Abstract
Brain metastatic papillary thyroid carcinomas (PTCs) are afflicted with unfavorable prognosis; however, the underlying molecular genetics of these rare metastases are virtually unknown. In this study, we compared whole transcript microarray expression profiles of a BRAF mutant, brain metastasis from a PTC, including its technical replicate (TR), with eight non-brain metastatic PTCs and eight primary brain tumors. The top 95 probe sets (false discovery rate (FDR) p-value < 0.05 and fold change (FC) > 2) that were differentially expressed between the brain metastatic PTC, including the TR, and both, non-brain metastatic PTCs and primary brain tumors were in the vast majority upregulated and comprise, e.g. ROS1, MYBPH, SLC18A3, HP, SAA2-SAA4, CP, CCL20, GFAP, RNU1-120P, DMBT1, XDH, CXCL1, PI3, and NAPSA. Cytokines were represented by 10 members in the top 95 probe sets. Pathway and network analysis (p-value < 0.05 and FC > 2) identified granulocytes adhesion and diapedesis as top canonical pathway. Most significant upstream regulators were lipopolysaccharide, TNF, NKkB (complex), IL1A, and CSF2. Top networks categorized under diseases & functions were entitled migration of cells, cell movement, cell survival, apoptosis, and proliferation of cells. Probe sets that were significantly shared between the brain metastatic PTC, the TR, and primary brain tumors include CASP1, CASP4, C1R, CC2D2B, RNY1P16, WDR72, LRRC2, ZHX2, CITED1, and the noncoding transcript AK128523. Taken together, this study identified a set of candidate genes and biofunctions implicated in, so far nearly uncharacterized, molecular processes of a brain metastasis from a PTC.
Keywords: Papillary thyroid carcinoma, brain metastasis, microarray expression profiling, cytokines, diapedesis
Introduction
Approximately 80% of all thyroid carcinomas are histologically classified as papillary thyroid carcinomas (PTCs). PTCs harbor in 40-70% of the cases a BRAF V600E mutation that renders BRAF in a constitutively active formation and has shown to confer vulnerability to protease inhibitors [1]. In thyroid cancer, the V600E mutation is frequently associated with unfavorable features including multifocality, lymph node involvement, and distant metastasis. We reported earlier on the comparison between V600E positive and negative PTCs and normal thyroid samples, revealing that V600E positive PTCs are comparably afflicted with more unfavorable features; however, both V600E positive and negative PTCs exhibited quite closely related expression profiles when compared to normal thyroid samples [2].
Common sites of distant PTC metastases are lung and bone whereas metastases to the brain are rare. Frequency of brain metastases varies between 0.15-1.3% of all diagnosed thyroid carcinomas [3]. Brain metastases can occur even after years of the primary thyroid tumor with an approximately 10 years difference of patients’ mean age; however, aggressive subtypes of PTC as tall cell variants are likely to metastasize earlier than other histological subtypes [4]. Prognosis for patients with intracranial brain metastasis of a PTC is in general poor and depends on factors like histology type and the presence of other distant metastases [5]. In approximately 10% of patients who die from thyroid carcinoma reveal to have a brain metastasis at autopsy [6].
Symptoms implicative for a brain metastasis include headache, nausea, seizure, polyuria, and various neuronal impairments such as ocular motor vision dysfunction. Asymptotic brain metastases from thyroid cancer are very rare and can be incidentally detected by 131I treatment. Treatment options include surgery, radiation, and systemic therapy. Surgical resection of a brain metastasis originating from differentiated thyroid cancer may improve prognosis of patients [6]. Sorafenib which is a dual-active inhibitor that inhibits the mitogen activated protein kinase pathway has been successfully applied to treat a patient with a brain metastasis originating from a follicular thyroid carcinoma [7]. It is proposed that the brain metastatic process is a multistep process involving migration, intravasation, circulation, arrest, extravasation, and settlement/invasion of the brain microenvironment [8,9].
In the present study, using whole transcript microarrays, we analyzed the expression profile of a brain metastasis from a PTC and compared it with those from non-brain metastatic, stage III and IV PTCs and with primary brain tumors. Our objectives were to identify molecular biomarkers that are specifically related to the brain metastatic PTC and that support unraveling molecular mechanisms underlying this deteriorative disease.
Material and methods
Tumor samples
In the core expression analysis we studied samples from one brain metastatic PTC, eight non-brain metastatic, stage III and IV PTCs and eight primary brain tumors. The specimens were derived from patients who were treated surgically in the period between 2010 and 2015 at the King Abdulaziz University Hospital, Jeddah, and the King Faisal Specialist Hospital and Research Center (KFSH&RC), Jeddah. Histopathological diagnosis was performed by a team of pathologists (JM, AJ, and FG) on established criteria. The brain metastasis was initially identified as a left frontal brain tumor in a middle-aged (< 45 years) old female without any known chronic illness and the tumor was treated by surgery. Histopathological examination sustained the diagnosis of a metastatic PTC and subsequent sonography demonstrated thyroid nodules and the primary, stage II [10], multifocal PTC and lymph node metastases were resected. This study was approved by the Research Ethics Committee of the King Abdulaziz University, Faculty of Medicine, #358-10, #976-12, and the Institutional Review Board of the KFSH&RC, #IRB2010-07.
Immunohistochemistry
Antibodies (Ventana Medical Systems, Tucson, AZ) employed for immunohistochemistry (IHC) consisted of thyroid transcription factor (TTF-1) (clone 8G7G3/1), thyroglobulin (clone 2H11/6E1), and proliferation marker ki-67 (clone 30-9) which is immunoreactive in the late G1, S, G2, and M phases of the cell cycle. Four μm sections of formalin-fixed and paraffin-embedded specimens were processed on an automated immunostainer (BenchMark XT, Ventana Medical Systems) according to the manufacturer’s protocols and using the ultraView Universal DAB Detection Kit for detection.
RNA and microarray processing
Isolation of total RNA and microarray sample processing were performed as described earlier [14,15]. In brief, the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) was employed to assess RNA integrity and integrity number was > 5 in samples used for the differential expression analysis between then brain metastatic PTC, non-brain metastatic PTCs, and primary brain tumors. The NanoDrop ND1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) was utilized to determine RNA concentration. All RNA samples were processed using the Ambion WT Expression Kit (Life Technologies, Austin, TX), the GeneChip WT Terminal Labeling and Controls Kit (Affymetrix, Santa Clara, CA), and the Affymetrix GeneChip Hybridization, Wash and Stain Kit. The RNA from the brain metastasis was processed twice to generate a technical replicate (TR). All processed samples were hybridized to Affymetrix Human Gene 1.0 ST GeneChip arrays. This type of microarray interrogates with a set of 764,885 probes 36.079 annotated reference sequences (NCBI build 36). The microarrays were scanned on a GeneChip Scanner 3000 7G and probe cell intensity data (CEL) files were generated using the GeneChip Command Console Software (AGCC).
Sample selection for microarray expression study
Sample selection criterion was to identify probe sets which are significant to the brain metastatic process. Therefore, eight non-brain metastatic, stage III and IV PTCs as advanced thyroid tumors and eight selected primary brain tumors were chosen for differential expression analysis. For selection, analysis of variance (ANOVA) in Partek Genomics Suite version 6.6 (Partek Inc., MO) was employed and performed on the imported CEL files and carried out between the comparison group, consisting of the brain metastatic PTC and its TR, and each of 40 primary brain tumors for which CEL files were available from our microarray data sets. Eight primary brain tumors with meningioma histology revealed to have the comparably lowest numbers of differentially expressed probe sets with the brain metastatic PTC and its TR (p-value < 0.05 and FC > 2) and therefore were taken, after assessing sample bias, as a group for differential gene expression analysis which included also, besides the brain metastatic PTC and its TR, the eight non-brain metastatic PTC group. Sample bias was assessed by replacing the eight primary brain tumors with comparably lowest numbers of differentially expressed probe sets with those eight brain tumors having the highest number of differentially expressed probe sets. The ANOVA result of the assessment showed that approximately 90% of the significantly differentially expressed probe sets were shared between both configurations.
Gene expression core analysis
In Partek Genomics Suite quality assessment of microarray experiments was performed on basis of QC metrics tables and QC graphical reports on the imported CEL files. The saved distribution file of 18 quantile normalized samples was used for quantile normalization of subsets of the 18 CEL files, i.e. 17 CEL files consisting of eight non-brain metastatic PTCs, eight primary brain tumors and either the brain metastatic PTC or its TR. All lists of differentially expressed probe sets were generated by applying ANOVA and using either a p-value < 0.05 and a fold change (FC) > 2 or using, where indicated, the more stringent criterion of the false discovery rate (FDR) p-value (step-up method) < 0.05, and fold change (FC) > 2. Principal component analysis was employed to illustrate overall variance in gene expression between samples or groups of samples. Average linkage hierarchical clustering was performed by using Spearman’s correlation as a similarity matrix. Venn diagrams were generated to display intersecting or non-intersecting groups of differentially expressed probe sets. The Gene Ontology (GO) enrichment tool was employed in the gene expression workflow to group significantly expressed genes into functional categories. The gene enrichment score utilizes the Fisher’s Exact test to determine the level of differential gene expression in a functional category. PANTHER version 10 which combines GO annotations and a phylogenetic tree model for inferring gene functions was primarily used for protein classification [16]. The generated microarray data set has been deposited at the NCBI Gene Expression Omnibus under accession number GSE66463 and it includes renormalized samples from non-brain metastatic PTCs and primary brain tumors of submissions GSE54958 [2,17] and GSE77259 [11].
Functional network and pathway analysis
The Ingenuity Pathways Analysis software (IPA; build version 338830M) (Ingenuity Systems, Redwood City, CA) was utilized to interpret biological significance of expression data [18]. IPA workflow comprises core and functional analysis and structural data categorization. The Ingenuity Knowledge Base was used as a reference data set. Direct and indirect molecular relationships were included in the analysis settings. Fisher’s exact test p-values indicate the significance of relationships between the analyzed data set molecules and the functional frameworks prebuilt or generated de novo by IPA. Where specified, the Molecule Activity Predictor was utilized to predict expression effects/coherence of expression effects of a molecule on other pathway or network molecules. The canonical pathway workflow was employed to identify molecules from the uploaded data set that are co-expressed in a directional, up- to downstream, pathway. The overlap percentage of uploaded molecules matching to a canonical pathway determines its overrepresentation and ranking. Upstream regulators analysis was employed to explain how differences in target gene expression are regulated by upstream molecules and what kind of biological activities are involved. The activation z-score predicts the activation states of regulators. The diseases & functions network analysis was employed to explore relationships and effects between the uploaded data set and downstream processes. The activation z-score measures in how far downstream processes are either up- or downregulated.
Semiquantitative RT-PCR and BRAF mutational analysis
Expression of eight genes was assessed by semiquantitative RT-PCR using housekeeping gene B2M as reference. Primer sequences of the genes are listed in Table 1 and in case of B2M have been reported previously [11]. RT-PCR were performed in 20 µL volumes containing each 2 µL buffer mix, 0.1% 2-mercaptoethanol, 0.0125% BSA, 3 mM MgCl2, 10 nmol of each dNTP, 5 pmol forward primers, 5 pmol reverse primers, 1.25 units GoTaq DNA Polymerase (Promega, Madison, WI), and 200 ng of second strand cDNA template that was generated using the Ambion WT Expression Kit. No cDNA template was included in the negative control. A modified standard PCR protocol was used as reported earlier [11]. Gel band densities of selected reactions were measured using ImageJ 1.49v [12] and then the ratio to the internal reference B2M was calculated. DNA extraction and BRAF mutational screening for the non-brain metastatic PTCs was reported earlier (GSE54958) and for the brain metastatic PTC was performed within the present study as described by us [13].
Table 1.
Primer sequences used for semiquantitative RT-PCR
| Gene | Forward primer sequence (5’-3’) | Reverse primer sequence (5’-3’) | Product size (bp) |
|---|---|---|---|
| ROS1 | ATTGTGGAGAGTTGCACG | ATGACCTTGCCATCTGTG | 205 |
| MYBPH | CCAGTCCAGCAATGATGG | AGGCTTAGTGGCTGTGGA | 202 |
| SLC18A3 | GCGAAGACGTGAAGATCG | ATGGCTATGCCAGACGTG | 240 |
| HP | CTACACCCTAACTACTCC | ACATACTTCAGATGGTCAGT | 184 |
| CP | ACCCTGATAACACCACAG | AAGGTCCGATGAGTCCTG | 185 |
| CCL20 | ACCATGTGCTGTACCAAG | ATGTCACAGCCTTCATTGG | 178 |
| GFAP | GCTTTGCCAGCTACATCG | ATTGTCCCTCTCAACCTC | 193 |
| SAA2 | TTTTCGTTCCTTGGCGAG | CTCTCTACAGTTCAGCTG | 192 |
aCGH analysis
aCGH study of the brain metastatic PTC was performed according to the manufacturer’s protocol (Agilent Technologies, Santa Clara, CA). In brief, tumor DNA was extracted by using the QIAmp DNA FFPE tissue kit (Qiagen, Hilden, Germany). Subsequently, 500 ng of tumor DNA and 500 ng fragmented Agilent, gender-matched reference DNA were labeled using ULS-Cy3 and ULS-Cy5 reagents (Genomic DNA ULS labeling kit), respectively. The hybridization mixture, containing 5 µg human Cot-1 DNA, 1 µL 100x blocking reagent, 2x Hi-RPM hybridization buffer, and both labeled DNA solutions was hybridized for 40 hrs at 65°C and 20 rpm to a 180K SurePrint G3 Human CGH microarray. The type of microarray contains 170,334 distinct biological features. Subsequently, the microarray was washed stringently, then scanned using the SureScan High resolution scanner, and the generated TIFF image analyzed, utilizing CytoGenomics v3.0.6 software (Agilent Technologies) according to its user guide (G1662-90046). Confidence of differential expression of biological features is based on p-values that are generated using the log ratios of the Cy3 to Cy5 channel signals and the log ratio errors which are a measure for the significance of computed log ratios.
Results
Hematoxylin-eosin staining revealed a characteristic papillary architecture of the brain metastatic PTC (Figure 1A). Nuclear immunoreactivity for TTF-1 and cytoplasmic immunoreactivity for thyroglobulin demonstrated origin of the brain metastasis from a PTC (Figure 1B, 1C). Proliferation marker Ki-67 showed immunoreactivity in about 20% of the tumor cells (Figure 1D). Five of the non-brain metastatic PTCs and the brain metastatic PTC exhibited a BRAF V600E mutation. An aCGH analysis of the brain metastasis identified a number of whole and partial chromosomal gains including +1q, +5, +7, +12p12pter, +17p, +18p, and +20q (Figure 2).
Figure 1.
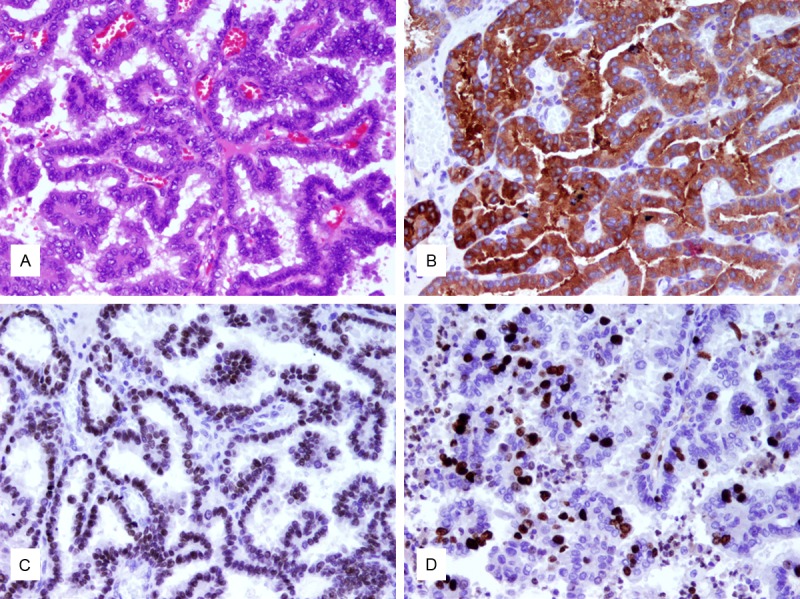
The brain metastatic PTC. A. Hematoxylin-eosin staining reveals papillary architectures covered by cuboidal to columnar epithelium. Cellular crowding is visible and neoplastic cells demonstrate nuclear clearing and nuclear grooves. B. TTF-1 staining as a marker for thyroid or lung origin exhibits strong nuclear staining. C. Thyroglobulin shows strong cytoplasmic staining. D. Proliferation marker Ki67 is positive in 20% of cells (original magnification, x400).
Figure 2.
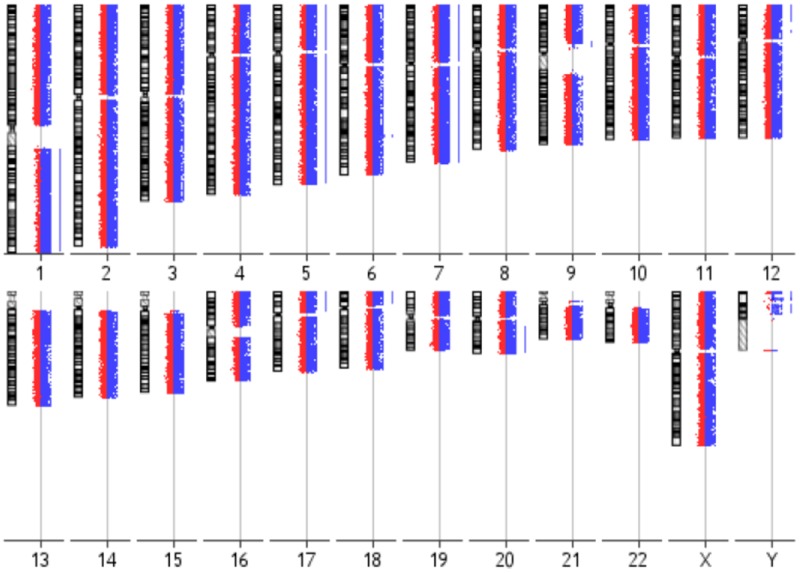
aCGH karyogram of the brain metastatic PTC. Each chromosome ideogram is shown with the green/red fluorescent profiles representing oligonucleotide features. Bars to the right of the profiles indicate gains affecting chromosomes/chromosomal regions including 1q, 5, 7, 12p12pter, 17p, 18p, and 20q.
Genes differentially expressed in the brain metastasis and the TR vs. non-brain metastatic PTCs and primary brain tumors
A detailed microarray expression analysis was performed on a rare brain metastasis from a PTC, including its TR, eight non-brain metastatic, stage III and IV PTCs and eight primary brain tumors (Table 2). The primary brain tumors were of meningioma histology and selected as described under Material and methods. Similarity of expression profiles of all 18 samples is illustrated by a distance related scale in a PCA 3D scatter plot (Figure 3). Hierarchical cluster analysis separately groups the brain metastatic PTC and the TR, from non-brain metastatic PTCs, and primary brain tumors (Figure 4). The 18 tumor samples served as basis to generate four differentially expressed comparison groups, i.e. brain metastatic PTC vs. non-brain metastatic PTCs, brain metastatic PTC (TR) vs. non-brain metastatic PTCs, brain metastatic PTC vs. primary brain tumors, and brain metastatic PTC (TR) vs. primary brain tumors. The top 95 differentially expressed probe sets, that primarily distinguish the brain metastatic PTC from both, non-brain metastatic PTCs and primary brain tumors were those that intersect with all four comparison groups (Figure 5A, Supplement Table 1). The 95 probe sets were in the vast majority comparably upregulated in the brain metastatic PTC. The encoded proteins/molecules represent various functional categories. These include the cytokines CCL20, CXCL1, CCL13, OSM, CXCL8, IL6, IL17C, CXCL2, IL1B, and IL32; proteins with receptor activity, e.g. ROS1, FPR2, IL1R2, and SIGLEC6; cytoskeletal and related proteins, e.g. MYBPH, GFAP, KRT17, ACTBL2, and TUBB3; enzymes including proteases and their inhibitors, e.g. XDH, PI3, NAPSA, FSTL5, SPINK14, TMPRSS3, and MMP7; and transporter proteins, e.g. SLC18A3, CP, CNTNAP5, AQP5, and SLC6A14. Molecules of various categories were, e.g.HP, SAA1, SAA2, SAA2-SAA4, DMBT1, ZER1, HSPA6, OTX2, RRAD, RAB7B, MIR146A, SBSN, and FAM83A. Eight differentially expressed genes, i.e. ROS1, MYBPH, SLC18A3, HP, CP, CCL20, and GFAP were subjected to semiquantitative RT-PCR that substantially confirmed microarray expression data (Figure 6).
Table 2.
Demographic data and clinicopathological tumor features
| Case | Histology1 | Age | Gender2 | Tumor location3 | Tumor stage |
|---|---|---|---|---|---|
| TM-1039-11 | Conventional PTC | 52 | F | Jugular vein wall | IV |
| TM-989-11 | FVPTC | 71 | F | Skull, bone | IV |
| TM-602-11 | Conventional PTC | 59 | F | Thyroid bed, lung | IV |
| TM-187-11 | Conventional PTC | 56 | M | Lymph node, perithyroidal extension | IV |
| TM-892-11 | Conventional PTC | 56 | M | Thyroid bed | IV |
| TM-279-08 | Conventional PTC | 77 | M | Cervical and retropharyngeal lymph nodes | III |
| TM-301-12 | FVPTC | 51 | F | No recurrence/metastasis | III |
| TM-824-12 | Conventional PTC | 55 | F | Extrathyroidal extension | IV |
| Jed51_MTT | Conventional PTC | 44 | F | Brain, left frontal | II |
| Jed04_MN | Transitional meningioma | 57 | F | Left tentoral | I |
| Jed13_MN | Meningothelial meningioma | 50 | F | Left frontal | I |
| Jed49_MN | Atypical meningioma with brain invasion | 51 | F | Posterior right subfalcine | II |
| Jed57_MN | Transitional meningioma | 48 | F | Left parasagittal | I |
| Jed64_MN | Fibroblastic meningioma | 49 | F | Right frontal | I |
| Jed70_MN | Fibroblastic meningioma | 73 | F | Right temporal | I |
| Jed88_MN | Secretory meningioma | 43 | M | Sub-occipital | I |
| Jed92_MN | Meningothelial meningioma | 49 | F | Olfactory groove | I |
FVPTC, follicular variant of PTC;
F, female, M, male;
Tumor location, location of PTC recurrence/metastasis and location of primary brain tumors including one recurrence (Jed49_MN).
Figure 3.
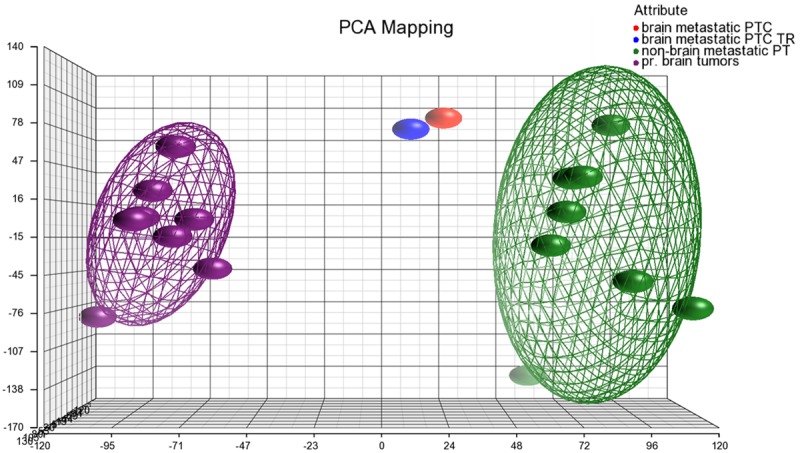
PCA 3D scatter plot as a dimensional, distance-related measure to display similarity of expression profiles of samples that are indicated by dots. Bounding ellipsoids are included for non-brain metastatic PTCs and primary brain tumors. Colors for samples/groups of samples as indicated in the color scheme legend.
Figure 4.
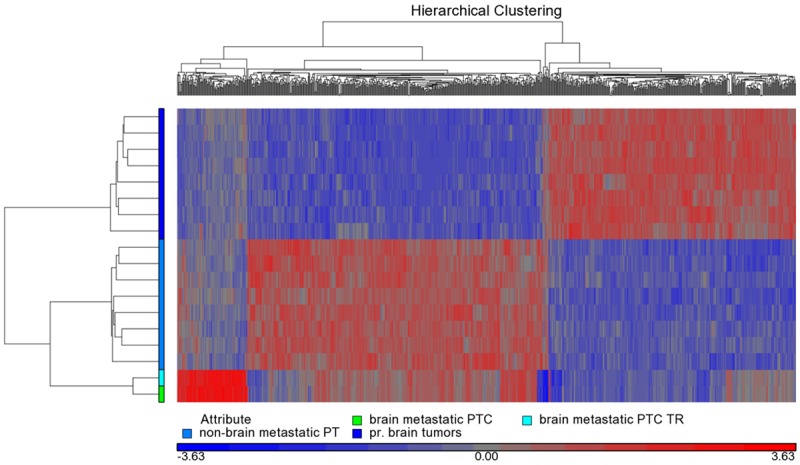
Hierarchical cluster analysis between the brain metastatic PTC, the TR, non-brain metastatic PTCs, and primary brain tumors. The brain metastatic PTC and the TR are clustering separately from the two other groups. Analysis is based on 796 differentially expressed probe sets (p-value < 0.0005). Colors for samples/ groups of samples as indicated in the color scheme legend.
Figure 5.
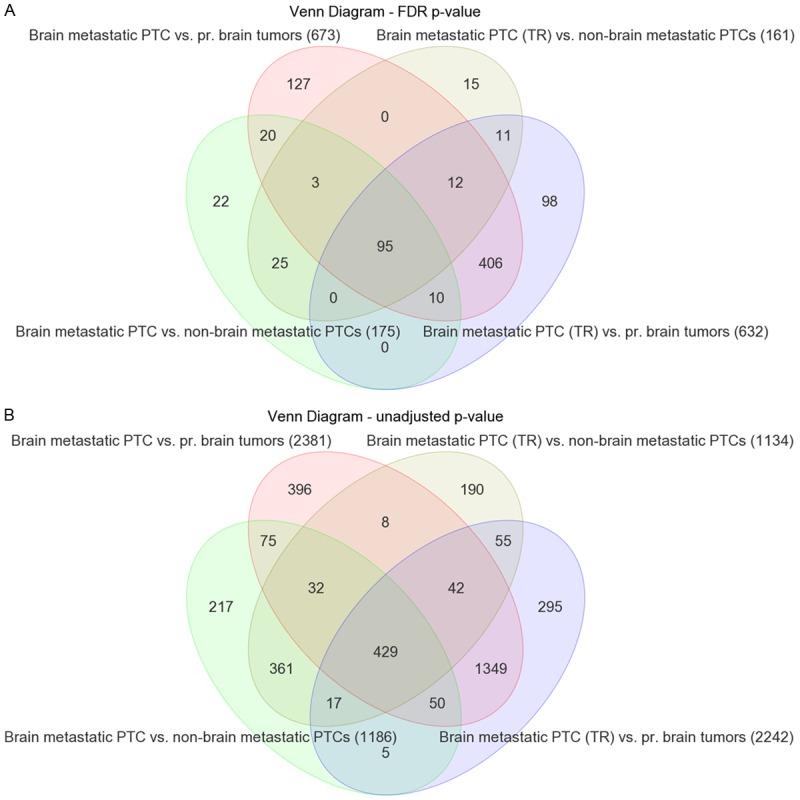
Venn diagrams displaying differentially expressed probe sets that intersect or non-intersect between different comparison groups. A. Applying a FDR p-value < 0.05 and FC > 2.0 results in 95 differentially expressed probe sets (Supplement Table 1) that intersect between the four comparison groups brain metastatic PTC vs. non-brain metastatic PTCs, brain metastatic PTC (TR) vs. non-brain metastatic PTCs, brain metastatic PTC vs. primary brain tumors, and brain metastatic PTC (TR) vs. primary brain tumors. The 95 probe sets include 90 genes represented by one probe set each, 2 genes represented by two probe sets each, and one unmapped transcript. B. Applying the unadjusted p-value < 0.05 and FC > 2.0 results in 429 differentially expressed probe sets that intersect between the comparison groups as outlined under A (Supplement Table 1). Numbers of probe sets that are differentially expressed in each comparison group are given in parentheses.
Figure 6.
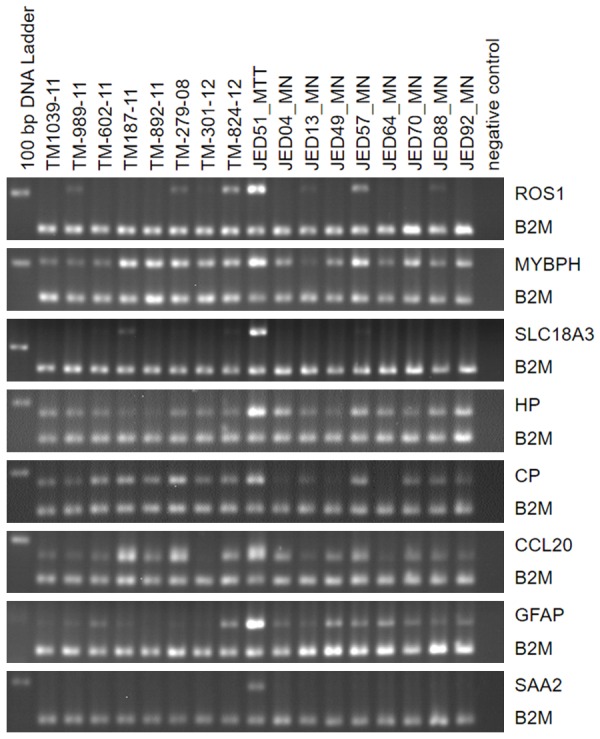
Semiquantitative RT-PCR for ROS1, MYBPH, SLC18A3, HP, CP, CCL20, GFAP, and SAA2 in the case series of eight thyroid tumors (TM-1039-11, TM-989-11, TM-602-11, TM-187-11, TM-892-11, TM-279-08, TM-301-12, and TM-824-12), the brain metastatic thyroid tumor (Jed51_MTT) and eight primary brain tumors (Jed04_MN, Jed13_MN, Jed49_MN, Jed57_MN, Jed64_MN, Jed70_MN, Jed88_MN, and Jed92_MN). The genes rank among the top differentially expressed probe sets (Supplement Table 1). The PCR products of the interrogated genes have a size between 178-240 bp (Table 1). Product size of the internal reference gene B2M is 156 bp. Relative band densities of the PCR products for all eight genes compared to B2M is comparably highest for Jed51_MTT. Some cases as e.g. Jed57_MN express considerable amounts of MYBPH (~55%, compared to Jed51_MTT), or TM-279-08 express considerable amounts of CP (~69%) and CCL20 (~64%). Lane 1 contains the DNA ladder with the 200 bp band visible in the image fields and lane 19 contains the negative control reactions.
Biofunctional prediction analysis
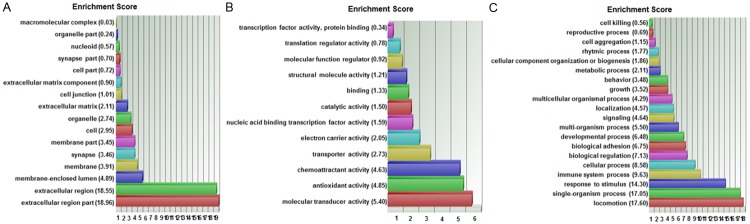
Biofunctional analysis was performed on the set of 429 differentially expressed probe sets (p-value < 0.05 and FC > 2) (Figure 5B, Supplement Table 1). Functional groups that were significantly overrepresented in different ontology categories are illustrated in a GO enrichment analysis (Figure 7). In the cellular component domain, the categories extracellular region part, extracellular region, and membrane-enclosed lumen were prevalent. In the molecular function domain, the most significantly related categories were molecular transducer activity, antioxidant activity, and chemoattractant activity. In the biological process domain, the prevalent categories were locomotion, single-organism process, and response to stimuli. The top canonical pathway was entitled granulocyte adhesion and diapedesis, illustrating molecular processes implicated in transendothelial migration (Table 3, Figure 8). Other top pathways were likewise related to inflammatory response and diseases. Network analysis identified as most significantly related upstream regulators lipopolysaccharide, the nuclear NFkB (complex), and the cytokines TNF, Il1A, and CSF2, otherwise known as GM-CSF (Table 3, Figure 9). Top networks under the category diseases & functions were entitled migration of cells, cell movement, cell survival, apoptosis, and proliferation of cells (Table 3, Figure 10).
Figure 7.
GO enrichment analysis for the 429 differentially expressed probe sets. The functional categories are ranked according to their p-values. A. The categories extracellular region part, extracellular region, and membrane-enclosed lumen were most overrepresented in the cellular component domain. B. The most overrepresented categories in the molecular function domain were molecular transducer activity, antioxidant activity, and chemoattractant activity. C. The dominant categories in the biological process domain were locomotion, single-organism process, and response to stimulus.
Table 3.
Top pathways and networks based on 429 differentially expressed probe sets
| Category | Brain metastatic PTC vs. non-brain metastatic PTCs | Brain metastastic PTC vs. primary brain tumors | ||
|---|---|---|---|---|
|
| ||||
| p-value | Overlap [%]/activation z-score | p-value | Overlap [%]/activation z-score | |
| Top canonical pathways | ||||
| Granulocyte adhesion and diapedesis | 1.38E-09 | 11.4 | 1.38E-09 | 11.4 |
| Agranulocyte adhesion and diapedesis | 8.24E-07 | 8.9 | 8.24E-07 | 8.9 |
| Role of IL-17A in psoriasis | 2.79E-06 | 38.5 | 2.79E-06 | 38.5 |
| LXR/RXR activation | 3.21E-06 | 10.0 | 3.21E-06 | 10.0 |
| Acute phase response signaling | 4.04E-06 | 8.4 | 4.04E-06 | 8.4 |
| Top upstream regulators | ||||
| Lipopolysaccharide | 1.81E-23 | 6.382 | 1.81E-23 | 6.734 |
| TNF | 2.82E-23 | 5.799 | 2.82E-23 | 6.102 |
| NFkB (complex) | 2.99E-22 | 5.477 | 2.99E-22 | 5.477 |
| IL1A | 4.46E-20 | 4.668 | 4.46E-20 | 5.016 |
| CSF2 | 4.12E-18 | 4.729 | 4.12E-18 | 4.729 |
| Diseases & functions | ||||
| Migration of cells | 9.77E-14 | 5.694 | 9.77E-14 | 4.989 |
| Cell movement | 1.48E-13 | 5.982 | 1.48E-13 | 5.298 |
| Cell survival | 3.24E-13 | 5.493 | 3.24E-13 | 5.203 |
| Apoptosis | 6.63E-13 | -0.923 | 6.63E-13 | -2.397 |
| Proliferation of cells | 1.22E-12 | 4.191 | 1.22E-12 | 4.063 |
Figure 8.
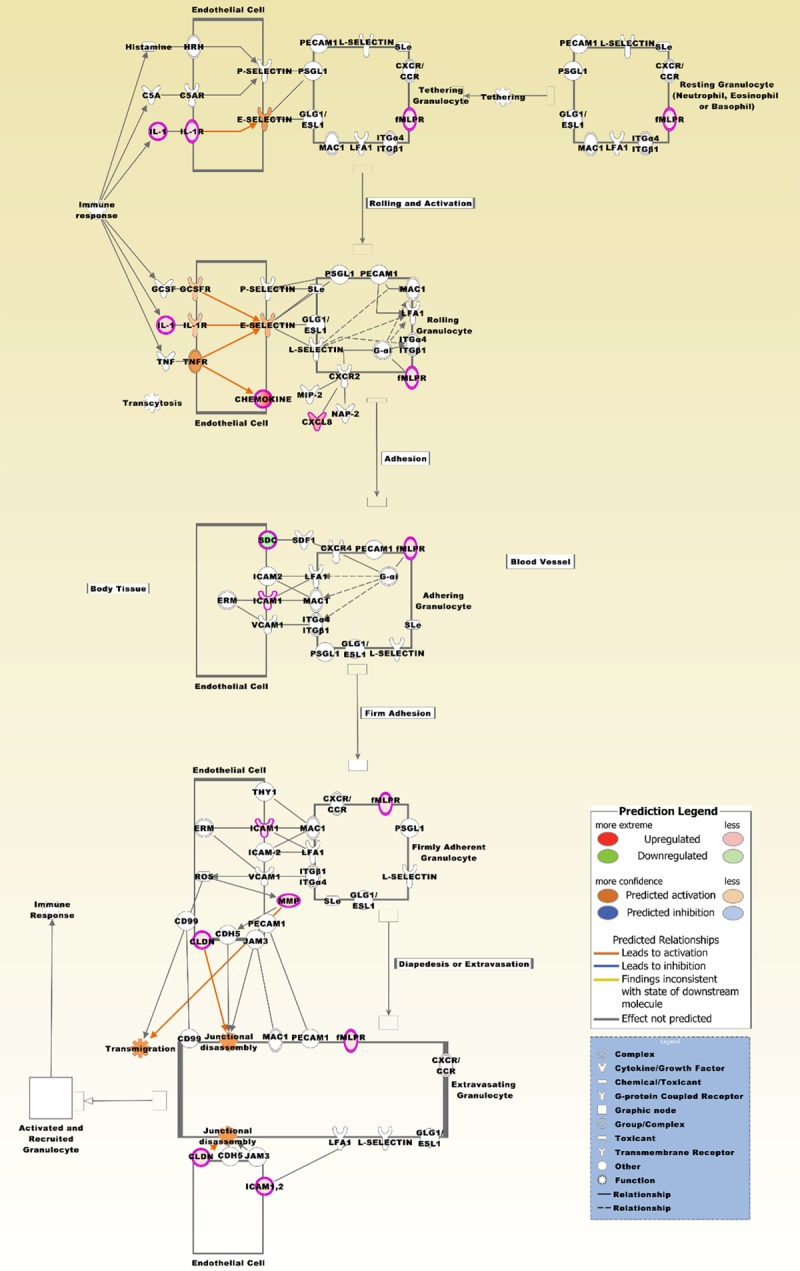
The top canonical pathway is entitled granulocyte adhesion and diapedesis. The pathway is based on the list of 429 differentially expressed probe sets that intersect between the four comparison groups (Figure 2B), and displaying the expression values derived from the two comparison groups, brain metastatic PTC vs. non-brain metastatic PTCs and brain metastatic PTC (TR) vs. non-brain metastatic PTCs (Supplement Table 1). Molecules that are comparably upregulated in the brain metastatic PTC include CCL2, CCL13, CCL18, CCL20, CCL22, CXCL1, CXCL2, CXCL8, CXCL14, CX3CL1, CLDN10, FPR2, IL1A, IL1B, IL1R2, ICAM1, and MMP7. SDC2 is comparably downregulated. A number of molecules from the probe sets are displayed under their group specific names, e.g. chemokines, CLDN, fMLPR, IL-1, MMP, and SDC. This pathway was also most significantly associated with the 95 probe sets (p-value 5.51E-08).
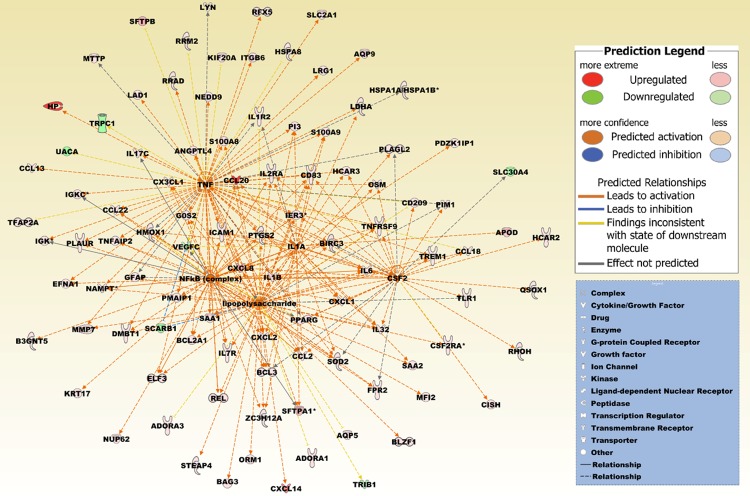
Figure 9.
A merged network based on the top five upstream regulators lipopolysaccharide, nuclear NFkB (complex) and the cytokines TNF, IL1A, and CSF2. IL1A is also included in the differentially expressed probe set list. Increased activation is predicted by a z-score > 2 (Table 3). The network is based on the list of 429 differentially expressed probe sets that intersect between the four comparison groups (Figure 5B), and reflecting the expression values derived from the two comparison groups, brain metastatic PTC vs. non-brain metastatic PTCs and brain metastatic PTC (TR) vs. non-brain metastatic PTCs (Supplement Table 1). Upregulated molecules include ADORA1, ADORA3, ANGPTL4, APOD, AQP5, AQP9, B3GNT5, BAG3, BCL2A1, BCL3, BIRC3, BLZF1, CCL13, CCL18, CCL2, CCL20, CCL22, CD209, CD83, CISH, CSF2RA, CX3CL1, CXCL1, CXCL14, CXCL2, CXCL8, DMBT1, EFNA1, ELF3, FPR2, G0S2, GFAP, HCAR2, HCAR3, HMOX1, HP, HSPA1A/HSPA1B, HSPA8, ICAM1, IER3, IGK, IGKC, IL17C, IL1A, IL1B, IL1R2, IL2RA, IL32, IL6, IL7R, ITGB6, KIF20A, KRT17, LAD1, LDHA, LRG1, LYN, MFI2, MMP7, MTTP, NAMPT, NEDD9, NUP62, ORM1, OSM, PDZK1IP1, PI3, PIM1, PLAGL2, PLAUR, PMAIP1, PPARG, PTGS2, QSOX1, REL, RFX5, RHOH, RRAD, RRM2, S100A8, S100A9, SAA1, SAA2, SFTPA1, SFTPB, SLC2A1, SOD2, STEAP4, TFAP2A, TLR1, TNFAIP2, TNFRSF9, TREM1, and ZC3H12A. Downregulated molecules include SCARB1, SLC30A4, TRIB1, TRPC1, UACA, and VEGFC. The pathway was overlaid with the Molecule Activity Predictor to precalculate further molecular effects, as outlined in the prediction legend.
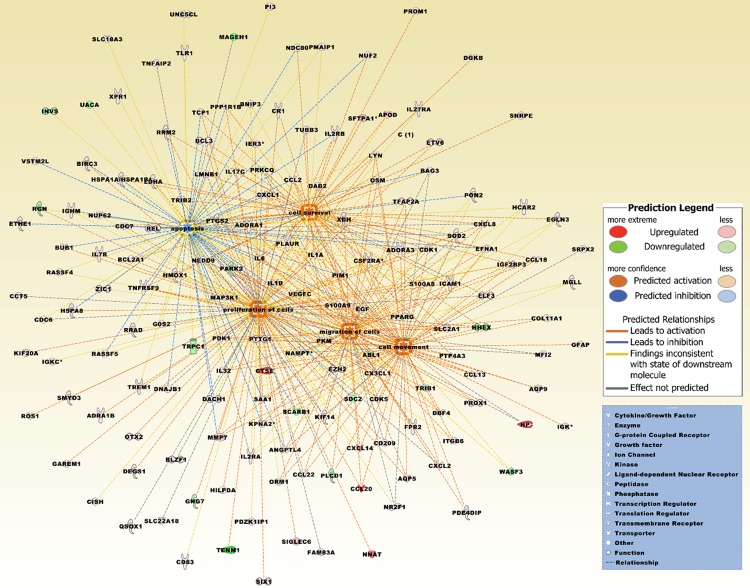
Figure 10.
The top five merged networks under the category diseases & functions were entitled migration of cells, cell movement, cell survival, apoptosis, and proliferation of cells. Increased activation is predicted by a z-score > 2 (Table 3). The network is based on the list of 429 differentially expressed probe sets that intersect between the four comparison groups (Figure 5B), and reflecting the expression values derived from the two comparison groups, brain metastatic PTC vs. non-brain metastatic PTCs and brain metastatic PTC (TR) vs. non-brain metastatic PTCs (Supplement Table 1). Upregulated molecules include ADORA1, ADORA3, ADRA1B, ANGPTL4, APOD, AQP5, AQP9, BAG3, BCL2A1, BCL3, BIRC3, BLZF1, BNIP3, BUB1, CCL13, CCL18, CCL2, CCL20, CCL22, CCT5, CD209, CD83, CDC6, CDC7, CDK1, CDK5, CISH, COL11A1, CR1, CSF2RA, CTSE, CX3CL1, CXCL1, CXCL14, CXCL2, CXCL8, DAB2, DACH1, DBF4, DEGS1, DGKB, DNAJB1, EFNA1, EGF, EGLN3, ELF3, ETHE1, ETV6, EZH2, FAM83A, FPR2, G0S2, GAREM1, GFAP, HCAR2, HILPDA, HMOX1, HP, HSPA1A/HSPA1B, HSPA8, ICAM1, IER3, IGF2BP3, IGHM, IGK, IGKC, IL17C, IL1A, IL1B, IL27RA, IL2RA, IL2RB, IL32, IL6, IL7R, ITGB6, KIF14, KIF20A, KPNA2, LDHA, LMNB1, LYN, MAP3K1, MFI2, MGLL, MMP7, NAMPT, NDC80, NEDD9, NNAT, NR2F1, NUF2, NUP62, ORM1, OSM, OTX2, PDE4DIP, PDK1, PDZK1IP1, PI3, PIM1, PKM, PLAUR, PMAIP1, PON2, PPARG, PPP1R1B, PROM1, PROX1, PTGS2, PTP4A3, PTTG1, QSOX1, RASSF4, RASSF5, REL, ROS1, RRAD, RRM2, S100A8, S100A9, SAA1, SFTPA1, SIGLEC6, SIX1, SLC18A3, SLC22A18, SLC2A1, SMYD3, SNRPE, SOD2, SRPX2, TCP1, TFAP2A, TLR1, TNFAIP2, TNFRSF9, TREM1, TRIB2, TUBB3, UNC5CL, VSTM2L, XDH, XPR1, and ZIC1. Downregulated molecules include ABL1, GNG7, HHEX, INVS, MAGEH1, PARK2, PLCD1, PRKCQ, RGN, SCARB1, SDC2, TENM1, TRIB1, TRPC1, UACA, VEGFC, and WASF3. The pathway was overlaid with the Molecule Activity Predictor to precalculate further molecular effects, as outlined in the prediction legend.
Genes shared between the brain metastasis and primary brain tumors
To identify candidate genes that may support the brain metastasis to adapt and expand in the brain microenvironment, we selected (FDR p-value < 0.05 and FC > 2.0) in our data set for those probe sets that intersect between the comparison groups, brain metastatic PTC vs. non-brain metastatic PTCs, brain metastatic PTC (TR) vs. non-brain metastatic PTCs, and primary brain tumors vs. non-brain metastatic PTCs. Furthermore, we excluded from the intersecting probe sets those that were included in the list of 429 probe sets. The resulting probe sets include CASP1, CASP4, and C1R which were comparably higher expressed and CC2D2B, RNY1P16, WDR72, LRRC2, ZHX2, CITED1, and the noncoding and uncharacterized transcript AK128523 which were comparably lower expressed in the brain metastatic PTC.
Discussion
In contrast to melanoma, breast and lung cancer, which contribute to the majority of cancers with brain metastasis, brain metastatic PTCs are rare and presence of a PTC brain metastasis prior to detection of the primary thyroid tumor is exceptionally rare [19]. We present to our knowledge the first whole transcript expression profile of a brain metastasis from a PTC. Critical steps of the brain metastatic process as transmigration through the blood brain barrier (BBB) are not fully explored yet and depend on a number of factors as tissue type of tumor cells, cellular interactions and the time interval needed for extravasation [8,9]. By comparing the expression profile of a brain metastasis with profiles of advanced stage, non-brain metastatic PTCs and with profiles of statistically sorted primary brain tumors, we selected for those differentially expressed candidate genes that are most probably related to in vivo characteristics of the brain metastatic process. The BRAF mutational status of PTC metastases to the brain has only been reported for a small number of cases providing limited information to the clinical behavior of these metastases [20,21]. A meta-analysis revealed that distant metastases from PTCs are not significantly associated with the BRAF mutational status [22]; however, a positive BRAF mutational status was more frequently observed in PTCs with a brain metastasis in comparison to other metastatic sites as soft tissue, lung, and retroperitoneum [20].
Cytokines
CCL20 is a ligand for CCR6 and its expression is a proinflammatory signal, known to be upregulated in a number of cancer types. In SW1736 thyroid cancer cells, migration and invasion was induced by activation of CCR6 through CCL20 [23]. In addition, CCL20 stimulated expression and secretion of MMP3 and the nuclear translocation and activation of NFkB which is known to be implicated in a variety of cellular functions including cytokine activation and exertion of pro-tumorigenic activities. Of notice, upregulation of CCL20 in endothelial cells that line specific zones of the BBB has been associated with a support for autoreactive T cells to cross the BBB regionally [24]. CCL20 was also included in the top scoring gene list derived from a comparison of parental breast cancer cell lines CN34 and MDA231 with their derivatives possessing brain metastatic behavior [25]. CXCL1 and CXCL2 are neutrophil chemoattractants. Overexpression of both chemokines in breast cancer cells increased survival of cancer cells at the metastatic sites [26]. In a rat model of soman-induced status epilepticus, CXCL1 expression was induced in neurons and endothelial cells [27]. An in vitro and in vivo study demonstrated that CCL2 expression from breast cancer cells and stroma attracts inflammatory monocytes that promotes metastasis of breast tumors [28]. CCL13 is the ligand of chemokine receptors CCR1, CCR2, CCR3, and CCR5. Although CCL13 is a critical factor in inflammatory diseases [29] its functions in cancer is not well defined. OSM is structurally related to IL6 and was found to inhibit thyroid peroxidase gene expression in porcine thyroid cell culture [30]. CXCL8, IL17, and IL6 were among a number of cytokines that were detected with increased levels upon neuroinflammatory stimulus in a 3D microfluidic chip model of the human BBB [31]. Furthermore, in SW480 and Caco-2 cells, proliferation, invasion, or epithelial-mesenchymal transition could be synergistically fostered by CXCL8 and CCL20 via the PI3K/AKT and ERK1/2 pathways [32]. GBM cells which were selected in a lung metastasis assay showed increased expression of IL6, IL8 and other factors known to be associated with unfavorable prognosis [33]. IL17C is preferentially expressed from non-immune cells and its expression was found to be comparably upregulated in the CNS of mice with induced autoimmune encephalomyelitis [34,35]. In an in vitro BBB model, treatment with recombinant IL1B abolished BBB integrity by suppressing astrocytic SHH production and furthermore, led to increased synthesis of CCL2, CCL20, and CXCL2 in astrocytes [36]. The proinflammatory cytokine IL32, alias NK4, is an established cancer associated factor that was demonstrated e.g. in cell culture experiments where in a splice variant of IL32 promoted migration and invasion of breast cancer cells via the downstream VEGF/STAT3 pathway [37].
Proteins with receptor activity
IL1R2 is a decoy receptor that interferes with the binding of IL1R1 to IL1A and IL1B. The receptor is known to be expressed in various cancer types [38]. ROS1 encodes a receptor tyrosine kinase with yet unknown ligand and it exhibits conserved relationship with ALK and the leukocyte tyrosine kinase LTK. Among other genes, upregulation of ROS1 was detected in GBM clones that were resistant to the EGFR inhibitor Gefitinib [39]. FPR2 is a G protein-coupled chemoattractant receptor [40]. In vitro and in vivo studies revealed that glioma cells overexpressing FPR showed increased motility and were more invasive than non-FPR overexpressing glioma cells [41]. SIGLEC6 is a cell membrane receptor containing an extracellular immunoglobulin domain for sialic acids binding and an intracellular immunoreceptor tyrosine-based inhibitory motif. Expression of SIGLEC6 in choriocarcinoma-derived BeWO cells resulted in decreased apoptosis and enhanced proliferation and invasiveness [42].
Cytoskeletal and related proteins
MYBPH is a cytoskeletal protein that is involved in a number of cellular processes; however, its functions are not fully explored yet. Of notice, in vitro experiments demonstrated that MYBPH expression is positively regulated by TTF-1 and furthermore, MYBPH overexpression impairs actomyosin organization and acts as a negative factor for cell motility and migration of collective cells but not of single cells [43]. This latter observation can be of relevance in the context that the brain metastatic cells in our study harbored expression signatures related to granulocyte/agranulocyte adhesion and diapedesis processes. GFAP is an intermediate filament protein that is an established marker for a number of glial tumors [44]. Overexpression of KRT17 has been found in an IHC study to be associated with lymph node metastasis in PTCs [45]. Within the cytoskeleton structures, actin filaments form dynamic networks that can rapidly reorganize, determine cytokinesis, cell shape, and are mandatory for cell adhesion, migration, and invasion [46]. Overexpression of the ubiquitously expressed gene ACTBL2, alias kappa-actin, is associated with unfavorable prognosis in hepatocellular carcinoma and its induced overexpression in hepatoma cells led to enhanced serum-independent cell proliferation and anchorage-independent colony formation [47]. Expressional dataset analysis detected overexpression of the beta tubulin protein TUBB3 in brain metastases of breast cancers but not in the primary tumors [48]. Furthermore, knockdown of TUBB3 in brain metastatic breast cancer cells reduced their metastatic capacity in mice which led to increased survival of the rodents.
Enzymes including proteases and their inhibitors
XDH encodes a molybdoflavin enzyme that catalyzes the oxidative reactions of hypoxanthine to xanthine and subsequently of xanthine to uric acid. The role of the enzyme in cancer has not been thoroughly elucidated; however, XDH expression has been identified by mass spectrometry in bovine brain capillary endothelial cells (BCECs) exhibiting functional BBB properties that were induced by co-cultured glial cells, whereas mono-cultured BCECs with impaired BBB properties did not express detectable XDH [49]. PI3, also known as elafin, is a serine protease inhibitor that is synthesized and secreted locally at the site of injury in response to primary cytokines such as IL1 and TNF [50]. In an experimental anti-angiogenesis in vivo model system, PI3 was upregulated in GBM and its upregulation was found to be associated with poor prognosis in GBM patients [51]. NAPSA encodes an aspartic protease that is used as an IHC marker for subclassification of lung tumors. In thyroid cancer, IHC expression of NAPSA was detected in a subset of poorly differentiated thyroid carcinomas and anaplastic carcinomas but only in a vast minority of PTCs [52]. FSTL5 is an enzyme modulator of the follistatin family and the secretory glycoprotein was identified in a microarray expression and IHC study as a marker of unfavourable prognosis in different subtypes of medulloblastomas [53]. The function of SPINK14 in cancer is barely known but the protease inhibitor was listed among the top downregulated genes in HepG2 cells upon treatment with the EZH2 inhibitor GSK343 [54]. TMPRSS3 is a type II transmembrane serine protease that, when overexpressed in ovarian A2780 cells, promotes their proliferation, invasion, and migration and an IHC study revealed that TMPRSS3 expression is an unfavorable prognostic factor in breast cancer patients [55,56]. MMP7 is a secreted matrix metalloproteinase. In vitro assays in prostate cancer cells demonstrated that the proteinase sufficiently digests perlecan, a core component of basement membranes, and by this facilitates cell invasion [57].
Transporter proteins
The vesicular acetylcholine transporter SLC18A3 encodes a transmembrane molecule that loads acetylcholine into secretory vesicles in presynaptic nerve terminals. An RT-PCR assay demonstrated that induction of differentiation of human neuroblastoma cell line LAN-5 by retinoic acid resulted in a number of deregulated genes under which SLC18A3 was shown to be initially upregulated [58]. CP owns ferroxidase activity and it is implicated in supporting iron transport across the BBB. In an in vitro BBB model system, expression of CP in C6 glioma cells was stimulated by IL1B and IL6 that were expressed from neighboring brain microvascular endothelial cells [59]. According to microarray expression data, CNTNAP5, which is a member of the neurexin family and associated with potassium channels is comparably high expressed in a number of brain cancers including medulloblastoma, pilocytic astrocytoma, and cerebellar tumors [60]. In prostate cancer, the water channel protein AQP5 was identified as an unfavorable prognostic marker and cell culture experiments indicated that siRNA knockdown of AQP5 inhibited proliferation and migration of prostate cancer cells [61].
Molecules of various categories
HP has been found with increased serum levels by quantitative protein analysis in patients with brain metastatic lung cancer [62]. Furthermore, in vitro and in vivo studies identified HP as a key mediator of metastatic cell homing, fostering invasion, migration, including transendothelial migration, and proliferation of cancer cells [63]. SAA proteins are the major acute phase response molecules and considered as a family of apolipoproteins. SAA2-SAA4 is transcripted as a readthrough between the adjunct SAA2 and SAA4 genes. Overexpression of isoforms SAA1 and SAA2 was found in sera of lung cancer patients and in lung carcinoma cells that metastasized and settled in the lung of a mouse model [64]. Furthermore, recombinant SAA1 differentially affected cell migration and invasion in two tested glioma cell lines [65]. DMBT1 is a member of the scavenger receptor cysteine-rich superfamily and it was initially identified as a gene deleted in a number of medulloblastoma and GBM samples and cell lines [66]. Detailed in vitro and in vivo studies established DMBT1 as a critical vascular extracellular matrix protein that promotes endothelial cell adhesion, migration, proliferation, and angiogenesis [67]. ZER1, that was in our study comparably downregulated in the brain metastatic PTC, encodes a subunit of an E3 ubiquitin ligase complex. A microarray study in bladder cancer identified, among other genes, upregulation of ZER1 at tumor stage Ta-T1; however, its downregulation at stage T1-T2 [68]. Upon heat induction, heat shock protein HSPA6 has been identified in centrioles that are known to have critical functions in cellular polarity and migration processes during neuronal differentiation [69]. OTX2 encodes a homeobox transcription factor and is frequently gained in non-Shh/non-Wnt medulloblastomas. In vitro and in vivo experiments demonstrated that OTX2 overexpression promotes cell proliferation, whereas its knockdown reduces tumorigenicity of cells [70]. RRAD is a member of the (Rem/Rem2, Gem, Kir) RGK subfamily of the Ras family of small GTPases, sharing in its GTPase domain a ~35% identity with HRAS and other small GTPases. Overexpression of RRAD in primary GBM is associated with poor prognosis [71]. Furthermore, cell culture experiments in GBM LN229 cells demonstrated that RRAD, by inducing EGF, enhances STAT3 activation. Furthermore, immunoprecipitation showed that RRAD physically interacts with EGFR and in vitro and in vivo studies demonstrated that RRAD enhances colony formation and cell migration as well as tumor growth in nude mice. Downregulation of RRAD by siRNA treatment resulted in loss of expression of the stemness-regulating genes OCT4, NANOG, and SOX2 as well as in sensitizing of otherwise temozolomide-resistant LN229 clones. The small GTPase Rab7b is known to be involved in retrograde transport of a number of sorting receptors and implicated in cell migration by actin cytoskeletal remodelling [72]. In a spinal cord injury rat model, MiR146A was one of the key miRNAs that was upregulated at the injury site over a longer time period and cell culture experiments indicated that MIR146A is a regulator of inflammation in glial cells [73,74]. SBSN is known as an invasion promoting protein that was identified in a proteomics study in the secretome of a GBM cell line that does not express EGFR and PTEN and in GBM derivate cells that have induced expression of both, EGFR and PTEN [75]. In mouse tumor endothelial cells, siRNA depletion of SBSN inhibited migration and tube formation capacity [76]. The functional properties of FAM83A are less known, however, its overexpression in breast cancer cells increased proliferation and invasion and augmented EGFR resistance to tyrosine kinase inhibitors [77]. Moreover, independently of an upstream EGF/EGFR activation, FAM83A overexpression resulted in activation of CRAF and PI3K p85 which are known upstream activators of ERK1/2 and AKT. Conversely, FAM83A depletion by siRNA resulted in delayed tumor growth of a xenograft tumor model and metadata analysis demonstrated that high FAM83A expression correlates with unfavorable prognosis in breast cancer patients.
Probe sets differentially expressed in the brain metastatic PTC, the TR, and primary brain tumors vs. non-brain metastatic PTCs
C1R is a serine protease of the complement system and an activator of C1S that is known to enhance tumorigenic invasion and migration by extracellular matrix disintegration [78]. CASP1 and CASP4 are effector caspases that are involved in caspase activation and regulation of inflammatory processes [79]. Comparably highest RNA expression levels for WDR72, which is a critical factor in calcium transport, have been found in kidney and thyroid tissues which may support our results showing lower expression of WDR72 in the brain metastatic PTC and primary brain tumors [80]. The function of CC2D2B, especially in cancer, is virtually unknown; however, microarray expression data indicated that CC2D2B is a top upregulated gene in PTCs which may also be a support for our results of comparably lower expression of the factor in the brain metastasis and primary brain tumors [60]. Similarly, CITED1 which encodes a CREB-binding protein/p300-interacting transactivator was identified in a microarray expression study as an upregulated gene in PTCs [81]. ZHX2 is a zinc finger and homeobox containing factor. Induced expression of ZHX2 in xenograft tumors resulted in reduced tumor growths [82].
In conclusion, whole transcript expression profiling identified candidate genes including a number of cytokines, and biofunctions, notably the granulocyte adhesion and diapedesis pathway, associated with a brain metastasis of a PTC, suggesting an inherited expression profile related to the brain metastatic process.
Acknowledgements
This study was supported by King Abdulaziz City for Science and Technology (KACST) grant AT-32-98 and KACST Technology Innovation Center in Personalized Medicine grant 13-CIPM-07. We thank Alaa Alamdi, Reem Alotibi, Nadia Baqtian, Ohoud Subhi, Manal Shabaat, Ishaq Khan, Mona Al-Omary, Nawal Madkhali, Lobna Siraj Mira, Maha Al-Quaiti, Fai Ashgan, Alaa Ghazi Almasri, Sabrin Bukhari, and Shireen Hussain for excellent technical assistance.
Disclosure of conflict of interest
None.
Abbreviations
- aCGH
array comparative genomic hybridization
- BBB
blood brain barrier
- FC
fold change
- FDR
false discovery rate
- GO
gene ontology
- GBM
glioblastoma multiforme
- IHC
immunohistochemistry
- FVPTC
follicular variant of PTC
- PCA
principal component analysis
- PTC
papillary thyroid carcinoma
- TR
technical replicate
Supporting Information
References
- 1.Zecchin D, Boscaro V, Medico E, Barault L, Martini M, Arena S, Cancelliere C, Bartolini A, Crowley EH, Bardelli A, Gallicchio M, Di Nicolantonio F. BRAF V600E is a determinant of sensitivity to proteasome inhibitors. Mol Cancer Ther. 2013;12:2950–2961. doi: 10.1158/1535-7163.MCT-13-0243. [DOI] [PubMed] [Google Scholar]
- 2.Schulten HJ, Alotibi R, Al-Ahmadi A, Ata M, Karim S, Huwait E, Gari M, Al-Ghamdi K, Al-Mashat F, Al-Hamour O, Al-Qahtani MH, Al-Maghrabi J. Effect of BRAF mutational status on expression profiles in conventional papillary thyroid carcinomas. BMC Genomics. 2015;16(Suppl 1):S6. doi: 10.1186/1471-2164-16-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dadu R, Cabanillas ME. Optimizing therapy for radioactive iodine-refractory differentiated thyroid cancer: current state of the art and future directions. Minerva Endocrinol. 2012;37:335–356. [PMC free article] [PubMed] [Google Scholar]
- 4.Tahmasebi FC, Farmer P, Powell SZ, Aldape KD, Fuller GN, Patel S, Hollis P, Chalif D, Eisenberg MB, Li JY. Brain metastases from papillary thyroid carcinomas. Virchows Arch. 2013;462:473–480. doi: 10.1007/s00428-013-1394-4. [DOI] [PubMed] [Google Scholar]
- 5.Tsuda K, Tsurushima H, Takano S, Tsuboi K, Matsumura A. Brain metastasis from papillary thyroid carcinomas. Mol Clin Oncol. 2013;1:817–819. doi: 10.3892/mco.2013.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiu AC, Delpassand ES, Sherman SI. Prognosis and treatment of brain metastases in thyroid carcinoma. J Clin Endocrinol Metab. 1997;82:3637–3642. doi: 10.1210/jcem.82.11.4386. [DOI] [PubMed] [Google Scholar]
- 7.Shen Y, Ruan M, Luo Q, Yu Y, Lu H, Zhu R, Chen L. Brain metastasis from follicular thyroid carcinoma: treatment with sorafenib. Thyroid. 2012;22:856–860. doi: 10.1089/thy.2011.0419. [DOI] [PubMed] [Google Scholar]
- 8.Singh M, Manoranjan B, Mahendram S, McFarlane N, Venugopal C, Singh SK. Brain metastasis-initiating cells: survival of the fittest. Int J Mol Sci. 2014;15:9117–9133. doi: 10.3390/ijms15059117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilhelm I, Molnar J, Fazakas C, Hasko J, Krizbai IA. Role of the blood-brain barrier in the formation of brain metastases. Int J Mol Sci. 2013;14:1383–1411. doi: 10.3390/ijms14011383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. AJCC Cancer Staging Manual. New York: Springer; 2010. [Google Scholar]
- 11.Schulten HJ, Hussein D, Al-Adwani F, Karim S, Al-Maghrabi J, Al-Sharif M, Jamal A, Al-Ghamdi F, Baeesa SS, Bangash M, Chaudhary A, Al-Qahtani M. Microarray Expression Data Identify DCC as a Candidate Gene for Early Meningioma Progression. PLoS One. 2016;11:e0153681. doi: 10.1371/journal.pone.0153681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulten HJ, Salama S, Al-Mansouri Z, Alotibi R, Al-Ghamdi K, Al-Hamour OA, Sayadi H, Al-Aradati H, Al-Johari A, Huwait E, Gari M, Al-Qahtani MH, Al-Maghrabi J. BRAF mutations in thyroid tumors from an ethnically diverse group. Hered Cancer Clin Pract. 2012;10:10. doi: 10.1186/1897-4287-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulten HJ, Al-Mansouri Z, Baghallab I, Bagatian N, Subhi O, Karim S, Al-Aradati H, Al-Mutawa A, Johary A, Meccawy AA, Al-Ghamdi K, Al-Hamour O, Al-Qahtani M, Al-Maghrabi J. Comparison of microarray expression profiles between follicular variant of papillary thyroid carcinomas and follicular adenomas of the thyroid. BMC Genomics. 2015;16(Suppl 1):S7. doi: 10.1186/1471-2164-16-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulten HJ, Alotibi R, Al-Ahmadi A, Ata M, Karim S, Huwait E, Gari M, Al-Ghamdi K, Al-Mashat F, Al-Hamour O, Al-Qahtani M, Al-Maghrabi J. Effect of BRAF mutational status on expression profiles in conventional papillary thyroid carcinomas. BMC Genomics. 2015;16(Suppl 1):S6. doi: 10.1186/1471-2164-16-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44:D336–342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulten HJ, Al-Mansouri Z, Baghallab I, Bagatian N, Subhi O, Karim S, Al-Aradati H, Al-Mutawa A, Johary A, Meccawy AA, Al-Ghamdi K, Al-Hamour O, Al-Qahtani MH, Al-Maghrabi J. Comparison of microarray expression profiles between follicular variant of papillary thyroid carcinomas and follicular adenomas of the thyroid. BMC Genomics. 2015;16(Suppl 1):S7. doi: 10.1186/1471-2164-16-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kramer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Dhahri SF, Al-Amro AS, Al-Shakwer W, Terkawi AS. Cerebellar mass as a primary presentation of papillary thyroid carcinoma: case report and literature review. Head Neck Oncol. 2009;1:23. doi: 10.1186/1758-3284-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Osta H, Falchook G, Tsimberidou A, Hong D, Naing A, Kim K, Wen S, Janku F, Kurzrock R. BRAF mutations in advanced cancers: clinical characteristics and outcomes. PLoS One. 2011;6:e25806. doi: 10.1371/journal.pone.0025806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Capper D, Berghoff AS, Magerle M, Ilhan A, Wohrer A, Hackl M, Pichler J, Pusch S, Meyer J, Habel A, Petzelbauer P, Birner P, von Deimling A, Preusser M. Immunohistochemical testing of BRAF V600E status in 1,120 tumor tissue samples of patients with brain metastases. Acta Neuropathol. 2012;123:223–233. doi: 10.1007/s00401-011-0887-y. [DOI] [PubMed] [Google Scholar]
- 22.Tufano RP, Teixeira GV, Bishop J, Carson KA, Xing M. BRAF mutation in papillary thyroid cancer and its value in tailoring initial treatment: a systematic review and meta-analysis. Medicine (Baltimore) 2012;91:274–286. doi: 10.1097/MD.0b013e31826a9c71. [DOI] [PubMed] [Google Scholar]
- 23.Zeng W, Chang H, Ma M, Li Y. CCL20/CCR6 promotes the invasion and migration of thyroid cancer cells via NF-kappa B signaling-induced MMP-3 production. Exp Mol Pathol. 2014;97:184–190. doi: 10.1016/j.yexmp.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 24.Arima Y, Harada M, Kamimura D, Park JH, Kawano F, Yull FE, Kawamoto T, Iwakura Y, Betz UA, Marquez G, Blackwell TS, Ohira Y, Hirano T, Murakami M. Regional neural activation defines a gateway for autoreactive T cells to cross the blood-brain barrier. Cell. 2012;148:447–457. doi: 10.1016/j.cell.2012.01.022. [DOI] [PubMed] [Google Scholar]
- 25.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massague J. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PG, Manova-Todorova K, Leversha M, Hogg N, Seshan VE, Norton L, Brogi E, Massagué J. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150:165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson EA, Dao TL, Guignet MA, Geddes CE, Koemeter-Cox AI, Kan RK. Increased expression of the chemokines CXCL1 and MIP-1alpha by resident brain cells precedes neutrophil infiltration in the brain following prolonged soman-induced status epilepticus in rats. J Neuroinflammation. 2011;8:41. doi: 10.1186/1742-2094-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, Kaiser EA, Snyder LA, Pollard JW. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendez-Enriquez E, Garcia-Zepeda EA. The multiple faces of CCL13 in immunity and inflammation. Inflammopharmacology. 2013;21:397–406. doi: 10.1007/s10787-013-0177-5. [DOI] [PubMed] [Google Scholar]
- 30.Isozaki O, Tsushima T, Miyakawa M, Emoto N, Demura H, Arai M, Sato-Nozoe Y. Oncostatin M: a new potent inhibitor of iodine metabolism inhibits thyroid peroxidase gene expression but not DNA synthesis in porcine thyroid cells in culture. Thyroid. 1997;7:71–77. doi: 10.1089/thy.1997.7.71. [DOI] [PubMed] [Google Scholar]
- 31.Herland A, van der Meer AD, FitzGerald EA, Park TE, Sleeboom JJ, Ingber DE. Distinct Contributions of Astrocytes and Pericytes to Neuroinflammation Identified in a 3D Human Blood-Brain Barrier on a Chip. PLoS One. 2016;11:e0150360. doi: 10.1371/journal.pone.0150360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng XS, Li YF, Tan J, Sun B, Xiao YC, Fang XB, Zhang XF, Li Q, Dong JH, Li M, Qian HH, Yin ZF, Yang ZB. CCL20 and CXCL8 synergize to promote progression and poor survival outcome in patients with colorectal cancer by collaborative induction of the epithelial-mesenchymal transition. Cancer Lett. 2014;348:77–87. doi: 10.1016/j.canlet.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 33.Xie Q, Thompson R, Hardy K, DeCamp L, Berghuis B, Sigler R, Knudsen B, Cottingham S, Zhao P, Dykema K, Cao B, Resau J, Hay R, Vande Woude GF. A highly invasive human glioblastoma pre-clinical model for testing therapeutics. J Transl Med. 2008;6:77. doi: 10.1186/1479-5876-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waisman A, Hauptmann J, Regen T. The role of IL-17 in CNS diseases. Acta Neuropathol. 2015;129:625–637. doi: 10.1007/s00401-015-1402-7. [DOI] [PubMed] [Google Scholar]
- 35.Chang SH, Reynolds JM, Pappu BP, Chen G, Martinez GJ, Dong C. Interleukin-17C promotes Th17 cell responses and autoimmune disease via interleukin-17 receptor E. Immunity. 2011;35:611–621. doi: 10.1016/j.immuni.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Jin S, Sonobe Y, Cheng Y, Horiuchi H, Parajuli B, Kawanokuchi J, Mizuno T, Takeuchi H, Suzumura A. Interleukin-1beta induces blood-brain barrier disruption by downregulating Sonic hedgehog in astrocytes. PLoS One. 2014;9:e110024. doi: 10.1371/journal.pone.0110024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park JS, Choi SY, Lee JH, Lee M, Nam ES, Jeong AL, Lee S, Han S, Lee MS, Lim JS, Yoon do Y, Kwon Y, Yang Y. Interleukin-32beta stimulates migration of MDA-MB-231 and MCF-7cells via the VEGF-STAT3 signaling pathway. Cell Oncol (Dordr) 2013;36:493–503. doi: 10.1007/s13402-013-0154-4. [DOI] [PubMed] [Google Scholar]
- 38.Liu X, Min L, Duan H, Shi R, Zhang W, Hong S, Tu C. Short hairpin RNA (shRNA) of type 2 interleukin-1 receptor (IL1R2) inhibits the proliferation of human osteosarcoma U-2 OS cells. Med Oncol. 2015;32:364. doi: 10.1007/s12032-014-0364-2. [DOI] [PubMed] [Google Scholar]
- 39.Aljohani H, Koncar RF, Zarzour A, Park BS, Lee SH, Bahassi el M. ROS1 amplification mediates resistance to gefitinib in glioblastoma cells. Oncotarget. 2015;6:20388–20395. doi: 10.18632/oncotarget.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang J, Chen K, Gong W, Zhou Y, Le Y, Bian X, Wang JM. Receptor “hijacking” by malignant glioma cells: a tactic for tumor progression. Cancer Lett. 2008;267:254–261. doi: 10.1016/j.canlet.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang J, Chen K, Chen J, Gong W, Dunlop NM, Howard OM, Gao Y, Bian XW, Wang JM. The G-protein-coupled formylpeptide receptor FPR confers a more invasive phenotype on human glioblastoma cells. Br J Cancer. 2010;102:1052–1060. doi: 10.1038/sj.bjc.6605591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rumer KK, Post MD, Larivee RS, Zink M, Uyenishi J, Kramer A, Teoh D, Bogart K, Winn VD. Siglec-6 is expressed in gestational trophoblastic disease and affects proliferation, apoptosis and invasion. Endocr Relat Cancer. 2012;19:827–840. doi: 10.1530/ERC-11-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hosono Y, Yamaguchi T, Mizutani E, Yanagisawa K, Arima C, Tomida S, Shimada Y, Hiraoka M, Kato S, Yokoi K, Suzuki M, Takahashi T. MYBPH, a transcriptional target of TTF-1, inhibits ROCK1, and reduces cell motility and metastasis. EMBO J. 2012;31:481–493. doi: 10.1038/emboj.2011.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goyal R, Mathur SK, Gupta S, Goyal R, Kumar S, Batra A, Hasija S, Sen R. Immunohistochemical expression of glial fibrillary acidic protein and CAM5.2 in glial tumors and their role in differentiating glial tumors from metastatic tumors of central nervous system. J Neurosci Rural Pract. 2015;6:499–503. doi: 10.4103/0976-3147.168426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim HS, Lee JJ, Do SI, Kim K, Do IG, Kim DH, Chae SW, Sohn JH. Overexpression of cytokeratin 17 is associated with the development of papillary thyroid carcinoma and the presence of lymph node metastasis. Int J Clin Exp Pathol. 2015;8:5695–5701. [PMC free article] [PubMed] [Google Scholar]
- 46.Simiczyjew A, Mazur AJ, Popow-Wozniak A, Malicka-Blaszkiewicz M, Nowak D. Effect of overexpression of beta- and gamma-actin isoforms on actin cytoskeleton organization and migration of human colon cancer cells. Histochem Cell Biol. 2014;142:307–322. doi: 10.1007/s00418-014-1199-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang KW, Chou A, Lee CC, Yeh C, Lai MW, Yeh TS, Chen TC, Liang KH, Yeh CT. Overexpression of kappa-actin alters growth properties of hepatoma cells and predicts poor postoperative prognosis. Anticancer Res. 2011;31:2037–2044. [PubMed] [Google Scholar]
- 48.Kanojia D, Morshed RA, Zhang L, Miska JM, Qiao J, Kim JW, Pytel P, Balyasnikova IV, Lesniak MS, Ahmed AU. betaIII-Tubulin Regulates Breast Cancer Metastases to the Brain. Mol Cancer Ther. 2015;14:1152–1161. doi: 10.1158/1535-7163.MCT-14-0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cantu-Medellin N, Kelley EE. Xanthine oxidoreductase-catalyzed reduction of nitrite to nitric oxide: insights regarding where, when and how. Nitric Oxide. 2013;34:19–26. doi: 10.1016/j.niox.2013.02.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sallenave JM. The role of secretory leukocyte proteinase inhibitor and elafin (elastase-specific inhibitor/skin-derived antileukoprotease) as alarm antiproteinases in inflammatory lung disease. Respir Res. 2000;1:87–92. doi: 10.1186/rr18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saidi A, Javerzat S, Bellahcene A, De Vos J, Bello L, Castronovo V, Deprez M, Loiseau H, Bikfalvi A, Hagedorn M. Experimental anti-angiogenesis causes upregulation of genes associated with poor survival in glioblastoma. Int J Cancer. 2008;122:2187–2198. doi: 10.1002/ijc.23313. [DOI] [PubMed] [Google Scholar]
- 52.Chernock RD, El-Mofty SK, Becker N, Lewis JS Jr. Napsin A expression in anaplastic, poorly differentiated, and micropapillary pattern thyroid carcinomas. Am J Surg Pathol. 2013;37:1215–1222. doi: 10.1097/PAS.0b013e318283b7b2. [DOI] [PubMed] [Google Scholar]
- 53.Remke M, Hielscher T, Korshunov A, Northcott PA, Bender S, Kool M, Westermann F, Benner A, Cin H, Ryzhova M, Sturm D, Witt H, Haag D, Toedt G, Wittmann A, Schottler A, von Bueren AO, von Deimling A, Rutkowski S, Scheurlen W, Kulozik AE, Taylor MD, Lichter P, Pfister SM. FSTL5 is a marker of poor prognosis in non-WNT/non-SHH medulloblastoma. J. Clin. Oncol. 2011;29:3852–3861. doi: 10.1200/JCO.2011.36.2798. [DOI] [PubMed] [Google Scholar]
- 54.Liu TP, Hong YH, Tung KY, Yang PM. In silico and experimental analyses predict the therapeutic value of an EZH2 inhibitor GSK343 against hepatocellular carcinoma through the induction of metallothionein genes. Oncoscience. 2016;3:9–20. doi: 10.18632/oncoscience.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang D, Qiu S, Wang Q, Zheng J. TMPRSS3 modulates ovarian cancer cell proliferation, invasion and metastasis. Oncol Rep. 2016;35:81–88. doi: 10.3892/or.2015.4356. [DOI] [PubMed] [Google Scholar]
- 56.Rui X, Li Y, Jin F, Li F. TMPRSS3 is a novel poor prognostic factor for breast cancer. Int J Clin Exp Pathol. 2015;8:5435–5442. [PMC free article] [PubMed] [Google Scholar]
- 57.Grindel BJ, Martinez JR, Pennington CL, Muldoon M, Stave J, Chung LW, Farach-Carson MC. Matrilysin/matrix metalloproteinase-7 (MMP7) cleavage of perlecan/HSPG2 creates a molecular switch to alter prostate cancer cell behavior. Matrix Biol. 2014;36:64–76. doi: 10.1016/j.matbio.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guglielmi L, Cinnella C, Nardella M, Maresca G, Valentini A, Mercanti D, Felsani A, D’Agnano I. MYCN gene expression is required for the onset of the differentiation programme in neuroblastoma cells. Cell Death Dis. 2014;5:e1081. doi: 10.1038/cddis.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCarthy RC, Kosman DJ. Activation of C6 glioblastoma cell ceruloplasmin expression by neighboring human brain endothelia-derived interleukins in an in vitro blood inverted question markbrain barrier model system. Cell Commun Signal. 2014;12:65. doi: 10.1186/s12964-014-0065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li J, Wang Z, Chong T, Chen H, Li H, Li G, Zhai X, Li Y. Over-expression of a poor prognostic marker in prostate cancer: AQP5 promotes cells growth and local invasion. World J Surg Oncol. 2014;12:284. doi: 10.1186/1477-7819-12-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marchi N, Mazzone P, Fazio V, Mekhail T, Masaryk T, Janigro D. ProApolipoprotein A1: a serum marker of brain metastases in lung cancer patients. Cancer. 2008;112:1313–1324. doi: 10.1002/cncr.23314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aguado BA, Wu JJ, Azarin SM, Nanavati D, Rao SS, Bushnell GG, Medicherla CB, Shea LD. Secretome identification of immune cell factors mediating metastatic cell homing. Sci Rep. 2015;5:17566. doi: 10.1038/srep17566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sung HJ, Ahn JM, Yoon YH, Rhim TY, Park CS, Park JY, Lee SY, Kim JW, Cho JY. Identification and validation of SAA as a potential lung cancer biomarker and its involvement in metastatic pathogenesis of lung cancer. J Proteome Res. 2011;10:1383–1395. doi: 10.1021/pr101154j. [DOI] [PubMed] [Google Scholar]
- 65.Knebel FH, Albuquerque RC, Massaro RR, Maria-Engler SS, Campa A. Dual effect of serum amyloid A on the invasiveness of glioma cells. Mediators Inflamm. 2013;2013:509089. doi: 10.1155/2013/509089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mollenhauer J, Wiemann S, Scheurlen W, Korn B, Hayashi Y, Wilgenbus KK, von Deimling A, Poustka A. DMBT1, a new member of the SRCR superfamily, on chromosome 10q25.3-26.1 is deleted in malignant brain tumours. Nat Genet. 1997;17:32–39. doi: 10.1038/ng0997-32. [DOI] [PubMed] [Google Scholar]
- 67.Muller H, Hu J, Popp R, Schmidt MH, Muller-Decker K, Mollenhauer J, Fisslthaler B, Eble JA, Fleming I. Deleted in malignant brain tumors 1 is present in the vascular extracellular matrix and promotes angiogenesis. Arterioscler Thromb Vasc Biol. 2012;32:442–448. doi: 10.1161/ATVBAHA.111.239830. [DOI] [PubMed] [Google Scholar]
- 68.Fang ZQ, Zang WD, Chen R, Ye BW, Wang XW, Yi SH, Chen W, He F, Ye G. Gene expression profile and enrichment pathways in different stages of bladder cancer. Genet Mol Res. 2013;12:1479–1489. doi: 10.4238/2013.May.6.1. [DOI] [PubMed] [Google Scholar]
- 69.Khalouei S, Chow AM, Brown IR. Stress-induced localization of HSPA6 (HSP70B’) and HSPA1A (HSP70-1) proteins to centrioles in human neuronal cells. Cell Stress Chaperones. 2014;19:321–327. doi: 10.1007/s12192-013-0459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Adamson DC, Shi Q, Wortham M, Northcott PA, Di C, Duncan CG, Li J, McLendon RE, Bigner DD, Taylor MD, Yan H. OTX2 is critical for the maintenance and progression of classic medulloblastoma. Cancer Res. 2010;70:181–191. doi: 10.1158/0008-5472.CAN-09-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yeom SY, Nam DH, Park C. RRAD Promotes EGFR-Mediated STAT3 Activation and Induces Temozolomide Resistance of Malignant Glioblastoma. Mol Cancer Ther. 2014;13:3049–3061. doi: 10.1158/1535-7163.MCT-14-0244. [DOI] [PubMed] [Google Scholar]
- 72.Distefano MB, Kjos I, Bakke O, Progida C. Rab7b at the intersection of intracellular trafficking and cell migration. Commun Integr Biol. 2015;8:e1023492. doi: 10.1080/19420889.2015.1023492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Iyer A, Zurolo E, Prabowo A, Fluiter K, Spliet WG, van Rijen PC, Gorter JA, Aronica E. MicroRNA-146a: a key regulator of astrocyte-mediated inflammatory response. PLoS One. 2012;7:e44789. doi: 10.1371/journal.pone.0044789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Strickland ER, Hook MA, Balaraman S, Huie JR, Grau JW, Miranda RC. MicroRNA dysregulation following spinal cord contusion: implications for neural plasticity and repair. Neuroscience. 2011;186:146–160. doi: 10.1016/j.neuroscience.2011.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sangar V, Funk CC, Kusebauch U, Campbell DS, Moritz RL, Price ND. Quantitative proteomic analysis reveals effects of epidermal growth factor receptor (EGFR) on invasion-promoting proteins secreted by glioblastoma cells. Mol Cell Proteomics. 2014;13:2618–2631. doi: 10.1074/mcp.M114.040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alam MT, Nagao-Kitamoto H, Ohga N, Akiyama K, Maishi N, Kawamoto T, Shinohara N, Taketomi A, Shindoh M, Hida Y, Hida K. Suprabasin as a novel tumor endothelial cell marker. Cancer Sci. 2014;105:1533–1540. doi: 10.1111/cas.12549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee SY, Meier R, Furuta S, Lenburg ME, Kenny PA, Xu R, Bissell MJ. FAM83A confers EGFR-TKI resistance in breast cancer cells and in mice. J Clin Invest. 2012;122:3211–3220. doi: 10.1172/JCI60498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rutkowski MJ, Sughrue ME, Kane AJ, Mills SA, Parsa AT. Cancer and the complement cascade. Mol Cancer Res. 2010;8:1453–1465. doi: 10.1158/1541-7786.MCR-10-0225. [DOI] [PubMed] [Google Scholar]
- 79.Kajiwara Y, Schiff T, Voloudakis G, Gama Sosa MA, Elder G, Bozdagi O, Buxbaum JD. A critical role for human caspase-4 in endotoxin sensitivity. J Immunol. 2014;193:335–343. doi: 10.4049/jimmunol.1303424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 81.Huang Y, Prasad M, Lemon WJ, Hampel H, Wright FA, Kornacker K, LiVolsi V, Frankel W, Kloos RT, Eng C, Pellegata NS, de la Chapelle A. Gene expression in papillary thyroid carcinoma reveals highly consistent profiles. Proc Natl Acad Sci U S A. 2001;98:15044–15049. doi: 10.1073/pnas.251547398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yue X, Zhang Z, Liang X, Gao L, Zhang X, Zhao D, Liu X, Ma H, Guo M, Spear BT, Gong Y, Ma C. Zinc fingers and homeoboxes 2 inhibits hepatocellular carcinoma cell proliferation and represses expression of Cyclins A and E. Gastroenterology. 2012;142:1559–1570. e2. doi: 10.1053/j.gastro.2012.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.