Abstract
Expressed protein ligation is a valuable method for protein semisynthesis that involves the reaction of recombinant protein C-terminal thioesters with N-Cys containing peptides but the requirement of a Cys residue at the ligation junction can limit its use. Here we employ subtiligase variants to efficiently ligate Cys-free peptides to protein thioesters. Using this method, we have more accurately determined the effect of C-terminal phosphorylation on the tumor suppressor protein PTEN.
The ability to site-specifically install post-translational modifications, biophysical probes, unnatural amino acids, isotopic labels, and drug-like small molecules into proteins of any size offers enormous potential for both fundamental and applied biomedical research1–4. Semisynthesis using expressed protein ligation (EPL) has been exploited frequently in the construction of proteins containing diverse chemical modifications1,3. In standard EPL, a recombinant protein fragment fused to an intein is reacted with a thiol to generate the isolated recombinant protein C-terminal thioester5,6. An N-Cys containing synthetic peptide is then added to the protein thioester which undergoes transthioesterifcation followed by rearrangement to a conventional amide bond5,6 (Fig. 1a). While powerful, the scope of EPL is narrowed by the requirement of a Cys at the ligation junction. Cys is one of the least frequently encoded residues in proteins7, and this sharply confines the flexibility in the ligation position or requires the introduction of Cys at unnatural locations, sometimes perturbing function. Methods to overcome the Cys requirement8,9 in EPL are under development but have not yet proved robust.
Figure 1.
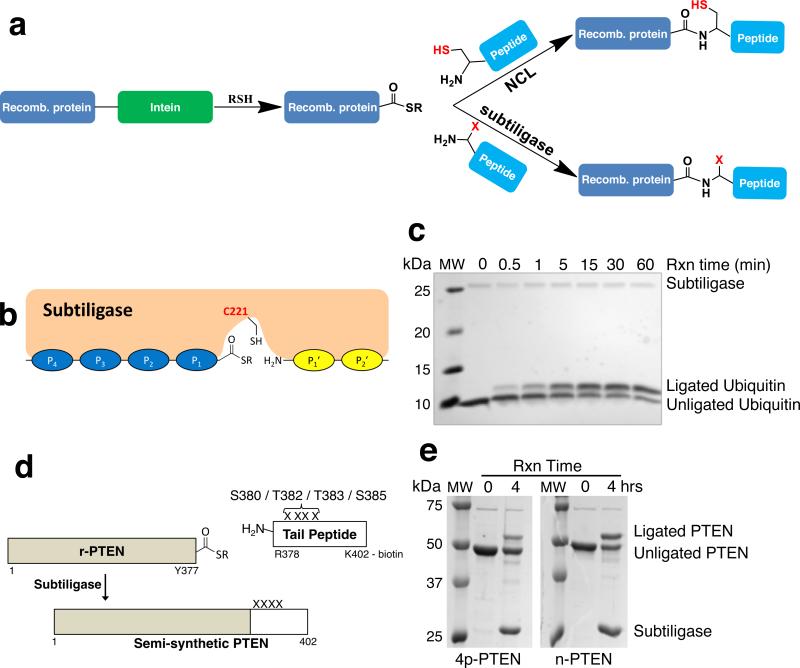
Subtiligase-catalyzed expressed protein ligation. (a) Schematic showing the intein-mediated generation of a protein thioester followed by ligation via native chemical ligation (NCL) or subtiligase. (b) Subtiligase is known to interact with 4 residues (P4 – P1) N-terminal to and 2 residues (P1′ – P2′) C-terminal to the ligation site. (c) Time-course analysis of a subtiligase-catalyzed ligation between G76Y ubiquitin thioester (P4 P1 = LY) and peptide 1 (P1′ P2′ = GL, see Supplementary Table 3). MW, molecular weight markers. (d) r-PTEN, consisting of residues 1 – 377 of PTEN, was ligated to a 25-mer peptide corresponding to residues 378 – 402 (tetra-phosphorylated 4p-tail or non-phosphorylated np-tail, see Supplementary Table 3) and also containing a biotin attached to the C-terminal Lys sidechain. (e) SDS-PAGE analysis of the results of 4-hour ligations between r-PTEN and either the tetra-phosphorylated or non-phosphorylated tail peptides. MW, molecular weight markers. Ligations with the tetra-phosphorylated and non-phosphorylated tails proceeded to ~30% and ~50% completion, respectively, possibly indicating that pSer380 in the P3′ position may interfere with the ligation reaction.
Subtiligase is an engineered peptide ligase derived from the protease subtilisin that contains mutations (S221C, P225A) which alter the mechanism to favor aminolysis over peptidase activity, facilitating the ligation of a peptide containing a donor C-terminal ester to an acceptor peptide containing an α-amine10–12. Subtiligase has been used in a variety of settings but has not been previously applied to ligations with recombinant protein thioester fragments, which we investigate here (Fig. 1a).
We initially examined subtiligase-catalyzed ligation with ubiquitin (Ub) C-terminal thioester, produced from an intein fusion protein after reaction with MESNA (Supplementary Fig. 1a). The extreme C-terminal residues of Ub are LRGG, and it is noteworthy that the reported preferences of subtiligase include a hydrophobic residue at the P4 position10–13 (Fig. 1b), which is Leu in Ub. A synthetic decapeptide (GLSGRGKGGK(biotin), 1 mM) was reacted with 100 μM Ub thioester, 7.5 μM subtiligase, 100 mM bicine at pH 8, and 5 mM Ca+2 at room temperature and the reaction was monitored by SDS-PAGE. As shown in Figure 1c, the ligation appeared to plateau by 60 min at ~50% ligated product with half-maximal conversion at 5 min. Mass spectrometry confirmed the correct structure of the product (Supplementary Fig. 2). As controls, it was shown that subtiligase was required for the transformation and standard Ub lacking a thioester functionality was unreactive under these conditions (Supplementary Fig. 1). Interestingly, recovered Ub showed that hydrolysis of the thioester was accelerated by subtiligase (Supplementary Fig. 3), which can account for the incomplete conversion at longer reaction times.
Prior data obtained with subtiligase and synthetic peptide oxyester ligations indicate that the P4, P1, P1′, and P2′ positions are the most influential for ligation efficiency10–13 (Fig. 1b). We studied Ub variants that contained a range of residues at the P4 (L,A,E) and P1 (Y,L,I,V,F,A,W,H,G) positions and a set of synthetic acceptor peptides with diversity introduced at the P1′ (G,A,R,F,M,S,Y,P,E) and P2′ (L,T,I,G) positions (Supplementary Table 3). As shown in Supplementary Figure 1 and Supplementary Table 1, the majority of the conversion efficiencies were in the range of 50-70%. Notable exceptions to these relatively good subtiligase-catalyzed Ub ligation efficiencies were when P4 or P1′ was Asp/Glu or when P1′ was Pro, previously identified as poor substrates for oxyester peptide ligations10–15.
In cases involving acidic residues at P1, QK subtiligase (E156Q, G166K) has been shown to improve oxyester peptide ligations10,13. We also designed Y217K subtiligase to complement acidic residues at the P1′ position (Supplementary Fig. 4a,b). In the case of Glu at P1, the conversion efficiency improved from 33% with standard subtiligase to ~50% with either QK or Y217K subtiligase (Supplementary Fig. 4c,d). In cases of Glu at P1′, the conversion efficiencies were markedly improved with Y217K subtiligase relative to the standard or QK forms (Supplementary Fig. 4c,d). These modified subtiligase forms thus broaden the scope of enzyme-catalyzed EPL.
To compare the efficiency of enzyme-catalyzed EPL to standard EPL, we performed the uncatalyzed reactions with an N-Cys-containing peptide substrate to a subtiligase-catalyzed reaction where the N-terminal residue was Gly (Supplementary Fig. 5a,b). The initial subtiligase reaction rate was approximately 3-fold faster than that of standard EPL (Supplementary Fig. 5c). These results underscore the rapid kinetics of enzyme-catalyzed EPL although further work will be needed to compare detailed rates across a range of substrates.
We assessed the generality of enzyme-catalyzed EPL with a series of glutathione S-transferase (GST) thioesters containing a range of C-terminal P4 (F,S,D) and P1 (Y,T,R,G,E) residues and a set of ligating decapeptides (Supplementary Table 3) containing varying P1′-P2′ residues (MT, GL, SI) (Supplementary Fig. 6). In general, as shown in Supplementary Fig. 6d and Supplementary Table 2, the efficiencies were similar to those of the Ub ligations. Interestingly, with Ser at P4, reaction was favorable when P1 was Tyr (conversions close to 60%) but not when P1 was Gly (<5%). However, Gly at P1 was acceptable when P4 was Phe with conversions of ~30%. Given the Ub thioester ligations described above, perhaps the most surprising result was that Glu at P1 was well-tolerated when P4 was Phe with conversions of ~60%. These results highlight how there can be an interplay among the C-terminal amino acids influencing ligation efficiency.
We next adopted enzyme-catalyzed EPL to determine the influence of C-terminal phosphorylation on the tumor suppressor lipid phosphatase PTEN. PTEN is phosphorylated on a cluster of four C-terminal S/T residues (S380, T382, T383, S385) in vivo by CK2 protein kinase, and these phosphorylations can inhibit the enzyme and induce an intramolecular conformational change in PTEN16,17. Conventional EPL has been used previously to generate tetraphosphorylated PTEN16. In this case, a Y379C mutation was introduced into PTEN since there is no nearby Cys for ligation. It has subsequently been discovered18 that Y379C PTEN behaves anomalously in cells. Thus, generating tetraphosphorylated PTEN without introducing a nearby Cys was desirable. The proximal natural sequence in this region, 374-PDHYRYS-380, suggested that positioning Y377 as the P1 residue and P374 as the P4 residue could be favorable for enzyme-catalyzed EPL. We thus generated the corresponding aa1-377 PTEN thioester (r-PTEN) and ligated it to a tetraphosphorylated (and unphosphorylated) peptide aa378-402 modified with a biotin at Lys402 (Fig. 1d,e and Supplementary Fig. 7), affording the desired semisynthetic tetraphosphorylated (Y379-4p-PTEN) and unphosphorylated (Y379-n-PTEN) products.
Western blot of Y379-4p-PTEN with an anti-phospho-PTEN Ab showed a ~4-fold more intense signal for the natural sequence compared with C379-4p-PTEN protein (Supplementary Fig. 7e), presumably because Y379 is important in the epitope. This enabled the use of the more natural 4p-PTEN to serve as a standard in determining the level of cellular C-terminal phosphorylation of endogenous PTEN isolated from mammalian cells. This revealed that the stoichiometry of tail phosphorylation of endogenous PTEN in mouse embryonic fibroblasts was ~72%, but the level of phosphorylation dropped after CK2 protein kinase inhibition (Fig. 2a) presumably due to the dynamic opposing actions of kinases and phosphatases19.
Figure 2.
Y379-4p-PTEN can be used as a standard to quantify PTEN phosphorylation in cell culture and shows lower activity and a more closed conformation compared to C379-4p-PTEN. (a) Representative Western blots showing a decrease in PTEN phosphorylation following treatment with a CK2 inhibitor. Fraction phosphorylated: Untreated, 0.72 ± 0.06; Treated, 0.33 ± 0.09 (n = 5 biological replicates, p = 0.0071, Student's t-test). Note that it has previously been shown that phosphorylation can stabilize cellular PTEN20, explaining the reduction in total PTEN after CK2 inhibition. (b) Activity of t-, Y379-n-, C379-4p-, and Y379-4p-PTEN toward 160 μM soluble di-C6 PIP3 with 60 mM or 200 mM NaCl. Values are the averages of 2 replicates and error bars represent the span of measurements. (c) Illustration of the open and closed conformations of PTEN and the change in accessibility of the phosphorylation sites to alkaline phosphatase. Red circles indicate phosphorylated residues. Cat., catalytic domain; C2, C2 domain; AP, alkaline phosphatase. (d) Y379C-4p-PTEN is more resistant to treatment with alkaline phosphatase than C379-4p-PTEN. Data are the average of two replicates for each PTEN construct. Representative Western blots show the decrease in phosphorylation with longer phosphatase treatment for both PTEN variants. C379-4p-PTEN half-life = 9.0 ± 2.2 min; Y379-4p-PTEN half-life = 112.1 ± 24.5 min (n = 2, p = 0.0273, Student's t-test).
The enzymatic activity of Y379-4p-PTEN was decreased compared with C379-4p-PTEN, which were both lower than the unphosphorylated and truncated (t-PTEN) forms (Fig. 2b), indicating that the Y379 residue may stabilize the closed, autoinhibited phosphorylated PTEN conformation16. Such autoinhibition was most marked at higher (200 mM) NaCl, suggesting that Y379-4p-PTEN is more resistant to conformational opening at high ionic strength compared with C379-4p-PTEN. Alkaline phosphatase has previously been employed to probe the conformation of C379-4p-PTEN16. We thus compared alkaline phosphatase mediated dephosphorylation of natively folded Y379-4p-PTEN to that of C379-4p-PTEN16, and this showed that the former had about a 10-fold slower rate of phosphate removal (Fig. 2c,d). These data suggest that Y379-4p-PTEN does indeed reside in a more tightly closed conformation relative to C379-4p-PTEN16. and can account for the anomalous cellular behavior of Y379 PTEN mutations18.
It would have been difficult to generate Y379-4p-PTEN without relying on enzyme-catalyzed EPL. The relatively broad flexibility of amino acid sequences around the ligation junction, the simplicity of the protocol, and the speed of the process combine to make enzyme-catalyzed EPL an attractive technique for protein semisynthesis.
Methods
B. Subtilis transformation with subtiligase
This was carried out analogously to the previously described methods10–13. E. coli–B. subtilis shuttle plasmid pPW 04 containing the pre-prosubtiligase sequence was purified from E. coli K12 ER1821 (NEB) and transformed into B. subtilis BG2864 (ΔaprE ΔnprE ΔflaA::kan, ATCC). B. subtilis was grown on LB agar + 25 μg/ml kanamycin. 2×YT media (5 ml) was inoculated with a single colony and grown overnight at 37°C in a spinning incubator. Cells were pelleted and resuspended in 5 ml bacillus medium A (80 mM K2HPO4, 45 mM KH2PO4, 15 mM (NH4)2SO4, 4 mM C6H5O7Na3, 5 mM MgCl2, 50 μg/ml tryptophan, 0.5% glucose, 0.02% amicase). 1 ml of this suspension was added to 50 ml medium A and grown at 37°C until 90 minutes after the culture had exited log phase growth. Then, 0.5 ml of this culture was added to 5 ml medium B (80 mM K2HPO4, 45 mM KH2PO4, 15 mM (NH4)2SO4, 4 mM C6H5O7Na3, 5 mM MgCl2, 2.5 μg/ml tryptophan, 0.5% glucose, 0.005% amicase). Next, 300 μl of this culture was added to tubes containing 2 μg of plasmid pPW 04 and grown at 37°C. After 2 hours, 1 ml 2×YT medium containing 0.5 μg/ml chloramphenicol was added, and the culture was grown for 1 additional hour. Culture was spun down (4000 × g, 2 min), resuspended in 150 μl 2×YT, and plated on LB agar containing 5 μg/ml chloramphenicol.
Site-directed mutagenesis of GST, ubiquitin, subtiligase, and PTEN
Supplementary Table 4 lists all primers used for cloning and mutagenesis. GST was cloned out of the pGEX-6P-1 vector. 1 μl of template was incubated with 200 nM forward and reverse primers and 2 mM dNTP mix (0.5 mM each dNTP) in 50 μl reaction buffer (20 mM Tris-HCl, 2 mM MgSO4, 10 mM KCl, 10 mM (NH4)2SO4, 0.1% Triton X-100, 0.1 mg/ml BSA, pH 8.8) along with 1 μl Pfu Ultra II polymerase (Agilent). PCR for mutagenesis was carried out as follows: 95°C for 30 sec, then 18 cycles of 95°C for 30 sec, 55°C for 1 min, and 68°C for 30 sec, followed by a final 2 min at 68°C, then 4°C for 5 min. Amplification of the GST gene was confirmed by agarose gel electrophoresis. The amplified GST gene was isolated from the agarose gel using a gel extraction kit (Qiagen) and the ends of the gene were trimmed using the NdeI and SmaI restriction enzymes (NEB). The gene was incubated with 1 unit/μl SmaI in NEBuffer 4 (50 mM potassium acetate, 20 mM Tris-acetate, 10 mM magnesium acetate, 1mM DTT, pH 7.9) for 2 hours at 25°C, then incubated with 1 unit/μl NdeI for 2 hours at 37°C. The pTYB2 vector was also digested in this way. To insert the gene into the vector, the digested gene and vector were incubated with 40 units/μl T4 ligase in T4 buffer (50 mM Tris-HCl, 10 mM MgCl2, 1 mM ATP, 10 mM DTT, pH 7.5) for 18 hours at 16°C. The completed ligation was transformed into chemically competent E. coli DH5α. The sequence was confirmed by Sanger sequencing. An overlooked SmaI site near the C-terminus of GST resulted in the truncation of the final 8 residues of GST and a single nucleotide frameshift of the intein-CBD relative to the GST. Quikchange mutagenesis was used to add Phe-Ala-Ala-Tyr to the C-terminus of GST and remove the frameshift. 1 μl of template was incubated with 200 nM forward and reverse primers and 2 mM dNTP mix in 50 μl reaction buffer along with 1 μl Pfu Ultra II polymerase. PCR for mutagenesis was carried out as follows: 95°C for 30 sec, then 18 cycles of 95°C for 30 sec, 55°C for 1 min, and 68°C for 7.5 min, followed by a final 5 min at 68°C, then 4°C for 5 min. After PCR, the reaction was incubated with 0.4 units/μl DpnI at 37°C for 1 hour to digest methylated DNA. The mutated plasmid was transformed into chemically competent E. coli DH5α. Quikchange mutagenesis was also used to further mutate the C-termini of GST and Ubiquitin, as well as mutate subtiligase and create r-PTEN from t-PTEN. Supplementary Table 2 shows the primers used for cloning and mutagenesis.
Subtiligase expression and purification
Parent and mutant subtiligase were expressed and purified using the following methods adapted from previous procedures10–13. Transformed B. subtilis was streaked out on LB agar + 10 μg/ml chloramphenicol. 2×YT media (5 ml) containing 10 μg/ml chloramphenicol was inoculated with 1 colony and grown overnight. 1 ml of overnight culture was added to 1 L 2×YT media containing 10 μg/ml chloramphenicol and grown at 37°C for 24 hours. Cells were pelleted and discarded and then the supernatant was slowly treated with 600 g (NH4)2SO4 while stirring on ice at 4°C. The solution was stirred for an additional hour during which a brown protein precipitation was observed. The mixture was centrifuged (10,000 × g, 30 min, 4°C) and the saved pellet was then re-dissolved in 75 ml sodium acetate buffer (25 mM CH3CO2Na, 5 mM DTT, pH 4.5). To this solution was added 300 ml ethanol, resulting in a pale brown precipitate, and the mixture was stirred for 30 minutes at 4°C. The mixture was then centrifuged (5000 × g, 15 minutes, 4°C) and the pellet was saved and re-dissolved in 50 ml sodium acetate buffer, then dialyzed thoroughly against the same buffer. After dialysis, the mixture was centrifuged (17,500 × g, 15 min, 4°C) and the pellet discarded. The supernatant was loaded onto a Mono S cation-exchange column (GE Healthcare Life Sciences). Protein was eluted in a 0 – 400 mM NaCl gradient in sodium acetate buffer, pH = 4.5. Fractions containing subtiligase were dialyzed overnight against 100 mM BICINE, 5mM DTT, pH 8.0, then concentrated to ~100 μM in an Amicon Ultra 10 kDa MWCO filter unit, flash frozen, and stored at −80°C. Protein was estimated to be greater than 95% by SDS-PAGE and the stock concentration was determined by comparison to bovine serum albumin standards stained with Coomassie blue.
GST expression and purification
GST was subcloned into a pTYB2 vector which contains the VMA intein from S. cerevisiae and subjected to insertional mutagenesis as described above, resulting in the appending of 4 residues (X-Ala-Ala-X) to the natural GST C-terminus followed by the intein. The GST plasmids were expressed in E. coli BL21. LB media (5 ml) containing 100 μg/ml carbenicillin was inoculated with a single colony and grown overnight at 37°C in a spinning incubator. 3.5 ml from overnight cultures was used to inoculate 1 L LB media containing 100 μg/ml carbenicillin, and the cultures were grown in shaker flasks at 37°C until OD600 = 0.7, then 1 ml 1M IPTG was added to induce expression and the cultures were incubated overnight while shaking at 16°C. Cells were pelleted and then resuspended in 40 ml lysis buffer (250 mM NaCl, 50 mM HEPES, 1 mM EDTA, 10% glycerol, pH = 7.5). Unused cell pellets were flash frozen and stored at −80°C until needed. E. coli cells were lysed by french press, the lysate was pelleted (17,500 × g, 15 minutes, 4°C), and the supernatant was loaded onto 5 ml of chitin resin (NEB). Resin was washed with 150 ml wash buffer (250 mM NaCl, 25 mM HEPES, 0.1% Triton X-100, pH = 7.5) then incubated overnight in cleavage buffer (250 mM NaCl, 50 mM HEPES, 300 mM MESNA, pH 7.5). To generate GST-biotin, 10 mM peptide containing a N-terminal cysteine and C-terminal Lys-biotin was also added to the cleavage buffer. The cleavage buffer was eluted from the resin, and the buffer was exchanged with 150 mM NaCl, 50 mM MES, pH 6.0 using an Amicon 10 kDa MWCO filter unit. The GST thioester proteins were concentrated to >1 mg/ml, flash frozen and stored at −80°C and appeared to be >95% pure by Coomassie-stained SDS-PAGE. Protein concentration was determined by SDS-PAGE referenced to bovine serum albumin standards using Coomassie staining.
Ubiquitin expression and purification
Human ubiquitin was C-terminally fused with Mxe intein into a pTXB1 vector and subjected to mutagenesis as described above to generate L73X and G76X residues. The ubiquitin plasmids were expressed in E. coli Rosetta(DE3). LB media (15 ml) containing 100 μg/ml ampicillin and 10 μg/ml Chloramphenicol was inoculated with a single colony and grown overnight at 37°C in a shaking incubator. 10 ml from overnight cultures was used to inoculate 1 L LB media containing 100 μg/ml ampicillin and 10 μg/ml Chloramphenicol, and the cultures were grown in shaker flasks at 37°C until OD600 = 0.7, then 1 ml 1M IPTG was added to induce expression and the cultures were further incubated for 3 hours at 37°C. Cells were pelleted and then resuspended in 20 ml lysis buffer (250 mM NaCl, 50 mM HEPES, 1 mM EDTA, 10% glycerol, pH = 7.5).
E. coli cells were lysed by french press, the lysate was pelleted (17,500 × g, 30 minutes, 4°C), and the supernatant was loaded onto 5 ml of chitin resin (NEB). Resin was washed with 150 ml wash buffer (250 mM NaCl, 25 mM HEPES, 0.1% Triton X-100, pH = 7.5) then incubated overnight in cleavage buffer (250 mM NaCl, 50 mM HEPES, 300 mM MESNA, pH 7.5). The cleavage buffer was eluted from the resin, and the buffer was exchanged with 150 mM NaCl, 50 mM MES, pH 6.0 using an Amicon 3.0 kDa MWCO filter unit. The ubiquitin thioester proteins were concentrated to 1 mM, flash frozen and stored at −80°C and appeared to be >95% pure by Coomassie-stained SDS-PAGE. Protein concentration was determined by SDS-PAGE referenced to bovine serum albumin standards using Coomassie staining.
Peptide synthesis
Peptides were synthesized either on a Prelude peptide synthesizer or PS3 peptide synthesizer from Protein Technologies using standard Fmoc-based solid phase peptide synthesis. Non-phosphorylated peptides were synthesized by double-coupling every residue. Fmoc groups were deprotected for five times, 10 minutes each with 20% piperidine in DMF. Coupling times were 1.5 hours. Biotinylated peptides were synthesized using N-ε-biotin-lysine Wang resin (Iris Biotech). Phosphorylated peptides corresponding to residues 378-403 of PTEN were synthesized by double-coupling residues 386-402 for 1.5 hours each, triple-coupling residues Asp384 and Asp381 for 1.5 hours each, and double coupling phosphorylated residues for 3 hours each. Phosphate groups were mono-protected by O-benzyl groups during the synthesis. All peptides were deprotected and cleaved from resin using reagent K (82.5:2.5:5:5:5 – trifluoroacetic acid:ethane dithiol:water:thioanisole:phenol) then purified using reverse-phase C18 HPLC and lyophilized. Peptide structures were confirmed using MALDI mass spectrometry and peptide concentrations were determined by amino acid analysis.
Ubiquitin and GST ligations
The standard ligation reaction conditions employed 40 μM protein-thioester, 3 mM biotinylated peptide, 0.5 μM subtiligase in a buffer containing 100 mM BICINE, 5 mM CaCl2, pH 8.0 for 90 minutes at 25°C before quenching with SDS loading dye. A non-subtiligase control was also included, as well as a zero time-point where the reaction was quenched prior to adding subtiligase. Aliquots (3 μl) of quenched reaction mixtures were run on 12% SDS-PAGE and detected by Coomassie blue staining or Western blot for ubiquitin or GST proteins respectively. For Western blot, after transfers to membranes using an iBlot system, the membranes were blocked overnight in 50 mg/ml BSA in TBS-T. Membranes were incubated with anti-biotin HRP-linked antibody (Sigma #A4541) at a 1:10,000 dilution for 1 hour, washed several times by TBS-T and developed with Amersham ECL Western blotting detection reagents (GE Healthcare) and imaged by a GeneSys imaging system for 4 minutes. Band intensities were quantified using ImageJ software (imagej.nih.gov). Ligations were performed at least twice, and each replicate showed similar yield (within 20%).
r-PTEN expression and purification
A pFastBac1 baculovirus vector containing the PTEN-intein-CBD fusion (PTEN aa1-377) was used to make bacmid and then baculovirus in SF-21 insect cells using standard methods16 which was then used to infect High Five insect cells with M.O.I. 1.0. After growing infected High Five cells in Express Five SFM Media (Gibco) for 48 hours at 27°C, they were pelleted (700 × g, 10 min, 4°C) and then resuspended in 1/20th the medium used for culture, pelleted again (discarding the supernatant), and then flash frozen and stored at −80°C. Resuspended cells from 200 ml culture were lysed in a 40 ml homogenizer in 30 ml lysis buffer (150 mM NaCl, 50 mM HEPES, 1 mM EDTA, 10% Glycerol, 0.1% Triton X-100) containing 3 dissolved protease tablets (Roche). The lysate was then centrifuged (17,500 × g, 40 min, 4°C) and the supernatant was added to a 10 ml bed of powdered cellulose (Sigma). After 1 hour of incubation at 4°C, the lysate was drained from the cellulose and then bound to 5 ml chitin resin (NEB). The resin was then washed with 150 ml washing buffer (250 mM NaCl, 25 mM HEPES, 0.1% Triton X-100, pH 7.5), incubated overnight in cleavage buffer (250 mM NaCl, 50 mM HEPES, 300 mM MESNA, pH 7.5). The cleavage buffer was eluted from the resin, and the buffer was exchanged with 150 mM NaCl, 50 mM MES, pH 6.5 using an Amicon 10 kDa MWCO filter unit. The PTEN thioester protein produced was shown to be >80% pure by Coomassie-stained SDS-PAGE and concentrated to greater than 1 mg/ml. If the r-PTEN protein thioester was not used within a day, it was flash frozen and stored at −80°C. For stable t-PTEN generation, 50 mM Cys was added to the cleavage buffer and it was otherwise handled as described above.
4p-PTEN ligation and purification
Under standard conditions (1 ml), a reaction mixture containing r-PTEN thioester (~1 mg/ml) C-terminal peptide (~10 mg/ml) in buffer (100 mM BICINE, 110 mM CaCl2, pH 8.0) was treated with subtiligase (~25 μM final). Note that higher subtiligase and increased Ca concentrations relative to the ubiquitin and GST cased appeared to improve the ligation yield, presumably because of the slower rate of this ligation. A 3 μl aliquot was saved as a negative control prior to adding subtiligase. The reaction was split into 150 μl aliquots, and incubated at 25°C. After 4 hours, 3 μl of the mixture was taken for SDS-PAGE analysis, and the remainder was injected onto a Superdex 75 size-exclusion column (GE Healthcare Life Sciences). Size exclusion chromatography was performed with a flow-rate of 0.5 ml/min in a buffer containing 500 mM NaCl, 50 mM Na2HPO4, 5 mM DTT, pH 7.0 and 0.3 ml fractions were collected and 5 μl of each fraction was analyzed by Coomassie-stained SDS-PAGE. Fractions containing ligated PTEN were combined and stored at 4°C overnight and then loaded onto 500 μl pre-blocked (10 mM biotin followed by 0.1 M Glycine, pH 2.8, followed by 500 mM NaCl, 50 mM Na2HPO4, 5 mM DTT, pH 7.0) mono-avidin resin (Thermo) on ice. Resin was washed sequentially with 5 ml of 500 mM NaCl, 50 mM Tris, 10 mM DTT, pH 8.0 for 1 hour followed by 5 ml of 1 M NaCl, 50 mM Tris, 10 mM DTT, pH 8.0 for another hour. Next, the resin was equilibrated with 5 ml 150 mM NaCl, 50 mM Tris, 10 mM DTT, pH 8.0 and then eluted with 10 mM biotin in the same buffer. The resin was incubated with 100 μl of elution buffer for 15 minutes at room temperature prior to an additional 500 μl of elution buffer. The purified semisynthetic protein was shown to be >90% pure by Coomassie-stained SDS-PAGE.
MEFs and Western blot analysis
Murine embryo fibroblasts (MEF) obtained from the Stivers lab at Johns Hopkins University or purchased from the ATCC (shown to be mycoplasma negative) were grown in DMEM, high glucose (Thermo Fisher) with 10% FBS at 37 °C, 5% CO2. Once the cells had reached ~70% confluency, the media was exchanged with DMEM with 2% FBS. Cells were then either treated with 50 μg/ml 4,5,6,7-tetrabromobenzotriazole (TBB) or DMSO. After 12-15 hours’ incubation at 37°C, cells were washed with PBS then lysed with RIPA buffer plus 1 mM PMSF. The lysate was pelleted at 15,000 × g (10 minutes, 4°C). The protein concentration in the supernatant was determined by BCA assay (Thermo Fisher). Supernatant with 50 μg of protein was run on a 10% SDS-PAGE gel along with Y379-4p-PTEN standards. Western blot membranes were incubated with either anti-phospho-PTEN antibody 44A7 (Cell Signaling #9549) or anti-PTEN antibody N-19 (Santa Cruz Biotech #sc-6818) at 1:1000 dilutions. Western blots were developed and analyzed as described above. Values reported in Fig. 2 are plus or minus standard error.
Assay of phospho-PTEN's sensitivity to alkaline phosphatase
This was adapted from previous methods.16 4p-PTEN (1 μg) was incubated with 5 μM calf intestine phosphatase (NEB) in 20 μl of reaction buffer (50 ng/μl PTEN) for a total of 4 hours. Reaction buffer consisted of 50 mM Tris, 10 mM BME, pH 8.0. Samples were taken from the reaction at various time points, diluted 10-fold in SDS loading dye and run on SDS-PAGE for Western blot analysis. 10 μl were loaded for the Cys-PTEN and 2 μl were loaded for the Tyr-PTEN. Blots were analyzed using anti-phospho-PTEN antibody (Cell Signaling #9554) at 1:1000 dilution. Images were analyzed using ImageJ software and Prism 6 software (Graphpad) was used to determine phosphorylation half-life fit to a standard exponential decay. Replicates with different preps showed similar half-lives (within 20%). Values reported in Fig. 2 are plus or minus standard error.
PTEN phosphatase activity assay
PTEN activity toward a water-soluble substrate (diC6-PIP3, from Avanti Polar Lipids) was determined by the evolution of inorganic phosphate as measured with a malachite green detection kit (R and D Biosystems). Assays were conducted as described previously16. Briefly, 0.5 – 1.5 μg PTEN was incubated with 160 μM diC6 PIP3 for 10 minutes in 25 μl reaction buffer (50 mM Tris, 10 mM BME, pH 8.0) at 30°C. Samples were quenched with malachite green reagent (R and D Biosystems) and absorbance was measured at 620 nm.
Statistics
All experiments were performed at least two times. As the relevant biological or technical replicates in almost every case gave conversions or rate constants that were within 30% of each other, duplicate experiments were believed to be sufficient to ensure confidence in these findings.
Supplementary Material
Acknowledgements
This work was supported by the NIH and the FAMRI foundation. We thank Y. Li for assisting with insect and mammalian cell culture. We thank the Stivers Lab (Johns Hopkins School of Medicine) for providing us with MEF cells. We thank A. Liu for assisting with the ubiquitin studies and we thank the other members of the Cole lab for valuable discussions and advice.
Footnotes
Contributions
S.H.H., N.C., J.W., and P.A.C. designed experiments. S.H.H., N.C., and Z.C. carried out experiments. S.H.H., N.C., Z.C., D.B., D.D., D.R.D., and Y.H. analyzed data. S.H.H., N.C., and P.A.C. drafted the manuscript and all authors contributed to editing it to produce the final version.
Competing financial interests
The authors declare no competing financial interests
References
- 1.Vila-Perelló M, Muir TW. Cell. 2010;143:191–200. doi: 10.1016/j.cell.2010.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun SB, Schultz PG, Kim CH. Chembiochem Eur. J. Chem. Biol. 2014;15:1721–1729. doi: 10.1002/cbic.201402154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Z, Cole PA. Curr. Opin. Chem. Biol. 2015;28:115–122. doi: 10.1016/j.cbpa.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmohl L, Schwarzer D. Curr. Opin. Chem. Biol. 2014;22:122–128. doi: 10.1016/j.cbpa.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 5.Muir TW, Sondhi D, Cole PA. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans TC, Benner J, Xu MQ. Protein Sci. Publ. Protein Soc. 1998;7:2256–2264. doi: 10.1002/pro.5560071103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooks DJ, Fresco JR, Lesk AM, Singh M. Mol. Biol. Evol. 2002;19:1645–1655. doi: 10.1093/oxfordjournals.molbev.a003988. [DOI] [PubMed] [Google Scholar]
- 8.Loibl SF, Harpaz Z, Seitz O. Angew. Chem. Int. Ed Engl. 2015;54:15055–15059. doi: 10.1002/anie.201505274. [DOI] [PubMed] [Google Scholar]
- 9.Machova Z, von Eggelkraut-Gottanka R, Wehofsky N, Bordusa F, Beck-Sickinger AG. Angew. Chem. Int. Ed Engl. 2003;42:4916–4918. doi: 10.1002/anie.200351774. [DOI] [PubMed] [Google Scholar]
- 10.Abrahmsén L, et al. Biochemistry. 1991;30:4151–4159. doi: 10.1021/bi00231a007. [DOI] [PubMed] [Google Scholar]
- 11.Chang TK, Jackson DY, Burnier JP, Wells JA. Proc. Natl. Acad. Sci. U. S. A. 1994;91:12544–12548. doi: 10.1073/pnas.91.26.12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson DY, et al. Science. 1994;266:243–247. doi: 10.1126/science.7939659. [DOI] [PubMed] [Google Scholar]
- 13.Braisted AC, Judice JK, Wells JA. Methods Enzymol. 1997;289:298–313. doi: 10.1016/s0076-6879(97)89053-2. [DOI] [PubMed] [Google Scholar]
- 14.Takeuchi Y, Satow Y, Nakamura KT, Mitsui Y. J. Mol. Biol. 1991;221:309–325. [PubMed] [Google Scholar]
- 15.Jain SC, Shinde U, Li Y, Inouye M, Berman HM. J. Mol. Biol. 1998;284:137–144. doi: 10.1006/jmbi.1998.2161. [DOI] [PubMed] [Google Scholar]
- 16.Bolduc D, et al. eLife. 2013;2:e00691. doi: 10.7554/eLife.00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masson GR, Perisic O, Burke JE, Williams RL. Biochem. J. 2016;473:135–144. doi: 10.1042/BJ20150931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen HN, et al. Oncogene. 2014;33:5688–5696. doi: 10.1038/onc.2013.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiao Y, Molina H, Pandey A, Zhang J, Cole PA. Science. 2006;311:1293–1297. doi: 10.1126/science.1122224. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, et al. J. Biol. Chem. 2016;291:14160–14169. doi: 10.1074/jbc.M116.728980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.