial cancer [3–10]. Although EXE is often prescribed to postmenopausal women with metastatic breast cancer following tumor progression on TAM, it is also a useful adjuvant for early hormone responsive breast cancer subsequent to 2–3 years initial TAM for a total duration of five years of hormone therapy [4, 11, 12]. The MAP.3 clinical trial examining long-term use in the risk reduction setting further suggests that EXE is a promising chemopreventative, as it is associated with a 65% decrease in incidence of invasive breast cancer (IBC) in high-risk postmenopausal women [13]. Recently completed studies imply that premenopausal women with mammary tumors may likewise derive benefit from ovarian function suppression (OFS) supplemented with daily EXE [14]. A meta-analysis of the SOFT and TEXT clinical trials in premenopausal women with hormone-responsive early breast cancer found decreased recurrence from combination treatment of EXE plus OFS compared to TAM with OFS [14]. On the basis of these observations, it appears that EXE is a safe, efficacious alternative to currently approved treatments with diverse applications in disease management.
EXE is structurally similar to androstenedione, a key substrate of aromatase [15]. Its potent therapeutic effect is mediated through so-called suicide inhibition of the enzyme [16]. EXE irreversibly binds to the active site of aromatase, disrupting androgen conversion into aromatic estrogens [16]. As aromatase is the predominant mechanism for estrogen biosynthesis in postmenopausal women, AIs afford clinical benefit by impeding the production of estrogens, which are known to exacerbate ER-positive mammary tumors [17–20]. Although EXE is highly effective in vivo, suppressing circulating estrogen levels in post-menopausal breast cancer patients by 98%, its complete metabolic pathway is as of yet uncharacterized nor is it known the extent to which genetic variation impacts EXE metabolism [21, 22]. The original manufacturer states that EXE (Aromasin®) is metabolized hepatically by CYP3A4 and aldo-keto reductases, of which there are 15 human members with varying tissue expression patterns [23–25].
The limited literature available describes 17β-dihydroexemestane (17β-DHE) as a prominent phase I EXE metabolite possessing both anti-aromatase and androgen agonistic activities in vitro [22, 26–30]. One study concluded that 17β-DHE concentrations were approximately 35–40% those of the parent drug in the plasma of healthy individuals taking EXE while another smaller study found that the amount of 17β-DHE relative to EXE in human plasma varied five-fold in a pool of only three participants [27, 31]. These observations support 17β-DHE as a major metabolite and highlight inter-individual variation in EXE metabolism. While drug disposition is undoubtedly multifactorial, it has been estimated that 20–95% of variability in drug response is attributable to genetic factors [32]. Discernment of relevant polymorphisms associated with varied EXE metabolism between individuals requires that enzymes participating in its biotransformation first be definitively identified.
Modification of the steroid scaffold at C17 is a well-documented phenomenon central to the regulation of human steroid hormone potency [33]. One early study predicted that EXE, like many steroids, is vulnerable to phase I metabolism at the carbonyl group occupying this position [34]. Carbonyl-reducing enzymes catalyze similar reactions despite contributions from two distinct protein phylogenies, the aldo-keto reductase (AKR) and short-chain dehydrogenase/reductase (SDR) superfamilies [35]. These enzymes play a prominent role in endogenous steroid metabolism by transforming ketosteroids into hydroxysteroid alcohols, altering their ligand affinities and rendering them available for conjugative reactions with phase II enzymes, such as the UGTs [36]. Reduction of EXE at C17 to form a reactive beta hydroxyl has since been confirmed and is known to facilitate additional metabolism by UGT2B17 and ultimately, excretion [22].
In total, twelve NADP(H)-dependent enzymes from the AKR and SDR superfamilies are believed to participate in carbonyl-containing xenobiotic reduction [35]. Of these enzymes, AKR1C1, 1C2, 1C3, and 1C4 (termed AKR1C1-4) as well as CBR1 are soluble, hepatically expressed and active against ketosteroids so it stands to reason that these reductases may be responsible for functionalizing the C17 carbonyl group of EXE to produce the 17β-DHE metabolite in human liver cytosol [35]. Numerous nonsynonymous polymorphisms have been described in hepatic xenobiotic-metabolizing enzymes, including the AKRs [37]. Naturally occurring variations in enzymes active in EXE metabolism could lead to differential metabolite production between individuals, potentially altering overall duration of exposure to antiestrogen therapy in vivo and thus, clinical outcomes. The present study seeks to clarify the metabolic pathway of EXE by identifying cytosolic hepatic reductases active in its biotransformation, as well as any phase I metabolites produced. For the first time, the functional consequences of common polymorphisms on reductase-mediated production of the active metabolite, 17β-DHE, are also explored.
Materials and Methods
Chemicals and materials
4-Androstene-3,17-dione, boldenone and testosterone used in exemestane and 17-dihydroexemestane synthesis were purchased from Hangzhou DayangChem Co. (Hangzhou, Zhejiang, China). All other reagents used in steroid synthesis were ACS grade or higher and purchased from Sigma-Aldrich (St. Louis, Missouri, US), Tokyo Chemical Industry Co. (Tokyo, Japan) or Thermo Fisher Scientific (Waltham, Massachusetts, US). Thin-layer chromatography plates from Bonna-Agela Technologies Inc. (Wilmington, DE, US) and silica columns from Yamazen Corp. (Osaka, Japan) were used for purification following synthesis. Codon-optimized pQE-T7 overexpression plasmids for wildtype AKR1C1-4, as well as CBR1, were purchased from Qiagen (Germantown, Maryland, US). The QuikChange II Site-Directed Mutagenesis Kit used to make variant reductase expression vectors was acquired from Agilent (Santa Clara, California, US). Oligonucleotides to prime site-directed mutagenesis were manufactured by Integrated DNA Technologies (Coralville, Iowa, US). Kanamycin, chlorophenicol, isopropyl β-D-1-thiogalactopyranoside (IPTG), and imidazole were obtained from Sigma-Aldrich. Pooled human liver cytosol collected from 50 individuals was procured from XenoTech (Lenexa, Kansas, US). Eighty-six percent of the donors were Caucasian, while 4% and 10% were African American or Hispanic, respectively. The NADPH regeneration system used for activity assays was purchased from Corning (Corning, New York, US). B-PER Complete protein extraction reagent, Halt EDTA-free protease inhibitor cocktail, Ni-NTA affinity purification columns, and Slide-a-Lyzer G2 dialysis cassettes (10 kDa MWCO) were purchased from Thermo Fisher Scientific. All solvents used for mass spectrometry were LC/MS grade and also obtained from Thermo Fisher Scientific. Luria broth base and 4–20% Tris-Glycine gels for SDS-PAGE were acquired from Invitrogen (Carlsbad, California, US). The BCA protein assay and silver staining kits used in protein purity assessment were purchased from Pierce (Rockford, Illinois, US).
Synthesis of EXE and DHE
Exemestane, 17α-dihydroexemestane (17α-DHE), and 17β-dihydroexemestane were synthesized on site at Washington State University to a purity of > 99%. EXE and 17β-DHE were prepared and authenticated in accordance with previously published methods [34, 38]. As depicted in the Supplemental Digital Content 1 schematic, 17α-DHE was derived from testosterone through stereospecific inversion of the Mitsunobu reaction, followed by removal of the carboxylic acid moiety, an unusual Mannich reaction, and 1,2-dehydrogenation [38, 39]. For a detailed description of the 17α-DHE synthesis scheme, refer to the text of Supplemental Digital Content 2. A Yamazen AI-580s flash chromatography system was employed to purify each compound following synthesis. Purity was determined by PDA spectrum (210 nm-400 nm) on an Acquity I Class UPLC platform from Waters (Milford, Massachusetts, USA). Prior to resuspension in ethanol and storage at −80°C, the structure of each steroid was verified by reviewing nuclear magnetic resonance spectra recorded on a Bruker AV300 instrument (Billerica, MA, US) with the kind permission of the Department of Chemistry at Gonzaga University (Spokane, WA).
Identification of nonsynonymous polymorphisms
Functional polymorphisms for the AKR1C subfamily and CBR1 were derived from the National Center for Biotechnology Information (NCBI) Variation Viewer using filters to search dbSNP and dbVar for any missense or nonsense gene variants arising from single nucleotide variations, insertions, deletions or frameshifts [40]. For the purpose of this study, common functional polymorphisms were those detected at a minor allele frequency (MAF) of > 1% in the human population according to the 1000 Genomes Project or GO-ESP datasets. Data regarding inter-ethnic differences in MAF for common ketosteroid reductase variants identified was extracted from NCBI’s 1000 Genomes Browser and organized into the table within Supplemental Digital Content 3.
Recombinant protein production
Overexpression vectors encoding common variant ketosteroid reductases (MAF > 0.01) were created from wildtype AKR1C and CBR1 plasmids via site-directed mutagenesis. Oligonucleotide sequences used to prime mutagenesis are provided in the table Supplemental Digital Content 4. Each expression vector was transformed into chemically competent BL21. Transformants were grown on kanamycin selection plates and confirmed via Sanger sequencing. Isolated bacterial colonies were scraped into 150-mL Luria broth supplemented with 7.5 mg kanamycin and 3.75 mg chlorophenicol and grown at 37°C for 16 h in a table-top shaker with gentle aeration (200 rpm). Seven-hundred and fifty mL fresh Luria broth containing 37.5 mg kanamycin and 18.75 mg chlorophenicol was inoculated with 120 mL of the overnight culture and grown for an additional 1.75 h to reach log-phase growth. Protein expression was induced by the addition of 0.5 mM IPTG followed by 3 h of growth at 37°C. BL21 were pelleted by centrifugation and lysed with 25 mL B-PER Complete with EDTA-free protease inhibitor cocktail.
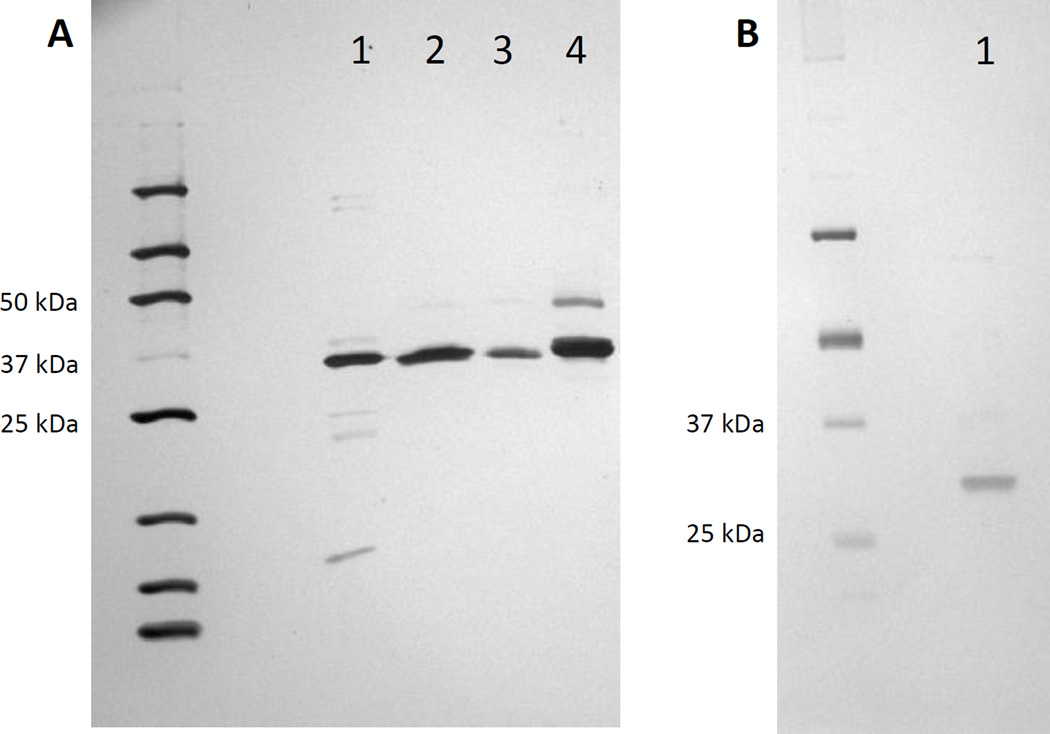
Polyhistidine-tagged recombinant protein was bound to Ni-NTA resin and then affinity purified through the sequential addition of increasing concentrations of imidazole suspended in PBS, pH 7.4. Purification columns were initially equilibrated with wash buffer containing 50 mM NaH2P04, 300 mM NaCl, and 10 mM imidazole, pH 8.0. Each bacterial lysate was mixed with equilibration buffer 1:1, loaded onto a dedicated column, and allowed to drain by gravity. Columns were incubated at room temperature for 30 min to promote maximum protein binding. Columns were then successively washed with 20 mM, 60 mM, 100 mM, and 250 mM imidazole. Each imidazole wash was allowed to drain from the resin bed completely by gravity. Flow-through was discarded. Recombinant enzyme was eluted using 500 mM imidazole wash buffer. Dialysis was performed against PBS for 12 h at 4°C. PBS was changed 90 min after initiating dialysis and then again after 3 h to ensure complete removal of imidazole prior to use in pharmacokinetic assays. 750 ng of each enzyme was subjected to SDS-PAGE on 4–20% Tris-Glycine polyacrylamide gel and all were found to be > 80% pure by silver staining (Figure 1).
Fig. 1.
Silver staining of purified recombinant reductases. Panel (A), AKR1C1; lane 1, AKR1C2; lane 2, AKR1C3; lane 3, AKR1C4; lane 4. Panel (B), CBR1; lane 1.
Metabolite Identification
EXE-derived metabolites were identified in 18-h enzymatic incubations with 200 µM EXE in PBS, pH 7.4 at 37°C. Each 50-µl incubation was supplemented with NADPH regeneration system (1.55 mM NADP+, 3.3 mM glucose-6-phosphate, 3.3 mM MgCl2, 0.5 µl of 40 U/ml glucose-6-phosphate dehydrogenase) and included 7.5 µg pooled human liver cytosol or 1.5 µg of recombinant wildtype reductase. Overnight incubations with 200 µM 17β-DHE and 17α-DHE were also performed under the same conditions to assess reversibility of EXE reduction. Following termination with ice-cold acetonitrile, each incubation was centrifuged at 4°C for 15 min at 13,200 g. Supernatant was collected and analyzed via ultra-pressure liquid chromatography coupled with tandem mass spectrometry using a Waters ACQUITY UPLC/MS/MS system equipped with a protective 0.2 µm in-line filter in series with a 1.7 µm ACQUITY UPLC BEH C18 column (2.1 mm × 50 mm, Ireland). The UPLC conditions used in these analyses are detailed in Table 1. Each supernatant was initially screened for the presence of EXE metabolites using UPLC/MS and electrospray ionization to detect positively charged molecular ions with m/z ranging from 100 to 450. To confirm the identity of peaks observed in the comprehensive scan, a second targeted UPLC/MS/MS scan was performed using mass transitions m/z 297.34 → 185.07, m/z 299.14 → 135.07, and m/z 299.20 → 135.13 to monitor EXE, 17α-DHE, and 17β-DHE, respectively. Desolvation temperature was 500°C with 800 L/h nitrogen gas used for drying. Collision energy was optimized at 25 V for EXE and 20 V for 17-DHE. 25 V cone voltage and 0.01 s dwell time resulted in high sensitivity detection of all three compounds. Retention times of metabolites observed in the enzymatic incubations were compared to EXE metabolite standards.
Table 1.
UPLC conditions for detection of phase I exemestane metabolites.
| Time (min) | % Aa | % B | % C | curve |
|---|---|---|---|---|
| initial | 70 | 20 | 10 | initial |
| 0.5 | 70 | 20 | 10 | 11 |
| 9 | 30 | 60 | 10 | 6 |
| 9.5 | 0 | 90 | 10 | 6 |
| 10 | 70 | 20 | 10 | 6 |
Mobile phase A, 0.1% formic acid; mobile phase B, 100% methanol; mobile phase C, 100% acetonitrile. Flow rate = 0.4 mL/min.
Enzyme kinetic assays
Varying concentrations of EXE (0.5–400 µM) were included as substrate for reductase-mediated DHE production in 2-h incubations. Each reaction was performed in PBS, pH 7.4 at 37°C in the presence of an NADPH regeneration system. One-thousand ng recombinant AKR1C protein was used per reaction while 250 ng recombinant CBR1 was used to avoid substrate depletion, which would have precluded Michaelis-Menten modeling. Similar incubations were conducted using 5 µg pooled human liver cytosol (HLC) in 90-min incubations. Protein concentration and incubation length fell within the linear range of reaction velocity curves for EXE reduction (data not shown). All reactions were terminated with cold acetonitrile, centrifuged at 4°C for 15 min at 13,200 g, and then dried for 2 h at ambient temperature. 20 µl of water/acetonitrile (1:1) was used to ensure complete resuspension prior to analysis. 17β-DHE formation was monitored via the UPLC/MS/MS method described above and quantitated against a reference curve of known concentrations.
Statistical analysis
Kinetic parameters were determined according to the Michaelis-Menten equation using GraphPad Prism 6 software (GraphPad Software, Inc., San Diego, California). VMAX values derived from kinetic assays were normalized to account for recombinant protein purity and expressed as picomoles·min−1·mg−1. All reported values represent the results of independent assays run in triplicate. The activity of each variant enzyme was compared to its respective wildtype using Student’s t-test or one-way ANOVA supplemented with Sidak’s test for multiple comparisons, as appropriate. A two-tailed p value < 0.05 was considered the threshold for statistical significance.
Results
Identification of cytosolic EXE metabolites
The predominant metabolite formed in overnight incubations of EXE with pooled human liver cytosol was 17β-DHE although a lesser amount of 17α-DHE was detected as well (Figure 2). When presented with 17α-DHE or 17β-DHE as substrate, neither exemestane nor secondary metabolite formation was observed in HLC incubations (data not shown). Recombinant AKR1C4 and CBR1 catalyzed reduction of EXE to both stereoisomers of DHE while only 17β-DHE formation was mediated by AKR1C1-3.
Fig. 2.
Identification of exemestane metabolites in overnight incubations. Panel (A), Mixed gender liver cytosol pooled from 50 human donors; panel (B), AKR1C1; panel (C), AKR1C2; panel (D), AKR1C3; panel (E), AKR1C4; and panel (F), CBR1. Peak 1, Exemestane; peak 2, 17β-dihydroexemestane; peak 3, 17α-dihydroexemestane.
Kinetic analysis of 17β-DHE formation
Assays monitoring reduction of EXE to 17β-DHE by purified wildtype protein suggest that AKR1C4 has the highest affinity for EXE (KM = 9.7 ± 1.9 µM) followed by AKR1C3, AKR1C2, AKR1C1, and CBR1 with KM values of 12.3 ± 1.1, 16.4 ± 0.6, 35.3 ± 3.8, and 265 ± 21 µM, respectively. The apparent KM for HLC was established as 55.7 ± 1.9 µM. Representative plots of 17β-DHE formation versus EXE substrate concentration are shown in Figure 3. 17β-DHE formation catalyzed by pooled HLC proceeded at a maximum rate of 21 ± 4.8 pmol·min−1·mg−1. CBR1 reduced EXE about 11-fold faster than AKR1C2 with an observed VMAX of 928 ± 244 pmol·min−1·mg−1. AKR1C2 (84.2 ± 15.6 pmol·min−1·mg−1) and AKR1C1 (71.1 ± 1.5 pmol·min−1·mg−1) formed 17β-DHE at similar maximum velocities. The VMAX for AKR1C3 was 36-fold higher than that of AKR1C4 (83.3 ± 15.4 versus 2.3 ± 0.6 pmol·min−1·mg−1). Intrinsic clearance as calculated by VMAX/KM was highest for AKR1C3 (6.8) followed closely by AKR1C2 (5.1), then CBR1 (3.5) and AKR1C1 (2.0). Recombinant AKR1C4 (0.24) displayed the lowest overall activity of wildtype enzymes assayed (Table 2).
Fig. 3.
Representative kinetics curves for the reduction of exemestane to 17β-dihydroexemestane. Panel (A), pooled human liver cytosol; panel (B), AKR1C1; panel (C), AKR1C2; panel (D), AKR1C3; panel (E), AKR1C4; and panel (F), CBR1.
Table 2.
Kinetic analysis of wildtype and variant ketosteroid reductases active against exemestane.
| Reductase enzyme or variant |
NCBI dbSNP Identifier |
1000 Genomes Project (MAF) |
GO-ESP (MAF) |
KM (µM) |
VMAX (picomoles·min−1·mg−1)a |
CLINT (nl·min−1·mg−1) |
|---|---|---|---|---|---|---|
| AKR1C1 | 35.3 ± 3.8 | 71.1 ± 1.5 | 2.0 | |||
| AKR1C2 | 16.4 ± 0.6 | 84.2 ± 15.6 | 5.1 | |||
| AKR1C2 Phe46Tyr | rs2854482 | 0.0649 | 0.07 | 13.5 ± 2.8 | 105.1 ± 6.3 | 7.8 |
| AKR1C3 | 12.3 ± 1.1 | 83.3 ± 15.4 | 6.8 | |||
| AKR1C3 His5Gln | rs12529 | 0.4203 | 0.43 | 17.9 ± 2.9 | 3.8 ± 0.1* | 0.21 |
| AKR1C3 Glu77Gly | rs11551177 | 0.0367 | 0.0503 | 13.9 ± 2.2 | 1.9 ± 0.2* | 0.14 |
| AKR1C3 Lys104Asn | rs12387 | 0.1518 | 0.1569 | 16.8 ± 1.0 | 6.4 ± 0.3* | 0.38 |
| AKR1C3 Pro180Ser | rs34186955 | 0.0086 | 0.0117 | 11.2 ± 3.1 | 3.6 ± 0.6* | 0.32 |
| AKR1C3 Arg258Cys | rs62621365 | 0.0325 | 0.001 | 75.8 ± 19.9* | 2.3 ± 0.1* | 0.03 |
| AKR1C4 | 9.7 ± 1.9 | 2.3 ± 0.6 | 0.24 | |||
| AKR1C4 Gly135Glu | rs11253043 | 0.027 | 0.0278 | 311.4 ± 75.7* | 19.1 ± 4.2* | 0.06 |
| AKR1C4 Ser145Cys | rs3829125 | 0.1028 | 0.1143 | 9.6 ± 2.4 | 5.9 ± 1.1 | 0.61 |
| AKR1C4 Leu311Val | rs17134592 | 0.1024 | 0.1143 | 11.9 ± 2.0 | 6.3 ± 0.8 | 0.53 |
| CBR1 | 265 ± 21 | 928 ± 244 | 3.50 |
All VMAX values were normalized to reflect the purity of recombinant enzymes assayed.
Denotes that a variant exhibited statistically significant deviations (p<0.001 in all cases) from the activity of its respective wildtype.
Impact of functional polymorphisms on EXE reduction
Screening the NCBI Variation Viewer for cytosolic ketosteroid reductase variants prevalent at 1% or greater yielded a single nonsynonymous polymorphism for AKR1C2, as well as three common variants in AKR1C4. Although six AKR1C3 allelic variants were identified, only five underwent kinetic screening to characterize 17β-DHE formation. AKR1C3 Glu36Term (rs1804062) was excluded as it encodes a truncated protein lacking functional co-factor and substrate binding domains. No polymorphisms matching our search criteria were detected for AKR1C1 or CBR1. KM and VMAX values were comparable between AKR1C2 and AKR1C2 Phe46Tyr (Table 2). Wildtype AKR1C3 demonstrated affinity for EXE similar to its variants His5Gln, Glu77Gly, Lys104Asn, and Pro180Ser. However, AKR1C3 Arg258Cys recombinant protein had roughly 6-fold lower affinity for EXE (KM = 12.3 ± 1.1 µM versus 75.8 ± 19.9 µM). Furthermore, all AKR1C3 functional variants assayed exhibited greatly diminished velocity of EXE reduction leading to sizable disparities from wildtype in intrinsic clearance. The KM was similar between wildtype AKR1C4 (9.7 ± 1.9 µM) and its Ser145Cys (9.6 ± 2.4 µM) and Leu311Val (11.9 ± 2.0 µM) variants. The experimental KM for AKR1C4 Gly135Glu using EXE as substrate (311.4 ± 75.7 µM) was 32-fold higher than that of wildtype, indicative of significantly lowered affinity resulting from amino acid substitution. No notable difference was observed in the VMAX of 17β-DHE formation between AKR1C4 and its Ser145Cys or Leu311Val allelic variants. Recombinant AKR1C4 Gly135Glu metabolized EXE 8-fold faster than its wildtype counterpart (VMAX = 19.1 ± 4.2 versus 2.3 ± 0.6 pmol·min−1·mg−1).
Discussion
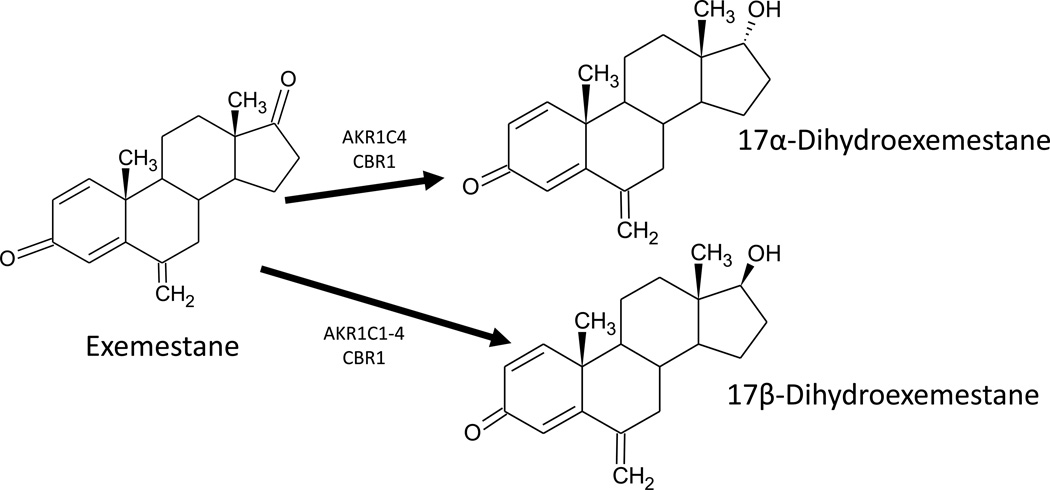
Studies examining phase I exemestane metabolism are scarce. However, the limited data available suggest that 17β-DHE is a major EXE metabolite produced via reduction of the C17 carbonyl moiety [26–29]. Although involvement of the aldo-keto reductase superfamily has previously been disclosed, specific enzymes catalyzing hepatic EXE metabolism have thus far been unidentified [23]. The present study confirms that C17 reduction of EXE by cytosolic carbonyl reductases to yield 17β-DHE is indeed a major metabolic pathway (Figure 4).
Fig. 4.
Schematic of in vitro exemestane metabolism by hepatic cytosolic AKR and SDR ketosteroid reductases.
Formation of an additional stereoisomer, 17α-DHE, was detected in concert with 17β-DHE in the current study. Oxidation of DHE to form EXE was not observed in incubations of human liver cytosol with an abundance of 17α- or β-DHE, suggesting that biotransformation of EXE by hepatic cytosolic reductases is irreversible (data not shown). A previous study exploring the ability of 17β-DHE to impede aromatase-mediated estrogen formation concluded that 17β-DHE inhibits aromatase with potency similar to its parent compound EXE [22]. 17α-DHE, on the other hand, appears to be a subsidiary phase I metabolite with no appreciable aromatase-inhibiting properties (unpublished observations). As an active EXE metabolite, 17β-DHE may contribute to the therapeutic mechanism of systemic estrogen deprivation and thus, influence in vivo efficacy of EXE in breast cancer treatment and prevention.
While CBR1, as well as all 1C members of the AKR superfamily reduce EXE to 17β-DHE, only AKR1C4 and CBR1 produced detectable quantities of 17α-DHE. Current literature strongly suggests that human AKR1C isoforms are remarkably functionally plastic [25]. The stereochemistry of hydroxylated AKR1C products is known to be dependent upon the initial docking position of substrate in the enzyme active site [41, 42]. Accommodation of multiple modes of substrate binding allows distinct hydroxysteroid products to be enzymatically derived from a single ketosteroid precursor, possibly explaining the production of both 17-DHE stereoisomers from parent EXE [25, 41, 42]. Furthermore, it has been reported that AKR1C4 reduces tibolone to both 3α- and 3β-hydroxytibolone in vitro using a single active site [41]. The same study suggests that AKR1C4 interconverts the two stereoisomers exhibiting dual oxidoreductase and epimerase activities [41]. Whether the production of 17α-DHE by recombinant AKR1C4 in the present study is attributable to innate promiscuity of the active site or epimerase activity is unknown.
Wildtype AKRs 1C2, 1C3, and 1C4 show similar affinity for EXE, while AKR1C1, which functions primarily as a 20-ketosteroid reductase, exhibited the lowest affinity of the AKRs assayed [43]. CBR1, a short-chain dehydrogenase reductase, displayed the lowest overall affinity for EXE but catalyzed 17β-DHE formation much more rapidly (11 to 403-fold) than recombinant AKR1C enzymes. The kinetic mechanism of AKRs is strictly ordered with NADP(H) cofactor binding prior to docking of substrate and dissociating only after its release [44–46]. Cofactor release as well as substrate reduction are thought to be the primary determinants of overall catalytic rate [47]. However, the kinetic mechanism of CBR1 may differ from that of the AKRs, which could account for the accelerated rate of CBR1-mediated EXE reduction.
Though our in vitro data suggest that intrinsic clearance of EXE is highest for AKR1C3 followed by AKR1C2 > CBR1 > AKR1C1 ≃ AKR1C4, the in vivo contribution of each enzyme to hepatic EXE metabolism is likely influenced by substrate availability, as well as relative expression. While CBR1 produces 17β-DHE quickly in vitro, it has low affinity for EXE (KM = 265 ± 21 µM) implying that it may play a lesser role at therapeutic doses given to patients. AKRs 1C1-4, on the other hand, display much higher affinity for EXE with KM values ranging from 9.7 ± 1.9 to 35.3 ± 3.8 µM. Quantitative RT-PCR data by Penning et al. suggests that transcripts encoding each AKR1C isoform are abundant and equally expressed in human liver, the principle site of EXE metabolism [23, 25]. AKR1C1-3 are also well expressed in breast tissue, which is a primary site of estrogen synthesis in postmenopausal women [25, 48]. These findings allude to the potential contributions of soluble AKR1C carbonyl reductases in converting a potent aromatase inhibitor to the active metabolite, 17β-DHE, in vivo.
Xenobiotic metabolizing enzymes, including the AKRs, are notoriously polymorphic with multiple high penetrance variants naturally found in the human populace [37]. Several AKR1C polymorphisms result in altered catalytic activity in vitro while other allelic variants are associated with increased risk of life-threatening pathologies [49–51]. In the present study, a subset of common cytosolic reductase variants differed from their wildtype counterparts with respect to affinity for EXE, as well as maximal velocity in its reduction to 17β-DHE.
AKR1C2 Phe46Tyr is similar to wildtype AKR1C2 in both affinity for EXE substrate and maximum velocity of 17β-DHE formation. While Bains et al. recorded reduction of the anti-cancer drugs daunorubicin and doxorubicin comparable to wildtype, tyrosine substitution at residue 46 decreased overall AKR1C2 activity against 1-acenaphthenol by approximately 30% [52]. Takahashi et al. reported a 40% decrease in VMAX for 5α-DHT reduction relative to wildtype enzyme [53]. These conflicting data suggest that deviations in catalytic activity could be substrate dependent. It has also been theorized that cofactor binding may be compromised in the Phe46Tyr variant leading to reduced enzymatic activity [49]. If this is indeed the case, it is feasible that use of supraphysiological concentrations of NADPH in the present study might mask moderately lowered cofactor affinity resulting in similar VMAX for EXE reduction by both wildtype and polymorphic AKR1C2 protein.
The AKR1C3 variants His5Gln, Glu77Gly, Lys104Asn, and Pro180Ser possess comparable affinity for EXE, but hydroxylation of the ketosteroid is sluggish compared to their wildtype counterpart, whose CLINT was 18–49-fold higher than that exhibited for the four AKR1C3 variants (Table 2). The Pro180Ser polymorphism occurs near α-helix 5 of the conserved (α/β)8 -barrel conformation characteristic of the aldo-keto reductase superfamily and has previously been associated with decreased VMAX using daunorubicin and doxorubicin as substrates for reduction [52, 54]. As it lies in close proximity to key cofactor binding residues (Ser166, Asp167, and Glu190), it has been proposed that NADPH binding may be adversely affected, thus lowering overall rates of catalysis [52].
The AKR1C3 Arg258Cys functional polymorphism is associated with a large (227-fold) decrease in CLINT and a substantial decrease in affinity for EXE substrate relative to wildtype (12.3 ± 1.1 versus 75.8 ± 19.9 µM), possibly because of its physical proximity to tryptophan residue 227 of the substrate binding pocket. This variant maps to α-helix 7 and is likewise near multiple residues of the cofactor binding pocket likely explaining decreased 17β-DHE production [54].
The catalytic tetrad of AKR1C3, in turn, is comprised of amino acid residues His117, Lys84, Tyr55, and Asp50 [54]. Substitution of negatively charged glutamic acid for nonpolar glycine at amino acid 77 decreases overall rates of EXE reduction by recombinant protein 44-fold ostensibly by altering the chemical and conformational environment near the catalytic tetrad (83.3 ± 15.4 pmole·min−1·mg−1 compared to 1.9 ± 0.2 pmole·min−1·mg−1). Results of a Swedish study found that Caucasian men heterozygous for the Glu77Gly genotype had lower serum testosterone levels than wildtype homozygotes, implying that this particular polymorphism may influence risk of androgen-dependent pathologies [55].
Our experimental results, as well as the locations of the His5Gln and Lys104Asn functional polymorphisms, suggest that significantly reduced VMAX for EXE reduction is mediated through changes in overall enzyme stability or folding rather than changes in EXE or cofactor binding. An inverse association between His5Gln and bladder cancer risk has previously been noted in subjects of the Spanish Bladder Cancer Study [56]. If His5Gln exhibits decreased catalysis for substrates other than EXE, these observations may be attributable to decreased bioactivation of polyaromatic hydrocarbons by AKR1C3, thus modifying exposure to genotoxic compounds promoting tumorigenesis [56]. At this time, additional data is needed to fully explicate the role of common AKR1C3 polymorphisms in attenuating or enhancing cancer risk.
While AKR1C4 Ser145Cys exhibited a slightly higher VMAX as compared to wildtype (5.9 +1.1 versus 2.3 + 0.6 pmole·min−1·mg−1, respectively), it had an affinity for EXE similar to wildtype with KM values of 9.6 ± 2.4 and 9.7 ± 1.9 µM, respectively. VMAX values for EXE reduction did not appreciably differ (6.3 ± 0.8 and 2.3 ± 0.6 pmol·min−1·mg−1), which is consistent with a previous study concluding that overall catalytic activity of the Ser145Cys variant is comparable to wildtype AKR1C4 using several test substrates [50].
However, the results of our current investigation contrast to prior reports describing altered catalytic activity in the common AKR1C4 Leu311Val and Gly135Glu functional polymorphisms. Valine substitution at amino acid 311 results in decreased activity against several substrates including daunorubicin, 1-acenaphthenol, as well as steroids androsterone and 5β-androstane-3α, 17β-diol [50, 52]. A mutagenesis study conducted by Matsuura et al. suggests that the leucine residue 311 is involved in substrate binding at the AKR1C4 active site, and valine substitution decreased overall activity by nearly 50% for several substrates [57]. Although several nearby amino acid residues (Asn306, Ala308, and Tyr310) contribute to substrate binding on the C-terminal loop of human AKRs, significant deviations from wildtype in EXE substrate affinity and overall velocity of catalysis were not observed in kinetic assays monitoring 17β-DHE formation [54]. AKR1C4 Gly135Glu, on the other hand, showed significantly less affinity for EXE (KM = 311.4 ± 75.7), and maximum velocity of its reduction (VMAX = 19.1 ± 4.2 pmol·min−1·mg−1) was 8-fold faster than wildtype AKR1C4. VMAX and KM values previously reported for AKR1C4 and the Gly135Glu variant do not differ for doxorubicin, daunorubicin, and 1-acenaphthenol substrates [52]. The extent to which functional polymorphisms impact catalysis is likely a complex interplay between the unique chemistry of each substrate and the specific nature of the amino acid deviations present. Thus, discrepancies between our observations and those reported in the literature may simply reflect a substrate-dependent effect on catalysis by variant AKR1C4 proteins.
In the present in vitro study, we attempted to clarify manufacturer claims regarding hepatic cytosolic EXE metabolism. Two isomers of DHE, the major phase I EXE metabolite, were identified in reactions with human liver cytosol, and five ketosteroid reductases were shown to be capable of catalyzing EXE reduction to the major metabolite 17β-DHE. Several functional polymorphisms in AKR1Cs resulted in altered rates of 17β-DHE formation compared to their respective wildtype enzymes as well as demonstrating substantially decreased affinity for EXE substrate. However, a limitation of the present study was the fact that the cytosolic reductases examined in our study were not over-expressed in a mammalian cell line system. Exemestane metabolism kinetics was performed using purified cloned reductase enzymes where protein mis-folding could potentially occur leading to possible artificial differences in protein instability, potentially affecting the kinetic activities observed.
Functional polymorphisms resulting in impaired reductase activity may partially underlie the varied response between individuals administered EXE by facilitating differential metabolite production. It has previously been suggested that 17β-DHE may contribute to the overall therapeutic mechanism of its parent compound [30]. Thus, it is feasible that genetic factors affecting conversion of EXE to its active metabolite could potentially impact circulating 17β-DHE levels, and in turn, influence overall clinical benefit. Albeit a well-established endocrine therapy, to our knowledge, this is the first time that the novel EXE metabolite 17α-DHE has been described in the literature. It is evident that many unanswered questions remain regarding EXE metabolism, and additional unidentified metabolites may yet exist. Although involvement of CYP3A4 is known, in vitro experiments by Kamdem et al. suggest that overall CYP450-mediated metabolism of exemestane may include contributions by additional isoforms [23, 58]. Therefore, future studies should examine the relative contributions of major hepatic CYP450s vs soluble cytosolic ketosteroids reductases to phase I metabolism of this drug. Genotype-phenotype correlative studies are also needed to determine whether functional AKR1C polymorphisms with deviant catalytic activity in vitro are associated with drug efficacy or incidence of adverse events in patients taking EXE for breast cancer treatment or prevention.
Supplementary Material
Acknowledgments
Source of Funding: None declared
These studies were graciously supported by Public Health Service (PHS) 1R01-CA164366-01A1 (National Cancer Institute) from the National Institutes of Health, U.S. Department of Health and Human Services to P. Lazarus.
Footnotes
Conflicts of interest
There are no conflicts of interest.
List of supplemental digital content
Supplemental Digital Content 1.ppt
Supplemental Digital Content 2.docx
Supplemental Digital Content 3.docx
Supplemental Digital Content 4.docx
References
- 1.Group USCSW. United States Cancer Statistics: 1999–2012 Incidence and Mortality Web-based Report. Atlanta: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute; [cited 2015]. Available from: www.cdc.gov/uscs. [Google Scholar]
- 2.Van Asten K, Neven P, Lintermans A, Wildiers H, Paridaens R. Aromatase inhibitors in the breast cancer clinic: focus on exemestane. Endocr Relat Cancer. 2014;21(1):R31–R49. doi: 10.1530/ERC-13-0269. [DOI] [PubMed] [Google Scholar]
- 3.Meier CR, Jick H. Tamoxifen and risk of idiopathic venous thromboembolism. Br J Clin Pharmacol. 1998;45(6):608–612. doi: 10.1046/j.1365-2125.1998.00733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coombes RC, Hall E, Gibson LJ, Paridaens R, Jassem J, Delozier T, et al. A randomized trial of exemestane after two to three years of tamoxifen therapy in postmenopausal women with primary breast cancer. N Engl J Med. 2004;350(11):1081–1092. doi: 10.1056/NEJMoa040331. [DOI] [PubMed] [Google Scholar]
- 5.Coombes RC, Kilburn LS, Snowdon CF, Paridaens R, Coleman RE, Jones SE, et al. Survival and safety of exemestane versus tamoxifen after 2–3 years' tamoxifen treatment (Intergroup Exemestane Study): a randomised controlled trial. Lancet. 2007;369(9561):559–570. doi: 10.1016/S0140-6736(07)60200-1. [DOI] [PubMed] [Google Scholar]
- 6.Fisher B, Costantino JP, Wickerham DL, Redmond CK, Kavanah M, Cronin WM, et al. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst. 1998;90(18):1371–1388. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 7.Fornander T, Rutqvist LE, Cedermark B, Glas U, Mattsson A, Silfversward C, et al. Adjuvant tamoxifen in early breast cancer: occurrence of new primary cancers. Lancet. 1989;1(8630):117–120. doi: 10.1016/s0140-6736(89)91141-0. [DOI] [PubMed] [Google Scholar]
- 8.Neven P, De Muylder X, Van Belle Y, Vanderick G, De Muylder E. Tamoxifen and the uterus and endometrium. Lancet. 1989;1(8634):375. [PubMed] [Google Scholar]
- 9.Rutqvist LE, Johansson H Stockholm Breast Cancer Study G. Long-term follow-up of the randomized Stockholm trial on adjuvant tamoxifen among postmenopausal patients with early stage breast cancer. Acta Oncol. 2007;46(2):133–145. doi: 10.1080/02841860601034834. [DOI] [PubMed] [Google Scholar]
- 10.Morales L, Timmerman D, Neven P, Konstantinovic ML, Carbonez A, Van Huffel S, et al. Third generation aromatase inhibitors may prevent endometrial growth and reverse tamoxifen-induced uterine changes in postmenopausal breast cancer patients. Ann Oncol. 2005;16(1):70–74. doi: 10.1093/annonc/mdi021. [DOI] [PubMed] [Google Scholar]
- 11.Kaufmann M, Bajetta E, Dirix LY, Fein LE, Jones SE, Zilembo N, et al. Exemestane is superior to megestrol acetate after tamoxifen failure in postmenopausal women with advanced breast cancer: results of a phase III randomized double-blind trial. The Exemestane Study Group. J Clin Oncol. 2000;18(7):1399–1411. doi: 10.1200/JCO.2000.18.7.1399. [DOI] [PubMed] [Google Scholar]
- 12.Walker G, Xenophontos M, Chen L, Cheung K. Long-term efficacy and safety of exemestane in the treatment of breast cancer. Patient Prefer Adherence. 2013;7:245–258. doi: 10.2147/PPA.S42223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goss PE, Ingle JN, Ales-Martinez JE, Cheung AM, Chlebowski RT, Wactawski-Wende J, et al. Exemestane for breast-cancer prevention in postmenopausal women. N Engl J Med. 2011;364(25):2381–2391. doi: 10.1056/NEJMoa1103507. [DOI] [PubMed] [Google Scholar]
- 14.Pagani O, Regan MM, Walley BA, Fleming GF, Colleoni M, Lang I, et al. Adjuvant exemestane with ovarian suppression in premenopausal breast cancer. N Engl J Med. 2014;371(2):107–118. doi: 10.1056/NEJMoa1404037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giudici D, Ornati G, Briatico G, Buzzetti F, Lombardi P, di Salle E. 6-Methylenandrosta-1,4-diene-3,17-dione (FCE 24304): a new irreversible aromatase inhibitor. J Steroid Biochem. 1988;30(1–6):391–394. doi: 10.1016/0022-4731(88)90129-x. [DOI] [PubMed] [Google Scholar]
- 16.Hong Y, Yu B, Sherman M, Yuan YC, Zhou D, Chen S. Molecular basis for the aromatization reaction and exemestane-mediated irreversible inhibition of human aromatase. Mol Endocrinol. 2007;21(2):401–414. doi: 10.1210/me.2006-0281. [DOI] [PubMed] [Google Scholar]
- 17.Labrie F. All sex steroids are made intracellularly in peripheral tissues by the mechanisms of intracrinology after menopause. J Steroid Biochem Mol Biol. 2015;145:133–138. doi: 10.1016/j.jsbmb.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 18.To SQ, Knower KC, Cheung V, Simpson ER, Clyne CD. Transcriptional control of local estrogen formation by aromatase in the breast. J Steroid Biochem Mol Biol. 2015;145:179–186. doi: 10.1016/j.jsbmb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Boudot A, Kerdivel G, Habauzit D, Eeckhoute J, Le Dily F, Flouriot G, et al. Differential estrogen-regulation of CXCL12 chemokine receptors, CXCR4 and CXCR7, contributes to the growth effect of estrogens in breast cancer cells. PLoS One. 2011;6(6):e20898. doi: 10.1371/journal.pone.0020898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sommer S, Fuqua SA. Estrogen receptor and breast cancer. Semin Cancer Biol. 2001;11(5):339–352. doi: 10.1006/scbi.2001.0389. [DOI] [PubMed] [Google Scholar]
- 21.Geisler J, King N, Anker G, Ornati G, Di Salle E, Lonning PE, et al. In vivo inhibition of aromatization by exemestane, a novel irreversible aromatase inhibitor, in postmenopausal breast cancer patients. Clin Cancer Res. 1998;4(9):2089–2093. [PubMed] [Google Scholar]
- 22.Sun D, Chen G, Dellinger RW, Sharma AK, Lazarus P. Characterization of 17-dihydroexemestane glucuronidation: potential role of the UGT2B17 deletion in exemestane pharmacogenetics. Pharmacogenet Genomics. 2010;20(10):575–585. doi: 10.1097/FPC.0b013e32833b04af. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aromasin Exemestane Tablets. [[cited 2015]]; Available from: http://www.pfizer.com/files/products/uspi_aromasin.pdf. [Google Scholar]
- 24.Penning TM. The aldo-keto reductases (AKRs): Overview. Chem Biol Interact. 2015;234:236–246. doi: 10.1016/j.cbi.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penning TM, Burczynski ME, Jez JM, Hung CF, Lin HK, Ma H, et al. Human 3alpha-hydroxysteroid dehydrogenase isoforms (AKR1C1-AKR1C4) of the aldo-keto reductase superfamily: functional plasticity and tissue distribution reveals roles in the inactivation and formation of male and female sex hormones. Biochem J. 2000;351(Pt 1):67–77. doi: 10.1042/0264-6021:3510067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lonning PE. Pharmacological profiles of exemestane and formestane, steroidal aromatase inhibitors used for treatment of postmenopausal breast cancer. Breast Cancer Res Treat. 1998;49(Suppl 1):S45–S52. doi: 10.1023/a:1006048722559. discussion S73-7. [DOI] [PubMed] [Google Scholar]
- 27.Evans TR, Di Salle E, Ornati G, Lassus M, Benedetti MS, Pianezzola E, et al. Phase I and endocrine study of exemestane (FCE 24304), a new aromatase inhibitor, in postmenopausal women. Cancer Res. 1992;52(21):5933–5939. [PubMed] [Google Scholar]
- 28.Mareck U, Geyer H, Guddat S, Haenelt N, Koch A, Kohler M, et al. Identification of the aromatase inhibitors anastrozole and exemestane in human urine using liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2006;20(12):1954–1962. doi: 10.1002/rcm.2545. [DOI] [PubMed] [Google Scholar]
- 29.Traina TA, Poggesi I, Robson M, Asnis A, Duncan BA, Heerdt A, et al. Pharmacokinetics and tolerability of exemestane in combination with raloxifene in postmenopausal women with a history of breast cancer. Breast Cancer Res Treat. 2008;111(2):377–388. doi: 10.1007/s10549-007-9787-1. [DOI] [PubMed] [Google Scholar]
- 30.Ariazi EA, Leitao A, Oprea TI, Chen B, Louis T, Bertucci AM, et al. Exemestane's 17-hydroxylated metabolite exerts biological effects as an androgen. Mol Cancer Ther. 2007;6(11):2817–2827. doi: 10.1158/1535-7163.MCT-07-0312. [DOI] [PubMed] [Google Scholar]
- 31.Corona G, Elia C, Casetta B, Diana C, Rosalen S, Bari M, et al. A liquid chromatography-tandem mass spectrometry method for the simultaneous determination of exemestane and its metabolite 17-dihydroexemestane in human plasma. J Mass Spectrom. 2009;44(6):920–928. doi: 10.1002/jms.1566. [DOI] [PubMed] [Google Scholar]
- 32.Kalow W, Tang BK, Endrenyi L. Hypothesis: comparisons of inter- and intra-individual variations can substitute for twin studies in drug research. Pharmacogenetics. 1998;8(4):283–289. doi: 10.1097/00008571-199808000-00001. [DOI] [PubMed] [Google Scholar]
- 33.Meier M, Moller G, Adamski J. Perspectives in understanding the role of human 17beta-hydroxysteroid dehydrogenases in health and disease. Ann N Y Acad Sci. 2009;1155:15–24. doi: 10.1111/j.1749-6632.2009.03702.x. [DOI] [PubMed] [Google Scholar]
- 34.Buzzetti F, Di Salle E, Longo A, Briatico G. Synthesis and aromatase inhibition by potential metabolites of exemestane (6-methylenandrosta-1,4-diene-3,17-dione) Steroids. 1993;58(11):527–532. doi: 10.1016/0039-128x(93)90029-m. [DOI] [PubMed] [Google Scholar]
- 35.Matsunaga T, Shintani S, Hara A. Multiplicity of mammalian reductases for xenobiotic carbonyl compounds. Drug Metab Pharmacokinet. 2006;21(1):1–18. doi: 10.2133/dmpk.21.1. [DOI] [PubMed] [Google Scholar]
- 36.Jin Y, Penning TM. Aldo-keto reductases and bioactivation/detoxication. Annu Rev Pharmacol Toxicol. 2007;47:263–292. doi: 10.1146/annurev.pharmtox.47.120505.105337. [DOI] [PubMed] [Google Scholar]
- 37.Penning TM, Drury JE. Human aldo-keto reductases: Function, gene regulation, and single nucleotide polymorphisms. Arch Biochem Biophys. 2007;464(2):241–250. doi: 10.1016/j.abb.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcos-Escribano A, BF A, Bonde-Larsen AL, Retuerto JI, Sierra IH. 1,2-Dehydrogenation of steroidal 6-methylen derivatives. Tetrahedron. 2009;65(36):7587–7590. [Google Scholar]
- 39.JM V. 2-(Prenyloxymethyl)benzoyl (POMB) group: a new temporary protecting group removable by intramolecular cyclization. Tetrahedron. 2007;63(45):10921–10929. [Google Scholar]
- 40.Variation Viewer [Internet] National Center for Biotechnology Information. U.S. National Library of Medicine; [cited 2015]. Available from: http://www.ncbi.nlm.nih.gov/variation/view/ [Google Scholar]
- 41.Steckelbroeck S, Jin Y, Oyesanmi B, Kloosterboer HJ, Penning TM. Tibolone is metabolized by the 3alpha/3beta-hydroxysteroid dehydrogenase activities of the four human isozymes of the aldo-keto reductase 1C subfamily: inversion of stereospecificity with a delta5(10)-3-ketosteroid. Mol Pharmacol. 2004;66(6):1702–1711. doi: 10.1124/mol.104.004515. [DOI] [PubMed] [Google Scholar]
- 42.Jin Y, Penning TM. Molecular docking simulations of steroid substrates into human cytosolic hydroxysteroid dehydrogenases (AKR1C1 and AKR1C2): insights into positional and stereochemical preferences. Steroids. 2006;71(5):380–391. doi: 10.1016/j.steroids.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 43.Jin Y, Mesaros AC, Blair IA, Penning TM. Stereospecific reduction of 5beta-reduced steroids by human ketosteroid reductases of the AKR (aldo-keto reductase) superfamily: role of AKR1C1-AKR1C4 in the metabolism of testosterone and progesterone via the 5beta-reductase pathway. Biochem J. 2011;437(1):53–61. doi: 10.1042/BJ20101804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Askonas LJ, Ricigliano JW, Penning TM. The kinetic mechanism catalysed by homogeneous rat liver 3 alpha-hydroxysteroid dehydrogenase. Evidence for binary and ternary dead-end complexes containing non-steroidal anti-inflammatory drugs. Biochem J. 1991;278(Pt 3):835–841. doi: 10.1042/bj2780835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neuhauser W, Haltrich D, Kulbe KD, Nidetzky B. NAD(P)H-dependent aldose reductase from the xylose-assimilating yeast Candida tenuis. Isolation, characterization and biochemical properties of the enzyme. Biochem J. 1997;326(Pt 3):683–692. doi: 10.1042/bj3260683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trauger JW, Jiang A, Stearns BA, LoGrasso PV. Kinetics of allopregnanolone formation catalyzed by human 3 alpha-hydroxysteroid dehydrogenase type III (AKR1C2) Biochemistry. 2002;41(45):13451–13459. doi: 10.1021/bi026109w. [DOI] [PubMed] [Google Scholar]
- 47.Cooper WC, Jin Y, Penning TM. Elucidation of a complete kinetic mechanism for a mammalian hydroxysteroid dehydrogenase (HSD) and identification of all enzyme forms on the reaction coordinate: the example of rat liver 3alpha-HSD (AKR1C9) J Biol Chem. 2007;282(46):33484–33493. doi: 10.1074/jbc.M703414200. [DOI] [PubMed] [Google Scholar]
- 48.Bulun SE, Lin Z, Imir G, Amin S, Demura M, Yilmaz B, et al. Regulation of aromatase expression in estrogen-responsive breast and uterine disease: from bench to treatment. Pharmacol Rev. 2005;57(3):359–383. doi: 10.1124/pr.57.3.6. [DOI] [PubMed] [Google Scholar]
- 49.Arthur JW, Reichardt JK. Modeling single nucleotide polymorphisms in the human AKR1C1 and AKR1C2 genes: implications for functional and genotyping analyses. PLoS One. 2010;5(12):e15604. doi: 10.1371/journal.pone.0015604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kume T, Iwasa H, Shiraishi H, Yokoi T, Nagashima K, Otsuka M, et al. Characterization of a novel variant (S145C/L311V) of 3alpha-hydroxysteroid/dihydrodiol dehydrogenase in human liver. Pharmacogenetics. 1999;9(6):763–771. [PubMed] [Google Scholar]
- 51.Lan Q, Mumford JL, Shen M, Demarini DM, Bonner MR, He X, et al. Oxidative damage-related genes AKR1C3 and OGG1 modulate risks for lung cancer due to exposure to PAH-rich coal combustion emissions. Carcinogenesis. 2004;25(11):2177–2181. doi: 10.1093/carcin/bgh240. [DOI] [PubMed] [Google Scholar]
- 52.Bains OS, Grigliatti TA, Reid RE, Riggs KW. Naturally occurring variants of human aldo-keto reductases with reduced in vitro metabolism of daunorubicin and doxorubicin. J Pharmacol Exp Ther. 2010;335(3):533–545. doi: 10.1124/jpet.110.173179. [DOI] [PubMed] [Google Scholar]
- 53.Takahashi RH, Grigliatti TA, Reid RE, Riggs KW. The effect of allelic variation in aldo-keto reductase 1C2 on the in vitro metabolism of dihydrotestosterone. J Pharmacol Exp Ther. 2009;329(3):1032–1039. doi: 10.1124/jpet.109.150995. [DOI] [PubMed] [Google Scholar]
- 54.Jez JM, Bennett MJ, Schlegel BP, Lewis M, Penning TM. Comparative anatomy of the aldo-keto reductase superfamily. Biochem J. 1997;326(Pt 3):625–636. doi: 10.1042/bj3260625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jakobsson J, Palonek E, Lorentzon M, Ohlsson C, Rane A, Ekstrom L. A novel polymorphism in the 17beta-hydroxysteroid dehydrogenase type 5 (aldo-keto reductase 1C3) gene is associated with lower serum testosterone levels in caucasian men. Pharmacogenomics J. 2007;7(4):282–289. doi: 10.1038/sj.tpj.6500419. [DOI] [PubMed] [Google Scholar]
- 56.Figueroa JD, Malats N, Garcia-Closas M, Real FX, Silverman D, Kogevinas M, et al. Bladder cancer risk and genetic variation in AKR1C3 and other metabolizing genes. Carcinogenesis. 2008;29(10):1955–1962. doi: 10.1093/carcin/bgn163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsuura K, Hara A, Deyashiki Y, Iwasa H, Kume T, Ishikura S, et al. Roles of the C-terminal domains of human dihydrodiol dehydrogenase isoforms in the binding of substrates and modulators: probing with chimaeric enzymes. Biochem J. 1998;336(Pt 2):429–436. doi: 10.1042/bj3360429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kamdem LK, Flockhart DA, Desta Z. In vitro cytochrome P450-mediated metabolism of exemestane. Drug Metab Dispos. 2011;39(1):98–105. doi: 10.1124/dmd.110.032276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.