Abstract
Recombinant adeno-associated viruses (rAAVs) are commonly used vehicles for in vivo gene transfer1-6. However, the tropism repertoire of naturally occurring AAVs is limited, prompting a search for novel AAV capsids with desired characteristics7-13. Here we describe a capsid selection method, called Cre-recombination-based AAV targeted evolution (CREATE), that enables the development of AAV capsids that more efficiently transduce defined Cre-expressing cell populations in vivo. We use CREATE to generate AAV variants that efficiently and widely transduce the adult mouse central nervous system (CNS) after intravenous injection. One variant, AAV-PHP.B, transfers genes throughout the CNS with an efficiency that is at least 40-fold greater than that of the current standard, AAV914-17, and transduces the majority of astrocytes and neurons across multiple CNS regions. In vitro, it transduces human neurons and astrocytes more efficiently than does AAV9, demonstrating the potential of CREATE to produce customized AAV vectors for biomedical applications.
rAAVs are the preferred vehicles for many in vivo gene transfer applications to non-dividing cell populations and are proving safe in clinical trials18-20. However, therapeutic applications have been limited by inefficient transduction of target cell populations. A prime example is gene transfer to the CNS; the transduction efficiency of currently available vectors in the mouse CNS after intravenous administration is at least an order of magnitude lower than in the liver21. Encouragingly, the transduction efficiency of rAAVs can be enhanced by creating AAV capsid libraries and selecting for variants with more desirable characteristics8-13. This approach, however, has not been successfully applied to the development of more effective gene delivery vehicles for global CNS transduction. Because the highly selective blood brain barrier (BBB) and cellular heterogeneity of the CNS present challenges for gene transfer to the CNS through the vasculature, we reasoned that it would be beneficial to provide selective pressure for capsids that cross the BBB and functionally transduce specific cell types. To meet these needs, we devised CREATE—a Cre recombination-dependent approach to selectively recover capsids that transduce predefined Cre expressing target cell populations (Fig. 1a).
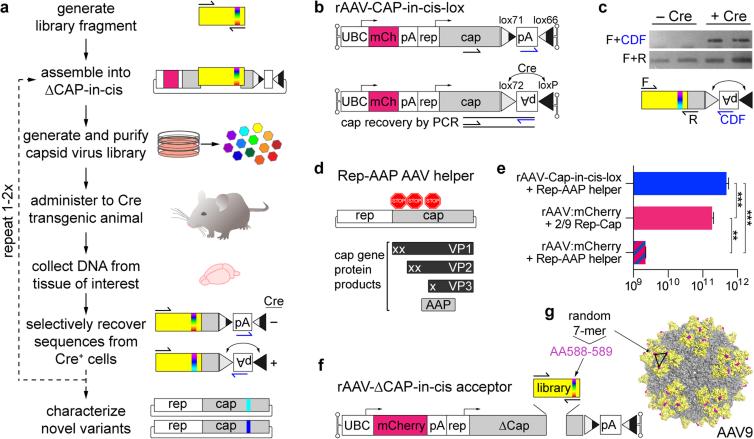
Figure 1. Cre-dependent recovery of AAV capsid sequences from transduced target cells.
(a) An overview of the CREATE selection process. PCR is used to introduce diversity (full visual spectrum vertical band) into a capsid gene fragment (yellow). The fragment is cloned into the rAAV genome harboring the remaining capsid gene (gray) and is used to generate a library of virus variants. The library is injected into Cre transgenic animals and PCR is used to selectively recover capsid sequences from Cre+ cells. (b) The rAAV-Cap-in-cis-lox rAAV genome. Cre inverts the polyadenylation (pA) sequence flanked by the lox71 and lox66 sites. PCR primers (half arrows) are used to selectively amplify Cre-recombined sequences. (c) PCR products from Cre recombination-dependent (top) and -independent (bottom) amplification of capsid library sequences recovered from two Cre+ or Cre− mice are shown. Schematics (bottom) show the PCR amplification strategies (see Supplementary Fig. 1 for details). (d) Schematic shows the AAV genes within the Rep-AAP AAV helper plasmid and the proteins encoded by the cap gene. Stop codons inserted in the cap gene eliminate VP1, VP2 and VP3 capsid protein expression. (e) DNase-resistant AAV vector genomes (vg) produced with the split AAV2/9 rep-AAP and rAAV-Cap-in-cis-lox genome (top) as compared to the vg produced with standard AAV2/9 rep-cap helper and rAAV-UBC-mCherry genome (middle) or with the AAV2/9 rep-AAP and rAAV-UBC-mCherry genome (bottom). N=3 independent trials per group; mean ± s.d.; **p<0.01, ***p<0.001; one-way ANOVA and Tukey multiple comparison test. (f) Cloning the 7-mer capsid library into the rAAV-ΔCap-in-cis vector. (g) The AAV9 surface model shows the location of the 7-mer inserted between AA588-589 (magenta). Sites encoded with the PCR-generated library fragment (AA450-592) are shown in yellow.
CREATE uses an rAAV capsid genome (rAAV-Cap-in-cis-lox) that couples a full-length AAV Cap gene, controlled by regulatory elements from the AAV Rep gene (Fig. 1b and Methods), with a Cre-invertible switch (Fig. 1b). By building capsid libraries within the rAAV-Cap-in-cis-lox backbone and delivering the virus libraries to animals with Cre expression in a defined cell population, the system enables the selective amplification and recovery of sequences that have transduced the target population (Fig. 1c). Because the rAAV-Cap-in-cis-lox genome lacks a functional Rep gene, Rep must be provided in trans for virus production. For this purpose, we modified an AAV2/9 rep-cap plasmid to eliminate capsid protein expression by inserting in-frame stop codons within the reading frame for each capsid protein, VP1-3 (Fig. 1d). These stop codons do not alter the amino acid sequence of the assembly-activating protein (AAP), which is expressed from an alternative reading frame within the Cap gene22. This split rAAV-Cap-in-cis-lox genome and Rep-AAP AAV helper system efficiently generates rAAV (Fig. 1e) and is the foundation of the CREATE selection platform, which enables capsid sequence recovery from genetically defined Cre-expressing cell populations within heterogeneous tissue samples (see Supplementary Fig. 1).
Within the rAAV-Cap-in-cis-lox acceptor genome we generated a library of AAV variants by inserting 7 amino acids (AA) of randomized sequence (7-mer) between AA588-589 (VP1 position) of the AAV9 capsid (Fig. 1f, g). To select for vectors that crossed the BBB and transduced cells throughout the CNS, we administered the capsid library intravenously into adult GFAP-Cre mice, which express Cre in astrocytes23. One week later, we isolated DNA from brain and spinal cord tissue and recovered capsid sequences by PCR from viral genomes that had undergone Cre-mediated recombination. We cloned the entire library of recovered Cap sequences back into the rAAV-Cap-in-cis-lox acceptor genome to generate the library GFAP1 and randomly chose 13 clones for sequencing. All tested sequences recovered from the GFAP1 library were unique, and therefore we used the GFAP1 plasmid library to generate a second virus library and performed an additional round of selection in GFAP-Cre mice. After the second selection, several variants were enriched (Supplementary Table 2) and showed enhanced CNS transduction (Supplementary Fig. 2a). We chose the most enriched variant, AAV-PHP.B, which represented 25% of recovered library sequences and encodes the 7-mer sequence TLAVPFK, for further tropism evaluation in vivo.
AAV-PHP.B and AAV9 capsids were used to package a single-stranded (ss) GFP reporter vector driven by the ubiquitous CAG promoter (ssAAV-CAG-GFP). Both AAV-PHP.B and AAV9 produced virus with similar efficiencies (Supplementary Fig. 2b). We next administered 1×1012 vector genomes (vg) of either vector to six-week-old mice by intravenous injection and assessed transduction by GFP expression three weeks later. Our data show that AAV-PHP.B transduced the entire adult CNS with high efficiency as indicated by imaging GFP immunohistochemistry (IHC) (Fig. 2a) or native eGFP fluorescence in several brain regions (Fig. 2b), the spinal cord (Fig. 2c) and retina (Fig. 2d). Using PARS-based CLARITY for whole body tissue clearing24, we imaged native eGFP fluorescence through cleared sections of tissue from the spinal cord (Fig. 2c), cortex and striatum (Fig. 2e). These 3D renderings (also see Supplementary Movies 1-3) further demonstrate the broad and efficient CNS transduction with the AAV-PHP.B vector. Outside the CNS, the cellular level tropism of AAV-PHP.B and AAV9 appeared similar in several organs, with the exception of the pancreas where the efficiency of transduction by AAV-PHP.B was reduced as compared with AAV9 (Fig. 2f).
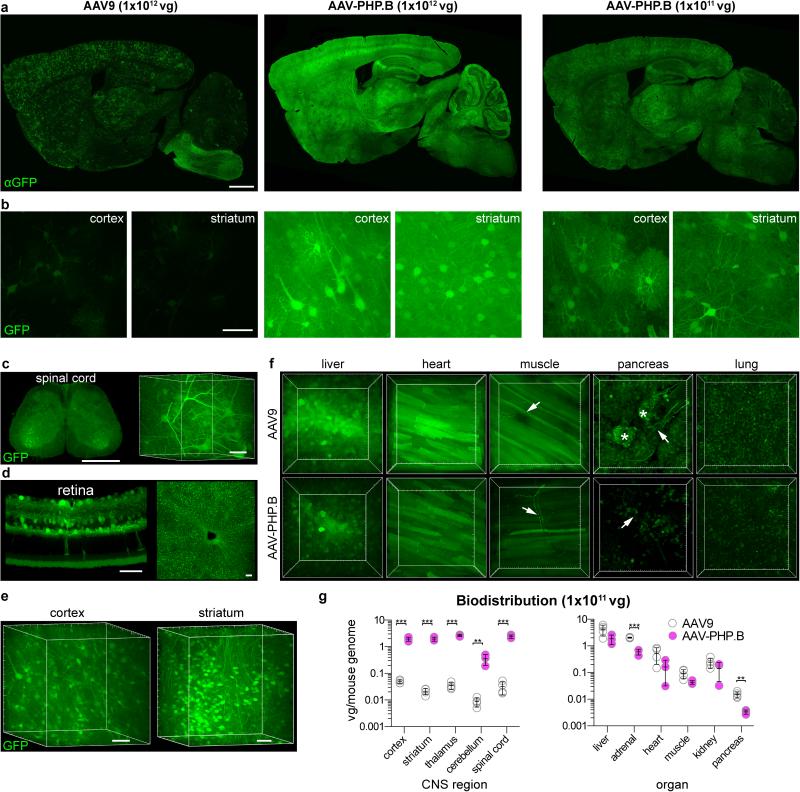
Figure 2. AAV-PHP.B mediates efficient gene delivery throughout the CNS after intravenous injection in adult mice.
(a-f) ssAAV9:CAG-GFP or ssAAV-PHP.B:CAG-GFP, at 1×1012 vg/mouse or 1×1011 (a, right), was intravenously injected into adult mice. Images show GFP expression 3 weeks after injection. (a) Representative images of GFP IHC in the brains of mice given AAV9 (left) or AAV-PHP.B (middle and right). (b) Native GFP fluorescence in the cortex (left) or striatum (right) in 50 μm maximum intensity projection (MIP) confocal images. (c) GFP fluorescence in the PARS-cleared24 lumbar spinal cord. (d) GFP fluorescence in the retina (left: 20 μm MIP, transverse section; right: whole-mount MIP). (e,f) GFP fluorescence in 3D MIP images of PARS-cleared tissue from AAV-PHP.B transduced cortex and striatum (e) and indicated organs from mice transduced with AAV9 (top) or AAV-PHP.B (bottom) (f). Arrows highlight GFP+ nerves. Asterisks in the image of the pancreas highlight GFP+ islet cells. Major tick marks in 3D projections are 100 μm. (g) AAV biodistribution in the indicated brain regions (top) and organs (bottom) 25 days after intravenous injection of 1×1011 vg into adult mice. N=3 for AAV-PHP.B and n=4 for AAV9; mean ± s.d.; **p<0.01, ***p<0.001, unpaired t tests corrected for multiple comparisons by the Holm-Sidak method. Scale bars: 1 mm (a, c (left)); 50 μm (b, c (right), d, e). Major tick marks in 3D projections in c, e, f are 100 μm.
To quantify the efficiency of gene transfer to the CNS and peripheral organs by AAV-PHP.B as compared with AAV9, we measured the number of viral genomes present in several brain regions and organs 25 days post-injection (Fig. 2g). AAV-PHP.B provided significantly greater gene transfer than AAV9 to each of the CNS regions examined: cortex (40-fold), striatum (92-fold), thalamus (76-fold), cerebellum (41-fold) and spinal cord (75-fold). Vector genome biodistribution outside the CNS showed that AAV-PHP.B transferred genes to the pancreas and adrenal gland less efficiently than AAV9 (Fig. 2g). No significant differences were found between the two vectors in the liver, heart, skeletal muscle and kidneys. When considered together with the CNS biodistribution data, in all CNS areas except the cerebellum, the number of viral genomes detected in mice treated with AAV-PHP.B was similar to that measured in the liver, an organ efficiently transduced by AAV921,25, and greater than that observed in other organs. In contrast, AAV9-mediated gene transfer to each of the examined CNS regions was at least 120-fold lower than in the liver. Therefore, although the tropism of AAV-PHP.B is not CNS specific, the enhanced gene transfer characteristics of this vector are CNS specific.
AAV9 preferentially transduces astrocytes when delivered intravenously to adult mice and non-human primates, but it also transduces neurons in several regions14,26. To examine the cell types transduced by AAV-PHP.B, we analyzed the co-localization of GFP with several cell-type markers. Owing to the highly efficient transduction, individual GFP expressing astrocytes were difficult to discern in mice that received 1×1012 vg AAV-PHP.B (Fig. 2a), but could be more easily identified morphologically by their compact, highly ramified processes in animals that received 10-fold less virus (Fig. 2a, b) and by co-localization with IHC for GFAP (Fig. 3a). In addition to astrocytes, AAV-PHP.B transduced CC1+ oligodendrocytes (Fig. 3b) and all neuronal subtypes examined, including NeuN+ throughout the brain (Fig. 3c) as well as midbrain tyrosine hydroxylase (TH)+ dopaminergic neurons (Fig. 3d), Calbindin+ cerebellar Purkinje cells (Fig. 3e), and several interneuron populations (Supplementary Fig. 3a-d). AAV-PHP.B also transduced CD31+ endothelial cells (Supplementary Fig. 3e) but did not appear to transduce Iba1+ microglia (Supplementary Fig. 3f,g). The paucity of GFP+ microglia seen after intravenous AAV-PHP.B delivery is consistent with previous reports of rare or nonexistent AAV-mediated gene expression in this cell population14,27-30. We have observed native GFP expression throughout the brain over a year after administration of AAV-PHP.B, suggesting that AAV-PHP.B can provide long-term, CNS-directed transgene expression (Supplementary Fig. 4).
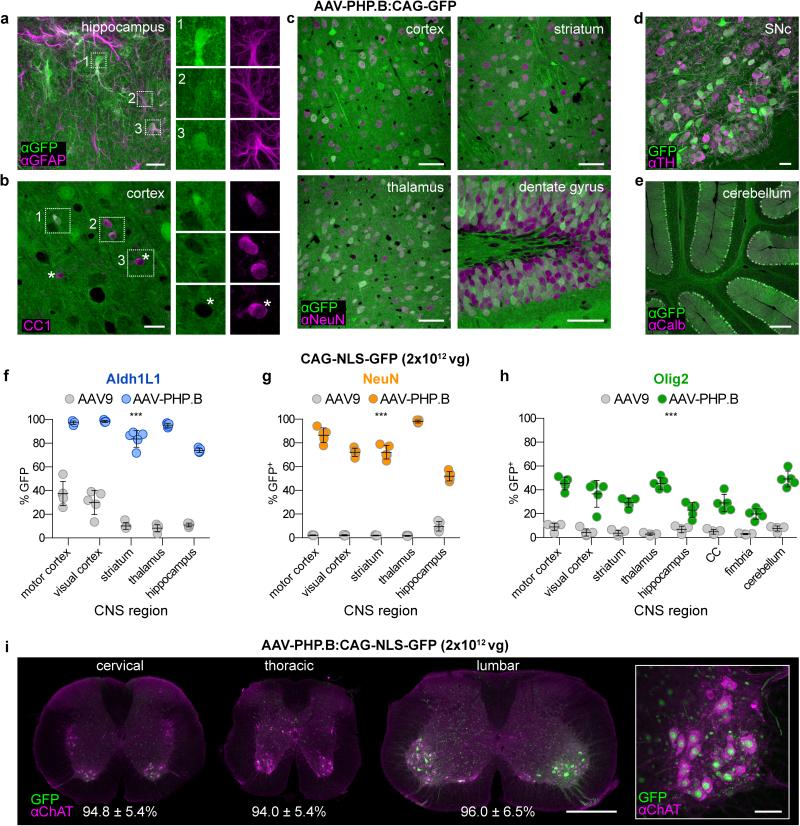
Figure 3. AAV-PHP.B transduces multiple CNS cell types more efficiently than AAV9.
(a-e) AAV-PHP.B transduces astrocytes, oligodendrocytes and neurons. Images show GFP expression 3 weeks after intravenous injection of 1×1011 (a) or 1×1012 (b-e) of ssAAV-PHP.B:CAG-GFP into adult mice. (a) MIP image of GFP (green) and GFAP (magenta) IHC in the hippocampus. (b) GFP and CC1 (magenta) IHC in the cortex. Numbered boxes highlight examples of double positive cells. Corresponding single-channel images are shown (right). Asterisks highlight cells without detectable GFP expression. (c) GFP and NeuN IHC in the indicated brain region. (d) MIP image of GFP fluorescence and TH IHC in the midbrain. (e) GFP and Calbindin (Calb) IHC in the cerebellum. (f-h) Quantification of the percentage of cells positive for NLS-GFP in the indicated brain region 3 weeks post injection. (i) AAV-PHP.B transduces ChAT+ spinal motor neurons. Images show native NLS-GFP fluorescence and IHC for ChAT in whole transverse spinal cord sections (left) or ventral horn MIP image (right). The percentage of ChAT+ neurons that expressed NLS-GFP in each spinal cord region is given ± the 95% confidence interval. For quantification, n=5 per group; mean ± s.d; All pairs of AAV9 vs AAV-PHP.B means were found to be different (***p<0.001) unpaired t tests corrected for multiple comparisons by the Holm-Sidak method. Scale bars: 20 μm (a,b,d), 50 μm (c,i, right), 200 μm (e) and 1 mm (i, left).
We next quantified the fraction of several cell types transduced by AAV-PHP.B as compared to AAV9. To facilitate reliable individual cell counting, we constructed a vector expressing a nuclear-localized GFP (NLS-GFP) under the control of the CAG promoter, ssAAV-CAG-NLS-GFP. A single injection of 2×1012 vg/mouse of ssAAV-PHP.B:CAG-NLS-GFP transduced the majority of Aldh1L1+ astrocytes (Fig. 3f and Supplementary Fig. 5a) and NeuN+ neurons (Fig. 3g and Supplementary Fig. 5b), as well as a modest fraction of Olig2+ oligodendrocyte lineage cells (Fig.3h and Supplementary Fig. 5c) across all brain regions examined. In all cases, AAV-PHP.B provided significantly enhanced transduction as compared to the same dose of AAV9. Notably, AAV-PHP.B also transduced over 94% of Chat+ motor neurons throughout the spinal cord (Fig. 3i), 91.4±1.6% of TH+ midbrain dopaminergic neurons (Supplemental Fig. 5d) and 91.7±5.8% of Calbindin+ Purkinje cells (n=5). In sum, adult intravenous administration of AAV-PHP.B efficiently targets multiple neuronal and glial cell types in the adult mouse.
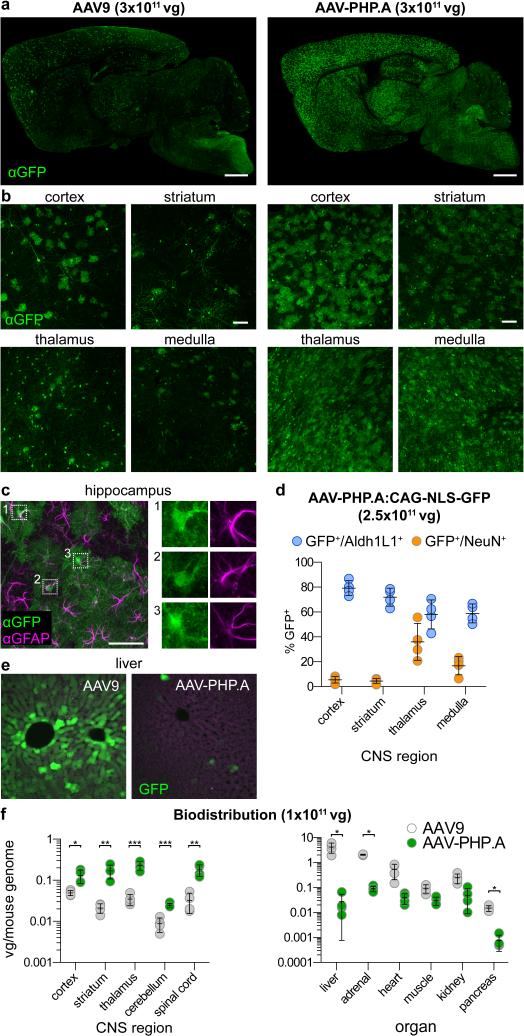
The method used to identify AAV-PHP.B only selects for transduction of the target cell population; it does not necessarily select for specificity. Nevertheless, in a separate trial in GFAP-Cre mice, we identified (after two rounds of in vivo selection) another AAV capsid variant, AAV-PHP.A, with the 7-mer sequence, YTLSQGW, that exhibits both more efficient and selective CNS astrocyte transduction (Fig. 4a-d), as well as reduced tropism for the liver (Fig. 4e, f) and other peripheral organs (Fig. 4f), as compared to AAV9. The increase in specificity for gene transfer to the CNS over the liver provided by AAV-PHP.A versus AAV9 is 400- to 1200-fold, resulting from a combination of enhanced adult CNS gene transfer (2.6- to 8-fold more depending on the specific region) and reduced liver gene transfer (152-fold). Two other variants enriched in this trial (Supplementary Fig. 2a) did not show enhanced GFP expression in CNS neurons or glia as compared with AAV9.
Figure 4. AAV-PHP.A exhibits efficient transduction of CNS astrocytes and reduced tropism for peripheral organs.
(a-c, e) Images show GFP expression 3 weeks after intravenous injection of 3×1011 of ssAAV9:CAG-GFP or ssAAV-PHP.A:CAG-GFP into adult mice. (a,b) Representative images of GFP IHC. (c) GFP IHC (green) and GFAP IHC (magenta) in the hippocampus. Numbered boxes highlight examples of GFP+/GFAP+ cells. Corresponding single-channel images are shown on the right. (d) 2.5×1011 vg of ssAAV-PHP.A:CAG-NLS-GFP was injected intravenously into adult mice. Graphs show quantification of the percentage of Aldh1L1+ (blue) and NeuN+ cells (orange) positive for NLS-GFP. (e) Native GFP expression in the liver (green) and tissue autofluorescence (magenta). (f) AAV biodistribution in the brain (top) and peripheral organs (bottom) 25 days after intravenous injection of 1×1011 vg into adult mice. (e, f) N=4 per group; mean ± s.d; *p<0.05, **p<0.01, ***p<0.001, unpaired t tests corrected for multiple comparisons by the Holm-Sidak method. Scale bars: 1 mm (a); 100 μm (b); 50 μm (c).
To determine whether AAV-PHP.A and AAV-PHP.B can also transduce human neural cells, we tested them on cortical neurons and astrocytes derived from human induced pluripotent stem cells (hiPSCs) using a 3D differentiation method31. HiPSC lines from two individuals were differentiated into 3D cerebral cortex-like structures (cortical spheroids), and maintained in vitro for up to 200 days. Aged cortical spheroids contain superficial and deep layer cortical excitatory neurons and up to 20% astrocytes31. In dissociated cortical spheroids that were exposed to the three viruses in monolayer, AAV-PHP.B more efficiently transduced both GFAP-expressing astrocytes and MAP2-expresing neurons in comparison with either AAV9 or AAV-PHP.A (Supplemental Fig. 5; two-way ANOVA p<0.01, n=3). In addition, all three viruses were capable of transducing intact 3D cortical spheroids (Supplementary Fig. 5c).
Using CREATE, we have developed new AAV variants that enable efficient widespread gene transfer to the adult mouse CNS after intravenous administration. An important advantage of this system is that it introduces selective pressure for capsids that mediate efficient intracellular trafficking and conversion of the single-stranded viral genome to persistent double-stranded DNA (dsDNA) forms necessary for long-term transduction (only the dsDNA genomes should serve as substrates for Cre). This additional selective pressure for functional capsids may have contributed to the identification of AAV-PHP.A and the AAV-PHP.B variants in independent trials after only two rounds of in vivo selection. In comparison, many previous in vivo and in vitro AAV capsid selection methods have applied 3-10 rounds of selection to identify capsid variants with enhanced properties9-13,32.
In principle, CREATE could be applied to discover AAV capsid variants that target defined, CRE-expressing cell types in any organ. Thus, it could be used not only in transgenic animals, as shown here, but also to develop AAV variants that target (i) specific Cre+ cell types in spheroid cultures31 or organoid cultures, (ii) cells made Cre+ in non-transgenic animals by, for example, viral injections that achieve population-, projection33-, or activity-based Cre expression34,35 or (iii) Cre+ human cells in human/mouse chimeric animals. Given the reported AAV tropism differences between animal models and humans11, selection schemes that use human Cre+ cells in vivo, cell-specific Cre expression in three-dimensional hiPSC-derived cellular models, or future Cre transgenic marmosets36 may be desirable for developing improved vectors for clinical applications. In addition, the success of AAV-based gene therapies, especially those requiring systemic delivery, can be hindered by the presence of neutralizing AAV capsid antibodies in the human population37-39. By using CREATE together with exposure of AAV libraries to pooled human sera, one could envision simultaneously selecting for capsids with retained or enhanced transduction characteristics that are also less susceptible to antibody-mediated neutralization.
A limitation of CREATE and other reported capsid selection methods is that it is difficult to predict, beyond an increase in target cell transduction efficiency, what characteristics the enriched variants will have before they are tested. In our two trials for astrocyte targeting, we identified several variants, AAV-PHP.B, B2, and B3 (Supplementary Fig. 2) that provide broad CNS transduction of both neurons and glia, and AAV-PHP.A that provides selectively more efficient astrocyte transduction. Identification of capsids with distinct properties from the same selection scheme was expected given that the recovery method we used selected for astrocyte transduction rather than for any specific intermediate step(s), e.g., brain vascular association, BBB transcytosis or astrocyte binding/internalization. Therefore, capsid variants that are more efficient at any of these intermediate steps should be recovered in our selection process. Indeed, by immunostaining for capsids we found that unlike AAV9, both AAV-PHP capsids readily localized to the brain vasculature shortly after intravenous administration (Supplementary Fig. 7a, b). In addition, by 24 hours post-administration, significantly more GFP-expressing cells were observed along the brain vasculature of mice that received AAV-PHP.B as compared with those that received AAV9 or AAV-PHP.A (Supplementary Fig. 7c, d). Considered together with the transduction characteristics of AAV-PHP.B and AAV-PHP.A in vivo (Figs. 2-4) and in human neural cultures (Supplementary Fig. 6), these data suggest that while both AAV-PHP vectors more efficiently associate with the brain vasculature, they may differ in subsequent cell type-specific entry or transport step(s).
In the future, CREATE could be used with next-generation sequencing to better predict the characteristics of the recovered sequences prior to testing the variants individually. Sequencing both the entire pool of variants recovered from the Cre-expressing target cells along with the unselected virus library should enable quantification of the relative extent of enrichment of each recovered sequence. Furthermore, sequencing capsids recovered from multiple Cre-expressing or Cre non-expressing populations could provide a means to perform both positive and negative selection in multiple cell types in a single experiment. Such in vivo-in silico selection approaches should increase the power of CREATE to enhance gene transfer to the CNS and other difficult-to-target cell populations.
METHODS
Plasmids
The rAAV-Cap-in-cis-lox genome plasmid contains three main elements flanked by AAV2 ITRs: (i) an mCherry expression cassette, which is comprised of a 398bp fragment of the human UBC gene upstream of the mCherry cDNA followed by a synthetic polyadenylation sequence40; (ii) the AAV9 capsid gene and regulatory sequences, which are comprised of the AAV5 p41 promoter sequence (1680-1974 of GenBank AF085716.1)41,42 and splicing sequences taken from the AAV2 rep gene; and (iii) a Cre-dependent switch, which is comprised of the SV40 polyadenylation sequence (pA) flanked by inverted lox71 and lox66 sites43 (Fig. 1b). The rAAV-Cap-in-cis-lox genome plasmid was further modified to introduce two unique restriction sites, XbaI and AgeI, within the capsid sequence. These sites flank the region (AA450-592) that is replaced by the randomized library fragment. The introduction of the XbaI site introduces a K449R mutation, which does not have an overt effect on vector production or transduction. The mutations required to insert the AgeI site are silent. For the rAAV-ΔCap-in-cis acceptor plasmid used for the capsid library cloning, the coding region between the XbaI and AgeI sites was removed to prevent virus production from the acceptor plasmid lacking the library fragment.
As a template for the library fragment, we PCR amplified the region spanning the XbaI and AgeI sites of the modified AAV9. This sequence was modified to remove a unique EarI restriction site and insert a unique KpnI site (both silent mutations) to create the xE fragment. The modified xE fragment was TA cloned into pCRII (Life Technologies) to generate pCRII-9Cap-xE. Eliminating the EarI site provided a second method that could be used, if necessary, to selectively digest contaminating (AAV9) capsid sequences recovered by PCR, but not digest the library-derived sequences. We did not find it necessary to use this digestion step. Using the rAAV-ΔCap-in-cis acceptor for cloning the libraries and taking standard PCR precautions (e.g., UV treating reagents and pipettors) was sufficient to prevent contamination.
The AAV2/9 REP-AAP helper plasmid was constructed by introducing five stop codons into the coding sequence of the VP reading frame of the AAV9 gene at VP1 AAs: 6, 10, 142, 148 and 216. The stop codon at AA216 was designed not to disrupt the coding sequence of the AAP protein, which is encoded within an alternative reading frame.
Several rAAV genomes were used in this study. Each is constructed within a single stranded (ss) rAAV genome with a reporter driven by the ubiquitous CMV-β-Actin-intron-β-Globin hybrid promoter (CAG). For simplicity, the vector descriptions have been abbreviated in the text. ssAAV-CAG-GFP refers to ssAAV-CAG-eGFP-2A-Luc-WPRE-SV40 polyA. ssAAV-CAG-NLS-GFP refers to ssAAV-CAG-NLS-GFP-WPRE-SV40 polyA, which was constructed by inserting the nuclear localization sequence PKKKRKV at both the N- and C-termini of GFP. ssAAV-CAG-mNeGreen-f refers to ssAAV-CAG-mNeonGreen-f-WPRE with a human growth hormone polyA signal. The mNeonGreen44 was modified with the membrane targeting (farnesylation and palmitoylation signals) sequence from c-Ha-Ras45.
Capsid library generation
The random 7-mer library fragment (inserted between amino acids 588 and 589) was generated by PCR using Q5 Hot Start High-Fidelity DNA Polymerase (NEB; M0493), primers XF and 7×MNN and pCRII-9Cap-xE as a template. A schematic showing the approximate primer binding sites and a table of the primer sequences are given in Supplementary Figure 1 and Supplementary Table 1, respectively. To generate the rAAV-based library, the PCR products containing the library and the XbaI- and AgeI-digested rAAV-ΔCap-in-cis acceptor plasmid were assembled using Gibson Assembly (NEB; E2611). The reaction products were then treated with Plasmid Safe (PS) DNase (Epicentre; E3105K) to digest any unassembled fragments and purified using a QIAquick PCR Purification Kit (Qiagen). This reaction typically yielded over 100 ng of assembled plasmid (as defined by the amount of DNA remaining after the PS DNase digestion step). 100 ng is sufficient to transfect ten 150mm tissue culture dishes at 10 ng/dish. Note, the libraries can also be constructed by ligation or Gibson Assembly and then amplified in E. coli, but bacterial transformation reduces the library diversity. By directly transfecting the assembled reaction products, the library diversity is limited instead by the number of successfully transfected HEK293 producer cells.
Virus production and purification
Recombinant AAVs were generated by triple transfection of 293T cells (ATCC) using polyethylenimine (PEI)46. Viral particles were harvested from the media at 72 hrs post transfection and from the cells and media at 120 hrs. Cell pellets were resuspended in 10mM Tris with 2mM MgCl2, pH 8, freeze-thawed three times, and treated with 100 U/mL Benzonase (Epicentre) at 37°C for at least 1 hr. Viral media was concentrated by precipitation with 8% polyethylene glycol 8000 (Sigma-Aldrich) with 500 mM sodium chloride47, resuspended in Tris-MgCl2, and then added to the lysates. The combined stocks were then adjusted to 500 mM NaCl, incubated at 37°C for 30 minutes, and clarified by centrifugation at 2000 × g. The clarified stocks were then purified over iodixanol (Optiprep, Sigma; D1556) step gradients (15%, 25%, 40% and 60%)48. Viruses were concentrated and formulated in phosphate buffered saline (PBS). Virus titers were determined by measuring the number of DNaseI-resistant vg using qPCR with linearized genome plasmid as a standard46.
For capsid library virus generation, two modifications were made to the above virus production protocol to reduce the production of mosaic capsids that could arise from the presence of multiple capsid sequences in the same cell. First, only 10 ng of the rAAV-Cap-in-cis library plasmid was transfected (per 150 mm dish) to increase the likelihood that most transfected cells only received one capsid variant sequence. Second, the virus was collected at 48 hrs (media) and 60 hrs (cells and media), rather than at 72 hrs and 120 hrs as described above, to minimize the secondary transduction of producer cells with rAAV library virus released into the medium.
Animals
All procedures were approved by the California Institute of Technology Institutional Animal Care and Use Committee (IACUC). GFAP-Cre mice expressing Cre under the control of the mouse GFAP promoter (012886)23 and C57Bl/6J mice (000664) were purchased from the Jackson Laboratory (JAX). Intravenous administration of rAAV vectors was performed by injecting the virus into the retro-orbital sinus. Mice were randomly assigned to groups of predetermined sample size. No mice were excluded from these analyses. Experimenters were not blinded to sample groups.
In vivo selection
For the selections in GFAP-Cre mice, 1×1011 vg of the capsid libraries were injected intravenously into adult Cre+ mice of either sex. Seven to eight days post-injection, mice were euthanized and the brain and spinal cord were collected. Vector DNA was recovered from one hemisphere of the brain and half of the spinal cord using 4-5 ml of Trizol (Life Technologies; 15596). To purify viral DNA, the upper aqueous fraction was collected according to the manufacturer's extraction protocol. We found that the aqueous fraction contains a significant portion of the viral DNA genomes as well as RNA. RNA was then digested by treatment with 1 uL of RNase A (Qiagen) at 37 °C overnight. Next, a two-step PCR amplification strategy was used to selectively recover Cap sequences from Cre-recombined genomes. The first amplification step preferentially amplifies Cre-recombined rAAV-Cap-in-cis-lox sequences using the primers 9CapF and CDF (see Supplementary Fig. 1). The PCR was performed for 20-26 cycles of 95°C for 20 sec, 60°C for 20 sec and 72°C for 30 sec using Q5 Hot Start High-fidelity DNA Polymerase. The PCR product was then diluted 1:10 or 1:100 and then used as a template for a second Cre-independent PCR reaction using primers XF and AR (Supplementary Fig. 1C). The second PCR generated the fragment that was cloned back into the rAAV-ΔCap-in-cis acceptor plasmid as described above. 1 μL of the Gibson Assembly reactions were then diluted 1:10 and transformed into SURE2 competent cells (Agilent; 200152) to generate individual clones for sequencing.
Variants that showed evidence of enrichment were cloned into an AAV Rep-Cap plasmid and transformed into DH5α competent cells (NEB). The novel AAV Rep-Cap variants, or AAV2/9 Rep-Cap as a control, were then evaluated using one of the reporter genomes described above.
Vector biodistribution
Six-week-old mice female C57Bl/6 mice were injected intravenously with 1×1011 vg of the ssAAV-CAG-GFP vector packaged into the indicated AAV capsid. Animals were randomly assigned to groups. 25 days after injection, the mice were euthanized and tissues and indicated brain regions were collected and frozen at −80°C. DNA was isolated from the tissue samples using Qiagen DNeasy Blood and Tissue kit. Vector genomes were detected using PCR primers that bind to the WPRE element and were normalized to mouse genomes using primers specific to the mouse glucagon gene. Absolute quantification was performed using serial dilutions of linearized plasmid standards of known concentration46. One randomly chosen animal injected with AAV-PHP.B was removed from the bio-distribution study for histological analysis.
Tissue preparation, immunohistochemistry and imaging
Mice were anesthetized with Nembutal and transcardially perfused with 0.1 M phosphate buffer (PB) at room temperature (RT) at pH 7.4 and then with freshly prepared, ice-cold 4% paraformaldehyde (PFA) in PB. Brains were post-fixed in 4% PFA overnight and then sectioned by vibratome or cryoprotected and sectioned by cryostat. IHC was performed on floating sections with primary and secondary antibodies in PBS containing 10% goat or donkey serum and 0.5% Triton X-100 (no detergent was used for GAD67 staining). Primary antibodies used were mouse anti-AAV capsid (1:20; American Research Products, 03-65158, clone B1), rabbit anti-GFP (1:1000; Invitrogen, A11122), chicken anti-GFP (1:1000; Abcam, ab13970), mouse anti-CC1 (1:200; Calbiochem, OP80), rabbit anti-GFAP (1:1000; Dako, Z0334), mouse anti-NeuN (1:500; Millipore, MAB377), rabbit anti-IbaI (1:500; Biocare Medical, CP290), mouse anti-Calbindin D28K (1:200; Sigma, CB-955), rabbit anti-Calretinin (1:1000; Chemicon, AB5054), mouse anti-GAD67 (1:1000; Millipore, MAB5406), guinea pig anti–MAP2 (1:1000; Synaptic Systems, 188004), mouse anti-Parvalbumin (1:1000; Sigma), Tyrosine Hydroxlyase (1:1000, Aves) and rabbit anti-CD31 (1:50; Abcam, ab28364). Primary antibodies incubations were performed for 16-24 hrs at RT. The sections were then washed and incubated with secondary Alexa-conjugated antibodies (1:1000; Invitrogen) for 2-16 hrs. For capsid detection with the B1 antibody that recognizes an internal epitope49, floating sections were treated with 2M HCl for 15 minutes at 37°C and then washed extensively with PBS prior to incubation with the primary antibody. For some images, the 16-bit green channel (GFP) gamma was adjusted to enable visualization (without oversaturation) of both low and high GFP-expressing cells present within the same field of view. In all cases, changes to gamma or contrast as well as microscope and laser settings remained consistent across sets of images. Images were taken with a Zeiss LSM 780 confocal microscope fitted with the following objectives: Fluar 5×/0.25 M27 Plan-Apochromat 10×/0.45 M27 (working distance 2.0 mm), Plan-Apochromat 25×/0.8 Imm Corr DIC M27 multi-immersion and LD C-Apochromat 40×/1.1 W Korr and Plan-Apochromat 100×/1.46 Oil DIC objectives. 3D MIP images and Supplementary Movies were generated with Imaris (Bitplane).
Quantification of cell type-specific transduction
Six-week-old female mice were randomly assigned to groups and injected intravenously with 2×1012 vg of ssAAV-CAG-NLS-GFP packaged into AAV9, AAV-PHP.B or AAV-PHP.A. Three weeks later, the mice were perfused and the brains were processed and immunostained for the indicated antigen as described above. The number of animals per group was pre-established; no animals were excluded from the analysis. Confocal single-plane images of the cell type-specific immunostaining and native NLS-GFP fluorescence were taken. To prevent bias, images were taken from the indicated matched regions identified by viewing only the cell type-specific immunostaining channel, rather than from GFP expression, prior to image acquisition. Likewise, cell counting was performed by first counting and marking each cell stained by the cell-specific antigen by viewing the IHC channel. Next those marked IHC+ cells that were positive for native GFP fluorescence were counted. Due to the stark transduction efficiency differences between capsids, the counting was not blinded by group.
Tissue clearing
Mice were perfused as above with 60-80 mL of ice-cold 4% PFA in PB at a flow rate of 14 mL per minute. The flow rate was then reduced to 2-3 mL/min and continued for 2 hrs at RT. The mice were then placed in individual custom-built chambers24 and perfused with 200 mL of recycling RT 4% acrylamide in PB at 2-3 mL/min overnight followed by a 2-hr perfusion flush with PB to remove residual polymers/monomers from the vasculature. The polymerization process was initiated by placing the chambers in a 42°C water bath and delivering, by perfusion (2-3 mL/min), of 200 mL of recycling, degassed PB containing 0.25% VA-044 initiator for 2-4 hrs. The mice were then perfused with 8% SDS in PB, pH 7.5 for 7 days. The SDS containing solution was refreshed two times during the 7 days and then flushed out by perfusion of 2 L of non-recirculating PB overnight. Cleared tissue samples were mounted in RIMS solution (refractive index of 1.46)24 for imaging.
Generation of cortical spheroids from human iPSC
Human cortical spheroids were generated from iPSC as previously described31. Briefly, iPSC lines derived from two healthy control individuals were grown on inactivated mouse embryonic fibroblast feeders in the following medium: DMEM/F12, Knockout Serum 20%, 1 mM non-essential amino acids (1:100), GlutaMax (1:200), β-mercaptoethanol (0.1 mM), penicillin and streptomycin (1:100) (Life Technologies). Cultures were regularly tested and maintained mycoplasma free. Colonies of iPSCs were detached intact with dispase (0.35 mg/ml, Invitrogen) and transferred into low-attachment plates in iPSC medium supplemented with dorsomorphin (5 μM, Sigma) and SB-431542 (10 μM, Tocris), and the medium was changed daily. On day six of in vitro differentiation, neural spheroids were transferred to NPC medium (Neurobasal A, B27 without vitamin A, GlutaMax (1:100), penicillin and streptomycin; Life Technologies), which was supplemented with EGF (20 ng/ml) and FGF2 (20 ng/ml) until day 24, and then supplemented with BDNF (20 ng/ml) and NT3 (20 ng/ml) from day 25 to 42. From day 43 onwards, cortical spheroids were maintained in NPC medium, which was changed every 4 days.
Dissociation and viral infection of cortical spheroids
For enzymatic dissociation and culture in monolayer, cortical spheroids at day 170-200 of in vitro differentiation (two independent neural differentiations of one iPSC line from one individual and one differentiation of an iPSC line from another individual) were incubated with Accutase (Innovative Cell Technologies) for 25 min at 37°C, washed three times with NPC media and gently triturated with a P-200 pipette. Cells were plated on poly-ornithine and laminin coated glass coverslip (15 mm) at ~300,000 cells/well and maintained in NPC media supplemented with BDNF (20 ng/ml) and NT3 (20 ng/ml) for the first 24 hrs, and then maintained in NPC media without growth factors.
Cultures grown on coverslips were infected with each of the viruses at a titer of 1×109 vg/well and fixed 5 days later with 4% paraformaldehyde (PFA) for 10 min. For immunocytochemistry, cells were permeabilized with 0.2% Triton X-100 for 10 min and blocked with 10% goat serum in PBS for 1 hr. Coverslips were then incubated with antibodies diluted in blocking solution for 2 hr. Nuclei were visualized with Hoechst 33258 (Life Technologies, 1:10,000).
Cells were imaged with a Zeiss M1 Axioscope using a 40× objective. The proportion of GFP+ cells co-labeled with either GFAP or MAP2 was quantified in images of 10 random fields per coverslip for each experimental condition. Results presented are the average of two separate dissociation and infection experiments.
To infect intact 3D cultures with AAVs, single human cortical spheroids at day 197 days of in vitro differentiation were transferred overnight into 1.5 ml Eppendorf tubes containing 6×109 vg/400 μl in NPC media, and were fixed 7 days later in 4% PFA overnight. Fixed spheroids were then transferred into 30% sucrose for 24 hrs, embedded in O.C.T. (Fisher Scientific) and cut at 14 μm sections. For immunohistochemistry, sections were blocked with 10% goat serum in PBS containing 0.3% Triton-X100 for 1 hr. Images were collected with a Leica TCS SP8 confocal microscope.
Supplementary Material
ACKNOWLEDGMENTS
This article and the naming of the novel AAV clones is dedicated to the memory of Paul H. Patterson (PHP) who passed away during the preparation of this manuscript. We wish to thank Laura Rodriguez and Pat Anguiano for administrative assistance, Alex Balazs and Stijn Cassenaer and the entire Gradinaru and Patterson laboratories for helpful discussions and Andrea Choe for helpful comments on the manuscript. We thank the U. Penn vector core for the AAV2/9 rep-cap plasmid, A. Balazs and David Baltimore for the AAV genome plasmid, and the Biological Imaging Facility, supported by the Caltech Beckman Institute and the Arnold and Mabel Beckman Foundation, for use of imaging equipment. This work was supported by the Hereditary Disease foundation and the Caltech-City of Hope Biomedical Initiative (to PHP), and by grants to VG: NIH Director's New Innovator IDP20D017782-01; NIH/NIA 1R01AG047664-01; Beckman Institute for CLARITY, Optogenetics and Vector Engineering Research; Gordon and Betty Moore Foundation through Grant GBMF2809 to the Caltech Programmable Molecular Technology Initiative. Work in the Gradinaru Laboratory is also funded by the following awards (to VG): NIH BRAIN 1U01NS090577; NIH/NIMH 1R21MH103824-01; Pew Charitable Trust; Kimmel Foundation; Human Frontiers in Science Program; Caltech-GIST; Caltech-City of Hope Biomedical Initiative. Work in the Pasca Laboratory is supported by a US National Institute of Mental Health (NIMH) 1R01MH100900 and 1R01MH100900-02S1, the NIMH BRAINS Award (R01MH107800), MQ Fellow Award and the Donald E. and Delia B. Baxter Foundation Scholar Award (to S.P.P.).
Footnotes
AUTHOR CONTRIBUTIONS
B.E.D designed and performed experiments, analyzed data, prepared figures and wrote the manuscript. P.L.P., B.P.S., S.K., A.B., and K.C. performed experiments, virus production and characterization. W.L.W. performed tissue processing and IHC. B.Y. assisted with tissue clearing and imaging. N.H. and S.P.P. performed the experiments with human cells, analyzed the data, and prepared the associated figure and text. V.G. helped with study design and data analysis, manuscript and figure preparation and supervised the project. All authors edited and approved the manuscript.
Competing Financial Interests
B.E.D. is listed as an inventor on a patent application relating to this work.
References
- 1.Kaplitt MG, et al. Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson's disease: an open label, phase I trial. Lancet. 2007;369:2097–2105. doi: 10.1016/S0140-6736(07)60982-9. doi:10.1016/S0140-6736(07)60982-9. [DOI] [PubMed] [Google Scholar]
- 2.Wu Z, Asokan A, Samulski RJ. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Molecular therapy : the journal of the American Society of Gene Therapy. 2006;14:316–327. doi: 10.1016/j.ymthe.2006.05.009. doi:10.1016/j.ymthe.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 3.High KH, Nathwani A, Spencer T, Lillicrap D. Current status of haemophilia gene therapy. Haemophilia : the official journal of the World Federation of Hemophilia 20 Suppl. 2014;4:43–49. doi: 10.1111/hae.12411. doi:10.1111/hae.12411. [DOI] [PubMed] [Google Scholar]
- 4.Borel F, Kay MA, Mueller C. Recombinant AAV as a platform for translating the therapeutic potential of RNA interference. Molecular therapy : the journal of the American Society of Gene Therapy. 2014;22:692–701. doi: 10.1038/mt.2013.285. doi:10.1038/mt.2013.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ojala DS, Amara DP, Schaffer DV. Adeno-associated virus vectors and neurological gene therapy. Neuroscientist. 2015;21:84–98. doi: 10.1177/1073858414521870. doi:10.1177/1073858414521870. [DOI] [PubMed] [Google Scholar]
- 6.Betley JN, Sternson SM. Adeno-associated viral vectors for mapping, monitoring, and manipulating neural circuits. Hum Gene Ther. 2011;22:669–677. doi: 10.1089/hum.2010.204. doi:10.1089/hum.2010.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartlett JS, Kleinschmidt J, Boucher RC, Samulski RJ. Targeted adeno-associated virus vector transduction of nonpermissive cells mediated by a bispecific F(ab'gamma)2 antibody. Nat Biotechnol. 1999;17:181–186. doi: 10.1038/6185. doi:10.1038/6185. [DOI] [PubMed] [Google Scholar]
- 8.Muller OJ, et al. Random peptide libraries displayed on adeno-associated virus to select for targeted gene therapy vectors. Nat Biotechnol. 2003;21:1040–1046. doi: 10.1038/nbt856. doi:10.1038/nbt856. [DOI] [PubMed] [Google Scholar]
- 9.Grimm D, et al. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. Journal of virology. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. doi:10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalkara D, et al. In vivo-directed evolution of a new adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous. Science translational medicine. 2013;5:189ra176. doi: 10.1126/scitranslmed.3005708. doi:10.1126/scitranslmed.3005708. [DOI] [PubMed] [Google Scholar]
- 11.Lisowski L, et al. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2014;506:382–386. doi: 10.1038/nature12875. doi:10.1038/nature12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maheshri N, Koerber JT, Kaspar BK, Schaffer DV. Directed evolution of adeno- associated virus yields enhanced gene delivery vectors. Nature biotechnology. 2006;24:198–204. doi: 10.1038/nbt1182. doi:10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- 13.Excoffon KJDA, et al. Directed evolution of adeno-associated virus to an infectious respiratory virus. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3865–3870. doi: 10.1073/pnas.0813365106. doi:10.1073/pnas.0813365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foust KD, et al. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nature biotechnology. 2009;27:59–65. doi: 10.1038/nbt.1515. doi:10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bevan AK, et al. Systemic gene delivery in large species for targeting spinal cord, brain, and peripheral tissues for pediatric disorders. Molecular therapy : the journal of the American Society of Gene Therapy. 2011;19:1971–1980. doi: 10.1038/mt.2011.157. doi:10.1038/mt.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maguire CA, Ramirez SH, Merkel SF, Sena-Esteves M, Breakefield XO. Gene therapy for the nervous system: challenges and new strategies. Neurotherapeutics. 2014;11:817–839. doi: 10.1007/s13311-014-0299-5. doi:10.1007/s13311-014-0299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray SJ, et al. Preclinical Differences of Intravascular AAV9 Delivery to Neurons and Glia: A Comparative Study of Adult Mice and Nonhuman Primates. Molecular therapy : the journal of the American Society of Gene Therapy. 2011;19:1058–1069. doi: 10.1038/mt.2011.72. doi:10.1038/mt.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maguire AM, et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. doi:10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nathwani AC, et al. Long-term safety and efficacy following systemic administration of a self- complementary AAV vector encoding human FIX pseudotyped with serotype 5 and 8 capsid proteins. Molecular therapy : the journal of the American Society of Gene Therapy. 2011;19:876–885. doi: 10.1038/mt.2010.274. doi:10.1038/mt.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaudet D, et al. Review of the clinical development of alipogene tiparvovec gene therapy for lipoprotein lipase deficiency. Atherosclerosis. Supplements. 2010;11:55–60. doi: 10.1016/j.atherosclerosissup.2010.03.004. doi:10.1016/j.atherosclerosissup.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 21.Pulicherla N, et al. Engineering Liver-detargeted AAV9 Vectors for Cardiac and Musculoskeletal Gene Transfer. Molecular therapy : the journal of the American Society of Gene Therapy. 2011;19:1070–1078. doi: 10.1038/mt.2011.22. doi:10.1038/mt.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sonntag F, Schmidt K, Kleinschmidt JA. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10220–10225. doi: 10.1073/pnas.1001673107. doi:10.1073/pnas.1001673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia ADR, Doan NB, Imura T, Bush TG, Sofroniew MV. GFAP-expressing progenitors are the principal source of constitutive neurogenesis in adult mouse forebrain. Nature neuroscience. 2004;7:1233–1241. doi: 10.1038/nn1340. doi:10.1038/nn1340. [DOI] [PubMed] [Google Scholar]
- 24.Yang B, et al. Single-Cell Phenotyping within Transparent Intact Tissue through Whole-Body Clearing. Cell. 2014;158:945–958. doi: 10.1016/j.cell.2014.07.017. doi:10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie J, et al. MicroRNA-regulated, Systemically Delivered rAAV9: A Step Closer to CNS- restricted Transgene Expression. Molecular therapy : the journal of the American Society of Gene Therapy. 2010;19:526–535. doi: 10.1038/mt.2010.279. doi:10.1038/mt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samaranch L, et al. Adeno-associated virus serotype 9 transduction in the central nervous system of nonhuman primates. Hum Gene Ther. 2012;23:382–389. doi: 10.1089/hum.2011.200. doi:10.1089/hum.2011.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dufour BD, Smith CA, Clark RL, Walker TR, McBride JL. Intrajugular vein delivery of AAV9-RNAi prevents neuropathological changes and weight loss in Huntington's disease mice. Molecular therapy : the journal of the American Society of Gene Therapy. 2014;22:797–810. doi: 10.1038/mt.2013.289. doi:10.1038/mt.2013.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartlett JS, Samulski RJ, McCown TJ. Selective and Rapid Uptake of Adeno-Associated Virus Type 2 in Brain. 1998 doi: 10.1089/hum.1998.9.8-1181. http://dx.doi.org/10.1089/hum.1998.9.8-1181. [DOI] [PubMed]
- 29.Wang H, et al. Widespread spinal cord transduction by intrathecal injection of rAAV delivers efficacious RNAi therapy for amyotrophic lateral sclerosis. Hum Mol Genet. 2014;23:668–681. doi: 10.1093/hmg/ddt454. doi:10.1093/hmg/ddt454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakrabarty P, et al. Capsid serotype and timing of injection determines AAV transduction in the neonatal mice brain. PLoS One. 2013;8:e67680. doi: 10.1371/journal.pone.0067680. doi:10.1371/journal.pone.0067680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasca AM, et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nature methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. doi:10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ying Y, et al. Heart-targeted adeno-associated viral vectors selected by in vivo biopanning of a random viral display peptide library. Gene therapy. 2010;17:980–990. doi: 10.1038/gt.2010.44. doi:10.1038/gt.2010.44. [DOI] [PubMed] [Google Scholar]
- 33.Wall NR, Wickersham IR, Cetin A, De La Parra M, Callaway EM. Monosynaptic circuit tracing in vivo through Cre-dependent targeting and complementation of modified rabies virus. Proc Natl Acad Sci U S A. 2010;107:21848–21853. doi: 10.1073/pnas.1011756107. doi:10.1073/pnas.1011756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawashima T, et al. Functional labeling of neurons and their projections using the synthetic activity-dependent promoter E-SARE. Nat Methods. 2013;10:889–895. doi: 10.1038/nmeth.2559. doi:10.1038/nmeth.2559. [DOI] [PubMed] [Google Scholar]
- 35.Guenthner CJ, Miyamichi K, Yang HH, Heller HC, Luo L. Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron. 2013;78:773–784. doi: 10.1016/j.neuron.2013.03.025. doi:10.1016/j.neuron.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Izpisua Belmonte JC, et al. Brains, genes, and primates. Neuron. 2015;86:617–631. doi: 10.1016/j.neuron.2015.03.021. doi:10.1016/j.neuron.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Marel S, et al. Neutralizing antibodies against adeno-associated viruses in inflammatory bowel disease patients: implications for gene therapy. Inflamm Bowel Dis. 2011;17:2436–2442. doi: 10.1002/ibd.21673. doi:10.1002/ibd.21673. [DOI] [PubMed] [Google Scholar]
- 38.Calcedo R, Vandenberghe LH, Gao G, Lin J, Wilson JM. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J Infect Dis. 2009;199:381–390. doi: 10.1086/595830. doi:10.1086/595830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boutin S, et al. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum Gene Ther. 2010;21:704–712. doi: 10.1089/hum.2009.182. doi:10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- 40.Levitt N, Briggs D, Gil A, Proudfoot NJ. Definition of an efficient synthetic poly(A) site. Genes and Development. 1989;3:1019–1025. doi: 10.1101/gad.3.7.1019. doi:10.1101/gad.3.7.1019. [DOI] [PubMed] [Google Scholar]
- 41.Chiorini JA, Kim F, Yang L, Kotin RM. Cloning and characterization of adeno- associated virus type 5. J Virol. 1999;73:1309–1319. doi: 10.1128/jvi.73.2.1309-1319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farris KD, Pintel DJ. Improved Splicing of Adeno-Associated Viral (AAV) Capsid Protein-Supplying Pre-mRNAs Leads to Increased Recombinant AAV Vector Production. Human gene therapy. 2008;19:1421–1427. doi: 10.1089/hum.2008.118. doi:10.1089/hum.2008.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Albert H, Dale EC, Lee E, Ow DW. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. The Plant journal : for cell and molecular biology. 1995;7:649–659. doi: 10.1046/j.1365-313x.1995.7040649.x. [DOI] [PubMed] [Google Scholar]
- 44.Shaner NC, et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods. 2013;10:407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hancock JF, Cadwallader K, Paterson H, Marshall CJ. A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. EMBO J. 1991;10:4033–4039. doi: 10.1002/j.1460-2075.1991.tb04979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gray SJ, et al. Production of recombinant adeno-associated viral vectors and use in in vitro and in vivo administration. Current protocols in neuroscience / editorial board, Jacqueline N. Crawley. 2011 doi: 10.1002/0471142301.ns0417s57. [et al.] Chapter 4, Unit 4 17, doi:10.1002/0471142301.ns0417s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ayuso E, et al. High AAV vector purity results in serotype- and tissue-independent enhancement of transduction efficiency. Gene therapy. 2010;17:503–510. doi: 10.1038/gt.2009.157. doi:10.1038/gt.2009.157. [DOI] [PubMed] [Google Scholar]
- 48.Zolotukhin S, et al. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene therapy. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. doi:10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
- 49.Wobus CE, et al. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J Virol. 2000;74:9281–9293. doi: 10.1128/jvi.74.19.9281-9293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.