Abstract
Ginsenoside F2 (F2), a protopanaxdiol type of saponin, was reported to inhibit human gastric cancer cells SGC7901. To better understand the molecular mechanisms of F2, an iTRAQ-based proteomics approach was applied to define protein expression profiles in SGC7901 cells in response to lower dose (20 μM) and shorter duration (12 hour) of F2 treatment, compared with previous study. 205 proteins were screened in terms of the change in their expression level which met our predefined criteria. Further bioinformatics and experiments demonstrated that F2 treatment downregulated PRR5 and RPS15 and upregulated RPL26, which are implicated in ribosomal protein-p53 signaling pathway. F2 also inhibited CISD2, Bcl-xl, and NLRX1, which are associated with autophagic pathway. Furthermore, it was demonstrated that F2 treatment increased Atg5, Atg7, Atg10, and PUMA, the critical downstream effectors of ribosomal protein-p53 signaling pathway, and Beclin-1, UVRAG, and AMBRA-1, the important molecules in Bcl-xl/Beclin-1 pathway. The 6 differentially abundant proteins, PRR5, CISD2, Bcl-xl, NLRX1, RPS15, and RPL26, were confirmed by western blot. Taken together, ribosomal protein-p53 signaling pathway and Bcl-xl/Beclin-1 pathway might be the most significantly regulated biological process by F2 treatment in SGC7901 cells, which provided valuable insights into the deep understanding of the molecular mechanisms of F2 for gastric cancer treatment.
1. Introduction
Gastric cancer is the fifth most common cancer and the third leading cause of cancer-related death worldwide. Annually it results in approximately 700,000 deaths [1]. Currently, chemotherapy has proved to decrease the rate of recurrence and improve overall survival; however, the drug resistance and serious toxic side effects largely reduce therapeutic efficacy and quality of life in patients [2, 3]. In recent years, compounds of natural products have caught wide attention due to their promising anticancer effects and minimal side effects [4–7]. Therefore, it is very necessary to develop new optimal anticancer agent from natural resource [3].
Ginsenosides, the major bioactive constituents in ginseng, have been demonstrated to exert potential anticancer ability [4, 5]. Exploration of ginsenoside as a new anticarcinogenic agent is of much interest [4–7]. Structural-function studies showed that the increased antitumor effect is implicated with the decrease of its sugar number [5]. Sugar moiety at C-6 significantly reduces the anticancer activities of ginsenosides. Ginsenoside F2 (see structure in Figure 1), a protopanaxdiol type ginsenoside with one sugar molecular at C-3 and one sugar molecule at C-20, has been shown to be potent in inhibiting tumorigenesis in several different cancers including gastric tumor and glioblastoma multiforme [6, 7]. Recently, our in vitro and in vivo studies demonstrated that ginsenoside F2 possesses anticancer effects in human gastric carcinoma cells SGC7901 [6]. However, the involved exact mechanisms of ginsenoside F2 on SGC7901 cancer cells at proteome level have not been systemically investigated.
Figure 1.
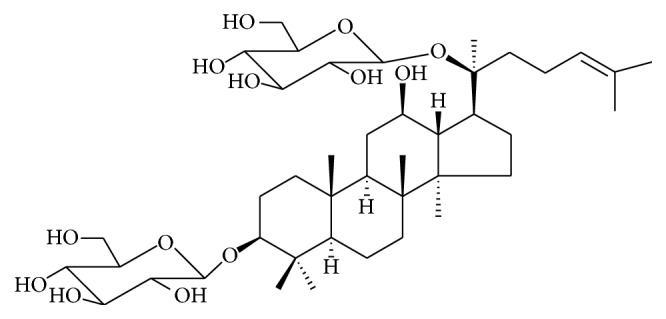
Structure of ginsenoside F2.
Advancements in the field of proteomics have made it possible to accurately monitor and quantitatively detect the changes of protein expression in response to drug treatment. The achieved data provide valuable insights into the molecular mechanisms of disease and help to identify therapeutic targets [8]. Isobaric tag for relative and absolute quantification (iTRAQ) is a robust mass spectrometry technique that allows quantitative comparison of protein abundance by measuring peak intensities of reporter ions released from iTRAQ-tagged peptides by fragmentation. iTRAQ with multiplexing capability up to eight distinct samples in a single experiment and relatively higher sensitivity has gained significant interest in the field of quantitative proteomics. In the present study, SGC7901 cells treated by lower dose and a shorter duration than that in previous report were analyzed by iTRAQ-based proteomics integrated with bioinformatics using Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Cluster of Orthologous Groups (COG) of proteins database. And network analysis was applied to identify critical molecules which are involved in anticancer mechanisms of ginsenoside F2 in gastric SGC7901 cells. General molecular biological techniques such as western blot were utilized for validation.
2. Materials and Methods
2.1. Reagents and Antibodies
Ginsenoside F2 was isolated previously from leaves of Panax ginseng by a series of chromatographic procedures [9]. Ginsenoside F2 has a molecular mass of 784 Da and was isolated with 98% purity. Primary antibodies of PRR5, CISD2, Bcl-2L, NLRX1, RPS15, RPL26, p53, PUMA, Beclin-1, UVRAG, AMBRA-1, mTOR, LC3-II, LC3-I, and β-actin together with all secondary antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA). The Atg5, Atg7, and Atg10 antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
2.2. Cell Culture and Treatment
SGC7901 cells were purchased from American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium (Hyclone) supplemented with 10% fetal bovine serum (FBS), 100 μg/mL streptomycin, and 100 μg/mL penicillin and grown at 37°C in 5% carbon dioxide.
2.3. Protein Preparation
In one of our recent reports [6], we have shown that the IC50 of ginsenoside F2 is in <50 μM in 24 hours. In order to characterize ginsenoside F2-related mechanism it is imperative to use samples that are at the early stages of ginsenoside F2 treatment. So, a lower dose than the IC50 (20 μM) and a shorter duration (12 hours in the study) were chosen in the study. The treated (20 μM) and untreated SGC7901 cells were suspended in the lysis buffer and sonicated in ice. The proteins were reduced with 10 μM DTT (final concentration) at 56°C for 1 h and then alkylated by 55 mM iodoacetamide (IAM) (final concentration) in the darkroom for 1 h. The reduced and alkylated protein mixtures were precipitated by adding 4x volume of chilled acetone at −20°C overnight. After centrifugation at 4°C, 30 000 ×g, the pellet was dissolved in 0.5 M triethylammonium bicarbonate (TEAB) (Applied Biosystems, Milan, Italy) and sonicated in ice. After centrifuging at 30000 ×g at 4°C, the supernatants were collected, and the total protein concentration was determined using a Bradford protein assay kit (BioRad, Hercules, CA, USA). The proteins in the supernatant were kept at −80°C for further analysis.
2.4. iTRAQ Labeling and SCX Fractionation
Total protein (100 μg) was taken out of each sample solution and then the protein was digested with Trypsin Gold (Promega, Madison, WI, USA) with the ratio of protein : trypsin = 30 : 1 at 37°C for 16 hours. iTRAQ labeling was performed according to the iTRAQ Reagents-8plex labeling manual (AB SCIEX, Madrid, Spain). Briefly, one unit of iTRAQ reagent was thawed and reconstituted in 24 μL isopropanol. iTRAQ labels 113 were used to label control sample separately, and 115 and 117 were used to label twice F2-treated samples for duplicated experiment. The peptides were labeled with the isobaric tags, incubated at room temperature for 2 h. The labeled peptide mixtures were then pooled and dried by vacuum centrifugation.
The mixed peptides were fractionated by strong cation exchange (SCX) chromatography on a LC-20AB HPLC Pump system (Shimadzu, Kyoto, Japan). The iTRAQ labeled peptide mixtures were reconstituted with 4 mL buffer A (25 mM NaH2PO4 in 25% acetonitrile, pH 2.7) and loaded onto a 4.6 × 250 mm Ul tremex SCX column containing 5 μm particles (Phenomenex). The peptides were eluted at a flow rate of 1 mL/min with a gradient of buffer A for 10 min, 5–60% buffer B (25 mM NaH2PO4, 1 M KCl in 25% acetonitrile, pH 2.7) for 27 min, and 60–100% buffer B for 1 min. The system was then maintained at 100% buffer B for 1 min before equilibrating with buffer A for 10 min prior to the next injection. Elution was monitored by measuring the absorbance at 214 nm, and fractions were collected at 1-minute intervals. The eluted peptides were pooled into 20 fractions, desalted with a Strata X C18 column (Phenomenex), and vacuum-dried. The cleaned fractions were then lyophilized again and stored at −20°C until analyzed by mass spectrometry.
2.5. LC-ESI-MS/MS Analysis Based on Q EXACTIVE
Each fraction was resuspended in buffer A (2% acetonitrile, 0.1% FA) and centrifuged at 20 000 ×g for 10 min. In each fraction, the final concentration of peptide was about 0.5 μg/μL. 10 μL supernatant was loaded on a LC-20AD nano-HPLC (Shimadzu, Kyoto, Japan) by the autosampler onto a 2 cm C18 trap column. Then, the peptides were eluted onto a 10 cm analytical C18 column (inner diameter 75 μm) packed in-house. The samples were loaded at 8 μL/min for 4 min; then the 44 min gradient was run at 300 nL/min starting from 2 to 35% B (98% acetonitrile, 0.1% FA), followed by 2-minute linear gradient to 80%, maintenance at 80% B for 4 min. Initial chromatographic conditions were restored in 1 min.
Data acquisition was performed with tandem mass spectrometry (MS/MS) in a Q EXACTIVE (Thermo Fisher Scientific, San Jose, CA) coupled online to the HPLC. Intact peptides were detected in the Orbitrap at a resolution of 70 000. Peptides were selected for MS/MS using high-energy collision dissociation (HCD) operating mode with a normalized collision energy setting of 27.0; ion fragments were detected in the Orbitrap at a resolution of 17500. In the octopole collision cell, the ten most intense peptide ions (charge states ≥ 2) were sequentially isolated to a maximum target value of 5 × 105 by pAGC and fragmented HCD. A data-dependent procedure that alternated between one MS scan and 15 MS/MS scans was applied for the 15 most abundant precursor ions above a threshold ion count of 20000 in the MS survey scan with a following Dynamic Exclusion duration of 15 s. The electrospray voltage applied was 1.6 kV. Automatic gain control (AGC) was used to optimize the spectra generated by the Orbitrap. A sweeping collision energy setting of 35 ± 5 eV was applied to all precursor ions for collision-induced dissociation. The AGC target for full MS was 3e6 and 1e5 for MS2. For MS scans, the m/z scan range was 350 to 2000 Da. For MS2 scans, the m/z scan range was 100–1800 Da. The iTRAQ experiments were performed as three technical replicates to gather reliable quantitative information.
2.6. Data Analysis
Raw data files acquired from the Orbitrap were converted into MGF files using Proteome Discoverer 1.2 (PD 1.2, Thermo) [5600 msconverter] and the MGF files were searched. Protein identifications were performed by using Mascot search engine (Matrix Science, London, UK; version 2.3.02) against database containing 143397 sequences.
For protein identification and quantification, a peptide mass tolerance of 20 ppm was allowed for intact peptide masses and 0.05 Da for fragmented ions, with allowance for one missed cleavage in the trypsin digests. Carbamidomethylation of cysteine was considered a fixed modification, and the conversion of N-terminal glutamine to pyroglutamic acid and methionine oxidation were considered variable modifications. All identified peptides had an ion score above the Mascot peptide identity threshold, and a protein was considered identified if at least one such unique peptide match was apparent for the protein. To reduce the probability of false peptide identification, only peptides at the 95% confidence interval by a Mascot probability analysis greater than “identity” were counted as identified. The quantitative protein ratios were weighted and normalized by the median ratio in Mascot. We set a 1.2-fold change as the threshold and a p value must be below 0.05 to identify significant changes.
2.7. Function Method Description
Functional annotations of the proteins were conducted using Blast2 GO program against the nonredundant protein database (NR; NCBI). The KEGG database (http://www.genome.jp/kegg/) and the COG database (http://www.ncbi.nlm.nih.gov/COG/) were used to classify and group these identified proteins.
GO is an international standardization of gene function classification system. It provides a set of dynamic updating controlled vocabulary to describe genes and gene products attributes in the organism. GO has 3 ontologies which can describe molecular function, cellular component, and biological process, respectively.
COG is the database for protein orthologous classification. Every protein in COG is supposed to derive from a same protein ancestor.
KEGG PATHWAY is a collection of manually drawn pathway maps representing our knowledge on the molecular interaction and reaction networks. Molecules are represented as nodes, and the biological relationship between two nodes is represented as an edge (line).
2.8. Western Blot
Western blot analyses were performed to confirm the presence of differentially expressed proteins. After the treatment of the indicated concentration of ginsenoside F2 (10, 20, and 40 μM) for 12 h, cells were harvested, washed with cold PBS (pH 7.4), and lysed with ice-cold lysis buffer (50 μM Tris-HCl, 150 μM NaCl, 1 μM EGTA, 1 μM EDTA, 20 μM NaF, 100 μM Na3VO4, 1%NP40, 1 μM PMSF, 10 μg/mL aprotinin, and 10 μg/mL leupeptin, pH 7.4) for 30 min and centrifuged at 12 000 ×g for 30 min at 4°C. The protein concentration of the clear supernatant was quantified using Bio-Rad Protein Assay Kit.
Approximately 30 μg of protein was loaded into a 10–15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE). Thereafter, proteins were electrophoretically transferred to nitrocellulose membrane and nonspecific sites were blocked with 5% skimmed milk in 1% Tween-20 (Sigma-Aldrich) in 20 μM TBS (pH 7.5) and reacted with a primary polyclonal antibody, PRR5, CISD2, Bcl-2L, NLRX1, RPS15, RPL26, p53, Atg5, Atg7, Atg10, LC3-II, LC3-I PUMA, Beclin-1, UVRAG, and mTOR and β-actin for 4 h at room temperature. After washing with TBS three times (5 min each), the membrane was then incubated with alkaline phosphatase-conjugated goat anti-rabbit secondary antibody. The signal was observed and developed with Kodak film by exposure to enhanced chemiluminescence (ECL) plus western Blotting Detection Reagents (Amersham Biosciences, Piscataway, NJ, USA).
2.9. Statistical Analysis
For cell-based assay, experiments were performed in duplicate and three independent experiments were performed. Western blot analyses of differential protein expressions were validated on cell lysates from three biological replicates. Statistical significance was analyzed using Student's t-test or ANOVA test by using GraphPad Prism v4.0 software (GraphPad Software, San Diego, CA, USA). Statistical significance is expressed as ∗∗∗ p < 0.001; ∗∗ p < 0.01; ∗ p < 0.05.
3. Results
3.1. Proteome Analysis
Human gastric carcinoma cells (SGC7901) are treated with ginsenoside F2 at a dose of 20 μM for 12 hours. The harvested proteins are used to perform iTRAQ for quantifying the difference of total 31853 peptides and 5411 proteins in SGC7901 cells with or without treatment. Finally, 205 proteins were screened out in terms of the change in their expression level which meet our predefined criteria of p < 0.05 with relative expression levels at least >1.2-fold (Table 1) or <0.83-fold (Table 2) (both 113/115 and 113/117) in ginsenoside F2-treated group compared with the control group. The protein properties, including pI, molecular weight (MW), and number of residues were calculated by Mascot. The results are highly reproducible in two individual experiments.
Table 1.
Differentially upregulated (>1.20-fold) proteins identified by iTRAQ in F2 treated SGC7901 cells.
| Rank # | Accession | Gene symbol (GN) | Definition (description) | Score | Mass | Cov% | Ration | COG function-description |
|---|---|---|---|---|---|---|---|---|
| Up 1 | sp|P07305-2 | H1F0 | Isoform 2 of histone H1.0 | 51 | 35582 | 13 | 2.11 | — |
| Up 2 | sp|P20962 | PTMS | Parathymosin | 503 | 15782 | 23.5 | 1.32 | — |
| Up 3 | tr|B8ZWD1 | DBI | Diazepam binding inhibitor, splice form 1A(2) | 121 | 15706 | 28.9 | 1.31 | Acyl-CoA-binding protein |
| Up 4 | sp|Q16576 | RBBP7 | Histone-binding protein RBBP7 | 877 | 55737 | 24.5 | 1.25 | FOG: WD40 repeat |
| Up 5 | sp|P46779-2 | RPL28 | Isoform 2 of 60S ribosomal protein L28 | 524 | 22107 | 27.6 | 1.35 | — |
| Up 6 | tr|B2R514 | — | cDNA, FLJ92300, Homo sapiens COP9 subunit 6 (MOV34 homolog, 34 kD) (COPS6), mRNA | 74 | 39068 | 20.2 | 1.22 | Predicted metal-dependent protease of the PAD1/JAB1 superfamily |
| Up 7 | tr|B3KY12 | — | cDNA FLJ46581 fis, clone THYMU3043200, highly similar to splicing factor 3A subunit 3 | 527 | 71859 | 22 | 1.24 | Splicing factor 3a, subunit 3 |
| Up 8 | sp|Q71DI3 | HIST2H3A | Histone H3.2 | 617 | 19694 | 26.5 | 1.40 | Histones H3 and H4 |
| Up 9 | tr|Q9P0H9 | RER1 | RER1 protein | 118 | 28927 | 22 | 1.26 | Golgi protein involved in Golgi-to-ER retrieval |
| Up 10 | tr|A8K3Q9 | — | cDNA FLJ76611, highly similar to Homo sapiens ribosomal protein L14 (RPL14), mRNA | 781 | 35114 | 25.9 | 2.24 | Ribosomal protein L14E/L6E/L27E |
| Up 11 | sp|Q9Y3A2 | UTP11L | Probable U3 small nucleolar RNA-associated protein 11 | 94 | 44174 | 21.7 | 1.30 | Uncharacterized conserved protein |
| Up 12 | tr|F2Z388 | RPL35 | 60S ribosomal protein L35 | 99 | 15372 | 32.3 | 1.35 | Ribosomal protein L29 |
| Up 13 | sp|Q9NZZ3 | CHMP5 | Charged multivesicular body protein 5 | 268 | 32218 | 21 | 1.42 | — |
| Up 14 | tr|B2R4D8 | — | 60S ribosomal protein L27 | 398 | 23061 | 36 | 1.28 | Ribosomal protein L14E/L6E/L27E |
| Up 15 | tr|M0QXF7 | C19orf10 | UPF0556 protein C19orf10 (fragment) | 265 | 11851 | 25 | 1.24 | — |
| Up 16 | tr|D3DV26 | S100A10 | S100 calcium binding protein A10 (annexin II ligand, calpactin I, light polypeptide (P11)), isoform CRA_b (fragment) | 134 | 27935 | 8.3 | 1.21 | — |
| Up 17 | tr|H7C2N1 | PTMA | Thymosin alpha-1 (fragment) | 117 | 18283 | 8.8 | 1.30 | — |
| Up 18 | tr|G2XKQ0 | — | Sumo13 | 60 | 14938 | 11.9 | 1.22 | Ubiquitin-like protein (sentrin) |
| Up 19 | tr|I3L1Y9 | FLYWCH2 | FLYWCH family member 2 | 99 | 19302 | 47.2 | 1.45 | — |
| Up 20 | tr|M0R210 | RPS16 | 40S ribosomal protein S16 | 1105 | 19391 | 57.4 | 1.27 | Ribosomal protein S9 |
| Up 21 | sp|O43715 | TRIAP1 | TP53-regulated inhibitor of apoptosis 1 | 82 | 12050 | 18.4 | 1.36 | — |
| Up 22 | sp|P49207 | RPL34 | 60S ribosomal protein L34 | 187 | 18684 | 20.5 | 1.66 | Ribosomal protein L34E |
| Up 23 | sp|Q92522 | H1FX | Histone H1x | 342 | 35250 | 25.4 | 1.33 | — |
| Up 24 | tr|J3KRX5 | RPL17 | 60S ribosomal protein L17 (fragment) | 795 | 27382 | 38.5 | 1.26 | Ribosomal protein L22 |
| Up 25 | sp|P02795 | MT2A | Metallothionein-2 | 104 | 9915 | 52.5 | 1.42 | — |
| Up 26 | tr|Q6FIE5 | PHP14 | PHP14 protein | 72 | 17301 | 8.8 | 1.27 | — |
| Up 27 | tr|A0PJ62 | RPL14 | RPL14 protein (fragment) | 536 | 21409 | 43.5 | 2.85 | Ribosomal protein L14E/L6E/L27E |
| Up 28 | tr|G3XAA2 | MAP4K4 | Mitogen-activated protein kinase kinase kinase kinase 4 | 142 | 156989 | 2.7 | 1.24 | Serine/threonine protein kinase |
| Up 29 | tr|C9JNW5 | RPL24 | 60S ribosomal protein L24 | 666 | 24642 | 32 | 1.67 | Ribosomal protein L24E |
| Up 30 | sp|Q13951 | CBFB | Core-binding factor subunit beta | 197 | 24461 | 18.1 | 1.20 | — |
| Up 31 | tr|D3DUE6 | N-PAC | Cytokine-like nuclear factor n-pac, isoform CRA_c | 219 | 76728 | 14.5 | 1.24 | 3-Hydroxyisobutyrate dehydrogenase and related beta-hydroxy acid dehydrogenases |
| Up 32 | tr|K7EKW4 | ISOC2 | Isochorismatase domain-containing protein 2, mitochondrial (fragment) | 130 | 21202 | 17.4 | 1.34 | Amidases related to nicotinamidase |
| Up 33 | sp|Q9NQ55-2 | PPAN | Isoform 2 of Suppressor of SWI4 1 homolog | 73 | 63713 | 10.7 | 1.37 | — |
| Up 34 | tr|B3KMF8 | — | cDNA FLJ10869 fis, clone NT2RP4001677 | 127 | 12398 | 27.7 | 1.28 | — |
| Up 35 | sp|P62424 | RPL7A | 60S ribosomal protein L7a | 613 | 42316 | 27.1 | 1.78 | Ribosomal protein HS6-type (S12/L30/L7a) |
| Up 36 | tr|B4E0X1 | — | Beta-2-microglobulin | 185 | 17093 | 13.1 | 1.25 | — |
| Up 37 | tr|H0Y7A7 | CALM2 | Calmodulin (fragment) | 735 | 24209 | 30.5 | 1.26 | Ca2+-binding protein (EF-Hand superfamily) |
| Up 38 | tr|J3KTJ8 | RPL26 | 60S ribosomal protein L26 (fragment) | 363 | 15545 | 34 | 1.24 | Ribosomal protein L24 |
| Up 39 | tr|B4DJM5 | — | cDNA FLJ61294, highly similar to keratin, type I cytoskeletal 17 | 326 | 21291 | 24.9 | 1.46 | — |
| Up 40 | sp|Q9Y3C1 | NOP16 | Nucleolar protein 16 | 79 | 27925 | 20.8 | 1.24 | — |
| Up 41 | sp|Q16543 | CDC37 | Hsp90 cochaperone Cdc37 | 384 | 57730 | 29.6 | 1.22 | — |
| Up 42 | sp|P16401 | HIST1H1B | Histone H1.5 | 801 | 42644 | 17.3 | 2.38 | — |
| Up 43 | sp|Q07866-3 | KLC1 | Isoform G of kinesin light chain 1 | 642 | 81828 | 23.9 | 1.24 | FOG: TPR repeat |
| Up 44 | tr|B4DKJ4 | — | cDNA FLJ57738, highly similar to translationally controlled tumor protein | 344 | 19250 | 32.4 | 1.28 | — |
Table 2.
Differentially downregulated (<0.83-fold) proteins identified by iTRAQ in F2 treated SGC7901 cells.
| Rank # | Accession | Gene symbol (GN) | Definition (description) | Score | Mass | Cov% | Ration | COG function-description |
|---|---|---|---|---|---|---|---|---|
| Down 1 | tr|F5H740 | VDAC3 | Voltage-dependent anion-selective channel protein 3 | 1114 | 39598 | 41.5 | 0.81 | — |
| Down 2 | sp|Q9H845 | ACAD9 | Acyl-CoA dehydrogenase family member 9, mitochondrial | 311 | 81512 | 21.9 | 0.69 | Acyl-CoA dehydrogenases |
| Down 3 | sp|Q969S9-2 | GFM2 | Isoform 2 of ribosome-releasing factor 2, mitochondrial | 153 | 94059 | 5.1 | 0.80 | Translation elongation factors (GTPases) |
| Down 4 | sp|P35908 | KRT2 | Keratin, type II cytoskeletal 2 epidermal | 338 | 76630 | 18.2 | 0.67 | Myosin heavy chain |
| Down 5 | tr|B7Z8A2 | — | cDNA FLJ51671, highly similar to prenylcysteine oxidase (EC 1.8.3.5) | 492 | 63740 | 23.8 | 0.83 | — |
| Down 6 | sp|Q9Y512 | SAMM50 | Sorting and assembly machinery component 50 homolog | 170 | 59339 | 18.6 | 0.76 | Outer membrane protein/protective antigen OMA87 |
| Down 7 | sp|Q6ZNW5 | GDPGP1 | GDP-D-glucose phosphorylase 1 | 118 | 45302 | 8.6 | 0.78 | — |
| Down 8 | sp|P51970 | NDUFA8 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8 | 72 | 25720 | 15.1 | 0.68 | — |
| Down 9 | tr|B4DRW0 | — | cDNA FLJ58125, highly similar to copper-transporting ATPase 1 (EC 3.6.3.4) | 102 | 61873 | 6.1 | 0.78 | Cation transport ATPase |
| Down 10 | tr|Q8NBW7 | KDELR1 | ER lumen protein retaining receptor | 51 | 20327 | 12.7 | 0.73 | ER lumen protein retaining receptor |
| Down 11 | tr|B2R6F5 | — | cDNA, FLJ92928, highly similar to Homo sapiens retinitis pigmentosa 2 (X-linked recessive) (RP2), mRNA | 59 | 47451 | 2.3 | 0.82 | — |
| Down 12 | tr|Q2VIN3 | — | RBM1 (fragment) | 1232 | 45756 | 26.8 | 0.81 | RNA-binding proteins (RRM domain) |
| Down 13 | sp|P14174 | — | Macrophage migration inhibitory factor | 608 | 13856 | 17.4 | 0.71 | — |
| Down 14 | tr|B2R6S4 | — | cDNA, FLJ93089, highly similar to Homo sapiens NCK adaptor protein 1 (NCK1), mRNA | 137 | 53755 | 18.3 | 0.83 | — |
| Down 15 | sp|Q16822 | PCK2 | Phosphoenolpyruvate carboxykinase [GTP], mitochondrial | 1795 | 78784 | 41.6 | 0.74 | Phosphoenolpyruvate carboxykinase (GTP) |
| Down 16 | tr|E9PM12 | TCIRG1 | V-type proton ATPase 116 kDa subunit a isoform 3 (fragment) | 63 | 25815 | 13.3 | 0.74 | Archaeal/vacuolar-type H+-ATPase subunit I |
| Down 17 | sp|Q2T9J0-2 | TYSND1 | Isoform 2 of peroxisomal leader peptide-processing protease | 96 | 43618 | 9.8 | 0.67 | — |
| Down 18 | tr|J3KPX7 | PHB2 | Prohibitin-2 | 1543 | 39466 | 51.8 | 0.82 | Membrane protease subunits, stomatin/prohibitin homologs |
| Down 19 | tr|Q8NCF7 | — | cDNA FLJ90278 fis, clone NT2RP1000325, highly similar to phosphate carrier protein, mitochondrial precursor | 517 | 48576 | 26.9 | 0.81 | — |
| Down 20 | tr|B4E0R0 | — | cDNA FLJ54220, highly similar to Long-chain-fatty-acid-CoA ligase 1 (EC 6.2.1.3) | 100 | 88560 | 6.2 | 0.74 | Long-chain acyl-CoA synthetases (AMP-forming) |
| Down 21 | tr|B3KRY3 | — | cDNA FLJ35079 fis, clone PLACE6005283, highly similar to lysosome-associated membrane glycoprotein 1 | 319 | 48851 | 11.1 | 0.79 | — |
| Down 22 | tr|B3KU09 | — | cDNA FLJ39034 fis, clone NT2RP7008085, highly similar to Homo sapiens ring finger protein 123 (RNF123), mRNA | 110 | 166029 | 2.4 | 0.78 | — |
| Down 23 | sp|Q9BVV7 | TIMM21 | Mitochondrial import inner membrane translocase subunit Tim21 | 86 | 35219 | 13.7 | 0.82 | — |
| Down 24 | sp|Q9UMY1 | NOL7 | Nucleolar protein 7 | 148 | 39504 | 12.5 | 0.78 | — |
| Down 25 | sp|Q9UNN8 | PROCR | Endothelial protein C receptor | 103 | 27909 | 15.1 | 0.80 | — |
| Down 26 | sp|Q86SF2 | GALNT7 | N-Acetylgalactosaminyltransferase 7 | 95 | 89410 | 9.9 | 0.81 | — |
| Down 27 | tr|I3L0U2 | PRSS21 | Testisin (fragment) | 115 | 27083 | 14.7 | 0.82 | Secreted trypsin-like serine protease |
| Down 28 | tr|B7ZLP5 | SAFB | SAFB protein | 557 | 121835 | 13 | 0.83 | — |
| Down 29 | tr|F2Z3N7 | TMEM106B | Transmembrane protein 106B | 135 | 12975 | 12.5 | 0.82 | — |
| Down 30 | tr|B7Z361 | — | Reticulon | 166 | 27838 | 12.2 | 0.76 | — |
| Down 31 | tr|H0Y6F2 | PRR5 | Proline-rich protein 5 (fragment) | 57 | 39929 | 2.3 | 0.78 | — |
| Down 32 | sp|Q7Z7E8 | UBE2Q1 | Ubiquitin-conjugating enzyme E2 Q1 | 92 | 54711 | 1.9 | 0.76 | — |
| Down 33 | tr|A8K4K9 | — | cDNA FLJ76169 | 146 | 42007 | 8.8 | 0.83 | — |
| Down 34 | sp|P13645 | KRT10 | Keratin, type I cytoskeletal 10 | 382 | 66321 | 21.6 | 0.55 | — |
| Down 35 | sp|Q8N5K1 | CISD2 | CDGSH iron-sulfur domain-containing protein 2 | 167 | 20364 | 26.7 | 0.81 | — |
| Down 36 | sp|Q8NI27 | THOC2 | THO complex subunit 2 | 282 | 241732 | 8.7 | 0.83 | — |
| Down 37 | tr|B4DEP8 | — | cDNA FLJ56960, highly similar to Homo sapiens phosphatidylinositol 4-kinase type II (PI4KII), mRNA | 127 | 61711 | 9.8 | 0.76 | — |
| Down 38 | sp|Q5BKZ1 | ZNF326 | DBIRD complex subunit ZNF326 | 145 | 78123 | 7.9 | 0.78 | — |
| Down 39 | tr|Q8IW24 | EXOC5 | Exocyst complex component 5 | 108 | 99962 | 9.3 | 0.82 | — |
| Down 40 | tr|B3KMG6 | — | cDNA FLJ10939 fis, clone OVARC1001065, highly similar to Homo sapiens MTERF domain containing 1 (MTERFD1), mRNA | 117 | 43225 | 9.8 | 0.76 | — |
| Down 41 | sp|Q8NBM4-2 | UBAC2 | Isoform 2 of ubiquitin-associated domain-containing protein 2 | 150 | 37306 | 18.1 | 0.83 | — |
| Down 42 | sp|Q8NGA1 | OR1M1 | Olfactory receptor 1M1 | 76 | 39512 | 2.2 | 0.69 | — |
| Down 43 | tr|E9PN17 | ATP5L | ATP synthase subunit g, mitochondrial | 366 | 11489 | 63.2 | 0.82 | — |
| Down 44 | tr|B2R686 | TGOLN2 | Trans-golgi network protein 2, isoform CRA_a | 166 | 61093 | 13 | 0.79 | — |
| Down 45 | tr|B4DIR5 | — | cDNA FLJ56026 | 51 | 143728 | 1.7 | 0.74 | — |
| Down 46 | tr|J3KS15 | ICT1 | Peptidyl-tRNA hydrolase ICT1, mitochondrial (fragment) | 169 | 26740 | 26 | 0.82 | Protein chain release factor B |
| Down 47 | tr|F5H0F9 | ANAPC5 | Anaphase-promoting complex subunit 5 | 72 | 98300 | 7.5 | 0.82 | — |
| Down 48 | tr|C8C504 | HBB | Beta-globin | 1233 | 20056 | 29.9 | 0.21 | — |
| Down 49 | tr|B2R921 | — | cDNA, FLJ94171, highly similar to Homo sapiens solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 (SLC25A15), nuclear gene encoding mitochondrial protein, mRNA | 53 | 39308 | 9 | 0.77 | — |
| Down 50 | sp|Q9Y613 | FHOD1 | FH1/FH2 domain-containing protein 1 | 255 | 141625 | 8.8 | 0.81 | — |
| Down 51 | sp|Q92643 | PIGK | GPI-anchor transamidase | 110 | 51592 | 10.9 | 0.77 | Glycosylphosphatidylinositol transamidase (GPIT), subunit GPI8 |
| Down 52 | tr|A4FTY4 | TXNRD2 | TXNRD2 protein | 331 | 41672 | 24.6 | 0.79 | Pyruvate/2-oxoglutarate dehydrogenase complex, dihydrolipoamide dehydrogenase (E3) component, and related enzymes |
| Down 53 | tr|D3DP46 | SPCS3 | Signal peptidase complex subunit 3 homolog (S. cerevisiae), isoform CRA_a | 147 | 24007 | 18.9 | 0.82 | — |
| Down 54 | sp|Q9Y5Q9 | GTF3C3 | General transcription factor 3C polypeptide 3 | 154 | 117216 | 7.8 | 0.79 | — |
| Down 55 | sp|P60468 | SEC61B | Protein transport protein Sec61 subunit beta | 192 | 11546 | 37.5 | 0.72 | — |
| Down 56 | sp|Q5RI15-2 | — | Isoform 2 of cytochrome c oxidase protein 20 homolog | 106 | 17682 | 20 | 0.83 | — |
| Down 57 | sp|Q9P206-2 | — | Isoform 2 of uncharacterized protein KIAA1522 | 146 | 128602 | 6.5 | 0.73 | — |
| Down 58 | sp|Q86YN1 | DOLPP1 | Dolichyldiphosphatase 1 | 64 | 28953 | 5.5 | 0.69 | Membrane-associated phospholipid phosphatase |
| Down 59 | sp|O00165-2 | — | Isoform 2 of HCLS1-associated protein X-1 | 111 | 34281 | 16 | 0.81 | — |
| Down 60 | tr|B4E303 | — | cDNA FLJ57449, highly similar to Notchless homolog 1 | 127 | 54134 | 16.5 | 0.82 | FOG: WD40 repeat |
| Down 61 | sp|O00194 | RAB27B | Ras-related protein Rab-27B | 56 | 29688 | 14.2 | 0.77 | GTPase SAR1 and related small G proteins |
| Down 62 | tr|B4DI41 | MBD1 | Methyl-CpG-binding domain protein 1 | 72 | 87409 | 1.8 | 0.80 | — |
| Down 63 | tr|B0UXB6 | ABHD16A | Abhydrolase domain-containing protein 16A | 129 | 73275 | 10.3 | 0.83 | Hydrolases of the alpha/beta superfamily |
| Down 64 | sp|Q5T8D3-2 | — | Isoform 2 of Acyl-CoA-binding domain-containing protein 5 | 148 | 64353 | 11.6 | 0.72 | Acyl-CoA-binding protein |
| Down 65 | tr|B4DNZ6 | GTF2H3 | General transcription factor IIH subunit 3 | 48 | 37020 | 4.5 | 0.79 | RNA polymerase II transcription initiation/nucleotide excision repair factor TFIIH, subunit TFB4 |
| Down 66 | sp|Q96FQ6 | S100A16 | Protein S100-A16 | 346 | 15197 | 22.3 | 0.83 | — |
| Down 67 | tr|B4DSE1 | — | cDNA FLJ55364, highly similar to CRSP complex subunit 6 | 55 | 84524 | 3.7 | 0.73 | — |
| Down 68 | tr|J3KNX9 | MYO18A | Unconventional myosin-XVIIIa | 157 | 282257 | 3.5 | 0.72 | Myosin heavy chain |
| Down 69 | tr|B4DMK6 | — | cDNA FLJ60055, highly similar to Rattus norvegicus Ssu72 RNA polymerase II CTD phosphatase homolog, mRNA | 51 | 23745 | 13.5 | 0.82 | RNA polymerase II-interacting protein involved in transcription start site selection |
| Down 70 | tr|G3V1A0 | TRAPPC4 | HCG38438, isoform CRA_b | 51 | 14838 | 20.5 | 0.81 | — |
| Down 71 | tr|B1AHA8 | HMOX1 | Heme oxygenase 1 (fragment) | 53 | 25525 | 15.5 | 0.83 | Heme oxygenase |
| Down 72 | sp|Q9Y3B3-2 | TMED7 | Isoform 2 of transmembrane emp24 domain-containing protein 7 | 193 | 24908 | 28.2 | 0.82 | — |
| Down 73 | tr|G3V1U5 | GOLT1B | Golgi transport 1 homolog B (S. cerevisiae), isoform CRA_c | 167 | 9121 | 20.3 | 0.77 | Membrane protein involved in Golgi transport |
| Down 74 | tr|B1PBA3 | — | SKNY protein | 148 | 109440 | 8.4 | 0.81 | — |
| Down 75 | sp|Q15061 | WDR43 | WD repeat-containing protein 43 | 138 | 91327 | 5.6 | 0.83 | FOG: WD40 repeat |
| Down 76 | tr|D3DUJ0 | AFG3L2 | AFG3 ATPase family gene 3-like 2 (yeast), isoform CRA_a (fragment) | 695 | 103842 | 21.2 | 0.83 | ATP-dependent Zn proteases |
| Down 77 | tr|B2RBL9 | — | cDNA, FLJ95582, highly similar to Homo sapiens breast cancer antiestrogen resistance 1 (BCAR1), mRNA | 204 | 104223 | 6 | 0.79 | — |
| Down 78 | sp|Q3SXM5-2 | — | Isoform 2 of inactive hydroxysteroid dehydrogenase-like protein 1 | 170 | 35499 | 13.5 | 0.83 | Short-chain dehydrogenases of various substrate specificities |
| Down 79 | sp|O43920 | NDUFS5 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 5 | 106 | 16388 | 11.3 | 0.74 | — |
| Down 80 | tr|H0YG20 | MAN1B1 | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase (fragment) | 155 | 90816 | 8.2 | 0.80 | — |
| Down 81 | tr|Q0KKI6 | — | Immunoglobulin light chain (fragment) | 66 | 28559 | 8.2 | 0.80 | — |
| Down 82 | sp|P62244 | RPS15A | 40S ribosomal protein S15a | 1521 | 18594 | 66.2 | 0.82 | Ribosomal protein S8 |
| Down 83 | tr|B4DL07 | — | cDNA FLJ53353, highly similar to ATP-binding cassette subfamily D member 3 | 398 | 92669 | 16.7 | 0.81 | ABC-type uncharacterized transport system, permease, and ATPase components |
| Down 84 | tr|B4DR67 | ALG5 | Dolichyl-phosphate beta-glucosyltransferase | 66 | 32213 | 10.9 | 0.81 | Glycosyltransferases involved in cell wall biogenesis |
| Down 85 | tr|Q9BTT5 | — | Similar to NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9 (39 kD) (fragment) | 189 | 45471 | 21 | 0.75 | Predicted nucleoside-diphosphate-sugar epimerases |
| Down 86 | tr|Q5U0H8 | — | Myelin protein zero-like 1 | 55 | 34725 | 4.8 | 0.74 | — |
| Down 87 | sp|Q5SY16 | NOL9 | Polynucleotide 5-hydroxyl-kinase NOL9 | 109 | 91782 | 7.4 | 0.79 | Predicted GTPase or GTP-binding protein |
| Down 88 | sp|O15173-2 | PGRMC2 | Isoform 2 of membrane-associated progesterone receptor component 2 | 620 | 30166 | 26.3 | 0.75 | — |
| Down 89 | sp|Q5VT52-3 | RPRD2 | Isoform 3 of regulation of nuclear pre-mRNA domain-containing protein 2 | 295 | 177879 | 4.5 | 0.82 | — |
| Down 90 | sp|Q8TC12 | RDH11 | Retinol dehydrogenase 11 | 494 | 41238 | 14.5 | 0.76 | Dehydrogenases with different specificities (related to short-chain alcohol dehydrogenases) |
| Down 91 | tr|B4DZ55 | — | cDNA FLJ52097, weakly similar to Homo sapiens transmembrane and tetratricopeptide repeat containing 1 (TMTC1), mRNA | 164 | 126875 | 10.1 | 0.79 | FOG: TPR repeat |
| Down 92 | tr|J3KQA9 | MTUS2 | Microtubule-associated tumor suppressor candidate 2 | 150 | 181383 | 0.6 | 0.77 | — |
| Down 93 | sp|Q96MG7 | NDNL2 | Melanoma-associated antigen G1 | 58 | 41645 | 7.6 | 0.72 | — |
| Down 94 | tr|H3BQH3 | KLHDC4 | Kelch domain-containing protein 4 (fragment) | 107 | 47359 | 10.7 | 0.83 | — |
| Down 95 | tr|J3KN00 | NDUFA13 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 | 258 | 28599 | 23.3 | 0.81 | — |
| Down 96 | sp|Q8NF37 | LPCAT1 | Lysophosphatidylcholine acyltransferase 1 | 708 | 67346 | 15.7 | 0.82 | 1-Acyl-sn-glycerol-3-phosphate acyltransferase |
| Down 97 | sp|Q9Y5P4-2 | COL4A3BP | Isoform 2 of collagen type IV alpha-3-binding protein | 82 | 81121 | 6.7 | 0.80 | — |
| Down 98 | tr|Q5T8U5 | SURF4 | Surfeit 4 | 418 | 22863 | 39.8 | 0.81 | Predicted membrane protein |
| Down 99 | sp|P26599-2 | PTBP1 | Isoform 2 of polypyrimidine tract-binding protein 1 | 570 | 69515 | 16.2 | 0.82 | — |
| Down 100 | sp|Q8NC56 | LEMD2 | LEM domain-containing protein 2 | 137 | 63423 | 7.4 | 0.76 | — |
| Down 101 | tr|Q2Q9H2 | G6PD | Glucose-6-phosphate 1-dehydrogenase (fragment) | 2165 | 64315 | 58.3 | 0.80 | Glucose-6-phosphate 1-dehydrogenase |
| Down 102 | sp|P21796 | VDAC1 | Voltage-dependent anion-selective channel protein 1 | 2340 | 38777 | 62.9 | 0.80 | — |
| Down 103 | tr|J3KNH7 | SENP3 | Sentrin-specific protease 3 | 88 | 73986 | 7.7 | 0.78 | Protease, Ulp1 family |
| Down 104 | sp|A6NHL2-2 | TUBAL3 | Isoform 2 of tubulin alpha chain-like 3 | 768 | 51287 | 11.8 | 0.79 | Tubulin |
| Down 105 | tr|B4DR71 | — | cDNA FLJ57078, highly similar to Homo sapiens opioid receptor, sigma 1 (OPRS1), transcript variant 1, mRNA | 63 | 18151 | 8.4 | 0.83 | — |
| Down 106 | sp|Q5JRA6-2 | MIA3 | Isoform 2 of melanoma inhibitory activity protein 3 | 415 | 249369 | 7.8 | 0.80 | — |
| Down 107 | tr|J9ZVQ3 | APOE | Apolipoprotein E (fragment) | 171 | 30543 | 12.2 | 0.79 | — |
| Down 108 | tr|G5E9V5 | MRPS22 | 28S ribosomal protein S22, mitochondrial | 224 | 49264 | 17.3 | 0.77 | — |
| Down 109 | tr|B7Z7X8 | ATL2 | Atlastin-2 | 112 | 76668 | 10.8 | 0.82 | — |
| Down 110 | sp|P54709 | ATP1B3 | Sodium/potassium-transporting ATPase subunit beta-3 | 243 | 39135 | 17.9 | 0.83 | — |
| Down 111 | tr|Q6IBK3 | SCAMP2 | SCAMP2 protein | 258 | 39155 | 9.7 | 0.81 | — |
| Down 112 | tr|A4LAA3 | ATRX | Alpha thalassemia/mental retardation syndrome X-linked | 129 | 374604 | 2.5 | 0.81 | Superfamily II DNA/RNA helicases, SNF2 family |
| Down 113 | sp|Q9UK59 | DBR1 | Lariat debranching enzyme | 203 | 72182 | 14.5 | 0.80 | — |
| Down 114 | tr|B4DI61 | — | cDNA FLJ58182, highly similar to protein CYR61 | 68 | 50414 | 6.4 | 0.70 | — |
| Down 115 | tr|H3BNF1 | CLN6 | Ceroid-lipofuscinosis neuronal protein 6 | 300 | 12918 | 20 | 0.80 | — |
| Down 116 | tr|E7ERK9 | EIF2B4 | Translation initiation factor eIF-2B subunit delta | 170 | 71199 | 8.8 | 0.79 | Translation initiation factor 2B subunit, eIF-2B alpha/beta/delta family |
| Down 117 | tr|H0Y8C3 | MTCH1 | Mitochondrial carrier homolog 1 (fragment) | 97 | 50964 | 12.9 | 0.81 | — |
| Down 118 | tr|B2RMV2 | CYTSA | CYTSA protein | 52 | 149539 | 2.5 | 0.79 | Ca2+-binding actin-bundling protein fimbrin/plastin (EF-hand superfamily) |
| Down 119 | tr|I3L1P8 | SLC25A11 | Mitochondrial 2-oxoglutarate/malate carrier protein (fragment) | 470 | 37200 | 35.5 | 0.83 | — |
| Down 120 | sp|Q8NBU5-2 | ATAD1 | Isoform 2 of ATPase family AAA domain-containing protein 1 | 124 | 40468 | 11.1 | 0.72 | ATPases of the AAA+ class |
| Down 121 | sp|Q9Y3E7 | CHMP3 | Charged multivesicular body protein 3 | 102 | 32415 | 14.4 | 0.83 | Conserved protein implicated in secretion |
| Down 122 | sp|P02763 | ORM1 | Alpha-1-acid glycoprotein 1 | 262 | 28288 | 20.4 | 0.80 | — |
| Down 123 | tr|Q53F51 | — | FGF intracellular binding protein isoform b variant (fragment) | 165 | 48798 | 12 | 0.83 | — |
| Down 124 | sp|Q3ZAQ7 | VMA21 | Vacuolar ATPase assembly integral membrane protein VMA21 | 241 | 12868 | 24.8 | 0.81 | — |
| Down 125 | tr|B2R6X8 | — | cDNA, FLJ93169, highly similar to Homo sapiens GPAA1P anchor attachment protein 1 homolog (yeast) (GPAA1), mRNA | 106 | 72151 | 7.6 | 0.80 | — |
| Down 126 | sp|Q9P0S9 | TMEM14C | Transmembrane protein 14C | 45 | 12774 | 8.9 | 0.70 | — |
| Down 127 | sp|P08779 | KRT16 | Keratin, type I cytoskeletal 16 | 630 | 57054 | 23.9 | 0.62 | — |
| Down 128 | sp|Q86UT6-2 | NLRX1 | Isoform 2 of NLR family member X1 | 75 | 110309 | 4.1 | 0.71 | — |
| Down 129 | tr|Q59E99 | — | Thrombospondin 1 variant (fragment) | 153 | 155789 | 3.4 | 0.68 | — |
| Down 130 | sp|Q8WXH0-2 | SYNE2 | Isoform 2 of nesprin-2 | 149 | 986758 | 1.1 | 0.82 | Ca2+-binding actin-bundling protein fimbrin/plastin (EF-hand superfamily) |
| Down 131 | sp|P78310-2 | CXADR | Isoform 2 of coxsackievirus and adenovirus receptor | 47 | 47491 | 3.8 | 0.74 | — |
| Down 132 | tr|B2R995 | — | Malic enzyme | 98 | 77738 | 5.8 | 0.83 | Malic enzyme |
| Down 133 | tr|Q5QP56 | BCL2L1 | Bcl-2-like protein 1 (fragment) | 98 | 21810 | 23.2 | 0.82 | — |
| Down 134 | tr|H0YK72 | SEC11A | SEC11-like 1 (S. cerevisiae), isoform CRA_a | 247 | 22018 | 16.5 | 0.81 | Signal peptidase I |
| Down 135 | tr|B4DDH8 | — | cDNA FLJ55184, highly similar to Homo sapiens leukocyte receptor cluster (LRC) member 4 (LENG4), mRNA | 137 | 54865 | 8.8 | 0.79 | Predicted membrane protein |
| Down 136 | sp|Q9UJS0-2 | SLC25A13 | Isoform 2 of calcium-binding mitochondrial carrier protein Aralar2 | 719 | 86824 | 17.5 | 0.82 | — |
| Down 137 | tr|A8KAK5 | — | cDNA FLJ77399, highly similar to Homo sapiens cofactor required for Sp1 transcriptional activation, subunit 2, 150 kDa (CRSP2), mRNA | 85 | 182987 | 3.4 | 0.82 | — |
| Down 138 | tr|H0YEF3 | RNASEH2C | Ribonuclease H2 subunit C (fragment) | 76 | 18856 | 25.3 | 0.77 | — |
| Down 139 | tr|Q5QNZ2 | ATP5F1 | ATP synthase F(0) complex subunit B1, mitochondrial | 406 | 27794 | 47.7 | 0.82 | — |
| Down 140 | sp|Q6UW68 | TMEM205 | Transmembrane protein 205 | 165 | 23294 | 15.9 | 0.82 | — |
| Down 141 | tr|B3KPJ4 | PHC2 | Polyhomeotic-like protein 2 | 193 | 59764 | 9.3 | 0.79 | — |
| Down 142 | tr|H0Y4D4 | ACAA1 | 3-Ketoacyl-CoA thiolase, peroxisomal (fragment) | 131 | 30218 | 12.7 | 0.78 | Acetyl-CoA acetyltransferase |
| Down 143 | tr|Q4G0F4 | POLRMT | DNA-directed RNA polymerase | 167 | 159664 | 4.6 | 0.81 | Mitochondrial DNA-directed RNA polymerase |
| Down 144 | tr|Q6FGZ3 | EPHX1 | EPHX1 protein (fragment) | 519 | 62281 | 14.9 | 0.77 | Predicted hydrolases or acyltransferases (alpha/beta hydrolase superfamily) |
| Down 145 | tr|B4DVN1 | — | cDNA FLJ52214, highly similar to DnaJ homolog subfamily B member 6 | 90 | 37740 | 8.6 | 0.70 | DnaJ-class molecular chaperone with C-terminal Zn finger domain |
| Down 146 | sp|Q92667-2 | AKAP1 | A-kinase anchor protein 1, mitochondrial | 66 | 111940 | 4.9 | ||
| Down 147 | sp|O00483 | NDUFA4 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 4 | 165 | 11855 | 46.9 | 0.83 | — |
| Down 148 | sp|Q9NTJ5 | SACM1L | Phosphatidylinositide phosphatase SAC1 | 179 | 77476 | 18.2 | 0.83 | Phosphoinositide polyphosphatase (Sac family) |
| Down 149 | tr|B3KVC5 | — | cDNA FLJ16380 fis, clone TLIVE2002882, weakly similar to imidazolonepropionase (EC 3.5.2.7) | 41 | 53582 | 3.3 | 0.83 | Imidazolonepropionase and related amidohydrolases |
| Down 150 | tr|B7ZLI5 | FAM98C | Family with sequence similarity 98, member C | 72 | 41696 | 9.5 | 0.68 | — |
| Down 151 | tr|B7Z6F5 | YIPF1 | Protein YIPF1 | 64 | 40866 | 2.7 | 0.61 | — |
| Down 152 | sp|Q6NVY1-2 | HIBCH | Isoform 2 of 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial | 101 | 46543 | 19.2 | 0.82 | Enoyl-CoA hydratase/carnitine racemase |
| Down 153 | tr|U3KQJ1 | POLDIP2 | Polymerase delta-interacting protein 2 | 282 | 46395 | 26.4 | 0.76 | Uncharacterized protein affecting Mg2+/Co2+ transport |
| Down 154 | tr|D6RGZ2 | THOC3 | THO complex subunit 3 | 172 | 12690 | 36.2 | 0.75 | — |
| Down 155 | tr|A0S0T0 | ATP6 | ATP synthase subunit a | 128 | 26896 | 4.4 | 0.78 | F0F1-type ATP synthase, subunit a |
| Down 156 | tr|G3V2U7 | ACYP1 | Acylphosphatase | 85 | 17520 | 14.7 | 0.80 | acylphosphatases |
| Down 157 | sp|Q9ULG6-2 | CCPG1 | Isoform 2 of cell cycle progression protein 1 | 79 | 93313 | 4.1 | 0.81 | — |
| Down 158 | tr|H7BXZ6 | RHOT1 | Mitochondrial Rho GTPase | 142 | 81600 | 5.9 | 0.77 | GTPase SAR1 and related small G proteins |
| Down 159 | sp|Q14151 | SAFB2 | Scaffold attachment factor B2 | 461 | 129824 | 13 | 0.83 | — |
| Down 160 | sp|Q96LD4 | TRIM47 | Tripartite motif-containing protein 47 | 138 | 75838 | 7.8 | 0.81 | — |
| Down 161 | tr|A8K2K2 | — | cDNA FLJ76494, highly similar to Homo sapiens GTPBP2 GTP-binding like protein 2 | 137 | 64767 | 11.7 | 0.83 | GTPase |
3.2. Classification of Differentially Expressed Proteins
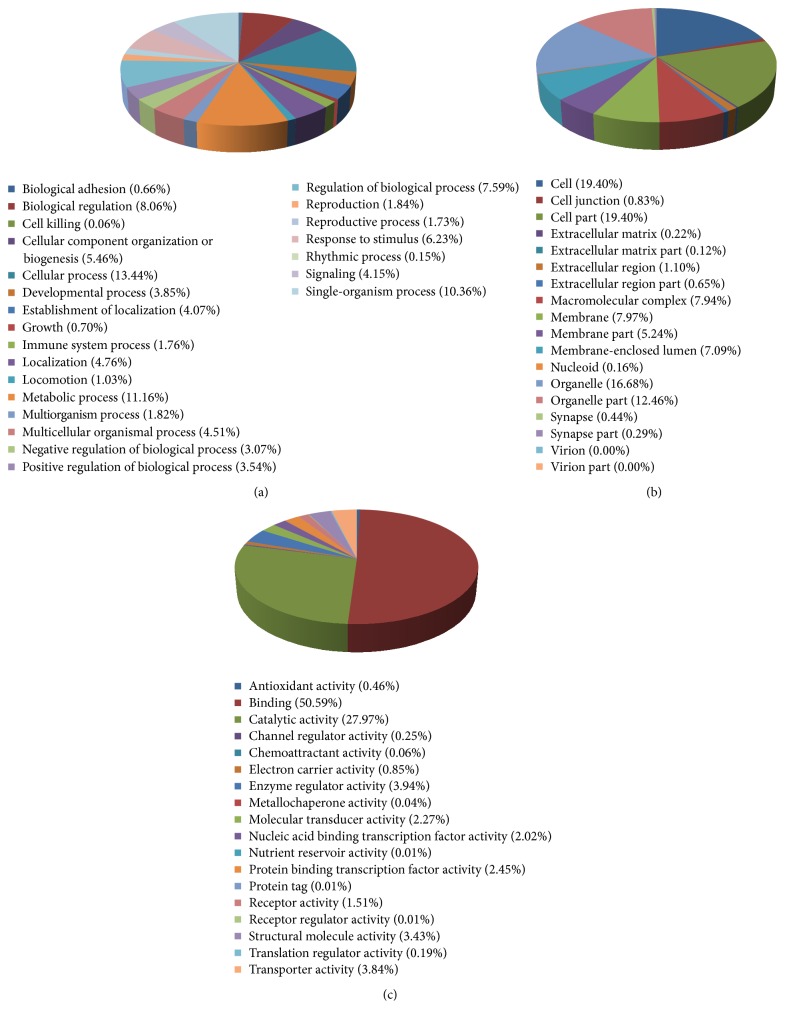
Firstly, screened proteins were functionally catalogued with GO and WEGO to three different groups (Figures 2 and 3(a)): biological process (BP), cellular component (CC), and molecular function (MF). As shown in Figure 2, the proteins are involved in BP including cellular process (13.44%), metabolic process (11.16%), single-organism process (10.36%), biological regulation (8.06%), and regulation of biological process (7.59%). The identified proteins separated according to CC include cell (19.40%), cell part (19.40%), organelle (16.68%), organelle part (12.46%), membrane (7.97%), and macromolecular complex (7.94%). MF of the proteins was classified and large groups were found to be binding (50.59%), catalytic activity (27.97%), enzyme regulator activity (3.94%), transporter activity (3.84%), and structural molecular activity (3.43%).
Figure 2.
Classification of identified proteins. (a) The biological processes (BPs), (b) cellular components (CCs), and (c) molecular functions (MFs) of the total identified proteins classified by GO database.
Figure 3.
WEGO (a) and COG (b) assay of the 205 differentially expressed proteins.
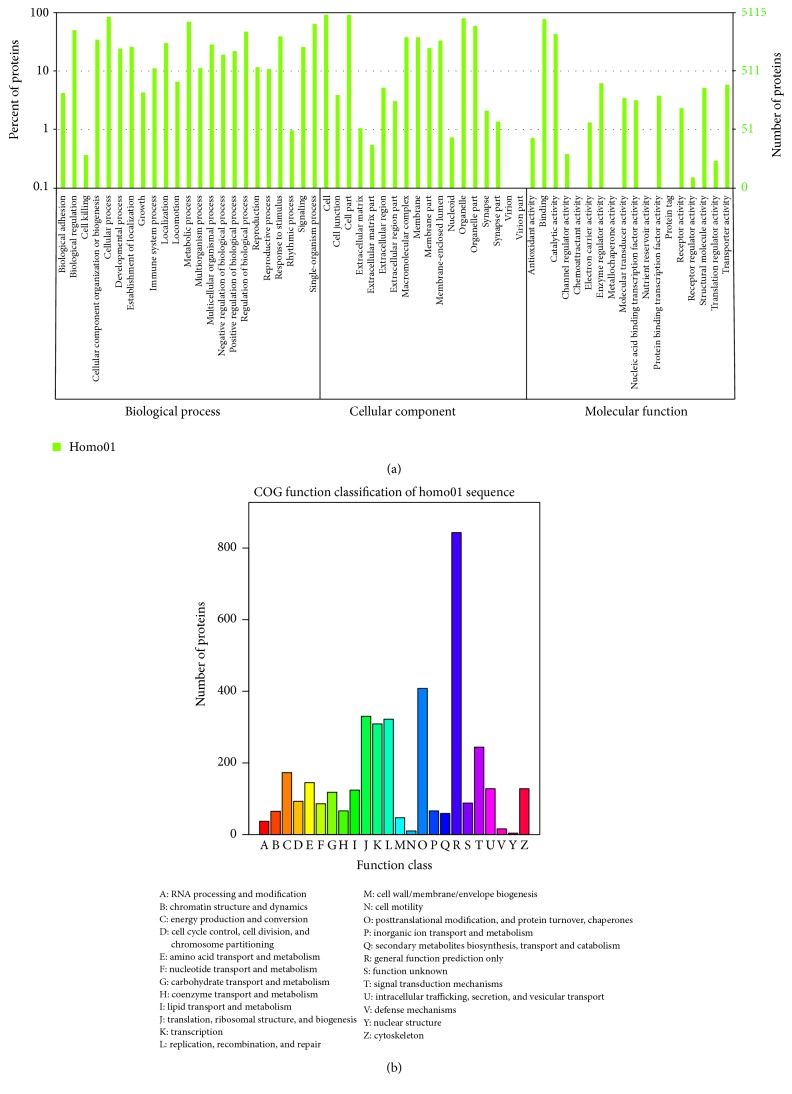
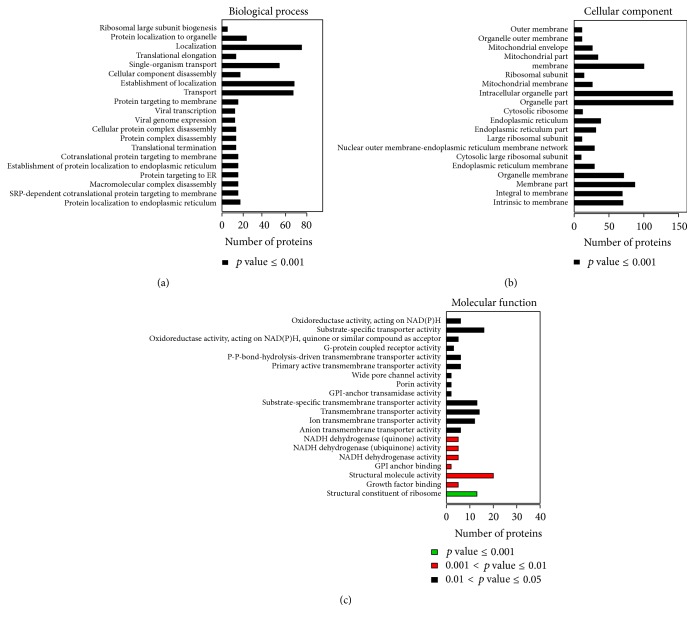
Further COG function classification revealed that posttranslational modification, protein turnover, and ribosomal structure biogenesis were major function of the screened 205 proteins (Figure 3(b)). In each category of BP, CC, and MF, top twenty proteins which generated bigger difference in response to ginsenoside F2 treatment are listed in Figure 4.
Figure 4.
GO annotation of the final selected differentially expressed proteins. The top 20 components for BP (a), CC (b), and MF (c) of the selected differentially expressed proteins are shown along with their enrichment score, represented as a p value.
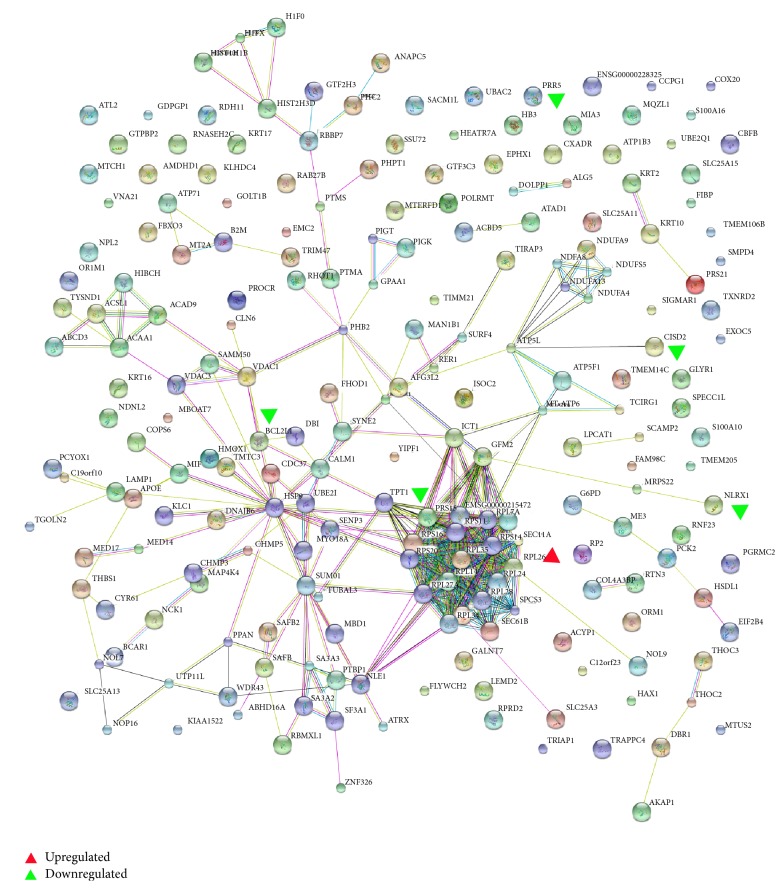
KEGG is a publicly available pathway database and could provide biologists excellent resources to attain a deeper understanding of biological mechanisms in response to different treatments. Protein analysis through KEGG indicated that 205 differentially expressed proteins were involved in 128 different pathways (data not shown). The connection degree between proteins is calculated by protein-protein interaction network analysis and the results are shown in Figure 5. Among these proteins, PRR5, RPS15, and RPL26 were found in ribosomal protein signaling pathway; CISD2, Bcl-xl, and NLRX1 were found in Beclin-1/Bcl-xL pathway. Therefore, PRR5, RPS15, RPL26, CISD2, Bcl-xl, and NLRX1 were selected for further validation and study in order to provide a comprehensive perspective for elucidating underlying molecular mechanisms of ginsenoside F2.
Figure 5.
The protein-protein interaction network of the differentially expressed proteins identified. Red triangle denotes upregulated proteins; green triangle denotes downregulated protein.
3.3. Western Blot Analysis
3.3.1. For Verification
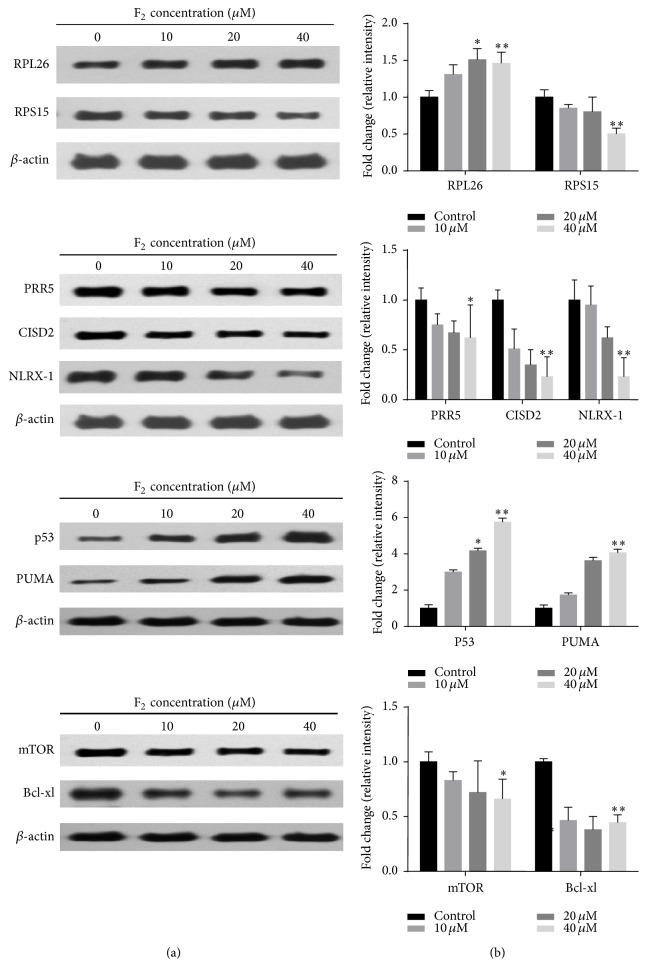
To validate the information obtained from the iTRAQ-based quantitative proteomics study and bioinformatics analysis, the screened proteins with strong response to ginsenoside F2 treatment were further confirmed by western blot. As shown in Figure 6, ginsenoside F2 significantly reduced protein expressions of PRR5, CISD2, Bcl-xl, NLRX1, and RPS15 (p < 0.01) and enhanced the expression of the RPL26 (p < 0.01) in SGC7901 cells in comparison with the treatment with vehicle control.
Figure 6.
Western blot validations of RPS15, RPL26, PRR5, CISD2, NLRX1, p53, PUMA, mTOR, and Bcl-xl in SGC7901 cells with different concentrations of ginsenoside F2. 1 × 106 SGC7901 cells are seeded in 6-well plate for overnight. On day 2, the cultured cells are treated with different concentration ginsenoside F2. 12 hours after treatment, the protein is prepared by lysating cells with RIPA buffer for performing western blot analysis. Left panel: the representative western blot analysis. β-actin was used as the loading control. Right panel: accumulated results show the relative protein density. Error bars represent means ± SEMs. Significant difference is expressed as ∗∗ p < 0.01, ∗ p < 0.05.
3.3.2. For Determining the Expression of Apoptosis and Autophagic Proteins
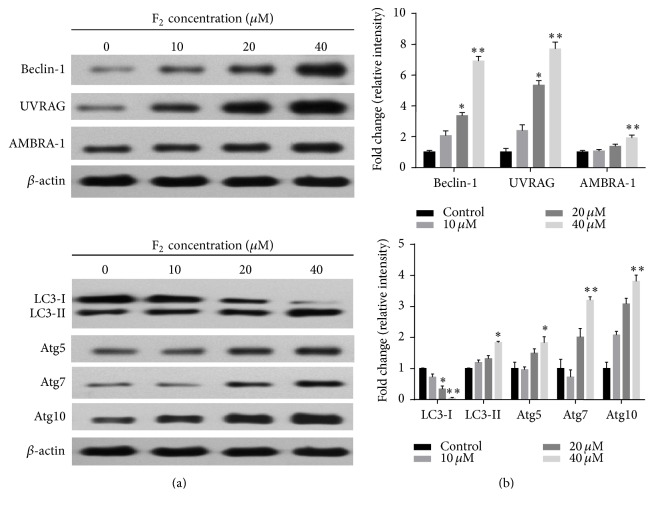
As shown in Figure 6, ginsenoside F2 suppressed the expression of mTOR and upregulated the expression of p53 in a dose-dependent manner. Atg5, Atg7, Atg10, PUMA, Beclin-1, UVRAG, and AMBRA-1 are known to be modulated by p53 or Bcl-xl signaling, which may trigger apoptosis or autophagy. Therefore, we proceeded to check the expressions of Atg5, Atg7, Atg10, PUMA, Beclin-1, UVRAG, and AMBRA-1. As shown in Figure 7, ginsenoside F2 upregulated the expressions of these proteins in a dose-dependent manner. LC3 is now widely used to monitor autophagy. During autophagy, the cytoplasmic form LC3-I is processed and recruited to phagophores, where LC3-II is generated by site-specific proteolysis and lipidation at the C-terminus. Thus, the amount of LC3-II positively correlates with the number of autophagosomes [10]. We examined the effect of F2 on LC3 conversion in SGC7901 cells. Western blot analysis showed that F2 treatment resulted in dose-dependent accumulation of LC3-II and reduction of LC3-I (Figure 7). The conversion of LC3-I to LC3-II suggested F2 treatment induces autophagy.
Figure 7.
Effect of ginsenoside F2 on the expression of Beclin-1, UVRAG, AMBRA-1, Atg5, Atg7, Atg10, LC3 I, and LC3-II. 1 × 106 SGC7901 cells are seeded in 6-well plate for overnight. On day 2, the cultured cells are treated with different concentration ginsenoside F2. 12 hours after treatment, the protein is prepared by lysating cells with RIPA buffer for performing western blot analysis. Left panel: the representative western blot analysis. β-actin was used as the loading control. Right panel: accumulated results show the relative protein density. Error bars represent means ± SEMs. Significant difference is expressed as ∗∗ p < 0.01, ∗ p < 0.05.
In the present study, combination of iTRAQ-based proteomics method with bioinformatics was used to identify critical molecules in SGC7901 cancer cells in response to ginsenoside F2 treatment. Ginsenoside F2 generated significant change of protein profile in SGC7901 cells. Some of them have been demonstrated to participate in either apoptosis or autophagy responses, suggesting that the antitumor mechanisms of ginsenoside F2 in SGC7901 cells are involved in both apoptosis and autophagy.
The current findings demonstrate that ginsenoside F2 impacts distinct signaling pathways and induces broad change in the protein profile of SGC7901 cells. Overall, 205 differentially expressed proteins were identified with ≥95% confidence in ginsenoside F2 treated group. Application of a ratio of 1.2-fold change as criteria resulted in 44 and 161 differentially abundant proteins in SGC7901 cells.
In our study, some proteins that were significantly altered by ginsenoside F2 show close relationship of protein-protein interaction (Figure 5). Ribosomal proteins, such as RPS15 and RPL26, exert critical roles in MDM2-p53 signal pathway [11, 12]. PRR5 [13], CISD2 [14], Bcl-xl [15], and NLRX1 [16, 17] have been reported to play a key role in the regulation of autophagy or apoptosis. The changes of these six potential proteins were verified by western blot analysis.
Ribosomal proteins (RPs) are considered to have diverse extra ribosomal functions, ranging from cell cycle progression to cell death and to malignant transformation and cellular metabolism [11]. Relevantly, a number of RPs have been shown to bind to MDM2, the inhibitor of p53 (murine double minute 2, and also HDM2 for its human ortholog), and inhibit MDM2 E3 ligase activity, leading to p53 stabilization and activation, then triggering apoptosis or autophagy [11]. Following the treatment of ginsenoside F2 in SGC7901 cells, the levels of RPL28, RPL34, RPL35, RPS16, RPL17, RPL14, RPL24, RPL7A, and RPL26 were increased, whereas that of RPS15 reduced. Although the functions of RPL28, RPL34, RPL35, RPS16, RPL17, RPL14, RPL24, and RPL7A have not been well studied, RPL26, a positive regulator of p53, was found to increase the translational rate of p53 mRNA by binding to its 50 untranslated region [12] and, in this case, MDM2 acts as an ubiquitin E3 ligase for ubiquitylation and degradation of RPL26 [18]. Thus, under the treatment of ginsenoside F2, the increased level of RPL26 indicated that RPL26 may inhibit MDM2 and subsequently activate p53. RPS15, identified as a direct p53 transcriptional target, was thought to activate p53 by repressing MDM2 activity [19]. Interestingly, in our study, the level of RPS15 reduced in SGC7901 followed by ginsenoside F2 treatment, suggesting that the roles of RPS15 and RPL26 involved in the anticancer mechanism of ginsenoside F2 are different, which warrant further investigation.
mTOR, existing in two multiprotein complexes, mTORC1 and mTORC2, regulates cell growth in response to a variety of cellular signals derived from growth factors and environmental stress [20]. mTORC2 is a kinase complex comprised of mTOR, PRR5, Rictor, mSin1, and mLST8/GbL. The expression level of PRR5 is correlated with that of mTORC2. Recent study showed that mTORC2 is implicated in actin cytoskeleton regulation, as well as phosphorylation of Akt [13]. Although TOR kinase has been largely attributed as a negative regulator of autophagy through TORC1, resent study indicated that mTORC2 was an independent positive regulator of autophagy during amino acid starvation [21]. In the present study, ginsenoside F2 decreased level of PPR5, indicated that ginsenoside F2 may inhibit the expression of PRR5, and consequently inhibited mTORC2.
Recent study indicated that p53 can be a positive or negative regulator of autophagy. In the nucleus, p53 may activate the AMPK pathway and inhibit the mTOR pathway, subsequently triggering autophagy. p53 may also transactivate multiple genes with proautophagic roles, including proapoptotic Bcl-2 proteins (Bax, PUMA) [22, 23]. In this network, PUMA induces the noncanonical autophagy pathway regulated via Atg5, Atg7, and Atg10. PUMA's initiation of autophagy promotes cytochrome c release, which then leads to apoptosis [22]. Interestingly, in our previous work, increasing level of cytochrome c and decreased mitochondrial transmembrane potential (MTP) were observed [6]. In present study, decreased expressions of PRR5 and RPL26 were found, which implied that ginsenoside F2 might trigger p53 signal pathway. It was reported that western blot analyses tended to show greater differential abundance compared with iTRAQ analyses [24]. Thus, the expressions of p53, Atg5, Atg7, Atg10, and PUMA were validated by western blot analyses. The increased level of Atg5 Atg7, Atg10, and PUMA and reduced level of P53 and mTORC2 suggested that ginsenoside F2 may initiate autophagy by ribosomal protein-p53 signaling pathway.
CISD2, also known as NAF-1, Miner1, Eris, and Noxp70, is a member of the 2Fe-2S cluster NEET family [25]. Our results showed that CISD2 was significantly decreased in ginsenoside F2 treated group, confirmed by western blot analysis. Recent work identified CISD2 as a Bcl-xl binding partner at a branch point between autophagy and apoptosis, life and death, under nutrient-deprived and oxidative stress conditions in vivo cells [25, 26]. Bcl-xl, also called Bcl-2L, is known to function through inhibition of the autophagy effector and tumor suppressor Beclin-1 [15]. CISD2 is required in this pathway for Bcl-xl to functionally antagonize Beclin-1-dependent autophagy. In our study, the expression of Bcl-xl decreased, confirmed by western blot analysis. Thus, CISD2 may be a Bcl-xl-associated cofactor that targets Bcl-2 for the autophagy pathway.
During initiation of autophagosome formation, after release from Bcl-xl, Beclin-1 functions as a platform by binding to class III PI3K/vacuolar protein sorting-34 (Vps34), UV-resistance-associated gene (UVRAG), activating molecule in Beclin-1-regulated autophagy (AMBRA-1) [15, 26, 27]. Previous studies have shown that binding of Beclin-1 to Bcl-2/Bcl-xl inhibits the autophagic function of Beclin-1, suggesting that Beclin-1 might have a role in the convergence between autophagy and apoptotic cell death [22]. For confirming the Beclin-1/Bcl-xl pathway, western blot was employed. The expressions of Beclin-1, UVRAG, and AMBRA-1 were increased, while Bcl-xl was decreased, which suggested that ginsenoside F2 may induce autophagy via Bcl-xl/Beclin-1 pathway.
NLRX1, a mitochondrial NOD-like receptor that amplifies apoptosis by inducing reactive oxygen species production, is an important component of TLR mediated inflammatory pathways [13, 16]. Recent evidence suggested that upregulated expression of NLRX1 may synergistically regulate metabolism and autophagy for highly invasive growth of the autophagy addicted MDA-MB-231 breast cancer cells [16]. And it acted as tumor suppressor by regulating TNF-α induced apoptosis and metabolism in cancer cells. In our iTRAQ results, expression of NLRX1 was significantly decreased in SGC7901 cells treated with ginsenoside F2. The phenomenon suggested different role of NLRX1 involved in the ginsenoside F2 treatment that may be different from that of published reports [16, 17], though the mechanism needs further research.
Mai et al. reported that F2 induces apoptotic cell death accompanied by protective autophagy in breast cancer stem cells [28]. In one of our previous studies, we found that F2 induces apoptosis by causing an accumulation of ROS and activating the apoptosis signaling pathway [6]. However, there was no report systemically comparing differently regulated proteins and building a network of F2-treated cancer cells at proteome level. In the current study, by the close look at cellular mechanisms at proteome level, we clearly identified the distinct pattern of cellular responses for the F2-treated cells, and 6 differentially regulated proteins were identified, which provide useful information on elucidating the anticancer mechanism of F2 to SGC7901 cells. Moreover, the integration of networks and pathway with the proteomic data enhanced our understanding of the functional relationship of proteome changes caused by the compound.
4. Conclusions
In conclusion, 44 upregulated proteins and 161 downregulated proteins were discovered by iTRAQ analysis in SGC7901 cells treated with lower dose and shorter duration of ginsenoside F2, compared with our previous study. 6 differentially abundant common proteins, PRR5, CISD2, Bcl-xl, NLRX1, RPS15, and RPL26, were confirmed by western blot analysis. Ribosomal protein-p53 signaling pathway and Bcl-xl/Beclin-1 pathway might be significantly regulated biological process by ginsenoside F2 treatment in SGC7901 cells. Although more work is required to find out the precise role of targeted proteins, our data lead to a better understanding of the molecular mechanisms of ginsenoside F2 for gastric cancer treatment.
Acknowledgments
This work was supported by the Natural Science Foundation of China (nos. 81573596, 81503191, 81274018, 81373946, and 81303221) and National High Technology Research and Development Plan of China (863 Plan) (2014AA022204).
Abbreviations
- iTRAQ:
Isobaric tag for relative and absolute quantification
- KEGG:
Kyoto Encyclopedia of Genes and Genomes
- COG:
Cluster of orthologous groups of proteins
- Go:
Gene Ontology
- FBS:
Fetal bovine serum
- SCX:
Strong cation exchange
- HCD:
High-energy collision dissociation
- AGC:
Automatic gain control
- NR:
Nonredundant protein database
- SDS-PAGE:
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- ECL:
Enhanced chemiluminescence
- BP:
Biological process
- CC:
Cellular component
- MF:
Molecular function
- RPs:
Ribosomal proteins
- MTP:
Mitochondrial transmembrane potential
- Vps34:
Vacuolar protein sorting-34
- UVRAG:
UV-resistance-associated gene
- AMBRA-1:
Activating molecule in Beclin-1-regulated autophagy.
Competing Interests
The authors declare that there is no conflict of interests.
References
- 1.Van Cutsem E., Sagaert X., Topal B., et al. Gastric cancer. The Lancet. 2016 doi: 10.1016/s0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 2.Niccolai E., Taddei A., Prisco D., Amedei A. Gastric cancer and the epoch of immunotherapy approaches. World Journal of Gastroenterology. 2015;21(19):5778–5793. doi: 10.3748/wjg.v21.i19.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Hagen P., Hulshof M. C. C. M., van Lanschot J. J. B., et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. The New England Journal of Medicine. 2012;366(22):2074–2084. doi: 10.1056/nejmoa1112088. [DOI] [PubMed] [Google Scholar]
- 4.Chen S., Wang Z., Huang Y., et al. Ginseng and anticancer drug combination to improve cancer chemotherapy: a critical review. Evidence-Based Complementary and Alternative Medicine. 2014;2014:13. doi: 10.1155/2014/168940.168940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi L.-W., Wang C.-Z., Yuan C.-S. American ginseng: potential structure-function relationship in cancer chemoprevention. Biochemical Pharmacology. 2010;80(7):947–954. doi: 10.1016/j.bcp.2010.06.023. [DOI] [PubMed] [Google Scholar]
- 6.Mao Q., Zhang P.-H., Wang Q., Li S.-L. Ginsenoside F2 induces apoptosis in humor gastric carcinoma cells through reactive oxygen species-mitochondria pathway and modulation of ASK-1/JNK signaling cascade in vitro and in vivo . Phytomedicine. 2014;21(4):515–522. doi: 10.1016/j.phymed.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 7.Shin J.-Y., Lee J.-M., Shin H.-S., et al. Anti-cancer effect of ginsenoside F2 against glioblastoma multiforme in xenograft model in SD rats. Journal of Ginseng Research. 2012;36(1):86–92. doi: 10.5142/jgr.2012.36.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao W., Zhou Y., Li Y., et al. iTRAQ-based proteomic analysis of combination therapy with taurine, epigallocatechin gallate, and genistein on carbon tetrachloride-induced liver fibrosis in rats. Toxicology Letters. 2015;232(1):233–245. doi: 10.1016/j.toxlet.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 9.Dou D., Wen Y., Weng M., et al. Minor saponins from leaves of Panax ginseng C.A. Meyer. Zhongguo Zhong Yao Za Zhi. 1997;22(1):35–37. [PubMed] [Google Scholar]
- 10.Hu X., Han W., Li L. Targeting the weak point of cancer by induction of necroptosis. Autophagy. 2007;3(5):490–492. doi: 10.4161/auto.4592. [DOI] [PubMed] [Google Scholar]
- 11.Wang W., Nag S., Zhang X., et al. Ribosomal proteins and human diseases: pathogenesis, molecular mechanisms, and therapeutic implications. Medicinal Research Reviews. 2015;35(2):225–285. doi: 10.1002/med.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takagi M., Absalon M. J., McLure K. G., Kastan M. B. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123(1):49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 13.Woo S.-Y., Kim D.-H., Jun C.-B., et al. PRR5, a novel component of mTOR complex 2, regulates platelet-derived growth factor receptor β expression and signaling. The Journal of Biological Chemistry. 2007;282(35):25604–25612. doi: 10.1074/jbc.m704343200. [DOI] [PubMed] [Google Scholar]
- 14.Chang N. C., Nguyen M., Germain M., Shore G. C. Antagonism of Beclin 1-dependent autophagy by BCL-2 at the endoplasmic reticulum requires NAF-1. The EMBO Journal. 2010;29(3):606–618. doi: 10.1038/emboj.2009.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S.-Y., Song X., Zhang L., Bartlett D. L., Lee Y. J. Role of Bcl-xL/Beclin-1 in interplay between apoptosis and autophagy in oxaliplatin and bortezomib-induced cell death. Biochemical Pharmacology. 2014;88(2):178–188. doi: 10.1016/j.bcp.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tattoli I., Carneiro L. A., Jéhanno M., et al. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-κB and JNK pathways by inducing reactive oxygen species production. EMBO Reports. 2008;9(3):293–300. doi: 10.1038/sj.embor.7401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia X., Cui J., Wang H. Y., et al. NLRX1 negatively regulates TLR-induced NF-κB signaling by targeting TRAF6 and IKK. Immunity. 2011;34(6):843–853. doi: 10.1016/j.immuni.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ofir-Rosenfeld Y., Boggs K., Michael D., Kastan M. B., Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Molecular Cell. 2008;32(2):180–189. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daftuar L., Zhu Y., Jacq X., Prives C. Ribosomal proteins RPL37, RPS15 and RPS20 regulate the Mdm2-p53-MdmX network. PLoS ONE. 2013;8(7) doi: 10.1371/journal.pone.0068667.e68667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson S. C., Rabinovitch P. S., Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493(7432):338–345. doi: 10.1038/nature11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vlahakis A., Powers T. A role for TOR complex 2 signaling in promoting autophagy. Autophagy. 2014;10(11):2085–2086. doi: 10.4161/auto.36262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang J. J., Di J. H., Cao H., Bai J., Zheng J. p53-Mediated autophagic regulation: a prospective strategy for cancer therapy. Cancer Letters. 2015;363(2):101–107. doi: 10.1016/j.canlet.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 23.Mathew R., Wadsworth V. K., White E. Role of autophagy in cancer. Nature Reviews Cancer. 2007;7(12):961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ralhan R., Desouza L. V., Matta A., et al. iTRAQ-multidimensional liquid chromatography and tandem mass spectrometry-based identification of potential biomarkers of oral epithelial dysplasia and novel networks between inflammation and premalignancy. Journal of Proteome Research. 2009;8(1):300–309. doi: 10.1021/pr800501j. [DOI] [PubMed] [Google Scholar]
- 25.Tamir S., Rotem-Bamberger S., Katz C., et al. Integrated strategy reveals the protein interface between cancer targets Bcl-2 and NAF-1. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(14):5177–5182. doi: 10.1073/pnas.1403770111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fimia G. M., Stoykova A., Romagnoli A., et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447(7148):1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 27.Liang C., Feng P., Ku B., et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nature Cell Biology. 2006;8(7):688–698. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 28.Mai T. T., Moon J. Y., Song Y. W., et al. Ginsenoside F2 induces apoptosis accompanied by protective autophagy in breast cancer stem cells. Cancer Letters. 2012;321(2):144–153. doi: 10.1016/j.canlet.2012.01.045. [DOI] [PubMed] [Google Scholar]