Abstract
The secreted axonal guidance molecular, Semaphorin-3F (SEMA3F), is widely expressed outside the central nervous system and inhibits tumor growth and metastasis. However, the role of SEMA3F expression in the osteosarcoma prognosis has not been elaborated. This study aimed to evaluate SEMA3F expression level in osteosarcoma and assess its prognostic value for patients. SEMA3F protein expression was detected by immunohistochemistry (IHC) in 112 cases of osteosarcoma. Kaplan-Meier analysis and Cox regression analysis were performed to evaluate the prognostic significance of SEMA3F. The results showed that the overall survival and metastasis-free survival of patients with negative SEMA3F expression were significantly shorter than patients with positive expression (both P<0.01). Multivariate Cox analysis identified SEMA3F expression as an independent prognostic factor to predict favorable overall survival and metastasis-free survival (both P<0.01). When endogenous SEMA3F expression was knocked down by siRNAs, cell proliferation, colony formation, migration and invasion in osteosarcoma cell lines were obviously promoted. Meanwhile, SEMA3F knockdown decreased E-cadherin expression but increased the expression of N-cadherin and β-Catenin, which indicated that SEMA3F could inhibit epithelial-mesenchymal transition (EMT). Mechanically, knockdown of SEMA3F inhibited GSK-3β protein expression and promoted the expression of β-Catenin and c-myc proteins. GSK-3β is a key upstream suppressor of β-Catenin and c-myc expression is an indicator of Wnt/β-Catenin activity. Therefore, these results suggest that down-regulated SEMA3F may promote EMT, migration, invasion and metastasis of osteosarcoma via activating Wnt/β-Catenin signaling. In conclusion, SEMA3F is downregulated and associated with prognosis of patients, indicating that SEMA3F may be a potential prognostic biomarker and therapeutic target for osteosarcoma.
Keywords: Osteosarcoma, SEMA3F, epithelial-mesenchymal transition (EMT), prognosis
Introduction
Osteosarcoma is the most commonly primary malignant tumors of bone and is a highly invasive neoplasms usually affecting the metaphysis of long bones in children and young adults [1]. The peak incidence of osteosarcoma usually occurs in the second and third decade of life [2]. Although treatment programs have been improved over the past decades, it is still a kind of tumor with a high mortality rate in children and adolescents. Although 5-year survival rates of approximately 50%-70% can be obtained by multimodal therapy, a large group of patients are still left with a poor prognosis as a result of lack of effective treatment patterns [3]. The shortage of a better molecular biomarker for prognosis estimation may lead to poor clinical response. Therefore, it is required for us to further understand the underlying molecular mechanisms and find a useful biomarker of osteosarcoma to predict prognosis.
The class 3 Semaphorin family is a group of secreted axon guidance molecules that plays crucial roles in neuronal development [4]. Recent evidence has reported that semaphorins including Semaphprin-3F (SEMA3F) are widely expressed outside the central nervous system and play important role in tumor angiogenesis, growth, and metastasis [5]. The human SEMA3F gene is located at 3p21.3, which is frequently lost in breast and lung cancers, implying its potential as a tumor suppressor gene [5]. Previous studies has shown that SEMA3F expression is significantly reduced in liver cancer, breast cancer, melanoma and ovarian cancer [6]. Recently, SEMA3F has been proved to perform an inhibitory role in tumorigenicity of lung [7], ovarian [8] and breast cancers [9]. However, the clinical significance of SEMA3F in osteosarcoma is not clear and its biological role in osteosarcoma remains to be further elucidated. In this study, we focused on investigation of SEMA3F expression and its clinical significance and biological function in osteosarcoma and also investigated which cell signaling pathway was regulated by SEMA3F in osteosarcoma. Our results suggest that down-regulated SEMA3F expression is associated with metastasis and poor prognosis.
Materials and methods
Ethics statement
The Medical Ethics Committee of the Xinqiao Hospital of Third Military Medical University has approved this protocol, and all patients provided written informed consent. All specimens were handled and stored anonymously according to ethical and legal standards.
Patients and tissue samples
Primary osteosarcoma samples were collected from 112 patients who had undergone surgical treatment for osteosarcoma with pathologic identification in Southwest Hospital and Xinqiao Hospital, Third Military Medical University from 1990 to 2013. Patients were enrolled in this retrospective study based on the following criteria: diagnosis of osteosarcoma with histopathological assessment, no prior anticancer treatment (radiotherapy or chemotherapy), and the availability of complete clinicopathologic and follow-up data. Tumor tissue specimens were grouped according to the sixth edition of the TNM classification of the International Union against Cancer (UICC).
Immunohistochemistry and evaluation
Immunohistochemical examination of SEMA3F was performed as described previously [10]. Briefly, after deparaffinising, rehydrating and blocking, the tissue sections of osteosarcoma were boiled in EDTA buffer (pH 8.0) for 15 min. The sections were then incubated with anti-SEMA3F (dilution 1:100; Millipore, USA) at 4°C overnight. Following incubation with secondary antibody (DAKO, Denmark), the sections were visualised using diaminobenzidine solution (DAKO) and lightly counterstained with haematoxylin. Negative control slides with the primary antibodies omitted were included for all assays.
The staining of SEMA3F was determined semi-quantitatively according to the intensity (0 = no staining, 1 = weak staining, 2 = moderate staining, 3 = strong staining) and the percentage of positive cells (0: none or <5%; 1: 5% to 20%; 2: 21% to 40%; 3: >40%). Scores of 0 to 2 were expected negative, and scores of 3 to 6 were expected positive. Cells were counted in at least three fields (at × 400 magnification) in the tumor areas. The scores were evaluated by two independent pathologists who were blinded to the pathological parameters and clinical outcomes of the patients.
Knockdown of SEMA3F
Small interfering RNAs (siRNAs) against SEMA3F were designed and purchased commercially from Genepharma (China) as follows: SEMA3F siRNA-1, 5’-AACTTCCTGCTCAACACAACC-3’; SEMA3F siRNA-2, 5’-AATCTTGCTCAAGGACGAGGA-3’; Scramble siRNA, 5’-GACTTCATAAGGCGCATGC-3’. These siRNAs were transfected into SaOS and MG-63 cells using Lipofectamine 2000 (Invitrogen) in the absence of serum according to the manufacturer’s instruction, when cells were reaching 40%~60% confluence. After 48 h, this medium was replaced with DMEM and cells were cultured for another 24 h. Then the cells were harvested to analyze the efficiency of SEMA3F knockdown.
Cell line and cell culture
The human osteosarcoma cell line, SaOS and MG-63 were purchased from the American Type Culture Collection (Rockville, USA). The osteosarcoma cell line SaOS and MG-63 were incubated in RPMI-1640 (Life Technologies, USA) supplemented with 10% fetal bovine serum (Gibco, USA). All cells were maintained in a humidified atmosphere containing 5% CO2 at 37°C.
Cell proliferation assay
Cell proliferation was performed by using Cell Counting Assay Kit-8 (CCK-8) (Dojindo, Japan) according to the manufacturer’s protocol. Briefly, 5 × 103 cells were harvested after 24, 48, 72 and 96 hours. 10 μL CCK-8 solution was added to each well, the cells were incubated for 1 hour, and the absorbance at 490 nm was measured by spectrophotometer (Bio-Rad, USA). All experiments were performed three times.
Wound healing migration and Matrigel invasion assays
For wound-healing migration assays, 1 × 105 cells were plated into in each well of 12-well plates. Three parallel lines were made in confluent cell cultures with a 200 ml tip. The wounds were observed at 0 and 48 h after scratching separately, and photographed via microscope at each time point. The distance between the two edges of the wound width was randomly quantified at 10 sites in all images. The average distance migrated by the cells was measured using a microscope calibrated with an ocular micrometer. The Matrigel invasion assay was performed with the BD BioCoatTM MatrigelTM Invasion Chamber (BD, USA) following the manufacturer’s instructions. SaOS (5 × 104 cell/plate) were seeded into the upper chamber of each well in serum-free medium, and complete medium was put into the bottom chamber. After incubated for 24 h, non-invading cells were removed by scrubbing from the upper surface of the membrane with cotton-tipped swabs. Following the processes of fixation in 100% methanol and staining in hematoxylin, the invading cells were counted in three images of each membrane under a microscope using a × 200 objective.
Western blotting
Western blotting was carried out by standard procedures as described previously. In brief, protein samples were separated on 10% (w/v) SDS gels and transferred onto PVDF membranes (Millipore, USA). The membranes were blocked with 5% milk proteins in TBST for 1 h at 37°C and then incubated with primary antibodies (SEMA3F, E-Cadherin, N-Cadherin, β-Catenin, GSK-3β, C-Myc, MMP-7, CD44, Abcam, USA) overnight at 4°C. The membranes were then incubated with HRP-conjugated anti-mouse secondary antibodies for 1 h at 37°C. Immunolabeling was detected using ECL reagent (Thermo Scientific, USA).
Statistical analysis
All group data are presented as mean ± SD. All statistical analyses were performed using SPSS version 13.0 statistical programs (SPSS, Chicago, IL). For all the analyses, a two-sided P value <0.05 was considered statistically significant. For the IHC analysis, the correlation of ASCL2 expression with the clinicopathologic data was analyzed using a Chi-square test. The prognostic values were evaluated by univariate Kaplan-Meier survival analysis and multivariate Cox proportional hazard model analysis. Overall survival (OS) was defined as the time from diagnosis until the date of either death or the last follow-up. For the metastasis-free survival (MFS) analysis, the duration was defined as the time from diagnosis until the occurrence of metastasis. If these patients had metastatic disease at diagnosis, this was considered time 0.
Results
Expression of SEMA3F mRNA in human non-metastatic and metastatic primary osteosarcoma samples
To examine the expression of SEMA3F mRNA in fresh osteosarcoma patient samples, we collected 12 primary tumor samples without metastasis and 14 primary tumor samples with metastasis. According to the qRT-PCR analysis, the expression levels of SEMA3F mRNA were found to be significantly decreased by 1.43-fold in the metastatic group compared with the non-metastatic group. The statistical analysis showed that the relative level of SEMA3F mRNA expression in metastatic primary osteosarcoma tissues (2.49±0.24) was remarkably lower (P<0.05) than that in non-metastatic tissues (3.57±0.15; Figure 1). These findings suggested that the lower expression level of SEMA3F in osteosarcoma was associated with metastasis.
Figure 1.
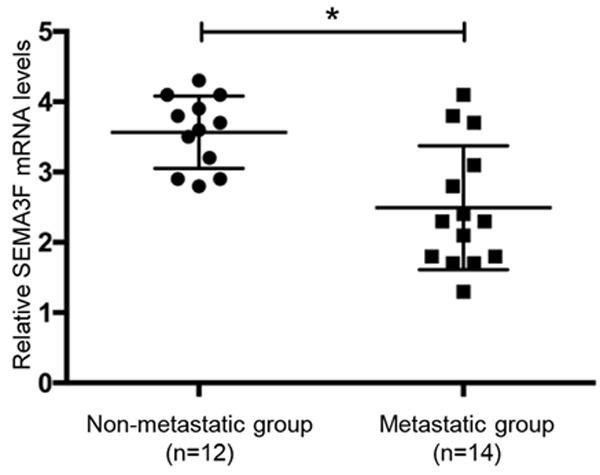
SEMA3F expression is in osteosarcoma tissues with or without metastasis. The SEMA3F mRNA levels were determined by real-time quantitative PCR. The data in the figure represent the averages ± SD of triplicate samples. *P<0.05.
Association between SEMA3F expression and the clinicopathologic features of osteosarcoma patients
To examine the expression of SEMA3F in human osteosarcoma patients, SEMA3F staining was positive in 76 osteosarcoma patients (67.9%). Microscopic observations indicated that SEMA3F was both expressed in the cytoplasm and nucleus of tumor cells (Figure 2). To elucidate the biologic significance, we investigated the association of patient clinicopathologic features and SEMA3F expression levels (Tables 1 and 2). Although positive staining of SEMA3F was present in both non-metastatic and metastatic tumors, the percentage of SEMA-3F-negative staining in osteosarcoma with metastases (39/64, 60.9%) was significantly higher than in the cases without metastases (25/64, 39.1%; P<0.05, Table 1). Positive SEMA-3F expression was more frequent in osteosarcoma tissues with low Enneking staging, and negative SEMA-3F expression with high Enneking staging (P<0.05). However, no significant difference was observed between the expression of SEMA-3F and patient gender, age, tumor location, histological classification, and histological grade.
Figure 2.
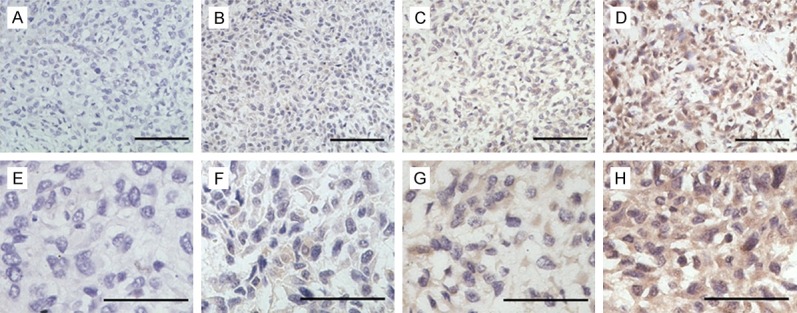
Representative immunohistochemical staining for SEMA3F in osteosarcoma tissues. Representative SEMA3F staining samples at low magnification at levels of 0, 1, 2, and 3 (A-D). Representative SEMA3F staining samples at high magnification at levels of 0, 1, 2, and 3 (E-H).
Table 1.
Correlation between SEMA3F expression and clinicopathologic factors of patients with osteosarcoma
| Variables | SEMA3F expression (N) | ||||
|---|---|---|---|---|---|
|
| |||||
| Total (N = 112) | Positive (76) | Negative (36) | X2 | P-values | |
| Gender | |||||
| Male | 67 (59.8%) | 42 | 25 | 2.044 | 0.215 |
| Female | 45 (40.2%) | 34 | 11 | ||
| Age | |||||
| ≤20 | 74 (66.1%) | 53 | 21 | 1.417 | 0.287 |
| ≥20 | 38 (33.9%) | 23 | 15 | ||
| Tumor Location | |||||
| Femur | 39 (34.8%) | 25 | 14 | 2.960 | 0.565 |
| Tibia | 28 (25.0%) | 17 | 11 | ||
| Humerus | 17 (15.2%) | 8 | 9 | ||
| Fibula | 15 (13.4%) | 7 | 8 | ||
| Others | 13 (11.6%) | 9 | 4 | ||
| Histological classification | |||||
| Osteoblastic | 67 (59.8%) | 36 | 31 | 3.598 | 0.165 |
| Chondroblastic | 36 (32.1%) | 13 | 23 | ||
| Others | 9 (8.1%) | 3 | 6 | ||
| Metastasis | |||||
| Yes | 64 (57.1%) | 25 | 39 | 3.258 | 0.071 |
| No | 48 (42.9%) | 27 | 21 | ||
| Histological grade | |||||
| I | 27 (24.1%) | 14 | 13 | 4.811 | 0.090 |
| II | 53 (47.3%) | 37 | 16 | ||
| III | 32 (28.6%) | 25 | 7 | ||
| Enneking staging | |||||
| I | 28 (25.0%) | 12 | 16 | 14.312 | 0.003 |
| II A | 46 (41.1%) | 31 | 15 | ||
| II B | 31 (27.7%) | 27 | 4 | ||
| III | 7 (6.2%) | 6 | 1 | ||
Abbreviation: SEMA3F, Semaphorin-3F.
Table 2.
Clinicopathologic patient characteristics and univariate survival analysis
| Variable | Patients (n = 112) | 5-y OS Rate (%) | P value | 5-y MFS Rate (%) | P value |
|---|---|---|---|---|---|
| Gender | |||||
| Male | 67 (59.8%) | 57.8% | 1.538 | 47.2% | 0.316 |
| Female | 45 (40.2%) | 53.2% | 43.6% | ||
| Age at diagnosis | |||||
| ≤20 | 74 (66.1%) | 54.3% | 0.621 | 37.8% | 0.217 |
| >20 | 38 (33.9%) | 60.1% | 42.3% | ||
| Tumor Location | |||||
| Femur | 39 (34.8%) | 62.3% | 0.458 | 54.2% | 0.625 |
| Tibia | 28 (25.0%) | 53.2% | 47.8% | ||
| Humerus | 17 (15.2%) | 59.4% | 50.7% | ||
| Fibula | 15 (13.4%) | 76.4% | 68.3% | ||
| Others | 13 (11.6%) | 68.9% | 58.5% | ||
| Histological classification | |||||
| Osteoblastic | 67 (59.8%) | 47.8% | 0.358 | 46.3% | 0.631 |
| Chondroblastic | 36 (32.1%) | 58.4% | 53.6% | ||
| Others | 9 (8.1%) | 71.2% | 67.2% | ||
| Metastasis | |||||
| Yes | 64 (57.1%) | 25.3% | <0.001 | 11.7% | <0.001 |
| No | 48 (42.9%) | 62.8% | 76.2% | ||
| Histological grade | |||||
| I | 27 (24.1%) | 63.2% | 0.012 | 65.2 | 0.026 |
| II | 53 (47.3%) | 52.7% | 37.8 | ||
| III | 32 (28.6%) | 26.4% | 27.4 | ||
| Enneking staging | |||||
| I | 28 (25.0%) | 86.2% | <0.001 | 76.5% | <0.001 |
| II A | 46 (41.1%) | 63.2% | 60.1% | ||
| II B | 31 (27.7%) | 26.3% | 23.7% | ||
| III | 7 (6.2%) | 2.8% | 0% | ||
| SEMA3F | |||||
| Negative | 36 (32.1%) | 41.3% | 0.013 | 37.8% | <0.001 |
| Positive | 76 (67.9%) | 76.2% | 64.5% | ||
Abbreviation: OS, Overall survival; MFS, Metastasis-free survival; SEMA3F, Semaphorin-3F.
Prognostic significance of SEMA3F expression in osteosarcoma patients
Follow-up information was available for all patients and the median period was 39 months (range: 5-106 months). During the follow-up period, cancer progression was found in 64 patients (57.1%). Kaplan-Meier curves were analyzed to show that SEMA3F high expression was statistically correlated with both favorable overall survival (OS, P<0.001) and disease-free survival (DFS, P<0.001) in osteosarcoma (Figure 3A, 3B). We then performed multivariate Cox regression analysis, which revealed that SEMA3F expression, histological grade and Enneking staging were indeed independent prognostic factors for OS and MFS (P<0.05; Table 3). These data indicated that SEMA3F may be a significant and novel biomarker for evaluating the prognoses of osteosarcoma patients.
Figure 3.
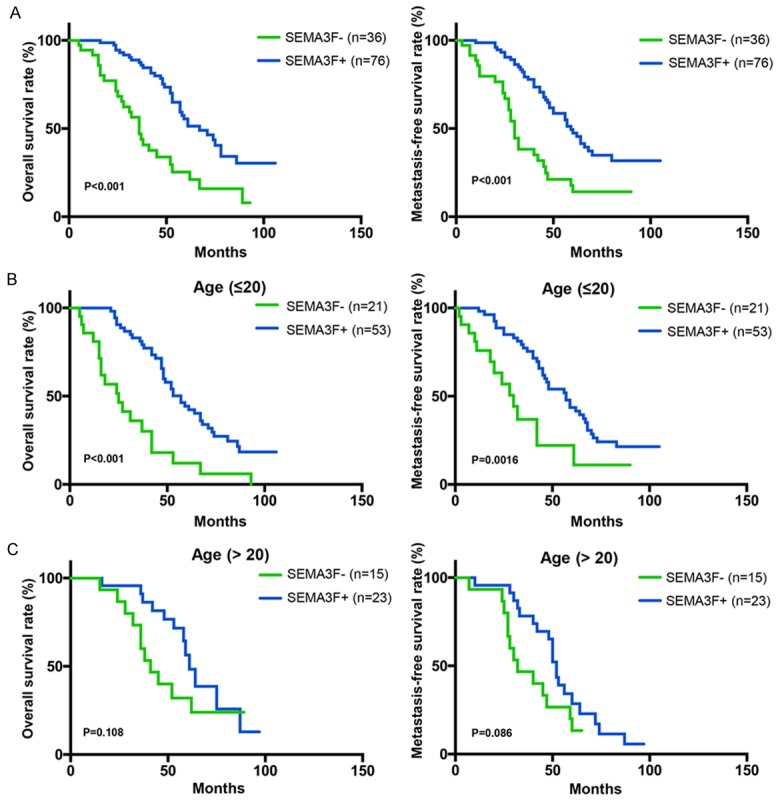
Negative SEMA3F immunohistochemical staining was associated with poor prognosis and survival. A. Overall survival (OS) and metastasis-free survival (MFS) of SEMA3F positive or negative patients was determined by Kaplan-Meier analysis. B. OS and MFS of SEMA3F positive or negative patients younger than 20 years old (≤20). C. OS and MFS of SEMA3F positive or negative patients elder than 20 years old (>20).
Table 3.
Multivariate analysis of factors associated with overall survival and metastasis-free survival
| Variable | HR (95% Cl) | P value |
|---|---|---|
| OS | ||
| SEMA3F(-vs+) | 2.153 (1.182-2.632) | 0.017 |
| Histological grade | 1.253 (0.843-1.284) | 0.032 |
| Enneking staging | 2.324 (1.274-3.852) | <0.001 |
| MFS | ||
| SEMA3F(-vs+) | 1.793 (0.873-3.194) | 0.018 |
| Histological grade | 2.632 (1.743-3.754) | 0.074 |
| Enneking staging | 4.643 (1.429-5.485) | <0.001 |
Abbreviation: SEMA3F, Semaphorin-3F.
As osteosarcoma is the second highest cause of cancer-related death in children and young adolescents [11], we next analyzed the OS and MFS in patients younger than 20 years old or elder than 20 years old. Kaplan-Meier analysis showed that young patients (≤20 years old) with negative SEMA3F expression had significantly worse OS and MFS than those with positive SEMA3F expression (Figure 3B; P<0.05). However, it was not significantly different in the patients elder than 20 years old between negative SEMA3F expression and positive expression (Figure 3C; P>0.05). It suggested that SEMA3F would be more accurate biomarker for evaluating OS and MFS of the patients within children and young adolescents (≤20 years old).
SEMA3F knockdown promotes proliferation and colony formation of osteosarcoma cells
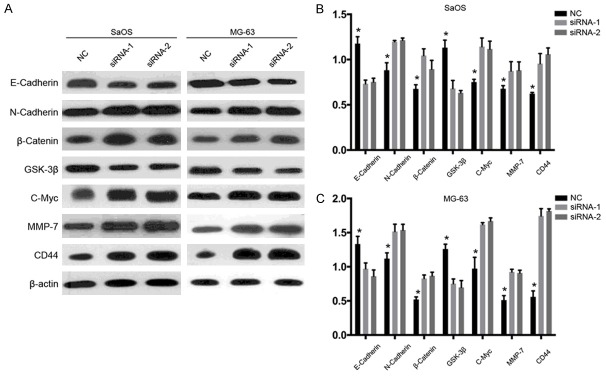
The above results suggested that downregulated SEMA3F is correlated with tumor progression of osteosarcoma and poor prognosis of patients. Thus, we explored whether knockdown of SEMA3F can promote growth of osteosarcoma cells. Two siRNAs were designed against SEMA3F and transfected into osteosarcoma cell lines SaOS and MG-63. Figure 4A showed that SEMA3F protein was downregulated in both SaOS and MG-63 cells transfected with the two siRNAs. CCK-8 assay (Figure 4B) showed that proliferation capacity of both SaOS and MG-63 cells transfected with siRNA-1 or -2 was significantly promoted compared with control group, indicating that downregulation of SEM3F could promote osteosarcoma cell proliferation. To confirm the effect of SEMA3F knockdown on the oncogenic growth of osteosarcoma cells, plate colony formation assay was also conducted on the two cells (Figure 4C). As expected, the colony numbers of the both cells with SEMA3F knockdown were remarkably more than those of the negative control cells (Figure 4D). These findings suggest that knockdown of SEMA3F enhances proliferation and survival ability of osteosarcoma cells.
Figure 4.
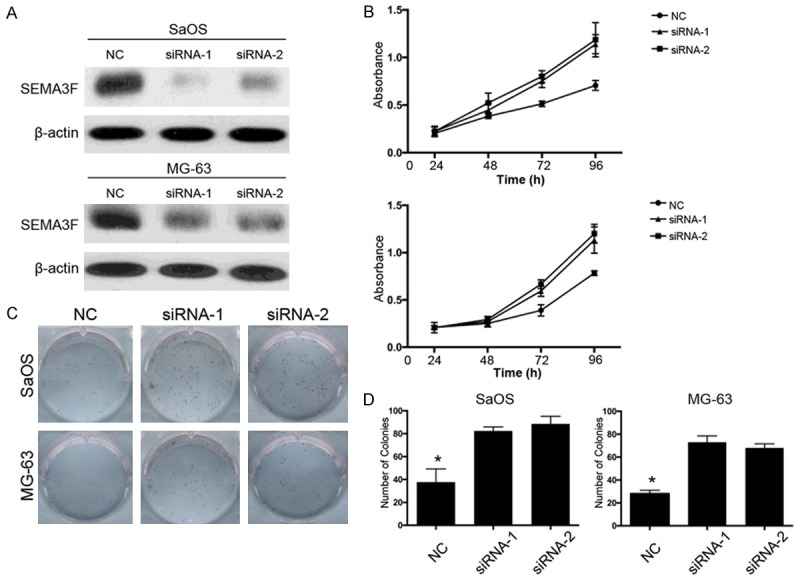
Knockdown of SEMA3F expression promotes the viability of osteosarcoma cells. A. The protein level of SEMA3F in SaOS and MG-63 cells after transfection of siRNAs against SEMA3F or scramble sequence was analyzed by Western blot. β-actin was used as the loading control. B. Growth curves of SaOS and MG-63 cells with transfected siRNAs or scramble sequence. Cell growth was determined by CCK-8. C, D. A colony-formation assay was performed to determine the oncogenic growth of SaOS and MG-63 cells transfected with siRNAs or scramble sequence. The data in the figure represent the averages ± SD of triplicate samples. *P<0.05 compared with the control group based on one-way ANOVA.
Downregulation of SEMA3F promotes migration and invasion of osteosarcoma cells
The above clinical results showed that SEMA3F down-expression was associated with metastasis, which suggested that SEMA3F could inhibit tumor progression and metastasis of osteosarcoma. It has been documented that cancer cell could metastasize only when they have the ability of migration and invasion. Therefore, we carried out in vitro experiments to demonstrate whether SEMA3F inhibits migration and invasion of osteosarcoma cells. Wound healing assay displayed that the downregulated SEMA3F expression by the two siRNAs led to higher migration rate in both osteosarcoma cell lines (Figure 5A-D). Transwell invasion assay also showed that the number of invaded cells were much more in SEMA3F knockdown cells compared with the control cells (Figure 5E-H). These results suggest that SEMA3F inhibited migration and invasion of osteosarcoma cells, while knockdown of SEMA3F promotes migration and invasion, which may lead to metastasis.
Figure 5.
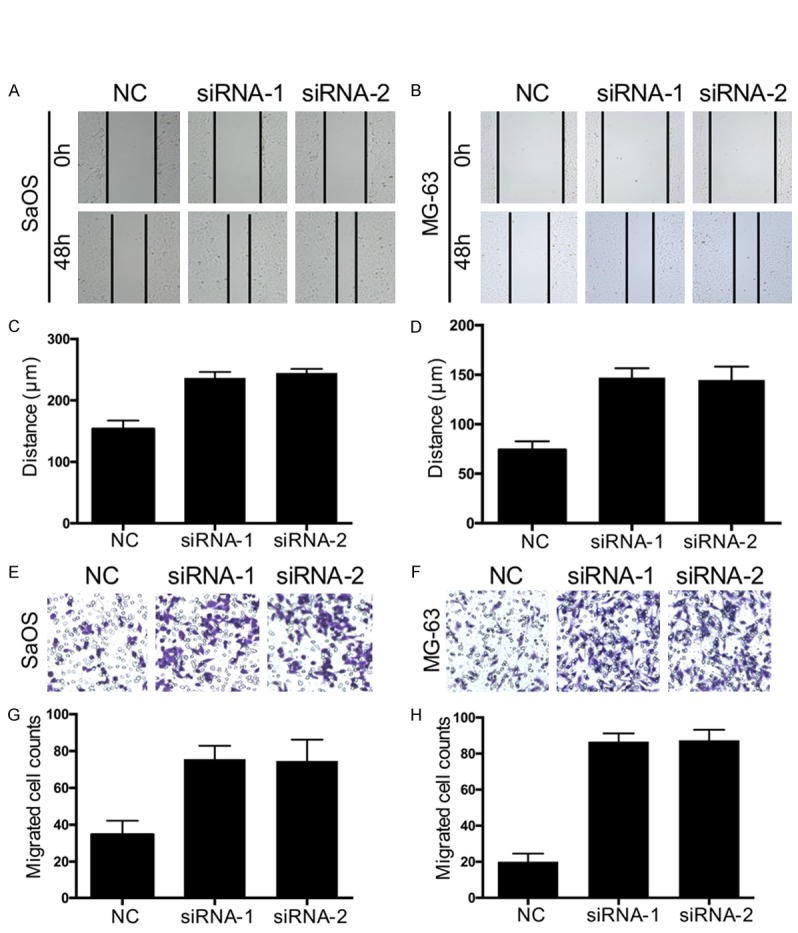
SEMA3F knockdown promotes migration and invasion of osteosarcoma cells. A-D. Wound healing assays were performed to determine the migratory abilities of si-SEMA3F-transfected SaOS and MG-63 cells. E-H. Transwell assays were performed to determine the migratory abilities of si-SEMA3F-transfected SaOS and MG-63 cells. The data in the figure represent the averages ± SD of triplicate samples. *P<0.05 compared with the control group based on one-way ANOVA.
SEMA3F knockdown increases epithelial-mesenchymal transition by Wnt/β-catenin pathway
Previous studies have demonstrated that before metastasis, cancer cells may have epithelial-mesenchymal transition (EMT), by which cancer cells lose cell-cell adhesion, epithelial markers, and apical-basolateral polarity, and acquire spindle-cell shape, motility and mesenchymal markers. Thus EMT was thought to facilitate cancer cell motility, invasion and metastasis. Combined with our results above, we hypothesize that SEMA3F can restrain EMT during its inhibition of migration and invasion. Thus, we further explored the markers of EMT with western blot: E-cadherin (epithelial marker), N-cadherin and β-catenin (mesenchymal markers). The results showed that SEMA3F knockdown repressed E-cadherin and upregulated N-cadherin and β-catenin (Figure 6A), which clearly indicated promotion of EMT in both osteosarcoma cells. Recent studies have shown that Wnt/β-catenin pathway is involved in cancer metastasis. β-catenin has been documented to be aberrantly accumulated in human tumors, which GSK-3β can induce ubiquitination and proteasomal degradation of β-catenin. In addition, c-myc expression is thought to be an indicator of Wnt/β-Catenin activity. Therefore, we simultaneously examined the expression of GSK-3β, c-myc and β-catenin when SEMA3F was knocked down. The results showed that GSK-3β expression was obviously decreased while c-myc and β-catenin expression was markedly elevated when SEMA3F was knocked down (Figure 5), which indicates that the activity of Wnt/β-catenin pathway was activated. These results suggest that down-regulated SEMA3F may promote EMT, migration, invasion and metastasis of osteosarcoma via activating Wnt/β-Catenin signaling.
Figure 6.
SEMA3F knockdown promotes EMT and activates Wnt/β-Catenin pathway in osteosarcoma cells. (A) Representative western blot images of E-cadherin, N-cadherin, β-catenin, GSK-3β, c-myc, MMP-7 and CD44 expression. The protein expression level of E-cadherin, N-cadherin, β-catenin, GSK-3β, c-myc, MMP-7 and CD44 in SaOS (B) and MG-63 (C) cells was determined. The data in the figure represent the averages ± SD of triplicate samples. *P<0.05 compared with the control group based on one-way ANOVA.
Discussion
Cancer metastasis is the major cause of mortality in osteosarcoma patients. Patients with lung or other bone metastases have a 5-year survival rate EMT causes an aggressive tumor phenotype, ultimately contributing to metastasis by direct local tissue invasion or distantly via blood vessels and lymphatics. Osteosarcoma is the most frequent primary malignant bone tumor and is a highly aggressive tumor that metastasizes primarily to the lungs [12]. Therefore, the study of biomarkers for osteosarcoma metastasis is important for improving the survival of patients.
SEM3F is originally identified as an axonal guidance factor in neural development, and is expressed outside central neural system. PlexinA1/PlexinA3 and neuropilion (NRP2) are co-receptors of SEMA3F. SEMA3F could bind to NRP2 with high affinity and PlexinA1/PlexinA3 is essential for signaling. The cytoplasmic region of PlexinA contain protein-protein interaction sites and a split GTPase-activating protein (GAP) domain, which the Rho and Ras families of small G protein. SEMA3F acts as a tumor suppressor in human breast cancer [9], melanoma [13], lung cancer [14] and fibrosarcoma [15]. It has been reported that SEMA3F inhibits the growth and metastasis of colorectal cancer [16]. In this study, we found that SAMA3F was expressed at a lower level in human osteosarcoma patients with metastasis compared with non-metastatic patients and that the negative SEMA3F expression in osteosarcoma tissues was significantly correlated with metastatic features and with shorter OS and MFS.
Patients outcome with osteosarcoma is varied greatly depending on patients age, histological classification, and Enneking stage. In our study, we found that the positive SEMA3F group had significantly more survival rates for OS and MFS than the negative SEMA3F group of osteosarcoma patients. Furthermore, it predicts significantly better outcome in the SEMA3F-positive patients younger than 20 years old. However, it is not significantly difference in the OS and MFS in the SEMA3F-positive patients elder than 20 years old. It suggested that SEMA3F would be more accurate biomarker for evaluating OS and MFS in the subgroups of patients within children and young adolescents (≤20 years old).
Although some studies on SEMA3F in cancer have been reported, little is known about the biological function and mechanism of SEMA3F in osteosarcoma. In this study, we found the extensive biological functions of SEMA3F in osteosarcoma cells, such as inhibition of proliferation, colony formation, migration, invasion and EMT of osteosarcoma cells. More important, we elucidate that SEMA3F downregulation promotes migration, invasion and EMT of osteosarcoma cells by activating Wnt/β-Catenin pathway. These results will help us understand how SEMA3F expression inhibits tumor progression, correlates with favorable prognosis in clinical patients with osteosarcoma and provide a potential molecular target for osteosarcoma therapy.
In conclusion, our results demonstrated that SEMA3F is down-expressed in osteosarcoma and is a potential prognostic biomarker of osteosarcoma, which provides additional information for guiding the therapeutic strategy. Our results warrant further studies on the detailed mechanisms by which SEMA3F inhibits tumor progression in osteosarcoma.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81472076, 81271982, 81301944 and 81401801).
Disclosure of conflict of interest
None.
References
- 1.Bielack S, Carrle D, Casali PG ESMO Guidelines Working Group. Osteosarcoma: ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann Oncol. 2009;20(Suppl 4):137–139. doi: 10.1093/annonc/mdp154. [DOI] [PubMed] [Google Scholar]
- 2.Damron TA, Ward WG, Stewart A. Osteosarcoma, chondrosarcoma, and Ewing’s sarcoma: National Cancer Data Base Report. Clin Orthop Relat Res. 2007;459:40–47. doi: 10.1097/BLO.0b013e318059b8c9. [DOI] [PubMed] [Google Scholar]
- 3.Bielack SS, Kempf-Bielack B, Delling G, Exner GU, Flege S, Helmke K, Kotz R, Salzer-Kuntschik M, Werner M, Winkelmann W, Zoubek A, Jurgens H, Winkler K. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. Oncol. 2002;20:776–790. doi: 10.1200/JCO.2002.20.3.776. [DOI] [PubMed] [Google Scholar]
- 4.Chen H, He Z, Bagri A, Tessier-Lavigne M. Semaphorin-neuropilin interactions underlying sympathetic axon responses to class III semaphorins. Neuron. 1998;21:1283–1290. doi: 10.1016/s0896-6273(00)80648-0. [DOI] [PubMed] [Google Scholar]
- 5.Tamagnone L. Emerging role of semaphorins as major regulatory signals and potential therapeutic targets in cancer. Cancer Cell. 2012;22:145–152. doi: 10.1016/j.ccr.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 6.Rehman M, Tamagnone L. Semaphorins in cancer: biological mechanisms and therapeutic approaches. Semin Cell Dev Biol. 2013;24:179–189. doi: 10.1016/j.semcdb.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Kusy S, Nasarre P, Chan D, Potiron V, Meyronet D, Gemmill RM, Constantin B, Drabkin HA, Roche J. Selective suppression of in vivo tumorigenicity by semaphorin SEMA3F in lung cancer cells. Neoplasia. 2005;7:457–465. doi: 10.1593/neo.04721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tse C, Xiang RH, Bracht T, Naylor SL. Human Semaphorin 3B (SEMA3B) located at chromosome 3p21.3 suppresses tumor formation in an adenocarcinoma cell line. Cancer Res. 2002;62:542–546. [PubMed] [Google Scholar]
- 9.Nasarre P, Kusy S, Constantin B, Castellani V, Drabkin HA, Bagnard D, Roche J. Semaphorin SEMA3F has a repulsing activity on breast cancer cells and inhibits E-cadherin-mediated cell adhesion. Neoplasia. 2005;7:180–189. doi: 10.1593/neo.04481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu XG, Chen L, Wang QL, Zhao XL, Tan J, Cui YH, Liu XD, Zhang X, Bian XW. Elevated expression of ASCL2 is an independent prognostic indicator in lung squamous cell carcinoma. J Clin Pathol. 2016;69:313–318. doi: 10.1136/jclinpath-2015-203025. [DOI] [PubMed] [Google Scholar]
- 11.Collins M, Wilhelm M, Conyers R, Herschtal A, Whelan J, Bielack S, Kager L, Kuhne T, Sydes M, Gelderblom H, Ferrari S, Picci P, Smeland S, Eriksson M, Petrilli AS, Bleyer A, Thomas DM. Benefits and adverse events in younger versus older patients receiving neoadjuvant chemotherapy for osteosarcoma: findings from a meta-analysis. J. Clin. Oncol. 2013;31:2303–2312. doi: 10.1200/JCO.2012.43.8598. [DOI] [PubMed] [Google Scholar]
- 12.Geller DS, Gorlick R. Osteosarcoma: a review of diagnosis, management, and treatment strategies. Clin Adv Hematol Oncol. 2010;8:705–718. [PubMed] [Google Scholar]
- 13.Bielenberg DR, Hida Y, Shimizu A, Kaipainen A, Kreuter M, Kim CC, Klagsbrun M. Semaphorin 3F, a chemorepulsant for endothelial cells, induces a poorly vascularized, encapsulated, nonmetastatic tumor phenotype. J Clin Invest. 2004;114:1260–1271. doi: 10.1172/JCI21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potiron VA, Sharma G, Nasarre P, Clarhaut JA, Augustin HG, Gemmill RM, Roche J, Drabkin HA. Semaphorin SEMA3F affects multiple signaling pathways in lung cancer cells. Cancer Res. 2007;67:8708–8715. doi: 10.1158/0008-5472.CAN-06-3612. [DOI] [PubMed] [Google Scholar]
- 15.Xiang R, Davalos AR, Hensel CH, Zhou XJ, Tse C, Naylor SL. Semaphorin 3F gene from human 3p21.3 suppresses tumor formation in nude mice. Cancer Res. 2002;62:2637–2643. [PubMed] [Google Scholar]
- 16.Zhou ZH, Rao J, Yang J, Wu F, Tan J, Xu SL, Ding Y, Zhan N, Hu XG, Cui YH, Zhang X, Dong W, Liu XD, Bian XW. SEMA3F prevents metastasis of colorectal cancer by PI3K-AKT-dependent down-regulation of the ASCL2-CXCR4 axis. J Pathol. 2015;236:467–478. doi: 10.1002/path.4541. [DOI] [PubMed] [Google Scholar]