Abstract
Acinetobacter baumannii had emerged as an important nosocomial and opportunistic pathogen worldwide. To assess the evolution of colistin resistance in A. baumannii and its effect on bacterial fitness, we exposed five independent colonies of A. baumannii ATCC 17978 to increasing concentrations of colistin in agar (4/5) and liquid media (1/5). Stable resistant isolates were analyzed using whole genome sequencing. All strains were colistin resistant after exposure to colistin. In addition to the previously reported lpxCAD and pmrAB mutations, we identified four novel putative colistin resistance genes: A1S_1983. hepA. A1S_3026, and rsfS. Lipopolysaccharide (LPS) loss mutants exhibited higher fitness costs than those of the pmrB mutant in nutrient-rich medium. The colistin-resistant mutants had a higher inhibition ratio in the serum growth experiment than that of the wild type strain in 100% serum. Minimum inhibitory concentration (MIC) results showed that the LPS-deficient but not the pmrB mutant had an altered antibiotic resistance profile. The compensatory mutations partially or completely rescued the LPS-deficient’s fitness, suggesting that compensatory mutations play an important role in the emergence and spread of colistin resistance in A. baumannii.
Keywords: Acinetobacter baumannii, colistin, resistance mechanism, fitness cost, whole genome sequencing
Introduction
Acinetobacter baumannii had emerged as an important nosocomial and opportunistic pathogen worldwide, especially in intensive care units (Peleg et al., 2008). A. baumannii treatment is difficult because the clinical strains generally show resistance to multiple antibiotics (Peleg et al., 2008). Because the extensively drug-resistant isolates demonstrated sensitivity only to colistin, colistin has become the only clinically useful agent against these pathogens (Falagas et al., 2005; Giske et al., 2008; Magiorakos et al., 2012). However, due to its low efficacy, colistin should be used in combination antibiotic therapy for the treatment of XDR A. baumannii (Durante-Mangoni et al., 2014).
Colistin is a polycationic antimicrobial peptide that targets the polyanionic bacterial lipopolysaccharide (LPS) of Gram-negative bacteria. There are two different colistin resistance mechanisms (Beceiro et al., 2014). The first involves the inactivation of the lipid A biosynthesis pathway and the complete loss of surface LPS. This loss might be caused by mutations in lpxC. lpxA, or lpxD. Isolates harboring these mutations showed high colistin minimum inhibitory concentrations (MICs), although their fitness cost was also high (Beceiro et al., 2014; Wand et al., 2015). The second resistance mechanism is mediated by the pmrAB two-component system. Mutations in pmrA and pmrB induce the activation of pmrC, which adds phosphoethanolamine (PEtn) to the hepta-acylated form of lipd A (Beceiro et al., 2011). The pmrB mutation did not result in growth retardation (Durante-Mangoni et al., 2015; Wand et al., 2015).
Antimicrobial pressure drives the evolution of antimicrobial resistance in bacteria (Baker et al., 2013), and resistance is often associated with reduced bacterial fitness. Epistasis can compensate for the fitness cost of these mutants (Andersson and Hughes, 2010). Epistasis occurs not only within the same resistance pathway but also in different resistance pathways. In fluoroquinolone resistance, the parC mutation had the greatest effect on increase fitness of low fitness strains that harbored mutations in gyrA and marR (Marcusson et al., 2009). Moreover, the cost of multiple resistance was smaller than expected during the evolution of multi-drug resistance (Trindade et al., 2009). Understanding the fitness cost of the evolution of resistance is important to control the spread of resistant bacteria (Baker et al., 2013).
To assess the evolution of A. baumannii colistin resistance, we evolved A. baumannii through continuous exposure to colistin in agar plates and liquid media. Four novel putative colistin resistance genes were identified in addition to lpxCAD and pmrAB. Fitness and resistance compensatory mutations appeared in the bacteria exposed to colistin.
Materials and Methods
Bacterial Isolates and Antimicrobial Susceptibility Testing
All bacteria used in this study were the ATCC 17978 strain and its laboratory-evolved mutants of A. baumannii. The MICs were determined by broth dilution using Etest (AB Biodisk, Solna, Sweden) on Mueller-Hinton (MH) agar. The results were interpreted according to the CLSI or EUCAST breakpoints (Clinical and Laboratory Standards Institute, 2014; European Committee on Antimicrobial Susceptibility Testing, 2015). The bacteria were cultured in Luria–Bertani (LB) medium at 37°C.
Laboratory-Evolved Colistin Resistant Mutants
Four independent single colonies of A. baumannii ATCC 17978 were grown overnight at 37°C. The in vitro serial passage experiments were performed using three methods. First, the bacteria were streaked on LB agar plates containing 10 mg/L colistin once each day for 15 days and then incubated at 37°C. Then, the bacteria were serially passaged for another 15 days in the presence of 50, 100, and 200 mg/L colistin. Second, the bacteria were streaked on LB agar plates containing 10 mg/L colistin once each day for 60 days and then incubated at 37°C. Third, the cultures were exposed to serially increasing concentrations of colistin, starting from 1/2 the MIC and doubling every day for 12 days. The overnight cultures were stored at -80°C prior to the experiments and analysis.
Whole Genome DNA Sequencing and Analysis
Bacteria from a single colony were cultured overnight at 37°C in MH broth. The genomic DNA was extracted using a QIAamp DNA minikit (Qiagen, Valencia, CA, USA) following the protocol of the manufacturer (Hua et al., 2012). Agarose gel electrophoresis and a NanoDrop spectrophotometer (ND-1000, ThermoFisher, Waltham, MA, USA) were used to determine the quality and quantity of the extracted genomic DNA. The 300 bp library used for Illumina paired-end sequencing was constructed from 5 μg of genomic DNA purified from the laboratory-evolved mutants. Mapping and SNP detection were performed using breseq (Deatherage and Barrick, 2014). The regions containing the detected SNPs were amplified by PCR. The PCR products were sent to Biosune (Biosune, Hangzhou, China) for Sanger sequencing. Duplications were detected using the CLC genomic workbench (8.02.2) and were verified by real-time PCR (Supplementary Table S2).
Fitness Cost and Serum Resistance Measurement
Four independent cultures per strain were grown overnight in MH (Mueller–Hinton), diluted 1:1000 [MH or fetal bovine serum (FBS)] and aliquoted into a flat-bottom 96-well plate in four replicates. The plate was agitated at 37°C. The OD600 of each culture was determined every 5 min for 16 h using a the BioTEK Synergy plate reader (Biotek, Winooski, VT, USA). The growth rate was estimated by an R script based on the OD600 curves (Li et al., 2015). Fitness data are shown in the Supplementary Table S1. Statistical analyses were performed using R 3.2.1 (R Development Core Team, 2015). Two-way analysis of variance (ANOVA) and TukeyHSD analysis were used to assess differences between the means, with a significant probability at a P value of ≤0.05.
Results
Mutations in Laboratory-Evolved Colistin Resistant Strains
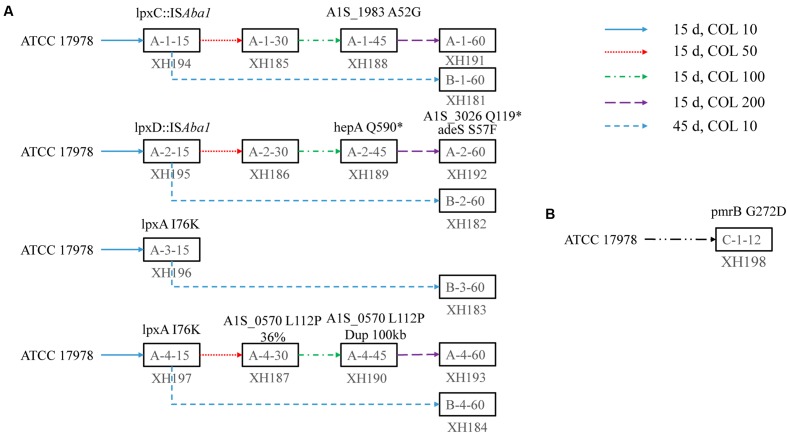
To investigate the colistin resistance mechanism in A. baumannii, we evolved A. baumannii ATCC 17978 in agar plates and liquid media with increasing or constant concentrations of colistin. A strain description and the MICs for colistin of the laboratory-evolved strains are listed in Supplementary Table S1. The MIC of ATCC 17978 for colistin was 0.19 mg/L. The MICs of the laboratory-evolved strains ranged from 6 mg/L to > 256 mg/L. The final isolates from each colony were sent for whole genome sequencing (WGS) to identify mutations. The mutations in the laboratory-evolved strains were detected via breseq, which mapped the sequence reads onto the ATCC 17978 reference genome. Then, the generation time each mutation appeared during the experiment was determined by PCR and sanger sequencing.
The mutations that occurred in the early stage of evolution were found in pmrB. lpxC. lpxA, and lpxD (Figure 1). Additional mutations were detected in A1S_1983. hepA (A1S_2462), A1S_3026. rsfS (A1S_0570), and adeS (A1S_1754) (Supplementary Tables S3 and S4).
FIGURE 1.
The evolutionary pathway of colistin resistance in Acinetobacter baumannii 17978 in solid agar and liquid medium. (A) Four colonies of A. baumannii ATCC 17978 were inoculated in a solid agar plate containing 10 mg/L colistin and serially passaged for 15 days. Then, one colony was continuously cultured with 10 mg/L colistin on agar plates for 45 days. Another colony was streaked on 50 mg/L colistin plates for a 15 days serial passage. Then, the bacteria were streaked onto 100 and 200 mg/L colistin plates for 15 days. The strains were stored at -80°C every 15 days. The final strains were sent for whole genome sequencing. The mutations were detected by breseq, which mapped the sequenced reads onto the reference genome. The mutation sites in the final strain were identified by PCR and Sanger sequencing. The mutations related to colistin resistance or fitness are labeled above the strain. (B) One lineage of A. baumannii 17978 was inoculated in Mueller Broth (MH) medium with increasing concentrations of colistin for 12 days. The final strains were sent for whole genome sequencing. The mutations were detected by breseq, which mapped the sequenced reads onto the reference genome. The mutations related to colistin resistance or fitness are labeled above the strain.
Colistin Resistance had a Fitness Cost
Among the mutations detected, A1S_1983. hepA. A1S_3026, and rsfS were hypothesized to be involved in colistin resistance. Conversely, the adeS mutation was not involved in colistin resistance (data not shown). The ratio of the A1S_0570 mutation in A-4-30 (XH187) was 36%, and this ratio increased to 100% in A-4-45 (XH190).
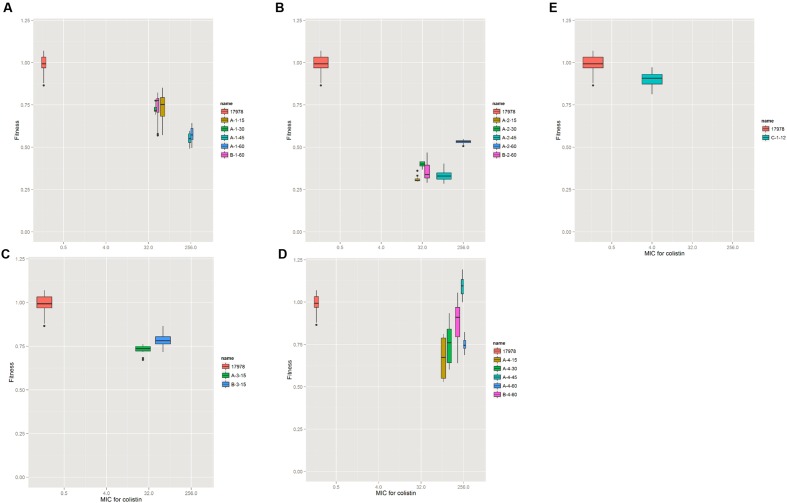
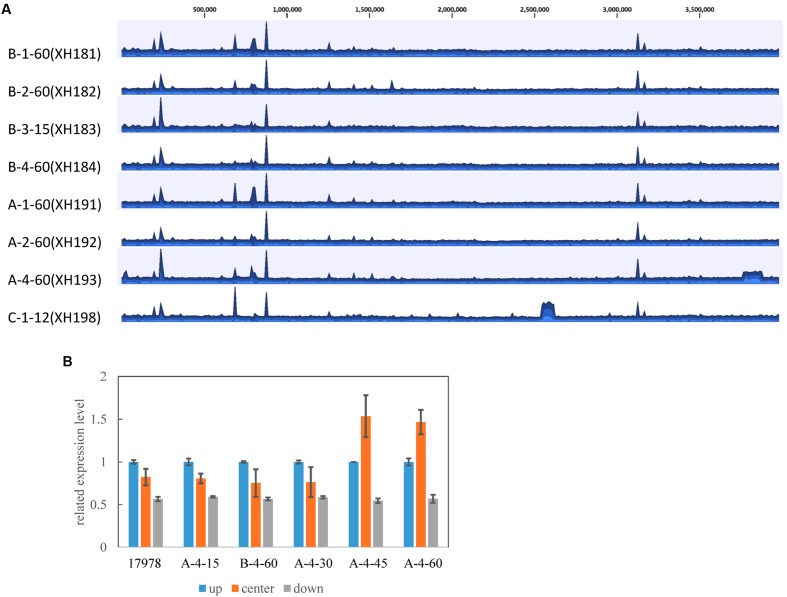
To investigate the effect of the mutations on A. baumannii fitness, we measured the growth rate of the laboratory-evolved strains as a proxy for fitness. The strains selected in the agar plates showed a high fitness cost in the first 15 selected generations, while increased resistance and fitness compensation were observed later in the later selection process (Figure 2). The evolution in colistin resistance and fitness was diverse among the four lineages. In lineage 1, no fitness compensation was observed while resistance to colistin was increased (Figure 2A). In lineage 2, the fitness was partially recovered with a mutation in A1S_3026, and the resistance to colistin increased (Figure 2B). Due to contamination, lineage 3 did not have enough isolates for analysis. In lineage 4, the fitness was completely recovered with a mutation in A1S_0570, while the resistance increased (Figure 2D). A 100 kb duplication occurred in A-4-45(XH193) and A-4-60(XH190; Figure 3), which showed a higher fitness compared to that of A-4-30(XH187). These results indicated that the duplication of the long chromosome fragment may play a role in the bacterial fitness. In addition, the growth of strains harboring the pmrB mutation was slightly retarded (Figure 2E).
FIGURE 2.
Resistance and fitness of the wild type and evolved strains. For the wild type and evolved strains, the relative fitness is shown as a function of the minimum inhibitory concentration (MIC) for colistin. Each box symbol represents one strain. The relative fitness was determined by measuring the growth rate in MH medium at OD600. The growth rate was calculated from the OD600 values during exponential growth by an R script. (A–D) Four colonies of A. baumannii strains evolved in solid agar plates. (E) The strain evolved in liquid media.
FIGURE 3.
(A) Coverage of colistin-resistant strains derived from A. baumannii ATCC 17978. Each plot shows the coverage for the alignment of each strain to the ATCC 17978 reference strain. The mean coverage for each strain is denoted by a curve (blue). Duplicated genomic segments appear as regions showing two or more mean coverages. (B) Confirmation of the 100 kb duplication in the colistin-resistant strains by RT-PCR. The genomic DNA was obtained from the strains and used as templates for RT-PCR. Three RT-PCR primers were designed upstream (up, A1S_3272), in the center (A1S_3302), and downstream (down, A1S_3366) of the duplication fragment. The error bars represent the SE from three replicates.
Lipopolysaccharide Affects Serum Resistance
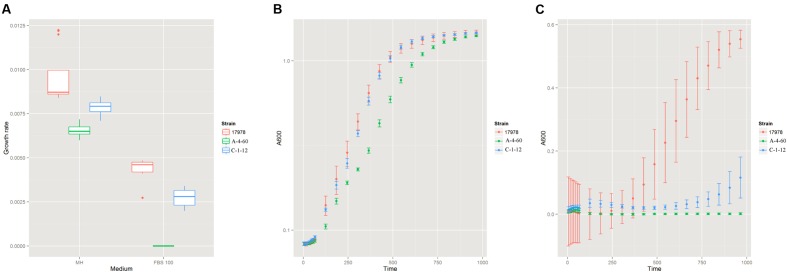
To investigate the effect of LPS on the serum resistance of A. baumannii, we measured the growth rate of the wild type strain (ATCC 17978), the LPS-deficient mutant (A-4-60, XH193), and the LPS-modified mutant (C-1-12, XH198) in FBS. There was significant difference in the growth rate of the wild type strain and LPS-deficient mutants in MH (Figure 4A). However, both the LPS-deficient mutant and LPS-modified mutant showed higher inhibition in FBS compared to that of the wild type strain. Both the LPS-deficient mutant and LPS-modified mutant exhibited a decrease in their final biomass (Figures 4B,C).
FIGURE 4.
(A) The growth rate of the wild type strain (ATCC 17978), the LPS (Lipopolysaccharide)-deficient mutant (A-4-60, XH193) and the LPS-modified mutant (C-1-12, XH198) in MH and fetal bovine serum (FBS). The wild type strain (ATCC 17978), LPS-deficient mutant (A-4-60, XH193), and LPS-modified mutant (C-1-12, XH198) were grown in MH and FBS at 37°C. The growth rates of the strains were determined by measuring the OD600 every 5 min and were estimated by an R script based on the OD600 curves. Each strain represents four biological and four technical replicates. The growth curves of the wild type strain (ATCC 17978), LPS-deficient mutant (A-4-60, XH193), and LPS-modified mutant (C-1-12, XH198) in MH (B) and FBS (C).
Colistin-Resistant Mutation Alters the Antibiotic Resistance Profiles
To examine the effect of the LPS mutation on antibiotic resistance, we determined the MICs of several antibiotics (including quinolones, aminoglycosides, tetracycline, and β-lactams). Table 1 shows the MICs of different antibiotics for A. baumannii ATCC 17978 and its colistin mutants. Overall, the changes in the MIC trends could be divided into two groups. The first group included seven strains that showed increased sensitivity to several types of antibiotics except colistin. All seven strains harbored mutations that disrupted the LPS biosynthesis pathway, leading to LPS loss. The second group contained only one strain: C-1-12 (XH198). This strain was resistant to all antibiotics except ampicillin and imipenem, and its colistin resistance mechanism involved an LPS modification. Thus, the LPS mutations resulted in differences in the antibiotic resistance profiles.
Table 1.
Antimicrobial susceptibilities of Acinetobacter baumannii ATCC 17978 and its colistin mutants.
| Minimum inhibitory concentration (MIC; mg/L)a | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Strain | Lineage | CI | GM | AK | MC | TGC | AM | MP | IP |
| 17978 | 0.25 | 0.75 | 2 | 0.75 | 0.38 | 48 | 0.38 | 0.38 | |
| XH191 | A-1-60 | 0.125 | 0.38 | 0.5 | <0.016 | 0.032 | 3 | 0.016 | 0.094 |
| XH181 | B-1-60 | 0.094 | 0.25 | 0.5 | <0.016 | 0.047 | 2 | 0.012 | 0.125 |
| XH192 | A-2-60 | 0.25 | 1 | 0.5 | 0.016 | <0.016 | 1 | 0.016 | <0.002 |
| XH182 | B-2-60 | 0.125 | 0.094 | 0.19 | 0.032 | 0.064 | 1 | 0.008 | 0.064 |
| XH183 | B-3-60 | 0.094 | 0.38 | 0.5 | 0.032 | 0.032 | 3 | 0.016 | 0.125 |
| XH193 | A-4-60 | 0.094 | 0.5 | 1 | 0.016 | 0.094 | 3 | 0.032 | 0.125 |
| XH184 | B-4-60 | 0.125 | 0.19 | 0.5 | 0.032 | 0.064 | 1.5 | 0.008 | 0.064 |
| XH198 | C-1-12 | 1 | 2 | 8 | 1 | 3 | 12 | 0.5 | 0.25 |
aCI, ciprofloxacin; GM, gentamicin; AK, amikacin; MC, minocycline; TGC, tigecycline; AM, ampicillin; MP, meropenem; IP, imipenem.
Discussion
In this study, we investigated the evolutionary pathway of A. baumannii that led to colistin resistance. Mutations in pmrB. lpxC. lpxA, and lpxD were detected early in the selection process. These results confirmed the findings of previous reports (Adams et al., 2009; Moffatt et al., 2010; Olaitan et al., 2014). In addition, four putative proteins, A1S_1983. hepA. A1S_3026, and rsfS were proposed to be involved in colistin resistance in this study. A1S_1983 is a putative exported protein. The ATP-dependent helicase HepA (A1S_2462) is a transcriptional regulator that activates transcription under stress conditions. The HepA mutation resulted in reduced exopolysaccharide coverage and increased retention of penicillin G compared to that of the wild type strain in E. coli strain; moreover, the mutant was more sensitive to norfloxacin, chloramphenicol, and gentamicin than the wild type (Lynch et al., 2007). A1S_3026 is a ribonuclease T2 family protein, and a mutation in this gene limits A. baumannii’s ability to colonize inanimate surfaces and bacterial mobility (Jacobs et al., 2014). RsfS (A1S_0570) is a ribosomal silencing factor that helps bacteria adapt to slow growth conditions (Hauser et al., 2012). The first protein is a membrane protein; its mutation would influence the integrity of the membrane, possibly leading to colistin resistance. The latter three proteins affected exopolysaccharide production, biofilm formation and bacterial adaptation, indicating new colistin resistance mechanisms. Recently, six genes (vacJ. pldA. ttg2C. pheS, and conserved hypothetical protein) were shown to have a role in colistin resistance (Thi Khanh Nhu et al., 2016). We also identified mutation in vacJ, but did not link the vacJ mutation to colistin resistance. This may be due to different mutations in vacJ in Nhu’s and our study. Moreover, these result also demonstrated the diversity of novel colistin resistance mechanism.
The LPS loss mutant showed a high fitness cost, which limited its spread in clinical environments. We identified two fitness compensatory mutations: A1S_3026 and RsfS in this study. These fitness compensatory mutations increase the fitness of the LPS loss mutant and promote its survival and spread in the clinical environment. However, we confirmed that the LPS-deficient mutants possessed an altered antibiotic resistance profile that was not observed in the LPS modification mutants. Colistin resistance in A. baumannii leads to changes in the antibiotic resistance profile (Moffatt et al., 2010; Lopez-Rojas et al., 2011b; Wand et al., 2015). The change in the antibiotic resistance profiles of the LPS-deficient mutants was caused by the increased permeability of the outer membrane to antibiotics (Moffatt et al., 2010). LPS in the outer membrane of A. baumannii acts as a highly selective permeability barrier (Nikaido, 2003). LPS loss removed the selective permeability of the barrier for antibiotics, whereas the LPS modification resulted in only minor changes in the permeability of the A. baumannii outer membrane. The pmrB mutant presented little or no change in susceptibility to these antibiotics (Wand et al., 2015). In this study, the pmrB mutant showed increased resistance to antibiotics. This may be due to changes in the permeability of the outer membrane by the pmrB mutation.
Previous studies have also demonstrated that the pmrB mutation resulted in no or a slight fitness cost (Durante-Mangoni et al., 2015; Wand et al., 2015). Our result confirmed these findings. These results explained why the LPS modification only resulted in a small change in the bacteria compared to the LPS loss. However, we did not determine the effect of colistin resistance-related mutations on the virulence of A. baumannii isolates in vivo. Colistin resistance was previously associated with lower in vivo fitness and reduced virulence in A. baumannii (Lopez-Rojas et al., 2011a).
In our study, LPS loss affected the growth rate of A. baumannii in nutrient-rich medium and serum. The loss of the O-ag capsule in Salmonella typhimurium was also reported to reduce the resistance to serum but did not affect the growth rate (Marshall and Gunn, 2015). The LPS loss was not shown to contribute to serum resistance in A. baumannii (Moffatt et al., 2013). However, our results did not support that conclusion. LPS modification decreased the growth rate of A. baumannii in serum and in nutrient-rich medium. The LPS modified mutation also contributed to serum resistance in A. baumannii. The conflict between Moffatt and our results may be caused by the different genetic backgrounds of ATCC 19606 used in Moffatt’s study and ATCC 17978 used in our study. Both of the LPS mutants showed a decreased the final biomass, whereas serum resistance was normal following incubation in serum for 30 min (Moffatt et al., 2013). The decreased biomass indicated that the serum resistance assay required further improvements.
Overall, colistin resistance A. baumannii rapidly developed. We not only confirmed the existence of a previously reported colistin resistance mechanism, but also identified four novel putative proteins involved in colistin resistance. The compensatory mutations partially or completely recovered the LPS-deficient mutant’s fitness, suggesting that compensatory mutations played an important role in the emergence and spread of colistin resistant A. baumannii.
Nucleotide Sequence Accession Numbers
The whole genome shotgun sequencing results for B-1-60 (XH181), B-2-60(XH182), B-3-15(XH183), B-4-60(XH184), A-1 -60(XH191), A-2-60(XH192), A-4-60(XH193), and C-1-12(XH198) have been deposited at DDBJ/EMBL/GenBank under the accession numbers MDWH00000000, MDWJ00000000, MDWK00000000, MDWL00000000, MDWG00000000, MDWI00000000, MDWF00000000, and MDWM00000000, respectively.
Author Contributions
XH and YY conceived and designed the study. XM, NW, XL, and KS performed the experiments. XH, ZZ, and YY performed data analysis and drafted the manuscript. All authors reviewed and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer YS and handling Editor declared their shared affiliation and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81230039, 31170130, and 81101285), the Natural Science Foundation of Zhejiang province, China (LY15H190004), the Science and Technology Project of Zhejiang Province (2016F50015), and the Zhejiang Province Medical Platform Backbone Talent Plan (2016DTA003).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01715/full#supplementary-material
References
- Adams M. D., Nickel G. C., Bajaksouzian S., Lavender H., Murthy A. R., Jacobs M. R., et al. (2009). Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 53 3628–3634. 10.1128/AAC.00284-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson D. I., Hughes D. (2010). Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 8 260–271. 10.1038/nrmicro2319 [DOI] [PubMed] [Google Scholar]
- Baker S., Duy P. T., Nga T. V., Dung T. T., Phat V. V., Chau T. T., et al. (2013). Fitness benefits in fluoroquinolone-resistant Salmonella Typhi in the absence of antimicrobial pressure. Elife 2:e01229 10.7554/eLife.01229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beceiro A., Llobet E., Aranda J., Bengoechea J. A., Doumith M., Hornsey M., et al. (2011). Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob. Agents Chemother. 55 3370–3379. 10.1128/AAC.00079-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beceiro A., Moreno A., Fernandez N., Vallejo J. A., Aranda J., Adler B., et al. (2014). Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob. Agents Chemother. 58 518–526. 10.1128/AAC.01597-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (2014). Performance Standards for Antimicrobial Susceptibility Testing: 24th Informational Supplement. CLSI Document M100-S24. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Deatherage D. E., Barrick J. E. (2014). Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151 165–188. 10.1007/978-1-4939-0554-6_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durante-Mangoni E., Del Franco M., Andini R., Bernardo M., Giannouli M., Zarrilli R. (2015). Emergence of colistin resistance without loss of fitness and virulence after prolonged colistin administration in a patient with extensively drug-resistant Acinetobacter baumannii. Diagn. Microbiol. Infect. Dis. 82 222–226. 10.1016/j.diagmicrobio.2015.03.013 [DOI] [PubMed] [Google Scholar]
- Durante-Mangoni E., Utili R., Zarrilli R. (2014). Combination therapy in severe Acinetobacter baumannii infections: an update on the evidence to date. Future Microbiol. 9 773–789. 10.2217/fmb.14.34 [DOI] [PubMed] [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing (2015). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 2.0. EUCAST; 2012. Basel: European Committee on Antimicrobial Susceptibility Testing. [Google Scholar]
- Falagas M. E., Bliziotis I. A., Kasiakou S. K., Samonis G., Athanassopoulou P., Michalopoulos A. (2005). Outcome of infections due to pandrug-resistant (PDR) Gram-negative bacteria. BMC Infect. Dis. 5:24 10.1186/1471-2334-5-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giske C. G., Monnet D. L., Cars O., Carmeli Y., ReAct-Action on Antibiotic Resistance. (2008). Clinical and economic impact of common multidrug-resistant gram-negative bacilli. Antimicrob. Agents Chemother. 52 813–821. 10.1128/AAC.01169-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser R., Pech M., Kijek J., Yamamoto H., Titz B., Naeve F., et al. (2012). RsfA (YbeB) proteins are conserved ribosomal silencing factors. PLoS Genet. 8:e1002815 10.1371/journal.pgen.1002815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X., Zhou H., Jiang Y., Feng Y., Chen Q., Ruan Z., et al. (2012). Genome sequences of two multidrug-resistant Acinetobacter baumannii strains isolated from a patient before and after treatment with tigecycline. J. Bacteriol. 194 6979–6980. 10.1128/JB.01887-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs A. C., Blanchard C. E., Catherman S. C., Dunman P. M., Murata Y. (2014). An ribonuclease T2 family protein modulates Acinetobacter baumannii abiotic surface colonization. PLoS ONE 9:e85729 10.1371/journal.pone.0085729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Liu L., Ji J., Chen Q., Hua X., Jiang Y., et al. (2015). Tigecycline resistance in Acinetobacter baumannii mediated by frameshift mutation in plsC, encoding 1-acyl-sn-glycerol-3-phosphate acyltransferase. Eur. J. Clin. Microbiol. Infect. Dis. 34 625–631. 10.1007/s10096-014-2272-y [DOI] [PubMed] [Google Scholar]
- Lopez-Rojas R., Dominguez-Herrera J., McConnell M. J., Docobo-Perez F., Smani Y., Fernandez-Reyes M., et al. (2011a). Impaired virulence and in vivo fitness of colistin-resistant Acinetobacter baumannii. J. Infect. Dis. 203 545–548. 10.1093/infdis/jiq086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rojas R., Jimenez-Mejias M. E., Lepe J. A., Pachon J. (2011b). Acinetobacter baumannii resistant to colistin alters its antibiotic resistance profile: a case report from Spain. J. Infect. Dis. 204 1147–1148. 10.1093/infdis/jir476 [DOI] [PubMed] [Google Scholar]
- Lynch S. V., Dixon L., Benoit M. R., Brodie E. L., Keyhan M., Hu P., et al. (2007). Role of the rapA gene in controlling antibiotic resistance of Escherichia coli biofilms. Antimicrob. Agents Chemother. 51 3650–3658. 10.1128/AAC.00601-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorakos A. P., Srinivasan A., Carey R. B., Carmeli Y., Falagas M. E., Giske C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18 268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- Marcusson L. L., Frimodt-Moller N., Hughes D. (2009). Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog. 5:e1000541 10.1371/journal.ppat.1000541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J. M., Gunn J. S. (2015). The O-antigen capsule of Salmonella enterica Serovar Typhimurium facilitates serum resistance and surface expression of FliC. Infect. Immun. 83 3946–3959. 10.1128/IAI.00634-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt J. H., Harper M., Harrison P., Hale J. D., Vinogradov E., Seemann T., et al. (2010). Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 54 4971–4977. 10.1128/AAC.00834-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt J. H., Harper M., Mansell A., Crane B., Fitzsimons T. C., Nation R. L., et al. (2013). Lipopolysaccharide-deficient Acinetobacter baumannii shows altered signaling through host Toll-like receptors and increased susceptibility to the host antimicrobial peptide LL-37. Infect. Immun. 81 684–689. 10.1128/iai.01362-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67 593–656. 10.1128/MMBR.67.4.593-656.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaitan A. O., Morand S., Rolain J. M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643 10.3389/fmicb.2014.00643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg A. Y., Seifert H., Paterson D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21 538–582. 10.1128/CMR.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2015). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Thi Khanh Nhu N., Riordan D. W., Do Hoang Nhu T., Thanh D. P., Thwaites G., et al. (2016). The induction and identification of novel Colistin resistance mutations in Acinetobacter baumannii and their implications. Sci Rep. 6:28291 10.1038/srep28291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trindade S., Sousa A., Xavier K. B., Dionisio F., Ferreira M. G., Gordo I. (2009). Positive epistasis drives the acquisition of multidrug resistance. PLoS Genet. 5:e1000578 10.1371/journal.pgen.1000578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wand M. E., Bock L. J., Bonney L. C., Sutton J. M. (2015). Retention of virulence following adaptation to colistin in Acinetobacter baumannii reflects the mechanism of resistance. J. Antimicrob. Chemother. 70 2209–2216. 10.1093/jac/dkv097 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.