Abstract
Studies have found that colorectal neoplasia differentially expressed (CRNDE) is related to cancer development. Herein, we found that the expression of CRNDE was increased in human hepatic carcinoma (HCC) tissues and cell lines. The ROC curve analysis illustrated CRNDE has a significant diagnostic value for HCC. At the same time, CRNDE promotes HCC cell proliferation, migration, and invasion in vitro. Quantitative real-time polymerase chain reaction (PCR) demonstrated that miR-384 was significantly downregulated in HCC tissues. Moreover, we indicated CRNDE negatively regulated miR-384 expression in HCC. In addition, we found that CRNDE accelerated the expression levels of NF-κB and p-AKT though inhibition of miR-384. Overall, these results suggested that CRNDE-miR-384 axis might be a promising therapeutic target for the treatment of HCC.
Keywords: lncRNA-CRNDE, miR-384, hepatocellular carcinoma, migration and invasion
Introduction
Hepatocellular carcinoma (HCC) is one of the most common tumors to impact human health, accounting for more than 5% of all cancers in the world [1,2]. It has been reported that HCC has become the second leading cause of cancer mortality in China [3]. Although surgical treatment is the most common and efficient method for treating HCC, the recurrence and metastasis rates are high. According to earlier reports, the 5-year survival rate after operation ranges between 36-50% [4]. HCC cell migration and invasion are very complex processes, including changes in multi-gene signatures. The early prevention of migration and invasion has become a primary topic in HCC research. Therefore, the molecular mechanisms of potential metastasis in HCC and the effective therapeutic targets require urgent exploration.
Long non-coding RNAs (lncRNAs), a new sort of non-coding RNA longer than 200 nucleotides, have no protein-coding capacity [5]. Numerous studies have indicated that lncRNAs regulate gene expressions through the processes of transcription regulation, post-transcription regulation, genomic imprinting, and chromatin modification [6,7]. There is already growing evidence that lncRNAs play major roles, not only in normal development but also in tumor genesis [8,9]. However, the biological function and mechanism of lncRNA-CRNDE are not clear in HCC. In this research, we measured the expression level of lncRNA-CRNDE in human hepatic carcinoma cell lines and tissues. We also demonstrated that lncRNA-CRNDE promoted HCC cell proliferation, migration, and invasion in vitro.
MicroRNAs (miRNAs), a class of endogenous non-coding RNA with the length of 20-25 nucleotides, can regulate the expression of massive target genes by targeting the homologous sequences of messenger RNAs (mRNAs) with the 3’-untranslated region (3’-UTR) to promote RNA degradation [10,11]. MiRNAs play key roles in the regulation of cell development process including proliferation, differentiation, metastasis, and apoptosis [12,13]. However, the mechanism of miR-384 action is not clear in HCC.
In this study, our results revealed a novel mechanism of lncRNA-CRNDE in tumorigenesis of HCC. We surmised that lncRNA-CRNDE can directly interact with miR-384 to promote HCC cell proliferation, migration, and invasion in vitro. Therefore, we speculated that the lncRNA-CRNDE, as a key regulator of gene expression, might be a promising therapeutic target for the treatment of hepatic carcinoma.
Materials and methods
Patients and clinical specimens
HCC tissues and matched adjacent noncancerous tissues samples were collected and written, informed consent was provided by patients in the Department of Hepatobiliary Surgery, Affiliated Hospital of Guizhou Medical University. According to World Health Organization (WHO), the HCC histological diagnosis was confirmed.
Cell culture
Seven HCC cell lines (MHCC97H, Hep3B, MHCC97L, BEL-7402, QGY-7703, HCCC9810, and HuH7), immortalized normal liver epithelial cell lines (THLE3), and human embryonic kidney 293T (HEK293T) cell lines were purchased from the Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China. MHCC97H, Hep3B, MHCC97L, BEL-7402, QGY-7703, HCCC9810, HuH7, and HEK293T were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (FBS, Invitrogen), 100 U/ml penicillin, and 1 μg/ml streptomycin (Invitrogen). THLE3 cells were cultured in bronchial epithelial growth medium (Clonetics Corporation, Walkersville, MD), including 5 ng/ml epithelial growth factor (EGF), 70 ng/ml phosphoethanolamine, and 10% FBS. All cells were cultured at 37°C with 5% CO2.
Lentiviral vector construction
The vectors carrying GFP, lncRNA-CRNDE, Luc, and shRNA-CRNDE were constructed. The packaging vectors (pCMV-VSVG, pMDLg/pRRE and pRSV-REV) and the lentivirus vectors were co-transfected in HEK293T cells, and then lentiviruses were concentrated and identified.
Transfection
MHCC97H cells were transfected with Lenti-GFP and Lenti-lncRNA-CRNDE. HuH7 cells were transfected with Lenti-shCRNDE, and Lenti- shLuc using 8 μg/mL polybrene (Sigma). G418 (Life Technologies, 0.8 mg/mL) was used to screen and establish the stable expression of cell lines. According to the manufacturer’s protocol, MHCC97H and HuH7 (2 × 105 cells/well) were transfected with 200 μl of mature miR-384 mimic (100 nM), mock, or inhibitor with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) for 72 hrs.
Reverse transcription and quantitative real-time reverse transcription PCR (qRT-PCR)
Total RNA was extracted using the TRIzol reagent (Invitrogen, CA, USA). Corresponding cDNA was synthesized using random primers and a RevertAid First Strand cDNA Synthesis kit (Thermo Fisher). SYBR-Green PCR Master Mix kit (Takara, Japan) and ABI 7500 Real-Time PCR System (Applied Biosystems) were used to detect the mRNA expression levels of related genes. GAPDH and U6 were chosen as internal loading controls. The sequences of GAPDH primers were 5’-GTCAGCCGCATCTTCTTTTG-3’ (sense) and 5’-GCGCCCAATACGACCAAATC-3’ (antisense); The primer sequences for U6 are: 5’-CTCGCTTCGGCAGCACA-3’ (the forward primer) and 5’-AACGCTTCACGAATTTGCGT-3’ (the reverse primer); The sequences of lncRNA-CRNDE primers were 5’-TGAAGGAAGGAAGTGGTGCA-3’ (sense) and 5’-TCCAGTGGCATCCTACAAGA-3’ (antisense); The sequences of miR-384 primers were 5’-TGTTAAATCAGGAATTTTAA-3’ (sense) and 5’-TGTTACAGGCATTATGAA-3’ (antisense); The sequences of NF-κB primers were 5’-AGTGTGGAGGCTGCCTTGCGAATG-3’ (sense) and 5’-TGGGCTTTCAAGACTGGAACGGTC-3’ (antisense); The sequences of p-AKT primers were 5’-GCAGCACGTGTACGAGAAGA-3’ (sense) and 5’-GGTGTCAGTCTCCGACGTG-3’ (antisense). All results were repeated three times. All results are shown as the mean ± SD of three independent experiments.
Western blot analysis
Cells were harvested in a lysis buffer containing a protease inhibitor cocktail (Roche Applied Science). The concentrations of total proteins were measured using a BCA protein assay kit (Thermo Fisher Scientific). The objective proteins were separated with 8% SDS polyacrylamide gel and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA). The membrane was incubated with primary antibodies including anti-rabbit-p-AKT (1:1000 dilutions, Santa Cruz, USA), anti-rabbit-AKT (1:1000 dilutions, Cell Signaling Technology, Beverly, MA, USA), anti-rabbit-NF-κB antibody (1:1000 dilutions, Cell Signaling Technology, Beverly, MA, USA), anti-rabbit-GAPDH antibody (1:4000 dilution, Cell Signaling Technology, Beverly, MA, USA). The enhanced chemiluminescence (ECL) substrate kit (Amersham Biosciences) and the enhanced chemiluminescence detection system (Amersham Biosciences) were used to detect the results.
Cell proliferation
The proliferate ability of HCC cells was measured using 3-(4, 5-dimethylthiazole-2-yl)-2, 5-diphenyl tetrazolium bromide (MTT). Next, 2 × 103 cells were seeded in a 96-well plate. At particular points in time, 20 μl MTT solutions (0.5 mg/ml, Sigma) were added to each well for 4 hrs at 37°C. The original culture was removed, and then 100 μl of dimethyl sulfoxide solutions (Sigma) were added to each well. The absorbance was detected by a Tecan plate reader at 490 nm. Each experimental condition was detected in quintuplicate. All experiments were performed in triplicate.
Migration and invasion assays
The migratory and invasive ability of HCC cells were assessed by migration assay with transwell (Corning Life Sciences, Bedford, MA) and the matrigel invasion (BD Biosciences, San Diego, CA, USA) assay, respectively. Cells were seeded to cell culture inserts (Corning Costar Corp) for incubation for 24 hrs at 37°C and then fixed using 4% paraformaldehyde. They were then stained using 0.1% crystal violet solution. The cells above the upper surface were cleared. The number of migratory cells was counted with the microscope.
Dual luciferase reporter assay
StarBase v2.0 (http://starbase.sysu.edu.cn/) was used to predict the binding sites between LncRNA-CRNDE and miR-384. HEK293T cells (5 × 104 cells/well) were co-transfected with the constructed plasmids using Lipofectamine 2000 (a renilla plasmid as internal reference) for 48 hrs. According to the manufacturer’s instructions, the luciferase activities were detected by Dual-Luciferase Reporter Assay kit (Promega).
Statistical analysis
Statistical significance was analyzed by Student’s t-test and one-way analysis of variance (ANOVA) using SPSS 15.0 software and GraphPad (GraphPad Prism Software, La Jolla, CA, USA). All results were shown as the means ± SD. P < 0.05 was considered statistically significant.
Results
The expression level of lncRNA-CRNDE was increased in human HCC cell lines and tissues
The qRT-PCR results show that the expression level of lncRNA-CRNDE was significantly increased in seven HCC cell lines (MHCC97H, Hep3B, MHCC97L, BEL-7402, QGY-7703, HCCC9810, and HuH7) compared with immortalized normal liver epithelial cell lines (THLE3) (Figure 1A). Moreover, we detect the expression of lncRNA-CRNDE in 87 pairs of HCC tissues compared to paired adjacent normal tissues by qRT-PCR (Figure 1B). As shown in Table 1, the expression level (ΔCt) of lncRNA-CRNDE was related to the clinicopathological factors in HCC tissues. The ROC curve was generated, and the area under the ROC curve of lncRNA-CRNDE was 0.699 (P < 0.0001), which implies that it may serve as an ideal biomarker for HCC diagnosis (Figure 1C). Taken together, these results demonstrated that lncRNA-CRNDE was upregulated in HCC.
Figure 1.
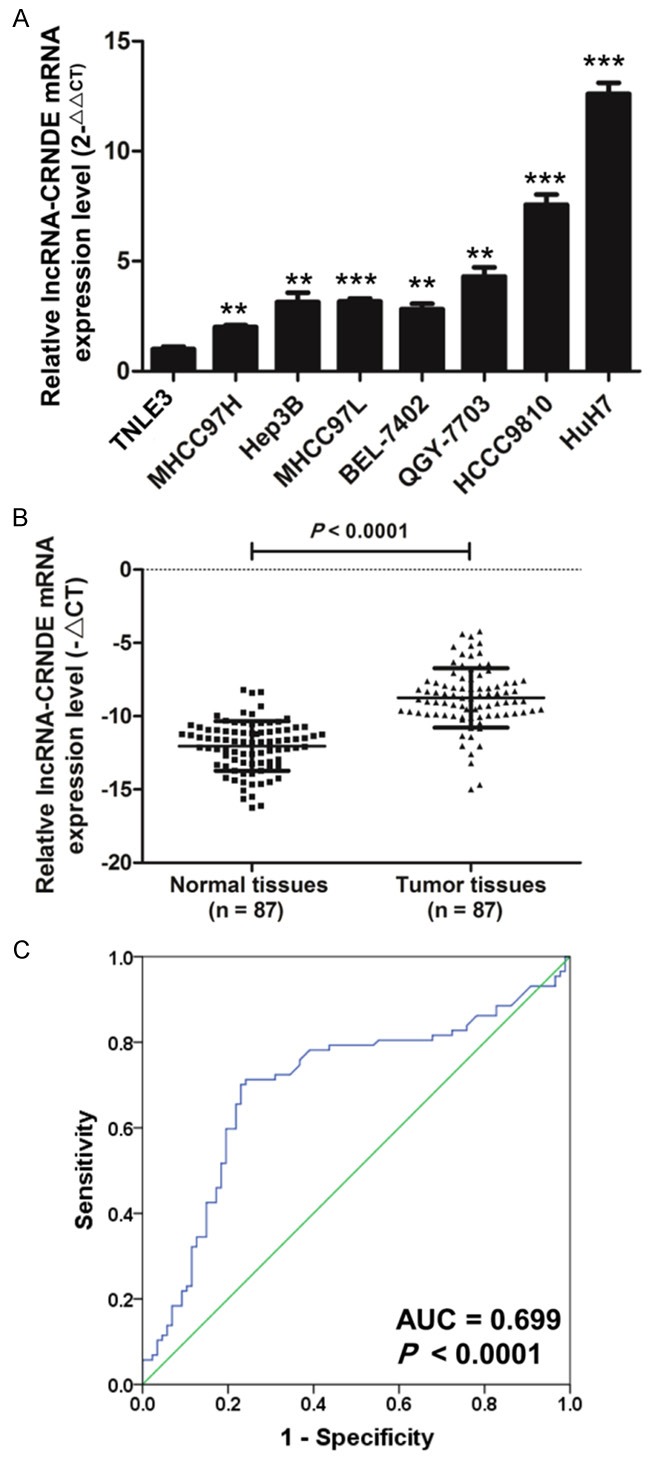
The expression level of lncRNA-CRNDE increases in human HCC cell lines and tissues. A. The expression level of lncRNA-CRNDE was detected by qRT-PCT in THLE3 and HCC cell lines (MHCC97H, Hep3B, MHCC977L, BEL-7402, QGY-7703, HCCC9810, and HuH7), U6 was used as an internal reference, (*P < 0.05, **P < 0.01, ***P < 0.001). B. The qRT-PCR was used to detect the mRNA expression level of lncRNA-CRNDE in 87 pairs of HCC tissues and paired adjacent normal tissues. C. The cut-off score of lncRNA-CRNDE expression was evaluated by a receiver operating characteristic curve (ROC) analysis.
Table 1.
The relationship of lncRNA-CRNDE and miR-384 expression levels (ΔCt) with clinicopathological factors in HCC tissues
| Characteristics | No. of patients (%) | LncRNA-CRNDE | miR-384 | ||
|---|---|---|---|---|---|
|
|
|||||
| Mean ± SD | P value | Mean ± SD | P value | ||
| Total no. of patients | 87 | ||||
| Age (year) | |||||
| > 60 | 38 (43.7) | 10.65 ± 2.23 | 0.09 | 14.11 ± 1.88 | 0.27 |
| ≤ 60 | 49 (56.3) | 11.87 ± 1.67 | 13.64 ± 1.62 | ||
| Gender | |||||
| Male | 35 (40.2) | 11.74 ± 2.12 | 0.08 | 14.96 ± 1.97 | 0.75 |
| Female | 52 (59.8) | 11.23 ± 2.32 | 14.82 ± 1.38 | ||
| Lymphatic metastasis | |||||
| N0 | 45 (51.7) | 11.93 ± 2.03 | 0.012* | 15.47 ± 1.84 | 0.007** |
| N1-N2 | 42 (48.3) | 9.65 ± 1.81 | 13.55 ± 1.49 | ||
| Distal metastasis | |||||
| M0 | 58 (66.7) | 11.58 ± 2.27 | 0.07 | 13.87 ± 1.80 | 0.47 |
| M1 | 29 (33.3) | 10.17 ± 1.94 | 14.47 ± 1.38 | ||
| TNM stage | |||||
| 0 & I & II | 49 (56.3) | 11.93 ± 2.03 | 0.012* | 14.87 ± 1.84 | 0.021* |
| III & IV | 38 (43.7) | 10.65 ± 1.81 | 13.15 ± 1.49 | ||
P < 0.05;
P < 0.01.
LncRNA-CRNDE promotes HCC cell proliferation, migration, and invasion in vitro
To investigate the biological function of lncRNA-CRNDE in the development process of HCC cells, we first detected the expression of lncRNA-CRNDE in MHCC97H cells transfected with lncRNA-CRNDE or the control by qRT-PCR. The results showed that lncRNA-CRNDE expression was significantly increased in MHCC97H cells transfected with lncRNA-CRNDE vector with lentivirus compared with the control (P < 0.001) (Figure 2A). Simultaneously, lncRNA-CRNDE expression was significantly decreased in HuH7 cells that were transfected with shRNA-CRNDE compared with the scramble (P < 0.001) (Figure 2B). Second, we measured the effect of lncRNA-CRNDE on the cell ability of HCC by MTT. As shown in Figure 2C, the overexpression of lncRNA-CRNDE promoted the proliferation of MHCC97H cells (P < 0.001). Meanwhile, the reduction of lncRNA-CRNDE markedly suppressed the proliferation of HuH7 cells (P < 0.001) (Figure 2D). Finally, we detected the effect of lncRNA-CRNDE on the ability of HCC cell migration and invasion by transwell assays. As shown in Figure 2E, the number of migratory and invasive MHCC97H cells was significantly increased when cells were transfected with the overexpression vector of lncRNA-CRNDE (P < 0.001). In addition, the number of migratory and invasive HuH7 cells were significantly decreased when transfected with shlncRNA-CRNDE (P < 0.001) (Figure 2F).
Figure 2.
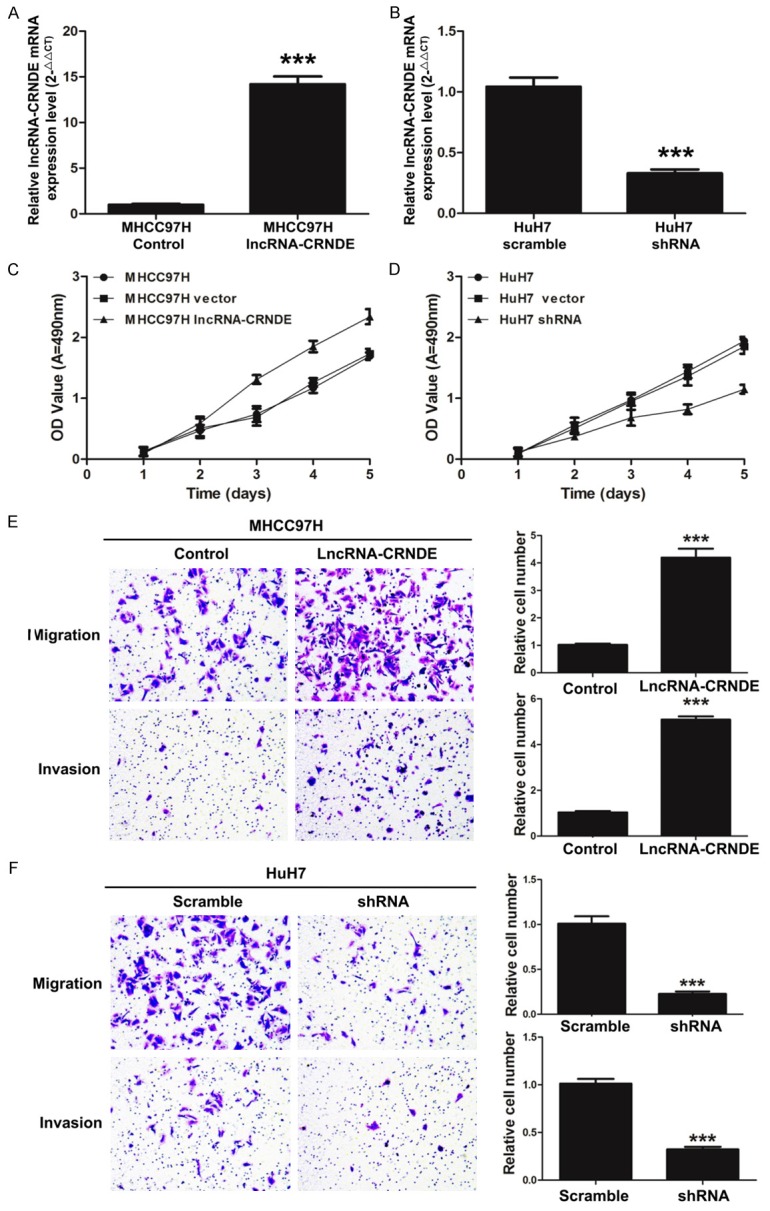
LncRNA-CRNDE promotes HCC cell proliferation, migration and invasion. A. The mRNA expression level of lncRNA-CRNDE was detected by qRT-PCR in MHCC97H cells transfected with lncRNA-CRNDE or the control (***P < 0.001). B. The mRNA expression level of lncRNA-CRNDE was detected by qRT-PCR in HuH7 cells transfected with shRNA-CRNDE or the scramble (***P < 0.001). C. The proliferation ability was measured by MTT assays in MHCC97H cells transfected with lncRNA-CRNDE or the control, (***P < 0.001). D. The silencing of lncRNA-CRNDE significantly inhibited cell proliferation. The proliferation ability was measured by MTT assays in HuH7 cells transfected with shRNA-CRNDE or the scramble (***P < 0.001). E. The overexpression of lncRNA-CRNDE significantly accelerated the migration and invasion ability of MHCC97H cells. Migrated and invasive cells were stained with crystal violet solution, and the quantification of migrated and invasive cells was shown; magnification 200 ×, ***P < 0.001. F. The silencing of lncRNA-CRNDE significantly inhibited migration and invasion ability of HuH7 cells. Migrated and invasive cells were stained with crystal violet solution, and the quantification of migrated and invasive cells was shown; magnification 200 ×, ***P < 0.001.
lncRNA-CRNDE negatively regulated miR-384 in human HCC
The expression level of miR-384 was analyzed by qRT-PCR in 87 pairs of HCC tissues and paired adjacent normal tissues to determine if miR-384 is associated with the development process of HCC and lncRNA-CRNDE expression. The expression level of miR-384 was significantly lower in HCC tissues compared to paired adjacent normal tissues (P < 0.001) (Figure 3A). A correlation analysis found that there was a negative correlation between miR-384 and lncRNA-CRNDE mRNA by qRT-PCR in 87 pairs of HCC tissues and paired adjacent normal tissues (R2 = 0.1335, P = 0.0005) (Figure 3B). Furthermore, we indicated that the expression level of miR-384 was significantly decreased in MHCC97H cells transfected with lncRNA-CRNDE compared with the control (P < 0.001), and the miR-384 expression was significantly increased when cells were transfected with miR-384 mimics again (P < 0.01) (Figure 3C). Simultaneously, we found that the expression level of miR-384 was significantly increased in HuH7 cells transfected with shRNA-CRNDE compared with the control (P < 0.01) and was significantly decreased when transfected with miR-384 inhibitors (P < 0.01) (Figure 3D). So, we further presumed that there is a binding site between miR-384 and lncRNA-CRNDE. StarBase v2.0 (http://starbase.sysu.edu.cn/) was used to predict the presumptive binding sites between LncRNA-CRNDE and miR-384. According to the results of the luciferase report assay, we showed that there was a significant decrease of the luciferase activities between the co-transfection of miR-384 mimics and wild-type lncRNA-CRNDE, with no changes in mutant lncRNA-CRNDE (Figure 3E). We also indicated that the proliferation ability was significantly increased in MHCC97H cells transfected with lncRNA-CRNDE compared with the control (P < 0.001), but it was significantly decreased in cells that were transfected with miR-384 mimics in combination (P < 0.001) (Figure 3F). Furthermore, we found that the proliferation ability of MHCC97H cells transfected with lncRNA-CRNDE was significantly decreased in HuH7 cells transfected with shRNA-CRNDE compared with the control (P < 0.001), yet, proliferation was significantly increased in cells transfected with miR-384 inhibitors in combination (P < 0.001) (Figure 3G). In short, these results showed that lncRNA-CRNDE promotes HCC cell proliferation though inhibition of miR-384.
Figure 3.
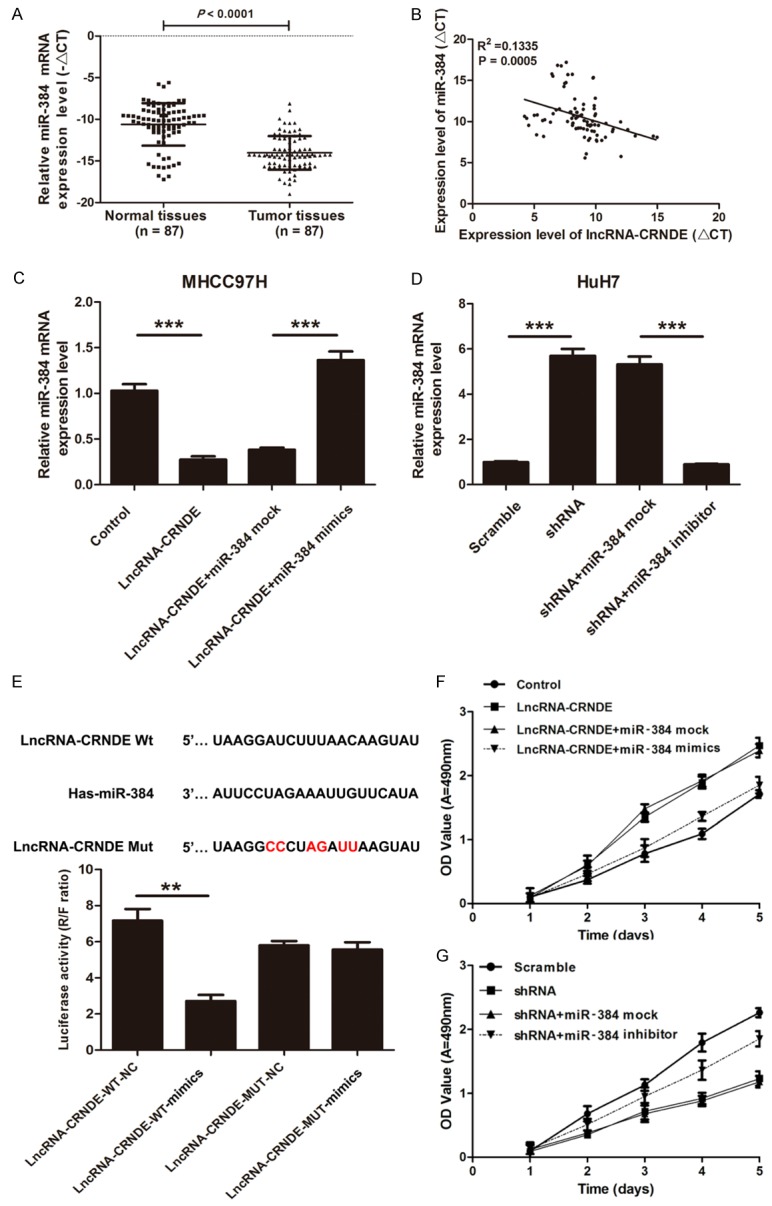
lncRNA-CRNDE negatively regulates miR-384 in human HCC. A. The qRT-PCR was used to detect the mRNA expression level of miR-384 in HCC tissues and paired adjacent normal tissues. The levels of transcript were normalized to U6 snRNA, (n = 87, ***P < 0.001). B. The correlation between lncRNA-CRNDE and miR-384 expression were measured (r = 0.546, P < 0.001). C. The mRNA expression level of miR-384 was measured by qRT-PCR in MHCC97H cells transfected with lncRNA-CRNDE or the control and in combination with miR-384 mock or mimics, (**P < 0.01, ***P < 0.001). D. The expression level of miR-384 was measured by qRT-PCR in HuH7 cells transfected with shRNA-CRNDE or the scramble and in combination with miR-384 mock or inhibitors, (**P < 0.01). E. The alleged binding sites of miR-384 on the lncRNA-CRNDE 3’UTR (Wt and Mut) were shown. The luciferase reporter assay was used to detect the activity of lncRNA-CRNDE in HEK 293T cells cotransfected with lncRNA-CRNDE (Wt or Mut) and miR-384 mimics or mock, (**P < 0.01, ***P < 0.001). F. The miR-384 mimics significantly decreased lncRNA-CRNDE-mediated cell proliferation. The proliferation ability was measured by MTT assays in MHCC97H cells transfected with lncRNA-CRNDE or the control, and then transfected with miR-384 mock or mimics, (***P < 0.001). G. The miR-384 inhibitors significantly promoted shRNA-CRNDE-mediated cell proliferation. The proliferation ability was measured by MTT assays in HuH7 cells transfected with shRNA-CRNDE or the scramble and then transfected with miR-384 mock or inhibitors (***P < 0.001).
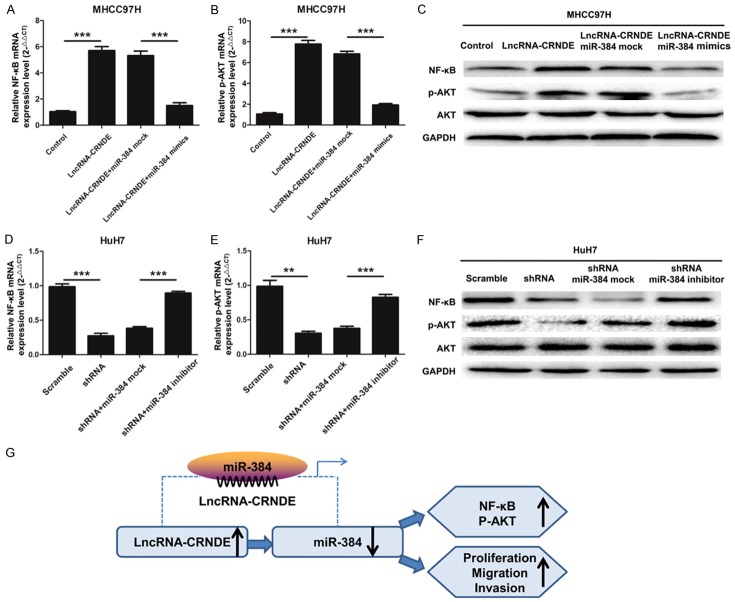
LncRNA-CRNDE regulates NF-κB and p-AKT expression
In order to study the molecular mechanism lncRNA-CRNDE in HCC, qRT-PCR and Western blot experiments were used to detect the mRNA and protein expression levels of NF-κB and p-AKT in HCC cells. As shown in Figure 4A, the qRT-PCR results indicated that the mRNA expression level of NF-κB was significantly up-regulated in MHCC97H cells transfected with lncRNA-CRNDE compared with the control (P < 0.001) and was down-regulated when cells were transfected with miR-384 mimics in combination (P < 0.001). As shown in Figure 4B, p-AKT has a similar reduction to NF-κB. Moreover, the protein levels of NF-κB and p-AKT also show this effect in transfected HCC cells (Figure 4C). As shown in Figure 4D, the qRT-PCR results indicated that the mRNA expression level of NF-κB was significantly down-regulated in HuH7 cells transfected with shRNA-CRNDE compared with the control (P < 0.001) and was significantly up-regulated in cells transfected with miR-384 inhibitors in combination (P < 0.001). As shown in Figure 4E, the mRNA expression level of p-AKT has a similar increasing trend. Likewise, the protein levels of NF-κB and p-AKT also have this effect in transfected HCC cells (Figure 4F). These data indicate that LncRNA-CRNDE accelerated the expression levels of NF-κB and p-AKT and promotes HCC tumorigenesis though inhibition of miR-384 (Figure 4G).
Figure 4.
LncRNA-CRNDE regulates NF-κB and p-AKT expression. (A) The mRNA expression level of NF-κB was measured by qRT-PCR in MHCC97H cells transfected with lncRNA-CRNDE or the control and then transfected with miR-384 mock or mimics, (***P < 0.001). (B) The mRNA expression level of p-AKT was measured by qRT-PCR in MHCC97H cells treated as A, (***P < 0.001). (C) The protein expression levels of NF-κB, AKT and p-AKT were detected by Western blotting in MHCC97H cells treated as A; GAPDH was used as a protein-loading control. (D) The mRNA expression level of NF-κB was measured by qRT-PCR in HuH7 cells transfected with shRNA-CRNDE or the scramble and then transfected with miR-384 mock or inhibitor, (**P < 0.01, ***P < 0.001). (E) The mRNA expression level of p-AKT was measured by qRT-PCR in HuH7 cells treated as D (**P < 0.01, ***P < 0.001). (F) The protein expression levels of NF-κB, AKT and p-AKT were detected by Western blotting in HuH7 cells treated as (D); GAPDH was used as a protein-loading control. (G) A diagrammatic sketch of lncRNA-CRNDE functions in HCC. LncRNA-CRNDE promoted HCC cell proliferation, migration, and invasion though inhibition of miR-384.
Discussion
By reasons of poor prognosis, HCC is the most common type of liver cancer and is the major leading cause of cancer mortality all over the world [14,15]. No completely effective treatment is available for primary liver cancer, however, the use of conventional radiotherapy and chemotherapy shows promise for prognosis [16,17]. To date, studies indicate that several signaling pathways that are associated with biological functions participated in the development and progression of HCC [18,19]. Particularly, the abnormally expressed proteins that are closely related to proliferation, migration, and invasion have been shown to be the main culprit in the formation process of liver cancer [20,21]. Therefore, analyzing potential biomarkers is the most direct and effective way to explore the molecular mechanism and function for HCC.
Non-coding RNAs (ncRNAs) are segmented into two types, small ncRNAs and long non-coding RNAs (lncRNAs). As a class of small ncRNAs, miRNAs regulate the post-transcriptional level of target mRNAs resulting in mRNAs degradation [22,23]. Some studies indicate that miR-122 [24], miR-372 [25], miR-375 [26] all participate in the process of liver cancer. LncRNA also has been shown to play important roles in gene regulation, such as epigenetic, transcriptional, posttranscriptional, and translational functions [27,28]. Numerous studies have indicated that lncRNAs can act as competitive endogenous RNA (ceRNA) and play key roles in the occurrence and development of copious diseases [29,30]. In this study, we found that the expression level of lncRNA-CRNDE was increased in human HCC. LncRNA-CRNDE promoted HCC cell proliferation, migration and invasion in vitro. Furthermore, we found that the upregulated lncRNA-CRNDE could act as a sponge for negatively regulated miR-384 and promoted HCC cells tumorigenesis.
NF-κB transcription factor, which acts as a regulator in cell survival, is involved in cancer development by encouraging a major inflammatory pathway activated during liver injury [31]. PI3K-Akt signaling pathway is regarded as a critical cause for tumorigenesis and plays major roles in cancer cell growth, survival, and proliferation. Members of the Akt pathway may become new targets for cancer treatment [32]. In our study, we first indicate that the lncRNA-CRNDE/miR-384 axis regulates these two important factors in HCC cell proliferation, invasion, and migration.
In conclusion, this study can be summarized by following major findings: 1) lncRNA-CRNDE was unregulated in HCC tissues and cells and therefore has a positive correlation with clinicopathological features. 2) lncRNA-CRNDE was negatively correlated with miR-384 and accelerated the expression levels of NF-κB and p-AKT though inhibition of miR-384.
Acknowledgements
This study was Supported by the National key clinical specialist construction Programs of China, Nation Health Office Medical Case, No. (2013) 544.
Disclosure of conflict of interest
None.
References
- 1.Woerns MA, Weinmann A, Schuchmann M, Galle PR. Systemic therapies in hepatocellular carcinoma. Dig Dis. 2009;27:175–188. doi: 10.1159/000218351. [DOI] [PubMed] [Google Scholar]
- 2.Spangenberg HC, Thimme R, Blum HE. Targeted therapy for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2009;6:423–432. doi: 10.1038/nrgastro.2009.86. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Hanazaki K, Kajikawa S, Shimozawa N, Mihara M, Shimada K, Hiraguri M, Koide N, Adachi W, Amano J. Survival and recurrence after hepatic resection of 386 consecutive patients with hepatocellular carcinoma. J Am Coll Surg. 2000;191:381–388. doi: 10.1016/s1072-7515(00)00700-6. [DOI] [PubMed] [Google Scholar]
- 5.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Morales DR, Thomas K, Presser A, Bernstein BE, van Oudenaarden A. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao W, An Y, Liang Y, Xie X. Role of HOTAIR long noncoding RNA in metastatic progression of lung cancer. Eur Rev Med Pharmacol Sci. 2014;18:1930–1936. [PubMed] [Google Scholar]
- 9.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ouyang L, Shi Z, Zhao S, Wang FT, Zhou TT, Liu B, Bao JK. Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012;45:487–498. doi: 10.1111/j.1365-2184.2012.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Serag HB, Mason AC. Rising incidence of hepatocellular carcinoma in the United States. N Engl J Med. 1999;340:745–750. doi: 10.1056/NEJM199903113401001. [DOI] [PubMed] [Google Scholar]
- 15.Lupberger J, Hildt E. Hepatitis B virus-induced oncogenesis. World J Gastroenterol. 2007;13:74. doi: 10.3748/wjg.v13.i1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Llovet JM. Updated treatment approach to hepatocellular carcinoma. J Gastroenterol. 2005;40:225–235. doi: 10.1007/s00535-005-1566-3. [DOI] [PubMed] [Google Scholar]
- 17.Fukuda S, Okuda K, Imamura M, Imamura I, Eriguchi N, Aoyagi S. Surgical resection combined with chemotherapy for advanced hepatocellular carcinoma with tumor thrombus: report of 19 cases. Surgery. 2002;131:300–310. doi: 10.1067/msy.2002.120668. [DOI] [PubMed] [Google Scholar]
- 18.Iizuka N, Oka M, Yamada-Okabe H, Mori N, Tamesa T, Okada T, Takemoto N, Hashimoto K, Tangoku A, Hamada K. Differential gene expression in distinct virologic types of hepatocellular carcinoma: association with liver cirrhosis. Oncogene. 2003;22:3007–3014. doi: 10.1038/sj.onc.1206401. [DOI] [PubMed] [Google Scholar]
- 19.Xu XR, Huang J, Xu ZG, Qian BZ, Zhu ZD, Yan Q, Cai T, Zhang X, Xiao HS, Qu J. Insight into hepatocellular carcinogenesis at transcriptome level by comparing gene expression profiles of hepatocellular carcinoma with those of corresponding noncancerous liver. Proc Natl Acad Sci U S A. 2001;98:15089–15094. doi: 10.1073/pnas.241522398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu TT, Hsieh YH, Hsieh YS, Liu JY. Reduction of PKCα decreases cell proliferation, migration, and invasion of human malignant hepatocellular carcinoma. J Cell Biochem. 2008;103:9–20. doi: 10.1002/jcb.21378. [DOI] [PubMed] [Google Scholar]
- 21.Saxena NK, Sharma D, Ding X, Lin S, Marra F, Merlin D, Anania FA. Concomitant activation of the JAK/STAT, PI3K/AKT, and ERK signaling is involved in leptin-mediated promotion of invasion and migration of hepatocellular carcinoma cells. Cancer Res. 2007;67:2497–2507. doi: 10.1158/0008-5472.CAN-06-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 23.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 24.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, Chen N, Sun F, Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He X, Chang Y, Meng F, Wang M, Xie Q, Tang F, Li P, Song Y, Lin J. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene. 2012;31:3357–3369. doi: 10.1038/onc.2011.500. [DOI] [PubMed] [Google Scholar]
- 27.Qiu MT, Hu JW, Yin R, Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol. 2013;34:613–620. doi: 10.1007/s13277-013-0658-6. [DOI] [PubMed] [Google Scholar]
- 28.Eades G, Zhang YS, Li QL, Xia JX, Yao Y, Zhou Q. Long non-coding RNAs in stem cells and cancer. World J Clin Oncol. 2014;5:134–141. doi: 10.5306/wjco.v5.i2.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859:169–176. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 30.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karin M. NF-κB as a critical link between inflammation and cancer. Cold Spring Harb Perspect Biol. 2009;1:a000141. doi: 10.1101/cshperspect.a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, Hennessy BT, Tseng H, Pochanard P, Kim SY. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]